Abstract
The Saccharomyces cerevisiae Ndt80 protein is the founding member of a class of p53-like transcription factors that is known as the NDT80/PhoG-like DNA-binding family. The number of NDT80-like genes in different fungi is highly variable and their roles, which have been examined in only a few species, include regulation of meiosis, sexual development, biofilm formation, drug resistance, virulence, the response to nutrient stress and programmed cell death. The protein kinase Ime2 regulates the single NDT80 gene present in S. cerevisiae. In this study we used a genetic approach to investigate whether the Aspergillus nidulans Ime2 homolog, ImeB, and/or protein kinases MpkC, PhoA and PhoB regulate the two NDT80-like genes (xprG and ndtA) in A. nidulans. Disruption of imeB, but not mpkC, phoA or phoB, led to increased extracellular protease activity and a defect in mycotoxin production similar to the xprG1 gain-of-function mutation. Quantitative RT-PCR showed that ImeB is a negative regulator of xprG expression and XprG is a negative regulator of xprG and ndtA expression. Thus, in contrast to Ime2, which is a positive regulator of NDT80 in S. cerevisiae, ImeB is a negative regulator as in Neurospora crassa. However, the ability of Ndt80 to autoregulate NDT80 is conserved in A. nidulans though the autoregulatory effect is negative rather than positive. Unlike N. crassa, a null mutation in imeB does not circumvent the requirement for XprG or NdtA. These results show that the regulatory activities of Ime2 and Ndt80-like proteins display an extraordinarily level of evolutionary flexibility.
Keywords: Aspergillus nidulans, XprG, Ndt80, Ime2, nutrient stress
The class of p53-like transcription factors that is known as the NDT80/PhoG-like DNA-binding family (http://pfam.xfam.org/family/PF05224) is found only in the unikont lineage, which includes animals, fungi and amoebozoa. The number of NDT80-like genes varies in different fungi and even varies within the same species/species complex (Katz et al. 2013; Katz and Kelly 2010). Most basidiomycetes appear to have no genes in this class whereas the Mucoromycotina fungi Mucor circinelloides and Rhizopus delemar have six and seven NDT80-like genes, respectively. Within the Ascomycota, the number of genes ranges from zero (Schizosaccharomyces pombe) to six (Fusarium oxysporum strain HDV247).
An analysis of all the Ndt80 homologs in a species has been completed in only three fungi, the haploid ascomycetes Saccharomyces cerevisiae, Aspergillus nidulans, and Neurospora crassa, which possess one, two, and three NDT80-like genes, respectively. In the budding yeast, S. cerevisiae, Ndt80 activates the transcription of more than 150 genes during the middle phase of meiosis (Chu et al. 1998). Ndt80 is required for completion of meiosis, which is triggered by nutrient limitation in yeast (Kupiec et al. 1997). Mutants lacking a functional copy of the NDT80 gene arrest during pachytene in meiosis I at the final nutritional checkpoint (Xu et al. 1995). The opportunistic pathogen Candida albicans is a diploid ascomycete that can switch between yeast and filamentous forms. Only one of the two or three (depending on strain) C. albicans NDT80-like genes has been characterized. CaNdt80 is required for antifungal drug resistance, hyphal growth, biofilm formation and virulence (Chen et al. 2004; Nobile et al. 2012; Sellam et al. 2010).
In the filamentous fungus, A. nidulans, one Ndt80-like protein (XprG) is a positive regulator that controls the response of a large number of genes to carbon starvation (Katz et al. 2013). Extracellular protease, mycotoxin and penicillin production are regulated by XprG (Katz et al. 1996, 2006, 2013). In addition, XprG regulates autolysis, a process involving hyphal fragmentation, and cell death induced by carbon starvation (Katz et al. 2013; Krohn et al. 2014). A second A. nidulans Ndt80-like protein (NdtA) has greater sequence similarity to Ndt80 and is required for sexual reproduction. The filamentous fungus, N. crassa, possesses two proteins (VIB-1 and NCU04729) that are more closely related to XprG and one that is more similar to NdtA (FSD-1) (Hutchison and Glass 2010). VIB-1 is required for expression of genes involved in heterokaryon-incompatibility programmed cell death and, like XprG, is a positive regulator of extracellular protease production (Dementhon et al. 2006; Hutchison and Glass 2010; Xiang and Glass 2002). Both VIB-1 and FSD-1 regulate formation of female sexual structures. The Δfsd-1 mutant is female sterile and defective in ascospore maturation. However, FSD-1 is not required for meiosis (Hutchison and Glass 2010). No phenotypic consequences have been discovered for deletion of NCU04729 (Hutchison and Glass 2010).
In S. cerevisiae, Ime2 is a positive regulator of Ndt80. Activation of NDT80 gene expression involves phosphorylation of the Sum1 repressor, which is bound to the NDT80 promotor, by Ime2 (reviewed in Winter 2012). There is also evidence that Ndt80 requires post-translational activation and that Ime2 plays a role in this step (Sopko et al. 2002; Benjamin et al. 2003). However, it is still not clear whether it is Ime2-dependent phosphorylation that is required for Ndt80 activity (Shubassi et al. 2003; Sopko and Stuart 2004; Wang et al. 2011). The regulation of Ndt80-like proteins by Ime2 homologs has been studied in only one filamentous fungus, N. crassa. In contrast to S. cerevisiae, N. crassa IME-2 is a negative regulator of vib-1 expression (Hutchison et al. 2012). Mutations in ime-2 have no effect on fsd-1 expression (Hutchison and Glass 2010) and no investigations into NCU04729 regulation have been reported. VIB-1 is phosphorylated at a site that matches the Ime2 consensus phosphorylation site. However, amino acid substitutions that were predicted to be phospho-null or phospho-mimetic had no effect on VIB-1-mediated programmed cell death (Hutchison et al. 2012).
In this study, we show that some aspects of the A. nidulans ImeB/XprG/NdtA regulatory pathway are similar to the N. crassa IME-2/VIB-1/FSD-1 pathway but others are not. Like IME-2, ImeB is a negative regulator of xprG expression. However, in N. crassa, ime-2 gene disruption suppresses the defects in extracellular protease production and heterokaryon-incompatibility induced cell death associated with the Δvib-1 mutation and the defect in female sexual development associated with the Δfsd-1mutation (Hutchison et al. 2012; Hutchison and Glass 2010). In contrast, in A. nidulans the requirement for XprG or NdtA is not circumvented by null mutations in imeB. We also show that XprG is a negative regulator of ndtA and xprG expression, though genetic evidence and transcriptional profiling indicate that XprG is usually a transcriptional activator. Thus, the ability of Ndt80-like proteins to regulate the transcription of NDT80-like genes is conserved in S. cerevisiae and A. nidulans even though in yeast the autoregulatory effect is positive whereas in A. nidulans it is negative. These results, coupled with the extreme variability in the number of NDT80-like genes, show that the regulatory activities of Ime2 and Ndt80-like proteins display an extraordinarily level of evolutionary flexibility.
Materials and Methods
Aspergillus strains and growth tests
The A. nidulans strains used in this study are listed in Table 1. The genetic techniques used to construct the strains listed in Table 1 have been described (Clutterbuck 1974). Growth tests were performed at 37° using Aspergillus minimal medium (Cove 1966). In media containing 1% glucose as a carbon source, nitrogen sources were added at a final concentration of 10 mM with the exception of skim milk or bovine serum albumin (BSA), which were used at 1%. In media that contained 1% skim milk or 1% BSA as a carbon source, 10 mM ammonium chloride was used as a nitrogen source. For media that contained skim milk, sodium deoxycholate (0.08%) was used to induce compact colony formation. Sexual development was initiated by growth on solid minimal medium containing sodium nitrate, proline, or alanine as a nitrogen source. After 3 days, air was excluded and the plates were incubated for a further 7–14 days before scoring and image capture using a Leica MZ6 stereomicroscope and Leica IC80 HD digital camera.
Table 1. List of Aspergillus nidulans strains used in this study.
| Strain | Genotypea | Source/Reference |
|---|---|---|
| A1313 | pyrG89; wA3; argB2; ΔnkuA::argB pyroA4; phoBΔ (phoB::pyrGAf); fwA1 chaA1 sE15 nirA14 | FGSC |
| A1338 | pyrG89; wA3; argB2; mpkCΔ (mpkC::pyrGAf); ΔnkuA::argB pyroA4; fwA1 chaA1 sE15 nirA14 | FGSC |
| A1357 | imeBΔ (imeB::pyrGAf); pyrG89; wA3; argB2; ΔnkuA::argB pyroA4; fwA1 chaA1 sE15 nirA14 | FGSC |
| MH2 | biA1; niiA4 | (Sandeman and Hynes 1989) |
| MH97 | pabaA1 yA1 acuE215 | (Sandeman and Hynes 1989) |
| MH11036 | pyroA4 ΔnkuA::argB; riboB2 | (Nayak et al. 2006) |
| MK85 | biA1; xprG1; niiA4 | (Katz et al. 2000) |
| MK414 | pabaA1 yA2; argB2; xprGΔ (xprG::argB) | (Katz et al. 2006) |
| MK422 | biA1; xprGΔ(xprG::argB) | (Katz et al. 2013) |
| MK481 | ndtAΔ (ndtA::pyroAAf); pyroA4 nkuA::argB; riboB2 | (Katz et al. 2013) |
| MK489 | hxkCΔ (hxkC::argB); niiA4 | This study |
| MK490 | biA1 acuE215; hxkDΔ3 (hxkD::argB); niiA4 riboB2 | This study |
| MK505 | ndtAΔ (ndtA::pyroAAf); pyroA4 nkuA::argB; prnΔ309 xprG2; niiA4 | (Katz et al. 2013) |
| MK552 | phoAΔ (phoA::pyroAAf); pyroA4 ΔnkuA::argB; riboB2 | This study |
| MK562 | biA ;veA+ | (Katz et al. 2015) |
| MK577 | pabaA1; phoAΔ (phoA::pyroAAf); pyroA4 ΔnkuA::argB; xprG1 | This study |
| MK578 | phoAΔ (phoA::pyroAAf); pyroA4 ΔnkuA::argB; xprGΔ (xprG::argB) | This study |
| MK582 | imeBΔ (imeB::pyrGAf) pabaA1 yA1 acuE215; ΔnkuA::argB; xprG1; fwA1 | This study |
| MK598 | imeBΔ (imeB::pyrGAf) biA1; argB2; ΔnkuA::argB; niiA4 chA1 | This study |
| MK601 | imeBΔ (imeB::pyrGAf) biA1; argB2; xprGΔ (xprG::argB) | This study |
| MK604 | argB2; pyroA4 ΔnkuA::argB; xprGΔ (xprG::argB) phoBΔ (phoB::pyrGAf) | This study |
| MK606 | xprG1 phoBΔ (phoB::pyrGAf) | This study |
| MK607 | phoBΔ (phoB::pyrGAf) | This study |
| MK608 | pabaA1; mpkCΔ (mpkC::pyrGAf) argB2; ΔnkuA::argB; xprGΔ (xprG::argB) | This study |
| MK609 | mpkCΔ (mpkC::pyrGAf) argB2; pyroA4 ΔnkuA::argB; nirA14 | This study |
| MK612 | mpkCΔ (mpkC::pyrGAf); xprG1 | This study |
| MK643 | imeBΔ (imeB::pyrGAf) biA1; argB2; xprGΔ (xprG::argB); veA+ | This study |
| MK645 | imeBΔ (imeB::pyrGAf) biA1; veA+ | This study |
| MK647 | imeBΔ (imeB::pyrGAf) ndtAΔ (ndtA::pyroAAf) biA1; pyroA4 ΔnkuA::argB; riboB2 nirA14 | This study |
| MK649 | imeBΔ (imeB::pyrGAf) ndtAΔ (ndtA::pyroAAf) biA1; argB2; pyroA4 ΔnkuA::argB; xprG2 niiA4 | This study |
The gene symbols are described in the Aspergillus Genome Database (http://www.aspgd.org/). FGSC, Fungal Genetics Stock Center
Protein kinase mutants
The A. nidulans genome contains two genes that have been designated phoA, AN8261, which encodes the cyclin-dependent protein kinase, and AN4055, which encodes a putative acid phosphatase. The entire AN8261 coding region (chromosome II coordinates 1,314,068-1,315,312; AspGD) was replaced with the Aspergillus fumigatus pyroA gene using a strategy similar to the one described in Nayak et al. (2006). Gene disruption was confirmed by PCR using the primers listed in Supporting Information, Table S1, and Southern blot analysis.
The A1313, A1338 and A1357 strains, which carry disruptions of the A. nidulans phoB (AN1867), mpkC (AN4668) and imeB (AN6243) genes, respectively, were obtained from the Fungal Genetics Stock Center (McCluskey et al. 2010). These strains were crossed to obtain kinase mutants that did not carry the sE15 mutation, which requires supplementation with methionine and, as a consequence, interferes with growth tests. The presence of mpkC:: pyrGAf (mpkCΔ) and phoB:: pyroAAf (phoBΔ) in segregants was confirmed by PCR using the primers listed in Table S1 as these mutants could not be scored based on growth morphology. The imeB::pyrGAf (imeBΔ) mutation results in reduced growth and compact colony morphology.
Assay for extracellular protease activity, sterigmatocystin, and autolysis
To measure production of extracellular proteases in response to carbon or nitrogen starvation, mycelia were grown in minimal medium containing glucose and ammonium tartrate, and then transferred to minimal medium containing no carbon source for 16 hr or no nitrogen source for 4 hr. Filtered culture medium was used in protease enzyme assays as described previously (Katz et al. 1996).
To measure production of sterigmatocystin (ST) in response to carbon starvation, mycelia were grown for 24 hr in minimal medium containing glucose and then transferred to minimal medium containing glucose for 24 hr or no carbon source for 24 or 48 hr. A volume of culture filtrate corresponding to 10 mg of mycelial dry weight was lyophilized and then resuspended in 1 ml of water. The ST was extracted with 1 ml of chloroform and then repeated with 0.5 ml of chloroform. After evaporation of the chloroform, the sample was resuspended in 25 µl of chloroform. ST was detected using a method described previously (Keller et al. 1994). A 5-µl sample of each extract was applied to aluminum-backed, silica thin layer chromatography sheets (Merck, Darmstadt, Germany) and separated using a mixture of benzene and glacial acetic acid (95:5). After drying, the plate was sprayed with 15% AlCl3 dissolved in 95% ethanol, baked at 65° for 15 min, and photographed under 365 nm UV illumination. ST (Sigma, St. Louis, MO) was used as a standard.
The progress of autolysis in submerged cultures following nutrient depletion was monitored as described previously (Katz et al. 2013). For each assay, six flasks containing 50 ml of minimal medium, 10 mM ammonium tartrate and vitamin supplements were each inoculated with 3 × 108 conidia and placed on an orbital shaker. Flasks were removed at 24 or 48 hr intervals, photographed and the weight of dried mycelium recorded. Each strain was assayed three times.
RNA extraction and qRT-PCR
Total RNA was extracted from mycelia transferred to medium containing glucose or no carbon source for 16 hr as described previously (Reinert et al. 1981) and treated with the Ambion Turbo DNA-free Kit (Invitrogen, AM1907, Carlsbad, CA) prior to quantification in a SpectraMax M2e Microplate Reader (Molecular Devices, M2E, Sunnyvale, CA). The primers used in qRT-PCR experiments were designed using the Primer3 program (http://frodo.wi.mit.edu/primer3/) and are listed in Table S1. Each primer pair was first tested with serial dilutions of RNA to determine the linear range of the qRT-PCR assays using the SuperScript III Platinum SYBR Green One-Step qRT-PCR Kits (Invitrogen, 11736). The experiments were performed using a Corbett CAS1200 liquid handling robot and Corbett Rotor-Gene 3000 real-time thermal cycler (QIAGEN, RG3000, Hilden, Germany). In the assays to determine relative transcript levels, 1 ng of total RNA was added to each reaction. A minimum of three independent RNA preparations were assayed.
Data availability Strains available upon request.
Results
Similarity of ImeB, MpkC, PhoA and PhoB to Ime2
The S. cerevisiae Ime2 protein kinase regulates the transcription and activity of the Ndt80 transcription factor. In contrast to S. cerevisiae, which possesses a single gene encoding an Ndt80-like transcription factor, A. nidulans possesses two genes encoding XprG and NdtA (Katz et al. 2006, 2013). Ndt80 shows greater similarity to NdtA (17.1% identity, E value 4.0e–09) than XprG (12.4% identity, E value 1.6). Like Ndt80, NdtA is required for sexual reproduction. We therefore considered the possibility that NdtA might be regulated by the A. nidulans homolog of Ime2 while XprG might be regulated by a different protein kinase. The four protein kinases that showed the greatest similarity to S. cerevisiae Ime2 are listed in Table 2. ImeB, at 781 amino acids in length, is similar in size to S. cerevisiae Ime2, which is 645 amino acids while MpkC, PhoB and PhoA are smaller. However, all four A. nidulans protein kinases show a high degree of similarity to the N-terminal half of Ime2 (Figure S1). The phenotype of the phoA1 deletion strain constructed by Bussink and Osmani (1998) showed some similarities with the xprG1 gain-of-function mutant, including an altered response to phosphate limitation and increased secretion of pigment (Katz et al. 2006). A mutant in which the entire phoA coding region was removed was constructed and deletion mutants for imeB, mpkC and phoB (De Souza et al. 2013) were obtained from the Fungal Genetics Stock Center (McCluskey et al. 2010).
Table 2. Aspergillus nidulans protein kinases showing highest similarity to Saccharomyces cerevisiae Ime2.
| Protein Kinase | No. of Identical Amino Acids | No. of Identical + Similar Amino Acids | Length of Protein (Amino Acids) | % Identity | E Value | Phenotype of Deletion Mutants |
|---|---|---|---|---|---|---|
| ImeB AN6243 | 201 | 445 | 736 | 40.6 | 1.0e–77 | Slow growth, abnormal sexual development, reduced production of sterigmatocystin (Bayram et al. 2009), cold sensitivity (De Souza et al. 2013) |
| MpkC AN4668 | 135 | 305 | 415 | 35.6 | 2.0e–29 | None noted (De Souza et al. 2013; Furukawa et al. 2005) |
| PhoA AN8261 | 115 | 255 | 366 | 29.2 | 8.0e–28 | Pigment secretion increased, conidiation decreased and sexual development increased in response to phosphate limitation (Bussink and Osmani 1998), lethal in combination with ΔphoB (Dou et al. 2003), NaCl sensitivity, marginal hydroxyurea sensitivity (De Souza et al. 2013) |
| PhoB AN1867 | 103 | 224 | 313 | 30.0 | 1.0e–27 | Lethal in combination with ΔphoA (Dou et al. 2003) |
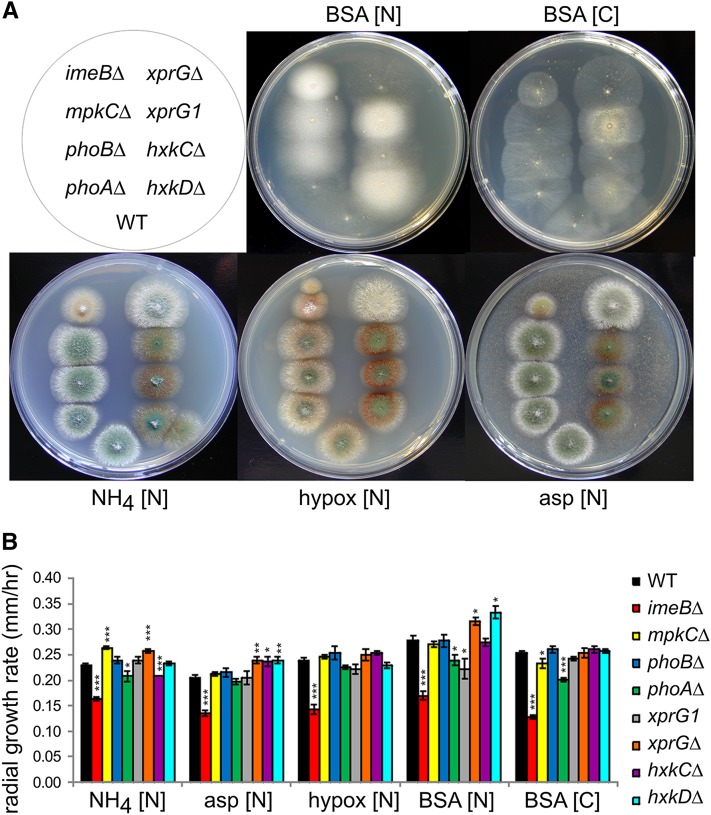
In A. nidulans, colony morphology is determined by the radial growth rate, density of hyphae, conidiation, and production of pigments. As reported previously, we found that the imeBΔ mutation results in reduced radial growth rate and compact colony morphology (Figure 1, Bayram et al. 2009; De Souza et al. 2013), and conidiation in the phoAΔ mutant is reduced (Bussink and Osmani 1998). The colony morphology of the mpkCΔ and phoBΔ mutants was indistinguishable from control strains on Aspergillus complete medium. The hxkC and hxkD genes encode noncatalytic, hexokinase-like proteins (Bernardo et al. 2007; Katz et al. 2000). Genetic evidence suggests that HxkC and HxkD are negative regulators of XprG. The hxkCΔ and hxkDΔ loss-of-function mutations and xprG1 gain-of-function mutations are associated with sparse hyphae and production of brown pigment on media containing nitrogen sources other than protein (Bernardo et al. 2007; Katz et al. 2006). No such growth defects were observed in the imeBΔ, mpkCΔ, phoAΔ, and phoBΔ mutants but the imeBΔ mutant displayed much denser hyphae due to stronger growth on media containing protein (BSA) as a nitrogen source, similar to the xprG1 and hxkDΔ mutants (Figure 1A).
Figure 1.
Phenotypic comparison of protein kinase mutants with strains carrying mutations in the xprG, hxkC and hxkD genes. Colony morphology (A) and radial growth rate (B) on media containing bovine serum albumin (BSA), ammonium tartrate (NH4), hypoxanthine (hypox) or aspartic acid (asp) as a nitrogen source [N] or BSA as a carbon source [C]. The full genotypes for the wild type (WT) (MH2), imeBΔ (MK598), mpkCΔ (MK609), phoBΔ (607), phoAΔ (MK552), xprGΔ (MK422), xprG1 (MK85), hxkCΔ (MK489) and hxkDΔ (MK490) strains are given in Table 1. Radial growth rate was measured between 20 and 44 hr after inoculation. For each strain, the average growth rate and standard error for three colonies grown on three separate plates are shown. An unpaired t-test was used to analyze the data. Values that differed significantly from the value for the WT strain are indicated with asterisks (* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001).
Extracellular protease production is elevated in the imeBΔ mutant
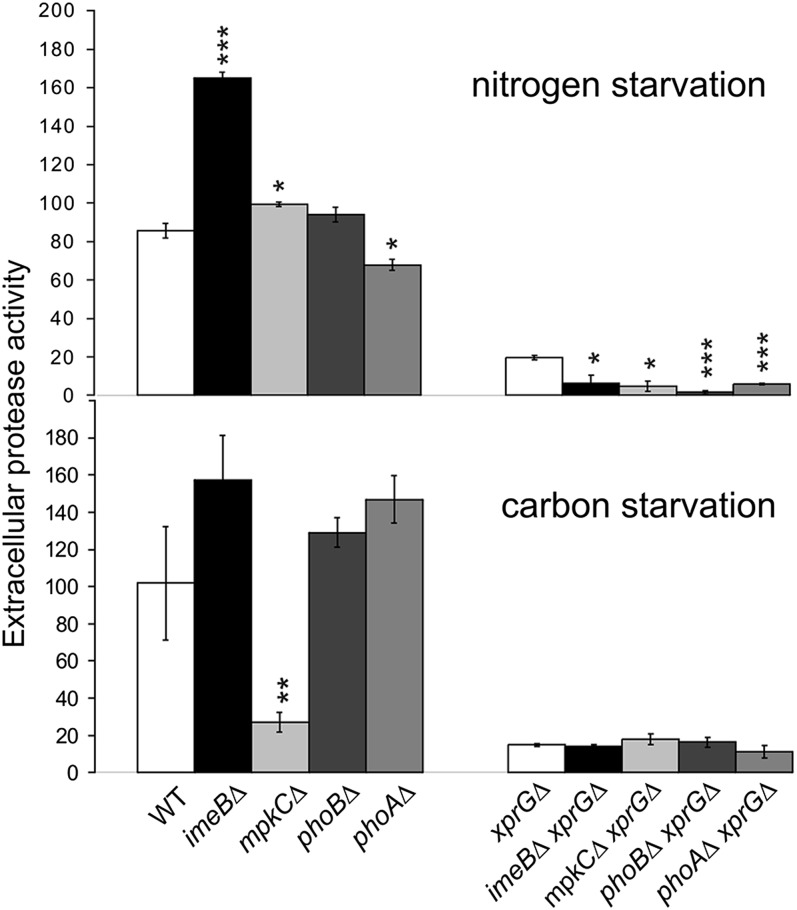
Mutations in xprG but not ndtA alter extracellular protease production (Katz et al. 2013). The xprG1 gain-of-function mutation increases extracellular protease production in response to carbon and nitrogen starvation whereas in loss-of-function mutants (e.g., xprGΔ), protease production is reduced (Katz et al. 1996, 2006, 2008). We examined the effect of the imeBΔ, mpkCΔ, phoAΔ, and phoBΔ mutations on extracellular protease levels and the interaction of the kinase mutations with the two types of xprG mutation using skim milk agar and protease enzymes assays (Figure S2 and Figure 2). These assays showed that the imeB mutation leads to an increase in extracellular protease levels in response to nitrogen limitation and suggested that ImeB might be a negative regulator of protease production. However, protease enzyme activity levels were very low in the imeBΔ xprGΔ double mutant. This result was unexpected. In N. crassa, protease deficiency due to mutations in the xprG homolog, vib-1, is suppressed by the Δime-2 mutation (Hutchison et al. 2012). Previous studies on the A. nidulans imeB gene have reported that some imeBΔ mutant phenotypes are expressed only in a veA+ genetic background (Bayram et al. 2009). Most laboratory strains of A. nidulans carry the veA1 point mutation, which allows asexual spore production in the absence of light (Kim et al. 2002). An imeBΔ xprGΔ veA+ strain was constructed to test whether imeBΔ was able to restore protease production in an xprGΔ mutant with a wild-type version of the VeA light sensor. When tested on solid medium containing skim milk, the phenotype of the imeBΔ xprGΔ veA+ and imeBΔ xprGΔ veA1 strains were indistinguishable—both were protease-deficient (Figure S2). Thus, it is likely that that the increase in protease production in the imeBΔ mutant is mediated by XprG.
Figure 2.
Extracellular protease enzyme activity in the protein kinase mutants. The effect of 4 hr of nitrogen starvation or 16 hr carbon starvation on extracellular protease activity was measured in protease enzyme assays using azocasein as a substrate. Protease activity was calculated as total absorbance units per gram (dry weight) of mycelium and is expressed in arbitrary units. The results are the average for a minimum of three cultures and standard errors are shown. An unpaired t-test was used to analyze the data. Values that differed significantly from the value for the WT strain (for the single mutants) or the xprGΔ strain (for the double mutants) are indicated with asterisks (* P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001). The full genotypes for the WT (MH2), imeBΔ (MK598), mpkCΔ (MK609), phoAΔ (MK552), phoBΔ (607), imeBΔ xprGΔ (MK601), mpkCΔ xprGΔ (MK608), phoAΔ xprGΔ (MK578), phoBΔ xprGΔ (604), and xprGΔ (MK422) strains are given in Table 1.
The phoBΔ deletion mutation did not alter protease production but, during nitrogen starvation, extracellular protease levels were significantly reduced in the phoAΔ mutant (Figure 2). When tested on skim milk agar, no changes in protease levels were detect in the mpkCΔ mutant (Figure S2), but in the enzyme assay protease activity in response to carbon starvation was much lower than in the wild-type strain (Figure 2). Discrepancies between the milk-clearing and protease enzyme assays could be due to the difference in growth conditions used in the two assays. In the enzyme assays no carbon/nitrogen source is provided whereas both milk and low molecular weight carbon/nitrogen sources are present in the skim milk agar.
Protein kinases do not play a role in autolysis but ImeB regulates mycotoxin synthesis
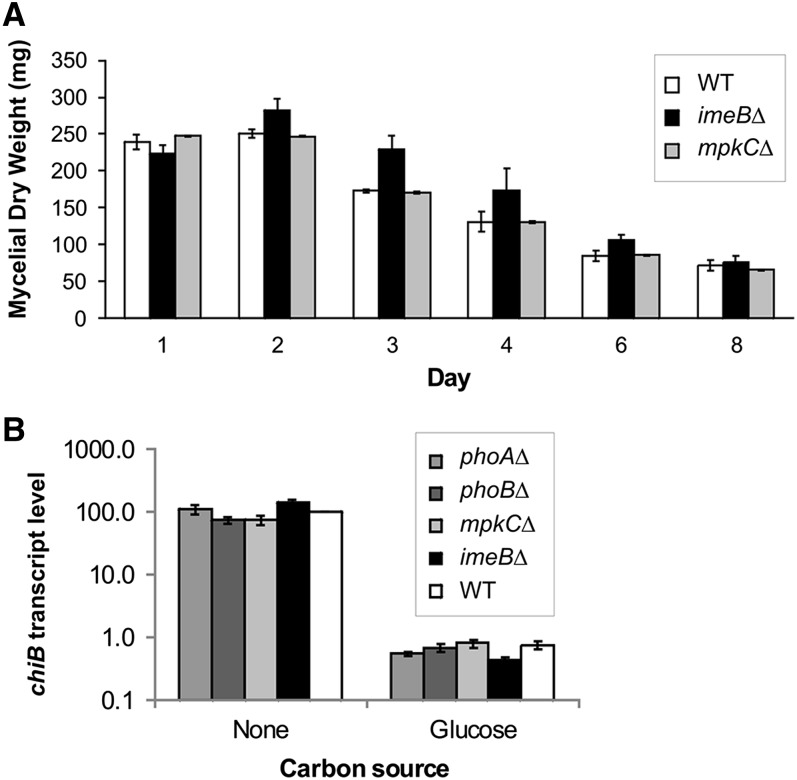
Autolysis, which occurs in stationary submerged cultures after carbon source depletion, is associated with an increase in extracellular protease and chitinase activity, loss of mycelial mass, accumulation of dark pigment, hyphal fragmentation and disintegration (Emri et al. 2004, 2005). Analysis of extracellular protease production indicated that ImeB could be a negative regulator of XprG and, as such, the imeBΔ mutant would be predicted to cause accelerated autolysis, similar to the xprG1 mutant. However, there was no evidence that autolysis occurred more rapidly in the imeBΔ mutant (Figure 3A). Autolysis was also examined in the mpkCΔ mutant, which showed low levels of extracellular protease in response to carbon starvation in protease enzyme assays. The loss of mycelial mass that occurs as a result of autolysis was not delayed in the mpkCΔ mutant as it is in the absence of XprG.
Figure 3.
Effect of protein kinase mutants on autolysis. (A) Loss of mycelial mass was monitored for 8 days in submerged cultures inoculated with 3 × 108 conidia. The average for the three experiments and standard errors are shown. The mycelial dry weight of the two mutants did not differ from the mass of the WT strain at each time point when the data were analyzed using an unpaired t-test. (B) Levels of the chiB transcript relative to actA mRNA levels. The chiB encoded chitinase is a marker of autolysis. The results are the average for three independent RNA preparations, each of which was assayed in duplicate. Transcript levels and standard errors, relative to the levels in the WT control during carbon starvation, are shown. Note the log scale on the x-axis. The data were analyzed using ANOVA after loge transformation. The 95% confidence intervals for all four mutants overlapped with the 95% confidence intervals for the WT strain. The strain numbers for the mutants is given in the legend of Figure 2, and the full genotypes are listed in Table 1.
Carbon starvation–induced autolysis is accompanied by increased expression of the chitinase gene, chiB (Yamazaki et al. 2007), therefore this gene can be used as a reporter of autolysis. Unlike the xprGΔ mutant, which has less than 20% of wild-type levels chiB transcript in carbon-starved mycelia, and the xprG1 mutant, which has elevated levels of chiB mRNA in nutrient-sufficient medium (Katz et al. 2015), none of the kinase mutants showed altered chiB transcript levels (Figure 3B).
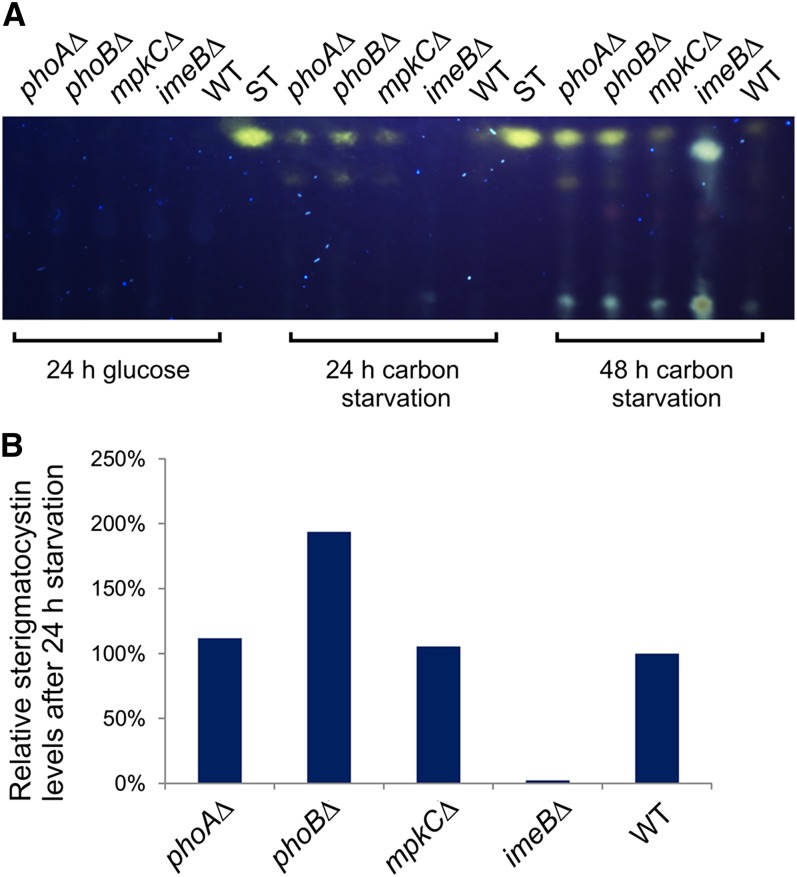
It has previously been reported that ImeB is required for production of the mycotoxin, sterigmatocystin, which is a precursor of aflatoxin (Bayram et al. 2009). We examined production of sterigmatocystin in response to carbon starvation in the kinase mutants (Figure 4). After 24 hr in carbon-free medium, sterigmatocystin was detected in the phoAΔ, phoBΔ and mpkCΔ mutants and control strain but not in the imeBΔ mutant. After 48 hr, a faint band corresponding in position to sterigmatocystin was visible in extracts from the imeBΔ mutant. These results are consistent with the findings of Bayram et al. (2009), in spite of the differences in culture conditions and genetic background, and show that ImeB-mediated regulation of mycotoxin production does not depend on the presence of the wild-type veA+ allele.
Figure 4.
Sterigmatocystin levels in the protein kinase mutants. (A) Sterigmatocystin extracted from culture medium containing glucose or no carbon source was analyzed using thin layer chromatography. Sterigmatocystin fluoresces yellow under ultraviolet light after treatment with AlCl3. Each sample was extracted from culture filtrate corresponding to 2 mg of mycelia (dry weight). In the 48-hr extract from the imeBΔ mutant, there is a bright blue band below the faint yellow sterigmatocystin band. Sterigmatocystin (ST) (Sigma) was applied as a standard. (B) Sterigmatocystin levels, relative to the WT strain, after 24 hr of carbon starvation. Sterigmatocystin levels were quantified using ImageJ software (Rasband 1997–2014). The strain numbers for the mutants are given in the legend of Figure 2 and the full genotypes are listed in Table 1.
XprG is a negative regulator and NdtA is a positive regulator of sexual development
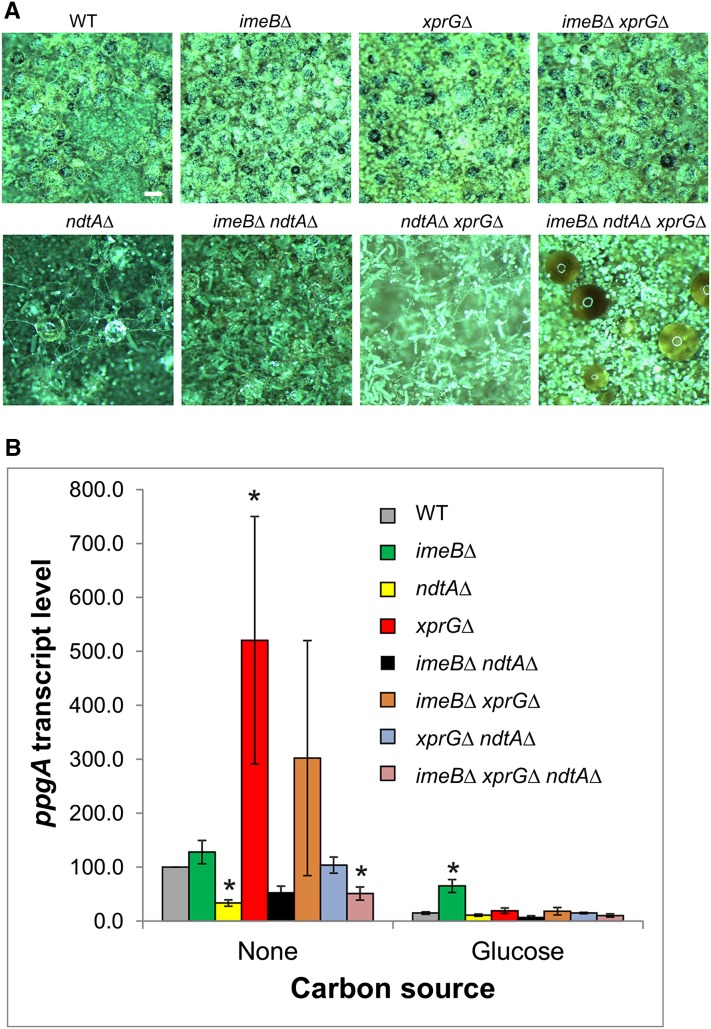
The genetic interactions between the imeBΔ and ndtAΔ mutations were examined to test whether ImeB is involved in regulating NdtA, the second member of the Ndt80/PhoG class of transcription factors. It has previously been shown that NdtA is required for development of sexual fruiting bodies (cleistothecia) in A. nidulans (Katz et al. 2013). As A. nidulans is self-fertile, the ability of imeBΔ, xprGΔ and ndtAΔ single, double and triple mutants to complete sexual development on media containing a variety of nitrogen sources was examined. The imeBΔ, xprGΔ, xprG1 and imeBΔ xprGΔ mutants formed large numbers of cleistothecia in the selfing assays but no cleistothecia were detected in the ndtAΔ, imeBΔ ndtAΔ, ndtAΔ xprGΔ or imeBΔ ndtAΔ xprGΔ mutants (Figure 5A and data not shown). Thus, neither imeBΔ nor xprGΔ are able to suppress the ndtAΔ developmental defect. Large clear spheres were observed in the selfing plates of the ndtAΔ mutants. Similar macroscopic aggregates of Hulle cells, which normally surround the cleistothecium, were seen in strains lacking the MAT1 or MAT2 mating type genes (Paoletti et al. 2007). In the imeBΔ ndtAΔ xprGΔ triple mutant, these aggregates were very large and dark in coloring (Figure 5A).
Figure 5.
Sexual reproduction in imeBΔ, ndtAΔ and xprGΔ mutants. (A) Formation of sexual fruiting bodies (cleistothecia). Mature cleistothecia, which are shiny opaque black spheres, are visible in the pictures in the top row. Each picture shows a 2 mm × 2 mm section of a colony. A white scale bar (200 µm) is shown in the WT picture. The white circles on the clear spherical structures in the second row of pictures are a reflection of the stereomicroscope lights. (B) Levels of the ppgA transcript, which encodes a putative sex pheromone, relative to actA mRNA levels. The results are the average for a minimum of three independent RNA preparations, each of which was assayed in duplicate. Transcript levels and standard errors, relative to the levels in the WT control during carbon starvation, are shown. The data were analyzed using ANOVA after loge transformation. The values marked with an asterisk are outside the 95% confidence intervals for the WT strain. The full genotypes of the WT (MH2), imeBΔ (MK598), ndtAΔ (MK481), xprGΔ (MK422), imeBΔ ndtAΔ (MK647), imeBΔ xprGΔ (MK601), xprGΔ ndtAΔ (MK505), and imeBΔ xprGΔ ndtAΔ (MK649) strains are given in Table 1.
The ppgA gene, which encodes a putative sex pheromone similar to S. cerevisiae α-factor, is upregulated during sexual development (Paoletti et al. 2007). Expression of ppgA and other genes involved in sexual development is increased in the xprGΔ mutant (Katz et al. 2013). To further investigate the role of ImeB in sexual reproduction, ppgA transcript levels were examined in imeBΔ, xprGΔ and ndtAΔ mutants using qRT-PCR (Figure 5B). The results show that ppgA expression is increased during carbon starvation and is dependent on NdtA. In the xprGΔ single mutant but not the xprGΔ ndtAΔ double mutant, carbon-starvation-induced ppgA expression is greatly increased. In nutrient-sufficient conditions, the imeBΔ mutation leads to increased ppgA transcript levels. However, there was no evidence that ImeB repressed ppgA expression during carbon starvation.
Regulation of ndtA and xprG expression
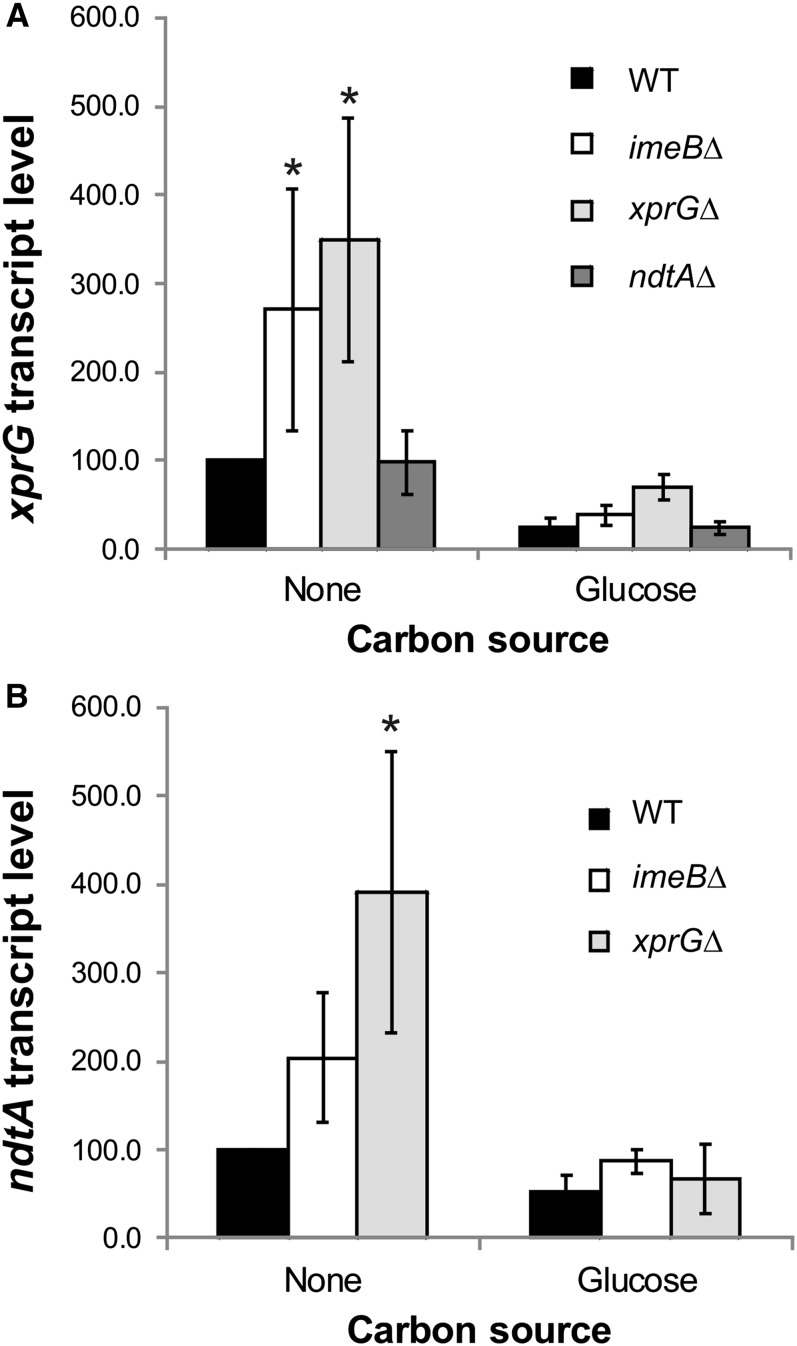
The elevated levels of the ppgA transcript observed in the xprGΔ mutant indicated that XprG might regulate ndtA expression or NdtA activity, whereas the XprG-dependent increase in extracellular protease activity seen in the imeB mutant suggested that ImeB could be involved in the regulation of XprG. We tested these hypotheses by measuring xprG and ndtA transcript levels (Figure 6). The binding sites for the primers used to measure xprG transcript levels are still present in the xprGΔ deletion mutation, which removes codons 248–344. The relative expression of both the xprG and ndtA genes was higher in carbon-starved mycelia than in mycelia that were not subjected to nutrient stress. In the imeBΔ mutant, xprG transcript levels were much higher than in the control strain and ndtAΔ mutant, but only in response to carbon limitation. Carbon-starvation-induced expression of ndtA was greatly increased in the xprGΔ mutant and to a much lesser extent (which was not outside the 95% confidence interval for the control strain) in the imeBΔ mutant. These results are consistent with a model in which ImeB is a negative regulator of xprG expression and XprG is a negative regulator of ndtA expression (Figure 7). NdtA does not appear to regulate xprG expression. In the xprGΔ mutant, carbon-starvation-induced xprG transcript levels are greatly increased, indicating that XprG has an autoregulatory function.
Figure 6.
Effect of the imeBΔ, ndtAΔ and xprGΔ mutation on xprG (A) and ndtA (B) transcript levels relative to actA mRNA levels. The results are the average for three to six independent RNA preparations. Transcript levels and standard errors, relative to the levels in the WT control during carbon starvation, are shown. The data were analyzed using ANOVA after loge transformation. The values marked with an asterisk are outside the 95% confidence intervals for the WT strain. The full genotypes of the WT (MH2), imeBΔ (MK598), ndtAΔ (MK481), and xprGΔ (MK422) strains are given in Table 1.
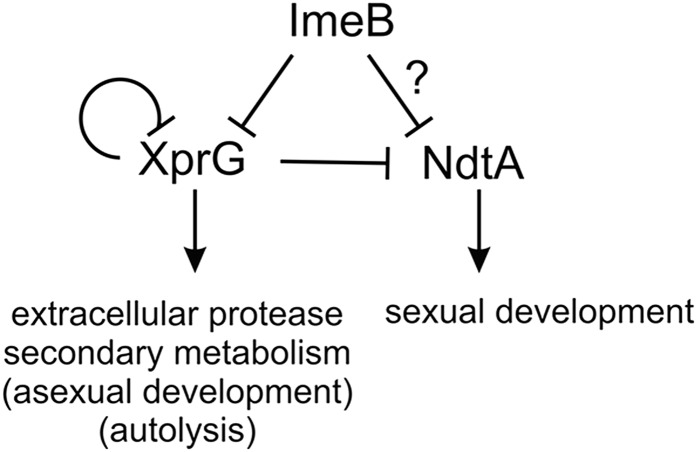
Figure 7.
Model for transcriptional control of the ImeB/XprG/NdtA regulatory pathway. ImeB is a negative regulator of xprG transcription and may also regulate ndtA expression. XprG is a negative regulator of ndtA transcript levels and also has a negative autoregulatory function. XprG plays a major role in the response to carbon starvation, including activation of genes encoding extracellular proteases, secondary metabolism (including the sterigmatocystin biosynthetic pathway), genes induced during autolysis, and genes that are upregulated during asexual development while NdtA is required for sexual development (Katz et al. 2013). There is no evidence that ImeB modulates xprG expression during autolysis or asexual development.
Discussion
The effect of the imeBΔ mutation in A. nidulans differs from similar mutations in S. cerevisiae and N. crassa. Ime2 is positive regulator of NDT80 expression in S. cerevisiae (Winter 2012). In contrast, A. nidulans ImeB and N. crassa IME-2 are negative regulators of xprG and vib-1 expression, respectively (Hutchison et al. 2012). Mutations in the N. crassa vib-1 gene have many phenotypic effects that are similar to loss-of-function mutations in the A. nidulans xprG gene. Both mutations affect programmed cell death, extracellular protease production and conidial pigmentation (Dementhon et al. 2006; Katz et al. 1996, 2006, 2013; Xiang and Glass 2002). The N. crassa Δime-2 mutation suppresses the Δvib-1 defect in heterokaryon-incompatibility induced programmed cell death and extracellular protease production in response to nitrogen starvation but wild-type conidial pigmentation is not restored (Hutchison et al. 2012). The Δime-2 mutation partially restores extracellular protease production even in a strain lacking all three NDT80-like genes (vib-1, fsd-1, and NCU4729) (Hutchison et al. 2012). The Δfsd-1 and Δvib-1 mutants are both defective in female sexual development. The Δime-2 mutation suppresses this defect in the Δfsd-1 mutant but not in the Δvib-1 mutant. The interaction between IME2- and NDT80 -like genes differs in A. nidulans. In contrast to the N. crassa Δime-2 mutation, which suppresses most Δvib-1 and Δfsd-1 defects, the A. nidulans imeBΔ mutation does not suppress the xprGΔ defect in extracellular protease production or the ndtAΔ defect in sexual reproduction. As the N. crassa Δime-2 mutation restores extracellular protease production in a strain lacking all three NDT80-like genes, IME-2 must regulate a parallel signaling pathway that does not exist in A. nidulans. Ime2 homologs have been shown to regulate a number of pathways that do not involve Ndt80-like transcription factors (Irniger 2011).
XprG plays a major role in the response to carbon starvation in A. nidulans (Katz et al. 2013). We have previously proposed that the common feature of Ndt80-like proteins is a role in nutrient sensing, and this may be the original role for this group of transcriptional activators (Katz et al. 2006, 2013). Is ImeB responsible for modulating XprG levels in response to nutrient stress? The imeBΔ mutant has increased levels of extracellular protease, particularly in response to nitrogen starvation, and increased levels of the xprG transcript in response to carbon starvation. However, ImeB does not appear to play a role in autolysis or the induction of chiB in response to carbon nutrient stress. Mutations in imeB do not increase extracellular protease production or xprG and ndtA expression in the absence of nutrient stress, so if ImeB is indeed involved in nutrient signaling it cannot act alone. Another protein must block transcription of these genes when nutrients are present. As previous studies (Katz et al. 2008) have shown that the CreA DNA-binding protein, which mediates carbon catabolite repression, may modulate XprG activity, it is a likely candidate.
In addition to ImeB, a number of other negative regulators of XprG have been identified. Genetic evidence suggests that the AtmA kinase modulates XprG activity (Krohn et al. 2014) and the hexokinase-like proteins HxkC and HxkD regulate XprG activity or expression (Bernardo et al. 2007; Katz et al. 2000).
In contrast to S. cerevisiae and N. crassa, sexual development in A. nidulans is not triggered by nutrient limitation and requires nutrient-sufficient conditions (Dyer and O’Gorman 2012). Yet, we have shown that mRNA levels for the ppgA sex pheromone gene and ndtA regulator of sexual reproduction are elevated in response to nutrient limitation. Thus, it appears that NdtA still retains the capacity to respond to nutrient stress. Whether this has any biological relevance is unknown. The imeBΔ mutation leads to increased ppgA transcript levels in nutrient-sufficient conditions. This result is consistent with the observation that, in N. crassa, the Δime-2 mutant produces abundant female sexual structures in nutrient-sufficient conditions that would normally repress sexual development (Hutchison and Glass 2010).
We have shown that ImeB is a negative regulator of xprG expression. Two observations suggest that ImeB may also regulate ndtA expression. 1) In the absence of nutrient stress, the xprGΔ mutation has no effect on ppgA expression. However, expression of the ppgA gene was increased in the imeBΔ mutant and this increase was NdtA-dependent. 2) A higher level of ndtA mRNA was detected in the imeBΔ mutant, though the level was not outside the 95% confidence interval of the control strain. As there is a higher level of the xprG transcript in the imeBΔ mutant, and XprG is a negative regulator of ndtA expression, we might expect to see a decrease in ndtA expression in an imeBΔ mutant rather than an increase if ImeB exerts no direct control over ndtA expression (i.e., acts only through XprG).
It has been reported that ImeB is required for inhibition of sexual development by light but that no defect was observed in A. nidulans strains carrying the veA1 mutation (Bayram et al. 2009). We have demonstrated that the imeBΔ mutation affects extracellular protease secretion, mycotoxin production, and transcript levels of the ppgA, ndtA and xprG genes in strains carrying the veA1 mutation. Therefore, ImeB must have some functions that are VeA-dependent and some that are VeA-independent.
Within the ascomycetes, there are two groups of Ndt80-like proteins, those that are similar to S. cerevisiae Ndt80, and those that are similar to XprG and VIB-1 (Hutchison and Glass 2010; Katz et al. 2013) (Figure S3; Larkin et al. 2007). It is clear from studies in A. nidulans and N. crassa that the proteins in the two groups have different functions (Hutchison et al. 2012; Hutchison and Glass 2010; Katz et al. 2013). In some fungal species, the number of Ndt80-like transcription factors has expanded (e.g., in F. oxysporum, R. delemar) and in others it has been reduced (e.g., in S. cerevisiae) or eliminated (e.g., in S. pombe and many basidiomycetes). Although, in most cases the function of the Ndt80-like proteins is not known, in the case of C. albicans, it is clear that expansion has been accompanied by the acquisition of new functions. The function of the C. albicans protein that is most closely related to S. cerevisiae meiosis-specific transcription factor Ndt80 (Q5A6P1, Figure S3) has not been reported. CaNdt80, which regulates genes involved in ergosterol biosynthesis, cell separation and hyphal development among many others, belongs to a novel Ndt80-like protein found only in the CTG clade of Saccharomycotina (Sellam et al. 2009, 2010). As CaNdt80 is required for virulence in C. albicans, it has been suggested that the gene duplication event which gave rise to CaNdt80 led to the ability of a number of fungi in the CTG clade to colonize mammalian hosts (Sellam et al. 2010). We have shown here that this extreme diversity in Ndt80-like proteins extends to the regulation of the genes encoding these proteins by Ime2 homologs and the ability of Ndt80-like proteins to regulate their own synthesis. Thus, the Ime2/Ndt80 signaling pathways display great flexibility in adapting to the lifestyle requirements of each species.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr Lisa Mascord for identification of mpkCΔ segregants, and the Fungal Genetics Stock Center (Kansas City, Missouri) for provision of Aspergillus nidulans strains.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.021378/-/DC1
Communicating editor: M. S. Sachs
Literature Cited
- Bayram O., Sari F., Braus G. H., Irniger S., 2009. The protein kinase ImeB is required for light-mediated inhibition of sexual development and for mycotoxin production in Aspergillus nidulans. Mol. Microbiol. 71: 1278–1295. [DOI] [PubMed] [Google Scholar]
- Benjamin K. R., Zhang C., Shokat K. M., Herskowitz I., 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17: 1524–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo S. M. H., Gray K.-A., Todd R. B., Cheetham B. F., Katz M. E., 2007. Characterization of regulatory non-catalytic hexokinases in Aspergillus nidulans. Mol. Genet. Genomics 277: 519–532. [DOI] [PubMed] [Google Scholar]
- Bussink H. J., Osmani S. A., 1998. A cyclin-dependent kinase family member (PHOA) is required to link developmental fate to environmental conditions in Aspergillus nidulans. EMBO J. 17: 3990–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. G., Yang Y. L., Shih H. I., Su C. L., Lo H. J., 2004. CaNdt80 is involved in drug resistance in Candida albicans by regulating CDR1. Antimicrob. Agents Chemother. 48: 4505–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., et al. , 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. [DOI] [PubMed] [Google Scholar]
- Clutterbuck, A. J., 1974 Aspergillus nidulans, pp. 447–510 in Handbook of Genetics, Vol. 1 Bacteria, Bacteriophages and Fungi, edited by R.C. King. Plenum Press, New York. [Google Scholar]
- Cove D. J., 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113: 51–56. [DOI] [PubMed] [Google Scholar]
- De Souza C. P., Hashmi S. B., Osmani A. H., Andrews P., Ringelberg C. S., et al. , 2013. Functional analysis of the Aspergillus nidulans kinome. PLoS One 8: e58008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dementhon K., Iyer G., Glass N. L., 2006. VIB-1 is required for expression of genes necessary for programmed cell death in Neurospora. Eukaryot. Cell 5: 2161–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou X. W., Wu D. L., An W. L., Davies J., Hashmi S. B., et al. , 2003. The PHOA and PHOB cyclin-dependent kinases perform an essential function in Aspergillus nidulans. Genetics 165: 1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer P. S., O’Gorman C. M., 2012. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol. Rev. 36: 165–192. [DOI] [PubMed] [Google Scholar]
- Emri T., Molnar Z., Pusztahelyi T., Pocsi I., 2004. Physiological and morphological changes in autolyzing Aspergillus nidulans cultures. Folia Microbiol. (Praha) 49: 277–284. [DOI] [PubMed] [Google Scholar]
- Emri T., Molnar Z., Pusztahelyi T., Varecza Z., Pocsi I., 2005. The FluG-BrlA pathway contributes to the initialisation of autolysis in submerged Aspergillus nidulans cultures. Mycol. Res. 109: 757–763. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Hoshi Y., Maeda T., Nakajima T., Abe K., 2005. Aspergillus nidulans HOG pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol. Microbiol. 56: 1246–1261. [DOI] [PubMed] [Google Scholar]
- Hutchison E. A., Glass N. L., 2010. Meiotic regulators Ndt80 and Ime2 have different roles in Saccharomyces and Neurospora. Genetics 185: 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison E. A., Bueche J. A., Glass N. L., 2012. Diversification of a protein kinase cascade: IME-2 is involved in nonself recognition and programmed cell death in Neurospora crassa. Genetics 192: 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S., 2011. The Ime2 protein kinase family in fungi: more duties than just meiosis. Mol. Microbiol. 80: 1–13. [DOI] [PubMed] [Google Scholar]
- Katz M. E., Kelly J. M., 2010. Glucose, pp. 291–311 in Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K., Ebbole D. ASM Press, Herndon, VA. [Google Scholar]
- Katz M. E., Flynn P. K., vanKuyk P., Cheetham B. F., 1996. Mutations affecting extracellular protease production in the filamentous fungus, Aspergillus nidulans. Mol. Genet. Genomics 250: 715–724. [DOI] [PubMed] [Google Scholar]
- Katz M. E., Masoumi A., Burrows S. R., Shirtliff C. G., Cheetham B. F., 2000. The Aspergillus nidulans xprF gene encodes a hexokinase-like protein involved in the regulation of the extracellular proteases. Genetics 156: 1559–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M. E., Gray K.-A., Cheetham B. F., 2006. The Aspergillus nidulans xprG (phoG) gene encodes a putative transcriptional activator involved in the response to nutrient limitation. Fungal Genet. Biol. 43: 190–199. [DOI] [PubMed] [Google Scholar]
- Katz M. E., Bernardo S. M., Cheetham B. F., 2008. The interaction of induction, repression and starvation in the regulation of extracellular proteases in Aspergillus nidulans: evidence for a role for CreA in the response to carbon starvation. Curr. Genet. 54: 47–55. [DOI] [PubMed] [Google Scholar]
- Katz M. E., Braunberger K., Yi G., Cooper S., Nonhebel H. M., et al. , 2013. A p53-like transcription factor similar to Ndt80 controls the response to nutrient stress in the filamentous fungus, Aspergillus nidulans. F1000 Res. 2: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M. E., Buckland R., Hunter C. C., Todd R. B., 2015. Distinct roles for the p53-like transcription factor XprG and autophagy genes in the response to carbon starvation. Fungal Genet. Biol. 83: 10–18. [DOI] [PubMed] [Google Scholar]
- Keller N. P., Kantz N. J., Adams T. H., 1994. Aspergillus nidulans verA is required for production of the mycotoxin sterigmatocystin. Appl. Environ. Microbiol. 60: 1444–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Han K., Kim K., Han D., Jahng K., et al. , 2002. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 37: 72–80. [DOI] [PubMed] [Google Scholar]
- Krohn N. G., Brown N. A., Colabardini A. C., Reis T., Savoldi M., et al. , 2014. The Aspergillus nidulans ATM kinase regulates mitochondrial function, glucose uptake and the carbon starvation response. G3 (Bethesda) 4: 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M., Byers B., Esposito R. E., Mitchell A. P., 1997. Meiosis and sporulation in Saccharomyces cerevisae, pp. 889–1036 in The molecular and cellular biology of the yeast saccharomyces, cell cycle and biology, edited by Pringle J. R., Broach J. R., Jones E. W. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., et al. , 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- McCluskey K., Wiest A., Plamann M., 2010. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J. Biosci. 35: 119–126. [DOI] [PubMed] [Google Scholar]
- Nayak T., Szewczyk E., Oakley C. E., Osmani A., Ukil L., et al. , 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Fox E. P., Nett J. E., Sorrells T. R., Mitrovich Q. M., et al. , 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148: 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M., Seymour F. A., Alcocer M. J., Kaur N., Calvo A. M., et al. , 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 17: 1384–1389. [DOI] [PubMed] [Google Scholar]
- Rasband, W. S., 1997–2014 U.S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/.
- Reinert W. R., Patel V. B., Giles N. H., 1981. Genetic regulation of the qa gene cluster of Neurospora crassa: induction of qa messenger ribonucleic acid and dependency on qa-1 function. Mol. Cell. Biol. 1: 829–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandeman R. A., Hynes M. J., 1989. Isolation of the facA (acetyl-Coenzyme A synthetase) and acuE (malate synthase) genes of Aspergillus nidulans. Mol. Genet. Genomics 218: 87–92. [DOI] [PubMed] [Google Scholar]
- Sellam A., Tebbji F., Nantel A., 2009. Role of Ndt80p in sterol metabolism regulation and azole resistance in Candida albicans. Eukaryot. Cell 8: 1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellam A., Askew C., Epp E., Tebbji F., Mullick A., et al. , 2010. Role of transcription factor CaNdt80p in cell separation, hyphal growth, and virulence in Candida albicans. Eukaryot. Cell 9: 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubassi G., Luca N., Pak J., Segall J., 2003. Activity of phosphoforms and truncated versions of Ndt80, a checkpoint-regulated sporulation-specific transcription factor of Saccharomyces cerevisiae. Mol. Genet. Genomics 270: 324–336. [DOI] [PubMed] [Google Scholar]
- Sopko R., Stuart D. T., 2004. Purification and characterization of the DNA binding domain of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80. Protein Expr. Purif. 33: 134–144. [DOI] [PubMed] [Google Scholar]
- Sopko R., Raithatha S., Stuart D., 2002. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 22: 7024–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chang C. Y., Wu J. F., Tung K. S., 2011. Nuclear localization of the meiosis-specific transcription factor Ndt80 is regulated by the pachytene checkpoint. Mol. Biol. Cell 22: 1878–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E., 2012. The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 76: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q., Glass N. L., 2002. Identification of vib-1, a locus involved in vegetative incompatibility mediated by het-c in Neurospora crassa. Genetics 162: 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Ajimura M., Padmore R., Klein C., Kleckner N., 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Yamazaki D., Takaya N., Takagi M., Ohta A., et al. , 2007. A chitinase gene, chiB, involved in the autolytic process of Aspergillus nidulans. Curr. Genet. 51: 89–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.