Abstract
Mutations in the Aristaless related homeodomain transcription factor (ARX) are associated with a diverse set of X-linked mental retardation and epilepsy syndromes in humans. Although most studies have been focused on its function in the forebrain, ARX is also expressed in other regions of the developing nervous system including the floor plate (FP) of the spinal cord where its function is incompletely understood. To investigate the role of Arx in the FP, we performed gain-of-function studies in the chick using in ovo electroporation, and loss-of-function studies in Arx-deficient mice. We have found that Arx, in conjunction with FoxA2, directly induces Sonic hedgehog (Shh) expression through binding to a Shh floor plate enhancer (SFPE2). We also observed that FoxA2 induces Arx through its transcriptional activation domain whereas Nkx2.2, induced by Shh, abolishes this induction. Our data support a feedback loop model for Arx function; through interactions with FoxA2, Arx positively regulates Shh expression in the FP, and Shh signaling in turn activates Nkx2.2, which suppresses Arx expression. Furthermore, our data are evidence that Arx plays a role as a context dependent transcriptional activator, rather than a primary inducer of Shh expression, potentially explaining how mutations in ARX are associated with diverse, and often subtle, defects.
Keywords: Spinal cord, neural tube, floor plate, development, Shh, Arx, FoxA2 and Nkx2.2
Introduction
Cell type specification is a dynamic process dependent on cell extrinsic and intrinsic signaling programs. The developing spinal cord serves as an excellent model system to study cell type specification. Many studies over the past decades have deduced that morphogenic gradients formed by several signaling molecules (e.g. sonic hedgehog and retinoic acid) initiate intrinsic transcription networks enabling first the specification of distinct progenitor cells and subsequently maintaining their identity (Davidson, 2002; Jessell, 2000). Understanding how the various factors in this process interact is crucial to unraveling the mechanisms underlying cell fate determination.
Sonic hedgehog (Shh) is a secreted protein with well-established roles in cell fate specification in the ventral spinal cord. It is first expressed in the mesodermally derived notochord and subsequently in the ventral midline of the developing neural tube (i.e., floor plate, FP). When Shh binds the transmembrane receptor, Patched (Ptc), it releases the inhibition of Smoothened (Smo), which then translocates to the cytoplasm and initiates a signaling cascade that results in the nuclear translocation of Gli (Gli1-3) transcription factors. Gli transcription factors bind specific cis-elements (GBSs; Gli binding sites) of downstream target genes to activate or repress their transcription (Briscoe et al., 2000; Dessaud et al., 2008). It is known that Shh stabilizes full-length Gli2 and Gli3 proteins in their activator forms (GliA); in the absence of ligand these bi-functional proteins undergo proteolysis and change to repressor forms (GliR) (Dessaud et al., 2008; B. Wang et al., 2000).
Shh signaling functions in a gradient to establish unique cell fates along the dorsal ventral axis of the developing spinal cord. In response to this morphogen gradient, transcription factors in responding cells are either induced or repressed to establish the p0, p1, p2, pMN, p3, and FP domains. In turn, each progenitor domain gives rise to a distinct neuronal (V3, MN, V2, V1 and V0), and non-neuronal (FP) subtypes (Dessaud et al., 2008; Jessell, 2000). Ventral neural tube development is not only dependent on its spatial concentration gradient of Shh, but also the timing and duration of the signaling (Chamberlain et al., 2008; Dessaud et al., 2008; 2007). Increasing levels and durations of Shh signaling direct progenitors to adopt progressively more ventral identities (Chamberlain et al., 2008; Dessaud et al., 2008; 2007). Furthermore, the interpretation of the Shh morphogen gradient into an intrinsic transcriptional network, rather than Shh gradient itself, has been shown to be responsible for differential spatial and temporal gene expression (Balaskas et al., 2012). Moreover, identification and characterization of the cis-regulatory modules (CRMs) of target genes operating downstream of Shh signaling, has clarified how different cells interpret their Shh signaling depending on their relative location in relation to the signaling source (Oosterveen et al., 2012; Peterson et al., 2012).
Specification of the most ventral cell type, the non-neuronal FP cells, is thought to be a sequential process. Initially, the presumptive FP cells, in response to notochord-derived Shh, express a set of transcription factors (e.g. FoxA2, Nkx2.2 and Nkx6.1) that are also expressed in adjacent progenitor cells (p3). Later the developing FP cells also begin expressing Shh and Arx, whereas Nkx2.2 expression is down-regulated and no longer detected in the presumptive FP but continues to be expressed in the adjacent p3 domain. Unlike other ventral neuronal subtypes, where high levels and longer duration of Shh signaling predicts more ventral identities, FP specification involves a biphasic response to Shh signaling. Initially, high levels of Shh signaling are required for FP specification (Ribes et al., 2010), however, maintenance of the FP is Shh signaling independent, although Shh continues to be expressed by FP cells. If Shh signaling is maintained during this time instead of down-regulated, FP cells convert their identity to ventral neural progenitors (Ribes et al., 2010).
Despite the down-regulation of Shh signaling in FP cells, Shh itself is not down-regulated, suggesting that the FP cells must maintain adequate levels of Shh production for the generation of other ventral cell types, and for functions such as a commissural axon chemoattraction (Bourikas et al., 2005). Paradoxically, the transcription factor FoxA2 is responsible for inducing Shh expression, while it simultaneously down-regulates Shh signaling to maintain FP identity and inhibit p3 fate.
Two enhancer regions have been identified in the regulatory regions of Shh that are responsible for spinal cord FP specific expression: Shh Floor Plate Enhancer 1 and 2 (SFPE1 and 2) (Epstein et al., 1999; Jeong and Epstein, 2003). SFPE1 activity is controlled in a FoxA2-independent manner. In contrast, SFPE2 activity is regulated by two elements, a Homeobox transcription factor Binding Site (HBS) and a FoxA2 binding site. Both are required for the full activity in the FP (Epstein et al., 1999; Jeong and Epstein, 2003). To date the homeodomain transcription factor(s) that binds to SFPE2 has not been identified.
The aristaless related homeodomain transcription factor (Arx) is the vertebrate homolog of Drosophila Aristaless (Miura et al., 1997). It is expressed in the developing brain including the cerebral cortex, basal ganglia, hypothalamus, thalamus, midbrain, and hindbrain (Colombo et al., 2004; Miura et al., 1997). Its expression is first detected at the 3 somite stage (~E8) in mouse embryos and it persists through early postnatal life (Colombo et al., 2004). Mutations ARX have been linked to morphological brain anomalies as well as multiple neurologic deficits in patients (Friocourt and Parnavelas, 2010; Kato et al., 2004; Kitamura et al., 2002; Mégarbané et al., 2011; Olivetti and Noebels, 2012; Sherr, 2003; Shoubridge et al., 2010; Strømme et al., 2002). Arx-deficient mice have intermediate progenitor cell proliferation defects in the forebrain resulting in small brains (Colasante et al., 2013; Kitamura et al., 2002). They also show aberrant migration and differentiation of interneurons in the ganglionic eminence and neocortex (Fulp et al., 2008; Kitamura et al., 2002; Marsh et al., 2009; Nasrallah et al., 2011). Furthermore, loss of Arx in mice, through conditional gene abrogation, results in structural brain anomalies, epilepsy, and neurocognitive phenotypes (Colasante et al., 2013; Fulp et al., 2008; Kitamura et al., 2002; Marsh et al., 2009).
Arx is also expressed in FP cells of the developing spinal cord, however its function in the FP has not been explored. Based on the observations that 1) Arx is expressed in FP cells during the period of Shh induction and 2) it is a homeodomain transcription factor, we hypothesized that Arx binds to the HBS of SFPE2 and induces Shh expression. To test our hypothesis, we performed both gain-of-function and loss-of-function experiments using the chick embryo and Arx deficient mice. We find Arx indeed binds the SFPE2 site and induces Shh expression in the presence of FoxA2. Furthermore, our data demonstrate that FoxA2 induces Shh via its activation domain, while Nkx2.2 represses FoxA2-induced Arx expression. These results support a model where Arx and FoxA2 participate in a feedback loop with Shh signaling, establishing a robust method to regulate the dynamic expression of Shh required for its multiple functions during spinal cord development.
Materials and Methods
Mice
Arx mutant mice (Fulp et al., 2008) were bred and maintained on C57Bl/6 background in according with an approved IACUC protocol at the Children’s Hospital of Philadelphia and Brigham and Women’s Hospital/Harvard Medical School. Arx−/y mouse embryos were generated by mating ArxF/+ with EIIacre male (The Jackson Laboratory stock number 003724). All genotyping was performed as previously described (Fulp et al., 2008).
DNA constructs
Arx, FoxA2, Nkx2.2 (human sequence for NKX2.2 was used but is referred throughout as Nkx2.2) and each deletion mutant, used for in ovo electroporation, were cloned into the pCIG vector (Megason and McMahon, 2002) that expresses eGFP under IRES, after PCR-amplification with the oligonucleotides as following: ArxF (5′-CGGAATTCCACCATGAGCAATCAGTACCAGGAAGAG-3′), Arx61F (5′-CGGAATTCCACCATGGAAAAAGCCATGCAAGGCTCCCCC -3′), Arx220F (5′-CGGAATTCCACCATGGGCGCCGAGGACGACGAGG -3′), Arx471mycR (5′-ACTTCAACGCGTCTACAGATCTTCTTCAGAAATAAGTTTTTGTTCCGCTGCTCCTAGAAAAGTGCTCAGACC-3′), ArxmycR (5′-ACTTCAACGCGTCGAGCTACAGATCTTCTTCAGAAATAAGTTTTTGTTCGCACACCTCCTTCCCCGTGCTG -3′), FoxA2FLAGF (5′-CGGAATTCCACCATGGATTACAAGGATGACGACGATAAGCTGGGAGCCGTGAAGATGGAA -3′), FoxA2R (5′-ACCGACGCGTTTAGGATGAGTTCATAATAGGCCTGGAGTACACTC-3′), FoxA2F52 (5′-CGGAATTCCACCATGGATTACAAGGATGACGACGATAAGGGCGGCGGTTCCGGCAACAT-3′), FoxA2R-418 (5′-AACCGACGCGTTTAGGAACCATAGCCCCCTGGGTAGTGC-3′), FoxA2D372-383F (5′-CCACCTGAAGCCCGAGCACCATTACTCGTCCGAGCAGCAACATCACCA -3′), Nkx2.2F (5′-CGGAATTCCACCATGGATTACAAGGATGACGACGATAAGATGTCGCTGACCAACACAAAGACGG-3′), Nkx2.2R (5′-AACCGACGCGTTCACCAAGTCCACTGCTGGGCCT-3′), Nkx2.2F113 (5′-CGGAATTCCACCATGGATTACAAGGATGACGACGATAAGGACAATGACAAGGAGACCCCGGGC -3′) and Nkx2.2R187 (5′-AACCGACGCGTTCACCGGGCGCGCTTCATCTTGTAG -3′). Arx MT is R332H, which does not bind to DNA due to mutation in homeodomain of Arx (Cho et al., 2012). The FoxA2 and Nkx2.2 constructs include a FLAG-tag embedded in the 5′ end of the oligonucleotide sequence. The FoxA2ΔA (52-418) deletion construct lacks the transcription activation domain (Pani et al., 1992). The FoxA2ΔI, internal deletion mutant which excludes amino acid 372–387 (TLE/Groucho binding site), was cloned into EcoRI and MluI of pCIG vector as described previously (J. C. Wang et al., 2000). Nkx2.2HD (aa113-187) (dominant negative mutant which contains only homeodomain) was constructed into pCIG as previously described (Watada et al., 2000). The PtcΔloop2, SmoM2, and ΔN-Gli3 constructs were all previously described (Lei et al., 2004; Lek et al., 2010; Tenzen et al., 2006). The deleted Arx DNA fragments, used for immunoprecipitation experiment, were subcloned into both EcoRI and XbaI digested pM vector (Clontech) after PCR amplification; ArxF (5′-CGGAATTCATGAGCAATCAGTACCAGGAAGAGG-3′), Arx221F (5′-CGGAATTCGGCGCCGAGGACGACGAGGAGGAG-3′), Arx321F (5′-CGGAATTCTCGGAGGAGGGGCTGCTGAAGCGC-3′), Arx471F (5′-CGGAATTCGCGGTGTTCCGCCACCCAGCCTTC -3′), Arx220R (5′-GCCCTCTAGACGTGCCACCACCCGCCGCGGGGGC -3′), Arx320R (5′-GCCCTCTAGAGTCGCTGCCGGCCGACAGGCACACG -3′), Arx470R (5′-GCCCTCTAGATGCTCCTAGAAAAGTGCTCAGACCC -3′) and ArxR (5′-GCCCTCTAGATTAGCACACCTCCTTCCCCGTGCTG -3′). For generation of the full length Arx protein in mammalian expression system, the FLAG-tagged Arx PCR product was subcloned into pcDNA3.1-d-TOPO (Invitrogen) with ArxFLAGF (5′-CACCATGAGCAATCAGTACCAGGAAGAGGGC-3′) and ArxFLAGR (5′-TTACTTATCGTCGTCATCCTTGTAATCGCACACCTCCTTCCCCGTGCTG-3′).
In ovo electroporation
Electroporation was performed as previously described (Briscoe et al., 2000; Lim et al., 2005). DNA was injected into the neural tube of Hamburger-Hamilton stage 10–12 (HH10-HH12) chick embryos at concentrations of 5.0 μg/μl in TE with 50 ng/μl Fast Green. For co-electroporation of low FoxA2 and ArxWT or ArxMT, FoxA2 expression construct was used in one tenth of Arx DNA. For all other co-electroporation, DNAs were mixed as 1:1 ratio. Approximately 48 h following electroporation, embryos were harvested and fixed in 4% paraformaldehyde for subsequent immunostaining.
Immunohistochemistry
All immunohistochemistry was performed as previously reported (Lim et al., 2005). Briefly, embryos were collected, fixed overnight in cold 4% paraformaldehyde, washed at 4°C in PBS, cryoprotected overnight at 4°C in PBS containing 30% sucrose, and frozen in OCT (Tissue-Tek). Twelve-micrometer sections at the spinal cord or hindbrain level were then cut for subsequent immunostaining. The primary antibodies used include: rabbit anti-FoxA2 (1:20, Epitomics), mouse anti-Shh (1:2000, Sigma), mouse anti-Shh (1:10, DSHB), rabbit anti-Shh (1:200, Santa Cruz Biotechnology), mouse anti-Nkx2.2 (1:5, DSHB), mouse anti-Pax7 (1:20, DSHB), rabbit anti-Nkx6.1 (1:5, DSHB), rabbit anti-Olig2 (1:5000, Epitomics, rabbit), mouse anti-Nkx2.2 (1:5, DSHB), mouse anti-Isl1/2 (1:20, DSHB), and rabbit anti-Arx (1:200, (Kitamura et al., 2002)). Nuclei were visualized with DAPI (1:30,000, Invitrogen) staining. Secondary antibodies conjugated with Alexa 488, 568, or 633 (1:200, Invitrogen) were utilized for visualization on a Leica DMR microscope equipped with epiflourescence and a Leica DFC345 FX camera. DAB and ABC kit (Vector Lab) were used for Shh, Alcam and Olig2 immunostaining. A minimum of 5 sections, from at least three embryos of each genotype, were examined in the mouse studies. Number of Olig2-positve cell was counted manually (E11.5) or automatically (E14.5) using Particle Analysis in ImageJ (Fiji).
Electromobility Shift Assay (EMSA), Chromatin Immunoprecipitation (ChIP), and Immunoprecipitation (IP)
EMSA was performed as previously described (Cho et al., 2012). The full length Arx protein was expressed in HEK293 transfected with pcDNA3.1-FLAG-Arx and purified with FLAG-beads (Sigma) in the TNE buffer (20 mM Tris-HCl [pH 7.4], 150 mM, NaCl, 0.5% Triton X-100, 5% glycerol). The purified Arx protein was applied to 5% polyacrylamide gel with double stranded oligonucleotides, SFPE2F (5′-CTTTATTGGATTTTAATTAGAAAATCCACACA-3′)/SFPE2R(5′-TGTGTGGATTTTCTAATTAAAATCCAATAAAG-3′) and mSFRP2F (5′-CTTTATTGGATTTTCCTTAGAAAATCCACACA-3′)/mSFRP2R(5′-TGTGTGGATTTTCTAAGGAAAATCCAATAAAG labeled by biotin (Integrated DNA Technologies). To detect the DNA/Arx complex, we utilized the LightShift Chemiluminescent EMSA kit (Thermo Scientific).
For ChIP assays, E9.5 mouse spinal cord tissues were used. After dissection and dissociation, the spinal cord tissues were fixed with 1% formaldehyde for 10 min at room temperature on a rotating rocker. ChIP was performed following the protocol for the EZ ChIP Immunoprecipitation Kit (EMD Millipore) using a rabbit anti-Arx and control rabbit antibody (Invitrogen). Primers spanning SFPE2 within intron2 of Shh gene, qSFPE2-R1 (5′-CCCGAGACTTGTGTGGATTT-3′) and qSFPE2-R2 (5′-TCCGAGGCTGTCTCCTATTTA-3′), primers −1kb away from upstream of SFPE2, −1kbF (5′-CGTAAGTCCTTCACCAGCTT-3′) and −1kbR (5′-CTCAACACCTGGTCTTTCTCTC-3′) were used for real time PCR of the immunoprecipitated DNA with SsoAdvanced Universal Supermix (Bio-rad). The primers, negR (5-ATGGTTGCCACTGGGGATCT-3′) and negR (5′-TGCCAAAGCCTAGGGGAAGA—3′) flanking genomic region between Gapdh and Cnap1 were used for negative control.
Immunoprecipitation experiment was performed as previously described with only slight modifications (Nasrallah et al., 2011). After co-transfected with FLAG-FoxA2 and Arx-Myc, HEK293T cells were lysed with TNE buffer (20 mM Tris-HCl [pH 7.4], 150 mM, NaCl, 0.5% Triton X-100, 5% glycerol) containing a protease inhibitor cocktail (Roche Applied Science). Anti-FLAG (M2) monoclonal antibody (Sigma) or anti-Myc antibody (Cell Signaling technology) was used for immunoprecipitation. For preclearance, lysates were incubated with protein G-conjugated beads (Invitrogen). Each primary antibody (2 μg) defined by the individual experiment was added, incubated at 4°C for 1 h, and then incubated with the protein G-conjugated beads for an additional hour. The beads were washed with TNE buffer twice and with TNE buffer containing 500 mM NaCl twice, followed by a final wash with TNE buffer. Bound proteins were eluted using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. To detect the associated protein in the immunoprecipitate, SDS-PAGE followed by Western blotting was performed using anti-Arx, anti-FLAG (M2, 1/500; Sigma) and anti-GAL4 (Clontech) antibody.
Results
Arx expression in the floor plate
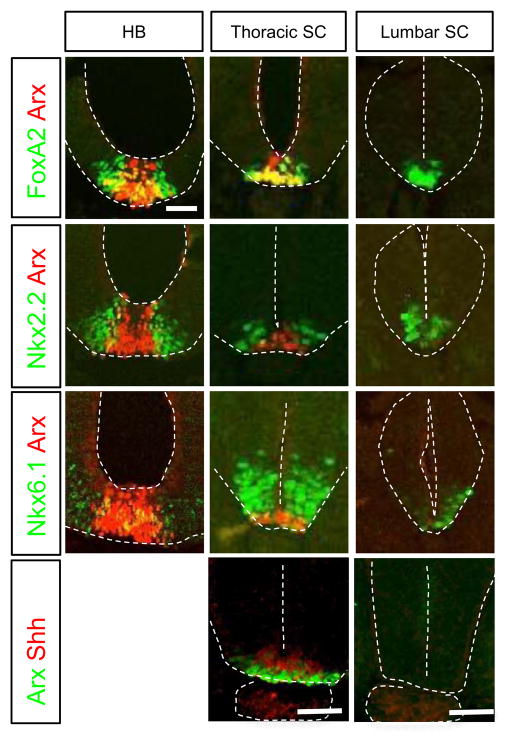
To establish the developmental time course of Arx expression in the developing chick spinal cord, we took advantage of the normal rostral to caudal gradient of neural tube development. In HH10 chick embryos, the neural tube is rostrally closed, however, a small segment remains open at the most caudal end. Arx is expressed in the rostral neural tube (hindbrain and thoracic level of the spinal cord) but it is not detected in the developmentally younger caudal neural tube (lumbar level of the spinal cord) (Fig 1). Arx expression is restricted to the ventral midline where it overlaps with FoxA2, although the FoxA2 expressing domain does extend to several cell bodies beyond that of Arx. In contrast, FoxA2, Nkx2.2 and Nkx6.1 are ventrally expressed along the entire rostral-caudal neural axis (Fig 1). As expected, Nkx2.2, and to a lesser extent Nkx6.1, are largely excluded from the presumptive FP at the more rostral levels of the neural tube; however both are expressed just lateral to the Arx expressing domain (Fig 1). The co-expression of FoxA2, Nkx2.2 and Nkx6.1 in the presumptive FP at the lumbar neural tube suggests that at the early stage, the FP identity is mixed with p3 identity, since the presumptive FP cells initially express markers that are shared with p3 and pMN progenitors such as Nkx2.2 and Nkx6.1, respectively. This data is consistent with the previous report of Shh expression in the FP (Ribes et al., 2010). Furthermore, we find Shh expression coincides with that for Arx (Fig 1). These results confirm that Arx is expressed late in ventral midline development, but in the definitive FP when p3 markers, such as Nkx2.2, are no longer expressed.
Fig. 1.
Cross-sections of embryonic chick neural tube (HH10) showing Arx, Nkx2.2, Nkx6.1, FoxA2 and Shh expression in the hindbrain (HB) and spinal cord (SC). The colored letters on the left side box indicate the antibodies used for immunohistochemistry. HB (hindbrain), thoracic SC, and lumber SC indicate the level of each section. Scale bar in the upper left panel is 50 μm for all images.
Arx positively regulates floor plate Shh induction in the presence of FoxA2
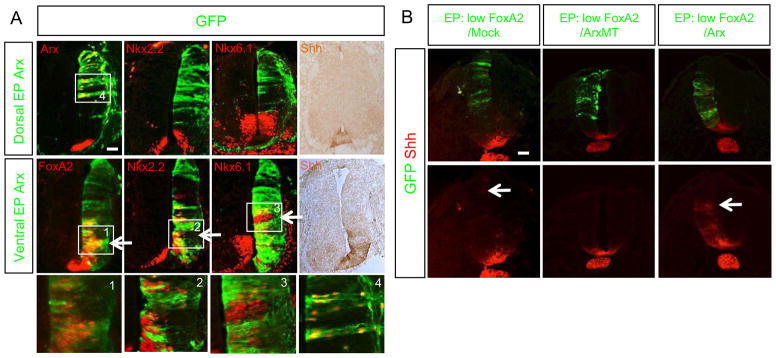
Arx expression in the FP coincides with the strong induction of Shh in these same cells (Fig. 1) (Ribes et al., 2010). Thus, we postulated that Arx might regulate Shh transcription in the FP. To test this assumption we electroporated an Arx expression construct into chick neural tubes and assayed for changes in gene expression. Targeting Arx to the dorsal neuroepithelium failed to induce Shh or the Shh downstream genes, Nkx2.2 and Nkx6.1 (Fig 2A). In contrast, when Arx was targeted to the ventral spinal cord, Shh as well as Shh downstream genes were induced (Fig 2A). These data suggest that Arx can induce Shh expression only in the presence of other ventral factor(s). Given that FoxA2 is also expressed in the FP and known to induce Shh (Ribes et al., 2010), we next tested whether FoxA2 might be the cooperating factor to function with Arx. When an Arx expression construct (WT) was co-electroporated in the dorsal spinal cord with low levels of a FoxA2 construct, the Shh gene was strongly induced in cell autonomous manner (Fig 2B). However, when a mutant Arx construct (MT) that harbors the R332H mutation in the homeodomain leading to loss of DNA binding activity (Cho et al., 2012) was co-electroporated with low levels of FoxA2, Shh was no longer induced (Fig 2B). In contrast, a low level of FoxA2 was not able to induce Shh expression or only did so slightly. Together these findings suggest Arx collaborates with FoxA2 to induce Shh.
Fig. 2.
Ectopic expression of Arx induces Shh and Shh-downstream genes in the presence of FoxA2. A) In ovo electroporation (EP) of Arx to the ventral or dorsal spinal cord analyzed for Nkx2.2, Nkx6.1, FoxA2, and Shh expression via immunostaining. Arx EP to the ventral neural tube induces FoxA2, Nkx2.2, Nkx6.1 and Shh (bottom panel), while dorsally targeted Arx does not (top panel). The electroporated plasmid expresses Arx and GFP, thus the presence of GFP reports the location of ectopic Arx expression. Boxed areas labeled 1–4 corresponded to high power images at bottom of A. B) Co-electroporation of low level of FoxA2 with ArxWT (wild type) (1:10 ratio in DNA concentration) induces Shh cell non-autonomously, even in the dorsal neural tube, whereas co-electroporations of low level of FoxA2 with Arx MT (non-DNA bound homeodomain mutant; R332H) or mock DNA do not. Arrows in A and B indicate the ectopic induction of Shh or Shh downstream target genes. Scale bars in A and B are 50 μm.
Arx can interact with FoxA2 and bind to the Shh enhancer, SFPE2
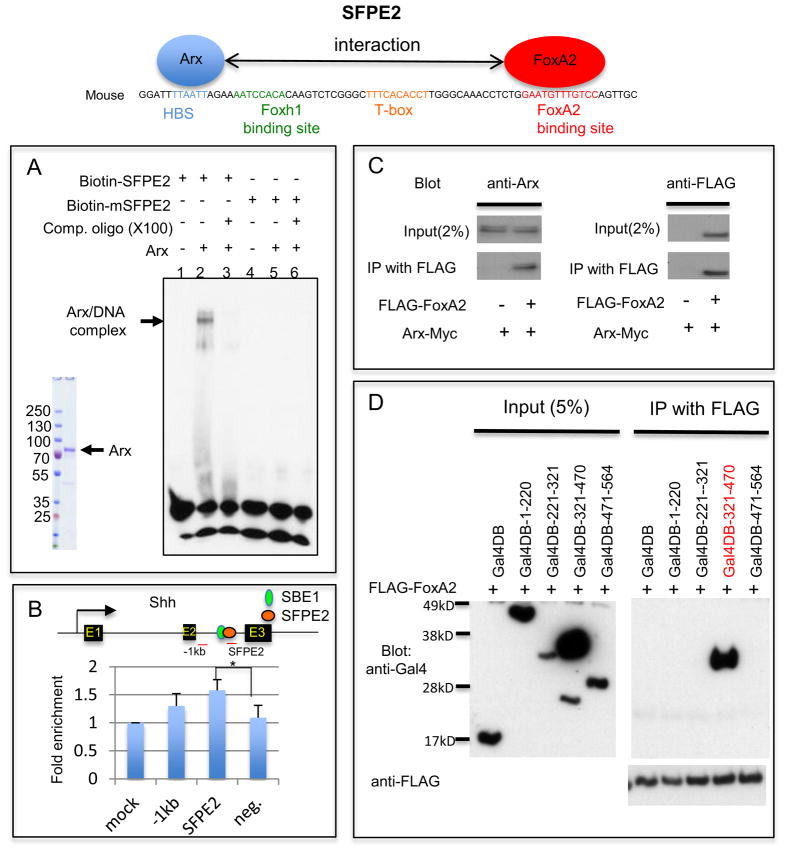
We next asked whether Arx modulates Shh expression through direct binding to its genomic regulatory sequence. Two enhancer regions have been reported for the FP expression of Shh: SFPE1 (Shh Floor Plate Enhancer1) and SFPE2 (Epstein et al., 1999; Jeong and Epstein, 2003). SFPE2 includes two sequence elements crucial for Shh induction; a homeodomain binding site (HBS) and a FoxA2 binding site separated by 51 base pairs (Jeong and Epstein, 2003). While FoxA2 binding in SFPE2 has been established, the homeodomain transcription factor(s) that binds to this HBS is unknown. We hypothesized that Arx, a paired-like homeodomain transcription factor, could bind to this cis-element and activate Shh expression in cooperation with FoxA2 in developing neural tube. In order to test this hypothesis, we performed electromobility shift assay (EMSA) and chromatin immunoprecipitation (ChIP). The Arx (Cho et al., 2012) bound directly to the HBS of SFPE2 and this binding exhibited specificity by cold competition, whereas a mutated SFPE2 fragment did not form a complex with Arx (Fig 3A). We verified this binding of Arx to the HBS in vivo by ChIP using embryonic spinal cords from E10.5 mouse and anti-Arx antibody. Our ChIP data confirmed Arx binding to the HBS of SFPE2 (Fig. 3B).
Fig. 3.
Arx binds to the HBS within Shh enhancer (SFPE2) and interacts with FoxA2. A) EMSA with the full-length Arx protein purified from transfected HEK293T cells and SFPE2 DNA fragments shows the DNA/protein complex (lane 2). The DNA/protein complex was reduced in the presence of unlabeled SFPE2 DNA competitor (lane 3). However, when the SFPE2 is mutated in the HBS (mSFPE2), it can no longer bind to Arx (lane 4–6). B) ChIP assay using developing spinal cord with either an anti-Arx antibody or control IgG. Enrichment of the PCR product in the immmunoprecipitated sample with Anti-Arx antibody was observed with three primer sets (one 1kb upstream region of SFPE2, a second in SFPE2 and the third a negative control which is in GAPDH locus) compared to control. (Note: ChIP was performed on whole spinal cords, of which less than 0.01% is FP.) Error bars correspond to SD (*, p=0.04, two-tailed, unpaired t-test). C) Arx directly interacts with FoxA2. HEK293T cells transfected with Arx-Myc and FoxA2-FLAG were used for immunoprecipitation (IP) experiments. IP with FLAG (FoxA2) antibody confirmed an interaction with Arx (Western blot with anti-Arx antibody). D) Arx interacts with FoxA2 through its homeodomain. HEK293T cells transfected with a series of Arx deletion mutants (aa 1–220, 221–321, 321–470 and 471–564 conjugated to Gal4DB) with FLAG-FoxA2, were used for IP. An Arx construct containing the homeodomain, Gal4DB-321-470 (red), was co-immunoprecipitated with FoxA2, whereas other constructs were not. The arrows identify the full-length protein of each mutant.
The HBS and FoxA2 binding site in the SFPE2 are very near each other (approximately 50bp apart) and FoxA2 is known to interact with other homeodomain transcription factors (Foucher et al., 2003; Rausa et al., 2003). This raised the possibility that Arx might physically interact with FoxA2. To test this possibility, we performed immunoprecipitation experiment using HEK293 cells, which were co-transfected with FLAG-FoxA2 and Arx-Myc. Arx was co-immunoprecipitated with FoxA2, confirming an interaction between Arx and FoxA2 (Fig. 3C). We further investigated which domain of Arx can interact with FoxA2, using a series of Arx deletion constructs, each fused with Gal4DB for nuclear targeting. A construct containing the homeodomain, Gal4DB-321-470, was co-immunoprecipitated with FoxA2, whereas constructs that did not contain the homeodomain were not co-immunoprecipitated (Fig 3D). These data indicate that the Arx homeodomain is required for an interaction with FoxA2. Taken together, our data support a model where Arx binds to the HBS sequence in SFPE2 and interacts with FoxA2, which binds to an adjacent site in SFPE2, to directly induce Shh expression. We postulate the interaction with FoxA2 for Shh induction is cooperative.
The transcriptional activation domain (aristaless domain) of Arx is required for Shh induction
Arx is known to function both as a transcriptional repressor and activator (Colasante et al., 2009; Collombat et al., 2003; Fullenkamp and El-Hodiri, 2008; Fulp et al., 2008; McKenzie et al., 2007). It contains two strong repression domains (one in the octapeptide domain at the N-terminal and the other in the region, aa400-495) and one activation domain, the aristaless domain at the C-terminus.) (McKenzie et al., 2007). Recent studies indicate Arx functions predominantly as a transcriptional repressor (Colasante et al., 2009; Collombat et al., 2003; Fulp et al., 2008; Quille et al., 2011). The octapeptide domain has been shown to bind to Groucho/transducing-like enhancer of split (TLE) cofactor, and repress transcription (McKenzie et al., 2007). How Arx functions as a transcriptional activator, however, is poorly understood.
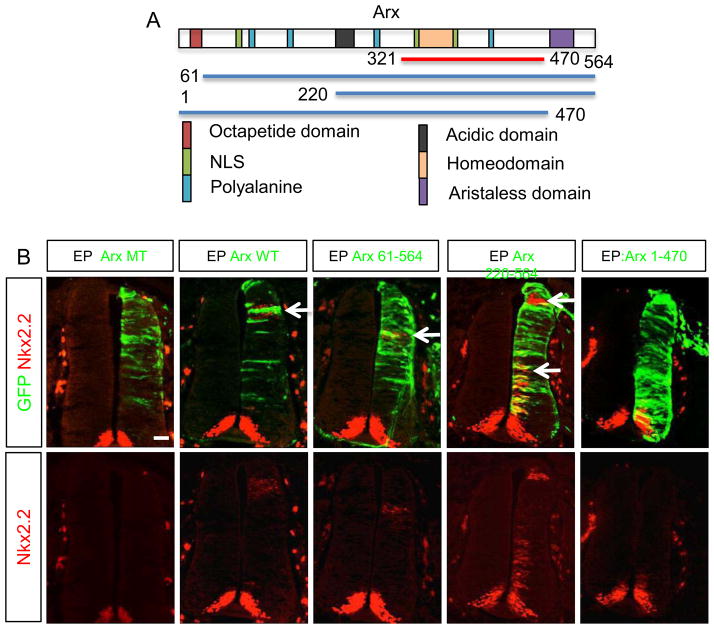
To determine whether the repression or the putative activation domain of Arx is crucial for Shh induction, we generated three Arx deletion constructs lacking the octapeptide domains for transcriptional repression (Arx 61–564 and Arx 220–564) and one lacking the C-terminal aristaless domain for transcriptional activation (Arx 1–470). All three mutants are expected to bind DNA and interact with FoxA2 since the homeodomain, which binds DNA, and the FoxA2 binding motif remains intact. These were independently co-electroporated with FoxA2 into the chick spinal cord (HH stage 10–12), and the expression of Nkx2.2, a downstream target gene of Shh, was assayed. While deletion of the known N-terminal repression domain (1–220 amino acids) did not affect Nkx2.2 expression, the Arx1-470 construct, which lacks the C-terminal aristaless domain failed to induce Nkx2.2 cell non-autonomously (Fig. 4). We believe the reason that Arx 220–564, with FoxA2, can better induce Nkx2.2 result from the loss of the repression domain in Arx. These data suggest that the C-terminal aristaless domain for the activation is required to induce Shh expression.
Fig. 4.
The transcriptional activation domain (aristaless domain) of Arx is required for induction of Nkx2.2. A) The schematic diagram depicts a full length Arx (top; rectangle) with the color-coded functional domains (e.g. NLS: nuclear localization signal), and Arx deletion mutants (bottom; lines) with beginning and ending amino acids indicated with numeric numbers (321–470 indicates the Arx fragment containing homeodomain used in Fig 3D). B) Electroporation of Arx or Arx deletion constructs to the chick spinal cords. Green fluorescence is an indicator of electroporated cells and Nkx2.2 immunostaining is shown in red. ArxMT means non-DNA bound homeodomain mutant (R332H). The arrows indicate examples of where Nkx2.2 induction was observed. Note that Arx 1–470 shows no Nkx2.2 induction. Scale bar is 50 μm for all images.
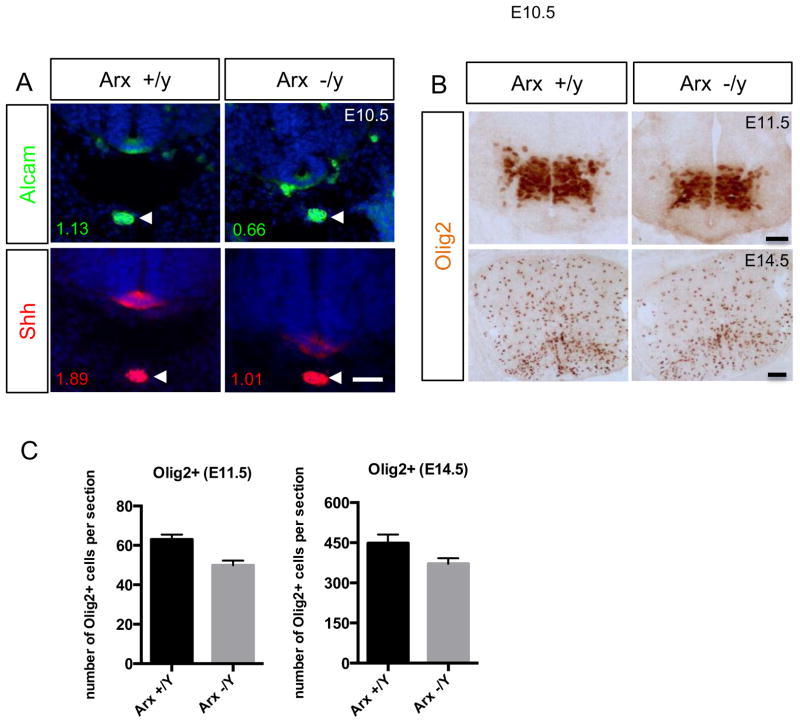
Shh expression is diminished in the spinal cord of Arx-deficient mice
Given our gain-of-function data in the chick embryo revealing Arx as a positive regulator of Shh induction, we next asked whether the endogenous loss of Arx would lead to a reduction in Shh expression and signaling. To test this, we examined the expressions of Shh and its downstream targets in the spinal cord of Arx−/y mice. The intensity and overall area of Shh immunostaining in the FP of Arx−/y (E10.5) mice was reduced when compared to control littermates (Fig. 5A). Although the change was rather mild, Shh level in the FP relative to that in the notochord was clearly reduced along the entire spinal cord in Arx−/y mice compared to control (0.5 in Arx+/y; 0.43 in Arx−/y) (n=3 mice). Similarly, the expression of FoxA2 and Alcam (another FP marker; (Schubert and Kaprielian, 2001) in the ventral spinal cord were all reduced (Fig. 5A and supplementary Fig. 1). These changes in gene expression level during early development persisted through later stages as shown with Olig2 (another Shh induced gene) immunostaining. Fewer Olig2 positive cells were detected in mutant spinal cords examined at E11.5 (labeling motor neuron; Roelink et al., 1994) and at E14.5 (labeling oligodendrocytes; Ligon et al., 2006; Rowitch, 2004) (Fig 5B). Notably, despite reduced levels of Nkx2.2 and FoxA2, likely due to reduction in Shh expression, we observed no change or expansion in the extent of the Nkx2.2 domain to the FP (supplementary Fig. 1), suggesting Arx does not repress Nkx2.2 expression. These data in Arx mutant mice are consistent with our findings in chick.
Fig. 5.
Shh expression in the FP is reduced in Arx-deficient mice. A) Shh immunostaining (bottom) in the FP of the spinal cord, but not in the notochord (arrowhead) is reduced in Arx−/y mice compared to Arx+/y. Note a similar reduction is detected in Alcam (another FP marker) immunostaining. The numbers below each image indicate the quantitative intensity of the signal in the FP relative to the signal in the notochord. The intensity is measured by area intensity of plot profile in Image J. B) Olig2, Shh induced gene, immunostaining show reduced expression in the mutant mouse spinal cord at E11.5 (top) and E14.5 (bottom). C) The quantification of the Olig2 immunostaining (representative images are shown in B) demonstrate that the number of Olig2+ cells is reduced in the mutant mouse spinal cord at E11.5 and E14.5 (n=3 mice per each). Error bars represent the SEM (p=0.0014 for E11.5 and p=0.086 for e14.5, two-tailed, unpaired t-test). Scale bars are 50 μm.
FoxA2 can induce Arx expression
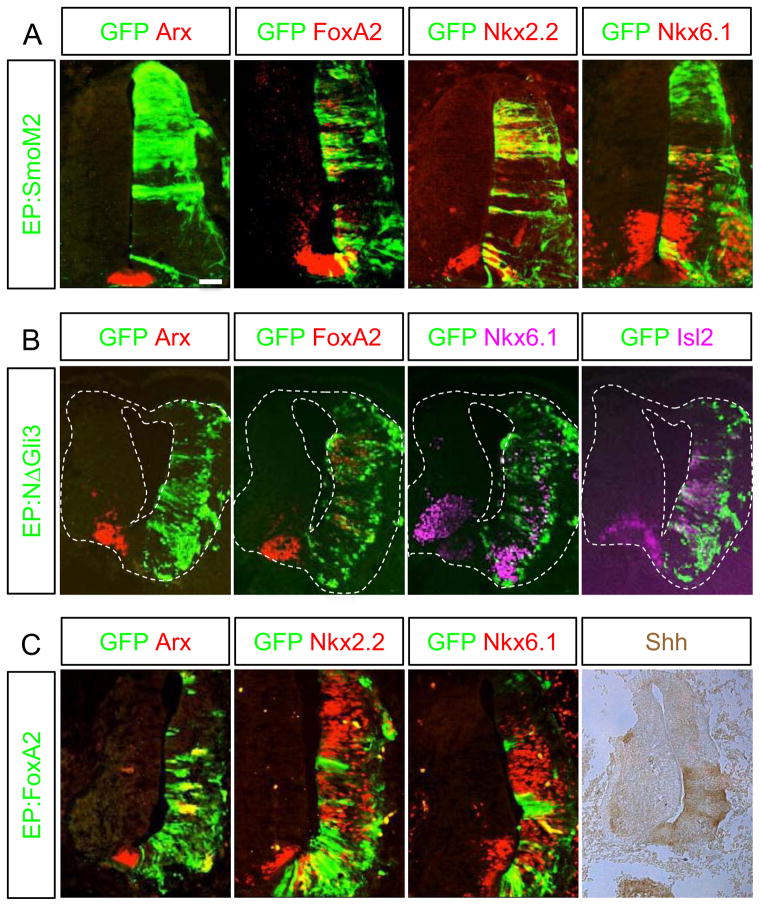
We next sought to determine how Arx expression was regulated in the FP. We introduced the constitutively active Shh receptor, SmoM2 (Xie et al., 1998), to the chick spinal cord (HH stage 10–12) by electroporation and examined the effect of ectopically induced Shh signaling. No induction of Arx was observed, while the known Shh target genes, Nkx6.1, Nkx2.2, and FoxA2, were ectopically induced (Fig. 6A). Next we electroporated the mutant Gli3 lacking its N-terminal repressor domain (ΔN-Gli3) (Lei et al., 2004), which should act as an activator of Shh signaling (Fig. 6B). ΔN-Gli3 disrupted neural tube structure on the electroporated side, suggesting that its effect is stronger than that of SmoM2, which did not affect neural tube morphology. Nonetheless, consistent with the SmoM2 result, forced expression of ΔN-Gli3 also did not induce Arx, while it successfully induced Nkx6.1, Isl2, and FoxA2 in cell-autonomous manner (Fig. 6B). Together, our data suggest that Shh signaling does not induce Arx in the FP.
Fig. 6.
Arx expression is induced by FoxA2 but not by Shh signaling. Electroporation of SmoM2 (A), ΔN-Gli3 (B) and FoxA2 (C) into the chick neural tube was followed by analysis for Arx, FoxA2, Nkx2.2, Nkx6.1 and Isl2 expression. FoxA2 electroporation induced the ectopic expression of Arx, Nkx2.2, Nkx6.1 and Shh. However, electroporation of SmoM2 or ΔN-Gli3, both activators of Shh signaling induced Nkx2.2 and Nkx6.1, or Nkx6.1 and Isl2, respectively but failed to induce Arx expression. Scale bar in A is 50 μm.
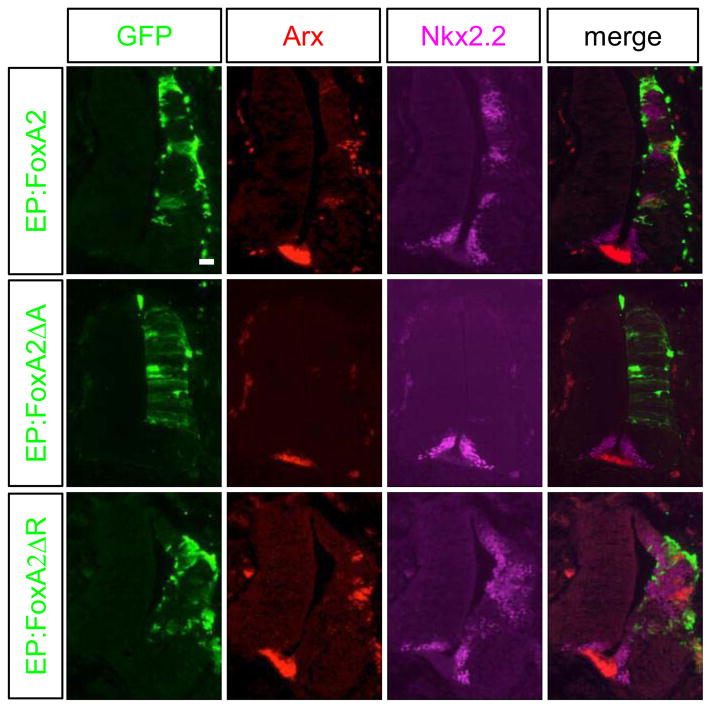
We next set out to examine the role of FoxA2 in Arx induction. FoxA2 has been previously reported to induce Arx (Ribes et al., 2010). We confirmed that electroporation of a FoxA2 expression construct into the chick spinal cord induced Arx cell autonomously, as well as Nkx2.2, Nkx6.1 and Shh (Fig 6C and Fig 7). Furthermore, co-electroporation of FoxA2 with either Ptch1Δloop2 or Gli3 in the chick spinal cord confirmed that FoxA2 induces Arx expression even when Shh signaling is blocked (Supplementary Fig 2). FoxA2 harbors both transcription activation and repression domains (Costa and Grayson, 1991; Qian and Costa, 1995; Rausa et al., 2003). Whether Arx induction by FoxA2 is mediated by its activation domain, or repression domain, is currently unknown. To determine which domain is responsible for Arx induction, we electroporated mutant FoxA2 constructs lacking either the activation or the repression domain. Forced expression of FoxA2 mutant construct lacking the activation domain (FoxA2ΔA) failed to induce Arx expression, whereas a construct lacking the repression domain (FoxA2ΔR) still induced Arx expression (Fig. 7). These results indicate that FoxA2 transcription activation domain is required for Arx induction.
Fig. 7.
The ability of FoxA2 to induce Arx expression is dependent on its activation domain. Constructs containing FoxA2, FoxA2ΔA (transcription activation domain deleted) or FoxA2ΔR (transcription repression domain deleted) were electroporated into the developing chick neural tube. Both the FoxA2 and the FoxA2ΔR expression constructs were able to induce the expression of Arx and Nkx2.2. In contrast, FoxA2ΔA failed to induce their expression. Scale bar in upper left panel is 50 μm.
Nkx2.2 can repress Arx expression
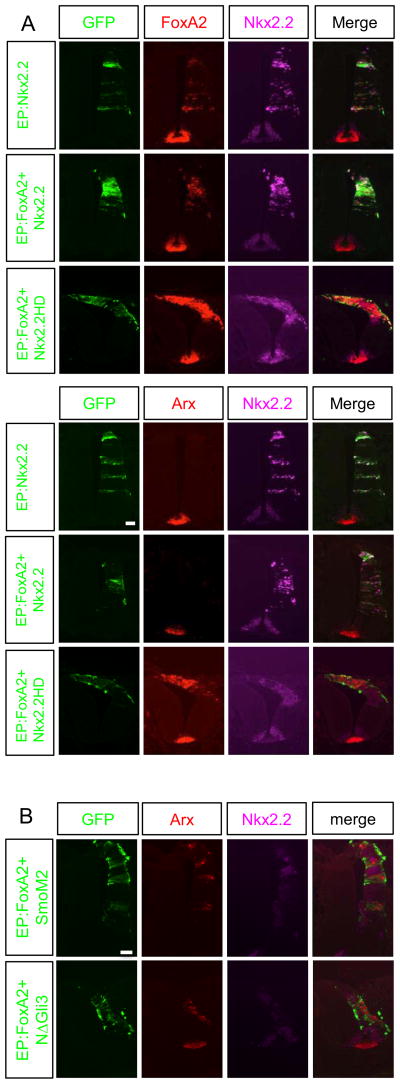
The fact that Shh induces FoxA2 and FoxA2 induces Arx expression, but Shh could not induce Arx, seemed incongruent. A further understanding of this apparent contradiction came through our observation of the functional relationships between Arx and Nkx2.2 (a Shh induced gene). Forced expression of FoxA2 can induce Nkx2.2 non-cell autonomously, while it induces Arx cell autonomously (Supplementary Fig. 3) (Ribes et al., 2010). Conversely, forced expression of Nkx2.2 induced FoxA2 expression (Fig. 8A; top left rows). Interestingly, this ectopic expression of FoxA2 by Nkx2.2 electroporation did not induce Arx (Fig. 8A; top right rows). Two possible explanations for this observation; 1) the FoxA2 levels induced by Nkx2.2 electroporation are not sufficient to induce Arx, or 2) Nkx2.2 might inhibit Arx induction. To distinguish these possibilities, we electroporated FoxA2 together with Nkx2.2 (DNA amount 1:1) to provide sufficient FoxA2 levels, but again did not detect Arx induction. This result suggests that the induction of Arx is not dependent on the level of FoxA2 expression. To determine if Nkx2.2 can repress Arx induction, we generated mutant form of Nkx2.2 that contains the homeodomain only (Nkx2.2HD), lacking the known transcriptional repression domain for co-electroporation with FoxA2 (Muhr et al., 2001; Watada et al., 2000). Furthermore, the repression domain in Nkx2.2 is required for Shh induction through a gene regulatory network. Nkx2.2 induces Shh, and therefore FoxA2, by repressing Pax6. Pax6 induces the Shh repressor Gli3 and thus Nkx2.2 induces Shh by repressing the expression of Gli3 (Lek et al., 2010). As we expected, the Nkx2.2HD mutant does not induce FoxA2 due to the loss of Nkx2.2 repression domain (Supplementary Fig. 4). Surprisingly, the FoxA2-induced Arx expression was no longer abolished when the Nkx2.2HD was used for FoxA2 co-electroporation, demonstrating that Nkx2.2 transcription repression function is required for abolishing FoxA2-induced Arx expression (Fig. 8A; bottom rows). Based on these observations, we argue that the reason Shh signaling does not induce Arx, despite the fact that it can induce FoxA2, is due to the Nkx2.2 related repression of Arx. This is consistent with the fact that Arx is not expressed in the early FP where Nkx2.2 is present, but only expressed in the later FP when Nkx2.2 is no longer expressed. Further evidence to support this mechanism is found in the results of our experiments ectopically expressing FoxA2. Arx and Nkx2.2 expression was always mutually exclusive (Supplementary Fig. 3). These data are similar to the mechanistic role Arx plays in pancreatic beta-cell. Nkx2.2 represses Arx in these cells to maintain beta cell identity. This repression is achieved through the recruitment of repression complex including Grg3, HDAC1 and DNMT3 (Papizan et al., 2012). We believe a similar repression mechanism may play a role in spinal cord FP cell specification.
Fig. 8.
Nkx2.2 represses Arx expression. A) Nkx2.2, FoxA2+Nkx2.2 or FoxA2 + Nkx2.2HD (homeodomain only) electroporation into the developing chick spinal cord. Nkx2.2 alone was able to induce FoxA2, but not Arx. The ability of FoxA2 to induce Arx is suppressed by forced expression of Nkx2.2 but not by the homeodomain only mutant form of Nkx2.2 (Nkx2.2HD). B) SmoM2 or ΔN-Gli3 co-electroporation with FoxA2 into the developing chick spinal cord. SmoM2 and ΔN-Gli3 failed to suppress the ability of FoxA2 to induce expression of Arx. Scale bars in upper left panel of A and B are 50 μm.
Interestingly, when SmoM2 or ΔN-Gli3 (activators of Shh signaling) was co-electroporated with FoxA2, they do not abolish FoxA2-induced Arx expression (Fig. 8B), suggesting that activated Shh signaling does not directly repress Arx induction. However, SmoM2 or ΔN-Gli3 can ectopically induce Nkx2.2, and Nkx2.2 positive cells do not express Arx (Fig. 8B), supporting our finding that Nkx2.2 represses FoxA2-induced Arx expression.
Discussion
Shh plays a critical role in spinal cord development as a morphogen for orchestrating cell type specification and maintenance, along with also functioning as a chemoattractant for commissural axon guidance (Bourikas et al., 2005; Dessaud et al., 2008). To serve these functions for normal spinal cord development, Shh expression requires tight spatial and temporal regulation. Although the function of Shh in spinal cord development has been intensively studied, the regulation of Shh expression in the FP is incompletely understood. To our knowledge, FoxA2 is the only known transcription factor shown to directly bind to one of the Shh enhancers and activate its expression in the spinal cord (Jeong and Epstein, 2003). Here we show that another transcription factor, Arx, in collaboration with FoxA2, directly induces Shh expression. It binds to the homeobox element, which is located adjacent to FoxA2 binding site of the Shh FP enhancer (SFPE2) that drives FP specific expression in the spinal cord. Our data support a model wherein Arx and FoxA2 physically interact while binding at the SFPE2 element and this cooperative binding regulates Shh transcription. Furthermore, FoxA2 induces Arx and this induction is repressed by Nkx2.2, suggesting a complex feedback loop exists between FoxA2, Arx and Shh signaling (Fig 9).
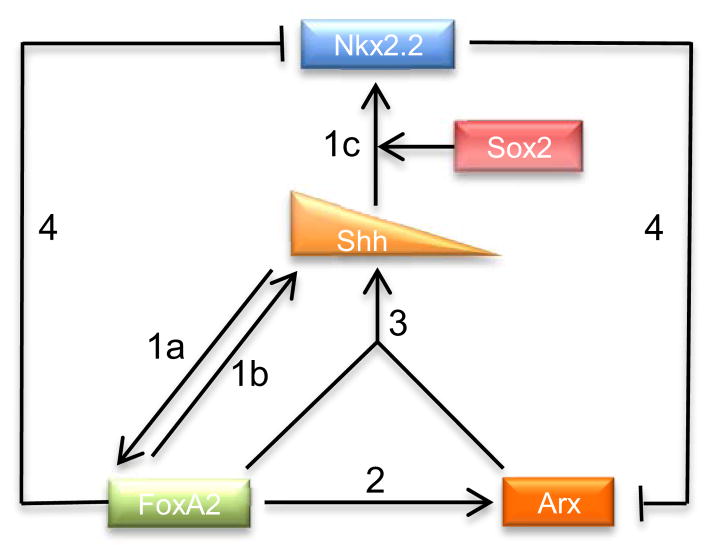
Fig. 9.
Schematic illustration of the proposed gene regulatory network in FP development. 1a) Shh from the notochord induces FoxA2 in the FP. 1b) FoxA2 from the FP in turn activates Shh induction in the FP. 1c) Shh also induces Nkx2.2, and Sox2 is required for this induction. 2) FoxA2 turns on Arx expression in the FP. 3) FoxA2 and Arx cooperatively activates Shh expression. 4a)–4b) FoxA2 represses Nkx2.2 and Nkx2.2 inhibits Arx expression, in a cell autonomous manner.
Arx collaborates with FoxA2 to regulate Shh expression in the floor plate
Shh expression along the anteroposterior axis of the mouse central nervous system is regulated by multiple enhancers. To date six enhancers distributed over 400kb, both upstream and downstream of the transcription initiation site, have been identified (Epstein et al., 1999; Jeong et al., 2006; Jeong and Epstein, 2003). Among these six enhancers, two Shh FP-specific enhancers, SFPE1 and SFPE2, are known to regulate Shh expression in the spinal cord. While SFPE1 is sufficient for directing Shh expression in the FP, SFPE2 works cooperatively with another enhancer, Shh brain enhancer 1 (SBE1), for its FP specific enhancer function. SFPE2 and SBE1 are intronic enhancers located adjacent to each other and within intron 2 of the Shh gene. Comparative sequence analysis among different species identified four highly conserved sequence elements (transcription factor binding sites) within SFPE2: HBS (homeobox transcription factor binding site), Foxh1 binding site, T-box binding site, and FoxA binding site. Further functional analysis in mice revealed that the HBS is required for driving Shh expression in the FP of the spinal cord and the FoxA binding site directs expression in the FP and notochord. In contrast, the Foxh1 binding site appears unnecessary and the T-box binding site drives repression. Interestingly, either HBS or FoxA binding site is not sufficient for SFPE2 activity on their own, but cooperative interaction is required between these two sites (Jeong and Epstein, 2003). No homeodomain protein has previously been identified that could bind to this HBS. Nkx2 or Nkx6 family members were considered candidates (Jeong and Epstein, 2003), however members of these families are not expressed in the FP when Shh gene is induced and thus are not considered good candidates. Our data in this study identifies Arx as the homeodomain transcription factor that binds to this HBS of the SFPE2, and in collaboration with FoxA2 regulates Shh gene expression in the spinal cord FP.
Considering the importance of Shh function in the spinal cord development, it is not surprising to find multiple and complex regulatory controls to direct and maintain precise levels of Shh expression. Since Shh expression in the ventral spinal cord is regulated by more than one enhancer, and SFPE1 (FoxA2 or Arx independent) alone is sufficient for Shh expression in the FP (Epstein et al., 1999), it is not surprising to observe only minimal changes in Shh expression in the Arx-deficient mice. In fact, even mild changes in Shh and downstream signaling in Arx-deficient mice suggest a significant involvement of Arx in a regulatory network of Shh expression. One sensitive measure of Shh signaling is to examine the downstream cell types generated by specific Shh concentrations, such as motor neuron- and oligodendrocyte progenitors (Allen et al., 2011; 2007; Dessaud et al., 2010; Ericson et al., 1996; Yu et al., 2013). Our data showing a mild reduction in motor neurons and oligodendrocytes (Olig2 positive cells) in Arx-deficient mice provides further evidence that loss of Arx is responsible for a perturbation in Shh signaling.
Arx is part of a complex gene regulatory network controlling the expression of intrinsic as well as extrinsic factors of the floor plate development
The FP specification involves the interplay between extrinsic signaling molecule, Shh, and intrinsic transcription factors such as FoxA2. Initially, Shh from the notochord can induce FoxA2 expression in the FP, and FoxA2 in turn induces Shh expression in the FP. Although the initial specification of the FP requires Shh signaling, the maintenance of the FP identity becomes no longer dependent on its signaling, as Ptch1, Hhip, Gli1, Gli3 and Nkx2.2 are directly or indirectly repressed by FP FoxA2 or GliR (Metzakopian et al., 2012; Peterson et al., 2012; Vokes et al., 2007), which results in attenuation of Shh signaling. While the FP cells down regulate Shh signaling to maintain their identity, Shh expression itself does not decrease in the FP, which can ensure enough production and secretion of Shh for the specification of adjacent cell populations such as V3 neurons and motor neurons from the p3 and pMN domains, respectively. Thus, it is important to understand how Shh expression can be maintained in the FP while Shh signaling is down regulated.
Our studies add Arx to the network of known transcription factors regulating and maintaining Shh expression in the FP. Conversely, whether Shh signaling can regulate Arx expression or not turns out to be a little more complicated. Our data show that activated Shh signaling, via SmoM2 or ΔN-Gli3 electroporation in the neural tube, does not induce Arx (Fig. 6). These results seem inconsistent with the previous report documenting Arx induction by Shh treatment in the naive neural plate explants (Ribes et al., 2010). This discrepancy suggests that the induction of Arx by Shh might have a temporal restriction. It is possible that at earlier stage (neural plate stage), Shh can induce Arx because Nkx2.2, which can repress Arx, is not present yet due to the absence of Sox2 which is necessary for induction of Nkx2.2 (Peterson et al., 2012). At a later stage (neural tube stage, which was used in our study), however, Shh signaling does not induce Arx since Nkx2.2 is available to repress Arx induction, as we showed in this study.
In summary, our studies have extended our understanding of the complex gene regulatory network involving intrinsic and extrinsic factors in FP development. We have provided evidence that Arx is a cooperating transcription factor with FoxA2 for Shh expression in the FP and that loss of Arx results in mild but significant decrease in Shh and Shh signaling. Our results suggest that the transcriptional co-activation function of ARX might be a key to understand the pathogenesis of human patients with ARX mutations. Given that many phenotypes in these patients are diverse and subtle, the result of this cooperative role of ARX in gene induction rather than a unique and independent role, provides a potential mechanistic insight into how this diverse spectrum of phenotypes could be observed.
Supplementary Material
Highlights.
Arx and FoxA2 collaborate in the floor plate of the neural tube to modulate Shh expression.
FoxA2 activates and Nkx2.2 represses Arx expression.
Arx plays a role in a gene regulatory network to control and maintain Shh expression and a proper morphogen gradient.
Acknowledgments
We thank Dr. M. P. Matise (Rutgers University) for Ptch1Δloop2, SmoM2 and ΔN-Gli3 expression constructs and Dr. D. Epstein (University of Pennsylvania) for discussion. This work was supported by NIH grant NS46616.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen BL, Song JY, Izzi L, Althaus IW, Kang JS, Charron F, Krauss RS, McMahon AP. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev Cell. 2011;20:775–787. doi: 10.1016/j.devcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BL, Tenzen T, McMahon AP. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 2007;21:1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. Gene Regulatory Logic for Reading the Sonic Hedgehog Signaling Gradient in the Vertebrate Neural Tube. Cell. 2012;148:273–284. doi: 10.1016/j.cell.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourikas D, Pekarik V, Baeriswyl T, Grunditz A, Sadhu R, Nardó M, Stoeckli ET. Sonic hedgehog guides commissural axons along the longitudinal axis of the spinal cord. Nat Neurosci. 2005;8:297–304. doi: 10.1038/nn1396. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Chamberlain CE, Jeong J, Guo C, Allen BL, McMahon AP. Notochord-derived Shh concentrates in close association with the apically positioned basal body in neural target cells and forms a dynamic gradient during neural patterning. 2008;135:1097–1106. doi: 10.1242/dev.013086. [DOI] [PubMed] [Google Scholar]
- Cho G, Nasrallah MP, Lim Y, Golden JA. Distinct DNA binding and transcriptional repression characteristics related to different ARX mutations. Neurogenetics. 2012;13:23–29. doi: 10.1007/s10048-011-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante G, Sessa A, Crispi S, Calogero R, Mansouri A, Collombat P, Broccoli V. Arx acts as a regional key selector gene in the ventral telencephalon mainly through its transcriptional repression activity. Developmental Biology. 2009;334:59–71. doi: 10.1016/j.ydbio.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Colasante G, Simonet JC, Calogero R, Crispi S, Sessa A, Cho G, Golden JA, Broccoli V. ARX Regulates Cortical Intermediate Progenitor Cell Expansion and Upper Layer Neuron Formation Through Repression of Cdkn1c. Cerebral Cortex. 2013 Aug 22; doi: 10.1093/cercor/bht222. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Mansouri A, Hecksher-Sorensen J, Serup P, Krull J, Gradwohl G, Gruss P. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17:2591–2603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E, Galli R, Cossu G, Gecz J, Broccoli V. Mouse orthologue of ARX, a gene mutated in several X-linked forms of mental retardation and epilepsy, is a marker of adult neural stem cells and forebrain GABAergic neurons. Dev Dyn. 2004;231:631–639. doi: 10.1002/dvdy.20164. [DOI] [PubMed] [Google Scholar]
- Costa RH, Grayson DR. Site-directed mutagenesis of hepatocyte nuclear factor (HNF) binding sites in the mouse transthyretin (TTR) promoter reveal synergistic interactions with its enhancer region. Nucleic Acids Research. 1991;19:4139–4145. doi: 10.1093/nar/19.15.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. A Genomic Regulatory Network for Development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Dessaud E, Ribes V, Balaskas N, Yang LL, Pierani A, Kicheva A, Novitch BG, Briscoe J, Sasai N. Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen sonic hedgehog. PLoS Biol. 2010;8:e1000382. doi: 10.1371/journal.pbio.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E, Yang LL, Hill K, Cox B, Ulloa F, Ribeiro A, Mynett A, Novitch BG, Briscoe J. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450:717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- Epstein DJ, McMahon AP, Joyner AL. Regionalization of Sonic hedgehog transcription along the anteroposterior axis of the mouse central nervous system is regulated by Hnf3-dependent and -independent mechanisms. Development. 1999;126:281–292. doi: 10.1242/dev.126.2.281. [DOI] [PubMed] [Google Scholar]
- Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell. 1996;87:661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- Foucher I, Montesinos ML, Volovitch M, Prochiantz A, Trembleau A. Joint regulation of the MAP1B promoter by HNF3beta/Foxa2 and Engrailed is the result of a highly conserved mechanism for direct interaction of homeoproteins and Fox transcription factors. Development. 2003;130:1867–1876. doi: 10.1242/dev.00414. [DOI] [PubMed] [Google Scholar]
- Friocourt G, Parnavelas JG. Mutations in ARX Result in Several Defects Involving GABAergic Neurons. Front Cell Neurosci. 2010;4:4. doi: 10.3389/fncel.2010.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullenkamp AN, El-Hodiri HM. The function of the Aristaless-related homeobox (Arx) gene product as a transcriptional repressor is diminished by mutations associated with X-linked mental retardation (XLMR) Biochem Biophys Res Commun. 2008;377:73–78. doi: 10.1016/j.bbrc.2008.09.116. [DOI] [PubMed] [Google Scholar]
- Fulp CT, Cho G, Marsh ED, Nasrallah IM, Labosky PA, Golden JA. Identification of Arx transcriptional targets in the developing basal forebrain. Human Molecular Genetics. 2008;17:3740–3760. doi: 10.1093/hmg/ddn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Epstein DJ. Distinct regulators of Shh transcription in the floor plate and notochord indicate separate origins for these tissues in the mouse node. Development. 2003;130:3891–3902. doi: 10.1242/dev.00590. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kato M, Das S, Petras K, Kitamura K, Morohashi KI, Abuelo DN, Barr M, Bonneau D, Brady AF, Carpenter NJ, Cipero KL, Frisone F, Fukuda T, Guerrini R, Iida E, Itoh M, Lewanda AF, Nanba Y, Oka A, Proud VK, Saugier-Veber P, Schelley SL, Selicorni A, Shaner R, Silengo M, Stewart F, Sugiyama N, Toyama J, Toutain A, Vargas AL, Yanazawa M, Zackai EH, Dobyns WB. Mutations of ARX are associated with striking pleiotropy and consistent genotype-phenotype correlation. Hum Mutat. 2004;23:147–159. doi: 10.1002/humu.10310. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K, Matsuo M, Kamijo SI, Kasahara M, Yoshioka H, Ogata T, Fukuda T, Kondo I, Kato M, Dobyns WB, Yokoyama M, Morohashi KI. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- Lei Q, Zelman AK, Kuang E, Li S, Matise MP. Transduction of graded Hedgehog signaling by a combination of Gli2 and Gli3 activator functions in the developing spinal cord. Development. 2004;131:3593–3604. doi: 10.1242/dev.01230. [DOI] [PubMed] [Google Scholar]
- Lek M, Dias JM, Marklund U, Uhde CW, Kurdija S, Lei Q, Sussel L, Rubenstein JL, Matise MP, Arnold HH, Jessell TM, Ericson J. A homeodomain feedback circuit underlies step-function interpretation of a Shh morphogen gradient during ventral neural patterning. Development. 2010;137:4051–4060. doi: 10.1242/dev.054288. [DOI] [PubMed] [Google Scholar]
- Ligon KL, Fancy SPJ, Franklin RJM, Rowitch DH. Olig gene function in CNS development and disease. Glia. 2006;54:1–10. doi: 10.1002/glia.20273. [DOI] [PubMed] [Google Scholar]
- Lim Y, Cho G, Minarcik J, Golden J. Altered BMP signaling disrupts chick diencephalic development. Mech Dev. 2005;122:603–620. doi: 10.1016/j.mod.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Marsh E, Fulp C, Gomez E, Nasrallah I, Minarcik J, Sudi J, Christian SL, Mancini G, Labosky P, Dobyns W, Brooks-Kayal A, Golden JA. Targeted loss of Arx results in a developmental epilepsy mouse model and recapitulates the human phenotype in heterozygous females. Brain. 2009;132:1563–1576. doi: 10.1093/brain/awp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie O, Ponte I, Mangelsdorf M, Finnis M, Colasante G, Shoubridge C, Stifani S, Gécz J, Broccoli V. Aristaless-related homeobox gene, the gene responsible for West syndrome and related disorders, is a Groucho/transducin-like enhancer of split dependent transcriptional repressor. Neuroscience. 2007;146:236–247. doi: 10.1016/j.neuroscience.2007.01.038. [DOI] [PubMed] [Google Scholar]
- Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Metzakopian E, Lin W, Salmon-Divon M, Dvinge H, Andersson E, Ericson J, Perlmann T, Whitsett JA, Bertone P, Ang SL. Genome-wide characterization of Foxa2 targets reveals upregulation of floor plate genes and repression of ventrolateral genes in midbrain dopaminergic progenitors. Development. 2012;139:2625–2634. doi: 10.1242/dev.081034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mégarbané A, Chouery E, Mignon-Ravix C, El Sabbagh S, Corbani S, Ghoch JA, Jalkh N, Mehawej C, Lévy N, Villard L. Ambiguous genitalia, microcephaly, seizures, bone malformations, and early death: a distinct MCA/MR syndrome. Am J Med Genet A. 2011;155A:1147–1151. doi: 10.1002/ajmg.a.33938. [DOI] [PubMed] [Google Scholar]
- Miura H, Yanazawa M, Kato K, Kitamura K. Expression of a novel aristaless related homeobox gene “Arx” in the vertebrate telencephalon, diencephalon and floor plate. Mech Dev. 1997;65:99–109. doi: 10.1016/s0925-4773(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Nasrallah MP, Cho G, Putt ME, Kitamura K, Golden JA. Differential effects of a polyalanine tract expansion in Arx on neural development and gene expression. Human Molecular Genetics. 2011 doi: 10.1093/hmg/ddr538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivetti PR, Noebels JL. Interneuron, interrupted: molecular pathogenesis of ARX mutations and X-linked infantile spasms. Curr Opin Neurobiol. 2012;22:859–865. doi: 10.1016/j.conb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterveen T, Kurdija S, Alekseenko Z, Uhde CW, Bergsland M, Sandberg M, Andersson E, Dias JM, Muhr J, Ericson J. Mechanistic differences in the transcriptional interpretation of local and long-range shh morphogen signaling. Dev Cell. 2012;23:1006–1019. doi: 10.1016/j.devcel.2012.09.015. [DOI] [PubMed] [Google Scholar]
- Pani L, Overdier DG, Porcella A, Qian X, Lai E, Costa RH. Hepatocyte nuclear factor 3 beta contains two transcriptional activation domains, one of which is novel and conserved with the Drosophila fork head protein. Mol Cell Biol. 1992;12:3723–3732. doi: 10.1128/mcb.12.9.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papizan JB, Singer RA, Tschen S, Dhawan S, Friel JM, Hipkens SB, Magnuson MA, Bhushan A, Sussel L. Nkx2.2 repressor complex regulates islet β-cell specification and prevents β-to-α-cell reprogramming. Genes Dev. 2012;25:2291–2305. doi: 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KA, Nishi Y, Ma W, Vedenko A, Shokri L, Zhang X, McFarlane M, Baizabal JM, Junker JP, van Oudenaarden A, Mikkelsen T, Bernstein BE, Bailey TL, Bulyk ML, Wong WH, McMahon AP. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 2012;26:2802–2816. doi: 10.1101/gad.207142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Costa RH. Analysis of hepatocyte nuclear factor-3 beta protein domains required for transcriptional activation and nuclear targeting. Nucleic Acids Research. 1995;23:1184–1191. doi: 10.1093/nar/23.7.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quille ML, Carat S, Quéméner-Redon S, Hirchaud E, Baron D, Benech C, Guihot J, Placet M, Mignen O, Férec C, Houlgatte R, Friocourt G. High-throughput analysis of promoter occupancy reveals new targets for Arx, a gene mutated in mental retardation and interneuronopathies. PLoS ONE. 2011;6:e25181. doi: 10.1371/journal.pone.0025181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausa FM, Tan Y, Costa RH. Association between hepatocyte nuclear factor 6 (HNF-6) and FoxA2 DNA binding domains stimulates FoxA2 transcriptional activity but inhibits HNF-6 DNA binding. Mol Cell Biol. 2003;23:437–449. doi: 10.1128/MCB.23.2.437-449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes V, Balaskas N, Sasai N, Cruz C, Dessaud E, Cayuso J, Tozer S, Yang LL, Novitch B, Marti E, Briscoe J. Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in the vertebrate neural tube. Genes Dev. 2010;24:1186–1200. doi: 10.1101/gad.559910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelink H, Augsburger A, Heemskerk J, Korzh V, Norlin S, Ruiz i Altaba A, Tanabe Y, Placzek M, Edlund T, Jessell TM. Floor plate and motor neuron induction by vhh-1, a vertebrate homolog of hedgehog expressed by the notochord. Cell. 1994;76:761–775. doi: 10.1016/0092-8674(94)90514-2. [DOI] [PubMed] [Google Scholar]
- Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Schubert W, Kaprielian Z. Identification and characterization of a cell surface marker for embryonic rat spinal accessory motor neurons. J Comp Neurol. 2001;439:368–383. doi: 10.1002/cne.1356. [DOI] [PubMed] [Google Scholar]
- Sherr EH. The ARX story (epilepsy, mental retardation, autism, and cerebral malformations): one gene leads to many phenotypes. Curr Opin Pediatr. 2003;15:567–571. doi: 10.1097/00008480-200312000-00004. [DOI] [PubMed] [Google Scholar]
- Shoubridge C, Fullston T, Gecz J. ARX spectrum disorders: making inroads into the molecular pathology. Hum Mutat. 2010;31:889–900. doi: 10.1002/humu.21288. [DOI] [PubMed] [Google Scholar]
- Strømme P, Mangelsdorf ME, Shaw MA, Lower KM, Lewis SME, Bruyere H, Lütcherath V, Gedeon AK, Wallace RH, Scheffer IE, Turner G, Partington M, Frints SGM, Fryns JP, Sutherland GR, Mulley JC, Gecz J. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet. 2002;30:441–445. doi: 10.1038/ng862. [DOI] [PubMed] [Google Scholar]
- Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, McMahon AP. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell. 2006;10:647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, Longabaugh WJR, Davidson EH, Wong WH, McMahon AP. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wang JC, Waltner-Law M, Yamada K, Osawa H, Stifani S, Granner DK. Transducin-like enhancer of split proteins, the human homologs of Drosophila groucho, interact with hepatic nuclear factor 3beta. J Biol Chem. 2000;275:18418–18423. doi: 10.1074/jbc.M910211199. [DOI] [PubMed] [Google Scholar]
- Watada H, Mirmira RG, Kalamaras J, German MS. Intramolecular control of transcriptional activity by the NK2-specific domain in NK-2 homeodomain proteins. Proc Natl Acad Sci USA. 2000;97:9443–9448. doi: 10.1073/pnas.97.17.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, de Sauvage FJ. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LMN, Mao M, Chan JR, Wu J, Lu QR. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.