Abstract
Purpose
MLH3, a MutL homolog protein in mammals playing a role in DNA mismatch repair, is associated with spermatogenesis and male infertility. The purpose of the present study was to investigate the association of the single-nucleotide polymorphism (SNP), rs 175080 in the MLH3 gene, with sperm parameters in a Greek population.
Methods
The study included 300 men of couples undergoing in vitro fertilization/intracytoplasmic sperm injection-embryo transfer (IVF/ICSI-ET) treatments (years 2011–2013). Genomic DNA was extracted from 300 peripheral blood samples, and conventional quantitative real-time PCR was performed for genotyping. Of them, 122 were from men used as “controls” and 178 from men used as “cases.” Allocation to the two groups was based on sperm concentrations (≥15 and <15 million/ml, respectively). Serum FSH, LH, estradiol, testosterone, and prolactin concentrations as well as sperm parameters were compared between three genotypes (GG, GA, and AA). Furthermore, the frequencies of these three genotypes were compared between “cases” and “controls.”
Results
Anthropometric parameters and hormonal values did not differ significantly between the three genotypes. Significantly lower sperm concentrations were found in men with the AA genotype as compared to men with the GG and GA genotypes (p < 0.001). The AA genotype had the lower progressive motility values as compared to the other two genotypes (p < 0.05). Also, there was a significantly different distribution of the frequencies of the three genotypes between “cases” and “controls” (p < 0.001).
Conclusions
It is suggested that the studied SNP in the MLH3 gene may be linked to oligozoospermia in Caucasian men of a certain area.
Keywords: MLH3 gene, Polymorphisms, Sperm parameters, Male infertility
Introduction
Infertility is generally accepted as the inability of a couple to achieve a pregnancy after at least 1 year of regular unprotected sexual intercourse. About 15 % of couples suffer from infertility with the male factor being responsible for almost half of the cases [1].
The routine use of genetic tests may diagnose a genetic factor of male infertility in about 15 % of cases, with karyotype abnormalities, Y chromosome microdeletions, and cystic fibrosis transmembrane conductance regulator (CFTR) gene mutation most commonly found [2].
Up to 2300 genes are involved in spermatogenesis and potentially each of them could be a specified testing in the diagnosis of male infertility. The currently considered idiopathic cases of infertility are possibly related to genetic and epigenetic factors, which may deteriorate spermatogenesis either alone or in combination with environmental factors. Accumulating evidence associates different polymorphisms in genes involved in spermatogenesis with male infertility.
MLH3 is a MutL homolog protein in mammals. It was found on 14q24.3 chromosome with a coding length of 4.3 kb [3]. Its basic role is in the DNA mismatch repair mechanism, while it has been proposed to play a distinct role in the meiotic recombination mechanism [4, 5]. Inactivation of the MLH3 gene has been suggested to play a role in both male and female infertility [3] since the presence of the MLH3 C2531T polymorphism leads to an increased risk for developing infertility [6]. Human MLH3 (hMLH3) gene may also be associated to spermatogenesis and male infertility, while MLH3-MLH1 pathway seems to play a key role in making crossovers during mammalian meiosis [7] since it appears that this pathway directs an unknown factor that resolves Holliday junction and intermediates into crossovers [8]. There are scanty data in humans, regarding the association between certain hMLH3 polymorphisms and male infertility [7, 9]; therefore, it is reasonable to assume that mutations and polymorphisms may impact negatively on spermatogenesis and subsequently on male fertility. Moreover, in a recent study, a decrease in the expression profile of the meiosis-involved mismatch repair genes was observed in patients with impaired spermatogenesis [10].
The aim of the present study was to investigate for the first time the association of single-nucleotide polymorphism (SNP) rs 175080 in the MLH3 gene with sperm parameters in the Caucasian race, such as in a Greek population, since it is one of the six polymorphisms in genes that is involved in DNA double-strand break repair and chromosome synapsis and associates with male infertility [11].
Materials and methods
A cohort of 300 men of infertile couples undergoing in vitro fertilization/intracytoplasmic sperm injection-embryo transfer (IVF/ICSI-ET) treatments in the years 2011 to 2013 inclusive were studied. All patients volunteered for the study and gave written informed consent, while Institutional Review Board approval of the study was also obtained.
The 300 men were divided into two groups, a group of 122 men serving as “controls” and a group of 178 men serving as “cases.” The allocation to the two groups was based on sperm concentrations, i.e., ≥15 and <15 million/ml, respectively. Genomic DNA was extracted from 300 peripheral blood samples obtained from all men, and conventional quantitative real-time PCR was performed for genotyping. Serum concentrations of FSH, LH, estradiol, testosterone, and prolactin were measured, while sperm parameters were analyzed. Three different genotypes, i.e., GG, GA, and AA were considered in the whole cohort.
DNA extraction
Genomic DNA extraction was performed with the use of the QIAamp DNA Blood Mini Kit (SafeBlood BioAnalytica SA, Greece). This kit purifies the genomic DNA in a rather automated procedure and processes sample sizes of up to 200 μl, with a preparation time of 20–40 min. The typical yield from 200 μl healthy whole blood is 4–12 μg, with an elution volume of 50–200 μl. Briefly, the procedure begins with the pipetting of 20 μl QIAGEN Protease (or proteinase K) and 200 μl of the sample into the bottom of a 1.5-ml microcentrifuge tube. The next step involves the addition of 200 μl Buffer AL to the previous mixture, and the whole mix is vortexed for 15 s. Following incubation at 56 °C for 10 min, the 1.5-ml microcentrifuge tube is centrifuged in order for the drops from the inside of the lid to be removed. Next, it is added 200 μl ethanol (96–100 %) to the sample and the mixture is vortexed for 15 s. The mixture is then centrifuged in order to remove drops from the inside of the lid. The produced mixture from the previous step is transferred to the QIAamp Mini spin column (in a 2-ml collection tube) without wetting the rim and is centrifuged at 6000×g (8000 rpm) for 1 min. After discarding the tube containing the filtrate, the QIAamp Mini spin column is placed in a clean 2-ml collection tube. The next step is to add 500 μl Buffer AW1 without wetting the rim, and the mixture is centrifuged at 6000×g (8000 rpm) for 1 min. Again, after discarding the tube containing the filtrate, the QIAamp Mini spin column is placed in a clean 2-ml collection tube and addition of 500 μl Buffer AW2 is followed. The whole mixture is centrifuged at full speed (20,000×g; 14,000 rpm) for 3 min. Last, the QIAamp Mini spin column is placed in a clean 1.5-ml microcentrifuge tube, while the collection tube containing the filtrate is discarded. Immediately after, 200 μl buffer AE is added, the mixture is incubated at room temperature (15–25 °C) for 1 min and is centrifuged at 6000×g (8000 rpm) for 1 min.
Single-nucleotide polymorphism
The single-nucleotide polymorphism (SNP) rs 175080 in MLH3 gene was genotyped by TaqMan SNP genotyping assay (Applied Biosystems, Life Technologies Corp.) using the real-time PCR. The genotyping was performed to the protocol recommended by the manufacturer.
Hormone assays
All blood samples were centrifuged at 1000×g for 15 min, and serum was stored at −20 °C until assayed. All hormones were measured in duplicate in each blood sample. Measurement of FSH, LH, and prolactin in serum was performed using an immunoradiometric assay (FSH-IRMA, LH-IRMA, and PROLACTIN IRMA, respectively; Immunotech, Prague, Czech Republic), and the results are expressed as international units per liter, international units per liter, and nanograms per milliliter, respectively. Measurement of estradiol and testosterone in serum was performed using a radioimmunoassay (RIA ESTRADIOL and RIA Testosterone, respectively, Immunotech), and the results are expressed as picograms per milliliter and nanograms per milliliter, respectively. The lower limits of detection for FSH, LH, estradiol, testosterone, and prolactin were 0.17 and 0.16 IU/l, 9.6 pg/ml, 0.02 and 0.5 ng/ml, respectively, while inter- and intra-assay coefficients of variation were 8.2 and 4.0, 8.4 and 7.3, 14.5 and 14.4, 15.0 and 5.6 and 10.1 and 7.1 %, respectively.
Statistical analysis
Anthropometric parameters, hormone values, and sperm parameters were normally distributed (one sample Kolmogorov-Smirnov test), and statistical analysis was performed by one-way analysis of variance (ANOVA), followed by Bonferroni post hoc testing. Frequencies of the genotypes were compared with the chi-square test. A α level of 0.05 was used to determine statistical significance. Numeric values are expressed as mean ± SEM. The statistical software package used was SPSS v.20.
Results
There were no significant differences in anthropometric parameters and hormonal values between the three genotypes, i.e., GG (n = 61), GA (n = 178), and AA (n = 61) in the cohort of the 300 men (Table 1).
Table 1.
Anthropometric parameters and hormonal values in the three genotypes in the cohort of 300 men (values expressed as mean ± SEM)
| GG | GA | AA | p value | |
|---|---|---|---|---|
| N = 61 | N = 178 | N = 61 | ||
| Age (years) | 37.9 ± 0.7 | 38.4 ± 0.4 | 39.1 ± 0.7 | 0.499 |
| BMI (kg/m2) | 27.8 ± 0.5 | 27.8 ± 0.2 | 28.6 ± 0.5 | 0.235 |
| Smoking (yes) | 12 % | 32.3 % | 12 % | 0.740 |
| FSH (IU/l) | 6.3 ± 0.7 | 6.7 ± 0.4 | 8.1 ± 0.7 | 0.188 |
| LH (IU/l) | 4.3 ± 0.3 | 4.8 ± 0.2 | 5.1 ± 0.3 | 0.160 |
| Testosterone (ng/ml) | 5.0 ± 0.3 | 4.8 ± 0.1 | 4.8 ± 0.3 | 0.758 |
| Estradiol (pg/ml) | 45.0 ± 1.7 | 47.9 ± 1.1 | 48.7 ± 2.3 | 0.344 |
| Prolactin (ng/ml) | 7.1 ± 0.5 | 7.5 ± 0.2 | 8.0 ± 0.6 | 0.351 |
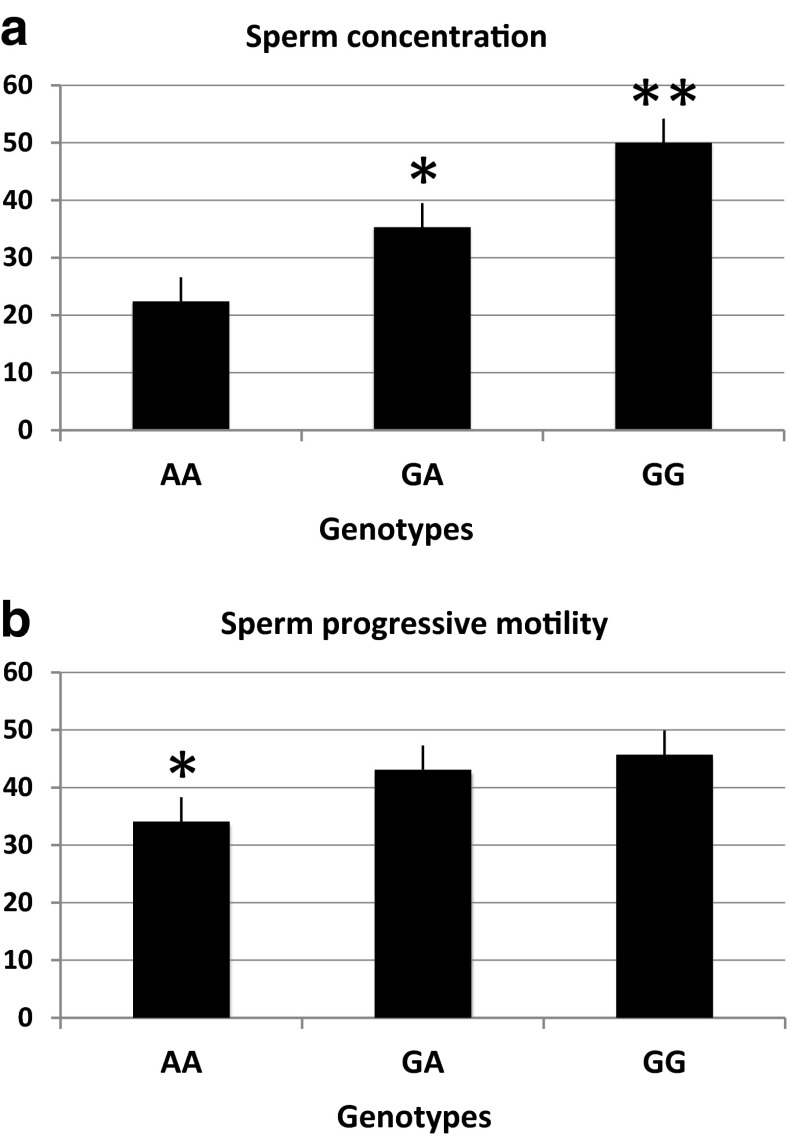
A significantly lower sperm concentration was found in men with the AA genotype (22.4 ± 4.2 mill/ml) as compared to men with the GG (50.0 ± 5.1 mill/ml, p < 0.001) and GA genotypes (35.3 ± 2.8 mill/ml, p < 0.05) (Fig. 1a).
Fig. 1.
a Sperm concentration (mean ± SEM) in the three genotypes. *p < 0.05 (difference between AA and GA) and **p < 0.001 (difference between AA and GG). b Progressive motility values (mean ± SEM) in the three genotypes. *p < 0.05 (difference between AA and GA and between AA and GG)
Men with the AA genotype had the lower progressive motility values (34.1 ± 3.3 %) as compared to men with the GG (45.7 ± 2.9 %) and GA (43.1 ± 1.7 %) genotypes (p < 0.05) (Fig. 1b). The other types of motility were comparable between the three groups.
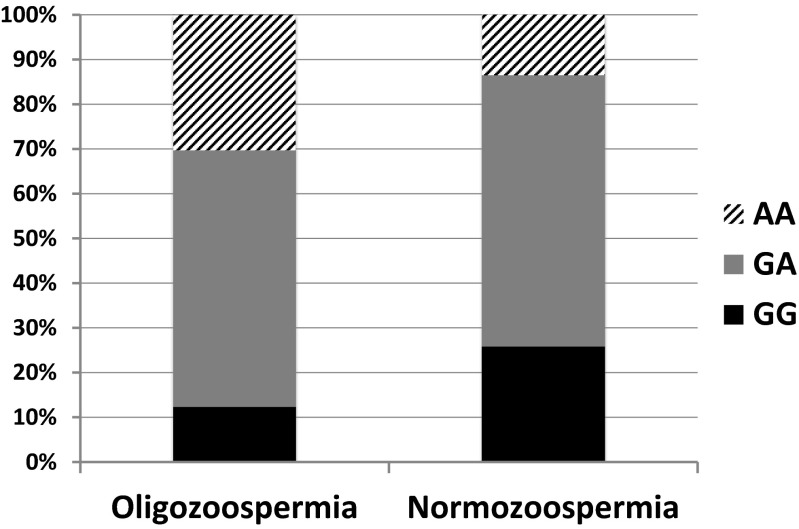
The men with sperm concentration <15 mill/ml were considered as “cases” while men with ≥15 mill/ml served as “controls.” The distribution of the frequencies of the three genotypes between “cases” and “controls” was significantly different (p < 0.001). In particular, in “cases,” the frequencies of GG, AG, and AA were 12.3, 57.4, and 30.3 %, while in “controls,” they were 25.8, 60.7, and 13.5 %, respectively (Fig. 2). The allele frequencies were for A 87.7 % in “cases” and 74.2 % in “controls” (p < 0.05), while for G, they were 70.5 % in “cases” and 86.0 % in “controls” (p < 0.05), respectively.
Fig. 2.
Frequencies of the three genotypes (AA, GA and GG) in men with oligozoospermia (cases) and in men with normozoospermia (controls). The “cases” show a lower frequency of GG and a higher frequency of AA than the “controls” (p < 0.001)
Discussion
Analyzing a total of 300 blood samples from men of infertile couples, we observed that the polymorphism rs 175080 of the MLH3 gene is associated with an increased rate of semen abnormalities. Specifically, the present data provide important findings of significantly lower sperm concentrations and lower progressive motility values in men with the AA genotype as compared to GG and GA genotypes, while the anthropometric parameters and hormonal values were comparable. These results suggest that the AA genotype at least regarding the parameters investigated in the present study significantly impact on spermatogenesis. Besides, a higher proportion of AA genotype and accordingly the allele A frequencies were found in oligospermic samples. It is probable that the A allele has a deteriorating effect in spermatogenesis, which is further amplified in a homozygotic condition.
The human MLH3 gene was first identified approximately in the year 2000, and it is located on 14q24.3 chromosome with a coding length of 4.3 kb, composed of 12 exons, of which exon 1 is 3.3 kb, accounting for 75 % of the coding region [3]. It is a member of a conserved protein family, which is involved in the DNA mismatch repair and meiotic recombination mechanism [4, 5]. Furthermore, MLH3 was found to interact with MLH1 [12]. This interaction seems to be essential for a direct role of the Mlh1-Mlh3 endonuclease activity in resolving recombination intermediates and in DNA mismatch repair [13]. The complex of Mlh1-Mlh3 also plays a role in DNA mismatch repair (MMR) by interacting with another complex, the Msh2-Msh3, in order to repair insertion and deletion mutations. In a recent study, an association was found between the SNP C2531T (P844L) mutation in the MLH3 gene and male infertility, proposing that this mutation may be a genetic risk factor for male infertility at least in Chinese people [7]. In the present study, it was found that the rs 175080 polymorphism of the MLH3 gene may impact on spermatogenesis reflected in abnormal sperm parameters in people from the Caucasian race. Specifically, the homozygotic mutation type (AA) is possibly related to a reduced ability of spermatozoa to obtain excellent motility, which is in concordance with the notion that this specific mutation affects the spermatogenesis. Moreover, mutations of the human MLH gene have been associated with spermatogenic failure or spermatogenic arrest [9]. It seems that Caucasian people that carry the present type of MLH mutation may be susceptible to disorders of sperm motility and concentration. In another recent study, involving azoospermic and oligozoospermic samples from three provinces of Iran and investigating the association between the SNP (rs28368082) in the exon 7 of SPO11 gene and male infertility, it was found a link between only azoospermic and not oligozoospermic parameters with this SNP [14]. This dissociation was attributed to differences in ethnic background and geographic variations.
The exact molecular mechanism via which this specific genetic mutation may negatively impact on male fertility is not yet clarified. Since male infertility is a multifactorial condition, it is likely that apart from the genetic background, the environment also plays a significant role. The MLH gene is essential for the repairing mechanism and the MLH1-MLH3 complex is a major contributor in making crossovers during mammalian meiosis. Therefore, it is probable that the specific mutation negatively affects the functionality of the repairing mechanism in a degree relative to the type of mutation. Furthermore, it is obvious that the homozygotic type of mutation (AA) does not completely eliminate the function of the repairing mechanism, suggesting that other compensatory molecular repairing mechanisms are functioning. Further research is needed to investigate this matter and the possible underlying molecular mechanism.
In conclusion, the present study demonstrates for the first time an association between the rs 175080 polymorphism of the MLH3 gene and the quality of sperm parameters in Caucasian people. Whether this polymorphism may also impact on male fertility needs further investigation.
Compliance with ethical standards
All patients volunteered for the study and gave written informed consent, while Institutional Review Board approval of the study was also obtained.
Footnotes
Capsule
It is suggested that the studied SNP in the MLH3 gene may be linked to oligozoospermia in Caucasian men of a certain area.
References
- 1.De Krester DM, Baker HW. Infertility in men: recent advances and continuing controversies. J Clin Endocrinol Metab. 1999;84:3443–50. doi: 10.1210/jcem.84.10.6101. [DOI] [PubMed] [Google Scholar]
- 2.Krausz C, Chianese C, Giachini C, Guarducci E, Laface I, Forti G. The Y chromosome-linked copy number variations and male fertility. J Endocrinol Invest. 2011;34(5):376–82. doi: 10.1007/BF03347463. [DOI] [PubMed] [Google Scholar]
- 3.Lipkin SM, Wang V, Jacoby R, Banerjee-Basu S, Baxevanis AD, Lynch HT, et al. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet. 2000;24(1):27–35. doi: 10.1038/71643. [DOI] [PubMed] [Google Scholar]
- 4.Cannavo E, Marra G, Sabates-Bellver J, Menigatti M, Lipkin SM, Fischer F, et al. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 2005;65:10759–66. doi: 10.1158/0008-5472.CAN-05-2528. [DOI] [PubMed] [Google Scholar]
- 5.Santucci-Darmanin S, Neyton S, Lespinasse F, Saunieres A, Gaudray P, Paquis-Flucklinger V. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum Mol Genet. 2002;11:1697–706. doi: 10.1093/hmg/11.15.1697. [DOI] [PubMed] [Google Scholar]
- 6.Pashaiefar H, Sheikhha MH, Kalantar SM, Jahaninejad T, Zaimy MA, Ghasemi N. Analysis of MLH3 C2531T polymorphism in Iranian women with unexplained infertility. Iran J Reprod Med. 2013;11(1):19–24. [PMC free article] [PubMed] [Google Scholar]
- 7.Xu K, Lu T, Zhou H, Bai L, Xiang Y. The role of MSH5 C85T and MLH3 C2531T polymorphisms in the risk of male infertility with azoospermia or severe oligozoospermia. Clin Chim Acta. 2010;411(1-2):49–52. doi: 10.1016/j.cca.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 8.Nishant KT, Plys AJ, Alani E. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics. 2008;179(2):747–55. doi: 10.1534/genetics.108.086645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrás C, Zhou XL, Sousa M, Lindblom A, Barros A. DNA mismatch repair gene hMLH3 variants in meiotic arrest. Fertil Steril. 2007;88(6):1681–4. doi: 10.1016/j.fertnstert.2007.01.063. [DOI] [PubMed] [Google Scholar]
- 10.Terribas E, Bonache S, García-Arévalo M, Sánchez J, Franco E, Bassas L, et al. Changes in the expression profile of the meiosis-involved mismatch repair genes in impaired human spermatogenesis. J Androl. 2010;31(4):346–57. doi: 10.2164/jandrol.109.008805. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Ding M, Ding X, Li T, Chen H. Six polymorphisms in genes involved in DNA double-strand break repair and chromosome synapsis: association with male infertility. Syst Biol Reprod Med. 2015;61(4):187–93. doi: 10.3109/19396368.2015.1027014. [DOI] [PubMed] [Google Scholar]
- 12.Kondo E, Horii A, Fukushige S. The interacting domains of three MutL heterodimers in man: hMLH1 interacts with 36 homologous amino acid residues within hMLH3, hPMS1 and hPMS2. Nucleic Acids Res. 2001;29(8):1695–702. doi: 10.1093/nar/29.8.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogacheva MV, Manhart CM, Chen C, Guarne A, Surtees J, Alani E. Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. J Biol Chem. 2014;289(9):5664–73. doi: 10.1074/jbc.M113.534644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghalkhani E, Sheidai M, Gourabi H, Noormohammadi Z, Bakhtari N, Malekasgar AM. Study of single nucleotide polymorphism (rs28368082) in SPO11 gene and its association with male infertility. J Assist Reprod Genet. 2014;31(9):1205–10. doi: 10.1007/s10815-014-0279-z. [DOI] [PMC free article] [PubMed] [Google Scholar]