RZFP34/CHYR1 functions as a ubiquitin E3 ligase and promotes stomatal movement in response to ABA and drought stress, during which RZFP34/CHYR1 activity is regulated by SnRK2.6-mediated phosphorylation.
Abstract
Abscisic acid (ABA) is a phytohormone that plays a fundamental role in plant development and stress response, especially in the regulation of stomatal closure in response to water deficit stress. The signal transduction that occurs in response to ABA and drought stress is mediated by protein phosphorylation and ubiquitination. This research identified Arabidopsis thaliana RING ZINC-FINGER PROTEIN34 (RZP34; renamed here as CHY ZINC-FINGER AND RING PROTEIN1 [CHYR1]) as an ubiquitin E3 ligase. CHYR1 expression was significantly induced by ABA and drought, and along with its corresponding protein, was expressed mainly in vascular tissues and stomata. Analysis of CHYR1 gain-of-function and loss-of-function plants revealed that CHYR1 promotes ABA-induced stomatal closure, reactive oxygen species production, and plant drought tolerance. Furthermore, CHYR1 interacted with SNF1-RELATED PROTEIN KINASE2 (SnRK2) kinases and could be phosphorylated by SnRK2.6 on the Thr-178 residue. Overexpression of CHYR1T178A, a phosphorylation-deficient mutant, interfered with the proper function of CHYR1, whereas CHYR1T178D phenocopied the gain of function of CHYR1. Thus, this study identified a RING-type ubiquitin E3 ligase that functions positively in ABA and drought responses and detailed how its ubiquitin E3 ligase activity is regulated by SnRK2.6-mediated protein phosphorylation.
INTRODUCTION
Plants have developed intrinsic mechanisms to cope with unfavorable environmental conditions. The phytohormone abscisic acid (ABA) plays a crucial role in plant responses to various abiotic stresses, especially drought stress, by inducing the closure of stomata, thus reducing water loss (Raghavendra et al., 2010; Osakabe et al., 2014). ABA also participates in seed maturation, dormancy, germination, and seedling growth (Koornneef et al., 1989; Leung and Giraudat, 1998; Finkelstein et al., 2002). While the complete cascade of ABA signaling has not been fully elucidated, significant progress has been achieved, especially with the identification of ABA receptors. In response to ABA, the PYR/PYL/RCAR (PYRABACTIN RESISTANCE/PYR-LIKE/REGULATORY COMPONENT OF ABA RECEPTOR) ABA receptors undergo a conformational change that favors their binding to protein phosphatase type 2Cs (PP2Cs). This binding relieves PP2Cs’ constitutive inhibition of SNF1-RELATED PROTEIN KINASE2 (SnRK2) activity, which functions as a major switch in downstream ABA signaling (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009; Santiago et al., 2009; Cutler et al., 2010).
SnRK2.6 (or SnRK2E), also known as OST1 (OPEN STOMATA1), along with SnRK2.2 (or SnRK2D) and SnRK2.3 (or SnRK2I), belong to Arabidopsis thaliana class 3 SnRK2s, whose kinase activity can be strongly activated by ABA and osmotic stress (Yoshida et al., 2006; Fujii et al., 2007). SnRK2.2 and SnRK2.3 regulate ABA responses in seed germination, dormancy, and seedling growth and are functionally redundant with SnRK2.6 (Fujii et al., 2007; Fujii and Zhu, 2009; Fujita et al., 2009; Nakashima et al., 2009). SnRK2.6 is expressed in guard cells and phosphorylates transcription factors, anion channels, and NADPH oxidase (RbohF), which synergistically regulates ABA-induced stomatal closure and plant drought tolerance (Mustilli et al., 2002; Kwak et al., 2003; Geiger et al., 2009; Lee et al., 2009; Sato et al., 2009; Sirichandra et al., 2009; Wege et al., 2014). ABA can be transported to guard cells in response to drought stress, inducing an efflux of ions, loss of turgor, and, thus, stomatal closure (Bray, 1997; Schroeder et al., 2001; Li et al., 2006; Mori et al., 2006). NADPH oxidase produces H2O2, a reactive oxygen species (ROS) that functions as a major endogenous signal molecule and is involved in high CO2 concentration-, UV-B-, darkness-, and ABA-regulated stomatal closure (Desikan et al., 2004; He et al., 2005; Kolla et al., 2007). H2O2 accumulation in guard cells also elevates cytosolic Ca2+ levels, conferring an integrated ABA-responsive signaling response in guard cells (McAinsh et al., 1996; Pei et al., 2000; Zhang et al., 2001a).
Ubiquitination, a type of protein posttranslational modification, is involved in many aspects of plant development and environmental response (Moon et al., 2004; Smalle and Vierstra, 2004; Dreher and Callis, 2007; Vierstra, 2009; Lee and Kim, 2011). Upon polyubiquitination, a substrate protein will undergo proteolysis by the 26S proteasome, thus providing a mechanism to fine-tune the abundance of key regulators in cells. The specificity of this modification is largely determined by the interaction of an ubiquitin E3 ligase with its substrate, which allows for the recognition, recruitment, and transfer of the 76-amino acid ubiquitin molecule to a lysine residue in the substrate protein (Moon et al., 2004; Smalle and Vierstra, 2004; Vierstra, 2009). Thus, E3 ligase functions as a crucial substrate recognition component in this pathway. Over 1400 genes in Arabidopsis encode ubiquitin (ubi) E3 ligases, among which 477 are potential RING (for Really Interesting New Gene) ubi E3 ligases that contain a RING domain structurally similar to a zinc finger (Stone et al., 2005; Vierstra, 2009). RING ubi E3 ligases play roles in cellular processes including phytohormone signaling, photomorphogenesis, self-incompatibility, flower development, senescence, and the regulation of plant growth and development in response to nitrogen limitation (Gray et al., 2001; Stirnberg et al., 2002; Xu et al., 2002; Guo and Ecker, 2003; McGinnis et al., 2003; Qiao et al., 2004; Mao et al., 2005; Peng et al., 2007). In response to ABA, the abundance of ABA receptors PYL4, PYL8, and PYL9 is regulated by DE-ETIOLATED1 and DDB1-ASSOCIATED PROTEIN1, which function as the substrate receptor for a CUL4-based, multisubunit RING ubi E3 ligase (Irigoyen et al., 2014). The basic leucine zipper transcription factor ABI5 can be ubiquitinated and degraded by a trans-Golgi network/cytosol-localized RING-type E3, KEEP ON GOING (KEG), which maintains a low level of ABI5 in the absence of ABA (Stone et al., 2006; Liu and Stone, 2013). KEG also mediates ABF1 and ABF3 degradation via the 26S proteasome (Chen et al., 2013). The B3-domain transcription factor ABI3, which functions as a downstream component of ABA signaling, is targeted by the RING ubi E3 ligase ABI3-INTERACTING PROTEIN2 (Zhang et al., 2005). Two RING ubi E3 ligases, DRIP1 and DRIP2, potentially mediate DREB2A protein degradation to fine-tune the abundance of this protein (Qin et al., 2008). SDIR1 positively regulates ABA signaling by affecting SDIRIP1 stability, the latter of which selectively regulates the expression of ABI5 (Zhang et al., 2007; Zhang et al., 2015). However, the function of a large number of RING ubi E3 ligases and the regulation of their activity by stress stimuli still remain to be elucidated.
Here, we found that Arabidopsis CHY ZINC-FINGER AND RING PROTEIN1 (CHYR1; previously identified as RING ZINC-FINGER PROTEIN34 [At-RZFP34]; Hsu et al., 2014), which encodes a protein containing both a CHY zinc-finger and a C3H2C3-type RING domain, was significantly inducible by ABA and drought. Seed germination and stomatal closure in chyr1-2 and chyr1-3 mutants were hyposensitive to ABA. Furthermore, the mutants exhibited reduced drought tolerance, suggesting that CHYR1 positively functions in the ABA response. In vitro, CHYR1 could self-ubiquitinate and form the Lys-48-type polyubiquitination chain. Importantly, SnRK2.6 interacted with the C-terminal region of CHYR1 and phosphorylated CHYR1 at the threonine residue 178 (Thr-178) in the RING domain, which probably enhances its ubi E3 ligase activity.
RESULTS
Identification of Arabidopsis RZFP34/CHYR1
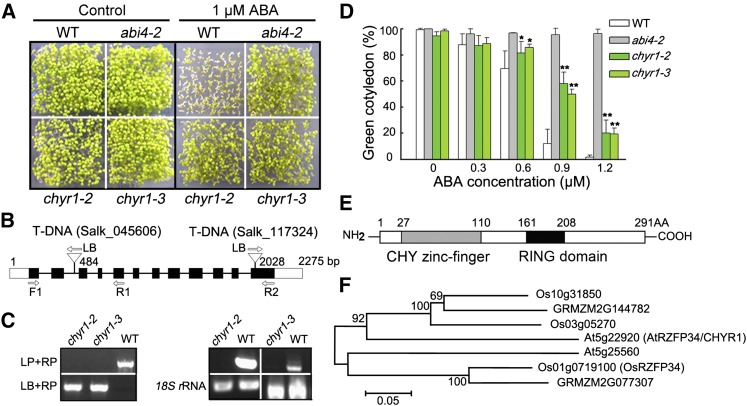
Homozygous T-DNA insertion mutants of a number of genes encoding RING domains, with ABA- and abiotic stress-responsive expression, were examined to gain a better understanding of the function of RING ubi E3 ligase in ABA and water stress responses. Two independent T-DNA insertion mutants of At5g22920 were identified that were, in comparison with the wild-type plants, consistently hyposensitive to exogenous ABA during seed germination (Figure 1A). At5g22920 encodes a protein containing a CHY zinc finger and a RING domain that was recently reported to be involved in stomatal closure and named At-RZFP34, based on orthology to Os-RSFP34 in rice (Oryza sativa; Hsu et al., 2014). Hsu et al. (2014) showed that another T-DNA mutant of At5g22920 (SALK_017562, which carries a T-DNA insertion in the gene promoter region, named atrzfp34) exhibited reduced stomatal aperture, which was complemented by transgenic expression of the rice protein sequence Os-RZFP34, but they did not further examine or characterize the function of the Arabidopsis gene. In addition, phylogenetic analysis showed that the closest Os-RZFP34 homolog in Arabidopsis is At5g25560 (Figure 1F in this work and Supplemental Figure 2 in Hsu et al., 2014). Thus, we cannot exclude the possibility that At5g22920 has a different function than Os-RZFP34. Therefore, we propose the alternate name CHYR1 for At5g22920 and note that the mutant described by Hsu et al. (2014) as atrzfp34 would also be known as chyr1-1 (SALK_017562). Here, we verified T-DNA insertions in lines chyr1-2 (SALK_045606) and chyr1-3 (SALK_117324) by genomic DNA amplification, which revealed insertions in the second intron and the last exon of CHYR1, respectively (Figures 1B and 1C). RT-PCR analysis showed that they are both CHYR1 loss-of-function mutants (Figure 1C). Green cotyledons of chyr1-2 and chyr1-3 were significantly higher than that of the wild type when the concentration of exogenous ABA was >0.6 µM, suggesting that the mutation in CHYR1 resulted in ABA hyposensitivity during seed germination (Figure 1D).
Figure 1.
ABA Hyposensitivity of chyr1-2 and chyr1-3 during Seed Germination and the Protein Sequence and Phylogenetic Analysis of CHYR1.
(A) Seed germination of wild-type and chyr1 mutants in response to ABA. Germination rates, defined by cotyledon greening, in wild-type, chyr1-2, chyr1-3, and abi4-2 seeds compared on MS medium with and without 1 μM ABA for 7 d. abi4-2, an ABA-insensitive mutant, was used as a control.
(B) Schematic structure of CHYR1 and the positions of the T-DNA insertion of chyr1-2 and chyr1-3. Black boxes denote exons, lines denote introns, and the untranslated regions are indicated with white boxes. The direction of the T-DNA left border (LB) is indicated with arrows.
(C) Left: Amplification of CHYR1 in genomic DNA from the wild type, chyr1-2, and chyr1-3. LP and RP are mutant-specific gene primers. Right: RT-PCR analysis of CHYR1 expression in the wild type and the mutants. Gene expression in chyr1-2 was checked using the F1 and R1 primer pair, and chyr1-3 was checked using the F1 and R2 primer pair (B).
(D) The percentage of cotyledon greening in wild-type and chyr1 mutants in response to different ABA concentrations. Values represent means ± sd (n = 3) from three biological replicates. Statistical significance between the wild type and chyr1 mutants was determined by a t test: *P < 0.05 and **P < 0.01.
(E) Schematic of the predicted protein domains of CHYR1. The gray and dark bars indicate the CHY zinc-finger and RING domains, respectively.
(F) Phylogenetic analysis of the seven CHYR1 homologs from Arabidopsis, rice, and maize. Bootstrap values from 1000 replicates are indicated at each node and the scale represents branch lengths.
CHYR1 consists of 291 amino acids with a CHY zinc-finger domain in the N-terminal region and a RING domain near the middle of the protein (Figure 1E). The CHY zinc-finger domain is named for the conserved amino acid motif “CxHY” present at its beginning and is a functionally uncharacterized protein domain (McDowall, 2007). The RING domain of CHYR1 is a C3H2C3-type zinc finger, based on the composition of the cysteine and histidine residues that coordinate two zinc ions (Supplemental Figure 1). Several proteins in different plant species share similarity with CHYR1, including in rice (Hsu et al., 2014) and maize (Zea mays) (Supplemental Figure 1). In addition, At5g25560 was identified as a paralog of CHYR1 in Arabidopsis, and both proteins retain their respective orthologs encoded in the rice and maize genomes (Figure 1F).
CHYR1 Expression Is ABA and Stress Inducible
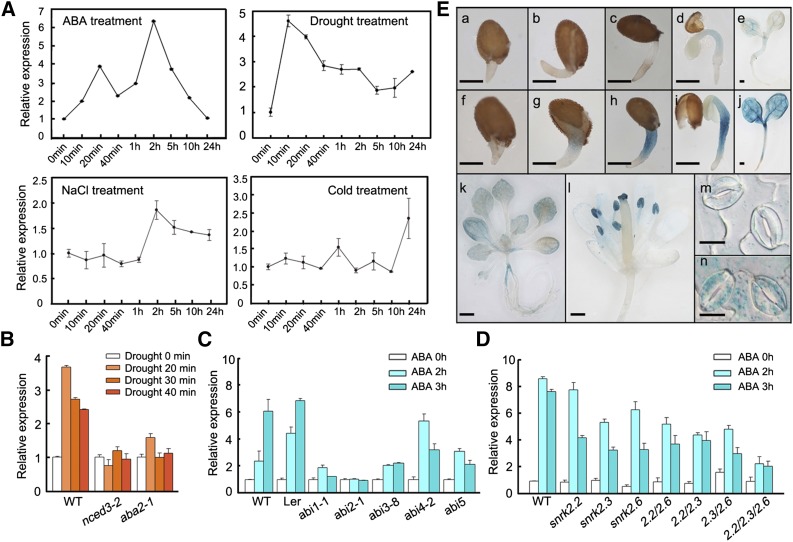
Three-week-old seedlings of wild-type Arabidopsis (Columbia) were treated with exogenous ABA and various abiotic stresses to investigate the expression of CHYR1. qRT-PCR analysis indicated that CHYR1 expression was significantly induced by ABA, reaching a maximum increase of ∼6-fold, relative to the controls, at 2 h after ABA treatment. CHYR1 expression was also significantly induced by drought stress within 10 min after the stress treatment was applied and then gradually declined and maintained at a level that was around 2-fold greater, relative to the control, over a 24-h period (Figure 2A). By contrast, CHYR1 was only mildly responsive to high salinity and low temperature stress, increasing by ∼2.0- to ∼2.5-fold relative to unstressed plants (Figure 2A). A 1.3-kb upstream DNA fragment of CHYR1 was fused with a GUS gene and the construct was introduced into Arabidopsis to study the tissue expression pattern of CHYR1. Histochemical staining was performed in different developmental stages of T2 plants. GUS activity during germination was only weakly detected in the radicals and vascular bundles of cotyledons (Figures 2Ea to 2Ee). However, in the presence of ABA, the intensity was dramatically increased, supporting the role of CHYR1 in ABA-inhibited seed germination (Figures 2Ef to 2Ej). At the rosette stage, a weak GUS signal was observed in leaves and in the stem section of the transition zone between the hypocotyl and root (Figure 2Ek). Stronger GUS expression was observed in the anthers and stigma of open flowers but was barely detected in petals (Figure 2El). Enhanced GUS expression was also detected in epidermal and guard cells after ABA treatment, consistent with the qRT-PCR analysis of gene expression (Figures 2Em and 2En). This observation suggests that CHYR1 may play a role in plant abiotic stress responses, especially ABA and drought responses.
Figure 2.
Expression Profile of CHYR1.
(A) Expression of CHYR1 in response to treatment with 100 μM ABA, drought, 250 mM NaCl, and 4°C. Total RNA was isolated from 3-week-old seedlings after the stress treatments. 18S rRNA transcript levels were used as an internal control for data normalization. Error bars indicate sd (n = 3).
(B) to (D) ABA induction of CHYR1 in the wild type and ABA-deficient (B), ABA-insensitive (C), and snrk2 (D) mutants. Three-week-old seedlings were subjected to drought stress or immersed in 100 μM ABA solution. 18S rRNA transcripts were used as an internal control for data normalization. Error bars indicate sd (n = 3). 2.2/2.3, 2.2/2.6, 2.3/2.6, and 2.2/2.3/2.6 in (D) denote the double mutants of snrk2.2 snrk2.3, snrk2.2 snrk2.6, and snrk2.3 snrk2.6 and the triple mutant of snrk2.2 snrk2.3 snrk2.6, respectively.
(E) Histochemical analysis of CHYR1 promoter activity in different tissues. (a) to (j) GUS activity in germinating seeds from radicle emergence to cotyledon greening, grown in MS medium without ([a] to [e]) or with 1 μM ABA ([f] to [j]). Bars = 250 μm. (k) and (l) GUS activity in 2-week-old seedlings and opened flowers. Bars = 500 μm. (m) and (n) GUS activity in guard cells with or without 100 μM ABA treatment for 3 h. Bars = 10 μm. All GUS staining patterns were obtained by observing at least 10 independent T2 transgenic lines.
The expression of CHYR1 was also examined in several ABA-related mutants to determine whether the observed ABA- and drought-inducible expression of CHYR1 is dependent on ABA biosynthesis and/or signal transduction. When two ABA biosynthesis mutants (nced3-2 and aba2-1) were subjected to dehydration stress, the stress-inducible expression of CHYR1 was almost abolished, suggesting that ABA biosynthesis is required to maintain the drought-inducible expression of CHYR1 (Figure 2B). In response to ABA, the induction of CHYR1 was also significantly diminished, relative to the wild type, in ABA signaling mutants such as abi1-1, abi2-1, and abi3-8 and was partially impaired in abi4-2 and abi5-1 (Figure 2C). Additionally, in the single, double, and triple mutants of SnRK2.2, SnRK2.3, and/or SnRK2.6, ABA-inducible expression of CHYR1 was inhibited to a variable extent, with the greatest level of inhibition observed in the snrk2.2 snrk2.3 snrk2.6 triple mutant (Figure 2D). Collectively, the data indicate that ABA- and drought-inducible expression of CHYR1 is dependent on both ABA biosynthesis and ABA signaling in plants.
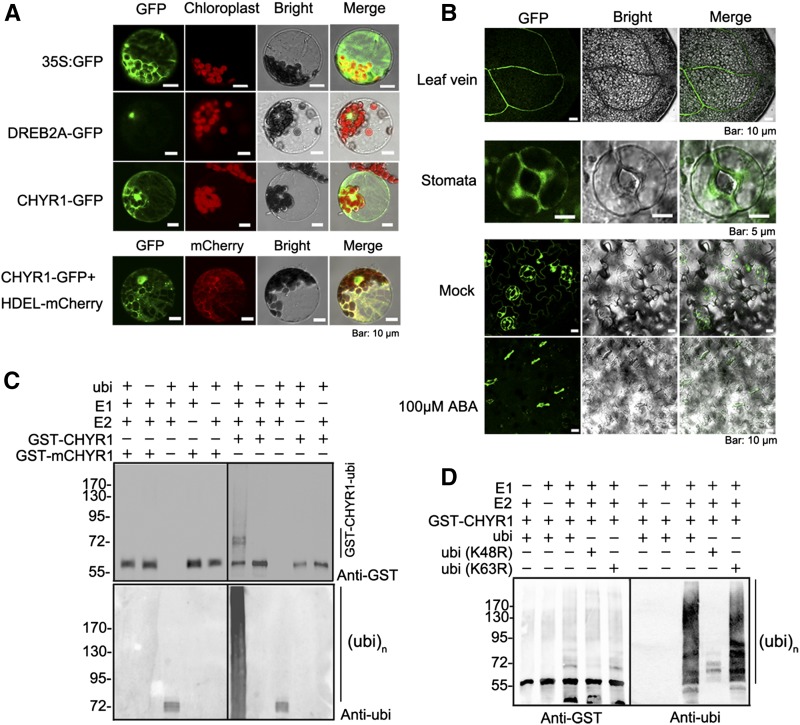
A 35S:CHYR1-GFP construct was transformed into Arabidopsis mesophyll protoplasts using a polyethylene glycol-mediated protocol (Yoo et al., 2007) to determine the subcellular localization of CHYR1 protein. The CHYR1-GFP fluorescence signal was localized in both the cytoplasm and nucleus (Figure 3A). The appearance of the GFP signal was reticular in the cytoplasm, similar to the distribution and appearance of the endoplasmic reticulum (ER). Arabidopsis protoplasts were then cotransformed with 35S:CHYR1-GFP and 35S:HDEL-mCherry plasmids (a fluorescence marker for ER localization). Colocalization of both CHYR1-GFP and HDEL-mCherry was clearly detected (Figure 3A). Therefore, it was concluded that CHYR1 was localized to the nucleus, cytoplasm, and ER. Additionally, the CHYR1-GFP protein in 35S:CHYR1-GFP stable transgenic plants was predominantly observed in vascular tissues and guard cells in plant leaves, which is consistent with the histochemical analysis of its promoter activity (Figure 3B).
Figure 3.
CHYR1 Protein Localization and Its Ubi E3 Ligase Activity.
(A) Subcellular localization of the CHYR1-sGFP fusion protein in plant cells. The 35S:GFP, 35S:DREB2A-GFP, and 35S:CHYR1-GFP constructs were transformed into Arabidopsis leaf protoplasts. DREB2A, a nuclear transcription factor, was used as a nuclear localization marker. HDEL is an ER retention signal. 35S:HDEL-mCherry was cotransformed with 35S:CHYR1-GFP to verify the ER localization of CHYR1.
(B) Vascular and stomatal localization of CHYR1-GFP in 35S:CHYR1-GFP transgenic plants. Ten-day-old seedlings of 35S:CHYR1-GFP transgenic plants were observed under a fluorescence microscope. CHYR1-GFP protein was clearly observed in leaf veins and stomata before and after 100 µM ABA treatment.
(C) In vitro self-ubiquitination assay of CHYR1. “+” and “−” denote the presence or absence of the components in each reaction mixture. The molecular weight of GST-CHYR1 and GST-mCHYR1 was ∼55 kD. Protein ubiquitination bands generated by GST-CHYR1 are indicated at the right and protein molecular mass markers are labeled at the left.
(D) CHYR1 mediates Lys-48-linked polyubiquitination. The purified ubi (K48R) and ubi (K63R) protein were used instead of the wild-type ubi protein.
CHYR1 Is a Ubi E3 Ligase and Mediates Lys-48-Linked Polyubiquitination
Since many RING domain-containing proteins possess ubi E3 ligase activity (Kraft et al., 2005; Stone et al., 2005), in vitro ubiquitination assays were performed using the purified GST-CHYR1 fusion protein to examine whether CHYR1 could function as a RING ubi E3 ligase. In addition, mutated protein (GST-mCHYR1) was generated in which the key residues Cys-182 and His-184 and -187 in the RING domain for Zn2+ ion chelating were mutated into Ser and two Tyr residues, respectively (Supplemental Figure 1). The GST-CHYR1 and GST-mCHYR1 proteins were each incubated with purified E1, E2, and His-ubi protein. The reaction products were analyzed by immunodetection using anti-GST and anti-ubi antibodies (Figure 3C). The formation of high molecular mass bands was detected only in the reactions containing GST-CHYR1, but not in those containing GST-mCHYR1. This was true using both anti-GST and anti-ubi detection. The reaction mixtures lacking any of the other components, E1, E2, GST-CHYR1, or His-ubi, failed to give positive results, except bands for ubiquitinated E2 obtained when GST-CHYR1 protein was absent (Figure 3C). These data clearly indicate that CHYR1 functions as a RING ubi E3 ligase in vitro and can mediate self-ubiquitination and the formation of a polyubiquitination chain. The results also demonstrate that the structure of the RING domain is essential for the E3 ligase activity.
To identify the type of polyubiquitination chain generated by CHYR1, the Lys-48 and -63 residues in the ubi protein were individually changed to Arg. These mutated proteins were employed in the reaction mixture instead of the wild-type ubi protein. The ubi E3 ligase activity of CHYR1 disappeared in the presence of the ubi (K48R) protein (Figure 3D). By contrast, the results obtained using the ubi (K63R) protein were comparable to those with the wild-type ubi protein (Figure 3D). Collectively, these data suggest that Lys-48 in the ubi protein is essential for CHYR1-mediated polyubiquitination chain formation. Therefore, CHYR1, in vitro, mainly mediates Lys-48-linked polyubiquitination.
CHYR1 Is a Positive Factor in Drought Response, ABA-Mediated ROS Production, and Stomatal Closure
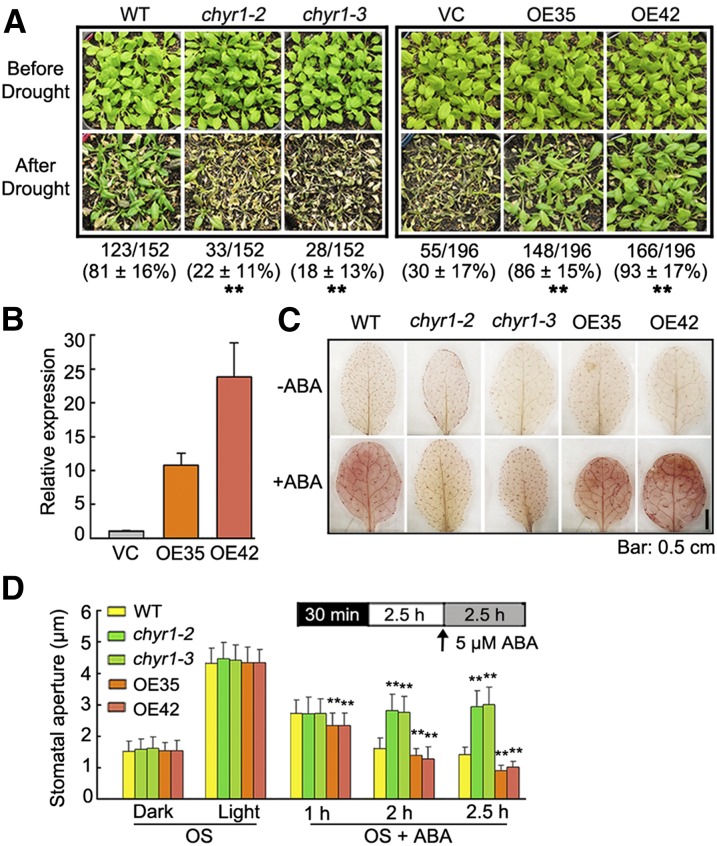
Since the expression of CHYR1 was inducible by ABA and drought stress (Figure 2A), it is plausible that CHYR1 plays a role in the ABA-dependent drought response. Thus, the drought tolerance of two mutants, chyr1-2 and chyr1-3, was compared with the wild type by subjecting 28-d-old seedlings, grown under well-watered conditions, to water stress by withholding irrigation for ∼14 d. Plants were then rewatered and survival rates were scored after 3 d of rewatering. As shown in Figure 4A, significantly lower survival rates were observed in the mutants (∼22% of chyr1-2 and 18% of chyr1-3) compared with wild-type plants (∼81%). Transgenic plants overexpressing CHYR1 were then generated using the enhanced cauliflower mosaic virus 35S promoter (35S:CHYR1), and their drought tolerance was compared with plants transformed with the empty vector control (VC). Approximately 45 T2 transgenic lines were obtained, and the morphology of all transgenic lines was similar to that of the VC lines. Two independent transgenic lines, 35S:CHYR1-35 and 35S:CHYR1-42, with enhanced CHYR1 expression, were used in further experiments (Figure 4B). When the 28-d-old transgenic seedlings were subjected to the drought test, only 30% of the VC plants were able to recover from the stress, whereas the survival rate of the 35S:CHYR1-35 and 35S:CHYR1-42 transgenic plants was ∼86 and 93%, respectively (Figure 4A). Similar results were obtained in repeated experiments, indicating that overexpression of CHYR1 improved plant tolerance to drought stress, while loss of function of CHYR1 compromised drought tolerance. Therefore, CHYR1 promotes plant drought tolerance.
Figure 4.
Phenotypic Analysis of chyr1 Mutants and 35S:CHYR1 Transgenic Plants in ABA and Drought Stress Response.
(A) Drought tolerance test of two independent 35S:CHYR1 transgenic lines (OE35 and OE42) and the chyr1 mutants. Twenty-eight-day-old seedlings were subjected to drought stress for 14 d and then rewatered when significant differences in wilting were observed. Representative photographs obtained from three independent experiments are shown. Values represent means ± sd from at least six biological replicates. Statistical significance between wild-type and chyr1 mutants; VC and the transgenic overexpression lines were determined by a t test: *P < 0.05 and **P < 0.01.
(B) Relative expression of CHYR1 in two 35S:CHYR1 transgenic lines and the empty vector control by qRT-PCR. Expression of CHYR1 in the wild type was defined as 1.0. Values represent means ± sd (n = 3) from three technical replicates.
(C) DAB staining indicates different levels of ABA-induced H2O2 production in leaves of the wild type, 35S:CHYR1, and the chyr1 mutants. The presence of H2O2 in the leaves is visualized as a red-brown color. Representative photographs are shown.
(D) Comparison of ABA-induced stomatal closure in the wild type, 35S:CHYR1, and chyr1 mutants. Leaves from 4-week-old plants were incubated in stomatal OS in the dark for 30 min and then exposed to the light for 2.5 h. ABA (5 μM) was added to the samples and stomatal closure was observed at 1, 2, and 2.5 h of ABA treatment. The data were obtained from ∼100 stomata. Values represent means ± sd (n > 100) from three to five biological replicates. Statistical significance between the wild type and chyr1 mutants or overexpression lines was by a t test: *P < 0.05 and **P < 0.01.
Since CHYR1 is expressed in stomata (Figure 3B) and overexpression of CHYR1 increased drought tolerance, we determined whether CHYR1 regulates ABA-dependent stomatal closure. Increased H2O2 levels have been reported to function as an early signaling molecule in response to ABA-induced signal transduction (Zhang et al., 2001b). ABA-induced ROS accumulation was compared in wild-type, chyr1-2, chyr1-3, 35S:CHYR1-35, and 35S:CHYR1-42 plants. Leaves of 4-week-old plants were treated with 0 or 100 μM ABA for 3 h, after which the leaves were incubated in 3,3′-diaminobenzidine (DAB) for 12 h. In the absence of ABA, no significant difference was observed among the leaves from the different lines (Figure 4C). In the presence of ABA, however, ABA-induced ROS production was impaired in chyr1-2 and chyr1-3 plants, while 35S:CHYR1 plants exhibited slightly enhanced ROS levels in comparison to wild-type plants. Stomatal movement in response to ABA was then analyzed in these plants. Detached leaves were incubated in a stomatal opening solution (OS) and exposed to dark and light conditions sequentially to synchronize the stomata (Wege et al., 2014). Stomatal closure or opening in chyr1 and 35S:CHYR1 plants was similar to the wild type under both light and dark conditions (Figure 4D). However, a clear difference was observed with the use of 5 μM ABA. The stomatal apertures in leaves of 35S:CHYR1 plants decreased more quickly than those of the wild type from 1 to 2.5 h of ABA treatment (Figure 4D). By contrast, the stomatal apertures in leaves of chyr1-2 and chyr1-3 remained in the range of 2.70 to 3.00 μm during the entire time course of the treatment. Collectively, these data indicate that CHYR1 positively regulates ABA-induced ROS production and stomatal closure.
CHYR1 Can Be Phosphorylated by SnRK2.6, Which Appears to Enhance CHYR1 Ubi E3 Ligase Activity
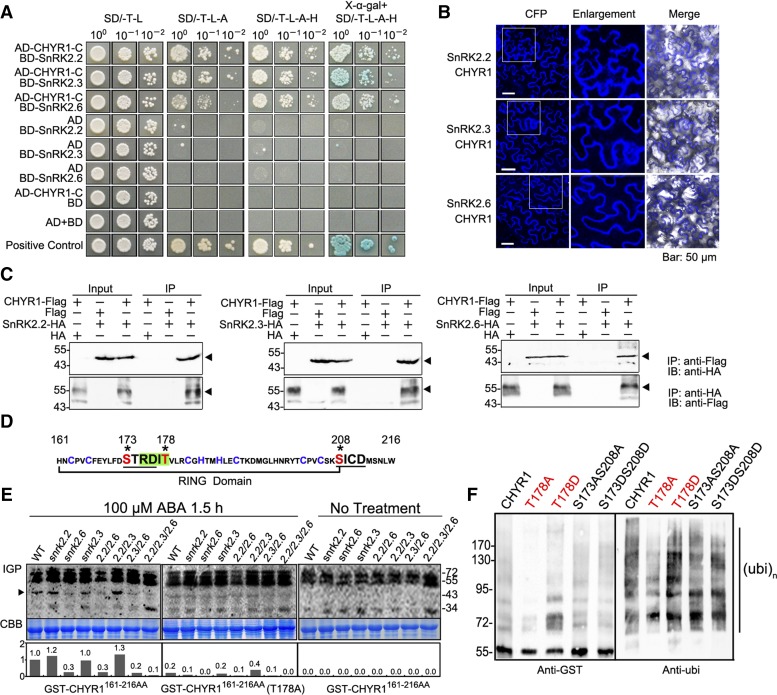
An in vivo affinity purification, using CHYR1-GFP as the bait protein, was performed to identify proteins that possibly interact with CHYR1. Total soluble protein was extracted from 17-d-old 35S:CHYR1-GFP and 35S:GFP (as a negative control) transgenic plants. Potential CHYR1-GFP interacting proteins were enriched and purified using anti-GFP agarose, and the resultant protein mixture was subjected to silver staining and immunoblot analysis (Supplemental Figure 2). Then, the samples were subjected to liquid chromatography and quadrupole time-of-flight mass spectrometry analysis. Proteins commonly detected in two replicate purifications were considered as potential CHYR1-GFP interacting proteins (Supplemental Data Set 1). Several peptides composed of amino acids representing a conserved region of a subgroup of SnRK2 kinases, SnRK2.2, SnRK2.3, SnRK2.6, and SnRK2.10, were identified among the potential interacting proteins. Considering the important role of this type of kinase in ABA and stress responses, the interactions of these proteins with CHYR1 were examined using a yeast two-hybrid system. A positive interaction with the C-terminal region of CHYR1 including the RING domain was detected for SnRK2.3, SnRK2.2, and SnRK2.6 but not SnRK2.10 (Figure 5A; Supplemental Figure 3A). Since the full CHYR1 had both autoactivation and autobinding activity in yeast, the yeast cells could not be used to evaluate the interaction of SnRK2s with full CHYR1 (Figure 5A). Therefore, the coding sequences of CHYR1 and SnRK2.2, SnRK2.3, and SnRK2.6 were inserted into pSCYNE and pSCYCE, respectively (Waadt et al., 2008). Bimolecular fluorescence complementation (BiFC) assays further verified that CHYR1 interacts with SnRK2.2, 2.3, and 2.6 in planta. Clear CFP fluorescence was observed in leaves coinfiltrated with Agrobacterium tumefaciens bacteria carrying the pSCYNE-CHYR1 and pSCYCE-SnRK2.2, -SnRK2.3, or -SnRK2.6 constructs (Figure 5B). The signal was observed in the cytoplasm but not in nuclei. However, no fluorescence was observed in the leaf samples infiltrated with agrobacteria carrying one gene construct in combination with an empty vector (Supplemental Figure 3B). The protein-protein interaction was further confirmed by coimmunoprecipitation detection, where either anti-Flag or anti-HA beads (which bind CHYR1-Flag or SnRK2s-HA, respectively) could successfully precipitate their respective counterpart, suggesting that CHYR1 physically interacts with SnRK2.2, SnRK2.3, and SnRK2.6 in plant cells (Figure 5C).
Figure 5.
CHYR1 Interacts with SnRK2.2, SnRK2.3, and SnRK2.6 and Is Regulated by Phosphorylation-Mediated by SnRK2.6.
(A) Yeast two-hybrid assays of CHYR1 and SnRK2.2, SnRK2.3, and SnRK2.6. Different concentrations of cotransformed yeast cells were dropped on synthetic dropout (SD) medium without tryptophan and leucine (SD/-T-L), without tryptophan, leucine, and adenine (SD/-T-L-A), without tryptophan, leucine, adenine, and histidine (SD/-T-L-A-H), or without tryptophan, leucine, adenine, and histidine with 20 mg/mL X-α-gal (X-α-gal+SD/-T-L-A-H).
(B) BiFC assays to evaluate interactions between pSCYNE-CHYR1 and pSCYCE-SnRK2.2, 2.3, and 2.6. Enlargements are 2.2-fold magnifications of the areas framed in the CFP field.
(C) Coimmunoprecipitation of CHYR1, SnRK2.2, SnRK2.3, and SnRK2.6 in BiFC assays. “Flag” and “HA” indicate the fusion tags in empty vector pSCYNE and pSCYCE, respectively. “+” or “−” denote the presence or absence of the protein in each sample. The black triangle indicates the target protein that was pulled down. The agarose beads for immunoprecipitation and antibodies for immunoblot detection are labeled at the right.
(D) Amino acid sequence of the CHYR1 RING domain used in IGP assays. The cysteine and histidine residues for Zn2+ binding are in blue. The SnRK2 phosphorylation motif is shaded in green, and the CKII phosphorylation motifs are underlined. The potential phosphorylated residues are in red and marked by stars.
(E) IGP assays of CHYR1161-216AA in the snrk2.2-, snrk2.3-, and snrk2.6-related mutants. Autoradiography of IGP is presented on the top, the Coomassie blue (CBB) staining indicates equal total protein loading, and the relative intensity of the 45-kD band in each lane (indicated by dark triangle) is shown on the bottom (the wild type in GST-CHYR1161-216AA is assigned as 1.0). The signals above 55 kD were considered to reflect the activity of CDPKs.
(F) In vitro ubiquitination assay of CHYR1 with mutations in potential phosphorylation residues. Left: Self-ubiquitination activity of the tested proteins, detected by anti-GST immunoblot analysis; right: formation of polyubiquitination chain, detected by anti-ubi immunoblot analysis.
The objective in our next line of investigation was to determine whether SnRK2 kinases are capable of phosphorylating CHYR1 protein. The kinase activity of SnRK2.2, SnRK2.3, and SnRK2.6 has been reported to be highly activated by ABA, which phosphorylates the Ser and/or Thr residue(s) in the conserved motif of RXXS/T of their substrates (Vlad et al., 2008; Wang et al., 2013). One RXXS/T motif, RDIT178, was present in the RING domain of CHYR1 (Figure 5D). Additionally, two phosphorylation motifs of Casein Kinase 2 (CKII), S/TXXD/E, S173TRD, and S208ICD, were also found in the RING domain of CHYR1 (Figure 5D). An in-gel kinase phosphorylation (IGP) assay, in which purified GST-CHYR1161-216AA (the RING domain) was imbedded in a gel, was used to determine if CHYR1 could serve as a substrate protein of SnRK2(s). Due to potential functional redundancy, total soluble protein extracts from 1.5-h ABA-treated wild type, single mutants of snrk2.2, snrk2.3, and snrk2.6, double mutants of snrk2.2 snrk2.3, snrk2.2 snrk2.6, and snrk2.3 snrk2.6, and the triple mutant of snrk2.2 snrk2.3 snrk2.6 were all checked for their ability to phosphorylate GST-CHYR1161-216AA. The protein extracts from snrk2.6 mutants (snrk2.6, snrk2.2 snrk2.6, snrk2.3 snrk2.6, and snrk2.2 snrk2.3 snrk2.6) exhibited much weaker phosphorylation signals in the ∼45-kD protein band (the molecular mass of SnRK2 kinases) than that observed in the protein extracts obtained from wild-type plants (Figure 5E). However, protein extracts prepared from the other mutants with intact SnRK2.6 provided a phosphorylation signal similar in intensity to that from wild-type protein extract (Figure 5E). These results indicate that the RING domain of CHYR1 can be phosphorylated and that the modification is predominantly mediated by SnRK2.6. When the Thr-178 amino acid was substituted with Ala in GST- CHYR1161-216AA (T178A), the corresponding phosphorylation signal in the IGP assay was reduced to 20 to ∼30% of that detected for CHYR1161-216AA, suggesting that phosphorylation of CHYR1 occurs at the Thr-178 residue (Figure 5E). To test whether the phosphorylation of CHYR1 by SnRK2.6 is ABA dependent, we applied protein extracts of snrk2-related mutants without ABA treatment to the IGP assay. We did not detect phosphorylation signals at the ∼45-kD region in either wild-type or snrk2-related mutants (Figure 5E). These results suggest that ABA signaling triggered the phosphorylation of CHYR1 by SnRK2.6.
The ability of the phosphorylation status of CHYR1 to influence its ubi E3 ligase activity was also examined, since the modification occurs directly in the RING domain, which is essential for its activity. GST-tagged wild-type CHYR1, CHYR1T178A (mimicking the unphosphorylated state), and CHYR1T178D (mimicking the phosphorylated state) were purified for use in the in vitro ubiquitination assay. Two other CHYR1 variants were also tested, in which the two putative CKII phosphorylation sites were also substituted by either Ala or Asp (CHYR1S173AS208A and CHYR1S173DS208D). Anti-GST immunoblot analysis indicated that the self-ubiquitination activity of CHYR1T187A was greatly diminished, whereas CHYR1T178D exhibited slightly enhanced activity. In the same experiment, CHYR1S173AS208A and CHYR1S173DS208D displayed comparable activity to the wild-type CHYR1 protein (Figure 5F). A similar pattern was observed in the anti-ubi immunodetection assay used to identify the generation of the polyubiquitination chain (Figure 5F). Collectively, the data suggest that CHYR1 can be phosphorylated by SnRK2.6 in response to ABA, and this phosphorylation appears to enhance CHYR1 ubi E3 ligase activity.
Overexpression of CHYR1T178A Interferes with CHYR1 Function in Drought and ABA Response
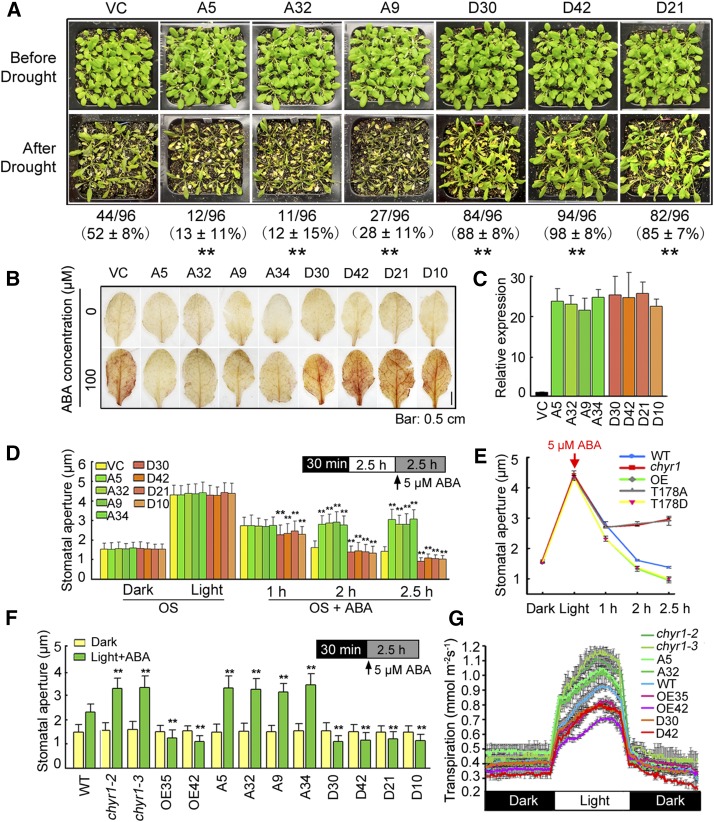
The nonphosphorylatable (CHYR1T187A) and phosphomimic (CHYR1T187D) forms of CHYR1 were overexpressed in Arabidopsis under the control of the 35S promoter to test whether or not phosphorylation of Thr-178 contributes to the function of CHYR1 in response to drought and ABA treatment. Four lines of each of 35S:CHYR1T178A (A5, A9, A32, and A34) and 35S:CHYR1T178D (D10, D21, D30, and D42) were used in the analysis, based on their increased level of CHYR1 transcript (Figure 6C). The drought tolerance of VC, 35S:CHYR1T178A, and 35S:CHYR1T178D plants was first analyzed. As shown in Figure 6A, the 35S:CHYR1T178A lines A5, A32, and A9 consistently exhibited a drought-sensitive phenotype, with a survival rate of 13, 12, and 28%, respectively. By contrast, the 35S:CHYR1T178D lines D30, D42, and D21 displayed significantly higher survival rates of 88, 98, and 85%, respectively. In parallel experiments, 52% of wild-type plants recover after rewatering, a survival level that was between that of 35S:CHYR1T178A and 35S:CHYR1T178D. ABA-inducible ROS production and stomatal closure were subsequently investigated in these plants. A DAB staining assay indicated significantly less ABA-induced H2O2 in leaves of 35S:CHYR1T178A transgenic plants compared with leaves of VC plants (Figure 6B). ABA-induced stomatal closure was also examined in these plants. Detached leaves from the VC, 35S:CHYR1T178A, and 35S:CHYR1T178D plants were incubated in a stomatal opening buffer in the dark for 30 min and then transferred to the light for 2.5 h. No obvious difference in stomatal aperture was observed among these plants (Figure 6D). However, when 5 μM ABA was added, the stomata of 35S:CHYR1T178D plants responded rapidly and started to close after 1 h of the treatment, while those of VC and 35S:CHYR1T178A plants did not show any evident changes. A clear difference in the stomatal aperture was observed after 2 and 2.5 h of the ABA treatment. The stomata in 35S:CHYR1T178D plants were more closed in response to ABA, whereas the stomata in 35S:CHYR1T178A were less closed (Figure 6D). Since ABA also inhibits light-induced stomatal opening, the stomatal movement in the dark-incubated leaf samples upon transfer to the light in the presence of 5 μM ABA was further examined. ABA-inhibited light-induced stomatal opening was enhanced in 35S:CHYR1T178D but impaired in 35S:CHYR1T178A (Figure 6F). When compared in parallel, it is important to note that 35S:CHYR1T178A was found to phenocopy the chyr1-2 and chyr1-3 mutants and that 35S:CHYR1T178D resembled the 35S:CHYR1 overexpressing plants (Figures 6E and 6F; Supplemental Figure 4). To address the concern that 35S:CHYR1T178A mimicking the chyr1 loss of function might be due to disruption of RING domain structure but not phosphorylation, we analyzed transgenic plants transformed by 35S:CHYR1S173AS208A. This mutated CHYR1 carries amino acid substitutions of Ser-173 and -208 to Ala. Both residues are in the RING domain. However, 35S:CHYR1S173AS208A showed a similar phenotype to 35S:CHYR1 in ABA-induced ROS production, stomatal movement, and plant drought tolerance (Supplemental Figure 5). These results indicate that not all amino acid substitutions in the RING domain affect the protein activity. Moreover, the leaf transpiration rate in these plants was monitored when plants grown at ∼60% RH for 8 weeks were transferred to 40% RH. Relative to wild-type plants, less water loss was observed in 35S:CHYR1 and 35S:CHYR1T178D plants, whereas more water loss was observed in chyr1-2, chyr1-3, and 35S:CHYR1T178A plants (Figure 6G).
Figure 6.
Analysis of 35S:CHYR1T178A and 35S:CHYR1T178D Transgenic Plants in Comparison to chyr1 Mutants and 35S:CHYR1 Plants.
(A) Drought tolerance of 35S:CHYR1T178A and 35S:CHYR1T178D plants. Water was withheld from 28-d-old plants for 14 d. A5, A32, and A9, and D30, D42, and D21 denote 35S:CHYR1T178A transgenic lines 5, 32, and 9, and 35S:CHYR1T178D transgenic lines 30, 42, and 21, respectively. Photographs were taken after 3 d of rewatering. Values represent means ± sd from three biological replicates. Statistical significance between VC and the transgenic lines was determined by a t test: *P < 0.05 and **P < 0.01.
(B) DAB staining of VC, 35S:CHYR1T178A, and 35S:CHYR1T178D leaves. A34 and D10 denote 35S:CHYR1T178A transgenic line 34 and 35S:CHYR1T178D transgenic line 10, respectively. The red-brown staining indicates H2O2 accumulation. Representative photos are shown. Bar = 0.5 cm.
(C) Relative expression of CHYR1 in the 35S:CHYR1T178A and 35S:CHYR1T178D lines analyzed by qRT-PCR. Expression of CHYR1 in the wild type was defined as 1.0. Values represent means ± sd (n = 3) from three technical replicates.
(D) ABA-induced stomatal closure in wild-type, 35S:CHYR1T178A, and 35S:CHYR1T178D plants. Leaves from 4-week-old plants were incubated in OS in the dark for 30 min. After exposure to light for 2.5 h, 5 μM ABA was added to the samples to observe the stomatal closure at 1, 2, and 2.5 h of ABA treatment. Data were obtained from ∼100 stomata for each sample. The error bars were obtained from different stomata. Values represent means ± sd (n > 100) from three to five biological replicates. Statistical significance between VC and the transgenic lines in each treatment was determined by a t test: *P < 0.05 and **P < 0.01.
(E) A comparison of ABA-induced stomatal closing among the wild type, the chyr1 mutants, and 35S:CHYR1, 35S:CHYR1T178A, and 35S:CHYR1T178D plants.
(F) ABA-inhibited light-induced stomatal opening in the wild type, the chyr1 mutants, and 35S:CHYR1, 35S:CHYR1T178A, and 35S:CHYR1T178D plants. Leaves from 4-week-old plants were incubated in OS for 30 min in dark and then transferred to the light in the presence of 5 μM ABA for 2.5 h. Data were obtained from ∼100 stomata for each sample. The error bars were obtained from different stomata. Values represent means ± sd (n > 100) from three to five biological replicates. Statistical significance between the wild type (comparable with VC) and chyr1 mutants or the transgenic lines in each treatment was determined by a t test: *P < 0.05 and **P < 0.01.
(G) Transpiration rates in 8-week-old plants grown under an 8-h-light/16-h-dark photoperiod were determined by gravimetric analysis. Water loss was measured as weight change at 5-min intervals over 24 h (means ± sd, n = 3).
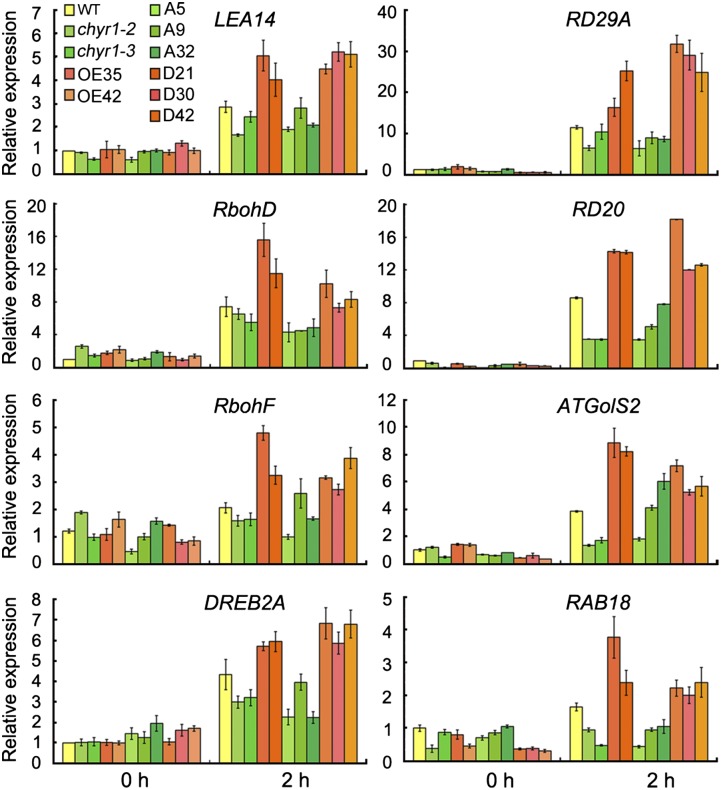
The expression of ABA- and/or drought-inducible genes was analyzed in chyr1-2, chyr1-3, 35S:CHYR1, 35S:CHYR1T178A, and 35S:CHYR1T178D transgenic plants by qRT-PCR. RAB18, LEA14, RD29A, RD20, DREB2A, and GolS2, which are well-known drought-responsive genes that function positively in drought tolerance, were induced to a greater extent 2 h after the onset of the dehydration treatment in the 35S:CHYR1 and 35S:CHYR1T178D plants and less induced in chyr1-2, chyr1-3, and 35S:CHYR1T178A plants, relative to the wild type (Figure 7). These expression patterns were consistent with and supported the observed difference in drought tolerance among these plants (Figure 6A). Similar changes were observed in the levels of expression of RbohD and RbohF, which are directly responsible for the production of H2O2 in guard cells in response to stress (Krieger-Liszkay et al., 2008). Taken together, these data suggest that CHYR1T178A can suppress the proper function of the endogenous CHYR1 and that the phosphorylation status of Thr-178 in CHYR1 is critical for the function of this protein in response to ABA and drought stress.
Figure 7.
Expression of Stress-Responsive Genes in the Wild Type, chyr1 Mutant, and 35S:CHYR1, and 35S:CHYR1T178A and 35S:CHYR1T178D Plants Subjected to Drought Stress.
Total RNA was obtained from 3-week-old seedlings treated by 2-h drought stress and analyzed by qRT-PCR using the gene-specific primers listed in Supplemental Table 1. The mean value of three replicates was normalized to the levels of ACTIN2. The gene expression in the wild type grown under non-stress conditions was taken as 1.0. Values represent means ± sd (n = 3) from three technical replicates.
DISCUSSION
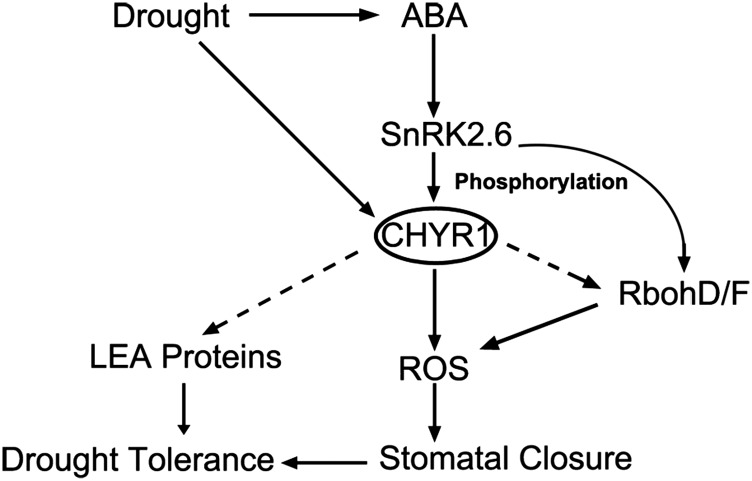
In this study, we identified an ABA- and drought-inducible gene, RZFP34/CHYR1, which encodes a RING ubi E3 ligase. CHYR1 is predominantly phosphorylated by SnRK2.6 at the Thr-178 in the RING domain, which appears to activate its ubi E3 ligase activity. Our analysis of CHYR1 gain-of-function plants, 35S:CHYR1 and 35S:CHYR1T187D, demonstrated that CHYR1 functions positively in ABA-induced ROS production, stomatal closure, and plant drought tolerance. These findings are supported by the enhanced expression of genes encoding NADPH oxidases (RbohD/F) and LEA (LATE EMBRYOGENESIS ABUNDANT) proteins in response to drought stress in the 35S:CHYR1 and 35S:CHYR1T178D plants (Figure 7). Under ABA and/or drought stresses, 35S:CHYR1T187D did not show a stronger phenotype than 35S:CHYR1, which suggests CHYR1 probably functions late in the signal response when the overexpressed CHYR1 is also activated. Overexpression of CHYR1T178A interfered with the activity and function of native CHYR1 protein, mimicking the phenotype of CHYR1 loss-of-function plants. By contrast, the S173AS208A mutation in CHYR1 did not change the protein activity and function in vitro or in vivo (Figure 5F; Supplemental Figure 5), indicating that Thr-178 is the key residue in modulating CHYR1 E3 activity, probably through SnRK2.6-mediated phosphorylation. Collectively, the data presented within this study establish CHYR1 as an ubi E3 ligase that is possibly phosphorylated by SnRK2.6 and promotes ABA and drought response in plants (Figure 8).
Figure 8.
Proposed Model for CHYR1 in ABA-Induced Stomatal Closure and Drought Tolerance.
CHYR1, a RING ubi E3 ligase, interacts with and is phosphorylated by SnRK2.6, which enhances ubi E3 ligase activity to promote ABA-induced ROS production and stomatal closure and plant drought tolerance.
In addition to the RING domain, CHYR1 also contains a CHY zinc-finger domain in the protein N terminus. The CHY zinc-finger domain, which was named for the conserved “CXHY” motif at the beginning of the domain, contains 12 cysteines and histidines that bind three zinc ions, forming a special zinc finger conformation (McDowall, 2007). The function of the CHY zinc-finger domain has not been characterized as yet, and only a few related yeast and human studies have been reported. In yeast, Host13p (HELPER OF TIM PROTEIN 13), which contains a CHY zinc-finger domain, assists small Tim (TRANSLOCASE OF THE INNER MEMBRANE) proteins to assemble into a 70-kD complex in the mitochondrial intermembrane space (Curran et al., 2004). In human, Pirh2 (P53-INDUCED PROTEIN WITH A RING-H2 DOMAIN), which also possesses a CHY zinc-finger and a RING-H2 domain, has been reported to negatively regulate p53 function through a physical interaction and ubiquitin-mediated proteolysis (Leng et al., 2003). Our phylogenetic analysis in plants revealed that CHYR1 orthologs are well conserved in different species, including Arabidopsis, rice, and maize, possibly indicating a conserved and indispensable function (Figure 1F).
Since several ubi E3 ligases have been reported to be involved in the ABA signaling pathway (Zhang et al., 2005; Gu and Innes, 2011; Lechner et al., 2011; Liu and Stone, 2013; Kim and Kim, 2013; Bueso et al., 2014), the molecular mechanism by which ubi E3 ligase activity is regulated is an important issue to elucidate. This study demonstrated that ubi E3 ligase CHYR1 is phosphorylated by SnRK2.6. Both CHYR1 and SnRK2.6 proteins are expressed in the vascular system and stomata in Arabidopsis, a finding that supports the potential interaction of these two proteins (Figure 3B; Mustilli et al., 2002). Phosphorylation of CHYR1 at the Thr-178 residue appears to activate its E3 ligase activity, based on the results obtained in the in vitro ubiquitination assays (Figure 5F). Plant U-Box 12 and 13 (PUB12 and PUB13) have been reported to be phosphorylated by BRI1 (BRASSINOSTEROID-INSENSITIVE1) ASSOCIATED KINASE1 to promote their association with an FLS2 (FLAGELLIN-SENSING2) substrate and, concomitantly, ubiquitination of FLS2, that can serve to attenuate FLS2 signaling in plant innate immunity responses (Lu et al., 2011). In that example, phosphorylation of PUB12 and PUB13 promotes the interaction of the proteins with their substrates. The direct phosphorylation of an ubi E3 ligase to regulate its ligase enzymatic activity has been studied in humans. In humans, ATM (ATAXIA-TELANGIECTASIA MUTATED)-mediated phosphorylation of the RING E3 ligase MDM2 (MURINE DOUBLE MINUTE2), which ubiquitinates the transcription factor p53, inhibits its E3 ligase activity by preventing oligomerization of the RING domain (Cheng et al., 2011). However, the activity of the human RING E3 ligase, Cbl-b (CASITAS B-LINEAGE LYMPHOMA-B), is activated rather than inhibited by phosphorylation on Tyr-363, blocking an autoinhibitory interdomain interaction (Kobashigawa et al., 2011). Recently, a plant protein kinase (ENHANCED DISEASE RESISTANCE1 [EDR1]) has been reported to negatively regulate the ubi E3 ligase activity of ARABIDOPSIS TOXICOS EN LEVDURA1 (ATL1) in the trans-Golgi network/early endosome and to influence stress responses initiated by ATL1-mediated ubiquitination events (Serrano et al., 2014). Our results demonstrated that phosphorylation deficiency of CHYR1 (i.e., CHYR1T178A) impaired ubi E3 ligase activity in vitro and disrupted gene function in vivo, suggesting that the Thr-178 residue is required for the proper functioning of CHYR1 (Figures 4 to 6). The unique, putative phosphorylation motif RXXS/T for SnRK2s is located in the RING domain of the CHYR1, and Thr-178 in this motif is close to the third cysteine residue of this domain. Thus, it is possible that the phosphorylation status of Thr-178 affects the structural conformation of the RING domain or its ability to incorporate zinc ions, which would affect CHYR1 ubi E3 ligase activity as detected in vitro (Figure 5F).
SnRK2.6 plays a key role in ABA-induced stomatal closure by directly phosphorylating and regulating the activity of ion channels, enzymes involved in H2O2 synthesis, and the induction of transcription factors (Wang et al., 2013). The H2O2 level increased in response to ABA in 35S:CHYR1 and 35S:CHYR1T178D transgenic plants, whereas it decreased in chyr1-2, chyr1-3, and 35S:CHYR1T178A (Figure 7). This finding was consistent with the results obtained for changes in the expression of RbohD and RbohF, which are key genes responsible for H2O2 production. These results suggest that CHYR1 may elevate ABA-induced ROS production in a manner that is dependent on SnRK2.6 phosphorylation. Previous results have demonstrated that inhibition of proteolysis can impair ABA-induced stomatal closure (Khanna et al., 2014). Some RING ubi E3 ligases have been reported to function in stomatal movement, but the specific regulation mechanism has been clarified only in a few studies (Bu et al., 2009; Ryu et al., 2010; Cho et al., 2011; Li et al., 2011). This research provides information that can assist in understanding the possible mechanism by which ubi E3 ligase modulates stomatal aperture.
The chyr1-2 and chyr1-3 mutants were hyposensitive, whereas 35S:CHYR1 transgenic plants were hypersensitive, to exogenous ABA during seed germination, relative to the wild type (Figure 1A). However, both 35S:CHYR1T178A and 35S:CHYR1T178D were hypersensitive to ABA during seed germination, similar to 35S:CHYR1 plants (Supplemental Figure 6). These findings indicate that Thr-178 phosphorylation status is not involved in CHYR1 function during seed germination. In regard to ABA-inhibited seed germination, the snrk2.6 single mutant does not display any obvious phenotype (Fujita et al., 2009). These results suggest that, besides functioning in ABA-induced stomatal closure and drought tolerance, CHYR1 may also play a role in seed germination that is not dependent on SnRK2.6-mediated protein phosphorylation. In the protein-protein interaction experiments, CHYR1 also interacted with SnRK2.2 and SnRK2.3, but it was not clearly phosphorylated by these two kinases in the IGP assay (Figure 5). The snrk2.2 snrk2.3 double mutant exhibits significant ABA hyposensitivity during seed germination (Fujita et al., 2009). Whether CHYR1 functions in ABA-inhibited seed germination through an interaction with SnRK2.2 and SnRK2.3 is unknown.
The chyr1-1 mutant (Salk_017562, previously called atrzfp), carrying a T-DNA insertion in the gene promoter, was reported to exhibit reduced (15%) stomatal apertures under non-stress conditions (Hsu et al., 2014). A similar phenotype was observed on our soil-grown chyr1-2 and chyr1-3 mutants, but their stomatal apertures can be opened to comparable in size with those in wild-type plants immersed in stomatal opening solution (Figure 4D). Thus, to examine stomatal movement to signal stimuli, we think it is important to synchronize the status of stomata before the onset of signal, especially when the initial aperture is different. When the soil-grown mutants in the transpiration experiment were examined, the stomatal response to low RH (from 60 to 40%) was found to be impaired, as indicated by the enhanced water loss (Figure 6G). Importantly, these phenotypes were reproducible in the 35S:CHYR1T178A transgenic plants and the response was opposite to those of the 35S:CHYR1 and 35S:CHYR1T178D plants (Figure 6). Collectively, these results suggest that loss of function of CHYR1 impairs both leaf stomatal aperture under normal soil-growth conditions and movement in response to ABA and/or stress stimuli. Whether the reduced opened stomata of the normal soil-grown plants can be attributed to inactive stomatal movement is unclear. The target protein of CHYR1 remains to be identified to fully resolve the function of CHYR1. The CHYR1-interacting proteins identified in the coimmunoprecipitation purification may provide important clues in this regard (Supplemental Data Set 1). The ability of CHYR1 to generate Lys-48-linked polyubiquitination was demonstrated (Figure 3D) and most likely facilitates 26S proteasome-mediated proteolysis. Further identification of CHYR1 substrate proteins and characterization of its activity in vivo will facilitate our understanding of the gene function. Collectively, our findings report that a CHY zinc-finger-containing RING ubi E3 ligase functions in responses to ABA and drought, probably through SnRK2.6-mediated phosphorylation.
METHODS
Plant Material and Transgenic Plant Construction
The Arabidopsis thaliana ecotype Col was used in this study. Seeds were grown on Murashige and Skoog (MS) medium plus 3% sucrose and 0.6% agar (pH 5.8). The 10-d-old seedlings were transferred into soil and were grown at 22°C in a 16-h-light/8-h-dark cycle. To study the effect of ABA on germination and cotyledon greening, seeds were planted on MS medium plates plus 1% sucrose and different concentrations of ABA (Sigma-Aldrich) in a growth chamber at 22°C under a 16-h-light/8-h-dark photoperiod with 60% relative humidity after 2 d of vernalization in darkness at 4°C. T-DNA insertion mutants chyr1-2 (SALK_045606) and chyr1-3 (SALK_117324) were obtained from the ABRC and verified to be homozygous by genotyping PCR using SALK_LBb1.3 and LP and RP primers from SIGnAL (SALK Institute Genomic Analysis Laboratory) (Figure 1B). The mRNA level of CHYR1 was observed in chyr1-2 and chyr1-3 by RT-PCR using the pair primers F1/R1 and F1/R2, respectively (Figure 1B). Mutants abi3-8 (Nambara et al., 2002), abi4-2 (SALK_080095), abi5-1 (SALK_013163), necd3-2 (GABI-Kat 129B08), aba2-1 (CS156), snrk2.2 (GABI-Kat 807G04), snrk2.6 (SALK_008068), and snrk2.3 (SALK_096546) are in Col, while abi1-1 (CS22) and abi2-1 (CS23) are in the Landsberg erecta background. A series of double and triple mutants of snrk2.2, snrk2.6, and snrk2.3 were constructed by genetic crosses and were screened using primers recommended by ABRC as described previously (Nakashima et al., 2009). To generate 35S:CHYR1, 35S:CHYR1T178A, 35S:CHYR1T178D, and 35S:CHYR1-GFP constructs, CHYR1 and mutated CHYR1 coding sequence were inserted into pGreenII and pGreenII-GFP vectors (Qin et al., 2008), respectively, by BamHI and XhoI sites. A 1.3-kb DNA fragment of CHYR1 upstream from start codon was constructed into pGreenII-GUS vector (Qin et al., 2008). All the constructed plasmids were introduced into Agrobacterium tumefaciens strains GV3101 and transformed into plants as described previously (Liu et al., 1998). Primers are listed in Supplemental Table 1.
Stress Treatment and qRT-PCR Analysis
Drought treatment was applied to 3-week-old seedlings by removing them from MS medium plates and desiccating them on Whatman 3MM paper on a clean bench for the indicated period of time. Low-temperature stress was applied to the seedlings growing on MS medium by transferring the plates into a 4°C incubator. NaCl and ABA treatment were applied by immersing the seedlings in 250 mM NaCl and 100 μM ABA, respectively. Total RNA was isolated from the seedlings after the stress treatments with Trizol reagent (Biotopped) and DNA was digested by RNase-free DNaseI. Two micrograms of total RNA was used for reverse transcription with M-MLV reverse transcriptase according to the supplier’s instructions (Promega). The RNA extraction and reverse transcription methods above were also applied for the RT-PCR in Figure 1. qRT-PCR analyses were performed with Applied Biosystems StepOne real-time PCR system using the SYBR Premix Ex Taq kit (Takara) and specific primers for PCR amplification. CHYR1 expression in response to different stresses and in various ABA-related mutants was calculated according to a standard curve calibration based on amplification of a dilution series of CHYR1 plasmid, according to the manufacturer’s protocol. 18S rRNA was used as an internal control for data normalization. The expression of different stress-responsive genes was compared based on the delta Ct method in normalization to ACTIN2. Each experiment was performed using at least three independent biological replicates. For each primer pair, the amplification efficiency was checked by a melting-curve analysis. The primers for RT-PCR and qRT-PCR are shown in Supplemental Table 1.
Phylogenetic Analysis
Protein homology searches were performed with the Phytozome program (http://www.phytozome.net/). Selected amino acid sequences were aligned using ClustalW software (http://workbench.sdsc.edu). The alignment is available as Supplemental Data Set 2. Phylogenetic trees were generated using the neighbor-joining method (Tamura et al., 2007) in MEGA software version 4. Bootstrap values were supported from 10,000 replicates. Branch length indicates divergence distance. Numbers on the branches designate percentage bootstrap support.
Histochemical GUS Staining
Different developmental stages and different parts of transgenic plants were collected and stained at 37°C in 1 mM Glc, 50 mM phosphate buffer, pH 7.0, and 10% methanol for ∼16 h (Qin et al., 2008). To remove chlorophyll after GUS staining, the plant tissues were immersed in 70% ethanol for several hours. Observation was conducted with a light microscope (SMZ1500; Nikon), and a camera (Fi1; Nikon) was used to take the digital images, with the corresponding Nikon NIS Elements D software.
Subcellular Localization
The 35S:CHYR1-GFP and 35S:GFP plasmids were transformed into Arabidopsis protoplasts by means of polyethylene glycol treatment (Yoo et al., 2007). Transformed protoplasts were observed using a fluorescence microscope (Leica TCS SP5). Images were analyzed with Image LAS-AF software. DREB2A-GFP and HDEL-mCherry were used as controls for nuclear and ER localization, respectively (Qin et al., 2008; Tian et al., 2013).
Protein Preparation and Immunoblot Analysis
The CHYR1 and ubi mutants were prepared using site-directed mutagenesis by primer extension and constructed into pGEX-4T-1 vector (GE Healthcare) and pET-28a vector (Novagen), respectively. GST-tagged CHYR1, mCHYR1, CHYR1T178A, CHYR1T178D, CHYR1S173AS208A, CHYR1S173DS208D, CHYR1161-216AA, CHYR1161-216AA (T178A), and His-tagged ubi, ubi (K48R), and ubi (K63R) were expressed in Rosetta plus Escherichia coli. The fusion proteins were purified by standard procedure using GST Sefinose Resin (Sangon Biotech) or nickle-coupled agarose (GenStar).
For immunoblot analysis, proteins were separated by SDS-PAGE in 8% acrylamide gel and electroblotted to polyvinylidene fluoride membrane (Millipore) at 360 V for 60 min. Bands were detected with ECL Prime Western Blotting Detection Reagent (GE Healthcare). Antibodies and the dilutions used in these experiments were as follows: anti-HA antibody (Covance; lot MMS-101R, 1:1000), anti-FLAG antibody (Sigma-Aldrich; lot F3165, 1:5000), anti-GST antibody (Santa Cruz Biotech; lot sc-138, 1:1000), antiubiquitin monoclonal antibody (Liu et al., 2010), and goat anti-mouse horseradish peroxidase-conjugated antibody (BPI; lot AbP71003-D-HRP, 1:10,000).
In Vitro Ubiquitination Assays and Protein Gel Blot Analysis
The ubiquitination assays were performed generally as described by Xie et al. (2002). Approximately 250 ng CHYR1-GST fusion protein was mixed with 100 ng wheat (Triticum aestivum) E1 (GI: 136632), 250 ng human E2 (UBCh5b), and 2 µg purified Arabidopsis ubiquitin (UBQ14, At4g02890) fused with the His tag. The reactions were performed in buffer containing 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, and 2 mM DTT. After incubation at 30°C for 2 h, the reactions were stopped with 2× SDS sampling buffer by boiling at 100°C for 5 min and analyzed by 8% SDS-PAGE and immunoblot analysis using a corresponding antibody.
Drought Phenotype Analysis
For the drought stress tolerance test, plants were grown for 28 d under normal conditions and subjected to water stress by withholding watering for 14 d (Qin et al., 2008). In each experiment, a small cup with 250 g soil (1:1 of black soil/vermiculite) was planted with 16 plants under the conditions of 16 h light/8 h dark. At least three independent experiments were performed. The plants were rewatered when significant differences in wilting were observed. Three days after rewatering, surviving plants were counted.
DAB Staining
DAB staining was used to monitor the accumulation of H2O2 (Thordal Christensen et al., 1997). Light-grown, 4-week-old rosette leaves were treated with 100 μM ABA for 3 h and immersed in freshly prepared 100 μg mL−1 DAB solution in the dark for 12 h. Stained leaves were fixed with a solution of 3:1:1 ethanol/acetic acid/glycerol and photographed.
Stomatal Movement Assay
Stomatal movement assays were performed as described previously (Ryu et al., 2010; Wege et al., 2014) with slight modification. Briefly, light-grown, 3-week-old rosette leaves were harvested and incubated in stomatal opening solution containing 10 mM KCl, 100 μM CaCl2, and 10 mM MES-KOH (pH 6.1). Subsequently, clear tape was applied to the ventral portion of the leaf blade to peel off the epidermal layer. After gently scraping off the mesophyll cells, the epidermal layer was mounted on glass slides, imaged using a Nikon 80i upright microscope, and measured using ImageJ software (Mao et al., 2005). For ABA-induced stomatal closure experiments, isolated leaves were incubated in stomatal OS for 30 min in the dark and 2.5 h in the light, and then 5 μM ABA was added. The stomatal apertures were measured at 1, 2, and 2.5 h of ABA treatment. For ABA inhibition of light-induced stomatal opening, leaves were incubated in OS buffer 30 min in the dark and 2.5 h in the light in the presence of 5 μM ABA and then stomatal apertures were measured after 2.5 h. Stomatal aperture values are means from at least 100 stomata, measured in five plants per treatment.
Leaf Transpiration Rate
Whole-plant leaf transpiration rate was determined by a gravimetric method as described previously (Yoo et al., 2010). Individual plant was grown in a little container in 8-h-light/16-h-dark photoperiod incubator with ∼60% RH for ∼8 weeks. Before measurement, each container was covered with polyethylene wrap completely to prevent evaporation from the soil surface. About two to three plants were placed on a balance and weighed every 5 min over 24 h, and the RH of the laboratory bench was ∼40%. After measurement, total leaf areas were determined by ImageJ software. Transpiration (mmol water m−2 s−1) was calculated based on gravimetric water loss rate and leaf area data.
Affinity Purification of CHYR1-Interacting Proteins
Affinity purification and subsequent mass spectrometry analysis were performed as described previously (Nishimura et al., 2010) with some modifications. 35S:CHYR1-GFP and 35S:GFP transgenic seedlings were grown on solid MS medium supplemented with 3% sucrose for 17 d. Samples of ∼4 to 6 g fresh weight were harvested, ground to fine powder in liquid nitrogen, and resuspended in 2× extraction buffer (50 mM Na-phosphate, pH 7.4, 150 mM NaCl, 0.1% Nonidet P-40, 1 mM DTT, and 1× protease inhibitor cocktail tablets [Roche]; 8 to 10 mL). The homogenate was centrifuged at 20,000g for 20 min at 4°C, and the complete supernatant was incubated with GFP agarose beads (MBL) for 2 h at 4°C. The beads were washed on a spin column (Pierce) with wash buffer (50 mM Na-phosphate, pH 7.4, 150 mM NaCl, and 0.1% Nonidet P-40) at least five times and then eluted with elution buffer (0.1 M glycine-HCl, pH 3.0, and 1× protease inhibitor cocktail tablets [Roche]). The elutions were neutralized with one-fifth to approximately one-tenth volume of 300 mM NH4HCO3 and denatured in 1% Rapigest (Waters), 20 mM Tris-HCl (pH 8.0), and 10 mM DTT at 37°C for 4 h. Subsequently, iodoacetamide (50 mM final concentration) was added to alkylate samples at room temperature in the dark for 1 h. The total mixtures were applied to ultrafitration with 20 mM NH4HCO3 and then one-tenth volume was analyzed by silver staining. The rest was digested with trypsin (2 µg trypsin/100 µg total protein) at 37°C for 16 to ∼18 h. The resulting mixtures were freeze-dried for analysis by liquid chromatography-tandem mass spectrometry (Absciex Quadrupole-TOF 5600+). Proteins identified in the CHYR1-GFP samples but not in the GFP samples are considered possible CHYR1-interacting proteins.
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was performed using the MatchMaker GAL4 Two-Hybrid System 3 (Clontech) according to the manufacturer’s manual. The full-length coding regions of SnRK2.2, SnRK2.3, and SnRK2.6 were individually ligated into pGBKT7 to make the bait plasmid vectors. The CHYR1 sequence encoding the C-terminal region containing the conserved RING finger domain was fused into pGADT7 vector as prey. For the interaction test, each bait construct was cotransformed with each prey construct in the AH109 yeast strain, plated on SD/-Trp-Leu medium, and grown at 30°C for 3 d. Then, the transformants were tested on SD screening medium at 30°C for 5 d. Empty vectors were cotransformed as negative controls. The primers used were listed in the Supplemental Table 1.
BiFC Assays
The Agrobacterium strains GV3101 containing the BiFC vectors pSCYCE and pSCYNE (Waadt et al., 2008) were infiltrated as described (Waadt and Kudla, 2008). Infiltration was performed on leaves of 1- or 1.5-month-old tobacco (Nicotiana benthamiana) plants and observed 3 to 5 d later using a fluorescence microscope (Leica TCS SP5). Images were analyzed with Image LAS-AF software.
Coimmunoprecipitation and Immunoblot Analysis
The coinfiltrated parts of N. benthamiana leaves were ground in the same volume of 2× extraction buffer (50 mM Na-phosphate, pH 7.4, 150 mM NaCl, 0.1% Nonidet P-40, 1 mM DTT, 50 µM MG132, and 1× protease inhibitor cocktail tablets [Roche]) and centrifuged at 20,000g for 20 min. HA agarose (MBL) or FLAG agarose (MBL) was added to the cell lysates, incubated at 4°C for 2 h, and washed at least five times with wash buffer (50 mM Na-phosphate, pH 7.4, 150 mM NaCl, and 0.1% NP-40). The agarose beads were recovered by centrifugation at 5000g for 1 min and then SDS sample buffer was added. The samples were boiled for 10 min before immunoblot analysis.
In-Gel Kinase Phosphorylation Assay
The in-gel kinase phosphorylation assay was performed as previously described (Zhang et al., 1993). In brief, the total proteins of each snrk2 mutant were extracted in extraction buffer (50 mM HEPES-KOH, pH 7.5, 5 mM EDTA, 5 mM EGTA, 2 mM DTT, 25 mM NaF, 1 mM Na3VO4, 50 mM β-glycerophosphate, 20% glycerol, 2 μg/mL leupeptin, 2 μg/mL pepstatin A, and 2 mM PMSF). The samples were then incubated with 2× sample loading buffer at 70°C for 10 min to complete the sample preparation. Protein extracts (50 µg) were electrophoresed on 12.5% SDS-PAGE gels embedded with 0.1 mg CHYR1161-216AA or CHYR1161-216AA (T178A) purified protein in the separating gel as a kinase substrate. After electrophoresis, the SDS was removed from the gel by washing with washing buffer (25 mM Tris-HCl, pH 7.5, 0.5 mM DTT, 0.1 mM Na3VO4, 5 mM NaF, 0.5 mg/mL BSA, and 0.1% Triton X-100) three times for 30 min each at room temperature. The proteins were then renatured in 25 mM Tris-HCl, pH 7.5, 1 mM DTT, 0.1 mM Na3VO4, and 5 mM NaF at 4°C overnight with three changes of the buffer. The gels were incubated at room temperature in reaction buffer (25 mM Tris-HCl, pH 7.5, 2 mM EGTA, 12 mM MgCl2, 1 mM DTT, and 0.1 mM Na3VO4) for 30 min. Phosphorylation was performed for 1.5 h at room temperature in 30 mL reaction buffer that contained 200 nM ATP and 50 µCi [γ-32P]ATP (3000 Ci/mmol). The reaction was stopped with a solution of 5% trichloroacetic acid (w/v) and 1% sodium pyrophosphate (w/v) at room temperature with five changes. The gel was dried, subjected to autoradiography, and scanned using a Typhoon 9410 gel and blot imager.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: wheat E1 (GI: 136632), human E2 (UBCh5b, GI: 1145689), ubiquitin (UBQ14, At4g02890), CHYR1 (At5g22920), SnRK2.2 (At3g50500), SnRK2.3 (At5g66880), SnRK2.6/OST1 (At4g33950), LEA14 (At1g01470), RD29A (At5g52310), RD20 (At2g33380), DREB2A (At5g05410), ATGolS2 (At1g56600), RAB18 (At1g43890), RbohD (At5g47910), and RbohF (At1g64060). Additional sequences used in phylogenetic analysis were as follows: At5g25560 (Arabidopsis); Os10g31850, Os03g05270, and Os01g0719100 (Oryza sativa); and GRMZM2G144782 and GRMZM2G077307 (Zea mays).
Supplemental Data
Supplemental Figure 1. Sequence Alignment of CHYR1 Homologs.
Supplemental Figure 2. Affinity Purification of CHYR1-GFP-Interacting Protein for LC-MS/MS Analysis.
Supplemental Figure 3. Yeast Two-Hybrid Assay of CHYR1 and SnRK2.10 and Negative Controls in BiFC Assays of CHYR1 and SnRK2.2, SnRK2.3, and SnRK2.6.
Supplemental Figure 4. Representative Photos for Stomatal Aperture of Wild-Type, chyr1 Mutants, 35S:CHYR1, 35S:CHYR1T178A, and 35S:CHYR1T178D Plants under ABA Treatment.
Supplemental Figure 5. Phenotype Analysis of 35S:CHYR1S173AS208A Transgenic Plants.
Supplemental Figure 6. ABA Sensitivity of 35S:CHYR1, 35S:CHYR1T178A, and 35S:CHYR1T178D Transgenic Plants during Seed Germination.
Supplemental Table 1. PCR Primer Sequences Used for This Research.
Supplemental Data Set 1. List of Proteins Identified in the Mass Spectrometry Analysis.
Supplemental Data Set 2. Text File of the Alignment Used in the Phylogenetic Analysis Shown in Figure 1F.
Supplementary Material
Acknowledgments
We thank Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for kindly providing us with the in vitro ubiquitination components and ubiquitin antibody for the assays. We thank June M. Kwak (University of Maryland) for providing us with the snrk2.2/2.3/2.6 mutant seeds. This research was supported by grants from the National Basic Research Program of China (2012CB114302-4) and the National Natural Science Foundation of China (31171163 and 31300995).
AUTHOR CONTRIBUTIONS
S.D. and B.Z. performed the experiments and analyzed the data together. F.Q. designed the experiments. S.D., B.Z., and F.Q. wrote the article.
Glossary
- ABA
abscisic acid
- ROS
reactive oxygen species
- ER
endoplasmic reticulum
- DAB
3,3′-diaminobenzidine
- BiFC
bimolecular fluorescence complementation
- IGP
in-gel kinase phosphorylation
- VC
empty vector control
- MS
Murashige and Skoog
- OS
opening solution
Footnotes
Articles can be viewed online without a subscription.
References
- Bray E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2: 48–54. [Google Scholar]
- Bu Q., Li H., Zhao Q., Jiang H., Zhai Q., Zhang J., Wu X., Sun J., Xie Q., Wang D., Li C. (2009). The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol. 150: 463–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso E., Rodriguez L., Lorenzo-Orts L., Gonzalez-Guzman M., Sayas E., Muñoz-Bertomeu J., Ibañez C., Serrano R., Rodriguez P.L. (2014). The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 80: 1057–1071. [DOI] [PubMed] [Google Scholar]
- Chen Y.T., Liu H., Stone S., Callis J. (2013). ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 75: 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Cross B., Li B., Chen L., Li Z., Chen J. (2011). Regulation of MDM2 E3 ligase activity by phosphorylation after DNA damage. Mol. Cell. Biol. 31: 4951–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.K., Ryu M.Y., Seo D.H., Kang B.G., Kim W.T. (2011). The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid-mediated drought stress responses. Plant Physiol. 157: 2240–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran S.P., Leuenberger D., Leverich E.P., Hwang D.K., Beverly K.N., Koehler C.M. (2004). The role of Hot13p and redox chemistry in the mitochondrial TIM22 import pathway. J. Biol. Chem. 279: 43744–43751. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Desikan R., Cheung M.K., Clarke A., Golding S., Sagi M., Fluhr R., Rock C., Hancock J., Neill S. (2004). Hydrogen peroxide is a common signal for darkness- and ABA-induced stomatal closure in Pisum sativum. Funct. Plant Biol. 31: 913–920. [DOI] [PubMed] [Google Scholar]
- Dreher K., Callis J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99: 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Gampala S.S., Rock C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (Suppl): S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.Y., Cutler S.R., Sheen J., Rodriguez P.L., Zhu J.K. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P.E., Zhu J.K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Zhu J.K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 106: 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50: 2123–2132. [DOI] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K.A., Romeis T., Hedrich R. (2009). Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 106: 21425–21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276. [DOI] [PubMed] [Google Scholar]
- Gu Y., Innes R.W. (2011). The KEEP ON GOING (KEG) protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE 1 (EDR1) protein to TGN/EE vesicles. Plant Physiol. 155: 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677. [DOI] [PubMed] [Google Scholar]
- He J.M., Xu H., She X.P., Song X.G., Zhao W.M. (2005). The role and the interrelationship of hydrogen peroxide and nitric oxide in the UV-B-induced stomatal closure in broad bean. Funct. Plant Biol. 32: 237–247. [DOI] [PubMed] [Google Scholar]
- Hsu K.H., Liu C.C., Wu S.J., Kuo Y.Y., Lu C.A., Wu C.R., Lian P.J., Hong C.Y., Ke Y.T., Huang J.H., Yeh C.H. (2014). Expression of a gene encoding a rice RING zinc-finger protein, OsRZFP34, enhances stomata opening. Plant Mol. Biol. 86: 125–137. [DOI] [PubMed] [Google Scholar]
- Irigoyen M.L., et al. (2014). Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 26: 712–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Li J., Tseng T.S., Schroeder J.I., Ehrhardt D.W., Briggs W.R. (2014). COP1 jointly modulates cytoskeletal processes and electrophysiological responses required for stomatal closure. Mol. Plant 7: 1441–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Kim W.T. (2013). The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol. 162: 1733–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashigawa Y., Tomitaka A., Kumeta H., Noda N.N., Yamaguchi M., Inagaki F. (2011). Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc. Natl. Acad. Sci. USA 108: 20579–20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolla V.A., Vavasseur A., Raghavendra A.S. (2007). Hydrogen peroxide production is an early event during bicarbonate induced stomatal closure in abaxial epidermis of Arabidopsis. Planta 225: 1421–1429. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C.J., Hilhorst H.W., Karssen C.M. (1989). In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol. 90: 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.S., Deng X.W., Callis J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139: 1597–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger-Liszkay A., Fufezan C., Trebst A. (2008). Singlet oxygen production in photosystem II and related protection mechanism. Photosynth. Res. 98: 551–564. [DOI] [PubMed] [Google Scholar]
- Kwak J.M., Mori I.C., Pei Z.M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D., Schroeder J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E., Leonhardt N., Eisler H., Parmentier Y., Alioua M., Jacquet H., Leung J., Genschik P. (2011). MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev. Cell 21: 1116–1128. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Kim W.T. (2011). Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol. Cells 31: 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Lan W., Buchanan B.B., Luan S. (2009). A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 106: 21419–21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng R.P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J.M., Lozano G., Hakem R., Benchimol S. (2003). Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112: 779–791. [DOI] [PubMed] [Google Scholar]
- Leung J., Giraudat J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 199–222. [DOI] [PubMed] [Google Scholar]
- Li H., Jiang H., Bu Q., Zhao Q., Sun J., Xie Q., Li C. (2011). The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol. 156: 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Assmann S.M., Albert R. (2006). Predicting essential components of signal transduction networks: a dynamic model of guard cell abscisic acid signaling. PLoS Biol. 4: e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2013). Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J. Biol. Chem. 288: 20267–20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Mao J., Zhang Y.C., Sang Y., Li Q.H., Yang H.Q. (2005). From The Cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl. Acad. Sci. USA 102: 12270–12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh M.R., Clayton H., Mansfield T.A., Hetherington A.M. (1996). Changes in stomatal behavior and guard cell cytosolic free calcium in response to oxidative stress. Plant Physiol. 111: 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall, J. (2007). Protein of the Month: Zinc fingers. http://www.ebi.ac.uk.
- McGinnis K.M., Thomas S.G., Soule J.D., Strader L.C., Zale J.M., Sun T.P., Steber C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Parry G., Estelle M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori I.C., Murata Y., Yang Y., Munemasa S., Wang Y.F., Andreoli S., Tiriac H., Alonso J.M., Harper J.F., Ecker J.R., Kwak J.M., Schroeder J.I. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol. 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A.C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Suzuki M., Abrams S., McCarty D.R., Kamiya Y., McCourt P. (2002). A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161: 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363. [DOI] [PubMed] [Google Scholar]
- Nishimura N., Sarkeshik A., Nito K., Park S.Y., Wang A., Carvalho P.C., Lee S., Caddell D.F., Cutler S.R., Chory J., Yates J.R., Schroeder J.I. (2010). PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61: 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Osakabe K., Shinozaki K., Tran L.S. (2014). Response of plants to water stress. Front. Plant Sci. 5: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z.M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734. [DOI] [PubMed] [Google Scholar]
- Peng M., Hannam C., Gu H., Bi Y.M., Rothstein S.J. (2007). A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 50: 320–337. [DOI] [PubMed] [Google Scholar]
- Qiao H., Wang F., Zhao L., Zhou J., Lai Z., Zhang Y., Robbins T.P., Xue Y. (2004). The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 16: 2307–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra A.S., Gonugunta V.K., Christmann A., Grill E. (2010). ABA perception and signalling. Trends Plant Sci. 15: 395–401. [DOI] [PubMed] [Google Scholar]
- Ryu M.Y., Cho S.K., Kim W.T. (2010). The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol. 154: 1983–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Dupeux F., Round A., Antoni R., Park S.Y., Jamin M., Cutler S.R., Rodriguez P.L., Márquez J.A. (2009). The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668. [DOI] [PubMed] [Google Scholar]
- Sato A., Sato Y., Fukao Y., Fujiwara M., Umezawa T., Shinozaki K., Hibi T., Taniguchi M., Miyake H., Goto D.B., Uozumi N. (2009). Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 424: 439–448. [DOI] [PubMed] [Google Scholar]
- Schroeder J.I., Kwak J.M., Allen G.J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410: 327–330. [DOI] [PubMed] [Google Scholar]
- Serrano I., Gu Y., Qi D., Dubiella U., Innes R.W. (2014). The Arabidopsis EDR1 protein kinase negatively regulates the ATL1 E3 ubiquitin ligase to suppress cell death. Plant Cell 26: 4532–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C., Gu D., Hu H.C., Davanture M., Lee S., Djaoui M., Valot B., Zivy M., Leung J., Merlot S., Kwak J.M. (2009). Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583: 2982–2986. [DOI] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590. [DOI] [PubMed] [Google Scholar]
- Stirnberg P., van De Sande K., Leyser H.M. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Williams L.A., Farmer L.M., Vierstra R.D., Callis J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z., Wei Y., Collinge D.B. (1997). Subcellular localization of H2O2 in plants, H2O2 accumulation in papillae and hypersensitive response during barley-powdery mildew interaction. Plant J. 11: 1187–1194. [Google Scholar]
- Tian L., Dai L.L., Yin Z.J., Fukuda M., Kumamaru T., Dong X.B., Xu X.P., Qu Q. (2013). Small GTPase Sar1 is crucial for proglutelin and α-globulin export from the endoplasmic reticulum in rice endosperm. J. Exp. Bot. 64: 2831–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Vlad F., Turk B.E., Peynot P., Leung J., Merlot S. (2008). A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 55: 104–117. [DOI] [PubMed] [Google Scholar]
- Waadt, R., and Kudla, J. (2008). In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc. 2008: pdb.prot4995. [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Wang X., Duan C.G., Tang K., Wang B., Zhang H., Lei M., Lu K., Mangrauthia S.K., Wang P., Zhu G., Zhao Y., Zhu J.K. (2013). RNA-binding protein regulates plant DNA methylation by controlling mRNA processing at the intronic heterochromatin-containing gene IBM1. Proc. Natl. Acad. Sci. USA 110: 15467–15472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege S., De Angeli A., Droillard M.J., Kroniewicz L., Merlot S., Cornu D., Gambale F., Martinoia E., Barbier-Brygoo H., Thomine S., Leonhardt N., Filleur S. (2014). Phosphorylation of the vacuolar anion exchanger AtCLCa is required for the stomatal response to abscisic acid. Sci. Signal. 7: ra65. [DOI] [PubMed] [Google Scholar]
- Xie Q., Guo H.S., Dallman G., Fang S., Weissman A.M., Chua N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C.Y., Pence H.E., Jin J.B., Miura K., Gosney M.J., Hasegawa P.M., Mickelbart M.V. (2010). The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22: 4128–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshida R., Umezawa T., Mizoguchi T., Takahashi S., Takahashi F., Shinozaki K. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281: 5310–5318. [DOI] [PubMed] [Google Scholar]
- Zhang H., Cui F., Wu Y., Lou L., Liu L., Tian M., Ning Y., Shu K., Tang S., Xie Q. (2015). The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27: 214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Jin C.D., Roux S.J. (1993). Casein kinase Il-type protein kinase from pea cytoplasm and its inactivation by alkaline phosphatase in vitro. Plant Physiol. 103: 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N.H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19: 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Miao Y.C., An G.Y., Zhou Y., Shangguan Z.P., Gao J.F., Song C.P. (2001a). K+ channels inhibited by hydrogen peroxide mediate abscisic acid signaling in Vicia guard cells. Cell Res. 11: 195–202. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang L., Dong F., Gao J., Galbraith D.W., Song C.P. (2001b). Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 126: 1438–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang C., Li Y., Zheng N., Chen H., Zhao Q., Gao T., Guo H., Xie Q. (2007). SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.