Abstract
The development and application of ribosome profiling has markedly advanced our understanding of ribosomes and mRNA translation. The experimental approach, which relies on deep sequencing of ribosome-protected mRNA fragments generated by treatment of polyribosomes with exogenous nucleases, provides a transcriptome-wide assessment of translation. The broad application of ribosome profiling has been slowed by the complexity and expense of the protocol. Here, we provide a simplified ribosome profiling method that uses micrococcal nuclease to generate ribosome footprints in crude cellular extracts, which are then purified simply by size selection via polyacrylamide gel electrophoresis. This simplification removes the laborious or expensive purification of ribosomes that has typically been used. This direct extraction method generates gene-level ribosome profiling data that are similar to a method that includes ribosome purification. This protocol should significantly ease the barrier to entry for research groups interested in employing ribosome profiling.
Introduction
The development of ribosome profiling stands as one of the most prominent recent advances in the field of translation, bringing the study of translation solidly into the genomics era (1). This experimental approach, which relies on deep sequencing of the mRNA fragments generated by nuclease treatment of translating ribosomes, and which details the location of the ribosome on the mRNA at the time of digestion (2), has greatly advanced our understanding of ribosome biochemistry and translational regulation. Prominent examples include transcriptome-wide definition of ribosome elongation rates (3), assignment of new coding sequences within viral RNAs (4), assessment of protein chaperone functions (5), and dissection of translational gene expression programs (5–7).
The broad application of ribosome profiling has been hindered, however, by the expense, complexity, and lengthiness of the protocol. Successful application of ribosome profiling typically requires cell lysis, nuclease digestion, purification of ribosomes, extraction of the ribosome-protected mRNA fragments (RPFs), rRNA depletion, and deep sequencing library preparation. Early protocols required upwards of a week to execute all of these steps, and required specialized equipment, such as an ultracentrifuge and density gradient fractionation system (8). More recently, commercial kits have been developed (9) that rely on size exclusion chromatography rather then ultracentrifugation to isolate ribosomes. This approach remains somewhat expensive: around US$250 per sample, not including sequencing, and reduces the time requirement only modestly.
Here, we present a protocol that allows for acquisition of high-quality ribosome profiling data while substantially reducing time requirement and expense. Instead of purification of ribosomes, RNA is directly extracted following nuclease digestion of crude cell extracts, and RPF isolated by gel electrophoresis without further need for rRNA depletion. Time- and labor-intensive RNA precipitation steps have also been minimized throughout the protocol. These simplifications are enabled by the use of micrococcal nuclease (MNase) in place of the more common RNAse I. Under the reaction conditions specified, MNase activity can be precisely controlled, and as a consequence, the rRNA is not markedly degraded. Due to the abundance of ribosomes, the relative stability of their binding to mRNA, and the relatively unique footprint size, the RPFs can then be purified by gel electrophoresis-based size selection alone.
We estimate the cost of this simplified method from cell lysis to library completion at around $50 per sample, an 80% reduction relative to currently available kits, and the protocol can reasonably be completed within two days. These simplifications make ribosome profiling similar in complexity and cost to MNase-seq, a routinely employed procedure in many research groups to map the position of histones on DNA (10–12).
Methods
Cell growth and lysis
Ribosome profiling has been performed in a large variety of cells and tissues, and this protocol can accommodate many inputs. Pre-treatment of cells with cycloheximide, generally used to stall ribosomes prior to lysis and stabilize polyribosomes, should be avoided due to artifacts that it can introduce (13). If using animal tissue as input, flash freezing in liquid nitrogen followed by physical disruption in lysis buffer is a preferred method (8). Regardless of the source of the biological material, the lysates are treated identically in the downstream protocol. The following text describes the protocol for use with adherent, tissue culture cells.
-
(1)
Grow cells to desired confluence. Approximately 5 million cells is generally sufficient for this protocol.
-
(2)
Aspirate media and wash cells twice with 1.5 mL ice cold PBS.
-
(3)
With the second PBS wash still on the plate, remove cells from plate surface using a cell scraper, then pipet into a 1.5 mL tube on ice.
-
(4)
Pellet cells by centrifugation for 2 min at 6,000 rpm in a refrigerated tabletop centrifuge.
-
(5)
Aspirate supernatant, then resuspend cells in 300 μL ice cold lysis buffer. We generally use a lysis buffer consisting of 1% NP-40, 200 mM KOAc, 25 mM K-HEPES pH 7.2, 10 mM MgCl, and 4 mM CaCl2. However, many non-ionic detergents are functional substitutes. Resuspend cells thoroughly by gentle trituration. Inclusion of calcium in the lysis buffer is required for the subsequent nuclease treatment.
-
(6)
Incubate cells on ice in lysis buffer for 5 min.
-
(7)
Pellet cell debris by centrifugation for 5 min at 9,000 rpm using a refrigerated tabletop centrifuge. Collect supernatant, taking care not to disturb the pellet.
Nuclease digestion
A variety of nucleases are effective in generation of RPFs. The two most common choices are RNase I and MNase. MNase generally leaves the ribosome more intact following digestion and yields a more discrete ribosome footprint (14–16), but has a bias to cut to the 5′ of A (17). For this reason, MNase is effective and appropriate for assessing translational efficiency of mRNAs and general ribosome positioning (16), but for applications requiring subcodon resolution, MNase should not be used. Because the method described here does not rely on the purification of ribosomes nor depletion of rRNA prior to RPF isolation, it is necessary to use MNase to obtain high-quality deep sequencing libraries. Because the sensitivity of rRNA to nuclease digestion can vary between species (14, 16), it is important for researchers to independently verify that a clear, discrete RPF is produced by this protocol before proceeding to large-scale experiments.
If performing mRNA-seq in parallel, which will allow for definition of translational efficiency when paired with ribosome profiling, a third of the lysate may be separated prior to nuclease digestion and analyzed at this point by one of many available protocols or kits.
Adjust KOAc concentration of lysate to 100 mM by 1:1 dilution with water.
Add MNase (from a 2 mg/ml stock in 50% glycerol, 1 mM EDTA pH 7, 10 mM Tris pH 8, 50 mM NaCl) to a final concentration of 10 ug/ml.
Incubate for 30min at 37 °C
Footprint isolation by direct post-digestion RNA purification
-
(8)
To the completed digestion reaction, add 1.5 volumes of GT buffer (4 M guanidinium thiocyanate, 25 mM sodium citrate pH 7.0, 0.5% N-lauryl sarcosine, 5 mM EDTA), gently mix, and add 0.75 volumes sodium citrate pH 4.5 buffer saturated phenol, and 5 ul 2-mercaptoethanol. Alternatively, Trizol or other commercially available phenol/chloroform RNA extraction products are acceptable substitutes.
-
(9)
Vortex thoroughly (e.g., 1 min)
-
(10)
Add 0.2 volumes chloroform and vortex 30 s.
-
(11)
Centrifuge 10min at 12,000 rpm using a refrigerated tabletop centrifuge
-
(12)
Transfer the upper phase to a new tube and add 1 volume isopropanol, 0.1 lysate volumes 3 M NaOAc pH 5.2, and 2 μL GlycoBlue or other co-precipitant.
-
(13)
Precipitate 30 min −20 °C, then centrifuge (12,000 rpm) for 15 min at 4 °C.
-
(14)
Decant supernatant, wash the pellet 2x with cold 80% ethanol, then air dry for 10 min or until ethanol is gone.
PNK treatment and footprint isolation
MNase digestion produces a 3′ phosphate and a 5′ hydroxyl fragment. Treatment with polynucleotide kinase is used to reverse the phosphate position, creating a 3′ hydroxyl and a 5′ phosphate, making fragments compatible with most deep sequencing library preparation methods. Following PNK treatment, RPFs are purified by gel electrophoresis. We resuspend the RNA pellet directly in a PNK reaction buffer and then purify the RPF directly following PNK treatment to avoid the RNA precipitation step that often must follow PNK treatment.
Resuspend the pellet in 10 μl PNK solution (1X PNK Buffer, 10 mM ATP, 1 U/uL (1 ul) PNK (NEB).
Incubate at 37 °C for 30 min.
Add 10ul RNA loading dye (80% formamide, 1 mg/ml xylonol blue, 1mg/ml bromophenol blue, 10 mM EDTA).
Prepare the following gel in mini-gel format: 8M urea, 15% acrylamide (29:1 acrylamide:bis), 1X TBE. Warm and shake to solubilize the urea.
To 5ml gel solution, add 20 μl 10% ammonium persulfate and 4μl TEMED. Load onto 0.75 mM mini-gel with no stacking gel. Use 10-well combs and 1XTBE as a running buffer.
Wash urea out of the wells with running buffer using a syringe, then immediately load the 20 μl footprinting reaction. Alongside it, run a ladder or oligonucleotide mobility marker.
Run the gel until the blue dye has left the gel (~1h at 400V).
Put the gel into 25ml 1X TBE in a small dish. Add 1:10000 SybrGreen and incubate for 5 min with shaking. If working with a sample of particularly low abundance, use SybrGold, as the increased sensitivity may be useful in visualizing the band.
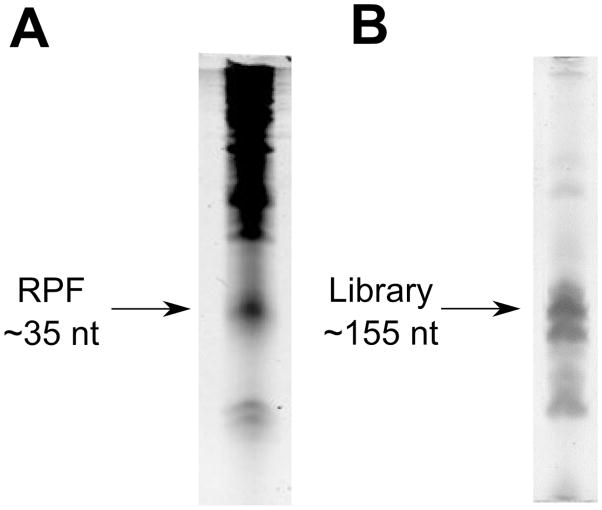
Under UV light, cut out the ~35nt band (Fig. 1A) with a clean razor blade and place it into a 1.5 mL tube.
To the tube, add 400μl 400 mM NaOAc pH 5.2.
Freeze the slice 10min −80 °C.
Thaw the gel slice at 95 °C for 5 min, then crush thoroughly using a pestle or pipet tip.
Extract the RNA from the slice with 3 cycles of (5min 95 °C, 20min vortexing or rocking)
Load the extracted gel slice onto a SpinX column (Corning) and centrifuge for 10 min maximum speed on a tabletop centrifuge. Alternatively, pellet acrylamide by centrifugation at maximum speed for 10 min and collect supernatant, then repeat to remove remaining acrylamide. This method will result in some sample loss, but does not require a SpinX column.
Add 1mL ethanol, 1ul GlycoBlue and incubate for 1 h −20 °C.
Pellet the RPF by centrifugation for 15 min at maximum speed in a tabletop centrifuge at 4 °C. Decant the supernatant and wash the pellet twice with cold 80% ethanol, centrifuging for 5 min after each wash.
Let pellet dry for 10min.
Resuspend in 7ul water.
Figure 1. Purification of ribosome-protected fragment and deep sequencing libraries.
A) The typical result of a ribosome profiling gel electrophoresis purification, as visualized with SybrGreen. MNase generates a RPF that is quite discrete and easily separable form other RNAs. B) Gel electrophoresis purification of the ribosome profiling deep sequencing library. Care must be taken when excising the band below the library, which represent primer dimers.
Library construction
The RNA fragment in this form is compatible with a large variety of deep sequencing library construction techniques. We prefer the NEBNext Small RNA Library Prep Set for Illumina for its simplicity and economic efficiency. A common alternative is an RNA cirularization approach that is commonly employed for ribosome profiling library generation (18). Each of these construction approaches carries particular ligation and sequencing biases, and the approach chosen should be selected with care depending on application. Below, we discuss our most commonly used approach.
We find that reactions of half the manufacturer’s specified volumes are effective. Use 3 μl of the ribosome footprint to prepare a library according to the NEBNext Small RNA Library Prep Set instructions. For PCR amplification, we typically use 14 cycles, although this number will vary depending on input levels.
To purify the deep sequencing library, prepare a 1.5 mm-thick 10% acrylamide minigel containing 1X TBE (and no urea). Magnetic bead-based methods of library purification are not desirable here because of the small difference in size between the cDNA library and primer dimers.
Prepare samples for loading using a 5x loading dye containing 75% glycerol, 0.1% SDS, 1 mg/ml xylenol blue, 1mg/ml bromophenol blue. Run gel at 250 V until yellow band has nearly run off the gel.
Stain the gel with SybrGold as above and excise the cDNA library (~155 nt) using a razor blade under UV illumination (Fig. 1B). Place the gel slice into 375 mM NaOAc pH 5.2. Crush the gel slice and incubate it overnight at 4 °C. Do not boil the sample, as it will result in denaturation of the dsDNA.
Centrifuge the crushed gel solution through a SpinX column or remove acrylamide by centrifugation as above.
Add 1mL ethanol, 2μl GlycoBlue. Precipitate 1h −20 °C.
Spin 15min max speed 4 °C. Wash 2x with cold 80% ethanol.
-
Let pellet dry for 10min.
Resuspend in 10 uL water.
This sample is now ready for Illumina HiSeq deep sequencing protocols.
Computational analysis
Generic mRNA-seq analysis tools are not compatible with ribosome profiling analysis because of the unique quantification requirements (for example, quantification of ribosome profiling typically only considers reads within the coding sequence when measuring levels of translation). Some tools online such as GWIPS-viz (19) offer a useful set of tools for ribosome profiling analysis, and a detailed pipeline for ribosome profiling analysis has been described previously (8). Each of these approaches carries its own set of complexities, and an important future area of progress will be the simplification of ribosome profiling data analysis.
Results and Discussion
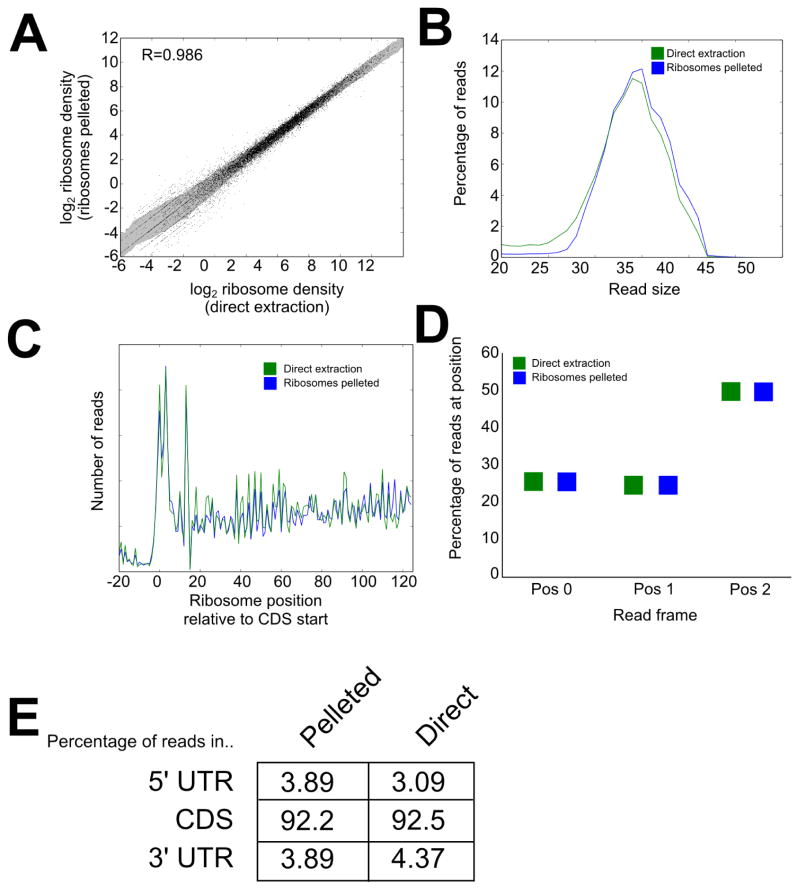
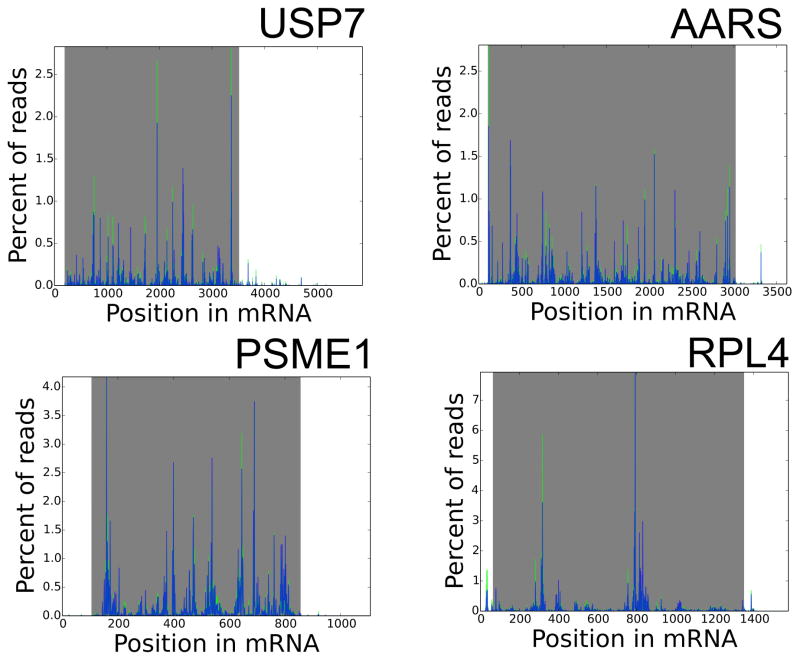
We performed ribosome profiling on immortalized human lymphocytes by the direct extraction method described above and, in parallel, a more standard method that we have used extensively (7, 15) that includes purification of ribosomes by centrifugation through a sucrose cushion, but is otherwise identical. Deep sequencing libraries were prepared and sequenced, generating ~40 million reads per sample. Reads were trimmed of adapters using Cutadapt (20), then mapped using Bowtie (21) allowing no mismatches. We compared the two approaches to assess the validity and performance of the direct extraction method. The read density of RPFs per mRNA was very well conserved, close in performance to technical replicates (1), indicating that the direct extraction method assesses transcriptome-wide translational levels with strong performance (Fig. 2A). The distribution of read length is also very close between the two approaches, centering around 35 nt (Fig. 2B), each of which is somewhat larger than the footprint yielded by digestion by RNase I (8). The distribution of ribosomes along mRNAs is also well recapitulated (Fig. 2C). Each approach yields a distribution of ribosomes with the expected 3 nt periodicity (Fig. 2D) and a strong enrichment of reads in annotated coding sequences (Fig. 2E). There was, however, an increase of ~25% in reads mapping to 3′ UTRs in the direct extraction method, perhaps representing the presence of some RNA binding protein footprints or an increased sensitivity of this method for ribosomes in the 3′ UTR (16). Investigators should consider both of these possibilities when assessing the cause of ribosome footprint detection in non-canonical locations. We also compared the distribution of ribosomes along several individual mRNAs in the two methods of RPF generation (Fig. 3), and observed very similar patterns.
Figure 2. Comparison of direct extraction and ribosome purification methods.
Ribosome profiling libraries were generated from human lymphocytes using the direct extraction method or with inclusion of ribosome purification by pelleting through a sucrose cushion. A) Quantification of ribosome association per mRNA in each sample, normalized by coding sequence length and library size. The shaded area represents ± 1 SD. B) Distribution of read sizes between the two methods. Both approaches generate ribosome footprints with sizes near 35 nt. C) Metagene profile of the distribution of ribosomes relative to the start codon across all mRNAs. D) Quantification of the three-nucleotide periodicity in the direct extraction and ribosome pelleting methods across the entire transcriptome. E) Distribution of reads between the coding sequence and UTRs in each method, each showing a strong enrichment for reads in the coding sequence.
Figure 3. Distribution of ribosomes on individual mRNAs.
The number of ribosome reads at each position along selected mRNAs is shown, with libraries prepared by direct extraction in green and libraries prepared including ribosome pelleting in blue. Shaded area represents the coding sequence.
The data derived from the direct extraction method differed from the protocol that included ribosome purification in the percentage of reads that mapped to our reference mRNA transcriptome: 33.4% mapped when including ribosome pelleting, whereas 26.8% mapped when using direct extraction. Additionally, a substantial number of reads (38.4% for pelleted ribosomes, 13.2% for direct extraction) mapped to rRNA, mostly towards the 3′ end of the 28S rRNA. Each method yielded around 60% of its reads mappable to the genome (61.0% for pelleted ribosomes, 58.1% for direct extraction). Overall, this tradeoff of speed and simplicity in exchange for a modest reduction (~20%) in read depth is one that will likely be worth making in many experiments.
The simplicity of this protocol may offer an advantage beyond its ease of use: the reduced number of steps involved may increase its reproducibility. A recent survey of public ribosome profiling datasets found a troublingly dramatic magnitude of variation between experiments, even those that used ostensibly identical methods on the same cell types (22). To this point, the yield of RPF following ribosome purification is often quite variable in our hands, likely depending on the efficiency of ribosome pellet resuspension, which can be difficult. In contrast, the direct extraction method has proven to be quite consistent in its yields. Decreasing the number of points where small differences may arise between individual researchers performing these methods may help to decrease experimental variability, ultimately helping to distinguish noise from biological differences.
By simplifying and shortening this protocol while reducing its expense, ribosome profiling becomes more viable for large-scale studies. We also hope that these simplifications serve to democratize the method, making it accessible to more research groups that lack the biochemical expertise involved in carrying out most previous protocols. As the community aims to standardize ribosome profiling protocols (22), the protocol we discuss here may become one of several employed by researchers: this MNase/direct extraction protocol can serve as a rapid, robust snapshot of transcriptome-wide translational efficiency, while other more laborious protocols can provide more detailed biochemical data regarding ribosome elongation and dynamics.
Ribosome profiling can be performed without inclusion of a ribosome purification step
Gene-level translation data acquired without ribosome purification closely match data generated by methods that include ribosome purification
This simplification makes ribosome profiling simple, faster, and less expensive to perform
Acknowledgments
We thank the Shenolikar and Nicchitta laboratories for helpful feedback, as well as the anonymous peer reviewers for their thoughtful critiques. This work was funded by an Individual Research Grant (NMRC/GMS/1252/2010) and Translational Clinical Research Flagship Award entitled “National Parkinson’s Disease Translational Clinical Research Programme” from National Medical Research Council of Singapore and Duke/Duke-NUS Collaborative Grant from Ministry of Health, Singapore (to SS), a Duke/Duke-NUS Research Collaboration Project funded by the Singapore Ministry of Health (to SS and CVN), and a grant from the NIH (GM101533, CVN),
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. The EMBO journal. 1988;7(11):3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingolia NT, Lareau LF, Weissman JS. Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell. 2011 doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingolia NT, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell reports. 2014;8(5):1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreev DE, et al. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife. 2014;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485(7396):55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reid DW, Chen Q, Tay AS, Shenolikar S, Nicchitta CV. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell. 2014;158(6):1362–1374. doi: 10.1016/j.cell.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature protocols. 2012;7(8):1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeberg L, Kuersten S, Syed F. Isolate and sequence ribosome-protected mRNA fragments using size-exclusion chromatography. Nat Meth. 2013;10(5) [Google Scholar]
- 10.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nature genetics. 2010;42(4):343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132(5):887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Gerashchenko MV, Gladyshev VN. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic acids research. 2014;42(17):e134. doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid DW, Nicchitta CV. Primary Role for Endoplasmic Reticulum-bound Ribosomes in Cellular Translation Identified by Ribosome Profiling. The Journal of biological chemistry. 2012;287(8):5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miettinen TP, Bjorklund M. Modified ribosome profiling reveals high abundance of ribosome protected mRNA fragments derived from 3′ untranslated regions. Nucleic acids research. 2015;43(2):1019–1034. doi: 10.1093/nar/gku1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dingwall C, Lomonossoff GP, Laskey RA. High sequence specificity of micrococcal nuclease. Nucleic acids research. 1981;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafner M, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17(9):1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michel AM, et al. GWIPS-viz: development of a ribo-seq genome browser. Nucleic acids research. 2014;42(Database issue):D859–864. doi: 10.1093/nar/gkt1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17(1):10–12. [Google Scholar]
- 21.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connor P, Andreev D, Baranov P. Surveying the relative impact of mRNA features on local ribosome profiling read density in 28 datasets. 2015 doi: 10.1038/ncomms12915. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]