A cytochrome P450 in the CYP76 family modulates linalool emission and linalool oxide (including lilac compounds) formation in Arabidopsis, making flowers repellent rather than attractive to insects.
Abstract
The acyclic monoterpene alcohol linalool is one of the most frequently encountered volatile compounds in floral scents. Various linalool oxides are usually emitted along with linalool, some of which are cyclic, such as the furanoid lilac compounds. Recent work has revealed the coexistence of two flower-expressed linalool synthases that produce the (S)- or (R)-linalool enantiomers and the involvement of two P450 enzymes in the linalool oxidation in the flowers of Arabidopsis thaliana. Partially redundant enzymes may also contribute to floral linalool metabolism. Here, we provide evidence that CYP76C1 is a multifunctional enzyme that catalyzes a cascade of oxidation reactions and is the major linalool metabolizing oxygenase in Arabidopsis flowers. Based on the activity of the recombinant enzyme and mutant analyses, we demonstrate its prominent role in the formation of most of the linalool oxides identified in vivo, both as volatiles and soluble conjugated compounds, including 8-hydroxy, 8-oxo, and 8-COOH-linalool, as well as lilac aldehydes and alcohols. Analysis of insect behavior on CYP76C1 mutants and in response to linalool and its oxygenated derivatives demonstrates that CYP76C1-dependent modulation of linalool emission and production of linalool oxides contribute to reduced floral attraction and favor protection against visitors and pests.
INTRODUCTION
The acyclic monoterpene alcohol linalool (3,7-dimethyl-1,6-octadiene-3-ol) is one of the most frequently encountered floral volatile compounds, occurring widely in both monocots and dicots (Raguso and Pichersky, 1999; Knudsen et al., 2006). Its two enantiomeric forms, due to the chirality of its hydroxylated carbon, occur in different proportions in floral extracts from distinct plant species (Dötterl et al., 2006a; Parachnowitsch et al., 2013). Various cyclic pyranoid or furanoid oxygenated derivatives are most often present along with linalool, such as the furanoid lilac compounds (Figure 1). These compounds, which usually occur as complex mixtures of diastereomeric alcohols and aldehydes with different organoleptic properties, are important fragrance and flavor components. Volatile linalool oxides are prominent constituents of the scent or aroma of flowers and/or fruits, for example, from lilac (Syringa vulgaris) (Wakayama et al., 1973; Kreck et al., 2003), Clarkia breweri (Pichersky et al., 1994), kiwi (Actinidia arguta) (Matich et al., 2003, 2011), papaya (Carica papaya) (Flath and Forrey, 1977; Winterhalter et al., 1986), and grape (Vitis vinifera) (Williams et al., 1980; Luan et al., 2006), as well as tea (Camelia sinensis) leaves (Morita et al., 1994; Wang et al., 2000). A large proportion of linalool and its oxides is often present as bound, glycosidic conjugates in the plant tissues, the aglycones being released upon fruit maturation or tissue disruption (Heidlas et al., 1984; Flath et al., 1990; Raguso and Pichersky, 1999). In the case of grape and tea, these compounds are released upon manufacturing processes for the production of wines and semifermented or fermented teas (Williams et al., 1980; Wang et al., 2000). Convergent labeling experiments carried out with lilac (Burkhardt and Mosandl, 2003; Kreck et al., 2003) and kiwi flowers (Matich et al., 2007, 2011) or with papaya fruit (Schreier and Winterhalter, 1986) and grape berry mesocarp (Luan et al., 2006) demonstrated that lilac compounds, as well as other oxides, derive from linalool via successive oxidation steps, but the mechanism for their formation has not been elucidated.
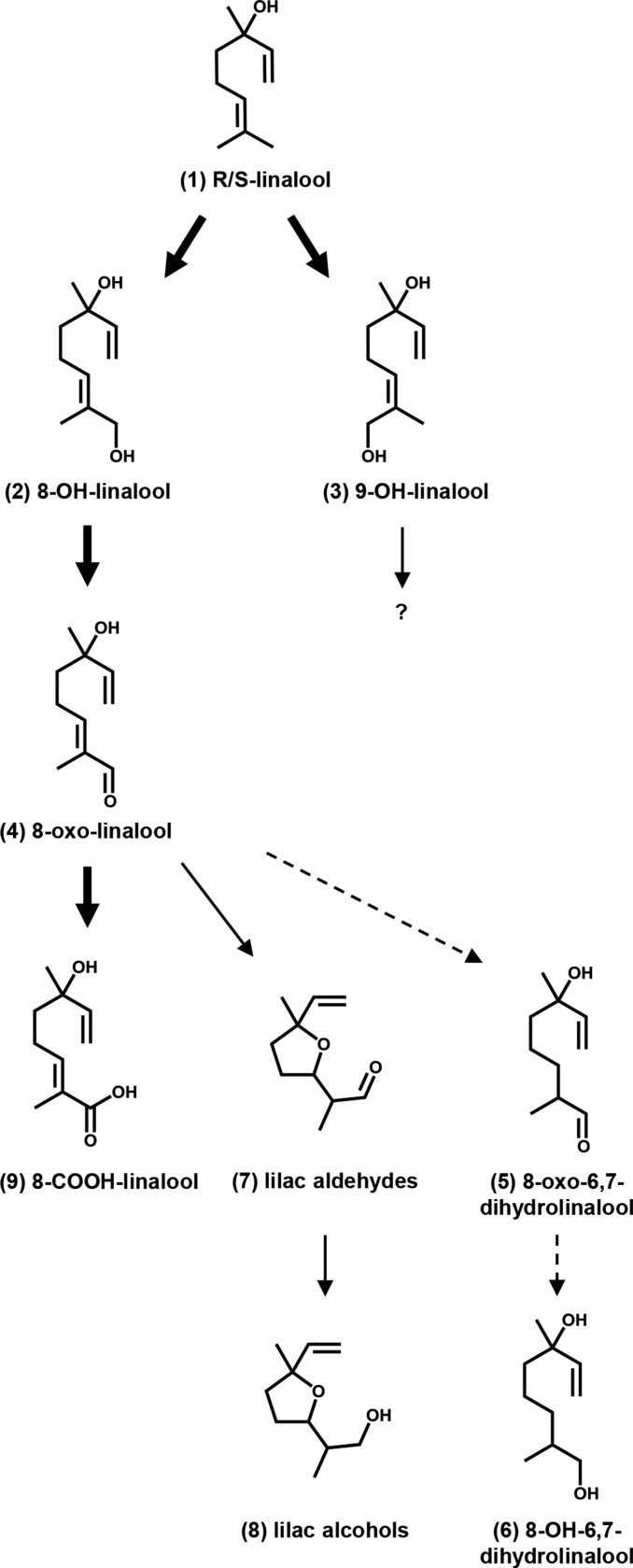
Figure 1.
Scheme Summarizing CYP76C1-Catalyzed Reactions.
Arrows indicate CYP76C1- and NADPH-dependent reactions in vitro. Mainstream CYP76C1-dependent pathway in vivo is shown in bold. Dashed arrows indicate that the formation of the compounds is not dependent on CYP76C1 in vivo. The minor product of primary linalool oxidation, 9-OH-linalool, was not available and thus not further tested as a substrate.
Arabidopsis thaliana flowers were long considered to be scentless and a poor model to study the metabolism of volatile compounds, until the sequencing of the plant genome revealed the existence of a quite broad family of terpene synthases and triggered investigation of the flower-emitted volatiles (Tholl and Lee, 2011). Initial analyses showed that sesquiterpenoids were predominant in the floral bouquet of Arabidopsis (Chen et al., 2003; Tholl et al., 2005). However, Chen et al. (2003) reported a flower-expressed terpene synthase (TPS14) producing (+)-(3S)-linalool. Rohloff and Bones (2005), in a more extensive analysis of the complex Arabidopsis floral volatile bouquet, then detected the presence of small amounts of linalool, as well as lilac aldehydes. Only recently have investigations based on coexpression of cytochrome P450 oxygenases with two terpene synthases started to reveal a complex floral linalool metabolism in the flowers of Arabidopsis (Ginglinger et al., 2013). This work revealed the coexistence of two flower-expressed linalool synthases, TPS14 and TPS10, producing (S)- and (R)-linalool enantiomers, respectively, and the role of two P450 enzymes, CYP76C3 and CYP71B31, in linalool oxidation. This study also suggested the presence of partially redundant P450 enzymes that may contribute to floral linalool metabolism. The hypothesis of the involvement of other P450 oxygenases in floral linalool metabolism was then further supported by a systematic functional exploration of the expression patterns and in vitro activities of the CYP76C3 paralogs present in Arabidopsis (Höfer et al., 2014), which revealed floral expression of CYP76C2 and CYP76C1, two other monoterpenol-metabolizing oxygenases. CYP76C1 emerged as the best candidate due to its high and selective linalool oxygenase activity, which is 10 times higher than the activity of CYP76C2. This work also indicated that CYP76C1 was mainly expressed in the inflorescence.
This led us to postulate that CYP76C1 might be a significant player in floral linalool metabolism and possibly in flower-insect interactions. Here, we provide evidence that CYP76C1 is the main linalool metabolizing oxygenase in Arabidopsis flowers. We demonstrate its prominent role in the control of linalool emission and in the formation of most linalool oxides detected in vivo, both as volatile and soluble conjugated compounds, including 8-hydroxy, 8-oxo, and 8-COOH-linalool, as well as lilac aldehydes and alcohols (Figure 1). Insect behavior on the CYP76C1 mutants and in response to linalool and its oxygenated derivatives demonstrates that CYP76C1, via controlling the emission of linalool and its conversion into more oxidized derivatives, contributes to reducing floral attraction of typical pollinator taxa and favors protection against floral antagonists and pests.
RESULTS
Gene Coexpression and Functional Screening Identify CYP76C1 as a Prime Candidate for Regulating Linalool Metabolism in Arabidopsis Flowers
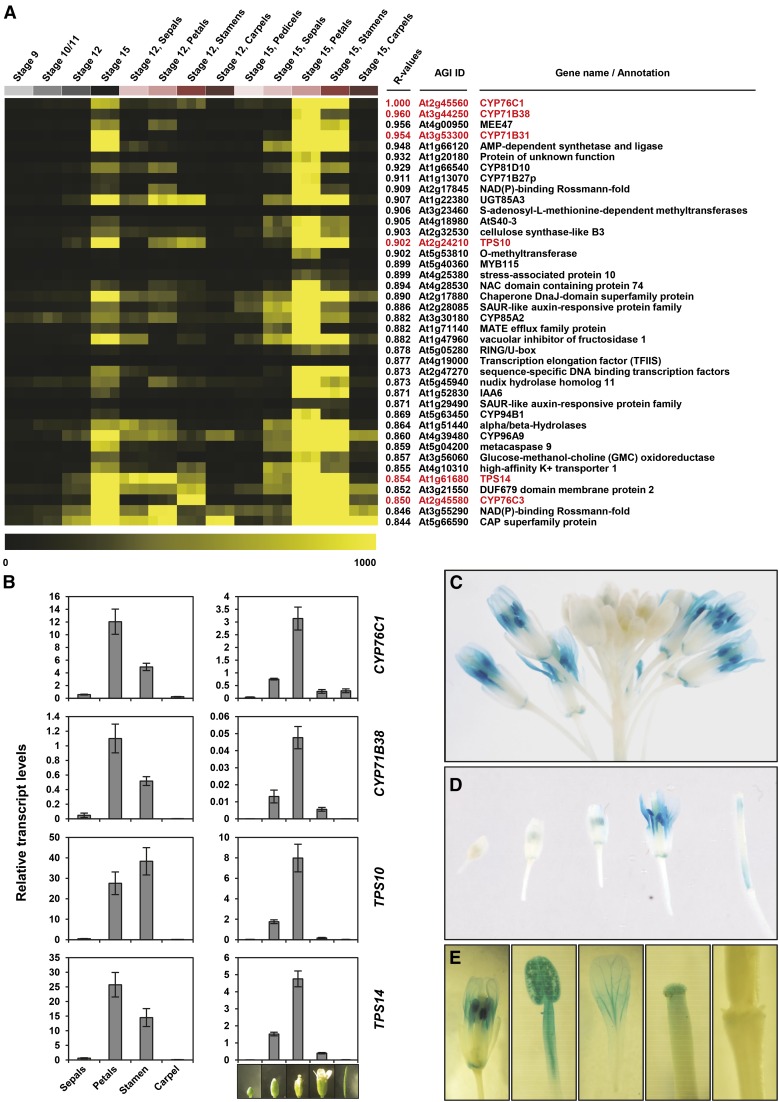
A thorough in silico analysis indicated that the expression of CYP76C1 was tightly coordinated in mature Arabidopsis flowers with the expression of genes involved in linalool metabolism. Figure 2A shows that CYP76C1 is expressed in the petals and anthers at anthesis and is tightly coregulated with TPS10 and TPS14, with high correlation coefficients of 0.902 and 0.854, respectively. It was also coexpressed with CYP71B31 and CYP76C3, encoding the two P450s recently shown to be active on linalool (Ginglinger et al., 2013). Coregulation of CYP76C1 with these genes involved in linalool metabolism (Figure 2B) and other enzymes potentially involved in this pathway (Supplemental Figure 1) was confirmed by qRT-PCR. The highest level of coregulation was found between CYP76C1 and CYP71B38 (Figure 2A), but expression of the latter was very low (Figure 2B). Therefore, CYP71B38 was not considered as a priority candidate for further investigations. Plants transformed with the CYP76C1 promoter fused to the β-glucuronidase gene (CYP76C1p:GUS) were used to obtain a more precise description of CYP76C1 activity. They revealed a transient and tissue-specific expression of CYP76C1 in filaments, anthers, stamen, and petals upon anthesis (Figures 2C to 2E), with some expression also detected in the stigma, petal abscission zone, and young siliques.
Figure 2.
CYP76C1 Expression and Coexpression Patterns in Arabidopsis Flowers.
(A) Expression heat map of the top 40 genes most closely coregulated with CYP76C1. Coexpression analysis was performed using the Expression Angler tool and the AtGenExpress Tissue Compendium data set (Toufighi et al., 2005). Heat map shows the expression levels in selected flowers tissues: flower stages 9, 10/11, 12, and 15 and petals, stamens, pedicels, and carpel at flower stages 12 and 15. Coexpressed genes are ranked according to their Pearson correlation coefficients with CYP76C1 (R values). Genes highlighted in red are known to be involved or putatively involved in linalool metabolism.
(B) Relative transcripts levels of CYP76C1, CYP71B38, TPS10, and TPS14 in flower organs (left panel) and during flower development (right panel). Relative transcript levels were determined by qRT-PCR using the EΔCt method, and the specific efficiency of each primer pair and normalization with four reference genes for which the stable expression level are known (Czechowski et al., 2005). Results represent the mean ± se of three biological replicates for the flower parts and four biological replicates for flower stages.
(C) to (E) GUS staining showing spatiotemporal floral expression of CYP76C1. Staining was 19 h for inflorescence (C), flowers at different stages (D), and parts of fully opened flowers (E).
The subcellular localization of the CYP76C1 protein was investigated via transient coexpression of CYP76C1:eGFP and TPS10:mRFP1 constructs in the leaves of Nicotiana benthamiana. This experiment indicated a localization of CYP76C1 at the endoplasmic reticulum contrasting with the patchy intrachloroplastic pattern observed for TPS10 (Supplemental Figure 2). The tridimensional imaging employed for this experiment better illustrated our previous observation (Ginglinger et al., 2013) that plastids appear wrapped in endoplasmic reticulum sheets, which would most likely favor P450 conversion of the plastid-emitted linalool.
CYP76C1 Sequentially Metabolizes Linalool and Its Successive Oxides to Form Lilac Aldehydes/Alcohols and 8-COOH-Linalool
CYP76C1 was previously shown to metabolize (−)-(3R)-linalool into 8-OH-linalool and small amounts of 9-OH-linalool (Höfer et al., 2014). To determine if CYP76C1 was selectively metabolizing R-linalool or both the (−)-(3R) and (+)-(3S) enantiomers, which are respectively produced by TPS10 and TPS14 (Ginglinger et al., 2013), the conversion of both of these enantiomers by yeast-expressed CYP76C1 was investigated (Supplemental Figure 3; Figure 3). Recombinant yeast microsomes metabolized the racemic mixture (Figure 3B) and pure enantiomers (Supplemental Figures 3B and 3C) to form a major metabolite identified as 8-OH-linalool (2) (Supplemental Figure 4A). Comparison of the catalytic parameters for the conversion of R- or S-linalool into 8-OH-linalool showed that both enantiomers were metabolized with very similar efficiencies (kcat/Km = 14.0 min−1 μM−1 for R-linalool and 9.8 min−1 μM−1 for S-linalool; Supplemental Figure 5). The other minor coeluting products (3/4) of linalool oxidation by CYP76C1 were identified as 9-OH-linalool (3) and 8-oxo-linalool (4) (Figure 3B; Supplemental Figures 3G, 4B, and 4C).
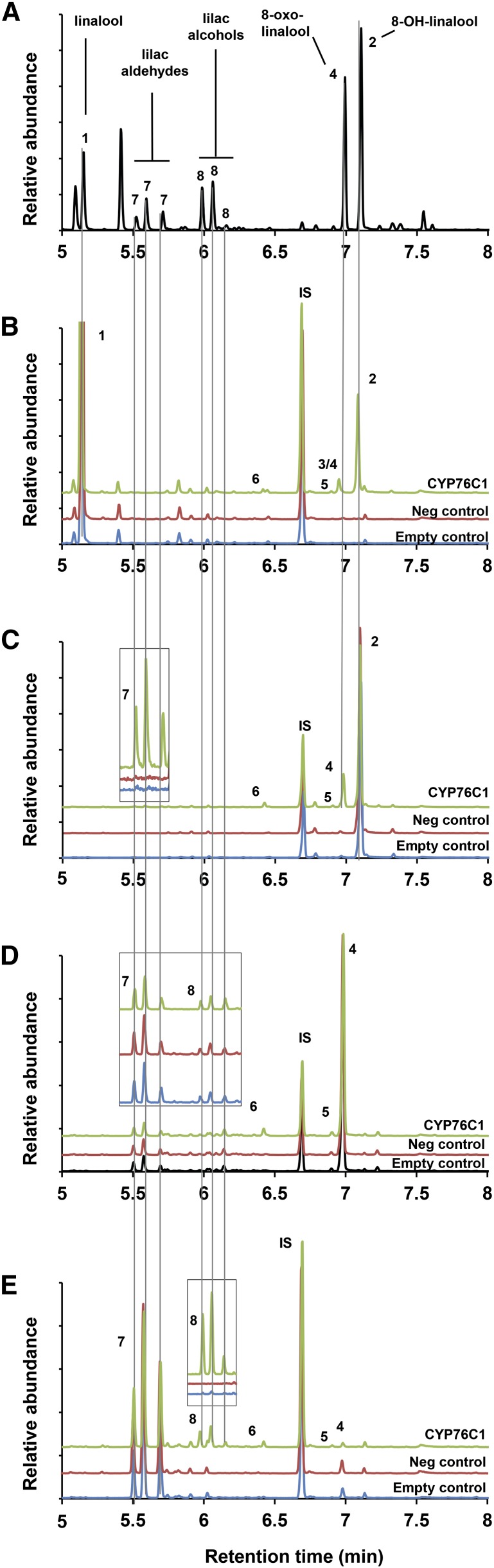
Figure 3.
Gas Chromatography Analysis of the Products Resulting from Yeast-Expressed CYP76C1 Activity on Linalool and Linalool Oxides.
GC-MS chromatograms of a mix of authentic standards (A) and of ethyl acetate extracts of the products from the conversion of a racemic mix of (R/S)-linalool (B), 8-OH-linalool (C), 8-oxo-linalool (D), and lilac aldehydes (E) by yeast-expressed CYP76C1. Microsomal membranes from the recombinant yeast expressing CYP76C1 (final [P450] ∼50 nM) or transformed with an empty vector (Empty control) were incubated for 15 min with 200 µM of substrate in presence or absence (neg. control) of NADPH. Chromatograms show the relative abundance of total ion current and the selected ions m/z 111 + 153 + 155 in the inserts. Compared with control, CYP76C1-dependent 8-oxo-linalool metabolism results in a decrease of 8-oxo-linalool and lilac aldehydes and a simultaneous increase in lilac alcohols (D). This suggested that the lilac aldehydes formed via conversion of 8-oxo-linalool might be converted rapidly into alcohols by CYP76C1 (E). Formation of lilac aldehydes from 8-oxo-linalool was confirmed performing a time-course experiment using low substrate and P450 concentrations (Supplemental Figure 7). (1) Racemic R/S-linalool, (2) 8-OH-linalool, (3) 9-OH-linalool, (4) 8-oxo-linalool, (5) 8-OH-6,7-dihydrolinalool, (6) 8-oxo-6,7-dihydrolinalool, (7) lilac aldehydes, and (8) lilac alcohols. IS, nonyl acetate used as internal standard for normalization. (2), (4), (7), and (8) were identified by comparison of RT and MS with those of authentic standards (Supplemental Figure 4). (3) was identified by comparison of its MS with MS of 8-OH-linalool (2) when separated from (4) on a HP-35ms column (Supplemental Figure 3). (5) and (6) were identified by NMR after reaction upscaling and purification by preparative GC (Supplemental Figure 6).
Another member of the CYP76 family, CYP76B6 from Madagascar periwinkle (Catharanthus roseus), was recently shown to metabolize geraniol and to further oxidize its hydroxylated product, 8-OH-geraniol into 8-oxo-geraniol (Höfer et al., 2013). The detection of 8-oxo-linalool in linalool conversion products thus suggested that CYP76C1 might also further metabolize 8-OH-linalool. When tested as a substrate of CYP76C1, 8-OH-linalool (2) was converted into 8-oxo-linalool (4) as the main product, along with two minor unknown compounds, (5) and (6), and small amounts of lilac aldehydes (7) (Figure 3C; Supplemental Figures 4D to 4F). The conversion of 8-OH-linalool into 8-oxo-linalool and four lilac aldehyde diastereoisomers was confirmed by NMR after upscaling the reaction (Supplemental Figures 6A and 6B).
Considering the presence of lilac compounds in the reaction products, further metabolism of 8-oxo-linalool by CYP76C1 was then also investigated (Figure 3D). Since the systematic presence of lilac aldehydes and lilac alcohols in the standard of 8-oxo-linalool (Supplemental Figure 3E) suggested that 8-oxo-linalool and the lilac compounds were prone to autoxidation, a time-course experiment was performed. This experiment confirmed the transient formation by CYP76C1 of lilac aldehydes after 30 s and 1 min of incubation followed by their decline in prolonged incubations (Supplemental Figure 7). This was explained by the capacity of the recombinant CYP76C1 to metabolize pure lilac aldehydes into several lilac alcohol diastereoisomers (Figure 3E; Supplemental Figure 4G). The minor products (5) and (6) were identified by NMR as 8-OH-6,7-dihydrolinalool and 8-oxo-6,7-dihydrolinalool, respectively (Supplemental Figure 6C).
Ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) analysis of the reaction products (Supplemental Figure 8) confirmed the formation of the products detected by gas chromatography (2, 8-OH-linalool; 3, 9-OH-linalool; 4, 8-oxo-linalool; 5, 8-OH-6,7-dihydrolinalool; 6, 8-oxo-6,7-dihydrolinalool; 7, lilac aldehydes; and 8, lilac alcohols). In addition, it revealed the formation of 8-COOH-linalool (9) from 8-oxo-linalool (Figure 4).
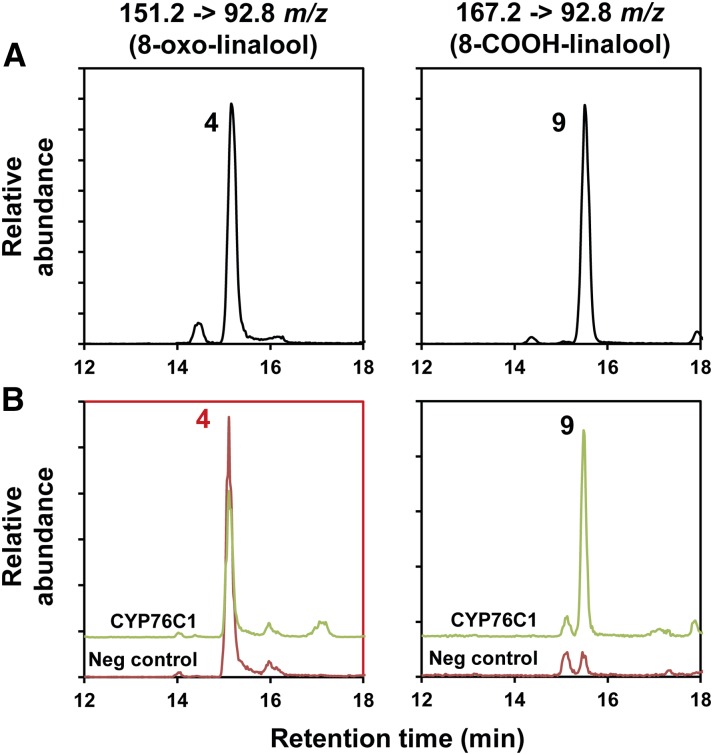
Figure 4.
Targeted UPLC-MS/MS Analysis of the Products Resulting from Yeast-Expressed CYP76C1 Activity on 8-Oxo-Linalool.
Samples were analyzed by UPLC-MS/MS using MRM. Left panels show a specific channel of MS/MS transition developed for the detection of 8-oxo-linalool (151.2 → 92.8 m/z). Right panels show a specific channel of MS/MS transition developed for the detection of 8-COOH-linalool (167.2 → 92.8 m/z).
(A) Mix of authentic standards.
(B) Methanol extract of the products from the conversion of 8-oxo-linalool by CYP76C1. Yeast microsomal membranes (final [P450] ∼100 nM) were incubated for 20 min with the substrate (200 µM) in the presence or absence (neg. control) of NADPH. Red panel shows the consumption of the substrate by CYP76C1 in presence of NADPH compared with the negative control. Minor amount of 8-COOH-linalool is detected in the negative control, probably due to 8-oxo-linalool autoxidation. 8-COOH-linalool was identified from its specific MS/MS and retention time compared with the authentic standard. More details and extended results are shown in Supplemental Figure 8. (4) 8-oxo-linalool and (9) 8-COOH-linalool.
Altogether, our data thus reveal an unexpectedly versatile activity of the recombinant CYP76C1 on linalool and its successive oxidized metabolites and suggest that it plays an important role in the formation of a broad range of linalool oxides, including 8-COOH-linalool and the cyclic volatile lilac compounds. The cascade of reactions catalyzed by CYP76C1 in vitro is illustrated in Figure 1.
CYP76C1 Is the Major Linalool Oxidase in Arabidopsis Flowers and Modulates the Emission of Linalool and Lilac Compounds
In order to investigate the physiological and ecological roles of CYP76C1 in Arabidopsis flowers, two independent homozygous insertion lines were selected (cyp76c1-1 and cyp76c1-2) and their genotype confirmed by RT-PCR (Supplemental Figure 9). In addition, a complemented/overexpression line was generated by transformation of the insertion mutant cyp76c1-1 with the full coding sequence of CYP76C1 under control of the CaMV 35S promoter (35S:CYP76C1) (Supplemental Figures 9B and 9C). Extensive gene expression analysis of the CYP76C1 insertion and complemented lines did not reveal any changes in expression for the genes known to be involved in linalool metabolism (TPS10, TPS14, CYP71B31, and CYP76C3) nor for P450s belonging to the same cluster as CYP76C1 and known to be active on linalool (CYP76C2, CYP76C3, and CYP76C4) (Höfer et al., 2014) (Supplemental Figure 9C). This ensured that any change in linalool metabolism would be the result of CYP76C1 activity.
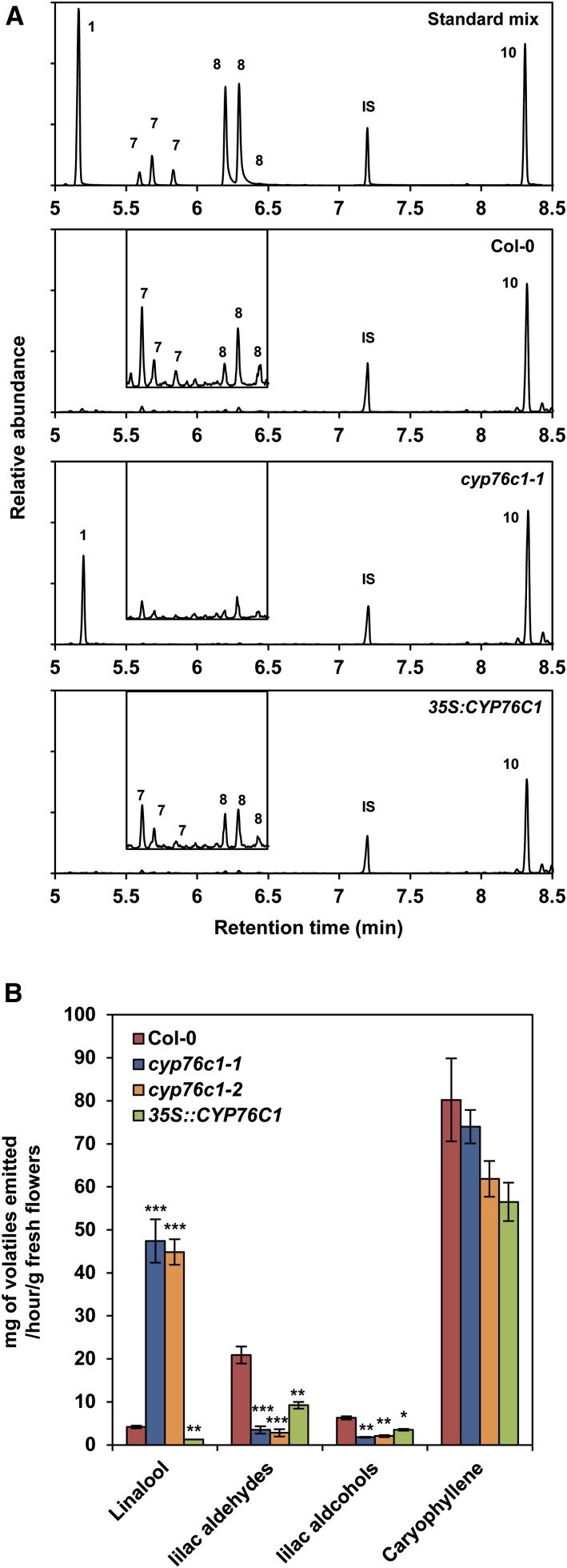
The headspaces collected from the inflorescences of the different lines were first evaluated by gas chromatography-mass spectrometry (GC-MS) analysis (Figure 5). Linalool emission was increased more than 10-fold in the insertion mutants compared with the wild type, reaching an emission rate of 47 ng h−1 g−1 fresh weight (FW), which was close to the emission rate of β-caryophyllene (up to 80 ng h1 g−1 FW), the most abundant volatile compound emitted by the flowers of Col-0 Arabidopsis. Conversely, overexpression of CYP76C1 resulted in barely detectable linalool emission compared with the wild type. In contrast, the emission of lilac aldehydes and lilac alcohols was reduced by ∼85 and 70%, respectively, in the cyp76c1 mutants compared with the wild type and was restored, although not to the wild-type level, in the complemented/35S:CYP76C1 line.
Figure 5.
Comparative GC-MS Analysis of the Flower Headspace of Wild-Type Arabidopsis, cyp76c1 Mutant, and 35S:CYP76C1 Lines.
(A) Targeted GC-MS chromatograms showing linalool, lilac compounds (inset), and caryophyllene emission based on selected ion currents 93 + 111 + 126 m/z (main panels) and 93 + 153 + 155 m/z (insets). Representative chromatograms are shown for an authentic standard mix of (1) linalool, (7) lilac aldehydes, (8) lilac alcohols, (10) caryophyllene, and (IS) nonyl acetate internal standard (top panel), as well as wild-type (Col-0), cyp76c1-1, and 35S:CYP76C1 lines.
(B) Quantification of headspace volatiles. Data are mean ± se of three biological replicates. Statistically significant differences relative to the wild type are indicated (Student’s t test: *P ≤ 0.05; **P > 0.01; ***P < 0.001).
Taken together, these results reveal a very active CYP76C1-dependent linalool oxidative metabolism that almost completely suppresses the emission of the linalool formed by TPS10 and TPS14 in wild-type Arabidopsis flowers. They also indicate that CYP76C1 largely determines the level of floral emission of linalool and linalool oxides. However, the amounts of lilac compounds emitted by the wild type do not mirror the amounts of linalool emitted by the cyp76c1 insertion lines, which could be explained by the retention of oxygenated and/or conjugated soluble metabolites in the wild-type flower tissues.
A Major Proportion of the CYP76C1-Derived Linalool Oxides Is Accumulated as Soluble Conjugates in Arabidopsis Flower Tissues
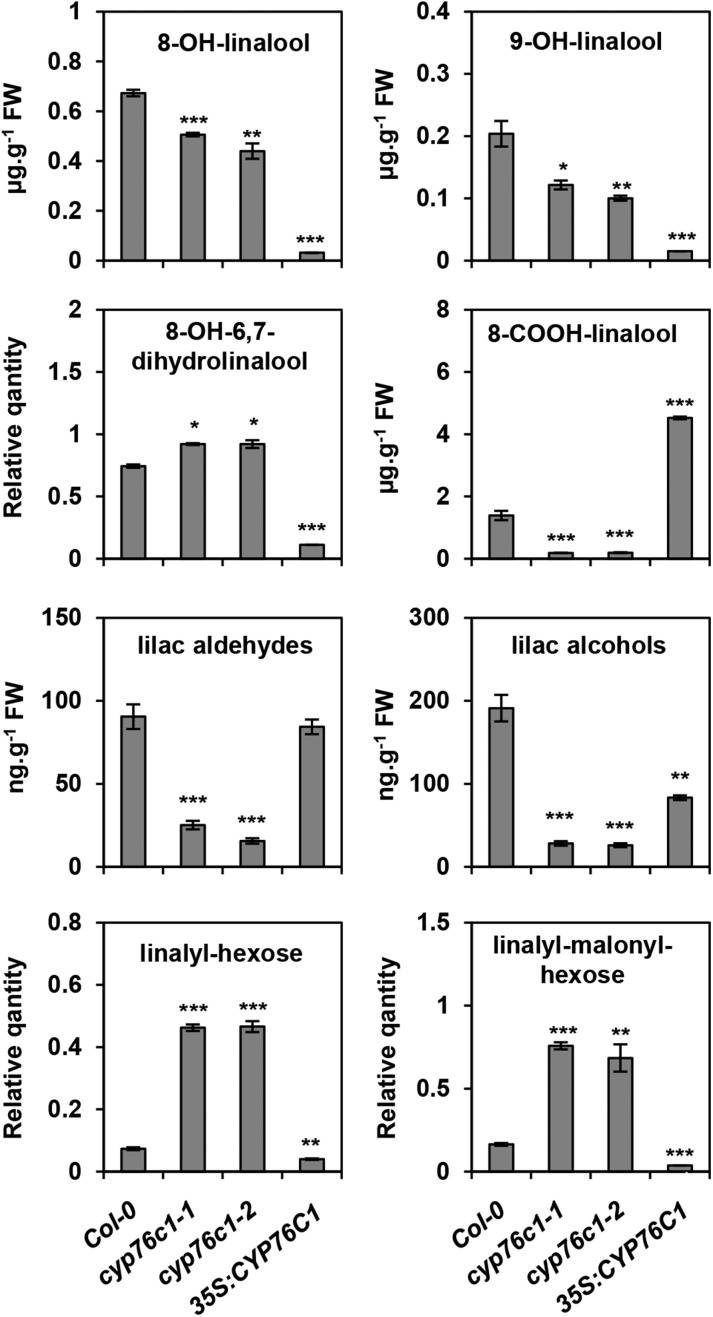
Linalool and its oxides have been reported to accumulate as free or conjugated compounds in flower tissues from various plant species including Arabidopsis (Aharoni et al., 2003; Ginglinger et al., 2013; Yang et al., 2013). Soluble linalool derivatives were thus quantified by targeted UPLC-MS/MS in the inflorescence of wild-type, cyp76c1, and complemented/35S:CYP76C1 lines (Figure 6; Supplemental Figure 10).
Figure 6.
Targeted UPLC-MS/MS Quantification of Linalool and Linalool Derivatives in the Flowers from Wild-Type and CYP76C1 Insertion and Complemented Lines.
Methanol extracts of the flowers were treated with β-glycosidase before UPLC-MS/MS analysis using the MRM method. The total amount of aglycones detected was quantified based on standard curves, except for lynalyl derivatives, which were only detected as conjugates and for 8-OH-6,7-dihydrolinalool, for which no standard was available. Data are mean ± se of three biological replicates. Statistically significant differences relative to the wild type are indicated (Student’s t test: *P ≤ 0.05; **P > 0.01; ***P < 0.001). Representative chromatograms are shown in Supplemental Figure 10.
The linalool conjugates were exclusively detected as putative linalyl-hexose and linalyl-malonyl hexoses, as previously observed (Ginglinger et al., 2013; Yang et al., 2013). They accumulated at up to 5 times higher concentrations in the cyp76c1 lines, and their levels were significantly reduced in the complemented/35S:CYP76C1 line compared with the wild type (Figure 6; Supplemental Figure 10A).
Conversely, all linalool oxides were detected as aglycones after deglycosylation. The levels of 8-OH-linalool and 9-OH-linalool, also formed by the CYP76C2 and CYP76C3 enzymes (Ginglinger et al., 2013; Höfer et al., 2014), were moderately reduced in the cyp76c1 flowers compared with the wild type, but almost completely abolished in the complemented/35S:CYP76C1 line. The levels of lilac aldehydes and alcohols conjugates were strongly reduced in the cyp76c1 flowers and partially restored in the complemented/35S:CYP76C1 line. Conversely, the levels of 8-COOH-linalool, which was almost absent in the cyp76c1 lines, strongly increased in complemented/35S:CYP76C1 plants compared with the wild type, which confirms the efficient CYP76C1-dependent conversion of 8-oxo-linalool into 8-COOH-linalool in the plant. Interestingly, whereas 8-COOH-linalool has not previously been detected in Arabidopsis flowers, it appears to be the most abundant linalool oxidation product, with amounts reaching 1.4 µg⋅g−1 of fresh weight in wild-type flower tissues. This concentration, estimated on the basis of the fresh weight of the whole inflorescence, was most likely much higher in the tissues specifically expressing CYP76C1, such as petal or anther filaments.
Although produced in vitro from the 8-oxo-linalool conversion by CYP76C1, the amounts of 8-OH-6,7-dihydrolinalool were slightly higher in the cyp76c1 mutants compared with the wild type and strongly reduced in the overexpressor line. A competing enzyme may thus be responsible for the formation of this metabolite in the plant.
Altogether, these results point to a major role of CYP76C1 in a complex and branched linalool metabolism leading to the accumulation of a variety of soluble conjugates in Arabidopsis flowers, with 8-COOH-linalool being the main product.
Transient Coexpression of TPS10/TPS14 and CYP76C1 in N. benthamiana Leaves Leads to Massive Accumulation of 8-COOH-Linalool but No Lilac Emission
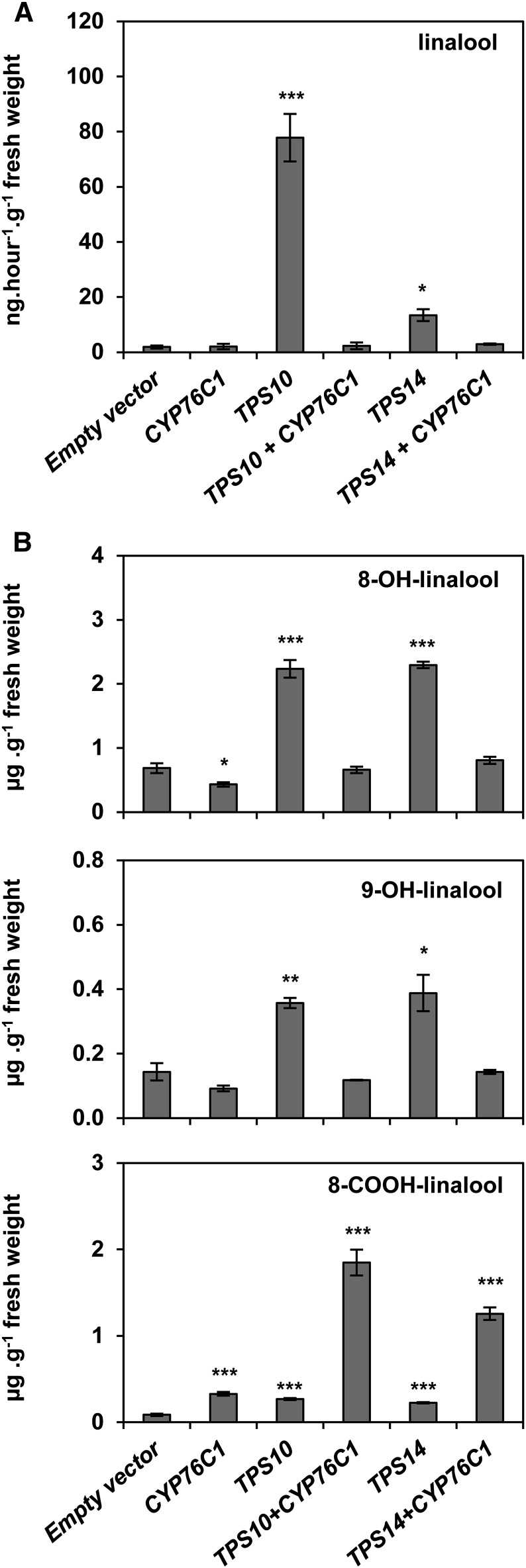
In an attempt to reconstruct the CYP76C1-dependent linalool pathway in a heterologous system, N. benthamiana leaves were transfected with vectors driving the expression of the enantiospecific linalool synthase TPS10 and TPS14 from Arabidopsis either alone or together with CYP76C1 (Figure 7). This experiment revealed the occurrence of an endogenous linalool-derived metabolism in N. benthamiana leaves resulting in low linalool emission (Figure 7A) and the accumulation of small amounts of 8- and 9-OH-linalool conjugates (Figure 7B). Minor amounts of 8-COOH-linalool conjugates were also detected in control leaves (Figure 7B). Expression of TPS10 or TPS14 alone led to increased emission of linalool and to increased accumulation of all linalool-derived metabolites.
Figure 7.
Heterologous Reconstitution of the Linalool-Derived Pathway by Transient Coexpression of Linalool Synthases and CYP76C1 in N. benthamiana Leaves.
N. benthamiana leaves were infiltrated with Agrobacterium tumefaciens transformed with an empty vector or with vectors driving the expression of CYP76C1, TPS10, or TPS14. Each gene was expressed alone or in the combinations TPS10/CYP76C1 or TPS14/CYP76C1. Five days postinfiltration, headspace volatiles were collected from the transformed leaves and analyzed by GC-MS. Simultaneously, β-glucosidase-treated methanol extracts from the leaves were analyzed by UPLC-MS/MS.
(A) Head-space analysis.
(B) Quantification by targeted UPLC/MS-MS of 8-OH-linalool, 9-OH-linalool, and 8-COOH-linalool, the only linalool-derived metabolites detected in the transformed leaves and for which the accumulation was modified depending on the enzymes coexpressed. Data are mean ± se of three biological replicates. Statistically significant differences relative to empty vector are indicated (Student’s t test: *P ≤ 0.05; **P > 0.01; ***P < 0.001).
Coexpression of each linalool synthase with CYP76C1 resulted in the restoration of the basal linalool emission observed in control leaves, confirming the CYP76C1-dependent metabolism of both linalool enantiomers produced by TPS10 and TPS14 in the plant (Figure 7A). Acceleration of the linalool oxidation cascade was further confirmed by the decrease in 8- and 9-OH-linalool levels in cotransfected leaves compared with leaves transformed with each TPS alone and by the concomitant accumulation of 8-COOH-linalool. The formation of other linalool-derived metabolites, such as the lilac compounds, or 8-OH-6,7-dihydrolinalool, was not detected in these experiments.
Reconstruction of the CYP76C1-dependent linalool pathway in N. benthamiana thus confirms that CYP76C1 efficiently converts both linalool enantiomers in vivo and that strong expression of CYP76C1 favors the production of 8-COOH-linalool.
CYP76C1-Dependent Modulation of Linalool and Linalool Oxide Production Affects the Behavior of Flower-Visiting Insects and Florivores
Engineered Arabidopsis plants transformed with the linalool/nerolidol synthase gene Fa-NES1 from strawberry (Fragaria ananassa) produce massive amounts of linalool and also accumulate glycosides of linalool oxides in leaves (Aharoni et al., 2003). In dual-choice tests, Fa-NES1 plants were shown to repel aphids. We thus tested the behavior of the aphid Myzus persicae on Arabidopsis flowers, expecting a preference for wild-type flowers over flowers of cyp76c1 mutants, which emit high levels of linalool. No significant difference was observed in aphid behavior in a dual-choice test between the flowers of wild-type and cyp76c1 lines (Supplemental Figure 11), possibly due to the preference of aphids for stems rather than flowers since they feed on phloem.
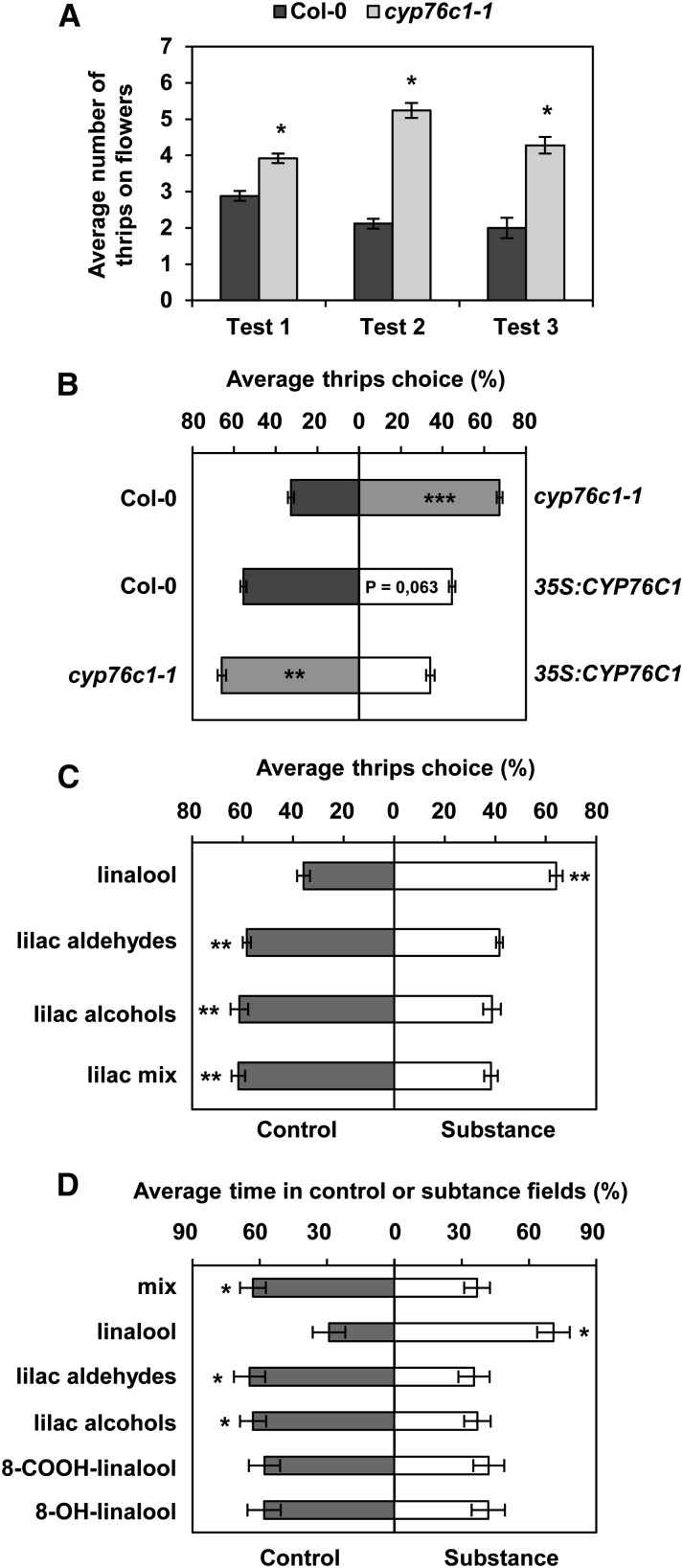
Arabidopsis is a self-pollinating plant that does not depend on pollinators to be fertilized. However, outcrossing events occur in natural populations due to visitation of flowers by insects such as solitary bees, hoverflies, and thrips (Jones, 1971; Snape and Lawrence, 1971; Hoffmann et al., 2003). The behavior of two of these insects, thrips (Frankliniella occidentalis) and hoverflies (Episyrphus balteus), was tested using cyp76c1 flowers and/or linalool oxides (Figure 8). Dual-choice tests performed on thrips demonstrated that they preferred to visit the flowers of the cyp76c1 mutants over the wild type (Figure 8A; Supplemental Figure 12). Thrips are antagonistic flower visitors described as being attracted by pure linalool (Koschier et al., 2000) and by linalool-emitting engineered chrysanthemum (Chrysanthemum morifolium) plants transformed with Fa-NES1, but repelled by linalool conjugates (Yang et al., 2013). To distinguish the role of volatiles from the influence of compounds stored in flowers the thrips could feed on, we conducted dual-choice tests in a Y-shaped olfactometer giving thrips the choice between volatiles from the flowers of the wild-type, cyp76c1, or complemented/35S:CYP76C1 lines using different combinations (Figure 8B). Thrips always preferred the volatiles from the plant emitting more linalool and less lilac compounds (cyp76c1-1 over the wild type, the wild type over complemented/35S:CYP76C1, and cyp76c1 over complemented/35S:CYP76C1). To discriminate between the influence of linalool and lilac compounds on this behavior, thrips preference was also tested using pure compounds (Figure 8C). In good agreement with their preference for cyp76c1 flowers emitting increased amounts of linalool and less lilacs compounds over the wild type, thrips were attracted by linalool, but they avoided lilac alcohols, lilac aldehydes, or an equimolar mix of both.
Figure 8.
Behavior of Flower Pollinators and Floral Antagonists on CYP76C1 Mutant Lines and Pure Compounds.
(A) Thrips dual-choice test between flowers from the wild type and cyp76c1-1 mutant. Results represent the average number of thrips (±se) on the flowers from five biological replicates and for three independent tests. Statistically significant differences relative to the wild type are indicated (Wilcoxon test: n = 5, *P ≤ 0.05).
(B) Thrips dual-choice test between volatiles from Col-0, cyp76c1, and 35S:CYP76C1 flowers performed in a Y-shaped arena of an olfactometer. Three headspace combinations were tested: Col-0 versus cyp76c1-1, Col-0 versus 35S:CYP76C1, and cyp76c1-1 versus 35S:CYP76C1. Results represent the average thrips’ choice + se from five independent replicates, each using different individual plants and for which choice of 30 to 40 individual thrips was scored. Statistically significant differences are indicated (paired t test: n = 5, **P < 0.01; ***P < 0.001).
(C) Thrips dual-choice test between pure compounds performed in Y-shaped arena of an olfactometer. Results represent the average thrips’ choice + se from six independent replicates and for which the choice of 30 individual thrips was scored. One hundred micrograms of each compound alone or 100 µg of both lilac compounds for the mix were used for each assay. Statistically significant differences are indicated (paired t test: n = 6, **P < 0.01).
(D) Hoverfly dual-choice test between pure compounds performed in a star-shaped arena of an olfactometer. Results represent the average percentage of time + se spent in substance or control fields for 30 individual hoverflies. One hundred micrograms of each compound alone or in mix were used for each assay. Statistically significant differences are indicated (paired t test: n = 25, **P < 0.05).
Hoverflies are known to be occasional pollinators of Arabidopsis in natural populations (Jones, 1971; Snape and Lawrence, 1971; Hoffmann et al., 2003). Their behavior was thus tested in a star-shaped olfactometer using pure compounds (Figure 8D). The syrphids spent significantly more time in fields supplied with linalool than in neutral fields serving as a control. Conversely, they avoided fields supplied with lilac compounds. Likewise, syrphids tended to avoid olfactometer fields supplied with 8-OH-linalool or 8-COOH-linalool, albeit not significantly. However, the neutral field was significantly preferred to fields supplied with an equimolecular mix of linalool and all of the linalool oxides (linalool, 8-OH-linalool, 8-COOH-linalool, lilac aldehydes, and lilac alcohols) tested separately (Figure 8D), which confirms that the attraction of linalool can be reduced/reversed by the presence of lilac compounds and other linalool oxides.
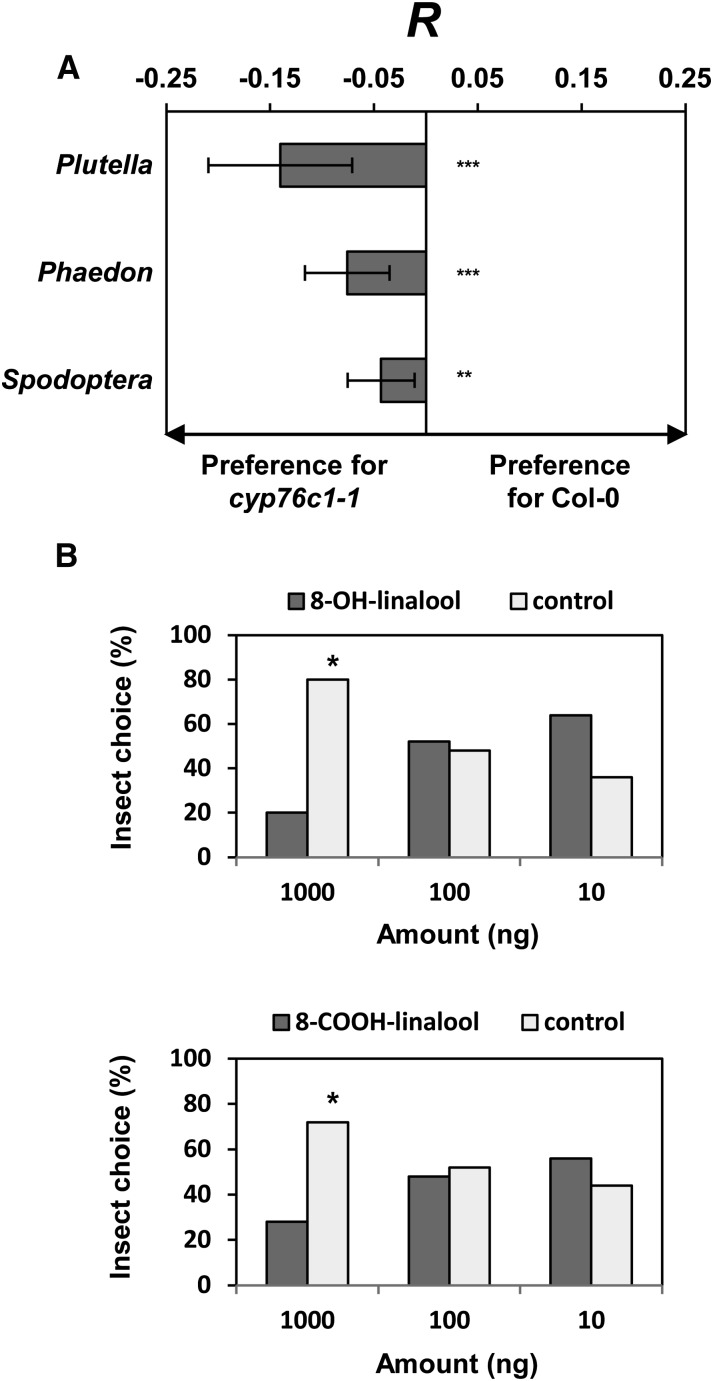
To test the hypothesis that the chemical display of Arabidopsis flowers represents an adaptation to defend against florivores instead of attracting pollinators, we tested the behavior of three florivorous insects, including the generalist Spodoptera littoralis (larvae) and the Brassicaceae specialists Plutella xylostella (larvae) and Phaedon cochleariae (adults), in dual-choice assays (Figure 9). All three of these insects significantly preferred feeding on cyp76c1-1 (Figure 9A), which may either result from a deterrent effect of the soluble linalool oxides that accumulate in wild-type flowers or attraction by linalool emitted from cyp76c1 flowers. This suggests an increase in flower fitness resulting from CYP76C1 expression due to reduced florivory. To test the effect of the soluble linalool oxides that accumulate in wild-type flowers, P. cochleariae feeding preference was then further tested on cabbage leaves treated with pure linalool or linalool oxides (Figure 9B; Supplemental Figure 13). The beetles significantly preferred to feed on control leaves over leaves treated with 1 μg g−1 FW of 8-OH-linalool or 8-COOH-linalool in dual-choice tests. No impact of linalool or lilac alcohols was detected in these choice assays (Supplemental Figure 13). Considering that 8-COOH-linalool is the most abundant linalool oxide that accumulates in wild-type flowers, with an estimated floral concentration around 1.4 µg g−1 FW, the response of the beetle to higher concentrations of the compounds or their glycosidic forms is not excluded. Our data are thus indicative of a deterrent effect or toxicity of soluble linalool oxides, such as 8-OH-linalool and 8-COOH-linalool, which accumulate in flower tissues as glycosides.
Figure 9.
Behavior of Florivores on Flowers of CYP76C1 Mutant Lines and Pure Compounds.
(A) Feeding preferences of larvae of P. xylostella and S. littoralis and adults of P. cochleariae for flowers from the wild type and cyp76c1-1 mutant in dual-choice test. Response index R represents the mean proportion consumed from cyp76c1-1 flowers minus the mean proportion of Col-0 flowers consumed in each case (±se) for 30 individuals per insect species. Negative values indicate a preference for the mutants. Statistically significant differences relative to the wild type are indicated (Student’s t test: n = 3, **P < 0.01; ***P < 0.001).
(B) Dual-choice test with adults of P. cochleariae feeding on cabbage leaves treated with pure 8-OH and 8-COOH-linalool. One gram of cabbage leave was treated with 10, 100, or 1000 ng of pure compounds dissolved in methanol or only with methanol (Control). Results represent the percentage mean choice of 30 individuals between treated or control leaves. Statistically significant differences relative to control are indicated (paired t test: n = 30, *P < 0.05).
The behavior of both pollinators and florivores on wild-type and mutant lines (or in the presence of pure compounds) thus supports the hypothesis that the expression of CYP76C1 reduces the attractiveness of Arabidopsis flowers by decreasing free linalool emissions but simultaneously increasing the production of volatile or conjugated lilac compounds and other linalool oxides that serve as defensive compounds against floral antagonists.
DISCUSSION
Linalool has been reported to be emitted by flowers of around 70% of the seed plant families (Knudsen et al., 2006). Linalool floral emission is often regarded as spatially and temporally governed by the expression of linalool synthases during flower development, but little information on the mechanism and role of its oxidative metabolism has been available, although sequestered and oxidized forms have been reported (Pichersky et al., 1994; Dudareva et al., 1996; Raguso and Pichersky, 1999; Lücker et al., 2001). Here, we describe how a single enzyme, CYP76C1, plays an essential role in linalool metabolism in Arabidopsis flowers by simultaneously tuning linalool emission and the production of its oxidized derivatives, which both affect (in different ways) the behavior of flower visiting insects.
CYP76C1 Is a Multifunctional and Pleiotropic Monoterpenol Oxidase
Our data provide clear evidence that the cytochrome P450 CYP76C1 is the major player in the oxidation of linalool in the flowers of Arabidopsis. CYP76C1 is expressed at a high level in flower tissues and catalyzes a cascade of oxidations on linalool and its oxidation products, producing soluble metabolites such as 8-COOH-linalool and the volatile cyclic derivatives lilac alcohols and lilac aldehydes (Figure 1). The prominent role of CYP76C1 in linalool metabolism is demonstrated by the massive emission of linalool and accumulation of linalool conjugates in cyp76c1 insertion lines, while the production of 8-COOH-linalool and lilac compounds is strongly reduced in these lines. The production of these compounds is largely recovered in the complemented line.
In vitro assays demonstrate that CYP76C1 is able to catalyze the whole oxidation cascade from linalool to lilac alcohols and 8-COOH-linalool. This cascade includes alcohol and aldehyde intermediates, some of which, such as alcohols, can be detected in the plant tissues, and others, such as the aldehyde, are too unstable or rapidly converted to final products. The cascade also includes some side products such as reduced derivatives resulting from the activity of other enzymes. A strictly parallel increase or decrease in the levels of all intermediates of the reaction cascade is not observed in Arabidopsis in vivo. In addition, transient expression of CYP76C1 was not sufficient to obtain the whole spectrum of metabolites in N. benthamiana. This partially results from the variable efficiency of CYP76C1 for catalyzing the different steps of the reaction cascade. It also reflects the existence of other enzymes, which participate in the biosynthesis of these compounds or in branching pathways in vivo. For example, 8- and 9-OH-linalool are also produced by CYP76C2 and CYP76C3 in Arabidopsis flowers (Ginglinger et al., 2013; Höfer et al., 2014), whereas in N. benthamiana leaves, oxidation of monoterpenols and monoterpenol oxides into the derived acids and glycosides was demonstrated by previous studies (Dong et al., 2013; Höfer et al., 2013).
Autoxidation of substrates, background metabolism by yeast enzymes, rapid conversion of the products by CYP76C1, and lack of pure enantiomers prevented the determination of the enzyme catalytic parameters for the reactions downstream of the formation of 8-OH-linalool. It was thus not possible to clearly identify potential bottlenecks in the reaction cascade. However, both the analyses of Arabidopsis complemented plants and transformed N. benthamiana indicate that CYP76C1 overexpression under a strong promoter favors direct formation of 8-COOH-linalool and decreases the accumulation of 8-OH-linalool, the precursor of 8-oxo-linalool, which was never detected in the plant. The conversion of 8-oxo-linalool into 8-COOH-linalool is thus likely to be faster in vivo than the upstream steps of the pathway. This is expected to reduce the pool of precursors required for the formation of lilac compounds, and fine-tuning of CYP76C1 expression might be required for effective production of lilac compounds. On the other hand, additional enzymes may be required for the effective formation of lilac alcohols in vivo. Oxydoreductases were recently found to enhance P450-catalyzed oxidation cascades in the biosynthesis of artemisinin (Paddon et al., 2013) and iridoids (Brown et al., 2015). Moreover, most linalool oxides are prone to autoxidation, in particular 8-oxo-linalool. Hence, a role of chemical conversion to form lilac compounds or of autoxidation to prevent 8-oxo-linalool accumulation in specific cellular environments such as photosynthetic tissues cannot be excluded.
Plant cytochrome P450s are usually regarded as enzymes with rather high substrate and regio/stereospecificities. CYP76C1 represents a counterexample. While this enzyme has a clear preference for linalool among other monoterpenols and is not metabolizing geraniol (Höfer et al., 2013, 2014), it is able to metabolize both R- and S-linalool with equal efficiencies, as well as a broad range of linalool oxides, in addition to phenylurea herbicides (Höfer et al., 2014). Comparison of its lilac products with a mixture of standards in which the eight isomeric forms of lilac aldehydes and lilac alcohols are present (racemic mixtures of the four diastereoisomers) suggests that CYP76C1 can generate the four lilac diastereoisomers, but not with the same efficiencies. Different proportions of lilac aldehydes or alcohols diastereoisomers are detected after conversion of racemic mixtures of 8-OH-linalool, 8-oxo-linalool, or lilac aldehydes. With respect to substrate preference, CYP76C1 radically differs from CYP76B6 from C. roseus, which oxidizes a broad range of monoterpenols (geraniol, nerol, linalool, and citronellol) with similar efficiencies, but never beyond the stage of the aldehyde, and cannot metabolize phenylurea (Höfer et al., 2013, 2014).
A broad substrate range seems to be a common feature of the CYP76 family. For example, the CYP76Ms of rice (Oryza sativa) evolved as multifunctional enzymes dedicated to the biosynthesis of labdane-related diterpenoid antifungal phytoalexins (Wang et al., 2012; Wu et al., 2013). This underscores the fast evolution and versatility of the CYP76 family and its specialization in different taxa to generate promiscuous enzymes, allowing fast adaption to ecological niches. This high enzyme versatility in the CYP76C subfamily seems to correlate with a high genetic variability (Höfer et al., 2014) and most likely favors the retention of gene duplicates.
CYP76C1 Sets the Balance between the Emission/Accumulation of Linalool and Linalool Oxides in the Flowers of Arabidopsis
By catalyzing linalool oxidation, CYP76C1 also reduces the emission of free linalool. The large increase in linalool emission and accumulation of linalyl conjugates upon gene inactivation demonstrates that CYP76C1 is the most abundant and active linalool metabolizing enzyme in the flowers of Arabidopsis. CYP76C1 diverts the free and conjugated linalool to volatile and soluble oxidation products, a large proportion of which is conjugated and stored in the plant tissues. Conjugation/deconjugation are thus expected to constitute other regulatory mechanisms for the storage and release of the bioactive oxygenated compounds. Indeed, a glycosyl transferase, UGT85A3, belonging to a family known to be active on monoterpenols and iridoids (Caputi et al., 2008; Nagatoshi et al., 2011) and an α-β hydrolase are coexpressed with CYP76C1. As previously discussed by Raguso and Pichersky (1999), some of the conjugates could be considered precursors of the free volatiles released upon flower maturation. However, the timing of TPS and P450 gene expression, which is quasisimultaneous with terpenoid emission, appears to exclude this possibility. The linalool oxide conjugates are thus expected to have a protective function, with aglycones potentially released upon attack by florivores or pathogens.
Unforeseen Complexity of Linalool-Derived Signaling in Arabidopsis Flowers
The two other linalool oxidases previously found to be expressed in the flowers of Arabidopsis, CYP76C3 and CYP71B31 (Ginglinger et al., 2013), led to the formation of 1,2-epoxylinalool, 4-OH-linalool, 8-OH-linalool, and different diastereomers of 5-OH-linanool as primary products in a transient plant expression assay, although their final products in Arabidopsis flowers were quantitatively too small for identification. Both CYP76C3 and CYP71B31 were mainly expressed in the upper segment of the anther filament and nectaries (with very low expression in petals), whereas CYP76C1 was expressed in filaments, anthers, stigma, and petals during anthesis. No CYP76C1 expression was detected in nectaries. This suggests the occurrence of local concentrations of specific compounds and fine-tuning of olfactory and gustatory cues in the different organs of Arabidopsis flowers. Considering the existence of other flower-expressed genes potentially encoding linalool/linalool oxide-metabolizing enzymes such as CYP76C2 and CYP71B38, further complexity in linalool oxides signaling in Arabidopsis is expected.
CYP76C1 Influences the Interactions of Flowers with Insects
Linalool in flower fragrance was long considered to function as a pollinator attractant (Borg-Karlson et al., 1996; Raguso and Pichersky, 1999; Reisenman et al., 2010). However, constitutive or induced/engineered linalool emission can also serve in plant defense (Junker et al., 2011; McCallum et al., 2011; Xiao et al., 2012), and linalool can also attract antagonistic flower visitors such as thrips (Koschier et al., 2000). In addition, herbivore-induced emission of linalool has been found to affect multitrophic interactions in several systems, where it attracts herbivores as well as their predators and parasitoids (Loughrin et al., 1995; Turlings et al., 1995; Xiao et al., 2012). Thus, it seems that the roles of linalool differ depending on the plant-insect interactions. Interestingly, Yang et al. (2013) reported that thrips were first attracted by linalool emitted by engineered chrysanthemum expressing linalool/nerolidol synthase from strawberry (Fa-NES1) but were ultimately repelled by linalool conjugates. The role of linalool conjugates and linalool oxides in ecological interactions is still a matter of debate (Raguso and Pichersky, 1999).
For self-compatible plants that do not depend on pollinators for successful reproduction, such as Arabidopsis, floral defenses may be more important than attractive displays to maximize the fitness of plant individuals. As the floral morphology of Arabidopsis does not provide any physical protection, we hypothesize that volatile or nonvolatile secondary metabolites produced by wild-type flowers may function as defenses against florivores (generalists and those specialized on Brassicaceae) and pollen thieves (Kessler et al., 2008; Junker and Blüthgen, 2010; Junker et al., 2010a).
In support for our hypothesis, we found that thrips and florivorous insects show strong preference for mutant flowers with increased linalool emission but reduced emissions of lilac aldehydes and lilac alcohols and lower concentrations of linalool oxides stored in the tissues. Bioassays with pure compounds further confirmed that preference for mutant flowers resulted from an attraction to linalool and from the lack of defensive volatiles and stored products (linalool oxides). It has been shown that lilac compounds are important in a specialized nursery pollination system for recognition by the noctuid moth nursery pollinator of white campion (Silene latifolia) (Dötterl et al., 2006b, 2007). Moreover, lilac aldehydes from Asimitellaria were shown to induce the nectaring behavior of long-tongued fungus gnats but are repellent for short-tongued fungus gnats in olfactometer tests (Okamoto et al., 2015). In our hands, both lilac aldehydes and lilac alcohols turned out to be repellent for a number of other insect taxa. This is in accordance with the dual function of individual volatiles and scent bouquets, with the obligate dependency of the moths to find their hosts for both resources and oviposition (Junker and Blüthgen, 2010; Schiestl, 2010) and the concurrent necessity for plants to protect reproductive tissues. In line with this, it is interesting to note that CYP76C1 is present as a pseudogene in the genome of the outcrossing relative Arabidopsis lyrata (Höfer et al., 2014).
Thus, we have shown how one gene/enzyme may be sufficient to turn an attractive display into a defensive phenotype using the attractive compound as substrate for the production of defensive metabolites. Interestingly, constitutively expressed CYP76C1 is localized specifically in the stamen (anthers and filament), petals, and top of the style of Arabidopsis flowers, which are the most valuable tissues for seed set and reproduction. Additionally, the role of CYP76C1 in flower defense against flower antagonists such as florivorous insects is supported by its temporally activated expression during flower opening and anthesis, the stages when flowers are most exposed and visited.
Prospect for Engineering Flower Scent, Aroma, Drug Production, and Plant Defense
Lilac aldehydes and alcohols form an important class of fragrance compounds, with low odor threshold and olfactory properties that depend on their complex stereochemistry (Kreck and Mosandl, 2003; Siska et al., 2014). Both lilac aldehydes and alcohols were first isolated from lilac flower oil (Wakayama et al., 1970, 1971; Wakayama and Namba, 1974) and later identified as fragrant components in Gardenia jasminoides flowers (Hattori et al., 1978), slender bog orchid (Platanthera stricta) (Patt et al., 1988), and Artemisia pallens (Misra et al., 1991). Lilac compounds, together with pyranoid and furanoid linalool oxides, are also produced in flowers and fruits of a broad range of plant species as diverse as papaya (Schreier and Winterhalter, 1986), grape (Luan et al., 2006), kiwi (Matich et al., 2003), and C. breweri (Pichersky et al., 1994). Their formation in these plants is expected to involve similar oxidative cascades. The demonstration of the role of CYP76C1 in multiple linalool oxidations is a first step in engineering lilac compounds. On the other hand, 8-COOH-linalool is a structural building block of compounds with high therapeutic potential, such as α,β-unsaturated monoterpene acid glucose esters, which are prevalent in the genus Eucalyptus (Goodger and Woodrow, 2011, 2013), and cytotoxic saponins from Acacia victoriae (Hanausek et al., 2001; Haridas et al., 2001) and Albizia lebbeck (Noté et al., 2015). Expression or modulation of CYP76C1 functional homologs in different plant species and microorganisms might thus be considered for applications as diverse as aroma enhancement, drug synthetic biology, engineering plant defense against herbivores, and enhancing/improving flower scent.
METHODS
Plant Growth
Arabidopsis thaliana Col-0 and Nicotiana benthamiana seeds were sown on a standard soil compost mixture in 7-cm-diameter pots and cultivated in growth chambers under white fluorescent lamps with a light intensity of 100 to 150 µmol m−2 s−1 at 22°C during the 12-h day period and 19°C during the 12-h night period for Arabidopsis and at 24°C during the 16-h day period and at 20°C during the 8-h night period for N. benthamiana.
Gene Coexpression Analysis and Quantification of Gene Expression
CYP76C1 expression and coexpression patterns were investigated using the ExpressionAngler tool (Toufighi et al., 2005). Quantification of gene expression was performed by qRT-PCR as previously described (Ginglinger et al., 2013; Höfer et al., 2014). Tissues and organs of Arabidopsis were harvested from at least five flowering plants and immediately frozen in liquid nitrogen. Total RNA was extracted by the classical lithium chloride-phenol protocol and treated with DNase I (Fermentas, Thermo Fisher Scientific) according to the manufacturer’s instructions. cDNAs were synthesized from 2 µg of total RNA with SuperScript III reverse transcriptase (Invitrogen, Life Technologies) in the presence of oligo(dT)18 primers (Fermantas, Thermo Fisher Scientific) and then 10-fold diluted. Primers used for each gene are provided in Supplemental Table 1. qRT-PCR reactions were prepared with a Biomek 3000 (Beckman Coulter) and contained 2 μL cDNA, 5 μL of LightCycler 480 SYBR Green I Master (Roche), and 250 nM of forward and reverse primers in a total volume of 10 µL. Samples were analyzed in triplicate on a LightCycler 480 II (Roche) with an amplification profile consisting of 95°C for 10 min and 40 cycles (95°C denaturation for 10 s, annealing at 60°C for 15 s, and extension at 72°C for 15 s), followed by a melting curve analysis from 55 to 95°C to verify the specificity of transcripts. Relative transcript levels were calculated using the EΔCt method (Pfaffl, 2001), corrected with the specific efficiency of each primer pair and normalized with four reference genes whose stable expression has been validated (Czechowski et al., 2005). Five biological replicates were used for the organs, three for the floral stages and four for flower parts.
Generation of Vector Constructs
Vector constructs were generated as previously described (Ginglinger et al., 2013; Höfer et al., 2014) with the USER cloning technique (New England Biolabs) according to Nour-Eldin et al. (2006). USER extensions were added to the primers (Supplemental Table 1) to integrate the appropriate sequences into the suitable plasmids. The CYP76C1 coding sequence was integrated into the yeast expression plasmid pYeDP60u2 and the plant expression vector pCAMBIA3300u (Höfer et al., 2013). CYP76C1 and TPS10 coding sequences with modified 3′ ends were respectively fused to the 5′ sequence of eGFP (Sagt et al., 2009) and mRFP (Campbell et al., 2002) according to Geu-Flores et al. (2007) as previously described (Ginglinger et al., 2013) and inserted into the plant expression vector pCAMBIA2300u. The 1-kb promoter region of CYP76C1 was integrated into the plant expression vector pBI101u as previously described (Ginglinger et al., 2013). Constructs were confirmed by sequencing. Plants and yeasts were transformed as previously described (Ginglinger et al., 2013; Höfer et al., 2014).
GUS Reporter Analysis of CYP76C1 Tissue-Specific Expression
A confirmed construct was introduced into the Agrobacterium tumefaciens strain GV3101 for Arabidopsis transformation by floral dip (Clough and Bent, 1998). Transformed lines were selected on plates containing kanamycin at 50 µg⋅mL−1. Three independent T1 kanamycin resistant lines were brought to T3 stable progeny by germination on kanamycin to obtain homozygous stable lines. GUS staining of the flowers was performed for 19 h after vacuum infiltration of 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) (Jefferson et al., 1987).
Subcellular Distribution of CYP76C1 and TPS10 Transiently Expressed in N. benthamiana Leaf Epidermal Cells Monitored by Confocal Microscopy
Constructs were transformed into the Agrobacterium LBA4404 strain (van der Fits et al., 2000). Twenty-day-old N. benthamiana leaves were cotransformed by agroinfiltration with a culture of equal density expressing the p19 protein of tomato bushy stunt virus (Voinnet et al., 2003) in a ratio of 1/1 (v/v) for transient expression as described by Bassard et al. (2012). Four days postinfiltration, leaf discs were excised for observation by confocal microscopy. Cell imaging was performed using a Leica TCS SP5X confocal laser scanning microscope equipped with a DM6000 microscope (Leica). Images were recorded using a 63× water immersion objective lens (HCX PL APO lambda blue 63× 1.2 water UV; Leica). Excitation/emission wavelengths were 488/495 to 530 nm for CYP76C1:eGFP construct, 548/560 to 600 nm for TPS10:mRFP1 construct, and 488/600 to 770 nm for chlorophyll fluorescence. The Z-stack image series were sequentially acquired using the resonant laser of the SP5X confocal system and the LAS Advanced Fluorescence version 2.7.3.9723 software (Leica). Images series were processed with ImageJ software version 1.49o (NIH; http://rsb.info.nih.gov/ij) or with Volocity version 6.3.0 (Perkin-Elmer). For ImageJ processing, a standard deviation Z-projection was prepared from images series, then contrasts were adjusted. For image processing by Volocity, noise was removed with the fine filter parameter, bleaching was corrected, and contrasts adjusted prior to 3D reconstruction with 3D opacity mode.
Synthesis of Linalool Oxides
Synthesis of racemic R/S-8-OH-linalool and racemic R/S-8-oxo-linalool was performed as previously described from racemic R/S-linalool (Ginglinger et al., 2013). Synthesis of racemic mixture of the four stereoisomers of lilac aldehydes (2-(5-methyl-5-vinyltetrahydrofuran-2-yl)propanol) was performed from (E)-2,6-dimethyl-6-hydroxy-2,7-octadienal following a slight modification of the procedure of Wilkins et al. (1993) and Wakayama et al. (1973). Dry NaH (172 mg, 7.13 mmol, 4 equiv.) was added to a stirred solution of racemic R/S-8-oxo-linalool (300 mg, 1.78 mmol, 1 equiv.) in anhydrous methanol (10 mL). The reaction mixture was stirred for 4 h, diluted with CH2Cl2 (30 mL), and washed with a saturated aqueous solution of NaCl (30 mL). The aqueous phase was extracted with CH2Cl2 (3 × 40 mL). The combined organic layers were dried over Na2SO4, filtrated, and concentrated under reduced pressure. Pure product was obtained by flash chromatography (PE/EtOAc 100/0 to 30/70) to afford the title compound as yellowish oil (127 mg, 0.75 mmol, 42%, four diastereoisomers). 1H NMR (CDCl3, 300 MHz) characterization of the title compound was as follows: δ = 9.72 (d, 1H, J = 2.3 Hz), 5.81 (d, 1H, J = 10.8 Hz), 5.11 (dd, 1H, J = 17.3, 1.4 Hz), 4.93 (dt, 1H, J = 10.4, 1.2 Hz), 4.04 to 4.14 (m, 1H), 2.38 to 2.47 (m, 1H), 1.62 to 2.07 (m, 4H), 1.24 (s, 3H), 1.01 (d, 3H, J = 7.0 Hz). 13C NMR (CDCl3, 75 MHz) characterization of the title compound was as follows: δ = 204.56, 204.52, 204.29, 144.21, 144.03, 143.45, 143.33, 111.76, 111.62, 111.58, 111.5, 83.40, 83.35, 83.13, 79.62, 79.17, 78.66, 78.46, 51.90, 51.71, 51.10, 50.74, 37.63, 37.43, 36.89, 36.62, 29.97, 29.36, 29.22, 28.65, 27.08, 26.76, 26.52, 26.14, 10.37, 10.29, 9.95, and 9.28. Rf: 0.78 (PE/EtOAc 70/30).
Synthesis of racemic mixture of the four stereoisomers of lilac alcohols (2-(5-methyl-5-vinyltetrahydrofuran-2-yl)propan-1-ol) was performed from the racemic mixture of the four stereoisomers of lilac aldehydes. NaBH4 (70 mg, 1.86 mmol, 4 equiv.) was added to a stirred solution of 2-(5-methyl-5-vinyltetrahydrofuran-2-yl)propanal (78 mg, 0.46 mmol, 1 equiv.) in anhydrous methanol (10 mL, 0.05 M). The reaction mixture was stirred for 30 min, subsequently diluted with Et2O (20 mL), and washed with NaCl saturated aqueous solution (20 mL). The aqueous phase was extracted with Et2O (3 × 20 mL). The combined organic layers were dried over Na2SO4, filtrated, and concentrated under reduced pressure. Pure product was obtained by flash chromatography (PE/EtOAc 100/0 to 30/70) to afford the title compound as yellowish oil (44 mg, 0.26 mmol, 56%, four diastereoisomers). 1H NMR (CDCl3, 300 MHz) characterization of the title compound was as follows: δ = 5.80 to 5.92 (m, 1H), 5.16 (dd, 1H, J = 17.0, 1.7 Hz), 4.98 (d, 1H, J = 10.6 Hz), 3.75 to 3.86 (m, 1H), 3.60 (m, 3H), 1.98 to 2.06 (m, 1H), 1.65 to 1.89 (m, 4H), 1.30 (s, 3H), 0.79 (d, 3H, J = 6.9 Hz). 13C NMR (CDCl3, 75 MHz) characterization of the title compound was as follows: δ = 144.57, 143.86, 143.76, 112.20, 111.97, 86.24, 85.84, 84.03, 83.89, 83.29, 82.72, 69.31, 69.06, 41.47, 41.35, 37.60, 37.29, 36.72, 31.49, 31.09, 27.57, 27.23, 27.06, 13.97, and 13.76 . Rf: 0.62 (PE/EtOAc 70/30).
Synthesis of a racemic mixture of 8-COOH-linalool [(E)-6-hydroxy-2,6-dimethylocta-2,7-dienoic acid] was performed following the procedure of Sharma and Chand (1996). A solution of NaClO2 (1.00 g, 11 mmol, 7.6 equiv.) and NaH2PO4 (1.00 g, 8.3 mmol, 5.7 equiv.) in distilled water (10 mL) was added dropwise to a stirred solution of (E)-6-hydroxy-2,6-dimethylocta-2,7-dienal (244 mg, 1.45 mmol, 1 equiv.) in a mixture of t-butanol (25 mL) and 2-methyl-2-butene (10 mL). The yellow solution was stirred overnight at room temperature and concentrated under a vacuum. The aqueous layer was diluted with water (30 mL) and extracted once with hexane (30 mL). The aqueous layer was titrated to pH 3.0 and extracted twice with Et2O (2 × 30 mL). The combined organic layers were washed with a saturated aqueous solution of NaCl, dried over Na2SO4, filtrated, and concentrated under vacuum, affording the title compound (232 mg, 1.26 mmol, 87%). 1H NMR (300 MHz, CDCl3) characterization of the title compound was as follows: 6.89 (td, 1H, J = 7.5, 1.6 Hz), 5.86 to 5.95 (m, 1H), 5.24 (dd, 1H, J = 17.3, 1.3 Hz), 5.10 (dd, 1H, J = 10.7, 0.9 Hz), 2.13 to 2.31 (m, 2H), 1.83 (s, 3H), 1.49 to 1.78 (m, 4H), 1.32 (s, 3H). 13C NMR (75 MHz, CDCl3): 173.4, 145.1, 144.7, 127.5, 112.7, 73.5, 40.8, 28.4, 24.0, and 12.3.
Yeast Expression and in Vitro Activity of CYP76C1 on Linalool and Linalool Oxides
Yeast expression of CYP76C1 was performed and quantified as previously described (Höfer et al., 2014). For GC-MS analysis of CYP76C1 activity on linalool and linalool oxides as substrates, assays were performed in 300 μL of 20 mM sodium phosphate buffer (pH 7.4) containing 100 µM of substrates, 1 mM NADPH, and adjusted amounts of P450 enzyme. After addition of NADPH, samples were incubated at 28°C and the reaction was stopped by addition of 600 μL ethyl acetate spiked with 10 µM nonyl acetate on ice. Samples were vortexed for 10 s and centrifuged at 4000g for 2 min. Four hundred microliters of the ethyl acetate phase was subsequently recovered and dried over anhydrous Na2SO4 (Sigma-Aldrich), rinsed with 100 μL ethyl acetate, and transferred into GC-MS vials.
For UPLC-MS/MS analysis of CYP76C1 activity, assays were performed in 500 μL of 20 mM sodium phosphate buffer (pH 7.4) containing 200 µM of substrates, 1 mM NADPH, and adjusted amounts of P450 enzyme. After addition of NADPH, samples were incubated at 28°C and the reaction was stopped by addition of 250 μL of methanol on ice. Samples were vortexed for 10 s and centrifuged at 5500g for 5 min at 4°C. Four hundred microliters of the supernatant was transferred to UPLC vials. For the determination of the kinetic parameters on R- and S-linalool by UPLC-MS/MS, assays were downscaled to a final volume of 200 µL, using R- and S-linalool concentrations ranging from 10 to 500 µM, 1 mM NADPH, and ∼2 nM P450s. Formation of products was quantified after 4.5 min of incubation. Kinetic parameters were deduced from Michaelis-Menten regression curves.
GC-MS Analysis
Capillary GC was performed on a Perkin-Elmer Clarus 680 gas chromatograph coupled to a Perkin-Elmer Clarus 600T mass spectrometer on a HP-5ms or HP-35ms column (30 m, 0.25 mm, 0.25 µm; Agilent Technologies) using splitless injection, 250°C injector temperature, and a temperature program of 0.5 min at 50°C, 20°C min−1 to 320°C, and 5 min at 320°C with a flow of 1.2 mL⋅min−1 of helium gas as vector. Products were identified based on their retention time and electron ionization mass spectra (70 eV, m/z 50 to 300) compared with those of authentic standards. The HP-35ms column was used to enable proper separation of 8-oxo-linalool and 9-OH-linalool.
NMR Characterization of Products
Standard in vitro enzyme assays were scaled up to a volume of 10 mL containing 1 mM of substrate. After a first incubation for 20 min at 28°C, a second aliquot of microsomes expressing CYP76C1 was added and incubated for another 20 min. The reaction was stopped by cooling the sample on ice. Membranes were pelleted by centrifugation at 5500g for 5 min. Products in the cleared buffer were extracted by solid phase extraction on SPE cartridges (Oasis HLB extraction cartridges; Waters) as previously described (Höfer et al., 2014). The products were eluted with CDCl3 prior to NMR analysis. Major products were directly analyzed by NMR as previously described (Höfer et al., 2014) on a 500-MHz Bruker Avance spectrometer (Bruker-Biospin) equipped with a 5-mm DCH dual cryoprobe with z-gradient operating at 500.13 MHz for 1H and 125.758 MHz for 13C. 1D 1H, 1H -1H COSY, edited 1H -13C HSQC, and 1H -13C HMBC were recorded for each sample, adding 1H-1H NOESY and 1D 13C when required.
Minor products from the 8-oxo-linalool transformation by yeast-expressed CYP76C1 were separated by a preparative GC Agilent 7890A GC instrument equipped with a HP-5 capillary column (30 m × 0.53 mm ID with 1.5-µm film; Agilent Technologies) connected to a flame ionization detector (Agilent) and a preparative fraction collector with a cryostatic trap cooler (PFC; Gerstel) as previously described (Ginglinger et al., 2013). NMR analysis of the collected products was conducted on a Bruker Avance 500 spectrometer equipped with a 5-mm TCI cryoprobe (5 mm) with z-gradient operating at 500.13 MHz for 1H and 125.76 MHz for 13C as previously described (Ginglinger et al., 2013).
Isolation of Insertion Mutant and Complemented/Overexpression Lines
Arabidopsis insertion lines cyp76c1-1 (SALK_010566) and cyp76c1-2 (SALK_001949) were selected from SALK and obtained from the Nottingham Arabidopsis Stock Center. Homozygous mutant lines were selected by PCR genotyping of genomic DNA extracted from young leaves using the primers provided in Supplemental Table 1. Absence of transcripts in the insertion lines was assessed by RT-PCR amplifying the full coding sequence and qRT-PCR in flower tissues as describe above.
To generate a complemented/overexpressed line (35S:CYP76C1), the plant expression vector pCAMBIA3300u harboring CYP76C1 was introduced into the Agrobacterium GV3101 strain before transformation of homozygous cyp76c1-1 plants by floral dip. The T1 progeny were screened by germination on phosphinothricin (BASTA), and several lines were screened by qRT-PCR for CYP76C1 expression in flower tissues. Selected T1 lines showing the highest expression were brought to T3 stable progeny by germination on BASTA. CYP76C1 overexpression was analyzed on T3 lines by RT-PCR and qRT-PCR in leaves and flowers as described above.
Headspace Volatile Collection from Arabidopsis Flowers and Transformed N. benthamiana
Collection of headspace volatiles was performed as previously described (Ginglinger et al., 2013). About 50 to 60 inflorescences from each line were used for the each sample collection with at least three biological replicates. Detached inflorescences were placed in 12-mL glass tubes filled with water and placed in 1-liter glass jars equipped with an inlet and an outlet. Volatiles were pumped out from the jar with a vacuum pump at ∼100 mL min−1 and trapped on a cartridge filled with 150 mg Tenax TA (20/35; Grace Scientific) at the outlet. A similar cartridge was placed at the inlet to ensure purification of the incoming air. Volatiles were sampled for 24 h. After volatile collection, flowers were cut from the inflorescences and weighed before further analysis of soluble compounds. Tenax cartridge desorption and volatile analysis were performed as described (Ginglinger et al., 2013) using a TurboMatrix 100 thermal desorber (Perkin-Elmer) and a Perkin-Elmer Clarus 680 equipped with a Perkin-Elmer Clarus 600T quadrupole mass spectrometer.
For headspace analysis of volatiles emitted from transformed N. benthamiana leaves, plant expression constructs carrying the coding sequences of CYP76C1, TPS10, or TPS14 were transformed into Agrobacterium LBA4404, which was used to infiltrate the N. benthamiana leaves as described previously (Ginglinger et al., 2013). Four days after infiltration, three infiltrated leaves per samples corresponding to ∼1.5 g of leaf material were detached and maintained in a 12-mL glass vial filled with water placed in a 1-liter glass jar. Leaf headspace was collected on Tenax cartridges as described above for 4 h. After volatile collection, leaves were weighed before further analysis of soluble compounds. Tenax cartridges were analyzed by GC-MS as described above.
Extraction and β-Glycosidase Treatment of Soluble Compounds
After headspace volatile collection from Arabidopsis flowers and N. benthamiana leaves, plant material was extracted with methanol for analysis of the soluble compounds. After weighing, samples were ground in 5 mL methanol, sonicated for 10 min, and kept at −20°C overnight. Methanol extracts were centrifuged 10 min at 5500g, and the supernatant was recovered and concentrated under argon to 500 µL. After addition of 1 volume of ultrapure water, samples were kept one night at −80°C to ensure precipitation of the chlorophylls. After centrifugation at 5500g for 10 min, the clear supernatant was split into two 500-μL samples and transferred into new vials. One of the vials was reduced to dryness under argon. The other was kept for analysis of the raw extract. The dried samples were treated overnight with β-glycosidase from almond (Sigma-Aldrich) in a final volume of 250 μL of sodium acetate buffer (150 mM) containing 6 units⋅mL−1 of enzymes. The next day, 250 μL of methanol was added to precipitate proteins, and after 10 min centrifugation at 5500g, the supernatant was transferred into a new tube.
UPLC-MS/MS Analysis of Soluble Compounds
UPLC analyses were essentially performed as previously described (Ginglinger et al., 2013) with minor changes using an Acquity UPLC system (Waters) coupled to a Quattro Premier XE mass spectrometer (Waters) equipped with an electrospray ionization source and an Acquity UPLC BEH C18 (100 × 2.1 mm, 1.7 µm; Waters) column and precolumn. The mobile phase consisted of (A) water and (B) methanol, both containing 0.1% formic acid. The run began with 2 min of 85% A. Then a linear gradient was applied to reach 100% B at 27 min, followed by an isocratic run using 100% B for 10 min. Return to initial conditions was achieved in 1 min followed by a conditioning of the column with 85% A for 2 min, with a total run time of 40 min. The column was operated at 35°C with a flow rate of 0.30 mL/min, injecting 4-μL samples. Nitrogen was used as the drying and nebulizing gas. The nebulizer gas flow was set to ∼50 L/h and the desolvation gas flow to 900 L/h. The interface temperature was set at 400°C and the source temperature at 135°C. The capillary voltage was set to 3.4 kV and the cone voltage to 15 V; the ionization was in positive mode. Low mass and high mass resolution were 15 for both mass analyzers, ion energies 1 and 2 were 0.5 V, entrance and exit potential were 50 V, and detector (multiplier) gain was 650 V.
For qualitative analysis and mass spectra determination, samples were analyzed with full scan in positive mode, with an energy cone of +15V on a range of 50 to 500 m/z and with a 0.2-s scan time. For quantitative analysis, multiple reaction monitoring mode (MRM)-specific MS/MS transitions, cone energy, and collision energy were determined for each compound using authentic standards. Analyses were performed using six MRM channels, each specific for a compound, with the specific tunes listed in Supplemental Table 2. For each channel, dwell time was 0.2 s and span was set up at 0.1 m/z. Data acquisition and analyses were performed with MassLynx and QuanLynx software version 4.1 (Waters).
Insect Behavior
Aphids (Myzus persicae) preference for either flowers from the wild-type or cyp76c1 insertion lines was tested in dual-choice assays. Detached inflorescences from Col-0 and cyp76c1 were placed on both sides of a Petri dish in a layer of agarose (1%). Ten individual aphids were then placed in the dish. The number of aphids on each inflorescence was scored for 48 h.
Thrips (Frankliniella occidentalis) behavior toward wild-type and mutant flowers was tested using dual-choice assays. Ten female adults for each assay were starved overnight and released the next day in a Petri dish containing (on both sides) one detached inflorescence from Col-0 and cyp76c1, each with its stem inserted in a layer of 1% agarose. The number of thrips on each inflorescence was scored every hour for 6 h. Evaluation is based on five biological replicates for each combination of lines and three independent assays.
Thrips preference for the volatiles emitted from wild-type and mutant flowers was studied in a y-shaped olfactometer as described by Junker et al. (2010b). All flower stems from each plant, bundled with Teflon tape, were wrapped in an oven bag. The bag was sealed with masking tape on the Teflon tape to avoid damaging the plant tissue. An inlet/outlet system was connected to the top of the oven bags. Inlet air was purified through charcoal and pushed into the bags with a pump at 100 mL min−1, thus pushing the volatiles emitted from the flowers through the outlet connected to one arm of the olfactometer arena. The two arms of the olfactometer arena were each supplied with the volatiles from the flowers of one of the two different lines tested per assay. Each arm was illuminated similarly to maintain uniform conditions for the thrips. About five thrips were released in the arena and their first choice for one of the arms was scored. After each trial, the volatile supply in the two arms was switched to avoid effects from side preferences. For each plant combination, the behavior of 30 to 40 individual thrips was tested in four independent replicates, each using different individual plants.
The thrips’ preference for pure compounds was tested in the same y-shaped olfactometer. The arm of the olfactometer defined as the substance arm was supplied with an air stream from a desiccator in which 100 µg of pure substance (racemic mix of R/S-linalool, mixture of lilac aldehyde diastereoisomers, mixture of lilac alcohol diastereoisomers, and equimolar mix of stereoisomeric lilac aldehydes and alcohols) dissolved in methanol was placed on a filter. The arm of the olfactometer defined as neutral was supplied with an air stream from a control desiccator in which the same amount of methanol was placed on a filter. About five thrips were released in the chamber, and their first choice for the neutral or substance arm was scored. After each trial, the volatiles supplying the arms were switched to avoid artifacts. For each compound, the behavior of up to 40 individual thrips was tested with six independent replicates. For each independent test, the pure substance was replaced with a new one.
The behavior of adult hoverflies (Episyrphus balteus) was tested with pure compounds in a star-shaped olfactometer described by Junker et al. (2010a). The fields of the olfactometer defined as substance fields were supplied as described above with 100 µg of pure compounds (racemic mix of R/S-linalool, racemic mix of R/S-8-OH-linalool, racemic mix of R/S-8-COOH-linalool, mixture of lilac aldehyde diastereoisomers, mixture of lilac alcohol diastereoisomer, and equimolar mix of all these compounds) in methanol placed on a filter. Hoverflies were placed one by one for 4 min in the olfactometer, and time spent in the neutral or substance fields was measured. Substances in the desiccator were replaced every 20 min.
The preference of the florivorous insects Plutella xylostella (L3 larvae), Phaedon cochleariae (adults), and Spodoptera littoralis (larvae) for flowers from the wild type and cyp76c1-1 mutants was tested using a dual-choice feeding test. Each set of five open Col-0 and five cyp76c1 flowers was offered on opposite sides of a Petri dish; flowers were placed into moist foam to keep them fresh for the duration of the experiment. One individual of either of three insect species mentioned above was placed in the Petri dish. The number of flowers (or flower parts) consumed by the insect was scored after 3 h. Each individual was used only once in order to prevent pseudo-replication.
The feeding preference of P. cochleariae (adults) on cabbage leaves supplied with linalool oxides and control leaves was tested using a dual-choice test. Small pieces of leaves were treated with 1 μL per 1 g FW methanol containing 10, 100, or 1000 ng µL−1 of pure compounds as described above (linalool, lilac alcohols, 8-COOH-linalool, or 8-OH linalool). Control leaves were treated in the same way with pure methanol. Leaves were offered in Petri dishes and one individual was allowed to choose between treatment and control. Within 10 min, the first choice (consumption) was scored.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TPS10 (At2g24210), TPS14 (At1g61680), CYP76C1 (At2g45560), and CYP71B38 (At3g53280).
Supplemental Data
Supplemental Figure 1. Extended CYP76C1 expression and coexpression analysis.
Supplemental Figure 2. CYP76C1 and TPS10 subcellular localizations.
Supplemental Figure 3. Extended GC-MS analysis of the products resulting from yeast-expressed CYP76C1 activity on linalool and linalool oxides.
Supplemental Figure 4. Mass spectra of the products formed by yeast-expressed CYP76C1 (in Figure 2 and Supplemental Figure 3).
Supplemental Figure 5. Determination of the catalytic parameters of the R- and S-linalool conversion by CYP76C1 into 8- and 9-OH-linalool.
Supplemental Figure 6. NMR characterization of linalool oxidation products formed by CYP76C1.
Supplemental Figure 7. Time-course production of lilac aldehydes from yeast-expressed CYP76C1 on 8-oxo-linalool.
Supplemental Figure 8. Targeted UPLC-MS/MS analysis of the products resulting from yeast-expressed CYP76C1 activity on linalool and linalool oxides.
Supplemental Figure 9. Genotyping of the Arabidopsis mutants.
Supplemental Figure 10. Targeted UPLC-MS/MS profiling of the linalool-derived metabolites present in the flowers of wild-type Arabidopsis and CYP76C1 mutants.
Supplemental Figure 11. Myzus persicae dual-choice test between flowers from the wild type and cyp76c1 insertion mutants.
Supplemental Figure 12. F. occidentalis dual-choice test between flowers from the wild type and cyp76c1-2 mutant.
Supplemental Figure 13. Dual-choice test of feeding with adults of Phaedon cochleariae on cabbage leaves treated with pure linalool and lilac alcohols.
Supplemental Table 1. PCR primer list.
Supplemental Table 2. Optimized MS/MS conditions for the UPLC analysis in multiple reaction monitoring of the selected linalool and linalool derivative compounds.
Supplementary Material
Acknowledgments
The data in this article were generated with the support of the European Community's Framework VII Program FP7/2007-2013 to the SMARTCELL project KBBE-2007-3-1-01. We thank Dana E. Seidel and Jessica Bossems for help in performing the behavioral bioassays, François Bernier and Léon Otten for critical reading of the article, and Raphael Lugan for training on UPLC-MS/MS. B.B. and D.W.-R. acknowledge the support of the European Fund for Regional Development in the Program INTERREG IVA Broad Region EU invests in your future. COST (European Cooperation in Science and Technology) of the Plant Engine Action (FA1006) supported two Short Term Scientific Missions (FA1006-10129 and FA1006-12615) relative to this project. Sascha Eilmus (Bayer) provided some of the insects used for the bioassays. R.R.J. acknowledges support of the Deutsche Forschungsgemeinschaft (DFG JU 2856/1-1). Imaging data were collected at the Center for Advanced Bioimaging Denmark, University of Copenhagen.
AUTHOR CONTRIBUTIONS
B.B. designed the experiments, generated constructs and mutants, did qRT-PCR expression analyses, analyzed enzyme expression in yeast and enzymatic assays, performed quantification of volatile emission, metabolites profiling, tested plant insect interactions, and wrote the article. R.R.J. designed experiments on insect behavior, discussed the results, and contributed to the writing of the article. K.L. supervised some the behavioral experiments. R.H. generated CYP76C1 constructs in yeast and insertion mutants in Arabidopsis. J.-F.G. generated constructs of TPS11 and TPS14 to transform N. benthamiana and gave support for sampling for qRT-PCR analyses. A.L. provided technical support. J.-E.B. supervised the subcellular colocalization and brought support for microscopy. R.C., D.E.S., and D.S.C.W. performed some of the behavioral experiments. R.C. and D.S.C.W. assisted in tests of plant-insect interactions. L.M., C.H., and M.M. synthesized the reference linalool oxides. L.A., C.P., F.B., B.V., and B.S. contributed to NMR identification of the products. D.W.-R. designed the project, supervised the discussions, and wrote the article.
Glossary
- UPLC-MS/MS
ultraperformance liquid chromatography-tandem mass spectrometry
- GC-MS
gas chromatography-mass spectrometry
- FW
fresh weight
- MRM
multiple reaction monitoring mode
Footnotes
Articles can be viewed online without a subscription.
References
- Aharoni A., Giri A.P., Deuerlein S., Griepink F., de Kogel W.J., Verstappen F.W.A., Verhoeven H.A., Jongsma M.A., Schwab W., Bouwmeester H.J. (2003). Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15: 2866–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassard J.-E., et al. (2012). Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 24: 4465–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg-Karlson , A.-K., Unelius, C.R., Valterova, I., and Nilsson, L.A. (1996). Floral fragrance chemistry in the early flowering shrub Daphne mezereum. Phytochemistry 41: 1477–1483. [Google Scholar]
- Brown S., Clastre M., Courdavault V., O’Connor S.E. (2015). De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl. Acad. Sci. USA 112: 3205–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt D., Mosandl A. (2003). Biogenetic studies in Syringa vulgaris L.: bioconversion of (18)O(2H)-labeled precursors into lilac aldehydes and lilac alcohols. J. Agric. Food Chem. 51: 7391–7395. [DOI] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99: 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi L., Lim E.-K., Bowles D.J. (2008). Discovery of new biocatalysts for the glycosylation of terpenoid scaffolds. Chemistry 14: 6656–6662. [DOI] [PubMed] [Google Scholar]
- Chen F., Tholl D., D’Auria J.C., Farooq A., Pichersky E., Gershenzon J. (2003). Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Miettinen K., Goedbloed M., Verstappen F.W.A., Voster A., Jongsma M.A., Memelink J., van der Krol S., Bouwmeester H.J. (2013). Characterization of two geraniol synthases from Valeriana officinalis and Lippia dulcis: similar activity but difference in subcellular localization. Metab. Eng. 20: 198–211. [DOI] [PubMed] [Google Scholar]
- Dötterl S., Burkhardt D., Jürgens A., Mosandl A. (2007). Stereoisomeric pattern of lilac aldehyde in Silene latifolia, a plant involved in a nursery pollination system. Phytochemistry 68: 499–504. [DOI] [PubMed] [Google Scholar]
- Dötterl S., Burkhardt D., Weissbecker B., Jürgens A., Schütz S., Mosandl A. (2006a). Linalool and lilac aldehyde/alcohol in flower scents. Electrophysiological detection of lilac aldehyde stereoisomers by a moth. J. Chromatogr. A 1113: 231–238. [DOI] [PubMed] [Google Scholar]
- Dötterl S., Jürgens A., Seifert K., Laube T., Weissbecker B., Schütz S. (2006b). Nursery pollination by a moth in Silene latifolia: the role of odours in eliciting antennal and behavioural responses. New Phytol. 169: 707–718. [DOI] [PubMed] [Google Scholar]
- Dudareva N., Cseke L., Blanc V.M., Pichersky E. (1996). Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8: 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flath R.A., Forrey R.R. (1977). Volatile components of papaya (Carica papaya l solo variety). J. Agric. Food Chem. 25: 103–109. [Google Scholar]
- Flath R.A., Light D.M., Jang E.B., Mon T.R., John J.O. (1990). Headspace examination of volatile emissions from ripening papaya (Carica papaya l solo variety). J. Agric. Food Chem. 38: 1060–1063. [Google Scholar]
- Geu-Flores F., Nour-Eldin H.H., Nielsen M.T., Halkier B.A. (2007). USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 35: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginglinger J.-F., et al. (2013). Gene coexpression analysis reveals complex metabolism of the monoterpene alcohol linalool in Arabidopsis flowers. Plant Cell 25: 4640–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodger J.Q.D., Woodrow I.E. (2011). α,β-Unsaturated monoterpene acid glucose esters: structural diversity, bioactivities and functional roles. Phytochemistry 72: 2259–2266. [DOI] [PubMed] [Google Scholar]
- Goodger, J.Q.D., and Woodrow, I.E. (2013). Oleuropeic and menthiafolic acid glucose esters from plants: Shared structural relationships and biological activities. Stud. Nat. Prod. Chem. 40: 427–452. [Google Scholar]
- Hanausek M., Ganesh P., Walaszek Z., Arntzen C.J., Slaga T.J., Gutterman J.U. (2001). Avicins, a family of triterpenoid saponins from Acacia victoriae (Bentham), suppress H-ras mutations and aneuploidy in a murine skin carcinogenesis model. Proc. Natl. Acad. Sci. USA 98: 11551–11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haridas V., Arntzen C.J., Gutterman J.U. (2001). Avicins, a family of triterpenoid saponins from Acacia victoriae (Bentham), inhibit activation of nuclear factor-kappaB by inhibiting both its nuclear localization and ability to bind DNA. Proc. Natl. Acad. Sci. USA 98: 11557–11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori R., Muraki S., Yoshida T. (1978). Chemical composition of absolute from gardenia flower. Agric. Biol. Chem. 42: 1351–1356. [Google Scholar]
- Heidlas J., Lehr M., Idstein H., Schreier P. (1984). Free and bound terpene compounds in papaya (Carica papaya, l) fruit pulp. J. Agric. Food Chem. 32: 1020–1021. [Google Scholar]
- Höfer R., Boachon B., Renault H., Gavira C., Miesch L., Iglesias J., Ginglinger J.-F., Allouche L., Miesch M., Grec S., Larbat R., Werck-Reichhart D. (2014). Dual function of the cytochrome P450 CYP76 family from Arabidopsis thaliana in the metabolism of monoterpenols and phenylurea herbicides. Plant Physiol. 166: 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfer R., Dong L., André F., Ginglinger J.-F., Lugan R., Gavira C., Grec S., Lang G., Memelink J., Van der Krol S., Bouwmeester H., Werck-Reichhart D. (2013). Geraniol hydroxylase and hydroxygeraniol oxidase activities of the CYP76 family of cytochrome P450 enzymes and potential for engineering the early steps of the (seco)iridoid pathway. Metab. Eng. 20: 221–232. [DOI] [PubMed] [Google Scholar]
- Hoffmann M.H., Bremer M., Schneider K., Burger F., Stolle E., Moritz G. (2003). Flower visitors in a natural population of Arabidopsis thaliana. Plant Biol. 5: 491–494. [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.E. (1971). The population genetics of Arabidopsis thaliana I. The breeding system. Heredity 27: 39–50. [Google Scholar]
- Junker R.R., Blüthgen N. (2010). Floral scents repel facultative flower visitors, but attract obligate ones. Ann. Bot. (Lond.) 105: 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker R.R., Gershenzon J., Unsicker S.B. (2011). Floral odor bouquet loses its ant repellent properties after inhibition of terpene biosynthesis. J. Chem. Ecol. 37: 1323–1331. [DOI] [PubMed] [Google Scholar]
- Junker R.R., Heidinger I.M.M., Blüthgen N. (2010a). Floral scent terpenoids deter the facultative florivore Metrioptera bicolor (Ensifera, Tettigoniidae, Decticinae). J. Orthop. Res. 19: 69–74. [Google Scholar]
- Junker R.R., Höcherl N., Blüthgen N. (2010b). Responses to olfactory signals reflect network structure of flower-visitor interactions. J. Anim. Ecol. 79: 818–823. [DOI] [PubMed] [Google Scholar]
- Kessler D., Gase K., Baldwin I.T. (2008). Field experiments with transformed plants reveal the sense of floral scents. Science 321: 1200–1202. [DOI] [PubMed] [Google Scholar]
- Knudsen J.T., Eriksson R., Gershenzon J., Stahl B. (2006). Diversity and distribution of floral scent. Bot. Rev. 72: 1–120. [Google Scholar]
- Koschier E.H., De Kogel W.J., Visser J.H. (2000). Assessing the attractiveness of volatile plant compounds to western flower thrips Frankliniella occidentalis. J. Chem. Ecol. 26: 2643–2655. [Google Scholar]
- Kreck M., Mosandl A. (2003). Synthesis, structure elucidation, and olfactometric analysis of lilac aldehyde and lilac alcohol stereoisomers. J. Agric. Food Chem. 51: 2722–2726. [DOI] [PubMed] [Google Scholar]
- Kreck M., Püschel S., Wüst M., Mosandl A. (2003). Biogenetic studies in Syringa vulgaris L.: synthesis and bioconversion of deuterium-labeled precursors into lilac aldehydes and lilac alcohols. J. Agric. Food Chem. 51: 463–469. [DOI] [PubMed] [Google Scholar]
- Loughrin J.H., Potter D.A., Hamilton-Kemp T.R. (1995). Volatile compounds induced by herbivory act as aggregation kairomones for the Japanese beetle (Popillia japonica Newman). J. Chem. Ecol. 21: 1457–1467. [DOI] [PubMed] [Google Scholar]
- Luan F., Mosandl A., Gubesch M., Wüst M. (2006). Enantioselective analysis of monoterpenes in different grape varieties during berry ripening using stir bar sorptive extraction- and solid phase extraction-enantioselective-multidimensional gas chromatography-mass spectrometry. J. Chromatogr. A 1112: 369–374. [DOI] [PubMed] [Google Scholar]
- Lücker J., Bouwmeester H.J., Schwab W., Blaas J., van der Plas L.H.W., Verhoeven H.A. (2001). Expression of Clarkia S-linalool synthase in transgenic petunia plants results in the accumulation of S-linalyl-beta-D-glucopyranoside. Plant J. 27: 315–324. [DOI] [PubMed] [Google Scholar]
- Matich A.J., Bunn B.J., Comeskey D.J., Hunt M.B., Rowan D.D. (2007). Chirality and biosynthesis of lilac compounds in Actinidia arguta flowers. Phytochemistry 68: 1746–1751. [DOI] [PubMed] [Google Scholar]
- Matich A.J., Comeskey D.J., Bunn B.J., Hunt M.B., Rowan D.D. (2011). Biosynthesis and enantioselectivity in the production of the lilac compounds in Actinidia arguta flowers. Phytochemistry 72: 579–586. [DOI] [PubMed] [Google Scholar]
- Matich A.J., Young H., Allen J.M., Wang M.Y., Fielder S., McNeilage M.A., MacRae E.A. (2003). Actinidia arguta: volatile compounds in fruit and flowers. Phytochemistry 63: 285–301. [DOI] [PubMed] [Google Scholar]
- McCallum E.J., Cunningham J.P., Lücker J., Zalucki M.P., De Voss J.J., Botella J.R. (2011). Increased plant volatile production affects oviposition, but not larval development, in the moth Helicoverpa armigera. J. Exp. Biol. 214: 3672–3677. [DOI] [PubMed] [Google Scholar]
- Misra L.N., Chandra A., Thakur R.S. (1991). Fragrant components of oil from Artemisia pallens. Phytochemistry 30: 549–552. [Google Scholar]
- Morita K., Wakabayashi M., Kubota K., Kobayashi A., Herath N.L. (1994). Glycoside precursor of tea aroma. 2. Aglycone constituents in fresh tea leaves cultivated for green and black tea. Biosci. Biotechnol. Biochem. 58: 687–690. [Google Scholar]
- Nagatoshi M., Terasaka K., Nagatsu A., Mizukami H. (2011). Iridoid-specific glucosyltransferase from Gardenia jasminoides. J. Biol. Chem. 286: 32866–32874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noté O.P., Jihu D., Antheaume C., Zeniou M., Pegnyemb D.E., Guillaume D., Chneiwess H., Kilhoffer M.C., Lobstein A. (2015). Triterpenoid saponins from Albizia lebbeck (L.) Benth and their inhibitory effect on the survival of high grade human brain tumor cells. Carbohydr. Res. 404: 26–33. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Hansen B.G., Nørholm M.H., Jensen J.K., Halkier B.A. (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T., Okuyama Y., Goto R., Tokoro M., Kato M. (2015). Parallel chemical switches underlying pollinator isolation in Asian Mitella. J. Evol. Biol. 28: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon C.J., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496: 528–532. [DOI] [PubMed] [Google Scholar]
- Parachnowitsch A.L., Burdon R.C.F., Raguso R.A., Kessler A. (2013). Natural selection on floral volatile production in Penstemon digitalis: highlighting the role of linalool. Plant Signal. Behav. 8: e22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patt J.M., Rhoades D.F., Corkill J.A. (1988). Analysis of the floral fragrance of Platanthera stricta. Phytochemistry 27: 91–95. [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E., Raguso R.A., Lewinsohn E., Croteau R. (1994). Floral scent production in Clarkia (Onagraceae). 1. Localization and developmental modulation of monoterpene emission and linalool synthase activity. Plant Physiol. 106: 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raguso R.A., Pichersky E. (1999). New perspectives in pollination biology: Floral fragrances. A day in the life of a linalool molecule: Chemical communication in a plant-pollinator system. Part 1: Linalool biosynthesis in flowering plants. Plant Species Biol. 14: 95–120. [Google Scholar]
- Reisenman C.E., Riffell J.A., Bernays E.A., Hildebrand J.G. (2010). Antagonistic effects of floral scent in an insect-plant interaction. Proc. Biol. Sci. 277: 2371–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohloff J., Bones A.M. (2005). Volatile profiling of Arabidopsis thaliana - putative olfactory compounds in plant communication. Phytochemistry 66: 1941–1955. [DOI] [PubMed] [Google Scholar]
- Sagt C.M.J., ten Haaft P.J., Minneboo I.M., Hartog M.P., Damveld R.A., van der Laan J.M., Akeroyd M., Wenzel T.J., Luesken F.A., Veenhuis M., van der Klei I., de Winde J.H. (2009). Peroxicretion: a novel secretion pathway in the eukaryotic cell. BMC Biotechnol. 9: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl F.P. (2010). The evolution of floral scent and insect chemical communication. Ecol. Lett. 13: 643–656. [DOI] [PubMed] [Google Scholar]
- Schreier, P., and Winterhalter, P. (1986). Precursors of papaya (Carica papaya, l) fruit volatiles. ACS Symp. Ser. 317: 85–98. [Google Scholar]
- Sharma M.L., Chand T. (1996). Allylic oxidation in terpenoids: Synthesis of(+/−)-E-linalool-1-oic acid, (+/−)-E-9-hydroxylinalool and (+/−)-7-hydroxyterpineol. Proc. Indian Acad. Sci. Chem. Sci. 108: 21–26. [Google Scholar]
- Siska P., Fodran P., Szolcsanyi P. (2014). Synthesis and olfactory properties of unnatural derivatives of lilac aldehydes. Tetrahedron 70: 6420–6427. [Google Scholar]
- Snape J.W., Lawrence M.J. (1971). The breeding system of Arabidopsis thaliana. Heredity 27: 299–302. [Google Scholar]
- Tholl D., Chen F., Petri J., Gershenzon J., Pichersky E. (2005). Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J. 42: 757–771. [DOI] [PubMed] [Google Scholar]
- Tholl D., Lee S. (2011). Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143, doi/10.1199/tab.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 43: 153–163. [DOI] [PubMed] [Google Scholar]
- Turlings T.C.J., Loughrin J.H., McCall P.J., Röse U.S.R., Lewis W.J., Tumlinson J.H. (1995). How caterpillar-damaged plants protect themselves by attracting parasitic wasps. Proc. Natl. Acad. Sci. USA 92: 4169–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L., Deakin E.A., Hoge J.H., Memelink J. (2000). The ternary transformation system: constitutive virG on a compatible plasmid dramatically increases Agrobacterium-mediated plant transformation. Plant Mol. Biol. 43: 495–502. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wakayama S., Namba S. (1974). Lilac aldehydes. Bull. Chem. Soc. Jpn. 47: 1293–1294. [Google Scholar]
- Wakayama S., Namba S., Hosoi K., Ohno M. (1973). The synthesis and the absolute configurations of lilac alcohols. New naturally occurring odorous ingredients of lilac flower. Bull. Chem. Soc. Jpn. 46: 3183–3187. [Google Scholar]
- Wakayama S., Namba S., Ohno M. (1970). Lilac alcohol-a and -b. New naturally occurring odorous ingredients. Bull. Chem. Soc. Jpn. 43: 3319. [Google Scholar]
- Wakayama S., Namba S., Ohno M. (1971). The odorous constituents of lilac flower oil. Nippon Kagaku Zassi 92: 256–259. [Google Scholar]
- Wang D., Yoshimura T., Kubota K., Kobayashi A. (2000). Analysis of glycosidically bound aroma precursors in tea leaves. 1. Qualitative and quantitative analyses of glycosides with aglycons as aroma compounds. J. Agric. Food Chem. 48: 5411–5418. [DOI] [PubMed] [Google Scholar]
- Wang Q., Hillwig M.L., Okada K., Yamazaki K., Wu Y., Swaminathan S., Yamane H., Peters R.J. (2012). Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J. Biol. Chem. 287: 6159–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins A.L., Lu Y., Tan S.T. (1993). Extractives from New Zealand honeys. 4. Linalool derivatives and other components from nodding thistle (Carduus nutans) honey. J. Agric. Food Chem. 41: 873–878. [Google Scholar]
- Williams P.J., Strauss C.R., Wilson B. (1980). Hydroxylated linalool derivatives as precursors of volatile monoterpenes of muscat grapes. J. Agric. Food Chem. 28: 766–771. [Google Scholar]
- Winterhalter P., Katzenberger D., Schreier P. (1986). 6,7-epoxy-linalool and related oxygenated terpenoids from Carica papaya fruit. Phytochemistry 25: 1347–1350. [Google Scholar]
- Wu Y., Wang Q., Hillwig M.L., Peters R.J. (2013). Picking sides: distinct roles for CYP76M6 and CYP76M8 in rice oryzalexin biosynthesis. Biochem. J. 454: 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Wang Q., Erb M., Turlings T.C.J., Ge L., Hu L., Li J., Han X., Zhang T., Lu J., Zhang G., Lou Y. (2012). Specific herbivore-induced volatiles defend plants and determine insect community composition in the field. Ecol. Lett. 15: 1130–1139. [DOI] [PubMed] [Google Scholar]
- Yang T., Stoopen G., Thoen M., Wiegers G., Jongsma M.A. (2013). Chrysanthemum expressing a linalool synthase gene ‘smells good’, but ‘tastes bad’ to western flower thrips. Plant Biotechnol. J. 11: 875–882. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.