Abstract
Like most eukaryotes, brown algae live in association with bacterial communities that frequently have beneficial effects on their development. Ectocarpus is a genus of small filamentous brown algae, which comprises a strain that has recently colonized freshwater, a rare transition in this lineage. We generated an inventory of bacteria in Ectocarpus cultures and examined the effect they have on acclimation to an environmental change, that is, the transition from seawater to freshwater medium. Our results demonstrate that Ectocarpus depends on bacteria for this transition: cultures that have been deprived of their associated microbiome do not survive a transfer to freshwater, but restoring their microflora also restores the capacity to acclimate to this change. Furthermore, the transition between the two culture media strongly affects the bacterial community composition. Examining a range of other closely related algal strains, we observed that the presence of two bacterial operational taxonomic units correlated significantly with an increase in low salinity tolerance of the algal culture. Despite differences in the community composition, no indications were found for functional differences in the bacterial metagenomes predicted to be associated with algae in the salinities tested, suggesting functional redundancy in the associated bacterial community. Our study provides an example of how microbial communities may impact the acclimation and physiological response of algae to different environments, and thus possibly act as facilitators of speciation. It paves the way for functional examinations of the underlying host–microbe interactions, both in controlled laboratory and natural conditions.
Introduction
Brown algae belong to the stramenopile lineage, a group of eukaryotes that has evolved independently of plants, animals and fungi for over one billion years (Cock et al., 2011). Among brown algae, Ectocarpus is a cosmopolitan genus of small filamentous species with a long history of research (Charrier et al., 2008) and has recently been established as a genomic and genetic model for this lineage (Peters et al., 2004; Cock et al., 2010). One of the particularities of Ectocarpus is its high capacity to acclimate to different environments, which has led to the development of a range of new approaches to study stress tolerance in this organism (for example, Dittami et al., 2009; Ritter et al., 2010; Tonon et al., 2011; Billoud et al., 2014). A key advantage of Ectocarpus for this type of study is that a number of different strains and ecotypes with different tolerance ranges towards abiotic factors are available. Particularly high tolerance ranges were observed with respect to salinity: while most Ectocarpus isolates are marine, at least one strain was isolated from a true freshwater habitat at Hopkins River Falls, Australia (West and Kraft, 1996). This freshwater strain (strain 371) is able to grow in both freshwater and seawater, adjusting its metabolism and cell wall structure to the corresponding conditions (Dittami et al., 2012; Meslet-Cladière et al., 2013; Torode et al., 2015). In addition, Ectocarpus strains related to the freshwater strain based on their internal transcribed spacer sequences, have been found in other ‘extreme' environments, such as on driftwood, or in areas of highly variable temperature (Bolton, 1983). The transition from marine environments to freshwater is a particularly rare event in brown algae, and only about 1% of brown algal species have colonized this habitat (Bold and Wynne, 1985). Although the exact taxonomic position of the freshwater strain and its relatives remains to be established, they do not belong to the genome-sequenced Ectocarpus siliculosus but constitute a sister species (Dittami et al., 2011) that has separated from E. siliculosus roughly 16 million years ago (Dittami et al., 2012).
So far, studies of acclimation to environmental changes in Ectocarpus, and in algae in general, have dealt primarily with the algae themselves, but very little is known about the reaction of the associated microbiome in response to these changes. Most, if not all, eukaryotes (including Ectocarpus) have evolved complex interactions with bacteria (Zilber-Rosenberg and Rosenberg, 2008). There are several examples where algal–bacterial associations benefit algal hosts, and have become an integral part of their biology (see Goecke et al., 2010; Hollants et al., 2013 for recent reviews): epi- and endophytic bacteria, for instance, are known to provide nitrate (Chisholm et al., 1996) or vitamins (Croft et al., 2006) to their hosts, thus increasing their fitness. Within the Ectocarpales, an absolute dependence on bacteria has been observed in E. fasciculatus as well as Pylaiella littoralis, both of which do not develop properly in axenic conditions, although algal growth can be restored by addition of the cytokinin kinetin to the culture medium (Pedersén 1968, 1973). In E. siliculosus, an uncultured bacterium closely related to the order of Rhizobiales, and tentatively named Candidatus ‘Phaeomarinobacter ectocarpi', is suspected to provide cytokinins as well as other compounds to the alga (Dittami et al., 2014a).
Here, we first generate a census of bacteria found in laboratory cultures of Ectocarpus, and then use one example (strain 371) to re-examine acclimation to low salinities focusing this time on the algal phycosphere. We show that, even in closed laboratory systems, bacterial community composition was clearly affected by changes in the culture conditions, and that the presence of bacteria positively affected the capacity of algae to acclimate to changing salinities. This underlines the importance of integrative approaches that consider both host and microbial symbionts to understand adaptation and acclimation in eukaryotes.
Materials and methods
Overview of experimental setup
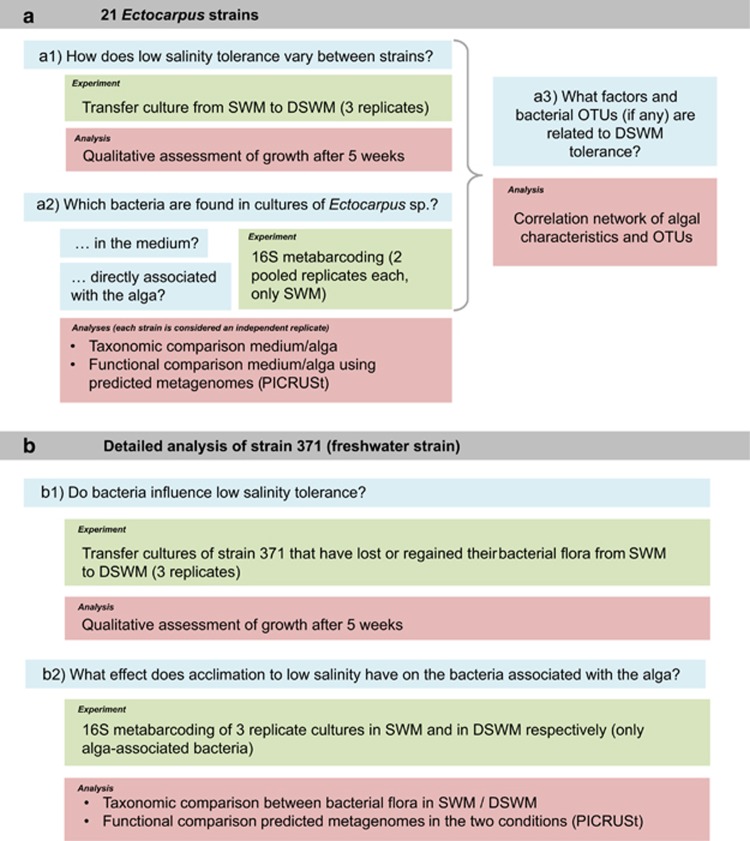
Two sets of experiments were carried out (Figure 1). A first set aimed to generate a census of the bacterial flora associated with 21 strains of Ectocarpus from different geographic origins, all cultured in standard seawater medium (SWM). It considered both bacteria attached to the alga and bacteria found in the culture medium, and was used to search for factors that correlated with low salinity tolerance in these strains. The second set of experiments focused on the freshwater strain 371, and examined the influence of the presence or absence of bacteria on the salinity tolerance of this strain, as well as the effect of acclimation to low salinity on the bacterial flora attached to the alga.
Figure 1.
Overview of the experimental setup. The two main experiments (a and b) are highlighted in gray, main scientific questions in blue and corresponding experiments and analyses in green and red, respectively.
Culture conditions and acclimation experiments
All 21 examined strains of Ectocarpus (Table 1) were obtained from BEZHIN ROSKO (http://www.bezhinrosko.com), and maintained in SWM or diluted SWM (DSWM) in 140 mm Petri dishes in a culture room at 13 °C. Seawater was collected offshore at Roscoff (48°46'40''N, 3°56'15''W), 0.45 μm filtered, diluted 1:20 with distilled water for DSWM and autoclaved. Both media were then enriched with autoclaved Provasoli nutrients (Starr and Zeikus, 1993), comprising sources of nitrate (270 μM), phosphate (glycerophosphate 15 μM), vitamins (vitamin B12 0.5 nM, thiamine 97 nM and biotin 1.4 nM) and trace metals (boron 31 μM, iron 3 μM, manganese 1.6 μM, zinc 127 nM, cobalt 28 nM). Light was provided at a photon flux density of 40 μmol m−2 s−1 for 12 h per day. All cultures were handled under laminar flow hoods, limiting their exposure to bacteria not present at the time of isolation. To determine the capacity of the 21 examined strains to acclimate to low salinity, cultures were transferred directly from SWM to DSWM, and their growth was monitored for 5 weeks. These experiments were carried out in biological triplicates. In parallel, 16S metabarcoding experiments were performed with all of these strains in SWM as described below. DNA from two replicate cultures was pooled and sequenced together. Each of these pools was considered as one independent replicate for which both algal and medium samples were examined.
Table 1. List of algal strains and samples used for 16S metabarcoding experiments.
| Name | Sex | Origin | Year | Isolator | ITS accession | Culture medium | Growth in DSWM |
Culture medium |
Alga |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. samples | Dsim | No. OTUs | No. samples | Dsim | No. OTUs | ||||||||
| 371 | Sp | Hopkins River, AUS | 1995 | J West | FR668844 | SWM | Yes | 1 | 0.15 | 43 | 3 | 0.13-0.16 | 74-84 |
| DSWM | Yes | 1 | 0.18 | 95 | 3 | 0.13-0.18 | 71-80 | ||||||
| Bft 1a | f | Beaufort, USA | 1978 | DG Müller | FR668733 | SWM | No | 1 | 0.34 | 39 | 1 | 0.19 | 61 |
| Bft 15b | m | Beaufort, USA | 1978 | DG Müller | FR668734 | SWM | No | 1 | 0.14 | 76 | 1 | 0.33 | 53 |
| Par 10b | f | Port Aransas, USA | 1978 | DG Müller | FR668791 | SWM | No | 1 | 0.50 | 51 | 1 | -c | 19c |
| PAr 10n_2 | f | Port Aransas, USA | 1978 | DG Müller | FR668792 | SWM | Yes | 1 | 0.14 | 72 | 1 | 0.12 | 48 |
| PAr 27a | m | Port Aransas, USA | 1978 | DG Müller | FR668795 | SWM | Yes | 1 | 0.20 | 53 | 1 | 0.12 | 54 |
| PAr 1027 | sp | Port Aransas, USA | 1978 | DG Müller | unpublished | SWM | No | 1 | 0.35 | 69 | 1 | 0.27 | 59 |
| Tam 2b | m | Tampa, USA | 1978 | DG Müller | FR668803 | SWM | Yes | 1 | 0.53 | 51 | 1 | 0.22 | 67 |
| Tam 12b | m | Tampa, USA | 1978 | DG Müller | FR668804 | SWM | No | 1 | 0.37 | 56 | 1 | 0.50 | 52 |
| Tam 18b | f | Tampa, USA | 1978 | DG Müller | FR668805 | SWM | Yes | 1 | 0.27 | 85 | 1 | 0.27 | 33 |
| Tam 19c | m | Tampa, USA | 1978 | DG Müller | FR668806 | SWM | C.d. | 1 | 0.24 | 59 | 1 | 0.20 | 67 |
| Grh 6c | sp | Grinhigen, SW | 1979 | DG Müller | FR668907 | SWM | C.d. | 1 | 0.17 | 40 | 1 | 0.39 | 39 |
| Kal 1c | sp | Killvik, SW | 1979 | DG Müller | FR668909 | SWM | Yes | 1 | 0.65 | 64 | 1 | 0.16 | 59 |
| Bri_1 | sp | Pill, UK | 2008 | AF Peters | unpublished | SWM | Yes | 1 | 0.37 | 56 | 1 | 0.70 | 68 |
| Bri_3 | sp | Pill, UK | 2008 | AF Peters | unpublished | SWM | Yes | 1 | 0.17 | 54 | 1 | 0.13 | 60 |
| DEV11_1a | m | Kingsbridge, UK | 2011 | AF Peters | unpublished | SWM | No | 1 | 0.43 | 49 | 1 | 0.21 | 60 |
| Ter11_1 | sp | Terenez, FR | 2011 | AF Peters | unpublished | SWM | No | 1 | 0.27 | 61 | 1 | 0.17 | 60 |
| Ter11_2 | sp | Terenez, FR | 2011 | AF Peters | unpublished | SWM | Yes | 1 | 0.20 | 68 | 1 | 0.09 | 33 |
| Rsa001 | sp | Muizenberg, RSA | 2004 | D Schroeder | unpublished | SWM | Yes | 1 | 0.31 | 52 | 1 | 0.70 | 35 |
| Rsa002 | Sp | Muizenberg, RSA | 2004 | D Schroeder | unpublished | SWM | Yes | 1 | 0.13 | 58 | 1 | -c | 31c |
| Ec32b | m | San Juan de Marcona, Peru | 1988 | AF Peters | genome | SWM | C.d. | 1 | 0.18 | 56 | 1 | 0.17 | 59 |
Abbreviations: DSWM, diluted seawater medium; f, female strain; ITS, internal transcribed spacer; m, male strain; OTU, operational taxonomic unit; sp, heterozygous sporophytes; SWM, seawater medium. The capacity of a strain to grow in DSWM was determined in a separate experiment with three biological replicates. ‘C.d.'=cell death, that is, cultures showed clear bleaching after 5 weeks of exposure.
This strain is likely to represent a species different from the remaining strains, despite high phenotypic similarity.
This strain is the genome-sequenced purely marine strain of E. siliculosus.
For these samples, less than 7891 sequences were obtained and no diversity-index was calculated.
For strain 371, which was originally isolated from freshwater (West and Kraft, 1996), freshwater tolerance experiments were carried out not only with standard non-antibiotic-treated cultures (I), but also with antibiotic-treated cultures (II), as well as antibiotic-treated cultures inoculated with 0.1% of medium from non-treated cultures of strain 371 (III), antibiotic-treated cultures inoculated with 0.1% of medium from non-treated cultures of the purely marine strain 32 (IV) and antibiotic-treated cultures inoculated with 0.1% of sterile (autoclaved) medium from non-treated cultures of strain 371 (V). Both antibiotic-treated (VI) and non-treated cultures (VII) were also transferred to fresh SWM as control. Antibiotic-treated cultures inoculated with medium from non-treated cultures were incubated under standard conditions (SWM) for 1 week prior to transferring algae to DSWM. Algal quantum yield (Fv/Fm), a fluorometric marker for the efficiency of photo system II, was determined 1 week after the transfer using a Walz Phyto-pulse amplitude modulation fluorometer (Walz, Effeltrich, Germany) and default parameters (Dittami et al., 2009). As this procedure incurred a risk of contamination with bacteria, separate cultures were used for these measurements and only one time point was sampled. These experiments were carried out in biological triplicates. Metabarcoding for strain 371 was performed in both SWM and DSWM (biological triplicates for algal samples, Table 1), and cultures were grown in the respective medium for at least 2 months prior to sampling.
Antibiotic treatments
Approximately 25 small algal filaments from either SWM or DSWM were placed in 90 mm Petri dishes with solid Zobell medium (Zobell, 1941) and antibiotic discs (Ampicillin 10 μg, Chloramphenicol 30 μg, Ciprofloxacin 5 μg, Erythromycin 15 μg, Kanamycin 30 μg, Penicillin G 10 μg, Polymyxine PB 50 μg, Rifampicin 30 μg, Tetracycline 30 μg and Vancomycin 30 μg). The Petri dishes were then incubated for 4 weeks in the aforementioned culture conditions, and the absence of bacterial contaminants verified microscopically (× 800 magnification, phase contrast). Random cultures were also selected for DAPI staining or for plating on Zobell medium. Filaments without detectable contamination were transferred to autoclaved (diluted) seawater without added Provasoli nutrients for 2 weeks, and then kept in SWM or DSWM for at least 4 weeks to allow algae to recover from the treatment. Only SWM, but not DSWM cultures, survived these treatments.
16S metabarcoding experiments and prediction of microbial metagenomes
Metabarcoding experiments were carried out as described in Supplementary File S1 for all samples listed in Table 1. Bacterial 16S V3-V4 rRNA gene sequences were amplified following Caporaso et al. (2012) (Supplementary Figure S2) and using the primers given in Supplementary Table S3. Approximately 5 × 106 pairs of raw sequence reads were generated on the Illumina (San Diego, CA, USA) MiSeq platform and processed as outlined in Supplementary Figure S4. We used the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) suite (Langille et al., 2013) to obtain an overview of the genomic and metabolic features represented by the bacterial communities in our samples. In this analysis, the mean Nearest Sequenced Taxon Index, calculated by PICRUSt to provide a reliability estimation of the metagenome predictions, was in a normal range for all samples (0.08±0.03 (s.d.), see Langille et al., 2013).
Statistical analyses and diversity assessment
All samples were subsampled to contain the same number of sequences after cleaning (7891), and samples with insufficient sequence coverage removed from the dataset (Table 1). To identify groups of similar samples, the Bray-Curtis index was used as a dissimilarity measure and non-metric multidimensional scaling (NMDS) of all data points was carried out. In addition, separate NMDS analyses were performed for only algal and only medium samples, and both NMDS plots subjected to a Procrustes comparison. These analyses were performed in R v2.15.2 (R Core Team, 2014), using the vegan 2.0–9 package (Oksanen et al., 2013).
To identify systematic differences in the taxonomic composition of algal and medium samples, we removed rare sequences (< 10 occurrences), then normalized counts using the total number of reads, and log2(x+1)-transformed the dataset to resemble a normal distribution. Each strain was considered as a biological replicate, and paired t-tests were carried out using STAMP 2.0 (Parks and Beiko, 2010). A Benjamini Hochberg correction for multiple testing was applied, limiting the overall false discovery rate to 5% (Benjamini and Hochberg, 1995). The same procedure was also used to compare functional KEGG categories inferred for each sample by PICRUSt (Langille et al., 2013).
Local bacterial diversity was assessed using Simpson's diversity index Dsimpson as calculated by mothur (Schloss et al., 2009). This index constitutes a robust measure of alpha-diversity that is biased towards the most abundant species, where 0 indicates maximal diversity and 1 indicates complete dominance by a single species or operational taxonomic unit (OTU) (Magurran, 2004).
Kendall correlations between the abundance of OTUs as well as other characteristics of the samples, namely year of isolation of the culture, life cycle stage (1=homozygous gametophyte or parthenosporophyte, 2=heterozygous sporophyte, see Charrier et al., 2008), capacity to grow in DSWM 5 weeks after transfer (1=visible growth, 0 =no growth, −1=bleaching/cell death), sample type (alga or medium) and diversity index, were calculated using the PAleontological STatistics software (PAST) version 3.01 (Hammer et al., 2001). Correlation networks across the different samples were generated using all correlations with P<0.001, a threshold corresponding to a false discovery rate of 3.4% according to Benjamini and Hochberg (1995). The resulting network was visualized using Cytoscape 3, and highly connected clusters identified using the MCODE plugin version 1.4.0.beta2 (Smoot et al., 2011).
Direct taxonomic comparisons of the microbial composition in the three replicate samples of strain 371 in SWM and in DSWM were carried out using t-tests as described for algal and medium samples above, except that samples were considered independent.
Data deposition
All custom scripts used in this study are available at http://sdittami.altervista.org/16Stools/. Raw Illumina reads were deposited at the European Nucleotide Archive under project accession number PRJEB5542.
Results
Each Ectocarpus strain has its unique microbiome
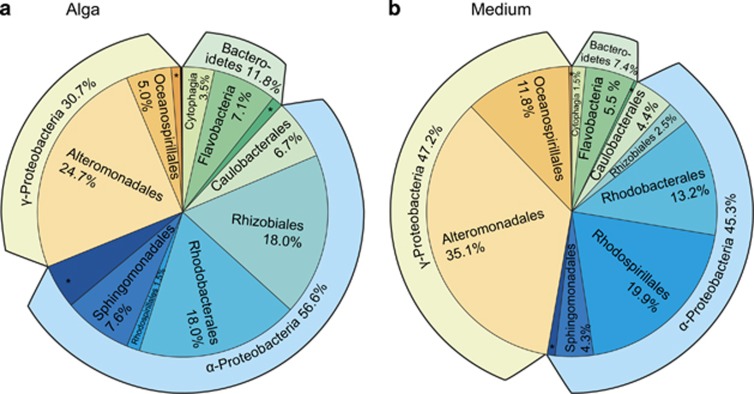
Using 16S metabarcoding, we examined how bacterial composition varied across different cultures of Ectocarpus, and which bacteria were attached to the algae or found in the medium. After data processing (Supplementary Figure S4), 1.8 million bacterial 16S rRNA gene reads with an average of 37 124±18 496 (s.d.) sequences per sample were obtained, corresponding to 438 OTUs at an identity threshold of 97%, with 33 to 85 OTUs per sample (Table 1). Intra-sample rarefaction curves (Supplementary Figure S5) were close to saturation, while inter-sample curves still increased when examining all 21 selected strains, indicating that the bacterial communities of algal strains differed with respect to the individual bacterial taxa found. On average, each algal strain contained 3.8 unique OTUs (that is, OTUs not found in other strains). Only a single OTU (identified as Roseobacter) was found in all algal samples, and three OTUs (Marinobacter, Alteromonas and a member of the OCS116 clade) were found in all medium samples (Supplementary Table S6). The taxonomic composition on higher levels (phylum to order), however, was similar across all samples. A vast majority of OTUs (92% in medium samples, 87% in algal samples) were identified as Proteobacteria (essentially Alpha- and Gammaproteobacteria), and the remaining sequences belonged almost exclusively to the phylum Bacteroidetes (Figure 2). No archaebacterial sequences were found in any of our samples, an observation that may at least partially be related to the fact that our primers targeted mainly eubacteria, and were predicted to amplify only ca. 60% of known archaebacterial species.
Figure 2.
Taxonomic distribution at the class and order levels of bacteria associated with the examined Ectocarpus strains (a) attached to the algae and (b) found in the culture medium. * indicates other or unknown bacteria. See Supplementary Table S6 for the complete dataset.
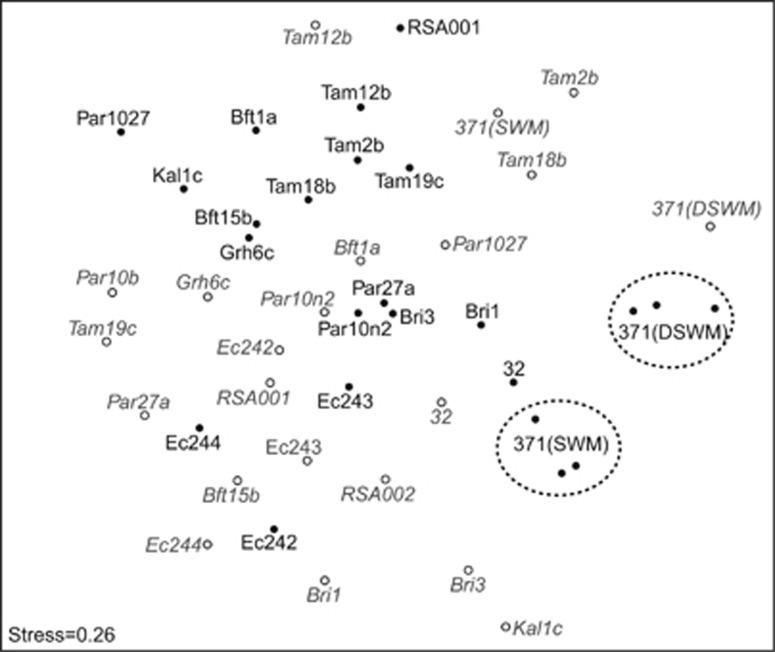
A comparison of dissimilarities between samples showed a rather homogenous distribution of strains and sample types, although we observed the triplicates of strain 371 in DSWM and SWM to form two distinct groups, reflecting the reproducibility of the experiments (Figure 3). There was a group of several algal samples isolated from Tampa, and microbial communities of all DSWM samples (both associated with the algae and found in the medium) could be separated from SWM samples. In a few but not all cases (especially Tam12b, Bft1a, Grh6c, 32, Ec243 and Ec244), samples corresponding to the medium were found not far from the algal samples, but a Procrustes analysis of NMDS plots generated separately for algal and medium samples revealed that this was not a significant general trend (r=0.38, P=0.12).
Figure 3.
NMDS analysis displaying the Bray–Curtis dissimilarity between bacterial communities from algal and medium samples. Solid black dots refer to algal samples, while gray outlined dots with labels in italics refer to medium samples. All data were resampled to contain 7981 reads per sample; two algal samples with low read coverage (RSA002 and Par10b) were discarded.
Differences between alga-attached and free-living bacteria
To further explore the observed absence of a clear separation of algal and medium samples in the NMDS plots, we performed direct comparisons of the two sample types on an OTU basis by paired t-tests using the different strains as biological replicates. Although our data show quantitative differences in the relative representation of OTUs in each strain, the low overlap between the bacterial communities of the 21 algal strains entails that these differences were usually strain-specific and thus not statistically significant. Moreover, qualitatively (that is, with respect to the mere presence or absence of OTUs), algal and medium samples of each strain were highly similar (Supplementary Table S6), reflecting the fact that algae and culture medium were in direct physical contact.
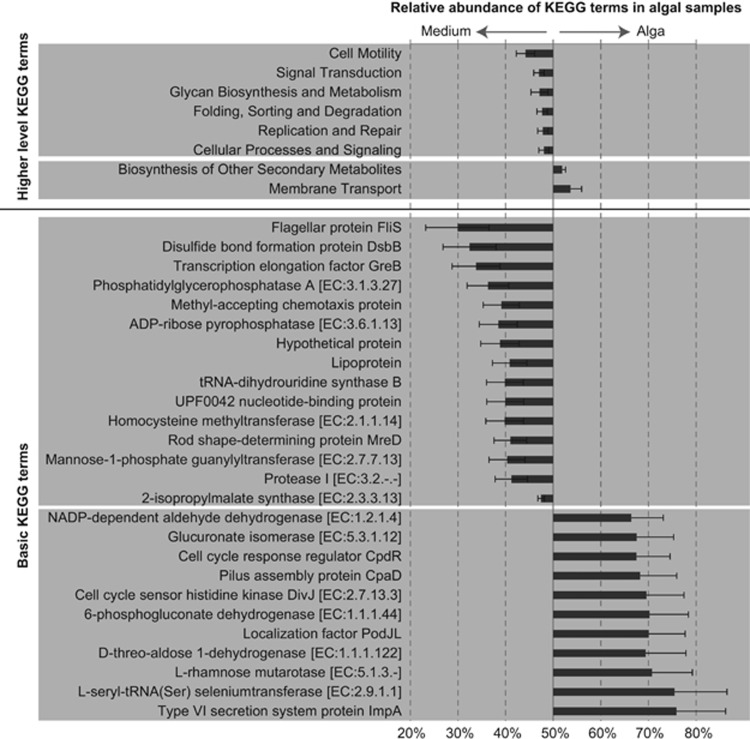
Despite the lack of a general pattern at the level of individual OTUs, we observed significant general differences between alga-attached and free-living bacteria on higher taxonomic levels across all 21 strains. As shown in Figure 2, Rhizobiales were ca. sevenfold more abundant attached to the algae, while Rhodospirillales were 13-fold more abundant in the medium. Moreover, an analysis of the predicted metabolic capacities encoded by the bacterial communities attached to algae and by free-living bacteria in the culture medium revealed a number of systematic differences (Figure 4). On a high hierarchical level (that is, KEGG level 2 in PICRUSt), membrane transport and biosynthesis of secondary compounds were predicted to be significantly more represented in alga-associated bacteria, while cell motility and several KEGG terms related to signaling and repair were predicted to be more represented in free-living bacteria. Specific examples for this are proteins belonging to Type VI secretion systems (bacteria associated with algae) and flagellar proteins (free-living bacteria).
Figure 4.
KEGG categories predicted to be differentially represented in bacterial communities found attached to the algae or free-living in the algal culture medium (false discovery rate<5%). Bars represent the relative abundance of a KEGG term in bacterial communities attached to the algae divided by its mean relative abundance in algal and medium samples (mean of 21 strains±s.e.). For example, 0% would denote a KEGG term predicted only in genomes of free-living bacteria, 100% a KEGG term predicted only in bacteria directly associated with the algae, and 50% a KEGG term predicted to be equally represented in both sample types. Metagenomes were inferred using PICRUSt.
The abundance of selected OTUs correlated with other algal and bacterial characteristics, including the strains' capacity to grow in low salinity
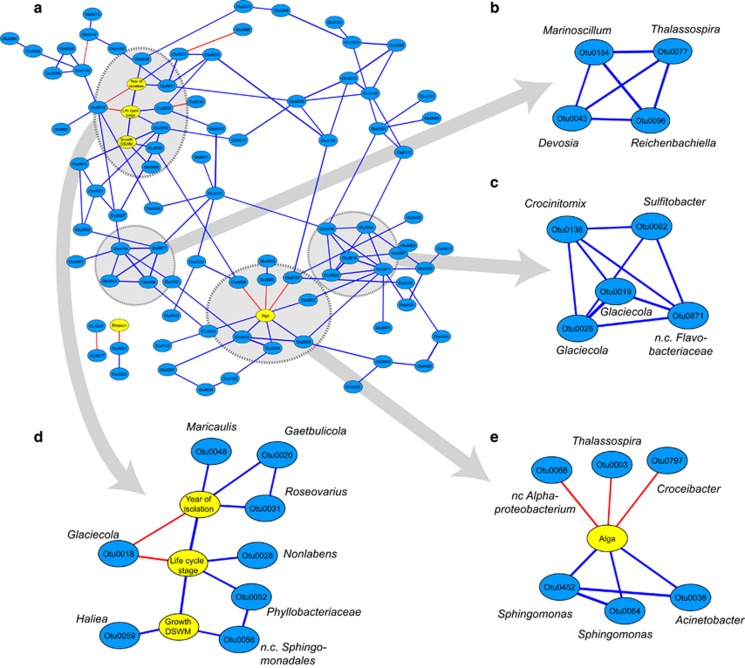
We built an association network to identify and visualize groups of OTUs from the 21 strains, the presence of which was significantly correlated (Kendall correlations, P<0.001, Figure 5a). This revealed two clusters of highly correlated OTUs in the network, the first (Figure 5b) comprising OTUs most abundant in strain 371 and in algal strains isolated from Tampa (Florida), and the second (Figure 5c) comprising OTUs characteristic for cultures originating from Port Aransas and Tampa.
Figure 5.
Correlation networks of OTUs (blue) and alga-related factors (yellow). Blue edges represent significant (P<0.001; false discovery rate<3.4%) positive correlations (Kendall-tau), red edges significant negative correlations. Panel a represents an overview of the network; panels b and c represent tightly connected groups as identified using the MCODE plugin in Cytoscape; panels d and e show sub-networks containing only the direct neighbors of selected alga-related factors; ‘n. c.'=non cultivated.
In addition to OTUs, we also observed three characteristics of the algal culture to be correlated, namely year of isolation, life cycle stage and capacity to grow in DSWM (Figure 5 and Table 1). Notably, sporophyte cultures (heterozygous phase) generally had a higher chance of tolerating a transfer from SWM to DSWM; at the same time, no clear links between freshwater tolerance and the origin of the 21 strains could be established. Furthermore, the presence of some OTUs was correlated with certain characteristics of the algal culture (Figure 5d). For instance, the presence of Haliea sp. and of an unclassified member of the Sphingomonadales was positively correlated with the capacity of cultures to grow in DSWM, and only three OTUs, Maricaulis, Gaetbulicola and Roseovarius, were significantly impacted by the age of the culture and most abundant in recently collected strains (2004–2011). The overall bacterial diversity found in our cultures, however, was not significantly correlated with either the age of the culture or its capacity of survival in low salinities and only one member of the Hoflea group (OTU0001) was found to be more abundant in communities with high diversity.
Lastly, three OTUs, two Sphingomonas and one Acinetobacter, were more frequently found attached to Ectocarpus, while three others (Croceibacter, Thalassospira and an unclassified Alphaproteobacterium) were more abundant in the culture medium (Figure 5e).
Bacteria facilitate algal growth under low salinities, but are impacted by changes in salinity
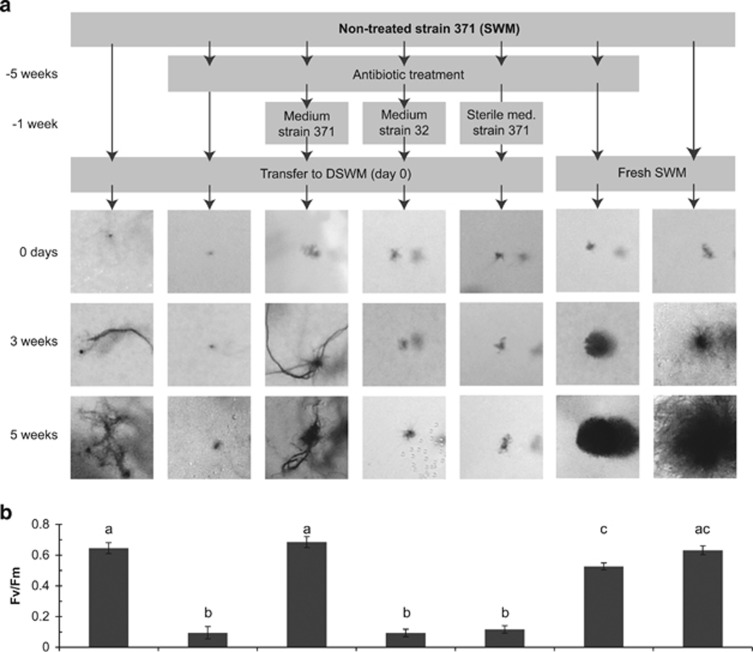
In our experiments focusing on strain 371 (Figure 1, part b), that is, the strain originally isolated from freshwater, we found that DSWM tolerance depended on the presence of bacteria in the culture medium: this strain did not survive antibiotic treatments when cultured in DSWM, and cultures treated with antibiotics in SWM did not survive the transfer to DSWM, even after several weeks of recovery. The only way we have found so far to re-establish the capacity of antibiotic-treated cultures of strain 371 to acclimate to DSWM was to inoculate them with a small quantity (that is, we tested 0.001 × volume) of culture medium from non-antibiotic-treated cultures (Figure 6a). The identity of the non-treated culture was important: medium from a purely marine strain of E. siliculosus, strain 32, did not restore growth, whereas medium from the freshwater strain 371 did. Furthermore, sterilizing the medium of xenic strain 371 by autoclaving prior to inoculation removed the beneficial effect on DSWM tolerance. Finally, inoculating cultures of the marine strain 32 with medium from strain 371 did not result in DSWM-tolerant cultures of strain 32 (not shown). These results were confirmed by Fv/Fm measurements, which were in a normal range (0.4–0.7) for growing cultures and <0.1 for cultures that did not exhibit growth (Figure 6b).
Figure 6.
(a) Growth of cultures of strain 371 after different treatments and transfer from SWM to DSWM. ‘Medium strain 371', ‘Medium strain 32' and ‘Sterile med. strain 371' refer to the inoculation of the antibiotic-treated culture of strain 371 with 0.1% (v/v) of medium from non-treated cultures of strains 371 and strain 32, respectively, where strain 32 is a purely marine strain of Ectocarpus and ‘sterile' medium was autoclaved prior to inoculation. All experiments were carried out in triplicates and yielded highly similar results, but only one replicate is shown here. Differences in morphology between algae cultivated in DSWM and SWM have been previously described (Dittami et al., 2012), and may be related to the described differences in the bacterial phycosphere. (b) Quantum yield (Fv/Fm) of cultures kept under identical conditions as in panel a, 1 week after the transfer to fresh medium. Means with different letters are significantly different (Tukey's HSD, P<0.05). Fv/Fm is a measure of photosystem II efficiency and rapidly decreases in response to stress.
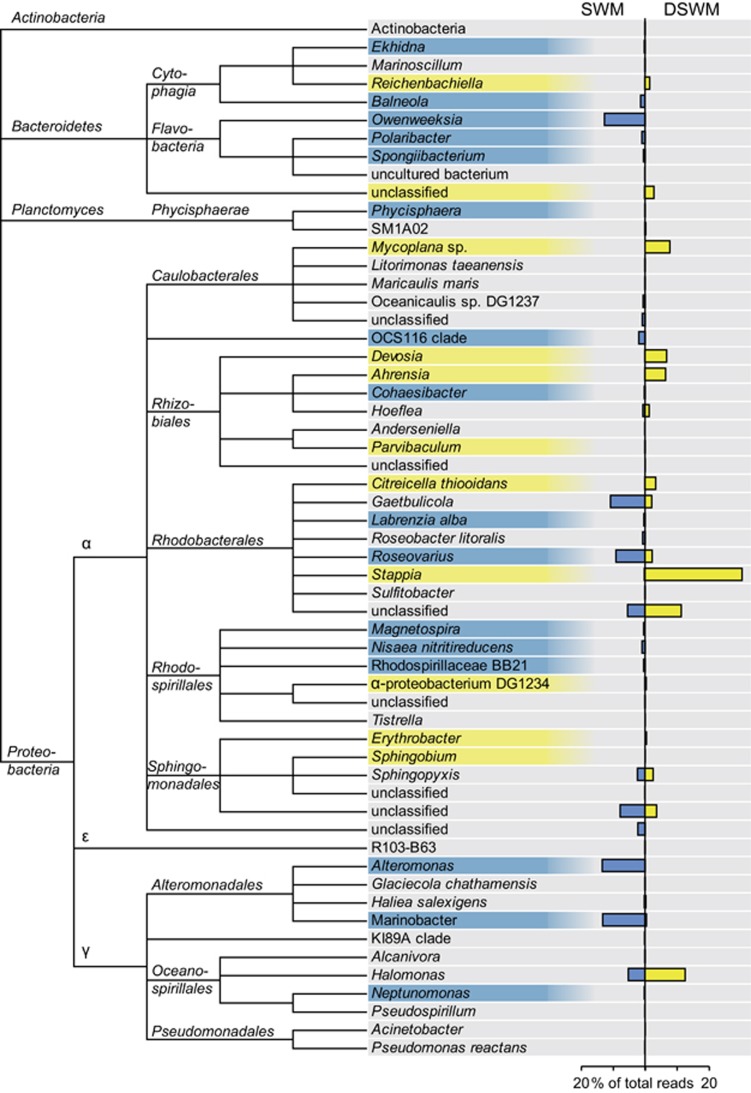
After having shown that bacteria have an effect on the capacity of algae to acclimate to DSWM, we were interested in the effect of algal acclimation to DSWM on the bacterial community. We therefore carried out a direct comparison of the relative composition of the community associated with strain 371 grown in SWM and DSWM (Figure 7). While numerous differences were observed at the genus level, most higher-level taxonomic groups were nearly equally abundant in both conditions. Exceptions here were Flavobacteria (in particular Owenweeksia) and Alteromonadales (especially Marinobacter and Alteromonas), which were both most abundant in SWM. The dominant genus in DSWM was Stappia sp., which accounted for 30.4% of the sequences in this condition. Interestingly, both OTUs positively correlated with growth in freshwater in the 21 examined strains (see Figure 5d), that is, Otu0056 (a non-classified member of the Spingomonadales) and Otu0059 (Haliea, Alteromonadales), did not exhibit significant differences in abundance between SWM and DSWM. In contrast to these taxonomic variations, a direct comparison of the KEGG categories associated with the bacterial communities in the two conditions did not reveal any significant differences (data not shown).
Figure 7.
Bacterial community composition associated with algae of strain 371 grown in DSWM and SWM. Taxonomic groups significantly overrepresented in DSWM or SWM (false discovery rate<5%, n=3) are highlighted in yellow or blue, respectively. Only taxonomic groups represented by at least 10 sequences were considered.
Discussion
It is well-established that seaweeds live in association with bacteria and other microbes, several of which have positive effects on algal growth and development (Wahl et al., 2012; Egan et al., 2013), and some seaweeds have been suspected to actively select specific bacterial communities (for example, Staufenberger et al., 2008). However, very little is known about these effects under changing environmental conditions and in small filamentous algae such as the brown algal model Ectocarpus. Here, we examined the bacterial communities associated with laboratory cultures of Ectocarpus and their behavior during the acclimation to a drastic environmental change: the transition from seawater to freshwater medium.
Conserved trends in phycospheres across lineages
The recruitment of plant microbiomes is thought to be governed by a combination of random processes (Hubbell, 2001) and selective properties related to the niche (Peterson et al., 2011), analogous to the competitive lottery model proposed for the colonization of coral reefs (Sale, 1979). Corresponding observations have been made, for example, during the formation of the phyllosphere in Arabidopsis (Maignien et al., 2014) or in biofilms on green algae (Burke et al., 2011). In both cases, strong taxonomic differences were observed between samples and over time, but functional equivalency of the bacterial communities was suspected within the pool of bacteria with the necessary properties to colonize the niche (referred to as guild). Our observations in laboratory cultures of Ectocarpus are consistent with this model: on a species or genus level, the composition of bacteria between algal cultures was highly variable, supporting the presence of random processes. However, compared with free-living bacteria in the medium, the genomes of bacteria predominantly attached to algae were enriched notably in membrane transport and secondary metabolism, and impoverished with respect to cell mobility. Although these comparisons were based purely on predictions made from 16S rRNA gene sequences, several of the observations are plausible: flagellar proteins, which were predicted to be less abundant in our algal samples, for example, are usually downregulated during biofilm formation (Guttenplan and Kearns, 2013), and may even be dispensable (Deligianni et al., 2010), while Type VI secretion systems, which were predicted to be more abundant in our algal samples, are transmembrane transporters frequently involved in pathogenesis but also suspected to function in other biotic interactions with hosts (Coulthurst, 2013). Thus, although the predicted overrepresented functional categories need to be interpreted with caution, we nevertheless consider our analysis to provide a good indication for the existence of functional enrichments in our system. This supports the existence of a guild of bacteria suitable to live in association with Ectocarpus, at least in a culture environment. Interestingly, as in field samples of Ulva (Burke et al., 2011), this guild comprised mainly alpha- and gammaproteobacteria, which together represented 91% of all sequences in our samples—far more than the approximately 50–70% typical for marine bacterioplankton (Pommier et al., 2007; Zinger et al., 2011).
These findings are important for three reasons. First, they suggest a high degree of convergence with respect to mechanisms governing algal–bacterial interactions in different systems: competitive lottery models may be applied to explain observations across different phyla (from the green lineage to stramenopiles), and in algae of different levels of morphological complexity. Second, traces of these mechanisms can still be detected in laboratory cultures that have been isolated from their natural environment for 2 to 35 years. This implies that the laboratory model Ectocarpus does not only provide a controlled platform that can be manipulated to gain functional understanding of the system, but that the forces governing bacterial recruitment in these cultures may still be similar to those encountered in the field. Lastly, these findings have consequences for our understanding of the biology of Ectocarpus cultures, which are discussed in the section below.
Acclimation of Ectocarpus to changing salinities requires changes in both the alga and its associated bacterial community
Bearing in mind that algae, including Ectocarpus as shown above, select a suitable bacterial flora and may depend on it to furnish nutrients, growth factors and vitamins, we were interested in how algal–bacterial interactions influence and are influenced by a radical change in salinity, that is, that from seawater to freshwater. It is plausible that such a strong abiotic change would alter not only the host but also the associated bacterial community, especially because this is a transition only rarely made by marine microbes (Logares et al., 2009). Here, we demonstrate that, in our system, this is indeed the case: not only does a change in salinity significantly modify dominance patterns in the phycosphere of the examined strain, we have also shown that the bacterial flora strongly affects the adaptive capacities of the system. This factor is still only rarely taken into consideration when studying adaptation and acclimation in eukaryotes.
So, what is necessary for an algal holobiont to be able to acclimate to freshwater? Clearly, factors that lie within the alga have a major role: our correlation analyses indicate that life cycle stages of the algae are linked significantly with growth in freshwater. Exclusively vegetative reproduction in the sporophyte life cycle stage has previously been reported in Fucus populations adapted to low salinities in the Baltic Sea (Tatarenkov et al., 2005), and the freshwater strain 371 is indeed a sporophyte that, to our knowledge, has never produced haploid gametophytes in a laboratory setting (see Charrier et al., 2008 for a review of the Ectocarpus life cycle). In connection with this, we have previously identified the alga's capacity to modify its cell wall composition, and in particular the degree of sulfation, as another possible factor for acclimation (Dittami et al., 2012; Torode et al., 2015). Here, we have shown that bacteria also have a role in algal acclimation, but so far, there was no indication that they produce compounds that specifically aid algae to acclimate to freshwater. A plausible alternative hypothesis is that bacteria provide essential services for the algae in general, such as the production of compounds that are crucial for algal growth. Thus, for an algal culture to acclimate to drastic environmental changes, it needs to be endowed with an associated microbiome that is able to do the same and continue to provide these services in the new conditions, or, alternatively, it needs to be able to recruit new bacteria to replace essential microbial functions. As our acclimation experiments were carried out in a closed system without external source of bacteria, only the former of the two possibilities was available. Dominance patterns in the bacterial flora changed significantly as a result of exposure to different salinities, but this did not result in a detectable modification of the community functions represented by the predicted metagenomes. Therefore, we propose that the modified bacterial community in DSWM cultures may still fulfill the same functions for the algae as in SWM. In the light of this hypothesis, we expected that, in a closed culture environment, algal strains with high bacterial diversity would have a higher chance of possessing a bacterial community capable of functioning in freshwater, yet no such correlation was observed. Instead, only the abundance of two OTUs was positively correlated with this factor. It may be coincidental that these OTUs exhibited similar relative abundances in DSWM and SWM cultures (Figure 7), that is, they, like the algae, thrived in both conditions, but they represent promising subjects for future targeted experiments.
Conclusion
Laboratory strains of small filamentous algae possess complex bacterial communities, which may exert beneficial effects on the growth and survival of the alga. On the basis of our culture experiments with the freshwater strain 371, we argue that, when an alga acclimates to environmental changes, this implies that the associated bacterial community needs to be able to follow the alga. This crucial factor is frequently overlooked when studying how an organism reacts to changes in the environment.
The freshwater strain 371 constitutes an excellent model to study the role of bacteria during algal acclimation. It can be cultured in a controlled environment, and bacteria induce a phenotype that is easily measured and of evolutionary interest, as drastic changes in habitat frequently lead to rapid evolutionary radiation events (Lee and Bell, 1999). One of the upcoming challenges is to understand the interactions between algae and bacteria in changing salinities on a functional and metabolic level and to examine their importance in the field. A possible approach towards elucidating these interactions is the use of metabolic networks to model potential metabolic fluxes between algae and bacteria during the acclimation process (Dittami et al., 2014b).
Acknowledgments
We are grateful to Catherine Leblanc, Christian Jeanthon, Dominique Davoult, Philippe Potin, Elianne Sirnæs Egge and Tristan Barbeyron for helpful advice on the experimental setup and analysis. We also thank Ludovic Delage for providing Zobellia galactanivorans DNA, Akira Peters (BEZHIN ROSKO) for providing algal strains, Declan Schroeder for allowing us to use the two South African algal strains, Laurence Dartevelle for performing antibiotic treatments on strain 371, Jonas Collén for critical reading of the manuscript, and the Biogenouest Genomics and Genomer platform core facility for technical support. This work benefited from the support of the French Government via the National Research Agency investment expenditure program IDEALG (ANR-10-BTBR-02-04) and additional support came from the region Brittany via the SAD (Stratégie d'Attractivité Durable) project COGEBRAL.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300. [Google Scholar]
- Billoud B, Nehr Z, Le Bail A, Charrier B. (2014). Computational prediction and experimental validation of microRNAs in the brown alga Ectocarpus siliculosus. Nucleic Acids Res 42: 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bold HC, Wynne MJ. (1985) Introduction to the Algae: Structure and Reproduction, 2nd ed. Prentice-Hall: Englewood Cliffs, New Jersey, USA. [Google Scholar]
- Bolton JJ. (1983). Ecoclinal variation in Ectocarpus siliculosus (Phaeophyceae) with respect to temperature growth optima and survival limits. Mar Biol 73: 131–138. [Google Scholar]
- Burke C, Thomas T, Lewis M, Steinberg P, Kjelleberg S. (2011). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J 5: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6: 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier B, Coelho SM, Le Bail A, Tonon T, Michel G, Potin P et al. (2008). Development and physiology of the brown alga Ectocarpus siliculosus: two centuries of research. New Phytol 177: 319–332. [DOI] [PubMed] [Google Scholar]
- Chisholm JRM, Dauga C, Ageron E, Grimont PAD, Jaubert JM. (1996). ‘Roots' in mixotrophic algae. Nature 381: 382–382. [Google Scholar]
- Cock JM, Peters AF, Coelho SM. (2011). Brown algae. Curr Biol 21: R573–R575. [DOI] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G et al. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465: 617–621. [DOI] [PubMed] [Google Scholar]
- Coulthurst SJ. (2013). The Type VI secretion system - a widespread and versatile cell targeting system. Res Microbiol 164: 640–654. [DOI] [PubMed] [Google Scholar]
- Croft MT, Warren MJ, Smith AG. (2006). Algae need their vitamins. Eukaryot Cell 5: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deligianni E, Pattison S, Berrar D, Ternan NG, Haylock RW, Moore JE et al. (2010). Pseudomonas aeruginosa cystic fibrosis isolates of similar RAPD genotype exhibit diversity in biofilm forming ability in vitro. BMC Microbiol 10: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittami SM, Barbeyron T, Boyen C, Cambefort J, Collet G, Delage L et al. (2014. a). Genome and metabolic network of ‘Candidatus Phaeomarinobacter ectocarpi' Ec32, a new candidate genus of Alphaproteobacteria frequently associated with brown algae. Front Genet 5: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittami SM, Eveillard D, Tonon T. (2014. b). A metabolic approach to study algal-bacterial interactions in changing environments. Mol Ecol 23: 1656–1660. [DOI] [PubMed] [Google Scholar]
- Dittami SM, Gravot A, Goulitquer S, Rousvoal S, Peters AF, Bouchereau A et al. (2012). Towards deciphering dynamic changes and evolutionary mechanisms involved in the adaptation to low salinities in Ectocarpus (brown algae). Plant J 71: 366–377. [DOI] [PubMed] [Google Scholar]
- Dittami SM, Proux C, Rousvoal S, Peters AF, Cock JM, Coppee J-Y et al. (2011). Microarray estimation of genomic inter-strain variability in the genus Ectocarpus (Phaeophyceae). BMC Mol Biol 12: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittami SM, Scornet D, Petit J-L, Ségurens B, Da Silva C, Corre E et al. (2009). Global expression analysis of the brown alga Ectocarpus siliculosus (Phaeophyceae) reveals large-scale reprogramming of the transcriptome in response to abiotic stress. Genome Biol 10: R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S, Harder T, Burke C, Steinberg P, Kjelleberg S, Thomas T. (2013). The seaweed holobiont: understanding seaweed-bacteria interactions. FEMS Microbiol Rev 37: 462–476. [DOI] [PubMed] [Google Scholar]
- Goecke F, Labes A, Wiese J, Imhoff J. (2010). Chemical interactions between marine macroalgae and bacteria. Mar Ecol Prog Ser 409: 267–299. [Google Scholar]
- Guttenplan SB, Kearns DB. (2013). Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev 37: 849–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø, Harper D, Ryan P. (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeontolia Electron 4. [Google Scholar]
- Hollants J, Leliaert F, De Clerck O, Willems A. (2013). What we can learn from sushi: a review on seaweed-bacterial associations. FEMS Microbiol Ecol 83: 1–16. [DOI] [PubMed] [Google Scholar]
- Hubbell SP. (2001) The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32). Princeton University Press: Princeton, NJ, USA. [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Bell M. (1999). Causes and consequences of recent freshwater invasions by saltwater animals. Trends Ecol Evol 14: 284–288. [DOI] [PubMed] [Google Scholar]
- Logares R, Bråte J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, Rengefors K. (2009). Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol 17: 414–422. [DOI] [PubMed] [Google Scholar]
- Magurran AE. (2004) Measuring Biological Diversity. Wiley: Oxford, UK. [DOI] [PubMed] [Google Scholar]
- Maignien L, DeForce EA, Chafee ME, Eren AM, Simmons SL. (2014). Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. MBio 5: e00682–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslet-Cladière L, Delage L, Leroux CJ, Goulitquer S, Leblanc C, Creis E et al. (2013). Structure/Function analysis of a type III polyketide synthase in the brown alga Ectocarpus siliculosus reveals a biochemical pathway in phlorotannin monomer biosynthesis. Plant Cell 25: 3089–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2013), Vegan: Community Ecology Package.
- Parks DH, Beiko RG. (2010). Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26: 715–721. [DOI] [PubMed] [Google Scholar]
- Pedersén M. (1968). Ectocarpus fasciculatus: marine brown alga requiring kinetin. Nature 218: 776–776.5690088 [Google Scholar]
- Pedersén M. (1973). Identification of a cytokinin, 6-(3 methyl-2-butenylamino) purine, in sea water and the effect of cytokinins on brown algae. Physiol Plant 28: 101–105. [Google Scholar]
- Peters AF, Marie D, Scornet D, Kloareg B, Cock JM. (2004). Proposal of Ectocarpus siliculosus (Ectocarpales, Phaeophyceae) as a model organism for brown algal genetics and genomics. J Phycol 40: 1079–1088. [Google Scholar]
- Peterson AT, Soberón J, Pearson RG, Anderson RP, Martínez-Meyer E, Nakamura M et al. (2011) Ecological Niches and Geographic Distributions (MPB-49). Princeton University Press: Princeton, NJ, USA. [Google Scholar]
- Pommier T, Canbäck B, Riemann L, Boström KH, Simu K, Lundberg P et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol 16: 867–880. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2014), R: A language and environment for statistical computing.
- Ritter A, Ubertini M, Romac S, Gaillard F, Delage L, Mann A et al. (2010). Copper stress proteomics highlights local adaptation of two strains of the model brown alga Ectocarpus siliculosus. Proteomics 10: 2074–2088. [DOI] [PubMed] [Google Scholar]
- Sale PF. (1979). Recruitment, loss and coexistence in a guild of territorial coral reef fishes. Oecologia 42: 159–177. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr RC, Zeikus JA. (1993). Utex - the culture collection of algae at the University of Texas at Austin: 1993 list of cultures. J Phycol 29: 1–106. [Google Scholar]
- Staufenberger T, Thiel V, Wiese J, Imhoff JF. (2008). Phylogenetic analysis of bacteria associated with Laminaria saccharina. FEMS Microbiol Ecol 64: 65–77. [DOI] [PubMed] [Google Scholar]
- Tatarenkov A, Bergström L, Jönsson RB, Serrão EA, Kautsky L, Johannesson K. (2005). Intriguing asexual life in marginal populations of the brown seaweed Fucus vesiculosus. Mol Ecol 14: 647–651. [DOI] [PubMed] [Google Scholar]
- Tonon T, Eveillard D, Prigent S, Bourdon J, Potin P, Boyen C et al. (2011). Toward systems biology in brown algae to explore acclimation and adaptation to the shore environment. Omics 15: 883–892. [DOI] [PubMed] [Google Scholar]
- Torode TA, Marcus SE, Jam M, Tonon T, Blackburn RS, Hervé C et al. (2015). Monoclonal antibodies directed to fucoidan preparations from brown algae. PLoS One 10: e0118366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M, Goecke F, Labes A, Dobretsov S, Weinberger F. (2012). The second skin: ecological role of epibiotic biofilms on marine organisms. Front Microbiol 3: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J, Kraft G. (1996). Ectocarpus siliculosus (Dillwyn) Lyngb. from Hopkins River Falls, Victoria–the first record of a freshwater brown alga in Australia. Muelleria 9: 29–33. [Google Scholar]
- Zilber-Rosenberg I, Rosenberg E. (2008). Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32: 723–735. [DOI] [PubMed] [Google Scholar]
- Zinger L, Amaral-Zettler LA, Fuhrman JA, Horner-Devine MC, Huse SM, Welch DBM et al. (2011). Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS One 6: e24570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobell CE. (1941). Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J Mar Res 4: 42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.