Abstract
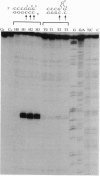
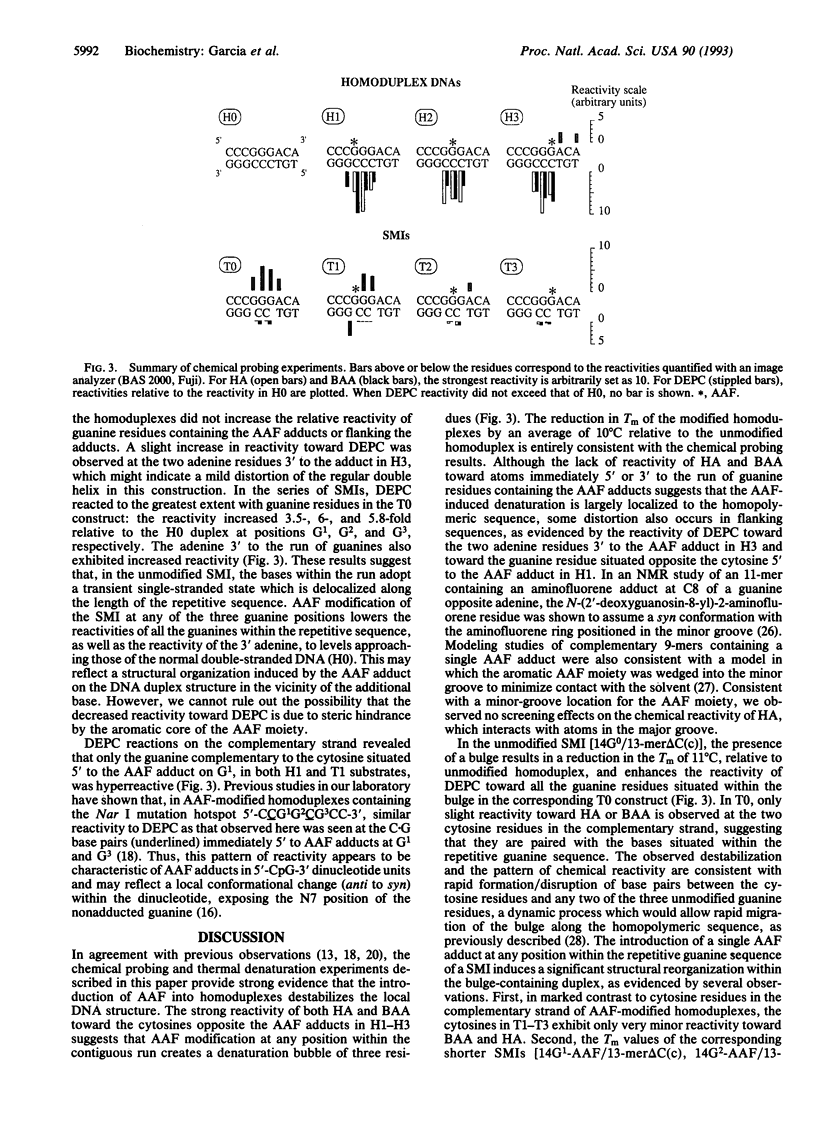
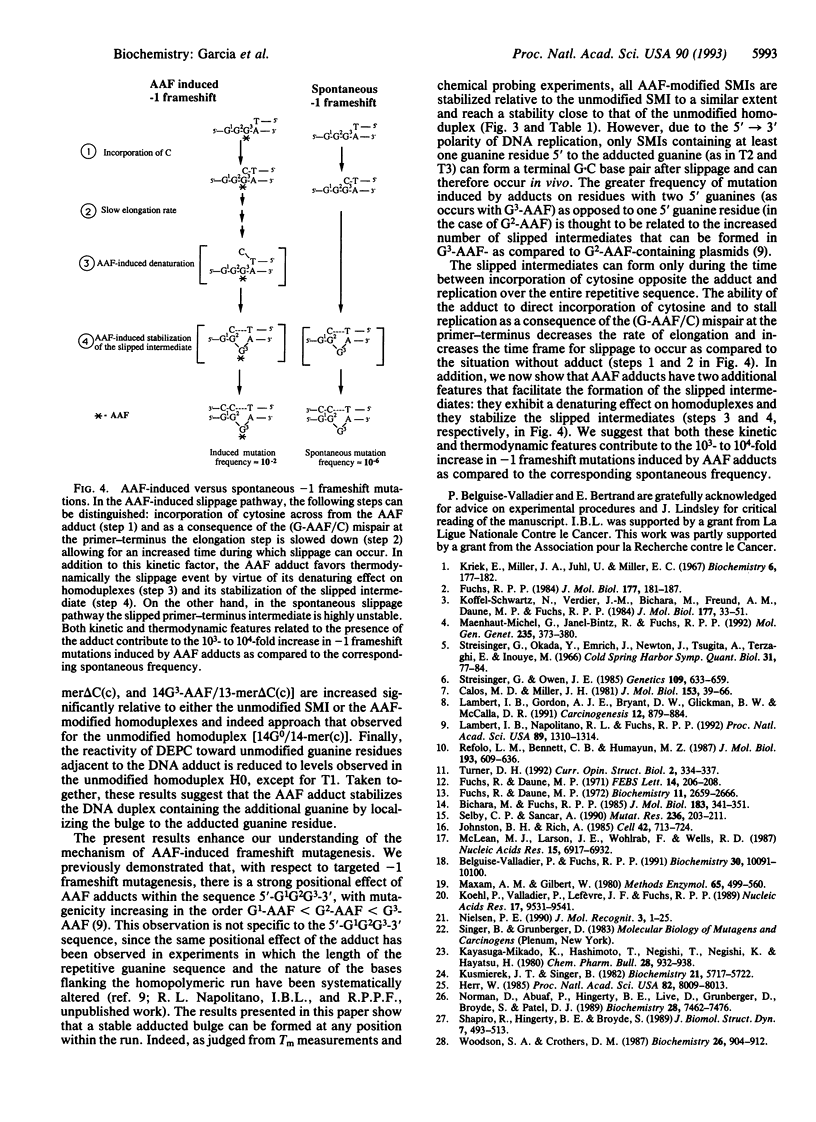
Chemical carcinogens such as the aromatic amide 2-acetylaminofluorene (AAF) are known to induce -1 frameshift mutation hotspots at repetitive sequences. This mutagenesis pathway was suggested to involve slipped intermediates formed during replication. To investigate the stability and structure of such intermediates we have constructed DNA duplexes containing single AAF adducts within a run of three guanine residues. The strand complementary to that bearing the AAF adducts contained either the wild-type sequence (homoduplexes) or lacked one cytosine directly opposite the run of guanines containing the AAF adduct and thus modeled the putative slipped mutagenic intermediates (SMIs). The melting temperature of AAF-modified homoduplexes or the unmodified SMI was reduced by approximately 10 degrees C relative to the unmodified homoduplex. Surprisingly, AAF adducts stabilized the SMIs as evidenced by an increase in melting temperature to a level approaching that of the unmodified homoduplex. The chemical probes hydroxylamine and bromoacetaldehyde were strongly reactive toward cytosine residues opposite the adduct in AAF-modified homoduplexes, indicating adduct-induced denaturation. In contrast, no cytosine reactivities were observed in the AAF-modified SMIs, suggesting that the two cytosines were paired with unmodified guanines. Use of diethyl pyrocarbonate to probe the guanine residues showed that all three guanines in the unmodified SMI adopted a transient single-stranded state which was delocalized along the repetitive sequence. However, when an AAF adduct was present, reduced diethyl pyrocarbonate reactivity at guanines adjacent to the adduct in AAF-modified SMIs reflected localization of the bulge to the adducted base. Our results suggest that AAF exerts a local denaturing and destabilizing effect within the homoduplex which is alleviated by the formation of a bulge. The stabilization by the AAF adduct of the SMIs may contribute to the dramatic increase in -1 frameshift mutation frequency induced by AAF adducts in repetitive sequences.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belguise-Valladier P., Fuchs R. P. Strong sequence-dependent polymorphism in adduct-induced DNA structure: analysis of single N-2-acetylaminofluorene residues bound within the NarI mutation hot spot. Biochemistry. 1991 Oct 22;30(42):10091–10100. doi: 10.1021/bi00106a005. [DOI] [PubMed] [Google Scholar]
- Bichara M., Fuchs R. P. DNA binding and mutation spectra of the carcinogen N-2-aminofluorene in Escherichia coli. A correlation between the conformation of the premutagenic lesion and the mutation specificity. J Mol Biol. 1985 Jun 5;183(3):341–351. doi: 10.1016/0022-2836(85)90005-1. [DOI] [PubMed] [Google Scholar]
- Fuchs R., Daune M. Physical studies on deoxyribonucleic acid after covalent binding of a carcinogen. Biochemistry. 1972 Jul 4;11(14):2659–2666. doi: 10.1021/bi00764a017. [DOI] [PubMed] [Google Scholar]
- Herr W. Diethyl pyrocarbonate: a chemical probe for secondary structure in negatively supercoiled DNA. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8009–8013. doi: 10.1073/pnas.82.23.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston B. H., Rich A. Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell. 1985 Oct;42(3):713–724. doi: 10.1016/0092-8674(85)90268-5. [DOI] [PubMed] [Google Scholar]
- Koehl P., Valladier P., Lefèvre J. F., Fuchs R. P. Strong structural effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Nucleic Acids Res. 1989 Dec 11;17(23):9531–9541. doi: 10.1093/nar/17.23.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffel-Schwartz N., Verdier J. M., Bichara M., Freund A. M., Daune M. P., Fuchs R. P. Carcinogen-induced mutation spectrum in wild-type, uvrA and umuC strains of Escherichia coli. Strain specificity and mutation-prone sequences. J Mol Biol. 1984 Jul 25;177(1):33–51. doi: 10.1016/0022-2836(84)90056-1. [DOI] [PubMed] [Google Scholar]
- Kriek E., Miller J. A., Juhl U., Miller E. C. 8-(N-2-fluorenylacetamido)guanosine, an arylamidation reaction product of guanosine and the carcinogen N-acetoxy-N-2-fluorenylacetamide in neutral solution. Biochemistry. 1967 Jan;6(1):177–182. doi: 10.1021/bi00853a029. [DOI] [PubMed] [Google Scholar]
- Lambert I. B., Napolitano R. L., Fuchs R. P. Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1310–1314. doi: 10.1073/pnas.89.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenhaut-Michel G., Caillet-Fauquet P. Effect of umuC mutations on targeted and untargeted ultraviolet mutagenesis in bacteriophage lambda. J Mol Biol. 1984 Jul 25;177(1):181–187. doi: 10.1016/0022-2836(84)90064-0. [DOI] [PubMed] [Google Scholar]
- Maenhaut-Michel G., Janel-Bintz R., Fuchs R. P. A umuDC-independent SOS pathway for frameshift mutagenesis. Mol Gen Genet. 1992 Nov;235(2-3):373–380. doi: 10.1007/BF00279383. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLean M. J., Larson J. E., Wohlrab F., Wells R. D. Reaction conditions affect the specificity of bromoacetaldehyde as a probe for DNA cruciforms and B-Z junctions. Nucleic Acids Res. 1987 Sep 11;15(17):6917–6935. doi: 10.1093/nar/15.17.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. E. Chemical and photochemical probing of DNA complexes. J Mol Recognit. 1990 Feb;3(1):1–25. doi: 10.1002/jmr.300030102. [DOI] [PubMed] [Google Scholar]
- Norman D., Abuaf P., Hingerty B. E., Live D., Grunberger D., Broyde S., Patel D. J. NMR and computational characterization of the N-(deoxyguanosin-8-yl)aminofluorene adduct [(AF)G] opposite adenosine in DNA: (AF)G[syn].A[anti] pair formation and its pH dependence. Biochemistry. 1989 Sep 5;28(18):7462–7476. doi: 10.1021/bi00444a046. [DOI] [PubMed] [Google Scholar]
- Refolo L. M., Bennett C. B., Humayun M. Z. Mechanisms of frameshift mutagenesis by aflatoxin B1-2,3-dichloride. J Mol Biol. 1987 Feb 20;193(4):609–636. doi: 10.1016/0022-2836(87)90344-5. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Structure and function of the (A)BC excinuclease of Escherichia coli. Mutat Res. 1990 Sep-Nov;236(2-3):203–211. doi: 10.1016/0921-8777(90)90005-p. [DOI] [PubMed] [Google Scholar]
- Shapiro R., Hingerty B. E., Broyde S. Minor-groove binding models for acetylaminofluorene modified DNA. J Biomol Struct Dyn. 1989 Dec;7(3):493–513. doi: 10.1080/07391102.1989.10508506. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985 Apr;109(4):633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson S. A., Crothers D. M. Proton nuclear magnetic resonance studies on bulge-containing DNA oligonucleotides from a mutational hot-spot sequence. Biochemistry. 1987 Feb 10;26(3):904–912. doi: 10.1021/bi00377a035. [DOI] [PubMed] [Google Scholar]