Abstract
Plants contain significant levels of natural phenolic compounds essential for reproduction and growth, as well as defense mechanisms against pathogens. Xanthomonas campestris pv. campestris (Xcc) is the causal agent of crucifers black rot. Here we showed that genes required for the synthesis, utilization, transportation, and degradation of 4-hydroxybenzoate (4-HBA) are present in Xcc. Xcc rapidly degrades 4-HBA, but has no effect on 2-hydroxybenzoate and 3-hydroxybenzoate when grown in XOLN medium. The genes for 4-HBA degradation are organized in a superoperonic cluster. Bioinformatics, biochemical, and genetic data showed that 4-HBA is hydroxylated by 4-HBA 3-hydroxylase (PobA), which is encoded by Xcc0356, to yield PCA. The resulting PCA is further metabolized via the PCA branches of the β-ketoadipate pathway, including Xcc0364, Xcc0365, and PcaFHGBDCR. Xcc0364 and Xcc0365 encode a new form of β-ketoadipate succinyl-coenzyme A transferase that is required for 4-HBA degradation. pobA expression was induced by 4-HBA via the transcriptional activator, PobR. Radish and cabbage hydrolysates contain 2-HBA, 3-HBA, 4-HBA, and other phenolic compounds. Addition of radish and cabbage hydrolysates to Xcc culture significantly induced the expression of pobA via PobR. The 4-HBA degradation pathway is required for full pathogenicity of Xcc in radish.
The members of genus Xanthomonas are economically important bacterial pathogens. These infect at least 124 monocotyledonous and 268 dicotyledonous plants and cause severe damage1. X. campestris pv. campestris (Xcc), the causal agent of black rot in crucifers, is the producer of xanthan gum and thus is of great commercial and biotechnological application value2. In addition, Xanthomonas is also a scientifically important bacterial pathogen. X. oryzae pv oryzae (Xoo), X. campestris pathovars, and X. axonopodis pathovars are currently recognized as three of the top 10 plant pathogenic bacteria in molecular plant pathology3.
A characteristic feature of Xanthomonas is the production of yellow, membrane-bound pigments called xanthomonadins4. These pigments are mixtures of unusual brominated, aryl-polyene esters5,6. A previous study conducted by Poplawsky and Chun7 has shown that xanthomonadin production in Xanthomonas is regulated by a diffusible factor (DF). Subsequent investigations showed that the DFs produced by Xcc and Xoo are 3-hydroxybenzoate (3-HBA) and 4-hydroxybenzoate (4-HBA)8,9. Our previous results showed that Xcc synthesizes 3-HBA and 4-HBA using the shikimate pathway product chorismate via the bifunctional chorismatase XanB210. 3-HBA and 4-HBA are further used as intermediates for xanthmonadin synthesis via the pig cluster, and for CoQ8 biosynthesis, respectively10. Further genomic analysis revealed that Xanthomonas strains also contain the putative genes for the transportation and degradation of 3-HBA and 4-HBA (Fig. 1; Supplementary Fig. S1). These findings suggest that the phytopathogen Xanthomonas might have evolved an extensive ability to metabolize 3-HBA and 4-HBA. The mechanistic details and biological significance of this phenomenon remain to be elucidated.
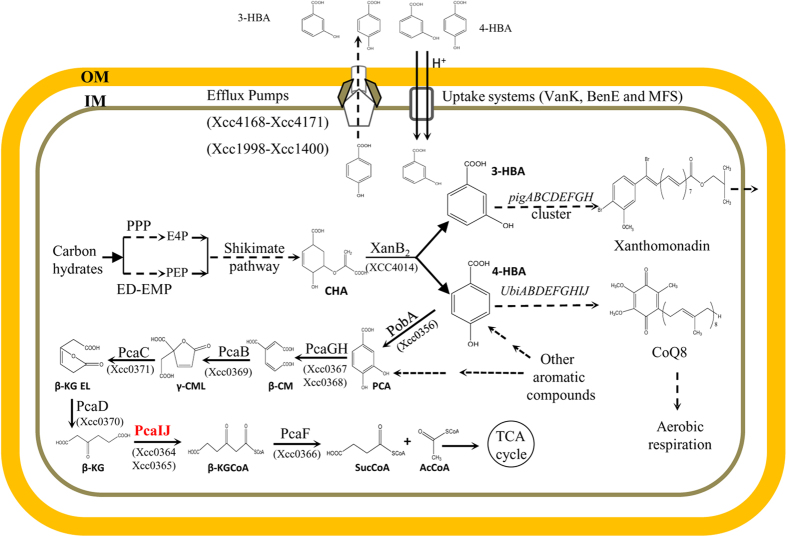
Figure 1. Schematic representation of a model of the synthesis, utilization, transportation, and degradation of 4-HBA in Xcc.
OM, outer membrane; IM, inner membrane; PPP, pentose phosphate pathway; ED-EMP, Entner–Doudoroff pathway and Embden-Meyerhof-Parnas pathway; TCA, tricarboxylic acid cycle; E4P, erythrose-4-phosphate; PEP, phosphoenol-pyruvate; CHA, chorismate; 3-HBA, 3-hydroxybenzoate; 4-HBA, 4-hydroxybenzoate; PCA, protocatechuate; β-CM, β-carboxy-cis,cis-muconate; γ-CML, γ-carboxymuconolactone; β-KG EL, β-ketoadipate enol-lactone; β-KG, β-ketoadipate; β-KGCoA, β-ketoadipyl-CoA; SucCoA, succinyl-CoA; AcCoA, acetyl-CoA; PobA, 4-hydroxybenzoate-3-monooxygenase; PcaGH, protocatechuate 3,4-dioxygenase; PcaB, β-carboxy-cis,cis-muconate cycloisomerase; PcaC, γ-carboxymuconolactone decarboxylase; PcaD, β-ketoadipate enol-lactonase; PcaLM, β-ketoadipate succinyl-CoA transferase; and PcaF, β-ketoadipyl-CoA thiolase.
Aromatic compounds constitute an important source of carbon and energy for soil-dwelling microorganisms and accumulate primarily as the result of the degradation of plant-derived molecules such as lignin11,12. Soil-dwelling microorganisms efficiently degrade a wide range of natural plant phenolic compounds, including 3-HBA and 4-HBA. The gentisate catabolic pathway has been described as the central route for 3-HBA degradation in some bacterial species13,14,15,16. Alternatively, 3-HBA could be degraded through the PCA catabolic pathway by the 3-HBA 4-hydroxylase, which is encoded by the mobA gene in Comamonas testosteroni KH12217. The PCA catabolic pathway, also called the PCA branches of the β-ketoadipate pathway, is a central catabolic route for aromatic compounds, which is widely distributed among taxonomically diverse bacteria and fungi18,19. PCA is a key central intermediate in bacterial degradation of diverse aromatic compounds, including 3-HBA, 4-HBA, and vanillate. PCA oxygenolytic ring-cleavage is catalyzed by PCA 3,4-dioxygenase (PcaGH) to generate 3-carboxy-cis,cis-muconate, which is converted into 4-carboxymuconolactone by 3-carboxy-cis,cis-muconate cycloisomerase (PcaB). 4-Carboxymuconolactone decarboxylase (PcaC) transforms 4-carboxymuconolactone into β-ketoadipate enol-lactone, which is then hydrolyzed by β-ketoadipate enol-lactone hydrolase (PcaD) into β-ketoadipate. The enzyme β-ketoadipate succinyl-CoA tranferase (PcaIJ) converts β-ketoadipate into b-ketoadipyl-CoA, which is finally transformed into succinyl-CoA and acetyl-CoA by β-ketoadipyl-CoA thiolase (PcaF)19. In some microorganisms, the PCA central pathway is involved in 4-HBA degradation. 4-HBA is hydroxylated by 4-HBA 3-hydroxylase, which is encoded by the pobA gene, to yield PCA in Pseudomonas, Burkholderia, Acinetobacter calcoaceticus, and Cupriavidus19,20,21,22. The resulting PCA is further metabolized via the PCA catabolic pathway.
The aims of this study were to characterize the 4-HBA degradation pathway and its biological significance in the model plant pathogen Xcc. This report described for the first time the genes and mechanism underlying 4-HBA degradation in plant pathogenic bacteria. This study demonstrated that the functional 4-HBA degradation pathway is required for full pathogenicity to Chinese radish and is probably involved in the plant-Xanthomonas interactions.
Results
Xcc genome contains a complete set of genes for 4-HBA metabolism
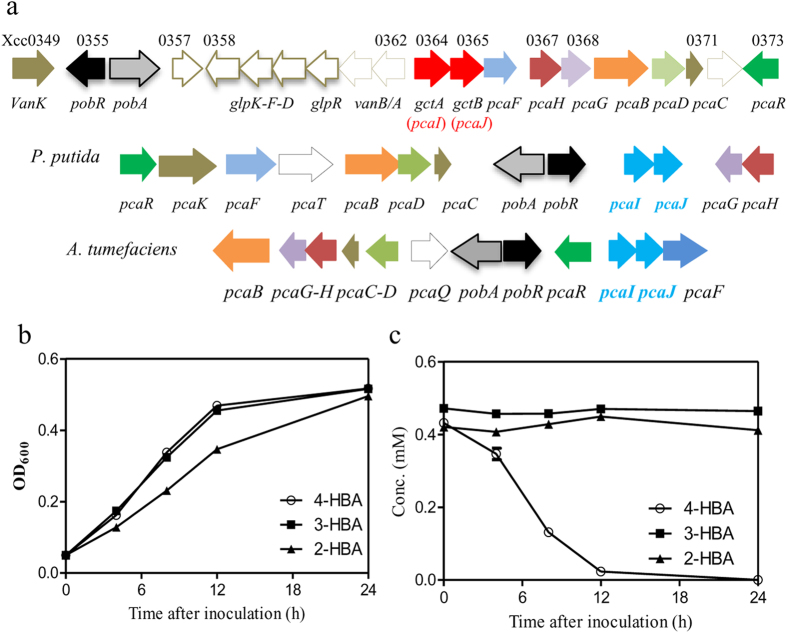
In the present study, we conducted a global comparative genome analysis of Xcc wild-type strain ATCC33913 to identify the genes involved in 3-HBA and 4-HBA metabolism. In addition to the previously characterized genes for 3-HBA and 4-HBA biosynthesis and utilization, we also identified a range of putative genes for 3-HBA and 4-HBA uptake, efflux pumping, and degradation (Fig. 1 and Supplementary Fig. S1). Among these, the products of the cluster Xcc4168-Xcc4171 are homologous to the previously identified 4-HBA efflux pump AaeXBA in Escherichia coli23 (Supplementary Fig. S1a). The gene cluster Xcc1398-Xcc1400 is homologous to the 4-HBA exporter, PP1271-PP1273, which encodes a multidrug efflux MFS transporter in Pseudomonas putida S1224 (Supplementary Fig. S1b). The protein product of the gene Xcc0349 is homologous to the characterized aromatic compound transporter BenK, VanK, or PcaK in P. putida or Acinetobacter sp. strain ADP125,26,27. The genes Xcc1685 and Xcc4153 encode an MFS transporter and benzoate transporter, BenE, respectively (Supplementary Fig. S2b). In particular, the Xcc genome also contains a superoperonic gene cluster (pca cluster hereafter) that harbored the gene pobA, which encodes a 4-HBA 3-monooxygenase and those for the β-ketoadipate pathway identified in P. putida and A. tumefaciens (Fig. 2a). These findings suggest that Xcc is a strain with an extensive ability to metabolize 4-HBA.
Figure 2. Xcc rapidly degrades 4-HBA.
(a) The 4-HBA degradation gene cluster in Xcc, Pseudomonas putida, and Agrobacterium tumefaciens. (b) Growth time course of Xcc in the presence of 2-HBA, 3-HBA or 4-HBA in XOLN medium. (c) Time course of 2-HBA, 3-HBA, and 4-HBA levels in the supernatant of the XC1 culture during growth in XOLN medium.
Xcc rapidly degrades 4-HBA
To further confirm whether the putative 4-HBA degradation pathway in Xcc was functional, 4-HBA was exogenously added into the XOLN cell cultures (OD600 = 0.1) at a final concentration of 0.5 mM. During growth, 4-HBA in the cultures was extracted and quantitatively analyzed by HPLC as previously described10. The results showed that the exogenous addition of 0.5 mM 4-HBA had little effect on Xcc growth (Fig. 2b; Supplementary Fig. S2b). The 4-HBA level in the culture rapidly decreased over time and a very low level of 4-HBA was detected in the culture after 12 h incubation (Fig. 2c). In contrast, when 3-HBA or 2-HBA was added to the same XOLN culture, its levels in the culture were relatively stable during growth (Fig. 2c), indicating that these were not degraded by Xcc.
The pca locus is responsible for 4-HBA degradation in Xcc
The pca locus consists of a total of 19 genes ranging from Xcc0355 to Xcc0373 (a 20-kb gene cluster from position 426,627 to 446,943 in the chromosome of Xcc strain ATCC33913). Among these, the product of Xcc0356 is highly homologous to PobA, which is a 4-HBA 3-monooxygenase that converts 4-HBA into PCA, whereas the product of Xcc0355 is homologous to the regulator PobR in the environmental bioremediation strains Pseudomonas, Burkholderia, Acinetobacter calcoaceticus, and Cupriavidus19,20,21,22. The products of Xcc366-Xcc0371 and Xcc0373 are homologous to PcaFHGBDC and PcaR in the well-characterized β-ketoadipate pathway in the strains Pseudomonas putida KT2440, A. tumefaciens, and Acinetobacter sp. strain ADP119,21. Therefore, Xcc0355, Xcc0356, Xcc366-Xcc0371, and Xcc0373 were renamed accordingly as pobA, pobR, pcaF, pcaH, pcaG, pcaB, pcaD, pcaC, and pcaR in the present study.
Previous studies have shown that when Xcc is grown in a rich medium, it produces and secretes 3-HBA and 4-HBA into the supernatant8,10. We hypothesized that disruption of the 4-HBA degradation pathway promotes the production and secretion of 4-HBA. To test this hypothesis, pobA was deleted or overexpressed in Xcc. The resulting two strains, i.e., ΔpobA and ΔpobA(pobA), and the wild-type strain XC1 were respectively grown in NYG medium and the level of 4-HBA in the culture supernatant was determined. Our results showed that deletion of pobA led to significantly higher level of 4-HBA in the supernatant than that observed in the wild-type strain (Fig. 3a). Overexpression of pobA in the strain ΔpobA resulted in a decrease in 4-HBA production to a level lower than that observed in the wild-type (Fig. 3a). To further confirm the role of pobA in 4-HBA degradation in Xcc, the same three strains were grown in an XOLN liquid medium supplemented with 0.5 mM 4-HBA. Wild-type strain XC1 and strain ΔpobA(pobA) rapidly degraded 4-HBA, whereas strain ΔpobA almost lost its activity (Fig. 3c). pobA deletion or overexpression had no effect on Xcc cell growth in XOLN supplemented with 0.5 mM 4-HBA (Supplementary Fig. S2). Furthermore, strains XC1 and ΔpobA(pobA) showed normal growth on the XOLN plate supplemented with 1.5 mM 4-HBA, whereas strain ΔpobA presented poor growth (Fig. 3e), indicating that PobA was involved in 4-HBA degradation.
Figure 3. PobA and PcaGH are involved in 4-HBA and PCA degradation in Xcc.
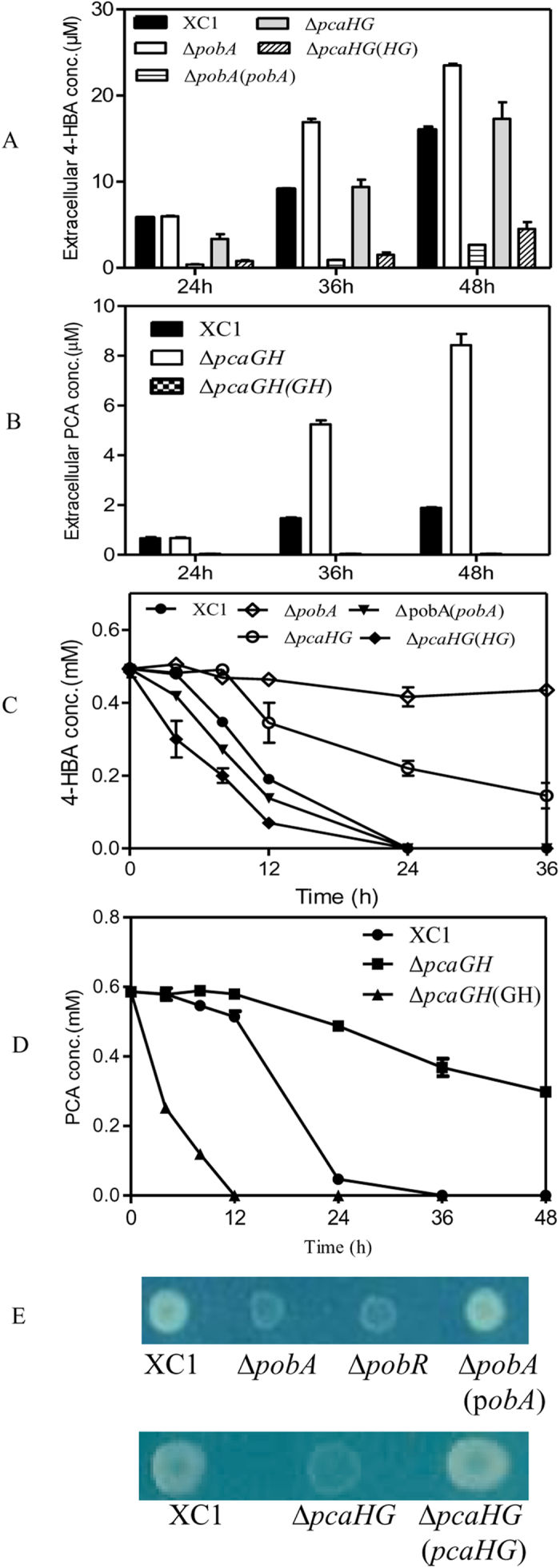
(A) Extracellular 4-HBA concentration of Xcc strains in NYG medium. (B) Extracellular PCA concentration of Xcc strains in NYG medium. (C) Time course of 4-HBA degradation of Xcc strains in XOLN medium with 0.5 mM 4-HBA. (D) Time course of PCA degradation of Xcc strains in XOLN with 0.5 mM PCA. (E) Growth of Xcc strains on an XOLN plate supplemented with 1.5 mM 4-HBA. Data are expressed as the means ± standard deviation of three independent assays.
pcaG and pcaH encode the α- and β-subunits of protocatechuate 3,4-dioxygenase, which acts to convert PCA into β-carboxy-cis,cis-muconate18. Deletion of pcaG and pcaH significantly increased both exogenous 4-HBA and PCA production in the supernatant of NYG cultures, which was restored by overexpression of pcaG and pcaH in the mutant (Fig. 3a,b). When grown in XOLN medium with 0.5 mM 4-HBA or 0.5 mM PCA, strain ΔpcaGH almost lost its ability to degrade PCA or 4-HBA (Fig. 3c,d). Wild-type strain XC1 showed normal growth in the XOLN plate supplemented with 1.5 mM 4-HBA, whereas strain ΔpcaGH presented poor growth (Fig. 3e). These findings confirmed that pcaG and pcaH were also involved in 4-HBA and PCA degradation.
The pca locus also contains two genes, Xcc0357 and Xcc0372, which encode hypothetical proteins, as well as the gene cluster Xcc0358–Xcc0363 (Fig. 2a). The products of Xcc0362 and Xcc0363 are predicted to be responsible for vanillic acid metabolism. Xcc0358–Xcc0361 was associated with glycerol uptake and catabolism. Deletion of these genes imparted minimal effects on exogenous 4-HBA levels, ability to degrade 4-HBA, and bacterial growth (Supplementary Fig. S3).
Xcc0364 and Xcc0365 encode a different form of β-ketoadipate-CoA transferase
In the β-ketoadipate pathway, β-ketoadipate succinyl-CoA transferase, which consists of a α-subunit (PcaI) and a β-subunit (PcaJ), is responsible for converting the β-ketoadipate into β-ketoadipyl-CoA18. In most β-ketoadipate pathway-containing bacterial species such as A. tumefaciens and A. baylyi, pcaI, pcaJ, and pcaF are usually transcribed within the same operon28. In the pca cluster of Xcc, genes encoding for β-ketoadipate succinyl-CoA transferase proteins (PcaIJ) were not detected (Fig. 2a). Two genes, Xcc0364 and Xcc0365, which were originally annotated as glutaconate CoA transferase subunits A (gctA) and B (gctB), were localized upstream of pcaF (Fig. 2a). The coding sequences of Xcc0364, Xcc0365, and PcaF overlapped by three base pairs, respectively, in the chromosome (Fig. 4a), which suggested that these were organized as a single transcriptional unit and were functionally associated. Domain organization analysis showed that Xcc0364, Xcc0365, PcaI, and PcaJ belong to the same SugarP_isomerase superfamily and contained the same CoA_trans domain (Supplementary Figs S4, S5), further supporting our hypothesis. However, the low amino acid sequence similarity of Xcc0364 and Xcc0365 with PcaI and PcaJ in A. tumefaciens (PcaI, 18.7%; PcaJ, 19.4%) and P. putida (PcaI, 16.7%; PcaJ, 15.3%) prevented their annotation as orthologs of PcaI and PcaJ. In addition, signature sequences (glycine cluster and SENG motif, respectively) typically present in PcaI and PcaJ of many species were absent or modified in the products of Xcc0364 and Xcc0365 (Supplementary Figs S4, S5). These findings suggest that Xcc0364 and Xcc0365 might be encoding a different form of β-ketoadipate-CoA transferase.
Figure 4. Xcc0364 and Xcc0365 are involved in 4-HBA degradation in Xcc.
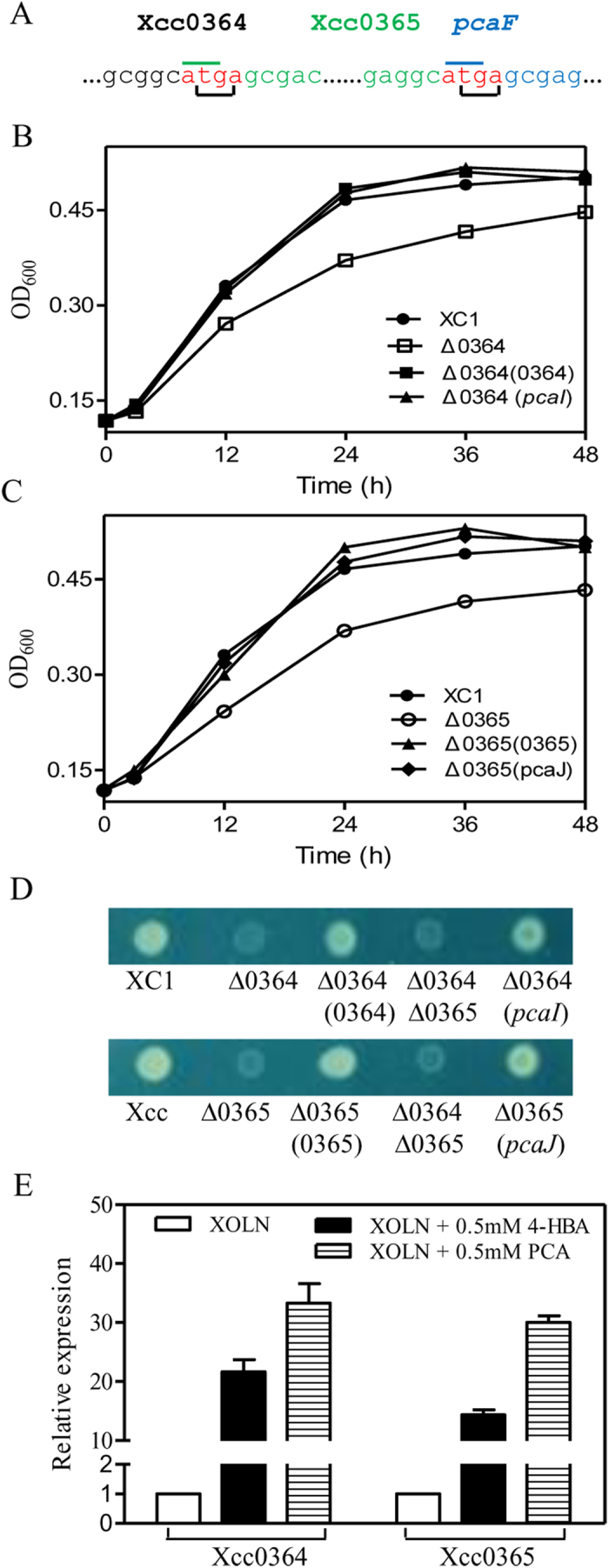
(A) Genetic organization of Xcc0364 and Xcc0365 in the Xcc genome. (B,C) Growth of Xcc strains in the XOLN medium supplemented with 1.5 mM 4-HBA. (D) Growth of Xcc strains on an XOLN plate supplemented with 2.5 mM 4-HBA. (E) Relative expression of Xcc0364 and Xcc0365 of XC1 strain in the presence of 0.5 mM 4-HBA or 0.5 mM PCA. Data are expressed as the means ± standard deviation of three independent assays.
To investigate whether Xcc0364 and Xcc0365 encode an β-ketoadipate succinyl-CoA transferase, we generated deletion and overexpression strains of the two genes, namely, ΔXcc0364 and ΔXcc0364(0364), and ΔXcc0365 and ΔXcc0365(0365). First, to determine whether β-ketoadipate accumulated from PCA metabolism in strains ΔXcc0364 or ΔXcc0365, we performed the Rothera test, which detects the presence of β-ketoadipate and thus indicates whether PCA has been metabolized to this pathway intermediate29. The wild-type strain XC1 exhibited a Rothera-negative phenotype in the presence of 0.1 mM PCA, whereas strains ΔXcc0364 or ΔXcc0365 were Rothera-positive, indicating the accumulation of β-ketoadipate. The strains overexpressing Xcc0364 or Xcc0365 also resulted in a Rothera-negative phenotype. These results suggest that Xcc0364 and Xcc0365 are involved in β-ketoadipate metabolism in Xcc.
Second, the growth of all strains was compared in XOLN liquid or solid media supplemented with 4-HBA. Wild-type strain XC1 and strains ΔXcc0364 (0364) or ΔXcc0365 (0365) showed better growth than strains ΔXcc0364 or ΔXcc0365 in liquid XOLN medium with 1.5 mM 4-HBA (Fig. 4b,c) or XOLN plate with 2.5 mM 4-HBA (Fig. 4d). A previous study has shown that genes pcaI and pcaJ in P. putida encode the α and β subunits of β-ketoadipate succinyl-CoA transferase18. The present study showed that pcaI-overexpressing strain ΔXcc0364 followed a similar growth pattern to that of wild-type strain XC1 (Fig. 4c,d). Similarly, pcaJ-overexpressing strain ΔXcc0365 displayed a similar growth pattern as that of wild-type strain XC1 (Fig. 4c,d).
Finally, qRT-PCR analysis showed that addition of PCA or 4-HBA to the XC1 XOLN culture at a final concentration of 0.5 mM significantly induced the expression of Xcc0364 and Xcc0365 (Fig. 4e). Taken together, we concluded that Xcc0364 and Xcc0365 encode subunits of a new form of β-ketoadipate succinyl-CoA transferase, and these two genes were renamed pcaI and pcaJ, respectively. Further genomic assessment revealed that the homologs of Xcc0364 and Xcc0365 were not only present in most of the genomes of Xanthomonas species deposited in the NCBI microbe genome database, but also present in the genomes of Lysobacter capsici, Pseudomonas aeruginosa PAO1, Pseudomonas knackmussii, Sinorhizobium meliloti, and Mesorhizobium loti, with high amino acid identity (>60%) (Supplementary Figs S6, S7).
pobA expression is significantly induced by 4-HBA via the transcriptional regulator PobR
The pca cluster in Xcc contains one gene, Xcc0355, which encodes an AraC-type transcriptional regulator, PobR (Fig. 5a). pobR is located adjacent to pobA, although its transcriptional orientation is in the opposite direction (Fig. 2a). PobR has been shown to be the activator for the 4-HBA degradation pathway in Acinetobacter sp. strain ADP130. To study the effect of pobR on the expression of the 4-HBA degradation gene in Xcc, we generated pobR deletion and overexpression strains ΔpobR and ΔpobR (pobR). The expression pattern of pobA in Xcc strains during growth in XOLN medium or XOLN medium supplemented with 4-HBA was determined by qRT-PCR analysis. When grown in XOLN medium, pobA expression was relatively low at 12 h and 24 h after inoculation, and significantly increased at 36 h after inoculation (Fig. 5b). Deletion of pobR significantly reduced the expression of pobA at 36 h after inoculation, whereas overexpression of pobR resulted in the upregulation of pobA (Fig. 5b). Addition of 4-HBA (0.1 mM or 0.5 mM) to the wild-type XC1 culture resulted in a 3.5 ∼ 6.0-fold increase in the expression of pobA, but not in the ΔpobR culture (Fig. 5c). Furthermore, our results showed that deletion of pobR almost abolished 4-HBA degradation activity, and overexpression of pobR in strain ΔpobR restored 4-HBA degradation activity to that of the wild-type level (Fig. 5d). These findings suggest that 4-HBA via the activator PobR induced the expression of pobA.
Figure 5. 4-HBA induces the expression of 4-HBA degradation genes via the regulator, PobR.
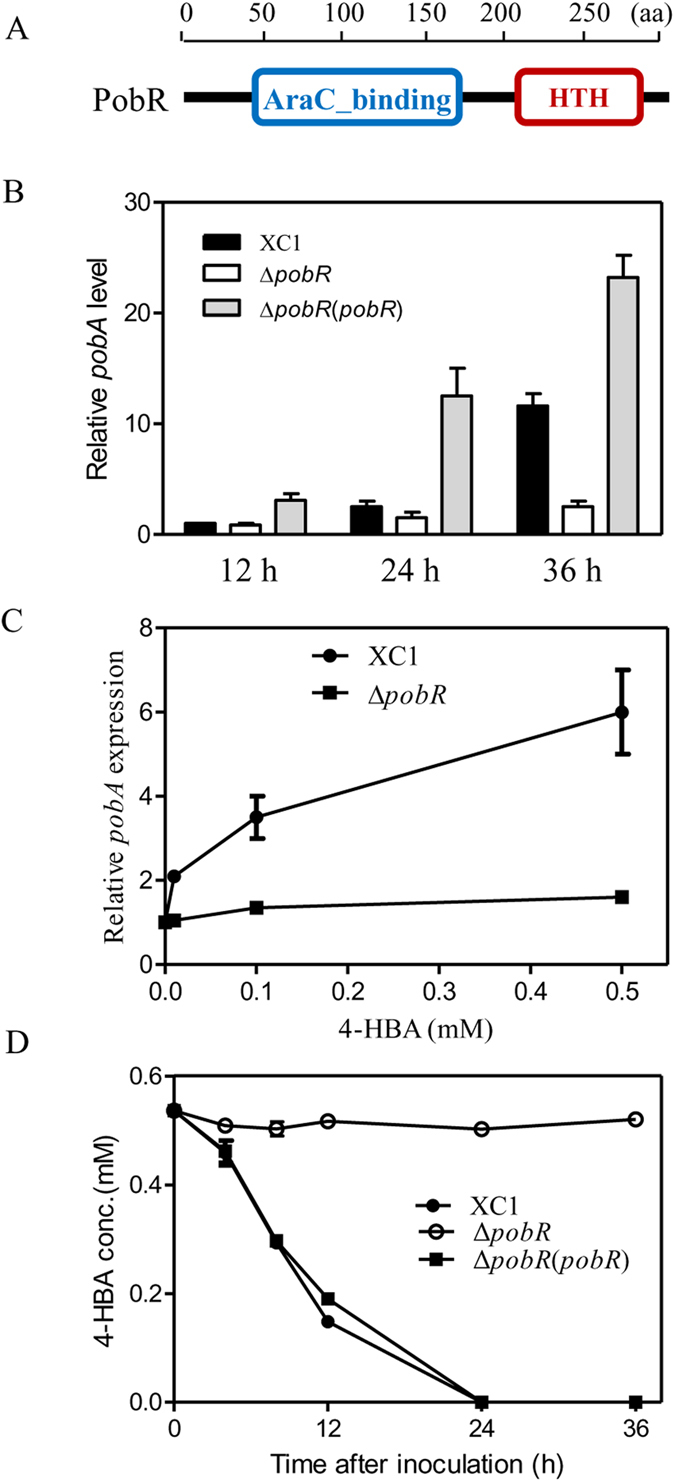
(A) Domain organization of PobR. (B) Time course of pobA expression in Xcc strains during growth. (C) Relative expression of pobA in strains XC1 and ΔpobR grown in the medium XOLN supplemented with 0.01 mM, 0.1 mM and 0.5 mM 4-HBA. (D) Time course of 4-HBA degradation of strains XC1, ΔpobR, and ΔpobR (pobR) in XOLN medium. Data are expressed as the means ± standard deviation of three independent assays.
Radish and cabbage hydrolysates induce pobA expression
Plants contain significant levels of natural phenolic compounds that play essential functions in plant reproduction and growth, as well as defense mechanisms against pathogens31. Phenolic acids are a major class of phenolic compounds, which mainly include hydroxybenzoic acids (e.g., gallic acid, 4-HBA, PCA, vanillic acid, and syringic acid) and hydroxycinnamic acids (e.g., ferulic acid, caffeic acid, p-coumaric acid, chlorogenic acid, and sinapic acid)32. We assumed that the 4-HBA degradation pathway in Xcc plays a role in detoxifying phenolic metabolites in the host during the infection. To test this hypothesis, radish and cabbage hydrolysates were prepared as described in Materials and Methods. Based upon the Folin-Ciocalteu method, the phenolic concentration within the radish and cabbage hydrolysates were 42.3 mg/g dry weight and 54.2 mg/g dry weight, respectively. The hydrolysate samples were added to the Xcc culture (OD600 = 0.8) in XOLN medium at three final phenolic concentrations, 1 mg/L, 10 mg/L, and 100 mg/L. After incubation for 3 h, the cells were collected for pobA gene expression analysis by qRT-PCR. Addition of radish or cabbage hydrolysates had little effect on Xcc growth (data not shown). The addition of the hydrolates at 10 mg/L or 100 mg/L phenolic compounds to the wild-type XC1 culture elicited a clear dose-dependent response in pobA expression (Fig. 6a). In contrast, the addition of the hydrolysates to the ΔpobR cultures had little effect on pobA expression (Fig. 6b).
Figure 6. Relative expression of pobA in the presence of radish hydrolates (RHs) or cabbage hydrolates (CHs).
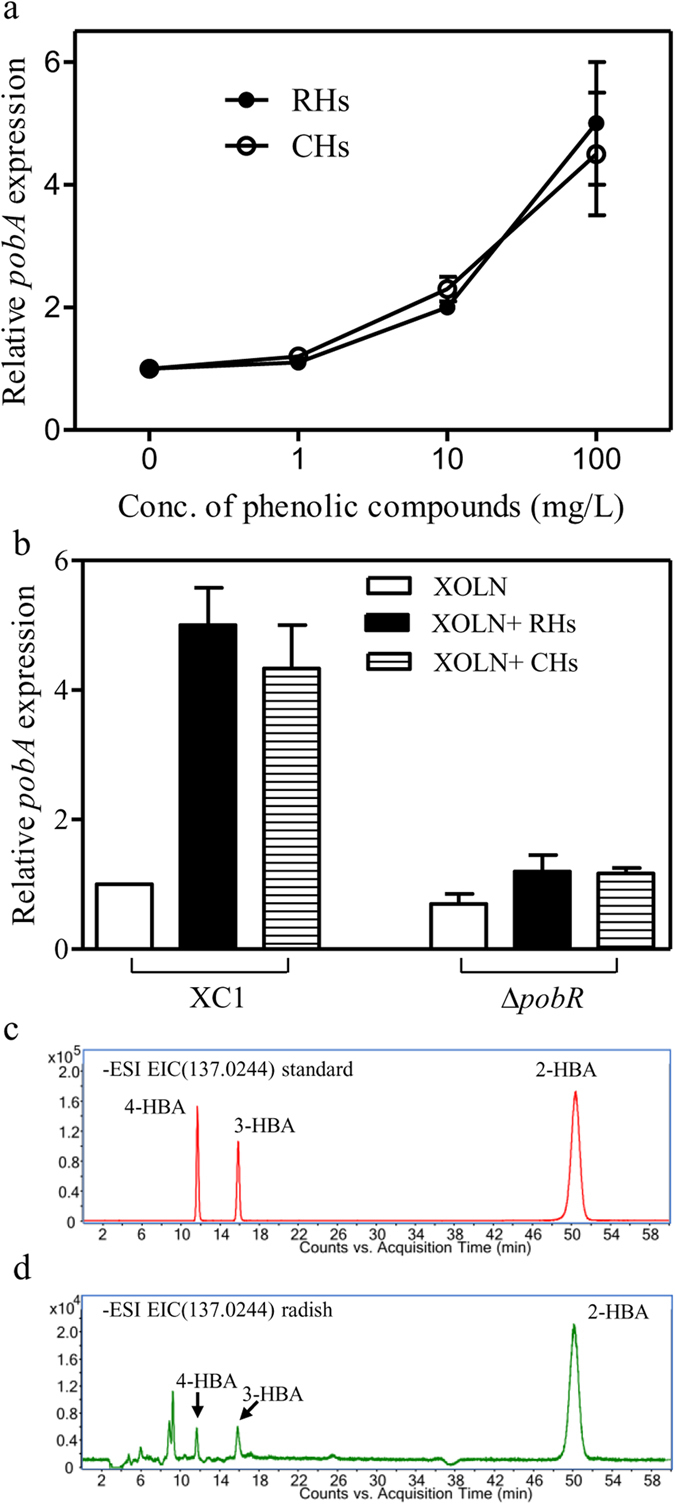
(a) The dose-dependent pobA expression in the wild-type strain XC1 in the presence of 0.1–100 mg/L phenolic compounds. (b) The relative pobA expression in the strains of XC1 and ΔpobR. (c) The extracted ion chromatograms of standards 2-HBA, 3-HBA and 4-HBA at 50 μM. (d) The extracted ion chromatograms of 2-HBA, 3-HBA and 4-HBA in radish hydrolysates. Total RNA was extracted from the cultures 3 h after addition of the hydrolates. The relative levels of pobA were determined by quantitative real-time RT-PCR. Data are expressed as the means ± standard deviation of three independent assays.
Furthermore, the phenolic compounds in radish and cabbage hydrolysates were extracted as previously described8. LC-MS analysis revealed that radish hydrolysates contain 2-HBA, 3-HBA, 4-HBA, and other uncharacterized phenolic compounds (Fig. 6c,d; Supplementary Fig. S9a). Based on the established standard curves (Supplementary Fig. S9b), the absolute concentration of 2-HBA, 3-HBA, and 4-HBA present in radish leaves was estimated to be 262 ng/g fresh weight, 114 ng/g fresh weight, and 122 ng/g fresh weight, respectively. Similar phenolic compounds pattern was also observed in cabbage hydrolysates (data not shown).
4-HBA degradation pathway is required for full pathogenicity in radish
In the present study, the production of virulence factors such as extracellular polysaccharide (EPS) and extracellular enzymes in mutant strains ΔpobA, ΔpcaG, and ΔpcaI were compared to those in wild-type strain XC1 using the rich medium NYG. Our results showed that deletion of pobA, pcaGH, or pcaI had minimal effects on the production of cellulase, amylase, protease, and EPS (Supplementary Fig. S8).
To determine the role of the 4-HBA degradation pathway on the pathogenicity of Xcc, mutant strains ΔpobA and ΔpcaGH were inoculated in Chinese radish. Our results showed that the lesion length of these mutant strains 2 weeks after inoculation ranged from 12.1 mm to 14.0 mm, which respectively were 15.0% to 26.5% less than the observed 16.5 mm in the wild-type strain XC1.
Discussion
The present study demonstrated that the phytopathogen Xcc contains a functional 4-HBA degradation pathway, which consists of 4-HBA hydroxylase (PobA) and the PCA branches of the β-ketoadipate pathway. 4-HBA degradation activity has been experimentally shown in P. putida, A. baylyi strain ADP1, A. tumefaciens, and C. necator JMP13430,33,34,35. Generally, the genes for 4-HBA degradation are organized, function, and are regulated in Xcc in a manner similar to those of the above strains, in particular, to that previously described in A. tumefaciens. However, the present study also revealed several unique features in the 4-HBA degradation mechanism in Xcc. First, the 4-HBA degradation genes in Xcc are organized in a more complicated superoperonic gene cluster. In A. tumefaciens, the two pca operons were clustered in close proximity, flanking the putative pobA gene (Fig. 2a). In P. putida, the genes for 4-HBA degradation were dispersed in three discrete regions (Fig. 2a). In Xcc, the pca genes were located in two discrete operons, with the 4-HBA catabolic genes about 9 kb away and the glycerol and vanillic acid catabolic genes in the intervening regions (Fig. 2a). The multioperonal grouping of genes may reflect their acquisition by horizontal transfer, as well as their evolution in concert by sequence exchange36. Although the present study showed that the genes for glycerol and vanillic acid catabolism in the superoperonic cluster are not required for 4-HBA degradation in Xcc (Supplementary Fig. S3), its biological significance requires further investigations. Second, a new form of β-ketoadipate succinyl-CoA transferase was involved in 4-HBA degradation in Xcc, which will be discussed in the next section. Third, the present study, for the first time, has shown that the expression of 4-HBA degradation genes was significantly induced by the hydrolysates of the host plants (Fig. 7), suggesting that the 4-HBA degradation pathway was involved in the interaction between the plant and Xanthomonas.
Figure 7. Virulence of pobA and pcaGH on Chinese radish.
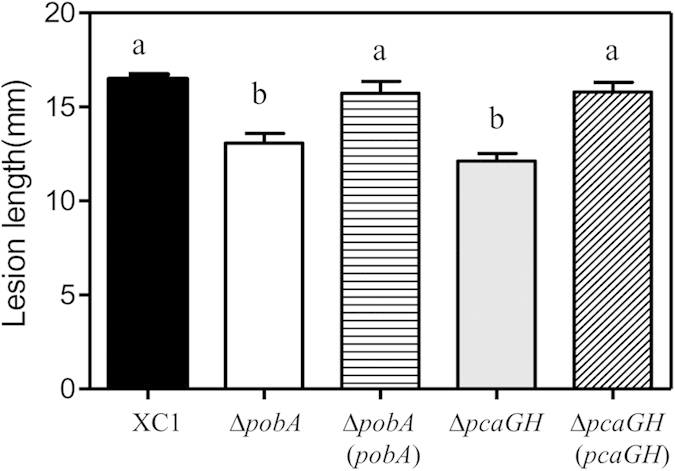
Virulence of the Xcc strains was tested by measuring lesion length after introducing bacteria into the vascular system of Chinese radish “Manshenhong” by leaf clipping. Values are expressed as the mean and standard deviation of triplicate measurements, each comprising 15 leaves. * and ** indicate significant differences between treatments (LSD at P = 0.05).
In bacterial species such as P. putida, A. baylyi, A. tumefaciens, and B. japonicum, the transfer of CoA to β-ketoadipate is catalyzed by β-ketoadipate succinyl-CoA transferase (PcaIJ). In the present study, by combining the Rothera test, expression profiles, and genetic data, we demonstrated that Xcc0364 and Xcc0365 have similar activity to PcaIJ and were required for 4-HBA and β-ketoadipate degradation in Xcc. Although the products of Xcc0364/Xcc0365 shared limited amino acid sequence identity to that of PcaIJ of A. tumefaciens and P. putida (Supplementary Figs S5, S6), these were highly homologous to those of SMB20587 (67%) and SMB20588 (60%), respectively, in S. meliloti. The latter two have been purified and shown to have β-ketoadipate succinyl-CoA transferase activity in vitro11. These findings strongly support that Xcc0364 and Xcc0365 encode the same form of β-ketoadipate succinyl-CoA transferase in S. meliloti. Therefore, the present findings are in good agreement with the previous assumption that at least two forms of β-ketoadipate succinyl-CoA transferase are present in bacteria11. The first one is present in the bacterial species such as A. baylyi, P. putida, A. tumefaciens, and B. japonicum, whereas the other one is present in Xanthomonas sp., Lysobacter sp., Pseudomonas aeruginosa, M. loti, and S. meliloti. The biological significance of the presence of two forms of β-ketoadipate succinyl-CoA transferase remains to be explored. It appears that in the course of evolution, natural selection has caused the β-ketoadipate pathway to assume a characteristic set of features or identity in different bacteria18. The new form of β-ketoadipate succinyl-CoA transferase encoded by Xcc0364 and Xcc0365 is present in most of the phytopathogens Xanthomonas. Whether these are related to specific lifestyles of Xanthomonas deserves further investigation.
Plants contain significant levels of natural phenolic compounds that play essential functions in the plant reproduction and growth, as well as defense mechanisms against pathogens31. In response to pathogenic attack, diverse broad spectrum antimicrobial substances are synthesized de novo by plants that accumulate rapidly at areas of pathogen infection. They may puncture the cell wall, delay maturation, disrupt metabolism or prevent reproduction of the pathogen in question37. Among these, glucosinolates and phenolics are well-known pathogen-induced metabolites of Brassicaceae family38,39. In addition, phytopathogens like Xcc may be exposed to a large amount of natural phenolic compounds derived from cell wall degradation during an infection. As a vascular pathogen, Xcc is normally restricted to the xylem tissues of infected plants. Within the xylem, Xcc multiplies and forms a microcolony, and then starts to produce various enzymes that would degrade the xylem walls for nutritional purpose40. The degradative enzymes not only cleave the cell wall to simple sugars, but also release lignin, which, when hydrolyzed, forms various types of aromatics such as 4-HBA, PCA, ferulic acid vanillic acid, and p-coumaric acid22. Several of these aromatics have been shown to be inhibitory towards fermentative microbes41,42. Some of them may influence the pathogen’s virulence machinery For instance, the classic immune hormone salicylic acid (2-HBA) has been shown to reduce virulence of A. tumefaciens by inhibiting the VirA/VirG two-component system43. In the opportunistic pathogen Pseudomonas aeruginosa, 2-HBA has been reported to reduce the production of several virulence factors including motility, biofilm formation and quorum sensing signal production44. Therefore, the ability of Xcc to survive phenolic compound stress is of critical importance for its successful colonization of host plants. The present study showed that Xcc contains a functional 4-HBA degradation system, which is required for full pathogenicity in radish (Fig. 7). The expression of the key gene pobA could be induced by 4-HBA or plant hydrolysates (Fig. 6). Therefore, the 4-HBA degradation system might be used to evade or subvert phenolic compound stress in Xcc. Similar results have also been reported in the Gram-positive Arthrobacter in the phyllosphere where the expression of cph genes for the degradation of pollutant 4-chlorophenol could be induced by natural phenolic compounds45. A similar 4-HBA degradation system is also present in other Xanthomonas species (Supplementary Figs S6, S7), suggesting that it could be a common strategy among phytopathogens. The detailed mechanisms on how 4-HBA degradation pathway contributes to the pathogenicity and plant-Xanthomonas interactions need to be further explored.
In addition to 2-HBA, 3-HBA and 4-HBA, plant phenolic compounds also include many other compounds like ferulic acid, vanillic acid, p-coumaric acid32. In bacteria, these compounds are initially transformed to a limited number of central intermediates, namely catechol and PCA. These intermediates are then channeled into two possible ring fission pathways, funneling them into the tricarboxylic acid cycle13,22. For example, ferulic acid is initially degraded to PCA via vanillic acid, whereas p-coumaric acid is degraded via 4-HBA in some Gram-negative bacteria46,47. These catabolic conversion steps required multiple genetic loci. The transformation of ferulic acid to vanillic acid involves an enoyl-CoA hydratase/aldolase, a vanillin dehydrogenase and a feruloyl coenzyme A synthase. Vanillic acid is then degraded to PCA by a demethylase encoded by two genes designated vanA and vanB22,47. The degradation of p-coumaric acid to 4-HBA also requires at least one locus that transforms ferulic acid to vanillic acid47. At lease two sets of vanA and vanB (Xcc0361-Xcc0362, Xcc0296-Xcc0297) were identified in the genome of Xcc strain ATCC33913. Interestingly, the former set is located within the pca cluster encoding 4-HBA degradation pathway (Fig. 2). These findings suggest that Xcc might degrade vanillic acid and other aromatic compounds via 4-HBA or PCA degradation pathway. Further genetic and functional identification of the molecular nature of diverse aromatic compounds degradation pathways in Xcc will not only help to elucidate the adaptation and virulence mechanism, but also provide a novel target for the development of Xcc-resistant crops.
Methods
Bacterial strains and growth conditions
The bacterial strains used in the present study are described in Supplementary Table S1. Xcc strain XC1 was grown in XOLN medium (5 g/L sucrose, 0.7 g/L K2HPO4, 0.2 g/L KH2PO4, 1 g/L (NH4)2SO4, 0.1 g/L MgCl2·6H2O, 0.01g/L FeSO4·7H2O, and 0.001 g/L MnCl2·4H2O, pH 7.15) or NYG medium (5 g/L peptone, 3 g/L yeast extract, and 2 g/L glycerol, pH 7.0) at 28 °C. E. coli strains were grown in LB medium at 37 °C. When required, rifampicin and kanamycin were added at final concentrations of 25 μg/mL and 50 μg/mL, respectively.
Construction of in-frame deletion mutants and complementation analysis
The Xcc wild-type strain XC1 was used as parental strains for the generation of deletion mutants, as previously described48. The primers used are listed in Supplementary Table S2. For complementation analysis, the target gene was PCR amplified and cloned into the MCS site of the expression plasmid pBBR1MCS2. The resulting construct was transferred into Xcc by triparental mating.
Extraction and quantitative analysis of 4-HBA and PCA by HPLC
4-HBA and PCA extraction and quantitative analysis were performed as previously described by Zhou et al.10. 4-HBA and PCA production was quantified using the peak area in HPLC elute. Commercially available 4-HBA and PCA (Sigma) were used as standards.
Rothera test for the detection of β-ketoadipate
Rothera test was conducted following the method described by Holding and Collee29, with minor modifications. Briefly, an overnight NYG culture was centrifuged and washed with XOLN medium. Cells were subcultured into the XOLN medium supplemented with 0.1 mM protocatechuate for overnight incubation at 30 °C. Cells were centrifuged and resuspended in 0.02 M Tris-HCl (pH 8.0) to an optical density (OD) of 1.0. Toluene (0.5 mL) was added to 2 mL of resuspended cells, which was then incubated at 30 °C with shaking for 1 h. After shaking, 1 g of (NH4)2SO4 was added to the mixture and then vortexed. One drop of a fresh aqueous sodium nitroprusside (1%) solution was then added, followed by the addition of 1 drop of concentrated NH3 (29%), and the mixture was vortexed. Development of a purple color within 5 min following the addition of NH3 was considered as a positive indication for the presence of β-ketoadipate.
Total RNA extraction and purification and qRT-PCR analysis
The total RNA of Xcc strains was extracted and purified using RNeasy Miniprep Kit (QIAGEN). Genomic DNA was removed by DNase I (QIAGEN). cDNA synthesis was conducting using PrimeScript RT Reagent Kit (TAKARA). RT-qPCR was performed in Mastercycler ep Realplex 4S (Eppendorf) with SYBR Premix EX Taq (TAKARA). Relative expression levels were calculated by using the 2−Δ Δ CT method, and the gene atpD was used as reference to normalize all samples and replicates.
Preparation of radish and cabbage hydrolysate samples and treatment of XC1 culture
A total of 1,000 g of radish (Raphanus sativus Manshenhong) or cabbage (Brassica oleracea L. Jingfeng) leaves were chopped into small pieces and blended in 200 mL of sterile water in an electric juicer (PHILIPS). The resulting samples were pretreated by adding NaOH at a final concentration of 1% (wt/vol) and by autoclaving at 121 °C for 15 min. After removing any remaining debris by passing the mixture through a filtration cloth, the filtrate was adjusted to a pH of 7.1 using hydrochloric acid (5 N) and lyophilized to generate a dry sample. The hydrolates were added to the XC1 cell culture at an OD600 = 0.8 at a range of final concentrations. After incubation for 3 h, the cells were collected for total RNA extraction and gene expression analysis. The phenolics present in the samples were quantified using the Folin-Ciocalteu reagent method49.
Quantitative analysis of 4-HBA and other phenolic compounds in radish and cabbage hydrolysates via LC-MS
We followed the previously described method by He et al.8 to extract 4-HBA and other phenolic compounds in plant hydrolysates. The resulting residues were dissolved in 500 μl of methanol. A three-microliter aliquote of extracted sample was then injected into the ultra-performance liquid chromatography coupled with mass spectrometry apparatus (Agilent UPLC1290-TOF-MS6230) under the following conditions: Agilent Zorbax XDB C18 reverse-phase (5 μm, 4.6 × 150 mm) eluted with methonal with 0.1% formic acid and H2O with 0.1% formic acid (30:70) at 0.4 ml/min. The MS analysis was performed under negative mode with a scanning range of m/z = 100–1700. The specific pseudo molecular ion (M-H)− of 2-HBA, 3-HBA, and 4-HBA were extracted at 137.0244. The retention time of 2-HBA, 3-HBA, and 4-HBA were 50.17 min, 15.88 min, and 11.68 min, respectively. The concentration of HBA molecules was quantified with a peak intensity (PI) of the specific extracted ion chromatogram (EIC) in the total ion chromatogram (TIC) according to the following formula: 2-HBA (μM) = 4.560 × 10−6 × PI + 0.404 with a R2 of 0.9994; 3-HBA (μM) = 2.643 × 10−5 × PI + 0.521 with a R2 of 0.9993; 4-HAB (μM) = 2.323 × 10−5 × PI + 0.181 with a R2 of 0.9998.
Determination of extracellular enzyme activity and EPS production and virulence testing
Determination of extracellular enzyme activity and EPS production was performed as previously described45. Xcc virulence in Chinese radish was estimated by leaf clipping. Fresh cell cultures were used to inoculate at an OD600 of 0.01. The lesion length was scored 14 days after inoculation. Fifteen leaves from each tested strain were inoculated. Each strain was tested in at least three separate experiments.
Additional Information
How to cite this article: Wang, J.-Y. et al. A functional 4-hydroxybenzoate degradation pathway in the phytopathogen Xanthomonas campestris is required for full pathogenicity. Sci. Rep. 5, 18456; doi: 10.1038/srep18456 (2015).
Supplementary Material
Acknowledgments
We thank Prof. Ping Xu for providing Pseudomonas putida strain. This work was financially supported by the research grants from the National Natural Science Foundation of China (No. 31272005 to HYW, No. 31301634 to ZL) and the Special Fund for Agro-Scientific Research in the Public Interest (No. 201303015 to HYW).
Footnotes
Author Contributions H.Y.W. and T.J.L. conceived and designed the experiments. W.J.Y., Z.L., C.B. and L.M. performed the experiments. H.Y.W., W.J.Y., Z.L. and T.J.L. analyzed the data. L.M. and T.H. contributed reagents & materials. S.S. and Z.W. performed LC-MS analysis. J.B.L. conducted the virulence. H.Y.W., W.J.Y. and Z.L. wrote the main manuscript text.
References
- Leyns F., De Cleene M., Swings J.-G. & De Ley J. The host range of the genus Xanthomonas. Bot Rev. 50, 308–356 (1984). [Google Scholar]
- Garcia-Ochoa F., Santos V. E., Casas J. A. & Gomez E. Xanthan gum: production, recovery, and properties. Biotechnol Adv. 18, 549–579 (2000). [DOI] [PubMed] [Google Scholar]
- Mansfield J. et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 13, 614–629 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr M. The Genus Xanthomonas. Pages 742-763 in: The Prokaryotes, Starr M., Stolp H., Trüper H., Balows A., and Schlegel H., eds Springer, Berlin Heidelberg (1981). [Google Scholar]
- Andrewes A. G., Jenkins C. L., Starr M. P., Shepherd J. & Hope H. Structure of xanthomonadin I, a novel dibrominated aryl-polyene pigment produced by the bacterium Xanthomonas juglandis. Tetrahedron Lett. 17, 4023–4024 (1976). [Google Scholar]
- Aririatu L. E. & Kester A. S. Isolation and characterization of the pigment esters of Xanthomonas juglandis (campestris). J Gen Microbiol. 131, 2047–2052 (1985). [Google Scholar]
- Poplawsky A. R. & Chun W. pigB determines a diffusible factor needed for extracellular polysaccharide slime and xanthomonadin production in Xanthomonas campestris pv. campestris. J Bacteriol. 179, 439–444 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. W. et al. Xanthomonas campestris diffusible factor is 3-hydroxybenzoic acid and is associated with xanthomonadin biosynthesis, cell viability, antioxidant activity, and systemic invasion. Molecular plant-microbe interactions. Mol Plant-Microbe Interact. 24, 948–957 (2011). [DOI] [PubMed] [Google Scholar]
- Zhou L. et al. The rice bacterial pathogen Xanthomonas oryzae pv. oryzae produces 3-hydroxybenzoic acid and 4-hydroxybenzoic acid via XanB2 for use in xanthomonadin, ubiquinone, and exopolysaccharide biosynthesis. Mol Plant-Microbe Interact. 26, 1239–1248 (2013). [DOI] [PubMed] [Google Scholar]
- Zhou L. et al. The diffusible factor synthase XanB2 is a bifunctional chorismatase that links the shikimate pathway to ubiquinone and xanthomonadins biosynthetic pathways. Mol Microbiol. 87, 80–93 (2013). [DOI] [PubMed] [Google Scholar]
- MacLean A. M., MacPherson G., Aneja P. & Finan T. M. Characterization of the beta-ketoadipate pathway in Sinorhizobium meliloti. Appl Environ Microbiol. 72, 5403–5413 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona M. et al. Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol Mol Biol Rev. 73, 71–133 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G., Boll M. & Heider J. Microbial degradation of aromatic compounds–from one strategy to four. Nat Rev Microbiol. 9, 803–816 (2011). [DOI] [PubMed] [Google Scholar]
- Ishiyama D., Vujaklija D. & Davies J. Novel pathway of salicylate degradation by Streptomyces sp. strain WA46. Appl Environ Microbiol. 70, 1297–1306 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. X., Liu H. & Zhou N. Y. MhbR, a LysR-type regulator involved in 3-hydroxybenzoate catabolism via gentisate in Klebsiella pneumoniae M5a1. Microbiol Res. 165, 66–74 (2010). [DOI] [PubMed] [Google Scholar]
- Chao H. & Zhou N. Y. GenR, an IclR-type regulator, activates and represses the transcription of gen genes involved in 3-hydroxybenzoate and gentisate catabolism in Corynebacterium glutamicum. J Bacteriol. 195, 1598–1609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromoto T. et al. Characterization of MobR, the 3-hydroxybenzoate-responsive transcriptional regulator for the 3-hydroxybenzoate hydroxylase gene of Comamonas testosteroni KH122-3s. J Mol Biol. 364, 863–877 (2006). [DOI] [PubMed] [Google Scholar]
- Harwood C. S. & Parales R. E. The beta-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 50, 553–590 (1996). [DOI] [PubMed] [Google Scholar]
- Romero-Silva M. J., Mendez V., Agullo L. & Seeger M. Genomic and functional analyses of the gentisate and protocatechuate ring-cleavage pathways and related 3-hydroxybenzoate and 4-hydroxybenzoate peripheral pathways in Burkholderia xenovorans LB400. PloS one. 8, e56038 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMarco A. A. & Ornston L. N. Regulation of p-hydroxybenzoate hydroxylase synthesis by PobR bound to an operator in Acinetobacter calcoaceticus. J Bacteriol. 176(14), 4277–84 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez J. I., Miñambres B., García J. L. & Díaz E. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol. 4, 824–841 (2002). [DOI] [PubMed] [Google Scholar]
- Bertani I., Kojic M. & Venturi V. Regulation of the p-hydroxybenzoic acid hydroxylase gene (pobA) in plant-growth-promoting Pseudomonas putida WCS358. Microbiology. 147, 1611–1620 (2001). [DOI] [PubMed] [Google Scholar]
- Van Dyk T. K., Templeton L. J., Cantera K. A., Sharpe P. L. & Sariaslani F. S. Characterization of the Escherichia coli AaeAB efflux pump: a metabolic relief valve? J Bacteriol. 186, 7196–7204 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef S., Ballerstedt H., Volkers R. J., de Winde J. H. & Ruijssenaars H. J. Comparative transcriptomics and proteomics of p-hydroxybenzoate producing Pseudomonas putida S12: novel responses and implications for strain improvement. Appl Microbiol Biotechnol. 87, 679–690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier L. S., Nichols N. N. & Neidle E. L. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J Bacteriol. 179, 5943–5946 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols N. N. & Harwood C. S. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J Bacteriol. 179, 5056–5061 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry M. T. et al. Genome-wide investigation of aromatic acid transporters in Corynebacterium glutamicum. Microbiology. 153, 857–865 (2007). [DOI] [PubMed] [Google Scholar]
- Trautwein G. & Gerischer U. Effects exerted by transcriptional regulator PcaU from Acinetobacter sp. strain ADP1. J Bacteriol. 183, 873–881 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding A. J. & Collee J. G. Routine biochemical tests, p. 1–32. In Norris J. R. & Ribbons D. W. (ed.), Methods in microbiology. Academic Press, London, United Kingdom (1971). [Google Scholar]
- DiMarco A. A., Averhoff B. & Ornston L. N. Identification of the transcriptional activator PobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J Bacteriol. 175, 4499–4506 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báidez A. G., Gómez P., Del Río J. A. & Ortuño A. Dysfunctionality of the xylem in Olea europaea L. Plants associated with the infection process by Verticillium dahliae Kleb. Role of phenolic compounds in plant defense mechanism. J Agric Food Chem. 55, 3373–3377 (2007). [DOI] [PubMed] [Google Scholar]
- Huang W. Y., Cai Y. Z. & Zhang Y. Natural phenolic compounds from medicinal herbs and dietary plants: potential use for cancer prevention. Nutr Cancer. 62, 1–20 (2009). [DOI] [PubMed] [Google Scholar]
- Romero-Steiner S., Parales R. E., Harwood C. S. & Houghton J. E. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J Bacteriol. 176, 5771–5779 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parke D. Acquisition, reorganization, and merger of genes: novel management of the the β-ketoadipate pathway in Agrobacterium tumefaciens. FEMS Microbiol Lett. 146, 3–12 (1997). [Google Scholar]
- Donoso R. A., Pérez-Pantoja D. & González B. Strict and direct transcriptional repression of the pobA gene by benzoate avoids 4-hydroxybenzoate degradation in the pollutant degrader bacterium Cupriavidus necator JMP134. Environ Microbiol. 13, 1590–1600 (2011). [DOI] [PubMed] [Google Scholar]
- Parke D. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens. J Bacteriol. 177, 3808–3817 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. & Ausubel F. M. Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA. 91, 8955–8959 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J. et al. Universally occurring phenylpropanoid and species-specific indolic metabolites in infected and uninfected Arabidopsis thaliana roots and leaves. Phytochemistry. 65, 691–699 (2004). [DOI] [PubMed] [Google Scholar]
- Abdel-Farid I. B., Kim H. K., Choi Y. H. & Verpoorte R. Metabolic characterization of Brassica rapa leaves by NMR spectroscopy. J Agric Food Chem. 55, 7936–7943 (2007). [DOI] [PubMed] [Google Scholar]
- Torres P. S. et al. Controlled synthesis of the DSF cell-cell signal is required for biofilm formation and virulence in Xanthomonas campestris. Environ Microbiol. 9, 2101–2109 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezeji T., Qureshi N. & Blaschek H. P. Butanol production from agricultural residues: Impact of degradation products on Clostridium beijerinckii growth and butanol fermentation. Biotechnol Bioeng. 97, 1460–1469 (2007). [DOI] [PubMed] [Google Scholar]
- Monnappa A. K., Lee S. & Mitchell R. J. Sensing of plant hydrolysate-related phenolics with an aaeXAB::luxCDABE bioreporter strain of Escherichia coli. Bioresour Technol. 127, 429–434 (2013). [DOI] [PubMed] [Google Scholar]
- Yuan Z. C. et al. The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc Natl Acad Sci USA. 104 11790–5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prithiviraj B. et al. Down regulation of virulence factors of Pseudomonas aeruginosa by salicylic acid attenuates its virulence on Arabidopsis thaliana and Caenorhabditis elegans. Infect Immun. 73 5319–5328 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheublin T. R. et al. Transcriptional profiling of Gram-positive Arthrobacter in the phyllosphere: induction of pollutant degradation genes by natural plant phenolic compounds. Environ Microbiol. 16, 2212–2225 (2014). [DOI] [PubMed] [Google Scholar]
- Toms A. & Wood J. M. The degradation of trans-ferulic acid by Pseudomonas acidovorans. Biochemistry. 9, 337–343 (1970). [DOI] [PubMed] [Google Scholar]
- Venturi V., Zennaro F., Degrassi G., Okeke B. C. & Bruschi C. V. Geneitics of ferulic acid bioconversion to protocatechuic acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology. 144, 965–973 (1998). [DOI] [PubMed] [Google Scholar]
- He Y. W. et al. Genome scale analysis of diffusible signal factor regulon in Xanthomonas campestris pv. campestris: identification of novel cell-cell communication-dependent genes and functions. Mol Microbiol. 59, 610–622 (2006). [DOI] [PubMed] [Google Scholar]
- Wolfe K. et al. Antioxidant activity of apple peels. J Agric Food Chem. 51, 609–614 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.