Abstract
The interplay of transcription and epigenetic marks is essential for oligodendrocyte cell (OPC) proliferation and differentiation during development. Here, we review the recent advances in this field and highlight mechanisms of transcriptional repression and activation involved in OPC proliferation, differentiation and plasticity. We also describe how dysregulation of these epigenetic events may affect demyelinating disorders, and consider potential ways to manipulate NG2 cell behavior through modulation of the epigenome.
1. Introduction
NG2 glial cells are traditionally defined as oligodendrocyte progenitors (OPC) receiving synaptic inputs and with the ability to respond to a variety of extracellular stimuli by proliferating, migrating, differentiating or modulating brain homeostasis and plasticity (Barres et al., 1994a; Demerens et al., 1996; Fannon et al., 2015; Hernandez and Casaccia, 2015; Nishiyama et al., 1999; Pringle et al., 1992; Raff et al., 1983; Tsai et al., 2009; Wake et al., 2011). These biological responses result from the integration of environmental signals with the intrinsic properties of the cells. The latter ones might evolve with age as progenitors in the neonatal period show different responsiveness than their adult counterparts, in terms of their biological properties, including fate-choice decisions, proliferation, migration or differentiation rates (Chari et al., 2003; Windrem et al., 2004; Wolswijk and Noble, 1989; Young et al., 2013). It is likely that these changes result from modifications of the epigenetic landscape over time.
Among well-defined epigenetic mechanisms this review will discuss: DNA methylation, chromatin modifications and remodeling and non-coding RNA.
DNA methylation is the only known epigenetic modification that directly modifies DNA components, by adding a methyl group at the C-5 position of cytosine residues at CpG dinucleotides (Eden and Cedar, 1994). This reaction is catalyzed by: the DNA maintenance methyltransferase DNMT1, which is responsible for the faithful transmission of DNA methylation from mother to daughter cells during replication and by the de novo methyltransferases DNMT3A and DNMT3B for the establishment of new methylation marks (Goll and Bestor, 2005; Lei et al., 1996; Okano et al., 1998). These enzymes are expressed in the CNS, where the DNA methylation level is higher than in any other tissues (Ono et al., 1993; Tawa et al., 1990). They have been shown to regulate survival and differentiation of neurons and astrocytes, while their role in the NG2 cells has not been thoroughly investigated (Fan et al., 2001; Noguchi et al., 2015; Takizawa et al., 2001; Wu et al., 2012b). DNA methylation at promoter regions is mainly associated with transcriptional repression, either by directly preventing the access of transcription factors to their binding sequence or by recruiting cofactors that modulate the chromatin environment (Schübeler, 2015; Smith and Meissner, 2013). Another modification of the DNA is the oxidation of 5-methylcytosine to 5-hydroxymethylcytosine (5-hmC) by the recently identified ten-eleven translocation (TET) enzymes, which are dynamically expressed in the oligodendroglial lineage (Branco et al., 2012; Tahiliani et al., 2009; Zhao et al., 2014). The low levels of 5-hmC initially found in the genome of embryonic stem cells led to the hypothesis that 5-hmC was only a short-lived intermediate associated with the removal of methyl groups from cytosine residues (Tahiliani et al., 2009). However, the abundance of 5-hmC in euchromatic regions, especially in the brain, suggested that it might also be an important epigenetic regulator of gene expression (Ficz et al., 2011; Münzel et al., 2010; Szulwach et al., 2011; Szwagierczak et al., 2010). Hydroxymethylation is characteristically enriched at gene bodies and transcription starting sites, where it has been associated with transcriptional activation and alternative splicing (Feng et al., 2015; Szulwach et al., 2011). In both human and mouse embryonic stem cells, hydroxymethylation enrichment at binding sites of pluripotency-associated transcription factors has been linked to regulation of cell lineage choice and differentiation (Ficz et al., 2011).
Histones H2A, H2B, H3 or H4 are protein components of the nucleosome, which defines the basic unit of chromatin. They can be subject to post-translational modifications including methylation, acetylation, sumoylation, phosphorylation, citrullination, ubiquitination, proline isomerization and ADP-ribosylation (Kouzarides, 2007). Addition or removal of these groups at specific amino acid residues on the tails of the histones, can either activate or repress gene expression (Jenuwein and Allis, 2001; Strahl and Allis, 2000). For example, acetylation of lysine 27 in histone H3 (H3K27ac) at active enhancers has been associated with transcriptional activation, whereas acetyl group removal by histone deacetylases (HDACs) is mainly linked to gene repression (Creyghton et al., 2010; Rada-Iglesias et al., 2011). Histone methylation marks, catalyzed by lysine-specific histone methyltransferase and arginine-specific histone methyltransferases, are also divided in two categories: methylation of lysine 4 in histone H3 (H3K4me1), usually enriched at enhancers, and dimethylation of arginine 3 in histone H4 (H4R3me2) are active histone marks, while trimethylation of lysines 9 or 27 in H3 (H3K9me3 and H3K27me3) are repressive marks, usually enriched in silenced genes (Di Lorenzo and Bedford, 2011; Mikkelsen et al., 2007; Shilatifard, 2006).
In addition to histone modifications, chromatin structure can also been rearranged by ATP-dependent chromatin remodelers that are characterized by nucleosomal sliding activity (Sohn et al., 2007). The SWI/SNF complex has first been identified in yeast, and several members of this family, including BRG1 (or SMARCA4) and BRM (or SMARCA2), have later been described in mammalian cells and also in NG2 cells (Bischof et al., 2015; Hargreaves and Crabtree, 2011; Ho and Crabtree, 2010; Yu et al., 2013). Chromatin remodeling has been associated to both activation and repression of gene expression (Clapier and Cairns, 2009).
Recently described non-coding RNA are another epigenetic mechanisms regulating gene expression, notably during brain development (Derrien et al., 2012; Qureshi and Mehler, 2012) and in neurological disorders (Esteller, 2011). In this family, the 20-25 nucleotide-long micro-RNAs (miRNA), which are processed by RNA polymerase II and DICER, can form a complex with Argonaute to induce mRNA silencing or degradation (Huntzinger and Izaurralde, 2011; Lee et al., 2004). By their post-transcriptional role, miRNA can affect several mRNA transcripts that are encoded at distant genome loci (Lim et al., 2005).
These epigenetic marks modify the organization or the condensation of the chromatin, thus altering its access to transcription factors and their ability to modulate gene expression (Bernstein et al., 2007; Li et al., 2007; Yao and Jin, 2014).
2. Regulation of NG2 cells proliferation
OPC proliferate in response to several mitogens, growth factors and cytokines (Barres et al., 1994b, 1996; Canoll et al., 1996; Diemel et al., 2003; Noble et al., 1988; Ohya et al., 2007), and also to electrical activity (Barres and Raff, 1993; Demerens et al., 1996; Gallo et al., 1996; Li et al., 2010). NG2 tend to proliferate more in white matter regions (i.e. corpus callosum) than in gray matter areas (Psachoulia et al., 2009; Young et al., 2013). OPC cell cycle time increases significantly with age: from a 24-48 hours duration in neonatal brains to 18 days in adult (2 month-old) and up to 70 days in aged (8 month old) murine brains (Clarke et al., 2012; Hughes et al., 2013; Psachoulia et al., 2009; Simon et al., 2011; Young et al., 2013). In addition, OPC derived from young or old brains display different degrees of responsiveness to treatment with equivalent mitogen concentrations (Lin et al., 2009; Ruffini et al., 2004; Shi et al., 1998), thereby supporting the existence of a cell intrinsic mechanism regulating proliferation which is affected by age.
2.1. Cell cycle regulation
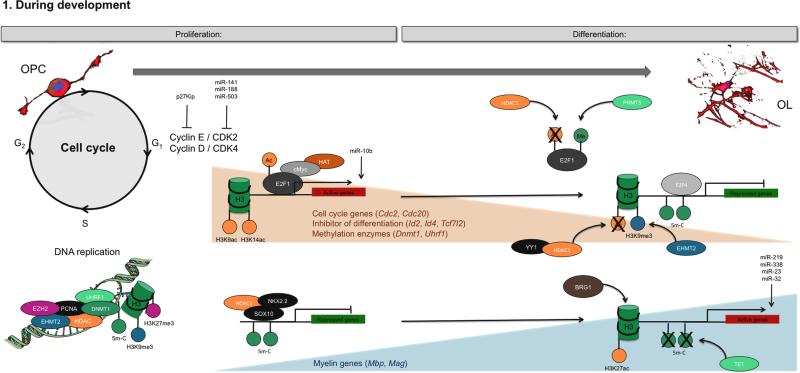
The decision of NG2 cells to enter the cell cycle or to arrest and start differentiating, is regulated at the G1/S transition, which is characterized by the phosphorylation of the retinoblastoma protein, and consequent release of the E2F1-mediated expression of S-phase genes (Huang et al., 2002; Magri et al., 2014a; Swiss and Casaccia, 2010; Swiss et al., 2011). The E2F family is divided into two main categories: the activating E2F1, E2F2, E2F3A and E2F3B, which bind exclusively to Rb, and the repressing E2F4 and E2F5, which can also interact with other members of the retinoblastoma family (e.g. p107 and p130) (Beijersbergen et al., 1994; Lees et al., 1993; Magri et al., 2014a; Swiss and Casaccia, 2010). The phosphorylation of the pocket proteins Rb, p107 and p130 is mediated by cyclin-dependent kinase (CDKs), which in turn can be activated by cyclins or inhibited by CDK inhibitors (Casaccia-Bonnefil et al., 1997, 1999; Hannon et al., 1993; Xiao et al., 1996). The cyclin E/CDK2 and cyclin D/CDK4 complexes have been identified as those responsible for cell cycle progression of NG2 cells (Belachew et al., 2002; Huang et al., 2002; Jablonska et al., 2007; Nobs et al., 2013). The detection of overall lower levels of expression of the cell cycle activating components in adult OPC compared to their neonatal counterparts, could in part explain the lower proliferative rate of adult OPC (Belachew et al., 2002; Young et al., 2013). Conversely, decreased levels of cell cycle inhibitors, such as p27Kip is associated with increased proliferation and expansion of the progenitor pool during developmental myelination and myelin repair (Casaccia-Bonnefil et al., 1997, 1999; Crockett et al., 2005; Durand et al., 1998; Tikoo et al., 1998).
E2F family proteins can be regulated by several mechanisms including lysine acetylation, arginine methylation and nuclear export. E2F1 acetylation has been described in non-oligodendroglial cell types, and associated with increased half-life, increased DNA binding and enhanced expression of target genes (Martínez-Balbás et al., 2000; Marzio et al., 2000). Although E2F1 acetylation has not been yet reported in NG2 cells it is intriguing to take into account the possibility that a histone deaceylase (i.e. HDAC1) with the ability to remove the acetyl groups from E2F1 (Marzio et al., 2000), has already been implicated as a factor that is necessary for oligodendrocyte (OL) differentiation (Marin-Husstege et al., 2002; Shen et al., 2008a). The stability of the E2F1 protein, in addition, is modulated by methylation on arginine residues by an enzyme previously shown to modulate OL differentiation in vitro (Huang et al., 2011) and called arginine methyltransferase 5 (PRMT5), which is expressed at high levels in progenitors and highly proliferative glioma cells (Cho et al., 2012; Zheng et al., 2013). Finally, it is important to take in consideration the fact that differentiation of NG2 cells is accompanied by E2F1 translocation from the nucleus to the cytoplasm, where it is degraded prior to reaching a late OL differentiation stage (Magri et al., 2014a). Importantly, while the levels of activating E2F family members decreases during the process of differentiation, the levels of repressive family members such as E2F4 remain constant (Magri et al., 2014a; Nygård et al., 2003; Panteleeva et al., 2007). The dynamic pattern of expression and the similarity of the DNA binding sequence for activating and repressive E2F family members in the oligodendrocyte lineage suggested a potential switch from activating to repressive protein complexes at target genes during OL differentiation (Magri et al., 2014a). Chromatin immunoprecipitation studies validated this hypothesis and revealed that E2F1 occupancy of cell cycle gene promoters (e.g. Ccne, Ccna2, Ccnb1, Cdc20) and epigenetic regulators (e.g. Dnmt1, Uhrf1) in proliferating NG2 progenitors, was replaced by E2F4-containing repressive complexes in OL (Magri et al., 2014a).
The oncogene transcription factor cMyc was also shown to be involved in cell cycle regulation of NG2 cells in response to mitogen stimulation (Dang, 1999; Magri et al., 2014b). In OPC, cMyc targeted several genes involved in cell cycle (e.g. Cdc2 or Cdc20), and it was progressively down-regulated during OL differentiation (Magri et al., 2014b). Consistent with the ability of cMyc to recruit histone acetyltransferases and modulate acetylation of histones over large chromatin domains (Guccione et al., 2006; Martinato et al., 2008; McMahon et al., 2000), NG2 cells -characterized by high levels of cMyc – also display global histone acetylation and high levels of gene targets (Magri et al., 2014b; Marin-Husstege et al., 2002). Silencing cMyc in OPC decreased histone acetylation (e.g. H3K9Ac), while increasing repressive histone methylation (e.g. H3K9me3), and favoring the initial compaction of peripheral nuclear chromatin and associated decrease of gene expression. Downregulated genes involved those modulating the cell cycle, such as Cdc2, which, in cMyc-silenced OPC was associated with lower H3K9ac and H3K14ac at the promoter (Magri et al., 2014b). Based on these data, we propose that cMyc might have a dual modality of regulation of NG2 cells proliferation by acting as DNA binding transcription factors on genes bearing its consensus sequence and also as a long-range chromatin modifier regulating histone acetylation.
2.2. DNA replication
DNA replication is another critical step of the cell cycle and the fidelity of replication is maintained by checkpoints that guarantee the quality control of DNA integrity, as well as the faithful transmission of the epigenetic marks (Budhavarapu et al., 2013; Probst et al., 2009). Both DNA and epigenetic marks duplication are coordinated by the proliferating cell nuclear antigen (PCNA) which recruits crucial players to the replication fork, including DNA polymerases, histone chaperone chromatin assembly factor 1 (cAF1 or cHAF1), HDACs, lysine methyltransferases, chromatin remodelers and DNA methyltransferases (Estève et al., 2006; Huen et al., 2008; Lu et al., 2013; Milutinovic et al., 2002; Stillman, 1986; Zhang et al., 2000). During cell division, the maintenance DNA methyltransferase DNMT1 is recruited by “Ubiquitin-like, containing PHD and RING Finger domains, 1” (UHRF1) to hemimethylated DNA, where it guarantees the duplication of DNA methylation marks from parents to daughter DNA strands (Bostick et al., 2007, 2007; Jacob et al., 2015; Knox et al., 2000; Milutinovic et al., 2003). The histone K27 methyltransferase “Enhancer of Zeste Homologue 2” (EZH2) has been shown to be necessary for DNA methylation of EZH2-target genes, at least at some genomic loci, like at the promoter of Myt1, which is involved in OL lineage cell function (McGarvey et al., 2007; Viré et al., 2006). During replication, EZH2 also binds to methylated H3K27me3 marks on the parent strand, to then replicate this mark on the new daughter strand (Hansen et al., 2008). Thus DNA methylation and histone K27 trimethylation are important epigenetic marks that guarantee the fidelity of transmission of genetic information from mother to daughter cell in highly proliferative populations.
2.3. Glioma proliferation
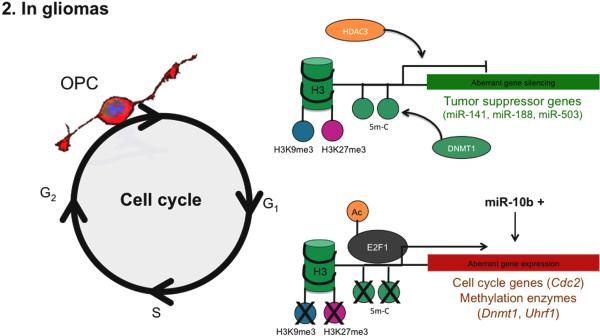
The disruption of the PCNA/DNMT1/UHRF1 complex is detected in conditions characterized by increased glial progenitor cells proliferation and lack of apoptosis (Hervouet et al., 2010). In the absence of a protein complex ensuring the transmission of correct genetic information and epigenetic marks during replication, the genomic contents and nuclear organization in daughter cells could have serious consequences and result in genomic instability and cancer. Indeed, glioma cells are largely characterized by aberrant DNA methylation and histone modifications and associated changes in chromatin organization (Brock et al., 2007; Felsberg et al., 2006; Kim et al., 2012; Long et al., 2013; Nair and Kumar, 2012; Uhlmann et al., 2003; Watanabe and Maekawa, 2010).
Extensive global DNA hypomethylation has been observed in several cancers, including gliomas, and often associated with aberrant activation of DNA coding and non-coding regions (Uhlmann et al., 2003; Watanabe and Maekawa, 2010). However, regional DNA hypermethylation at specific sites encoding for cell cycle inhibitors or tumor suppressor genes has also been observed in gliomas, further highlighting the complexity of the role of DNA methylation in tumorigenesis (Felsberg et al., 2006; Sharma et al., 2010).
Histone deacetylation and methylation have also been reported in glial tumors (Long et al., 2013; Zhu et al., 2013). Oligodendroglioma, for instance are characterized by histone hypoacetylation, and high levels of histone deacetylases (i.e. HDAC3). Acetate supplementation or inhibition of HDAC3 using siRNA have both been shown to suppress tumor cell proliferation in vitro (Kim et al., 2012). Alterations of repressive methylation marks at lysine residues K9 and K27 in histone H3 have also been detected in gliomas. More precisely, H3K9 and H3K27 methylation levels can be either low or high at different gene loci in glioblastomas, leading to aberrant expression of cell cycle genes or silencing of tumor suppressor genes respectively (Bender et al., 2013; Chan et al., 2013; Venneti et al., 2013a, 2013b). In addition, the loss of H3K9me3 and H3K27me3 repressive marks at DNMT1 promoter, associated with DNMT1 upregulation in gliomas, also suggests the synergistic role of DNA and histone methylation in tumor development (Rajendran et al., 2011).
Finally, miRNAs can function as either oncogenes or tumor suppressors, depending on their target genes. In glioblastomas, miR-10b is highly expressed and can modulate E2F1-mediated transcription, which leads to increased proliferation of NG2 cells (Teplyuk et al., 2015). In fact, E2F family proteins as well as their genes targets (e.g. Uhrf1, H2afz, Cdc2, Ccnd2) are up-regulated in gliomas, whereas silencing of E2f1 in glioma cells decreases also its targets expression (Magri et al., 2014a; Parr et al., 1997). The down-regulation of the oncogenic miR-10b in cells expressing high levels of the cell cycle inhibitor p21 has been shown to down-regulation of numerous E2F1 target genes and stabilization of cell cycle progression (Teplyuk et al., 2015). On the contrary, the tumor suppressors mirR-141, miR-188 and miR-503 have the ability to decrease the levels of cyclins and CDKs and further down-regulate the levels of E2F target genes in cancer cells, thereby promoting a beneficial effect (Cao et al., 2014; Wu et al., 2014; Xiao et al., 2013; Xue et al., 2014). Together these data identify modulation of E2F1 as a valuable therapeutic option to be further explored in proneural gliomas, which share several characteristics of OPCs.
Overall, we have discussed the complex regulation of proliferation in NG2 cells by transcription factors and epigenetic marks that can modulate each other expression or activity (Figure 1). Disruption of this complex regulatory network results in loss of homeostatic control and potentially explain tumorigenic transformation (Figure 2).
Figure 1.
Epigenetic mechanisms regulating NG2 cell proliferation and differentiation during development.
OPC proliferation is controlled by epigenetic marks that activate cell cycle genes expression and repress mature genes expression. During the S phase, genetic and epigenetic marks are also tightly duplicated. Combined epigenetic marks, especially deacetylation and methylation, lead to cell cycle genes and inhibitor of differentiation genes repression during OL differentiation, while acetylation and hypomethylation induce expression of myelin genes like Mag.
Figure 2.
Epigenetic mechanisms regulating NG2 cell proliferation in gliomas.
Increased levels of DNMT1 and HDAC3 result in aberrant gene silencing, such as tumor suppressor genes. In parallel, hypomethylation and overexpression of miR-10b maintain the expression of cell cycle genes, which keep the OPC in a proliferative stage.
3. Epigenetic mechanisms regulating NG2 cell differentiation
3.1. Necessary epigenetic regulatory machinery for NG2 cell differentiation
It is well-established that differentiation of NG2 cells is triggered by exit from the cell cycle and it is accompanied by dramatic changes of the transcriptional program, which include down-regulation of cell cycle regulatory molecules and differentiation inhibitors, followed by up-regulation of genes enriched in oligodendrocytes (Cahoy et al., 2008; Swiss et al., 2011; Zhang et al., 2014). RNA polymerase II ChIP-Sequencing revealed that start of NG2 cell differentiation is also accompanied by active transcription of genes involved in chromatin remodeling (Yu et al., 2013). Furthermore, chromatin condensation driven by heterochromatin formation can be detected at the periphery of the nucleus (Wu et al., 2012a). In light of these new findings, it is important to provide a new outlook of NG2 cell differentiation. Traditionally, NG2 cell differentiation is considered to be tightly controlled by a hierarchy of transcription factors (TFs) (Emery, 2010). Nevertheless, TFs alone cannot complete the task because genomic DNA is not linear or naked within a cell. Actually, it is compacted by histones into nucleosomes that are further assembled into a higher degree of structure inside the nucleus (Gorkin et al., 2014). Therefore, accessibility of the TFs to the genome is highly regulated and epigenetics is the mechanism that mediates this level of regulation (Chen and Weiss, 2014). In NG2 cells, many epigenetic enzymes are coupled with TFs to regulate the transcriptional program during differentiation. Loss of enzymatic activities results in severe attenuation of differentiation (Table 1). For instance, genetic ablation of Hdac1/2 in OPC resulted in failure of differentiation in vivo (Cunliffe and Casaccia-Bonnefil, 2006; Ye et al., 2009). In vitro, inhibition of HDAC enzymatic activity blocked differentiation in both rodent (Marin-Husstege et al., 2002; Swiss et al., 2011) and human OPCs (Conway et al., 2012). Similarly, the histone lysine methyltransferases “Euchromatic Histone-lysine N-MethylTransferase 2” (EHMT2 or G9a) and “Suppressor of variegation 3-9 homolog 1” (Suv39H1), are required for efficient differentiation (Liu et al., 2015). An important role of a histone arginine modifying enzyme, PRMT5, for OPC differentiation in vitro has also been reported (Huang et al., 2011). In addition, genetic deletion of an enzymatic component of the ATP-dependent SWI/SNF chromatin remodeling complex BRG1 in OPC stopped differentiation in vivo (Yu et al., 2013), although this finding has been challenged by a more recent report demonstrating that BRG1 is not essential for CNS myelination (Bischof et al., 2015). A detailed discussion of the epigenetic machinery regulating NG2 cell differentiation is presented below (Figure 1).
Table 1.
Epigenetic enzymes required for normal oligodendrocyte development.
| Enzyme | Modification | Required for | Experimental system |
|---|---|---|---|
| HDAC1/2 | Histone de-acetylation | Specification, differentiation and myelination | Primary rodent OPC cultures (Marin-Husstege et al., 2002; Wu et al., 2012a), human OPC culture (Conway et al., 2012), zebrafish (Cunliffe and Casaccia-Bonnefil, 2006) and conditional knockout mice (Olig1-Cre) (Ye et al., 2009) |
| HDAC11 | Histone de-acetylation | Differentiation | OPC cell line (Liu et al., 2009) |
| EZH2 | H3K27 methylation | Specification | Neural stem cell culture (Sher et al., 2008) |
| G9a/SUV39h1 | H3K9 methylation | Differentiation | Primary rodent OPC culture (Liu et al., 2015) |
| PRMT5 | Histone arginine symmetric methylation | Differentiation | Primary rodent OPC culture and glioma cell line (Huang et al., 2011) |
| BRG1 | Nucleosome remodeling | Differentiation and myelination | Primary rodent OPC culture and conditional knockout mice (Olig1-Cre) (Yu et al., 2013) |
| DICER | Process pre-microRNAs | Differentiation and myelination | Primary OPC cultures and conditional knockout mice (Plp-CreERT (Shin et al., 2009); Olig2-Cre and Cnp-Cre (Dugas et al., 2010); Olig1-Cre (Zhao et al., 2010)) |
3.2. Histone deacetylases (HDACs)
The requirement of HDAC1 (with HDAC2 being functionally redundant to HDAC1, Shen et al., 2008a; Wu et al., 2012a; Ye et al., 2009) for NG2 cell differentiation can be explained by the findings that HDAC1 is actively recruited to chromatin by different key TFs in order to repress gene expression through modulation of histone acetylation. For example, in OPC, it has been shown to interact with NKX2.2 (Ji et al., 2011; Wei et al., 2005) at the promoters of Mbp and Sirt2 genes and inhibits their expression. In addition, it can also interact with SOX10 (Liu et al., 2015). In the differentiated cells, however, HDAC1 no longer binds to SOX10 (Liu et al., 2015). Instead, it becomes an interacting partner of YY1, a TF essential for differentiation (He et al., 2007a). Recruitment of YY1/HDAC1 complex to the promoter of genes encoding for differentiation inhibitors (e.g. Tcf7l2 and Id4) resulted in loss of acetylated histone marks and transcriptional repression (He et al., 2007a). Supporting the importance of this mechanism was also the finding that a high percentage of down-regulated genes during OPC differentiation, were also modulated by type-I HDAC activity (Swiss et al., 2011). Global histone deacetylation mediated by HDAC1/2 was detected in OPC treated with extracellular factors promoting lineage progression (i.e. SHH) while increased acetylation was observed in cells treated with factors (i.e.BMP4) blocking oligodendrocyte differentiation (Wu et al., 2012a). Collectively, these findings support a ‘de-repression’ model where histone deacetylation is required for repressing the expression of differentiation inhibitors during NG2 cell differentiation, hence releasing the transcriptional inhibition from the promoters of myelin genes (Liu and Casaccia, 2010). The recent finding that inhibiting a major acetylated histone binding protein (BRD4) could promote NG2 cell differentiation (Gacias et al., 2014) further underscores the importance of the balance between histone acetylation and deacetylation for NG2 cell differentiation, with acetylation being favored at the progenitor stage and deacetylation required for differentiation.
In addition to modifying histones, HDACs are able to deacetylate also non-histone substrates. Notably, beta-catenin can be modified by acetylation that facilitates its interaction with TCF7L2 (Lévy et al., 2004). Since HDAC1 interacts with beta-catenin in OPC (Ye et al., 2009), it is possible that in this complex HDAC1 keeps beta-catenin in a de-acetylated state in addition to sequestering it from complexing with TCF7L2. Similarly, many other TFs found to be important for oligodendrocyte specification (e.g. GLI2) (Coni et al., 2013) and differentiation (e.g. YY1) (He et al., 2007b; Yao et al., 2001) are also regulated by acetylation/deacetylation. Additional factors which have been shown to be acetylated in other cell types (i.e. E2F1) (Martínez-Balbás et al., 2000) or have lysine residues which could play an important functional role (i.e. MYRF, OLIG1) have also the potential to be acetylated. Given the relevance of post-translational modification of TF in modulating their activity, the study of acetylation of non-histone substrates could be of high interest for future studies. In addition of the several HDAC family members expressed in the oligodendrocyte lineage (Broide et al., 2007; Shen et al., 2005; Tiwari et al., 2014), it is important to take into consideration that, besides HDAC1/2 other family members may play important functional roles including HDAC3 working in synergy with thyroid hormone T3 in promoting differentiation of Oli-neu cells (Castelo-Branco et al., 2014). This finding is actually not surprising considering that HDAC1/2 tend to form complexes with co-repressor proteins (e.g. Sin3a) (Grzenda et al., 2009) and a TF like YY1 (He et al., 2007a), HDAC3 is usually associated with the NCOR/SMRT co-repressor and a nuclear receptor (Perissi et al., 2010). Therefore, it is very likely that they may mediate different mechanisms in the same cells. While the functional significance of the HDAC1-Sin3a-YY1 has been demonstrated in the NG2 cells, further study is still needed to shed new light on the mechanism mediated by the HDAC3- NCOR/SMRT-nuclear receptor complex.
3.3 Histone methyltransferases (HMTs)
Two types of amino acid residues can be modified by methylation on the core histones-lysine and arginine-by two different groups of methyltransferases. Regarding lysine methylation, EZH2, the enzyme responsible for the repressive histone mark H3 lysine 27 trimethylation (H3K27me3), was the first one to be characterized in neural stem cells differentiated along the oligodendrocyte lineage, while its expression was decreased in neurons and astrocytes (Sher et al., 2008). It was described that EZH2 overexpression favored, while silencing it blunted, oligodendrocyte differentiation. This was consistent with the relatively unchanged genomic occupancy of EZH2 on the neuronal (e.g. Phox2b, Six1, Neurod2, Tlx3 and Otp) and astrocytic genes (e.g. Tal1) from neural stem cell differentiation into oligodendrocyte (Sher et al., 2012) and with the decreased EZH2 occupancy of oligodendrocyte lineage specific genes such as Olig2, Pdgfra, Nkx6.2 and Nkx 2.2, thereby suggesting that EZH2 is required for the early stages of oligodendrocyte differentiation from neural stem cells and may modulate cell fate decision of progenitors. A recent study confirmed these findings and identified another group of HMTs, the ones that are responsible for generating H3 lysine 9 trimethylation (H3K9me3), catalyzed by EHMT1 (also known as Glp) and EHMT2 (also known as G9a) and by Suv39H1, to be important for the subsequent stages of differentiation from NG2 cells to oligodendrocytes (Liu et al., 2015). In this study, the authors performed ChIP-Sequencing analysis for H3K9me3 and H3K27me3 in proliferating progenitors and their differentiated progeny and detected a global increase of H3K9me3 but not H3K27me3. At the progenitor stage 920 genes were shown to be uniquely repressed by H3K9me3 marks and more than 600 genes were uniquely repressed by H3K27me3 mark (Liu et al., 2015). Interestingly, as OPCs differentiated into OLs, the number of genes with H3K9me3 marks increased, in parallel with an up-regulation of EHMT2. The number of genes with H3K27me3 marks decreased, in parallel with the down-regulation of EZH2 in myelinating OL, compared to NG2 cells (Liu et al., 2015; Sher et al., 2008; Zhang et al., 2014). These data suggest that distinct repressive marks characterize diverse functional states. Repressed genes with unique H3K9me3 marks included those regulating membrane excitability in NG2 cells, such as clusters of potassium channel subunits (e.g. Kcnb1, Kcnb2, Kcnk1 and Kcnmb4). Accordingly, silencing the enzymes Ehmt2 and Suv39h1, responsible for generating this mark in NG2 cells altered the electrical properties of the cells. Additional genes regulated by both marks were transcription factors modulating neuronal fate, including Lhx1, Pax6, Grip1 and Dcx (Liu et al., 2015). In agreement, silencing Ehmt2 and Suv39h1 rather than Ezh2 impaired oligodendrocyte differentiation and altered expression of neuronal genes. Taken together, these results suggest that H3K27 HMT is required for oligodendrocyte lineage specification from neural progenitors to OPC, while the H3K9 HMT is critical for NG2 cell differentiation into OL.
Arginine residues in the tails of nucleosomal histones are also methylated by protein arginine methyltransferases (PRMTs) (Di Lorenzo and Bedford, 2011). In NG2 cells, a role of PRMT5 in regulating the transcriptional program during differentiation has been suggested (Huang et al., 2011). Considering its critical role in modulating E2F1-dependent gene expression in gliomas and as gatekeeper of “stemness” and survival in neural stem cells mediated by regulation of the mRNA splicing machinery (Bezzi et al., 2013), it would be interesting to determine whether it plays a similar role in NG2 cells and OLs, where alternative splicing events are known to occur in abundance (e.g. DM20/Plp1, immature/mature MBP transcripts, long/short MAG transcripts) (Zhang et al., 2014).
3.4. DNA methylation
In addition to histone deacetylation and H3 lysine 9 trimethylation, DNA methylation is another important epigenetic mechanism for gene repression, which might be important for developmental myelination. While DNA methylation has been studied extensively in neurons and astrocytes (Fan et al., 2001; Hutnick et al., 2009; Li et al., 1992; Milutinovic et al., 2003; Unterberger et al., 2006), it still remains poorly characterized in the oligodendrocyte lineage. During development, DNA methylation has been shown to be essential for stem cell differentiation into the neural lineage (Wu et al., 2012b), while demethylation has been reported to be associated with precocious astrogliogenesis (Fan et al., 2005; Wu et al., 2012b). Genetic ablation of Dnmts in the neuronal lineage has been linked either to defective survival in proliferating neuroblasts or defective neuronal plasticity in post-mitotic neurons (Feng et al., 2010; Hutnick et al., 2009). The concept of demethylation-associated expression of mature genes has been proposed for neuronal, astrocytic, and Schwann cell differentiation (Fan et al., 2001; Takizawa et al., 2001; Varela-Rey et al., 2014). A similar model could be envisioned for OLs as the TET proteins appear to be necessary for OPC differentiation and since the myelin gene Mag is specifically demethylated during differentiation (Grubinska et al., 1994; Zhao et al., 2014). However, a previous study performed in neonatal rats during developmental myelination, showed that injection of 5-azacytidine, a nucleoside inhibitor of DNMTs, resulted in an almost complete absence of myelin in the optic nerve (Ransom et al., 1985). When rats were allowed to recover from the chemical perturbation, myelinated fibers began to reappear, indicating that an oligodendrocyte population persisted but could not differentiate into myelin-forming cells. Taken together, these data suggest that, in the oligodendroglial lineage, DNA methylation is a complex mechanism, requiring both methylation and demethylation of specific genomic regions.
3.5. Nucleosome remodeling
Since genomic DNAs including coding and regulatory regions are packed into nucleosomes inside the nucleus, fully activating a gene requires sliding of the nucleosomes to expose the DNA regions that TFs or other transcriptional machineries can land on. It has been demonstrated that this process is energy-consuming (requiring the use of ATP) and mediated by a set of large protein complexes that are collectively termed as ATP-dependent chromatin remodeling complexes (Hargreaves and Crabtree, 2011). It has been shown that activation of genes by TF during NG2 cell differentiation depends on one of such mechanisms. Upon differentiation, the ATP-dependent SWI/SNF chromatin-remodeling enzyme BRG1 was recruited by OLIG2 to the active enhancer regions (defined by enrichment of H3 lysine 27 acetylation, H3K27ac) and this correlated with the expression of genes characteristic of the differentiated state (Yu et al., 2013). However, further studies are required to examine the genome-wide distribution of OLIG2 binding and whether it is changed upon loss of BRG1. This is of great importance considering a more recent finding from another group that demonstrated that ablation of Brg1 using NG2-cre and Cnp-cre is not essential for oligodendrocyte differentiation (Bischof et al., 2015). In addition, it would be of relevance to define whether the OLIG2/BRG1 complex contributes to the formation of a novel type of regulatory hubs (‘super-enhancers’) at clusters of enhancer regions that are cell type specific and developmentally regulated, as initially described in embryonic stem cells (Whyte et al., 2013). It is also conceivable that such a role could be played by SOX10 through its interaction with MED12 (Vogl et al., 2013), a component of the Mediator complex that functions by bridging enhancer regions with the basal transcription machinery at proximal promoter regions. In addition, given the current debate on the plastic and multipotential nature of NG2 cells, it would be of great relevance to map all the enhancer regions that retain the potential for transcriptional activation in these cells, by defining the ‘poised’ enhancers - which are enriched in H3K4me1, H3K4me2 and H3K27me3 marks and depleted of H3K4me3 and H3K27ac histone marks (Heinz et al., 2010). This would address a different question that mapping the active enhancers defined by the H3K27ac mark, as in the previous study (Yu et al., 2013), by uncovering the “multipotentiality” of the cells at a molecular level and provide novel insights into the controversial “multipotency” of NG2 cells (Richardson et al., 2011).
3.6. MicroRNAs play an additional role
An additional layer that adds to the complexity of epigenetic modulation of OL differentiation is the presence of microRNAs. The necessity of these molecules for differentiation was firstly revealed by the phenotype of mice with ablation of the gene encoding a key enzyme for microRNA maturation (DICER), which revealed microRNAs were not required for the specification of the oligodendrocyte lineage but are necessary for differentiation and myelination (Dugas et al., 2010; Shin et al., 2009; Zhao et al., 2010). Several microRNAs have been characterized in detail along with their mRNA targets. Specifically, miR-7a has been shown to safeguard from aberrant gene expression of Pax6 and Neurod4 and pro-oligodendrocytic genes during the specification of NG2 cells from neural stem cells (Shin et al., 2009). Similarly, miR-9 has been shown to prevent expression of Schwann cell specific transcripts, such as Pmp22 (Lau et al., 2008). On the other hand, miR-219, miR-338, miR-23 and miR-32 are much more abundant in differentiated cells and facilitate lineage progression towards myelinating oligodendrocytes. Specifically, miR-219 has been identified to be the most dramatically up-regulated microRNA during NG2 cell differentiation. It targets a wide range of mRNAs whose protein products constitute major obstacles to differentiation, including the mitogen receptor (Pdgfra) and multiple TFs (Sox6, Foxj3, Zfp238 and Hes5) (Dugas et al., 2010; Zhao et al., 2010). At later stages of differentiation, miR-219 targets the transcripts of a gene encoding a lipid metabolizing enzyme (Elovl7) whose overexpression would result in abnormal lipid accumulation in myelin (Shin et al., 2009). Similarly, miR-32 has also been shown to regulate the expression of an enzyme (Slc45a3) important for lipid metabolism in oligodendrocytes and myelin (Shin et al., 2012). Among other microRNAs enriched in differentiated NG2 cells, miR-338 seems to be functionally redundant to miR-219 (Zhao et al., 2010) while miR-23 is likely to play a different yet critical role in mature oligodendrocytes by suppressing the expression of a nuclear lamina component Lamin-B1, whose overdosing is the genetic cause of the an adult-onset demyelinating disease (Lin and Fu, 2009). Besides, it has been revealed more recently that over-expression of miR-23a in mice led to increased thickness of myelin in CNS, further supporting the importance of this microRNA for myelination (Lin et al., 2013).
3.7. Coordination between extrinsic signaling mechanisms and epigenetics
It should be noted that one of the most important reasons why epigenetic changes are considered as critical modulators of gene expression is because they are dynamic and with varying degrees of reversibility and with a longer lasting effect on transcription than TF alone. Some regulatory mechanisms are associated with turning on or off transcription in a cell, in response to the signals it receives. For example, it is well established that neurons utilize many of the epigenetic enzymes such as the histone acetyltransferase CBP to mediate their transcriptional response to synaptic activity and depolarizing stimuli (Riccio, 2010). In contrast, not enough studies have been performed in the NG2 cells to illustrate unambiguously how extrinsic signals can affect the epigenetic machinery. However, some findings are emerging to suggest such coordination may exist. Wu et al. showed that two important morphogens for oligodendrocyte development (SHH and BMP4) converge on regulating the histone state in oligodendrocyte progenitors. While SHH promoted oligodendrocyte differentiation and heterochromatin formation by decreasing global histone acetylation, BMP4 blocked this process by increasing histone acetylation globally and at astrocytic genes (Wu et al., 2012a). Interestingly, downstream of SHH signaling in the developing telencephalon, gene repression and activation has been reported to be modulated by BRG1, whose pleiotropic function is influenced by the developmental stage and brain region (Zhan et al., 2011). It would be of interest to see whether similar mechanisms exist in the NG2 cell lineage and such insight may be able to reconcile the findings that deletion of the BRG1 gene in different populations of the lineage results in different phenotypes on myelination (Bischof et al., 2015; Yu et al., 2013).
Further, many of the other aforementioned epigenetic mechanisms that are important for NG2 cells can be potentially regulated by hormonal ligands. It is well established that members of the nuclear receptor family (including the thyroid hormone receptor, Vitamin D receptor and Retinoid acid receptor) enter the cells and bind to their cognate receptor, thereby inducing an exchange of binding partners, from HDACs to HATs that would result in functional switch of the complex from repressive – in the absence of the ligand- to activating, in the presence of the hormone (Kato et al., 2011). The current finding that silencing the HMTs responsible for H3K9 methylation resulted in block of lineage maturation only in OPCs maintained in a proliferating condition (with the mitogens PDGFaa and bFGF) but not in the cells already induced to differentiate (with the thyroid hormone T3) suggests that in the NG2 cells also, the HMTs functions are likely linked to extracellular signals of the nuclear receptor superfamily (Liu et al., 2015).
Finally, recent studies have highlighted the importance of neuronal activity (Gibson et al., 2014) and social behavior (Liu et al., 2012) in modulating post-translational modifications of histones in NG2 cells in the adult brain. Using optogenetic stimulation of NG2 cells in the adult motor cortex in order to induce a motor behavior, Gibson and colleagues reported the subsequent occurrence of histone deacetylation and increased H3K9me3 methylation in response to neuronal stimulation. They further reported that pharmacological inhibition of histone deacetylation with the inhibitor trichostatin, prevented the occurrence of these histone marks and prevented the acquisition of the motor behavior (Gibson et al., 2014). Conversely, studies on mice subject to social isolation revealed decreased repressive H3K9me3 marks and the persistence of an immature chromatin state, which correlated with behavioral changes (i.e. a decrease in the social interaction time) and fewer myelinated fibers in the prefrontal cortex (Liu et al., 2012). Together these studies support the concept of modulation of epigenetic machinery by external stimuli.
4. Therapeutic approaches based on epigenomic modulators
4.1. Epigenomic approaches to remyelination failure
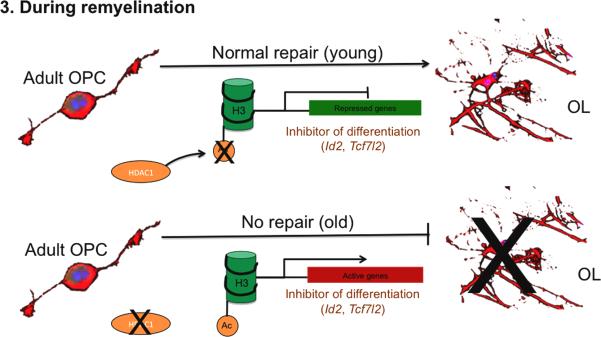
Consistent with their role in development, NG2 cells and the oligodendrocytes that they generate are extensively involved in the pathology of many CNS demyelinating disorders including multiple sclerosis (MS). Despite the many immunomodulatory treatments, a cure is still not available to repair the damage (Ransohoff et al., 2015). As a consequence, even when the immune attack or inflammation has receded, the demyelinated axons may not be properly remyelinated and therefore they remain exposed to axonal damage. In addition, a portion of progenitor cells, despite their presence around MS lesions, show a declining propensity to differentiate as the disease progresses and fail to generate enough OLs capable to remyelinate axons (Fancy et al., 2010). However, remyelination is a robust regenerative process in the mammalian CNS and NG2 cells are responsible for this process. In response to injury, NG2 cells are not only capable to proliferate and migrate to the lesions but also to differentiate into OLs and form new compact myelin sheaths wrapping around the demyelinated axons and leading to functional recovery. Repair can be promoted either by resident NG2 cells within the brain, or by cells from the subventricular zone. These cells may undergo similar steps as during postnatal development and produce oligodendrocytes (Huang and Franklin, 2012; Vidaurre et al., 2012). This suggests that endogenous myelin repair in MS patients is achievable. At the cellular level, it is known that many key mechanisms for remyelination, including the ones composed of epigenetic modulation, recapitulate those in development for regulating NG2 cell proliferation and differentiation (Fancy et al., 2011). For example, the type-I HDAC activities are required for both efficient developmental myelination and remyelination (Shen et al., 2008b). In agreement with this, increased immunoreactivity for acetylated histone H3 accompanied by high levels of transcriptional inhibitors of NG2 cell differentiation (i.e., TCF7L2, ID2, and SOX2) and HAT transcript levels (i.e., CBP, P300) have been observed in oligodendrocytes localized to the normal-appearing white matter (NAWM) of chronic MS lesions where remyelination is not sufficient in contrast to a low level of histone H3 acetylation in early MS lesions where remyelination is still efficient (Pedre et al., 2011). This implies that the status of H3 acetylation correlates with the differentiation state of the cells, similar to what happens during development. In addition, similar to the situation in MS, the ability of NG2 cells to regenerate myelinating oligodendrocyte also seems to decline with aging. This phenomenon can be at least partially explained by inefficient recruitment of HDACs in the aged but not young brains during remyelination (Shen et al., 2008b) (Figure 3). Therefore, it may be feasible to provide therapeutic interference in the NG2 cells to promote remyelination by modulating the epigenome. For instance, in light of a recent report that a small molecule inhibitor to one of the acetylated lysine binding domain of BRD4 is capable to tilt the balance of histone acetylation in NG2 cells to favor differentiation (Gacias et al., 2014), the acetylated histone binding protein BRD4 may become a druggable target for remyelination, considering a panel of different chemical inhibitors have been developed lately for this protein (Gacias et al., 2014; Shi and Vakoc, 2014).
Figure 3.
Epigenetic mechanisms regulating NG2 cell differentiation during repair.
Epigenetic modifications identified during development can be recapitulated, like the histone deacetylation, leading to gene repression (of inhibitor of differentiation). In chronic disease or in old animals, the lack of remyelination can be partially explained by the downregulation of HDAC1, thus the expression of inhibitors of differentiation.
4.2. Epigenomic approaches to program/reprogram the NG2 cells
It has been proven that transplantation of human NG2 cells has significant beneficial effects in several rodent models that mimic human neurological pathologies, including spinal cord injury (Volarevic et al., 2013) and the side effects of radiation therapy to brain tumors (Piao et al., 2015). Therefore, it is desirable to obtain human NG2 cells. A practical and feasible way to do so is the generation of human pluripotent stem cells (hPSCs). Although differentiation of hPSCs into NG2 cells (Douvaras et al., 2014; Stacpoole et al., 2013; Wang et al., 2013) is achievable, the process is still time-consuming (it takes from 95 days to 150 days). Human NG2 cells have the potential also to be obtained by direct reprogramming from other lineages (e.g. skin fibroblasts) and the pioneering results in murine cells are quite encouraging (Najm et al., 2013; Yang et al., 2013). However, the molecular identity of these cells is not entirely equivalent to the one of the native counterpart, since these reprogrammed cells express some Schwann cell markers and myelinate single axons as Schwann cells do (Najm et al., 2013; Yang et al., 2013), thereby suggesting insufficient epigenetic reprogramming. Therefore, derivation of NG2 cells in vitro still needs to be improved and it is predictable that the process could be enhanced by epigenetic manipulations, considering the many successful cases made in other lineage of cells (Zhang et al., 2012).
5. Concluding remarks
Several epigenetic mechanisms underlie the functions of NG2 cells by interacting with transcription factors and regulating both their proliferation and differentiation properties. It is foreseeable that with accumulating knowledge regarding the epigenetic landscape of OPC and its regulation, we will be able to answer many of the open questions regarding NG2 cells, especially their heterogeneity and plasticity. Recently, zinc-finger proteins or transcription activator-like effectors fused with chromatin-modifying enzymes or nanofiber-mediated microRNA have been used to modulate epigenetic modifications in vitro or in vivo (Diao et al., 2015; Heller et al., 2014; Mendenhall et al., 2013; Sanjana et al., 2012; Snowden et al., 2002). Such new tools could be used as strong therapeutic strategies in NG2 glial cells to specifically promote their remyelination capacities or unlock their multipotentiality in vivo.
Highlights.
- Epigenetic regulation includes DNA methylation, chromatin and non-coding RNA changes
- It affects the transcriptional landscape of NG2 glial cell
- Epigenetic changes of nuclear structural components define transitions in NG2 cells
- Epigenetic dysregulation can result in defective myelin repair or gliomas formation
- Modulation of epigenetic marks can be explored as therapeutic strategy for NG2 cells
Acknowledgements
Supported by NIH-NINDS R37-NS042925 and NSR01-NS52738.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barres BA, Raff MC. Proliferation of oligodendrocyte precursor cells depends on electrical activity in axons. Nature. 1993;361:258–260. doi: 10.1038/361258a0. [DOI] [PubMed] [Google Scholar]
- Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994a;120:1097–1108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- Barres BA, Raff MC, Gaese F, Bartke I, Dechant G, Barde YA. A crucial role for neurotrophin-3 in oligodendrocyte development. Nature. 1994b;367:371–375. doi: 10.1038/367371a0. [DOI] [PubMed] [Google Scholar]
- Barres BA, Burne JF, Holtmann B, Thoenen H, Sendtner M, Raff MC. Ciliary Neurotrophic Factor Enhances the Rate of Oligodendrocyte Generation. Mol Cell Neurosci. 1996;8:146–156. doi: 10.1006/mcne.1996.0053. [DOI] [PubMed] [Google Scholar]
- Beijersbergen RL, Kerkhoven RM, Zhu L, Carlée L, Voorhoeve PM, Bernards R. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 1994;8:2680–2690. doi: 10.1101/gad.8.22.2680. [DOI] [PubMed] [Google Scholar]
- Belachew S, Aguirre AA, Wang H, Vautier F, Yuan X, Anderson S, Kirby M, Gallo V. Cyclin-Dependent Kinase-2 Controls Oligodendrocyte Progenitor Cell Cycle Progression and Is Downregulated in Adult Oligodendrocyte Progenitors. J. Neurosci. 2002;22:8553–8562. doi: 10.1523/JNEUROSCI.22-19-08553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DTW, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The Mammalian Epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, Bonday ZQ, Guccione E. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 2013;27:1903–1916. doi: 10.1101/gad.219899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof M, Weider M, Küspert M, Nave K-A, Wegner M. Brg1-Dependent Chromatin Remodelling Is Not Essentially Required during Oligodendroglial Differentiation. J. Neurosci. Off. J. Soc. Neurosci. 2015;35:21–35. doi: 10.1523/JNEUROSCI.1468-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Estève P-O, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- Brock MV, Herman JG, Baylin SB. Cancer as a manifestation of aberrant chromatin structure. Cancer J. Sudbury Mass. 2007;13:3–8. doi: 10.1097/PPO.0b013e31803c5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1-11 in the rat brain. J. Mol. Neurosci. MN. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- Budhavarapu VN, Chavez M, Tyler JK. How is epigenetic information maintained through DNA replication? Epigenetics Chromatin. 2013;6:32. doi: 10.1186/1756-8935-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canoll PD, Musacchio JM, Hardy R, Reynolds R, Marchionni MA, Salzer JL. GGF/neuregulin is a neuronal signal that promotes the proliferation and survival and inhibits the differentiation of oligodendrocyte progenitors. Neuron. 1996;17:229–243. doi: 10.1016/s0896-6273(00)80155-5. [DOI] [PubMed] [Google Scholar]
- Cao T, Li H, Hu Y, Ma D, Cai X. miR-144 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting E2F3. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2014;35:10759–10764. doi: 10.1007/s13277-014-2017-7. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Tikoo R, Kiyokawa H, Friedrich V, Chao MV, Koff A. Oligodendrocyte precursor differentiation is perturbed in the absence of the cyclin-dependent kinase inhibitor p27Kip1. Genes Dev. 1997;11:2335–2346. doi: 10.1101/gad.11.18.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Hardy RJ, Teng KK, Levine JM, Koff A, Chao MV. Loss of p27Kip1 function results in increased proliferative capacity of oligodendrocyte progenitors but unaltered timing of differentiation. Development. 1999;126:4027–4037. doi: 10.1242/dev.126.18.4027. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco G, Lilja T, Wallenborg K, Falcão AM, Marques SC, Gracias A, Solum D, Paap R, Walfridsson J, Teixeira AI, et al. Neural stem cell differentiation is dictated by distinct actions of nuclear receptor corepressors and histone deacetylases. Stem Cell Rep. 2014;3:502–515. doi: 10.1016/j.stemcr.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K-M, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari DM, Crang AJ, Blakemore WF. Decline in rate of colonization of oligodendrocyte progenitor cell (OPC)-depleted tissue by adult OPCs with age. J Neuropathol Exp Neurol. 2003;62:908–916. doi: 10.1093/jnen/62.9.908. [DOI] [PubMed] [Google Scholar]
- Chen J, Weiss WA. When deletions gain functions: commandeering epigenetic mechanisms. Cancer Cell. 2014;26:160–161. doi: 10.1016/j.ccr.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E-C, Zheng S, Munro S, Liu G, Carr SM, Moehlenbrink J, Lu Y-C, Stimson L, Khan O, Konietzny R, et al. Arginine methylation controls growth regulation by E2F-1. EMBO J. 2012;31:1785–1797. doi: 10.1038/emboj.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Clarke LE, Young KM, Hamilton NB, Li H, Richardson WD, Attwell D. Properties and fate of oligodendrocyte progenitor cells in the corpus callosum, motor cortex, and piriform cortex of the mouse. J Neurosci. 2012;32:8173–8185. doi: 10.1523/JNEUROSCI.0928-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coni S, Antonucci L, D'Amico D, Di Magno L, Infante P, De Smaele E, Giannini G, Di Marcotullio L, Screpanti I, Gulino A, et al. Gli2 acetylation at lysine 757 regulates hedgehog-dependent transcriptional output by preventing its promoter occupancy. PloS One. 2013;8:e65718. doi: 10.1371/journal.pone.0065718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway GD, O'Bara MA, Vedia BH, Pol SU, Sim FJ. Histone deacetylase activity is required for human oligodendrocyte progenitor differentiation. Glia. 2012;60:1944–1953. doi: 10.1002/glia.22410. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crockett DP, Burshteyn M, Garcia C, Muggironi M, Casaccia-Bonnefil P. Number of oligodendrocyte progenitors recruited to the lesioned spinal cord is modulated by the levels of the cell cycle regulatory protein p27Kip-1. Glia. 2005;49:301–308. doi: 10.1002/glia.20111. [DOI] [PubMed] [Google Scholar]
- Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech. Dev. 2006;123:24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerens C, Stankoff B, Logak M, Anglade P, Allinquant B, Couraud F, Zalc B, Lubetzki C. Induction of myelination in the central nervous system by electrical activity. Proc Natl Acad Sci U A. 1996;93:9887–9892. doi: 10.1073/pnas.93.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao HJ, Low WC, Milbreta U, Lu QR, Chew SY. Nanofiber-mediated microRNA delivery to enhance differentiation and maturation of oligodendroglial precursor cells. J. Control. Release Off. J. Control. Release Soc. 2015 doi: 10.1016/j.jconrel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemel LT, Jackson SJ, Cuzner ML. Role for TGF-beta1, FGF-2 and PDGF-AA in a myelination of CNS aggregate cultures enriched with macrophages. J Neurosci Res. 2003;74:858–867. doi: 10.1002/jnr.10837. [DOI] [PubMed] [Google Scholar]
- Douvaras P, Wang J, Zimmer M, Hanchuk S, O'Bara MA, Sadiq S, Sim FJ, Goldman J, Fossati V. Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Rep. 2014;3:250–259. doi: 10.1016/j.stemcr.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Fero ML, Roberts JM, Raff MC. p27Kip1 alters the response of cells to mitogen and is part of a cell-intrinsic timer that arrests the cell cycle and initiates differentiation. Curr. Biol. CB. 1998;8:431–440. doi: 10.1016/s0960-9822(98)70177-0. [DOI] [PubMed] [Google Scholar]
- Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr. Opin. Genet. Dev. 1994;4:255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- Emery B. Regulation of oligodendrocyte differentiation and myelination. Science. 2010;330:779–782. doi: 10.1126/science.1190927. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Estève P-O, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Dev. Camb. Engl. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- Fancy S, Kotter M, Harrington E, Huang J, Zhao C, Rowitch D, Franklin R. Overcoming remyelination failure in multiple sclerosis and other myelin disorders. Exp Neurol. 2010 doi: 10.1016/j.expneurol.2009.12.020. [DOI] [PubMed] [Google Scholar]
- Fancy SPJ, Chan JR, Baranzini SE, Franklin RJM, Rowitch DH. Myelin regeneration: a recapitulation of development? Annu. Rev. Neurosci. 2011;34:21–43. doi: 10.1146/annurev-neuro-061010-113629. [DOI] [PubMed] [Google Scholar]
- Fannon J, Tarmier W, Fulton D. Neuronal activity and AMPA-type glutamate receptor activation regulates the morphological development of oligodendrocyte precursor cells. Glia. 2015 doi: 10.1002/glia.22799. [DOI] [PubMed] [Google Scholar]
- Felsberg J, Yan PS, Huang TH-M, Milde U, Schramm J, Wiestler OD, Reifenberger G, Pietsch T, Waha A. DNA methylation and allelic losses on chromosome arm 14q in oligodendroglial tumours. Neuropathol. Appl. Neurobiol. 2006;32:517–524. doi: 10.1111/j.1365-2990.2006.00759.x. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C, Le T, Ferguson D, Cahill ME, Li Y, et al. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat. Neurosci. 2015 doi: 10.1038/nn.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Gacias M, Gerona-Navarro G, Plotnikov AN, Zhang G, Zeng L, Kaur J, Moy G, Rusinova E, Rodriguez Y, Matikainen B, et al. Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression. Chem. Biol. 2014;21:841–854. doi: 10.1016/j.chembiol.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo V, Zhou JM, McBain CJ, Wright P, Knutson PL, Armstrong RC. Oligodendrocyte progenitor cell proliferation and lineage progression are regulated by glutamate receptor-mediated K+ channel block. J Neurosci. 1996;16:2659–2670. doi: 10.1523/JNEUROSCI.16-08-02659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson EM, Purger D, Mount CW, Goldstein AK, Lin GL, Wood LS, Inema I, Miller SE, Bieri G, Zuchero JB, et al. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science. 2014;344:1252304. doi: 10.1126/science.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic Cytosine Methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gorkin DU, Leung D, Ren B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell. 2014;14:762–775. doi: 10.1016/j.stem.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubinska B, Laszkiewicz I, Royland J, Wiggins RC, Konat GW. Differentiation-specific demethylation of myelin associated glycoprotein gene in cultured oligodendrocytes. J. Neurosci. Res. 1994;39:233–242. doi: 10.1002/jnr.490390302. [DOI] [PubMed] [Google Scholar]
- Grzenda A, Lomberk G, Zhang J-S, Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochim. Biophys. Acta. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall'Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007a;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Sandoval J, Casaccia-Bonnefil P. Events at the transition between cell cycle exit and oligodendrocyte progenitor differentiation: the role of HDAC and YY1. Neuron Glia Biol. 2007b;3:221–231. doi: 10.1017/S1740925X08000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EA, Cates HM, Peña CJ, Sun H, Shao N, Feng J, Golden SA, Herman JP, Walsh JJ, Mazei-Robison M, et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat. Neurosci. 2014;17:1720–1727. doi: 10.1038/nn.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez M, Casaccia P. Interplay between transcriptional control and chromatin regulation in oligodendrocyte progenitors. Glia. 2015 doi: 10.1002/glia.22818. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervouet E, Lalier L, Debien E, Cheray M, Geairon A, Rogniaux H, Loussouarn D, Martin SA, Vallette FM, Cartron P-F. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PloS One. 2010;5:e11333. doi: 10.1371/journal.pone.0011333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JK, Franklin RJM. Current status of myelin replacement therapies in multiple sclerosis. Prog. Brain Res. 2012;201:219–231. doi: 10.1016/B978-0-444-59544-7.00011-1. [DOI] [PubMed] [Google Scholar]
- Huang J, Vogel G, Yu Z, Almazan G, Richard S. Type II arginine methyltransferase PRMT5 regulates gene expression of inhibitors of differentiation/DNA binding Id2 and Id4 during glial cell differentiation. J. Biol. Chem. 2011;286:44424–44432. doi: 10.1074/jbc.M111.277046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Tang X-M, Cambi F. Down-regulation of the retinoblastoma protein (rb) is associated with rat oligodendrocyte differentiation. Mol. Cell. Neurosci. 2002;19:250–262. doi: 10.1006/mcne.2001.1077. [DOI] [PubMed] [Google Scholar]
- Huen MSY, Sy SM-H, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J. Biol. Chem. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes EG, Kang SH, Fukaya M, Bergles DE. Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat Neurosci. 2013;16:668–676. doi: 10.1038/nn.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Hutnick LK, Golshani P, Namihira M, Xue Z, Matynia A, Yang XW, Silva AJ, Schweizer FE, Fan G. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum Mol Genet. 2009;18:2875–2888. doi: 10.1093/hmg/ddp222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonska B, Aguirre A, Vandenbosch R, Belachew S, Berthet C, Kaldis P, Gallo V. Cdk2 is critical for proliferation and self-renewal of neural progenitor cells in the adult subventricular zone. J. Cell Biol. 2007;179:1231–1245. doi: 10.1083/jcb.200702031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob V, Chernyavskaya Y, Chen X, Tan PS, Kent B, Hoshida Y, Sadler KC. DNA hypomethylation induces a DNA replication-associated cell cycle arrest to block hepatic outgrowth in uhrf1 mutant zebrafish embryos. Development. 2015;142:510–521. doi: 10.1242/dev.115980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Ji S, Doucette JR, Nazarali AJ. Sirt2 is a novel in vivo downstream target of Nkx2.2 and enhances oligodendroglial cell differentiation. J. Mol. Cell Biol. 2011;3:351–359. doi: 10.1093/jmcb/mjr009. [DOI] [PubMed] [Google Scholar]
- Kato S, Yokoyama A, Fujiki R. Nuclear receptor coregulators merge transcriptional coregulation with epigenetic regulation. Trends Biochem. Sci. 2011;36:272–281. doi: 10.1016/j.tibs.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Kim W, Kim R, Park G, Park J-W, Kim J-E. Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation. J. Biol. Chem. 2012;287:5588–5599. doi: 10.1074/jbc.M111.328138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JD, Araujo FD, Bigey P, Slack AD, Price GB, Zannis-Hadjopoulos M, Szyf M. Inhibition of DNA methyltransferase inhibits DNA replication. J. Biol. Chem. 2000;275:17986–17990. doi: 10.1074/jbc.C900894199. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of Dynamically Regulated MicroRNA and mRNA Networks in Developing Oligodendrocytes. J. Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom K-H, Lee S, Baek SH, Kim VN. EMBO J. 2004;MicroRNA genes are transcribed by RNA polymerase II.23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees JA, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol. Cell. Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Lévy L, Wei Y, Labalette C, Wu Y, Renard C-A, Buendia MA, Neuveut C. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol. Cell. Biol. 2004;24:3404–3414. doi: 10.1128/MCB.24.8.3404-3414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li Q, Brus-Ramer M, Martin JH, McDonald JW. Electrical stimulation of the medullary pyramid promotes proliferation and differentiation of oligodendrocyte progenitor cells in the corticospinal tract of the adult rat. Neurosci Lett. 2010;479:128–133. doi: 10.1016/j.neulet.2010.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin S-T, Fu Y-H. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis. Model. Mech. 2009;2:178–188. doi: 10.1242/dmm.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Mela A, Guilfoyle EM, Goldman JE. Neonatal and adult O4(+) oligodendrocyte lineage cells display different growth factor responses and different gene expression patterns. J Neurosci Res. 2009;87:3390–3402. doi: 10.1002/jnr.22065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-T, Huang Y, Zhang L, Heng MY, Ptácek LJ, Fu Y-H. MicroRNA-23a promotes myelination in the central nervous system. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17468–17473. doi: 10.1073/pnas.1317182110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33:193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Hu Q, D'ercole AJ, Ye P. Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia. 2009;57:1–12. doi: 10.1002/glia.20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Dietz K, DeLoyht JM, Pedre X, Kelkar D, Kaur J, Vialou V, Lobo MK, Dietz DM, Nestler EJ, et al. Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat Neurosci. 2012;15:1621–1623. doi: 10.1038/nn.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Magri L, Zhang F, Marsh NO, Albrecht S, Huynh JL, Kaur J, Kuhlmann T, Zhang W, Slesinger PA, et al. Chromatin Landscape Defined by Repressive Histone Methylation during Oligodendrocyte Differentiation. J. Neurosci. Off. J. Soc. Neurosci. 2015;35:352–365. doi: 10.1523/JNEUROSCI.2606-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long PM, Tighe SW, Driscoll HE, Moffett JR, Namboodiri AMA, Viapiano MS, Lawler SE, Jaworski DM. Acetate supplementation induces growth arrest of NG2/PDGFRα-positive oligodendroglioma-derived tumor-initiating cells. PloS One. 2013;8:e80714. doi: 10.1371/journal.pone.0080714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585:2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Tian Y, Salwen HR, Chlenski A, Godley LA, Raj JU, Yang Q. Histone-lysine methyltransferase EHMT2 is involved in proliferation, apoptosis, cell invasion, and DNA methylation of human neuroblastoma cells. Anticancer. Drugs. 2013;24:484–493. doi: 10.1097/CAD.0b013e32835ffdbb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri L, Swiss VA, Jablonska B, Lei L, Pedre X, Walsh M, Zhang W, Gallo V, Canoll P, Casaccia P. E2F1 coregulates cell cycle genes and chromatin components during the transition of oligodendrocyte progenitors from proliferation to differentiation. J. Neurosci. Off. J. Soc. Neurosci. 2014a;34:1481–1493. doi: 10.1523/JNEUROSCI.2840-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri L, Gacias M, Wu M, Swiss VA, Janssen WG, Casaccia P. c-Myc-dependent transcriptional regulation of cell cycle and nucleosomal histones during oligodendrocyte differentiation. Neuroscience. 2014b;276:72–86. doi: 10.1016/j.neuroscience.2014.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PloS One. 2008;3:e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Balbás MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. E2F Family Members Are Differentially Regulated by Reversible Acetylation. J. Biol. Chem. 2000;275:10887–10892. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–5102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinovic S, Zhuang Q, Szyf M. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J. Biol. Chem. 2002;277:20974–20978. doi: 10.1074/jbc.M202504200. [DOI] [PubMed] [Google Scholar]
- Milutinovic S, Zhuang Q, Niveleau A, Szyf M. Epigenomic stress response. Knockdown of DNA methyltransferase 1 triggers an intra-S-phase arrest of DNA replication and induction of stress response genes. J Biol Chem. 2003;278:14985–14995. doi: 10.1074/jbc.M213219200. [DOI] [PubMed] [Google Scholar]
- Münzel M, Globisch D, Brückl T, Wagner M, Welzmiller V, Michalakis S, Müller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew. Chem. Int. Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Nair SS, Kumar R. Chromatin remodeling in cancer: a gateway to regulate gene transcription. Mol. Oncol. 2012;6:611–619. doi: 10.1016/j.molonc.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm FJ, Lager AM, Zaremba A, Wyatt K, Caprariello AV, Factor DC, Karl RT, Maeda T, Miller RH, Tesar PJ. Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nat Biotechnol. 2013;31:426–433. doi: 10.1038/nbt.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Chang A, Trapp BD. NG2+ glial cells: a novel glial cell population in the adult brain. J Neuropathol Exp Neurol. 1999;58:1113–1124. doi: 10.1097/00005072-199911000-00001. [DOI] [PubMed] [Google Scholar]
- Noble M, Murray K, Stroobant P, Waterfield MD, Riddle P. Platelet-derived growth factor promotes division and motility and inhibits premature differentiation of the oligodendrocyte/type-2 astrocyte progenitor cell. Nature. 1988;333:560–562. doi: 10.1038/333560a0. [DOI] [PubMed] [Google Scholar]
- Nobs L, Nestel S, Kulik A, Nitsch C, Atanasoski S. Cyclin D1 is required for proliferation of Olig2-expressing progenitor cells in the injured cerebral cortex. Glia. 2013;61:1443–1455. doi: 10.1002/glia.22533. [DOI] [PubMed] [Google Scholar]
- Noguchi H, Kimura A, Murao N, Matsuda T, Namihira M, Nakashima K. Expression of DNMT1 in neural stem/precursor cells is critical for survival of newly generated neurons in the adult hippocampus. Neurosci. Res. 2015 doi: 10.1016/j.neures.2015.01.014. [DOI] [PubMed] [Google Scholar]
- Nygård M, Wahlström GM, Gustafsson MV, Tokumoto YM, Bondesson M. Hormone-dependent repression of the E2F-1 gene by thyroid hormone receptors. Mol. Endocrinol. Baltim. Md. 2003;17:79–92. doi: 10.1210/me.2002-0107. [DOI] [PubMed] [Google Scholar]
- Ohya W, Funakoshi H, Kurosawa T, Nakamura T. Hepatocyte growth factor (HGF) promotes oligodendrocyte progenitor cell proliferation and inhibits its differentiation during postnatal development in the rat. Brain Res. 2007;1147:51–65. doi: 10.1016/j.brainres.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Ono T, Uehara Y, Kurishita A, Tawa R, Sakurai H. Biological significance of DNA methylation in the ageing process. Age Ageing. 1993;22:S34–S43. doi: 10.1093/ageing/22.suppl_1.s34. [DOI] [PubMed] [Google Scholar]
- Panteleeva I, Boutillier S, See V, Spiller DG, Rouaux C, Almouzni G, Bailly D, Maison C, Lai HC, Loeffler J-P, et al. HP1alpha guides neuronal fate by timing E2F-targeted genes silencing during terminal differentiation. EMBO J. 2007;26:3616–3628. doi: 10.1038/sj.emboj.7601789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr MJ, Manome Y, Tanaka T, Wen P, Kufe DW, Kaelin WG, Fine HA. Tumor-selective transgene expression in vivo mediated by an E2F-responsive adenoviral vector. Nat. Med. 1997;3:1145–1149. doi: 10.1038/nm1097-1145. [DOI] [PubMed] [Google Scholar]
- Pedre X, Mastronardi F, Bruck W, López-Rodas G, Kuhlmann T, Casaccia P. Changed histone acetylation patterns in normal-appearing white matter and early multiple sclerosis lesions. J. Neurosci. Off. J. Soc. Neurosci. 2011;31:3435–3445. doi: 10.1523/JNEUROSCI.4507-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Piao J, Major T, Auyeung G, Policarpio E, Menon J, Droms L, Gutin P, Uryu K, Tchieu J, Soulet D, et al. Human embryonic stem cell-derived oligodendrocyte progenitors remyelinate the brain and rescue behavioral deficits following radiation. Cell Stem Cell. 2015;16:198–210. doi: 10.1016/j.stem.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle NP, Mudhar HS, Collarini EJ, Richardson WD. PDGF receptors in the rat CNS: during late neurogenesis, PDGF alpha-receptor expression appears to be restricted to glial cells of the oligodendrocyte lineage. Development. 1992;115:535–551. doi: 10.1242/dev.115.2.535. [DOI] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- Psachoulia K, Jamen F, Young KM, Richardson WD. Cell cycle dynamics of NG2 cells in the postnatal and ageing brain. Neuron Glia Biol. 2009;5:57–67. doi: 10.1017/S1740925X09990354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff MC, Abney ER, Cohen J, Lindsay R, Noble M. Two types of astrocytes in cultures of developing rat white matter: differences in morphology, surface gangliosides, and growth characteristics. J Neurosci. 1983;3:1289–1300. doi: 10.1523/JNEUROSCI.03-06-01289.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran G, Shanmuganandam K, Bendre A, Muzumdar D, Mujumdar D, Goel A, Shiras A. Epigenetic regulation of DNA methyltransferases: DNMT1 and DNMT3B in gliomas. J. Neurooncol. 2011;104:483–494. doi: 10.1007/s11060-010-0520-2. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM, Hafler DA, Lucchinetti CF. Multiple sclerosis-a quiet revolution. Nat. Rev. Neurol. 2015;11:134–142. doi: 10.1038/nrneurol.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom BR, Yamate CL, Black JA, Waxman SG. Rat optic nerve: disruption of gliogenesis with 5-azacytidine during early postnatal development. Brain Res. 1985;337:41–49. doi: 10.1016/0006-8993(85)91607-5. [DOI] [PubMed] [Google Scholar]
- Riccio A. New endogenous regulators of class I histone deacetylases. Sci. Signal. 2010;3:pe1. doi: 10.1126/scisignal.3103pe1. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Young KM, Tripathi RB, McKenzie I. NG2-glia as multipotent neural stem cells: fact or fantasy? Neuron. 2011;70:661–673. doi: 10.1016/j.neuron.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffini F, Arbour N, Blain M, Olivier A, Antel JP. Distinctive properties of human adult brain-derived myelin progenitor cells. Am J Pathol. 2004;165:2167–2175. doi: 10.1016/S0002-9440(10)63266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Levanon EY, Hueske EA, Ambrose JM, Li JB. Activity-dependent A-to-I RNA editing in rat cortical neurons. Genetics. 2012;192:281–287. doi: 10.1534/genetics.112.141200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Liu A, Li J, Wolubah C, Casaccia-Bonnefil P. Epigenetic memory loss in aging oligodendrocytes in the corpus callosum. Neurobiol Aging. 2008a;29:452–463. doi: 10.1016/j.neurobiolaging.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Sandoval J, Swiss V, Li J, Dupree J, Franklin R, Casaccia-Bonnefil P. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008b;11:1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F, Rössler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells Dayt. Ohio. 2008;26:2875–2883. doi: 10.1634/stemcells.2008-0121. [DOI] [PubMed] [Google Scholar]
- Sher F, Boddeke E, Olah M, Copray S. Dynamic changes in Ezh2 gene occupancy underlie its involvement in neural stem cell self-renewal and differentiation towards oligodendrocytes. PloS One. 2012;7:e40399. doi: 10.1371/journal.pone.0040399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Marinovich A, Barres BA. Purification and characterization of adult oligodendrocyte precursor cells from the rat optic nerve. J Neurosci. 1998;18:4627–4636. doi: 10.1523/JNEUROSCI.18-12-04627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Shin D, Shin J-Y, McManus MT, Ptácek LJ, Fu Y-H. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann. Neurol. 2009;66:843–857. doi: 10.1002/ana.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Howng SYB, Ptáček LJ, Fu Y-H. miR-32 and its target SLC45A3 regulate the lipid metabolism of oligodendrocytes and myelin. Neuroscience. 2012;213:29–37. doi: 10.1016/j.neuroscience.2012.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Götz M, Dimou L. Progenitors in the adult cerebral cortex: cell cycle properties and regulation by physiological stimuli and injury. Glia. 2011;59:869–881. doi: 10.1002/glia.21156. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr. Biol. CB. 2002;12:2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- Sohn DH, Lee KY, Lee C, Oh J, Chung H, Jeon SH, Seong RH. SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J. Biol. Chem. 2007;282:10614–10624. doi: 10.1074/jbc.M610563200. [DOI] [PubMed] [Google Scholar]
- Stacpoole SRL, Spitzer S, Bilican B, Compston A, Karadottir R, Chandran S, Franklin RJM. High yields of oligodendrocyte lineage cells from human embryonic stem cells at physiological oxygen tensions for evaluation of translational biology. Stem Cell Rep. 2013;1:437–450. doi: 10.1016/j.stemcr.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Swiss VA, Casaccia P. Cell-context specific role of the E2F/Rb pathway in development and disease. Glia. 2010;58:377–390. doi: 10.1002/glia.20933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiss VA, Nguyen T, Dugas J, Ibrahim A, Barres B, Androulakis IP, Casaccia P. Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation. PLoS One. 2011;6:e18088. doi: 10.1371/journal.pone.0018088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song C-X, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Tawa R, Ono T, Kurishita A, Okada S, Hirose S. Changes of DNA methylation level during pre- and postnatal periods in mice. Differentiation. 1990;45:44–48. doi: 10.1111/j.1432-0436.1990.tb00455.x. [DOI] [PubMed] [Google Scholar]
- Teplyuk NM, Uhlmann EJ, Wong AH-K, Karmali P, Basu M, Gabriely G, Jain A, Wang Y, Chiocca EA, Stephens R, et al. MicroRNA-10b inhibition reduces E2F1-mediated transcription and miR-15/16 activity in glioblastoma. Oncotarget. 2015 doi: 10.18632/oncotarget.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikoo R, Osterhout DJ, Casaccia-Bonnefil P, Seth P, Koff A, Chao MV. Ectopic expression of p27Kip1 in oligodendrocyte progenitor cells results in cell-cycle growth arrest. J. Neurobiol. 1998;36:431–440. [PubMed] [Google Scholar]
- Tiwari S, Dharmarajan S, Shivanna M, Otteson DC, Belecky-Adams TL. Histone deacetylase expression patterns in developing murine optic nerve. BMC Dev. Biol. 2014;14:30. doi: 10.1186/1471-213X-14-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Macklin WB, Miller RH. Distinct modes of migration position oligodendrocyte precursors for localized cell division in the developing spinal cord. J Neurosci Res. 2009;87:3320–3330. doi: 10.1002/jnr.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann K, Rohde K, Zeller C, Szymas J, Vogel S, Marczinek K, Thiel G, Nürnberg P, Laird PW. Distinct methylation profiles of glioma subtypes. Int. J. Cancer J. Int. Cancer. 2003;106:52–59. doi: 10.1002/ijc.11175. [DOI] [PubMed] [Google Scholar]
- Unterberger A, Andrews SD, Weaver IC, Szyf M. DNA methyltransferase 1 knockdown activates a replication stress checkpoint. Mol Cell Biol. 2006;26:7575–7586. doi: 10.1128/MCB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Rey M, Iruarrizaga-Lejarreta M, Lozano JJ, Aransay AM, Fernandez AF, Lavin JL, Mósen-Ansorena D, Berdasco M, Turmaine M, Luka Z, et al. S-adenosylmethionine levels regulate the schwann cell DNA methylome. Neuron. 2014;81:1024–1039. doi: 10.1016/j.neuron.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. Zurich Switz. 2013a;23:558–564. doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Felicella MM, Coyne T, Phillips JJ, Gorovets D, Huse JT, Kofler J, Lu C, Tihan T, Sullivan LM, et al. Histone 3 lysine 9 trimethylation is differentially associated with isocitrate dehydrogenase mutations in oligodendrogliomas and high-grade astrocytomas. J. Neuropathol. Exp. Neurol. 2013b;72:298–306. doi: 10.1097/NEN.0b013e3182898113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurre OG, Liu J, Haines J, Sandoval J, Nowakowski R, Casaccia P. An integrated approach to design novel therapeutic interventions for demyelinating disorders. Eur. J. Neurosci. 2012;35:1879–1886. doi: 10.1111/j.1460-9568.2012.08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden J-M, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Vogl MR, Reiprich S, Küspert M, Kosian T, Schrewe H, Nave K-A, Wegner M. Sox10 cooperates with the mediator subunit 12 during terminal differentiation of myelinating glia. J. Neurosci. Off. J. Soc. Neurosci. 2013;33:6679–6690. doi: 10.1523/JNEUROSCI.5178-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volarevic V, Erceg S, Bhattacharya SS, Stojkovic P, Horner P, Stojkovic M. Stem cell-based therapy for spinal cord injury. Cell Transplant. 2013;22:1309–1323. doi: 10.3727/096368912X657260. [DOI] [PubMed] [Google Scholar]
- Wake H, Lee PR, Fields RD. Control of local protein synthesis and initial events in myelination by action potentials. Science. 2011;333:1647–1651. doi: 10.1126/science.1206998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Bates J, Li X, Schanz S, Chandler-Militello D, Levine C, Maherali N, Studer L, Hochedlinger K, Windrem M, et al. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell. 2013;12:252–264. doi: 10.1016/j.stem.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Maekawa M. Methylation of DNA in cancer. Adv. Clin. Chem. 2010;52:145–167. doi: 10.1016/s0065-2423(10)52006-7. [DOI] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R. Stage-specific expression of myelin basic protein in oligodendrocytes involves Nkx2.2-mediated repression that is relieved by the Sp1 transcription factor. J. Biol. Chem. 2005;280:16284–16294. doi: 10.1074/jbc.M500491200. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windrem MS, Nunes MC, Rashbaum WK, Schwartz TH, Goodman RA, McKhann G, Roy NS, Goldman SA. Fetal and adult human oligodendrocyte progenitor cell isolates myelinate the congenitally dysmyelinated brain. Nat Med. 2004;10:93–97. doi: 10.1038/nm974. [DOI] [PubMed] [Google Scholar]
- Wolswijk G, Noble M. Identification of an adult-specific glial progenitor cell. Development. 1989;105:387–400. doi: 10.1242/dev.105.2.387. [DOI] [PubMed] [Google Scholar]
- Wu J, Lv Q, He J, Zhang H, Mei X, Cui K, Huang N, Xie W, Xu N, Zhang Y. MicroRNA-188 suppresses G 1 /S transition by targeting multiple cyclin/CDK complexes. Cell Commun. Signal. CCS. 2014;12:66. doi: 10.1186/s12964-014-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Hernandez M, Shen S, Sabo JK, Kelkar D, Wang J, O'Leary R, Phillips GR, Cate HS, Casaccia P. Differential modulation of the oligodendrocyte transcriptome by sonic hedgehog and bone morphogenetic protein 4 via opposing effects on histone acetylation. J Neurosci. 2012a;32:6651–6664. doi: 10.1523/JNEUROSCI.4876-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Huang K, Yu J, Le T, Namihira M, Liu Y, Zhang J, Xue Z, Cheng L, Fan G. Dnmt3a regulates both proliferation and differentiation of mouse neural stem cells. J. Neurosci. Res. 2012b;90:1883–1891. doi: 10.1002/jnr.23077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Zhang W, Chen L, Chen F, Xie H, Xing C, Yu X, Ding S, Chen K, Guo H, et al. MicroRNA-503 inhibits the G1/S transition by downregulating cyclin D3 and E2F3 in hepatocellular carcinoma. J. Transl. Med. 2013;11:195. doi: 10.1186/1479-5876-11-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao ZX, Ginsberg D, Ewen M, Livingston DM. Regulation of the retinoblastoma protein-related protein p107 by G1 cyclin-associated kinases. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4633–4637. doi: 10.1073/pnas.93.10.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Niu Y-F, Huang J, Peng G, Wang L-X, Yang Y-H, Li Y-Q. miR-141 suppresses the growth and metastasis of HCC cells by targeting E2F3. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2014;35:12103–12107. doi: 10.1007/s13277-014-2513-9. [DOI] [PubMed] [Google Scholar]
- Yang N, Zuchero JB, Ahlenius H, Marro S, Ng YH, Vierbuchen T, Hawkins JS, Geissler R, Barres BA, Wernig M. Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol. 2013;31:434–439. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Jin P. Unlocking epigenetic codes in neurogenesis. Genes Dev. 2014;28:1253–1271. doi: 10.1101/gad.241547.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KM, Psachoulia K, Tripathi RB, Dunn SJ, Cossell L, Attwell D, Tohyama K, Richardson WD. Oligodendrocyte dynamics in the healthy adult CNS: evidence for myelin remodeling. Neuron. 2013;77:873–885. doi: 10.1016/j.neuron.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Shi X, Zhang Z, Chen Y, Wu JI. Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proc. Natl. Acad. Sci. U. S. A. 2011;108:12758–12763. doi: 10.1073/pnas.1018510108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li W, Laurent T, Ding S. Small molecules, big roles -- the chemical manipulation of stem cell fate and somatic cell reprogramming. J. Cell Sci. 2012;125:5609–5620. doi: 10.1242/jcs.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. Off. J. Soc. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Dai J, Ma Y, Mi Y, Cui D, Ju G, Macklin WB, Jin W. Dynamics of ten-eleven translocation hydroxylase family proteins and 5-hydroxymethylcytosine in oligodendrocyte differentiation. Glia. 2014;62:914–926. doi: 10.1002/glia.22649. [DOI] [PubMed] [Google Scholar]
- Zheng S, Moehlenbrink J, Lu Y-C, Zalmas L-P, Sagum CA, Carr S, McGouran JF, Alexander L, Fedorov O, Munro S, et al. Arginine methylation-dependent reader-writer interplay governs growth control by E2F-1. Mol. Cell. 2013;52:37–51. doi: 10.1016/j.molcel.2013.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wan H, Xue C, Jiang T, Qian C, Zhang Y. Histone deacetylase 3 implicated in the pathogenesis of children glioma by promoting glioma cell proliferation and migration. Brain Res. 2013;1520:15–22. doi: 10.1016/j.brainres.2013.04.061. [DOI] [PubMed] [Google Scholar]