Abstract
Centralized nervous systems (NSs) and complex brains are among the most important innovations in the history of life on our planet. In this context, two related questions have been formulated: How did complex NSs arise in evolution, and how many times did this occur? As a step towards finding an answer, we describe the NS of several representatives of the Xenacoelomorpha, a clade whose members show different degrees of NS complexity. This enigmatic clade is composed of three major taxa: acoels, nemertodermatids and xenoturbellids. Interestingly, while the xenoturbellids seem to have a rather ‘simple’ NS (a nerve net), members of the most derived group of acoel worms clearly have ganglionic brains. This interesting diversity of NS architectures (with different degrees of compaction) provides a unique system with which to address outstanding questions regarding the evolution of brains and centralized NSs. The recent sequencing of xenacoelomorph genomes gives us a privileged vantage point from which to analyse neural evolution, especially through the study of key gene families involved in neurogenesis and NS function, such as G protein-coupled receptors, helix-loop-helix transcription factors and Wnts. We finish our manuscript proposing an adaptive scenario for the origin of centralized NSs (brains).
Keywords: Xenacoelomorpha, acoela, cephalization, brain, nervous system, evolution
1. The phylogenetic placement of Xenacoelomorpha
Acoels are mostly marine bilateral worms with a simple body plan. They are triploblastic and acoelomate, with an outer epidermis of multiciliated cells and a single gut opening. Together with nemertodermatids, they constitute a taxon called Acoelomorpha, which is characterized by shared morphological features such as the epidermal ciliation, intestinal organization, certain glandular and sensory structures, and the limited presence of an extracellular matrix [1–7]. Recent phylogenetic analysis maintains that together with xenoturbellids [8], which form another taxon of considerably larger marine worms, they constitute the phylogenetic group Xenacoelomorpha [9–11]. This relationship is also supported by morphological similarities [1,3,7,12–18], for instance the ultrastructure of the ciliary tips and the system of epidermal ciliary rootlets [5,6,15,18].
However, historically, the phylogenetic relationship of the acoelomorph worms (and also Xenoturbella) with other metazoans has been a controversial issue. Traditionally, they were classified within the Platyhelminthes [19], but with the introduction of molecular characters in phylogenetic analysis, the acoelomorph relationships were reassessed and they were placed as the first offshoot of the Bilateria [20,21]. For this reason, acoels, for instance, were taken as good proxies for the complexity of an ancestral bilaterian animal [22]. This phylogenetic position was confirmed later on by deep phylogenomic analysis [10]. When the relationships between these animals seemed finally settled, a study using alternative methods and datasets suggested that Acoelomorpha instead represented a deuterostomian group with affinities to the Ambulacraria (the group formed by echinoderms and hemichordates) [9]. In all cited phylogenomic studies, the Acoelomorpha plus Xenoturbellida form what is now considered to be a clear monophyletic group: the above-mentioned Xenacoelomorpha.
Whether Xenacoelomorpha is actually placed as a basal bilaterian group or is included within Deuterostomia remains a matter of debate. However, the monophyletic nature of Xenacoelomorpha still allows us to study specific genomic and morphological changes within this group, irrespective of its metazoan affinities. We are especially interested in the Xenacoelomorpha nervous system (NS), as it seems to present certain evolutionary structural changes which have led to the establishment of a more elaborated brain architecture. In the following paragraphs, we will describe the structure of xenacoelomorph NSs, though we will pay special attention to the acoel species for they are the most studied so far.
2. Clarifying the used terminology
Before starting the comparison between the xenacoelomorphs' NSs, we need to clarify some terminology in order to avoid usual misunderstandings. We decided to follow (as in previous papers of our laboratory) the glossary for invertebrate neuroanatomy provided by Richter et al. [23] and the terminology used in Raikova's PhD thesis [24] (followed by her and other neuroanatomists). However, there are some terms that we should explain here. The major point to consider in this paper is what we understand by anterior nerve centralization, or a ‘brain-like structure’. Understanding that other authors might prefer other definitions, we use these terms here for any ensemble or ‘knot-like’ mass of nerve cells forming a defined anterior structure, without implying a necessary homology to the true brains of the so-called ‘higher’ animals. Specifically, only the anterior centralization that contains a central neuropile surrounded by a cortex of nerve cells will be here considered a brain or ‘true’ brain. In addition we should explain what we understand by a nerve net. We will be calling a nerve net the arrangement of the NS in the form of a diffuse plexus, with the shape of a network, formed by neural cell bodies plus neurites, and where the impulses travel through them in no preferential direction (see also [19, pp. 33–34, 83]). Sometimes we use the term nerve condensation to describe the concentration of a few neurons forming a cluster, but without the complexity of a ganglion. Finally, it is important to emphasize what we call neurite bundles. Neurite bundles are simple clusters of neurites arranged in parallel, giving rise to a bunch of tracks that extend longitudinally. They are not arranged in the form of a neuropile surrounded by a cellular cortex as occurs in a typical cord [6,23].
3. Comparative analysis of Xenacoelomorpha nervous system morphology
For a long time it has been clear that the use of different morphological characters allows us to generate hypotheses regarding the specific phylogenetic affinities of different clades. One set of characters that has been used regularly in this and other groups of animals is the neuroanatomical characters [25–28]. We have already shown elsewhere that the architecture of the NS within the Xenacoelomorpha shows different grades of NS morphological complexity [6] (see also fig. 1 in [29]). This feature of NS organization has made the Xenacoelomorpha an especially interesting group for the study of cephalization processes. However, an important question remains to be better explained: how are the different NSs specifically arranged in the members of this clade?
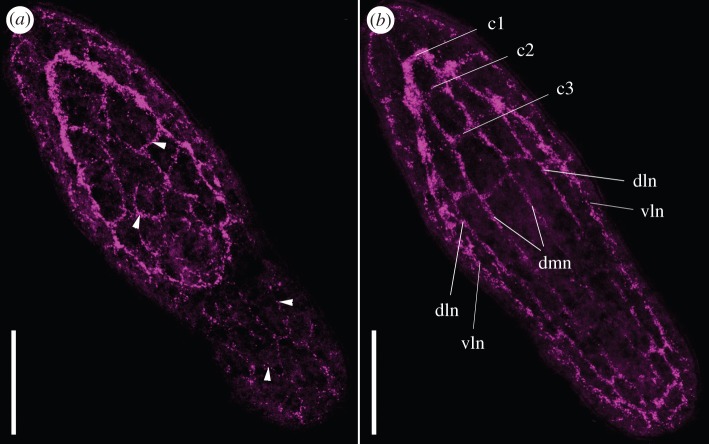
Figure 1.
Confocal microscopy stacks of S. roscoffensis juvenile nervous system stained with the synaptotagmin antibody. Anterior part is up. (a) Ventral view of the outer layers of a whole specimen. Arrowheads point to the thin commissures, part of the nerve net. (b) Dorso-central projection of the juvenile nervous system with the two lobes of the brain, connected by three commissures. c1, first brain commissure; c2, second brain commissure; c3, third brain commissure; dln, dorso-lateral neurite bundles; dmn, dorsomedial neurite bundles; vln, ventro-lateral neurite bundles. Scale bar: (a,b) 35 µm.
The sister group to all acoelomorphs, the Xenoturbellida [8,9,30], is represented by only two nominal species: Xenoturbella bocki [31] and Xenoturbella westbladi [32]; however, newly identified specimens have been collected from the Pacific [33]. These worms are notably larger than the acoelomorphs (up to 4 cm in length) but share with them a relatively simple morphology. Members of Xenoturbellida possess the simplest neuroanatomical organization and ultrastructure of the Xenacoelomorpha, and probably one of the simplest of all bilaterians. This NS is organized as a complete basal intraepidermal nerve net (fig. 1 in [29]) with interwoven neuronal fibres. Neither brain or brain-like structures as nerve rings are present; nor are neurite bundles or similar arrangements [18,31,34]. The presence of nerve processes is completely restricted to the epidermis and the subepidermal membrane complex (SMC) [18], with a lack of nerve structures in the subepidermal muscle layer and in the parenchyma [18,34]. The absence of neural structures below this layer and the lack of obvious direct musculature innervation led some authors to suggest that neuronal substances were transmitting individually from the basiepidermal nerve net to the muscle fibres, or that these animals relied on the use of muscular pacemakers [34]. The unique sensory organs found in Xenoturbella are the statocyst and the three sense-furrows (two lateral and one central) [31]. The statocyst consists of a vesicle formed by an outer cellular capsule and an internal layer of parietal cells. It is located in the anterior part of the body and inside the SMC [18,35] fenced by some neural projections that reach its periphery [31,35] (H. Nakano 2015, personal communication). Due to the composition and the structure of this organ, it has been speculated that it cannot work as a ‘true’ georeceptor; hence its function still remains unclear [35]. Detailed ultrastructural studies have detected that the nerve plexus is a little thickened in the periphery of the statocyst, however no accumulation of ganglionic cells have been observed [31,35,36]. The nerve net at the bottom of the two anterior side-furrows exceeds by two or three times its normal thickness, containing ganglion cells of considerable size [31].
A different degree of morphological complexity of the NS is observable in the sister group of the Xenoturbellida: the Acoelomorpha (Acoela plus Nemertodermatida) [11]. There we can see, in some of its constituent taxa, obvious neural aggregations, positioned in the most anterior part of the body. Nemertodermatida consists of nine species of marine worms. Their NSs have been studied using light microscopy in Nemertoderma westbladi [37,38]; by electron microscopy in Nemertoderma sp. [1] and Flagellophora apelti [39]; and in later studies through the use of GYIRFamide, FMRFamide, tyrosinated-tubulin and 5-HT immunoreactivity, for instance in N. westbladi and Meara stichopi [40–42]. According to those studies, most nemertodermatids’ NSs consist of a basiepidermal plexus with small neural masses surrounding the statocyst and arranged in defined structures [1,37,39,40]. The species described in most detail (N. westbladi, F. apelti and M. stichopi) present different degrees of NS centralization. Nemertoderma westbladi possesses a broad peripheral ring of neurites located anteriorly and outside the body wall musculature, with a higher density of processes and cell bodies in the dorsal side. Four neurite bundles start at the level of the ring structure: two lateral bundles and two ventral ones, interconnected by thin commissures; they run along the whole length of the body [37,41]. In the case of F. apelti, a peripheral nerve ring has been reported with a subepidermal neural mass that is exceptionally larger, bilobed, and placed caudally to the statocyst [39]. In M. stichopi, the arrangement of the NS is slightly different: an anterior commissure connects the only two longitudinal neurite bundles [40,42,43] (though Westblad only sees vaguely these neurite bundles) that run externally to the muscles and along the whole body length [42]. No brain or neural condensations were detected besides this anterior commissure. As is common to the anterior part of all xenacoelomorphs [1,31,34–36,40,41,43–47], the nemertodermatids also have a statocyst. In this taxon, the statocyst is divided in two chambers, and within each one there is a statolith ([1,36]; fig. 1 in [29]). Signs of a slight nerve concentration have been reported in the area of the statocyst [1,37–39]. Previous studies of nemertodermatids have shown that some outer neurons reach the basal lamina of the statocyst, but without penetrating it [36]. More recent immunohistochemistry observations in M. stichopi suggest that the statocyst is innervated [42].
The elaboration of a more centralized NS, within Xenacoelomorpha, is most clearly seen in the Acoela. In fact, the acoel NS is characterized by its high plasticity, which is evident in the very different neuronal arrangements that the acoel species show. A quick comparison of, for instance, acoels belonging to the most basal family (Diopisthoporidae) and those of the most divergent families within the class Crucimusculata reveals very striking differences in neural system arrangement [4,6,48–51]. Using the knowledge obtained from several studies, old and new, we are in a position to understand, for the first time, the evolution of the NS within this group (see also [6]), always in accordance with the latest available phylogeny of the Acoela [48]. In the following paragraphs, we summarize what is known on how the NS is organized in the different clades forming the Acoela.
The most basal family, Diopisthoporidae, possesses a NS formed by two ring commissures, the anterior being smaller than the posterior. In the case of Diopisthoporus gymnopharingeus, two ventral tracts connect both structures [49] (fig. 12 in [6]). Immunocytochemical studies carried out in Diopisthoporus longitubus show an extensively well-developed nerve net [24]. The anterior nerve centralization in D. longitubus is slightly different to that of D. gymnopharingeus, showing the second ring reduced to a semicircle on the dorsal side. Three pairs of longitudinal neurite bundles originate from the brain-like structure and run posteriorly, until they disappear within the nerve net [24]. The next branching family, Paratomellidae, is the sister group of Prosopharyngida plus Crucimusculata. Paratomella rubra, a representative of Paratomellidae, possesses two ring commissures connected by two tracts and also a concentrated network of cells in the area around the statocyst. Two neurite bundles extend along the whole length of the body [52,53] (fig. 12 in [6]). More clear evidence of evolutionary transformation is seen in the clade Prosopharyngida (including: Hallangiidae, Hofsteniidae and Solenofilomorphidae; fig. 12 in [6]). For instance, within Hofsteniidae, there are species with the NS located in different positions with respect to the epidermis: from a NS built as a basiepidermal plexus to a NS completely positioned below the epidermis, with some species showing some intermediate states (as in the case of Marcusiola tinga, where the dorsal neurite bundles are at the base of the epidermis while the rest is positioned clearly below it [54]). In the case of Hofstenia sp., the entire NS is located below the epidermis and consists of a wide ring of neurites, thickest in its dorsal part, with nerve fibres and somata that are partially located below the body wall musculature, and having some neurons surrounding the statocyst [6,55,56]. The NS in the Solenofilomorphidae is located below the body wall musculature and consists, close to the statocyst, of one to three commissures in the anterior part; these animals possess eight longitudinal neurite bundles that run along the body of the animal. The neurite bundles have two different spatial distributions: those that are clearly separated from the epidermis; and those that are associated with the epidermis (especially in the case of the dorsolateral and the ventrolateral neurite bundles) [6,57].
The sister group of Prosopharyngida is the clade Crucimusculata, which includes the acoels with complex, ganglionic, brains [24,29,50,51]. In the species of this clade, the NS becomes structurally more complex, forming a compact anterior bilobed brain with a cellular cortex and a dense central neuropile [6,29,50]. This anterior neuronal mass gives rise to a variable number of neurite bundles, which tend to be highly interconnected [6,24,29,50,51]. Over the last decade, several research groups, including ours, have studied some specific species belonging to Crucimusculata, such as Isodiametra pulchra and Symsagittifera roscoffensis, with the NS as their major research objective (fig. 12 in [6]). In §4, we revisit some of their findings and analyse the impact of the introduction of new tools on our current understanding of the acoel NS architecture.
4. The neuroanatomy of one acoel as revealed by a species-specific antibody
To shed light on the development and the neuroanatomy of acoel NSs, our laboratory recently obtained several species-specific antibodies, using cDNA sequences derived from our transcriptomic projects. Here, we describe in detail the expression pattern revealed using an antibody raised against the synaptic protein synaptotagmin [29], in both juvenile and adult stages of the acoel S. roscoffensis. As has been shown in previous studies, a high condensation of neural bodies and their processes characterizes the NS of S. roscoffensis. Confocal and electron microscopic studies have confirmed that, in juveniles, this neural mass fills most of the anterior third of the body (figure 1 and [50]). The so-called brain consists of a compact bilobed mass with a peripheral layer of neuronal cell bodies surrounding a central neuropile, all located around the statocyst, with two ocelli located laterally [50]. The NS contains also three pairs of neurite bundles that run along the major body axis: a dorsomedial pair (dmn), a dorsolateral pair (dln) and a ventrolateral pair (vln), all connected by numerous commissures that are arranged irregularly (figure 2c–e; fig. 2 in [29]) [50,51]. This compact (centralized) arrangement is preserved from the juvenile to the adult stages.
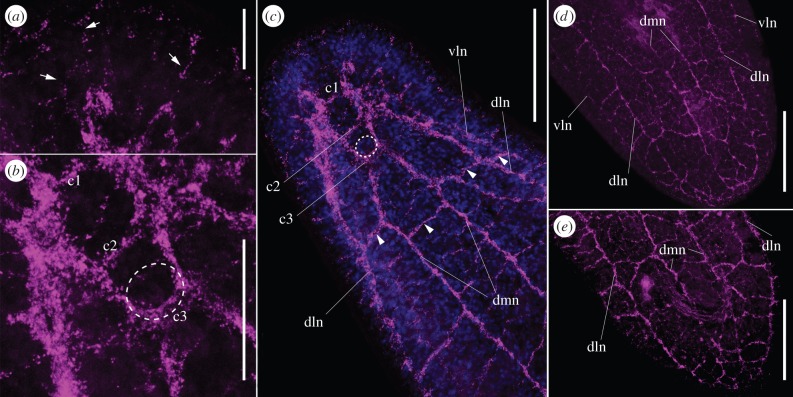
Figure 2.
Confocal microscopy stacks of S. roscoffensis adult nervous system stained with a specific antibody against synaptotagmin. Anterior part is up. (a) Anterior end of adult specimen detailed, thin reminiscences connect the brain to the epithelia (arrows). (b) Detailed projection of the brain; from the anterior to posterior we detect the three brain commissures (c1; c2; c3) and the position of the statocyst (shown here as a discontinuous circle). (c) Adult anterior part additionally stained with the nuclear marker DAPI. Five of the six neurite bundles are visible. Statocyst shown here as a discontinuous circle. Arrowheads point to the commissures connecting the neurite bundles. (d) Ventral view of the posterior end of the body. The three pairs of neurite bundles converge at the posterior end. (e) Dorsal view of the posterior end of the body. Note that the nervous system extends to the posterior end of the body. Refer to figure 1 for abbreviations. Scale bars: (a) 35 µm, (b) 65 µm and (c–e) 130 µm.
The granular pattern obtained by the anti-synaptotagmin antibody is explained by the fact that synaptotagmin is a highly conserved protein that acts as a calcium receptor located in synaptic vesicles [58]. The structures revealed in both adult and juvenile developmental stages coincide with those obtained using neuronal or neurotransmitter markers such as the anti-tubulin tyrosine antibody, or the anti-serotonin and RFamide antibodies [50,51]. However, our synaptotagmin antibody is most probably a pan-neuronally expressed epitope and exclusive to the NS, not labelling other structures such as those revealed with less-specific antibodies, for instance the anti-tyrosinated tubulin. On a more practical level, it allows us to perform immunochemistry and in situ hybridization protocols in the same specimen (unpublished results, 2014 and 2015).
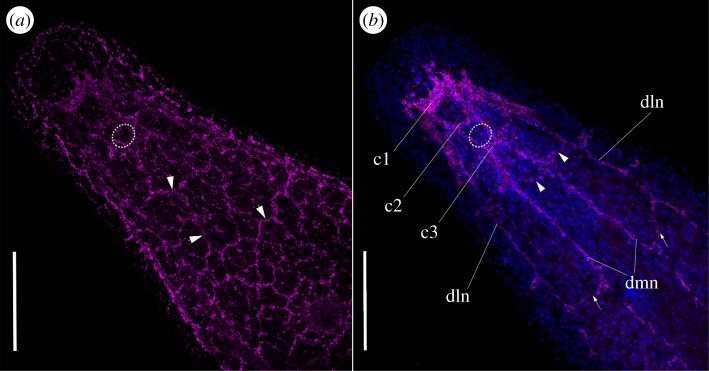
As revealed by the anti-synaptotagmin antibody, in both stages (adult and juvenile), the brain is a structure with clear bilateral symmetry: two triangular lobes interconnected by three commissures, named c1, c2 and c3 from the most anterior to the most posterior (figures 1b, 2b,c and 3b) [50]. Numerous thin fibres radiate from the brain towards the most anterior surface of the body (figure 2a). Posterior to the brain, the next five commissures connect the six neurite bundles at different positions along the body (figures 2c (arrowheads) and 3b (arrowheads)) [29]. The neurite bundles converge at the end of the body, and in the case of the adult, posterior to the male genital pore (figure 2d,e) [59]. According to the neural inmunohistochemical patterns obtained in previous studies [50,51] and now confirmed by our observations with the anti-synaptotagmin antibody, no major changes in the general arrangement of the NS occur from the young to the adult stages (figures 1 and 2), despite the obvious changes in the proportion of the whole body that the brain occupies [50].
Transmission electron microscopic analysis carried out in juvenile stages determined that the neurite bundles are fundamentally formed of neurites with the cell bodies dispersed along the neurite length, and specifically concentrated at the commissural junctions [50]. In agreement with this, double staining with the anti-synaptotagmin antibody and the nuclear marker DAPI shows a clear lack of nuclei in the major part of these bundles (E. Perea-Atienza 2015, personal observation). In the most anterior part, these bundles are thicker, revealing that they include more axons [50]. In the brain region, a prominent organ is located: the gravity-sensing statocyst (figures 2b,c and 3, discontinuous circle) [29,50,51]. As shown in previous immunochemical studies, with this methodology and antibody, we are not able to detect neurons directly innervating the statocyst [29,51]. It is important to point out here that all the analysis using histological sections showed the presence of neuronal concentrations surrounding the acoel statocyst [6,39,50,59–62], innervating the capsule [36]. As has been shown in previous studies using immunoreactivity against anti-tubulin tyrosine antibody, a peripheral nerve net formed by thin processes is visible in the epidermal layer, a feature also revealed with the anti-synaptotagmin antibody (figures 1 and 3a). The pattern is very similar in juvenile and adult stages. The neurons from this peripheral plexus establish a high number of synapses with neighbouring muscle fibres [50].
Figure 3.
Confocal microscopy projections of the anterior part of S. roscoffensis adult specimen revealing the nervous system stained with the specific antibody against synaptotagmin. (a) Ventral projections of numerous nerve connections (arrows) all giving rise to the nerve net. (b) Central projection evidencing the internal arrangement of the nervous system. Immediately posterior to the brain some commissures connect the neurite bundles (arrowheads). They are followed by other less prominent commissures (arrows). Refer to figure 1 for abbreviations. Statocyst shown here as a discontinuous circle. Scale bar: (a,b) 100 µm.
In the future, the use of species-specific antibodies will be extended to other species of the Acoela and later to other species of the Xenacoelomorpha, with the aim of providing higher resolution maps of neural architecture in the different members of this clade. The use of anti-synaptotagmin is just the first step of this project.
5. Molecular control of acoel neurogenesis
Very little is known concerning the control of neurogenesis in Nemertodermatida and Xenoturbellida. For this reason, this section focuses on the molecular control of neurogenesis in those acoel species that have been studied. Acoel development follows a special cleavage pattern called ‘duet cleavage’ [63]. The ectodermal cells (which give rise to epidermal and neural cells) are derived from the first, second and third duet micromeres of the 4-, 6- and 12-cell stage embryos, respectively. Later on in development, the progeny of those micromeres that will differentiate into neurons become internalized. As some studies suggest, these internalized neuron precursors express SoxB, a most likely pro-neural marker present in several bilaterian and non-bilaterian species [51,64,65].
Although much information is still lacking about the genes that trigger the initial steps of neural commitment and differentiation, most of our current knowledge corresponds to the analysis of anteroposterior patterning in the acoel NS. In all the species studied, only three Hox genes have been identified: one anterior, one central and one posterior [65–67]. As development proceeds, these genes seem to provide the needed positional information to the newborn neurons.
All three Hox genes are transcribed after gastrulation in the acoel Convolutriloba longifissura and they do it almost simultaneously, thus, showing no clear temporal colinearity. Nevertheless, they do present clear spatial nested patterns. The anterior Hox gene is expressed in two bilateral patches of cells in the animal hemisphere of the embryo, with the anterior boundary delimited by the expression domain of the Six3/6 orthologous gene [65], the latter being a gene involved in anterior ectoderm patterning in bilaterians and in the differentiation of the aboral pole in some cnidarians [68]. In the juvenile acoel, the anterior Hox is expressed in an anterior domain that extends posteriorly to the statocyst [65]. The central Hox gene is expressed in the embryo in a more posterior domain, surrounding the closed blastopore. Later on, it is expressed in two lateral bands along most of the body length. The posterior Hox gene is expressed in a restricted area at the most posterior end of the embryo. Unlike the other two, its expression does not seem to be limited to a specific germ layer [65,69]. Interestingly, Hox gene expression is maintained not only during regular development [65,66], but also during the budding of new specimens in asexually reproducing species [67].
A few other ‘classical’ neural genes have been studied in the acoel C. longifissura, for instance the orthologue of the posterior ParaHox gene, caudal/Cdx (expressed in many bilaterian neural domains [70]), which is also expressed anteriorly in juvenile neural structures, most probably in the cells that also express the neural genes NK2.1 and Otp [65,71].
From what we have described here it is clear that our understanding of the molecular control of neurogenesis in acoels (or in the xenacoelomorphs) is very limited. Efforts should be made to improve the characterization of many more genes involved in this process (see below). It is also necessary to stress that we completely lack information regarding the dorsoventral patterning of the acoel NS. This is an intriguing issue, for it has been observed that acoels do not have, apparently, an obvious preference for ventral or dorsal NS localization. However, in a preliminary study, and in the context of animal regeneration, it has been shown that the acoel Hofstenia miamia have differential expression of BMPs (bone morphogenetic proteins) and their antagonistic ADMPs (anti-dorsalizing morphogenetic proteins) along this secondary axis [72]. How the expression of these genes is related to the position of the neurite bundles can be, at present, just a matter of speculation. More expression and functional studies of these and other families of neural regulatory factors are urgently needed.
6. A genomics view of Xenacoelomorpha neurogenesis
Our laboratory's main research objective is to understand the process of neurogenesis and neural function in the different members of the Xenacoelomorpha. In particular, we aim at understanding how families of putative regulatory and effector neural genes have evolved within this clade, and how these changes relate to the inherent complexity of the NSs. In order to achieve this, we have started using the (mostly complete) genomes of the acoel S. roscoffensis and the xenoturbellid X. bocki. We focus our attention on the basic helix-loop-helix (bHLH) transcription factors, known regulators of NS development that control aspects such as neural differentiation or migration [73,74]; and on the G protein-coupled receptors (GPCRs), known transducers of signals from many sensory systems. Our interest in the latter group lies not only in their crucial and varied functions, but also in the fact that they are the largest and most diverse superfamily of transmembrane receptors [75,76], which allows us to assess gene complement changes (and families' diversification) within the clade and relate them to the different degrees of NS centralization observed in the constituent species.
A detailed study performed on those two groups of genes is already published [29]. Summarizing, and as shown in table 1, we found that, as a general pattern, the number of genes belonging to each family is lower in S. roscoffensis than in X. bocki: 18 bHLHs and 225 GPCRs were present in the former; 33 bHLHs and 258 GPCRs in the latter. This is consistent with, for instance, the number of Hox genes detected: three in acoels [66]; five in xenoturbellids [77]. Here, the complexity of the different families (we refer to the total number of genes per family plus the number of subfamilies present) in these two species is not directly related to the apparent structural complexity of acoel and Xenoturbella neural architectures. We observed that less complex neural architectures (the nerve net of Xenoturbella) seem to depend on richer complements of regulatory and effector genes (within, again, these specific families) than the complex/centralized structure of S. roscoffensis brain. It is important to point out that all these gene families were characterized in depth, with all members classified into subfamilies based on a thorough phylogenetic analysis, leaving little room for missing data [29]. Interestingly, our recent analysis of a different family, that of the Wnt ligands, has shown a similar pattern. Let us now take a closer look at this in the next paragraph.
Table 1.
Complement of genes of different families in Xenoturbella bocki and Symsagittifera roscoffensis. Data were published previously on the Hox genes [66,77] and on the bHLHs and GPCRs [29], and data on the Wnts are from this study.
| species | Hox | Wnts | bHLHs | GPCRs |
|---|---|---|---|---|
| X. bocki | 5 | 11 (12?) | 33 | 258 |
| S. roscoffensis | 3 | 5 | 18 | 225 |
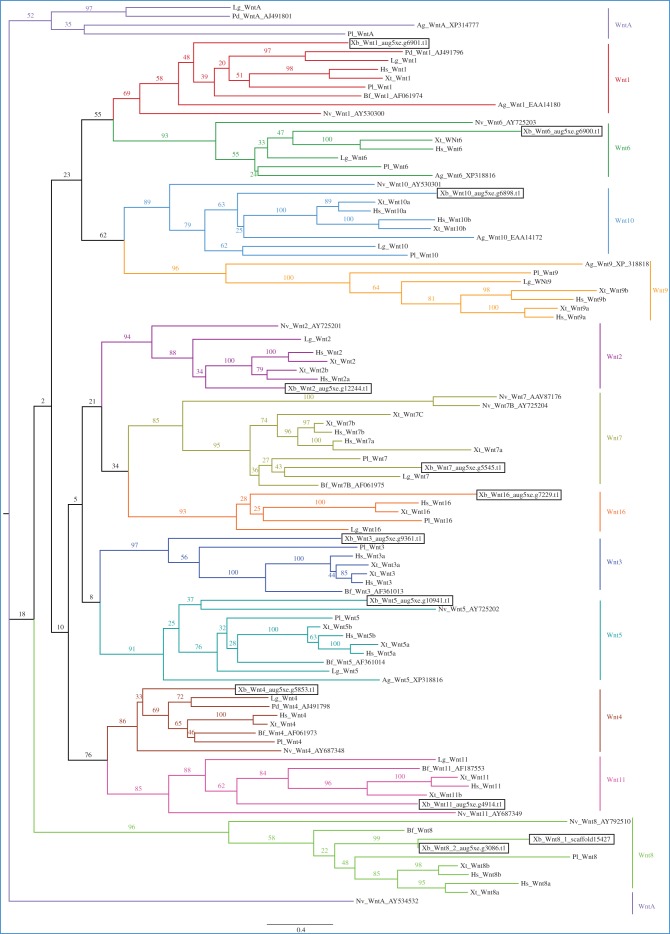
The Wnt glycoprotein signalling pathways play a crucial role in development by regulating aspects of cell fate determination, migration, polarity, neural patterning and organogenesis [78]. Furthermore, it has been observed that most animals use these signalling pathways to control their primary body axis, specifically the anteroposterior axis in all bilaterian animals [79]. We searched for Wnt domains in the predicted proteins obtained from the genomes of S. roscoffensis and X. bocki using the HMMER tool, as explained previously [29], and found that there are five different Wnt ligands in the genome of S. roscoffensis and 12 in that of X. bocki (table 1). We used phylogenetic analysis for subfamily classification. First, we aligned our sequences with the reference species: Lottia gigantea, Homo sapiens, Xenopus tropicalis, Paracentrotus lividus, Nematostella vectensis, Platynereis dumerilii, Anopheles gambiae and Branchiostoma floridae (Dr Jenifer C. Croce provided Wnt aligned protein sequences of the first four species; the rest were downloaded from the NCBI database). With this data, we reconstructed the phylogenetic trees (figure 4 and electronic supplementary material, figure S1) and obtained the results that are summarized in table 2. We were able to complete the analysis with good bootstrap values (more than 85 in most cases, except one of them that was 69) for all the X. bocki Wnts (we found clear orthologues of: Wnt1, Wnt2, Wnt3, Wnt4, Wnt5, Wnt6, Wnt7, Wnt8, Wnt10, Wnt11 and Wnt16) (figure 4). We found two putative Wnt8 orthologues; however, since they share a stretch of 35 amino acids it is possible that they correspond, in fact, to two predictions for the same Wnt protein. Intriguingly, it was impossible to classify with confidence the S. roscoffensis Wnts, since they do not clearly cluster with members of any known Wnt subfamily. When they do, their positions in the tree are only supported with excessively low bootstrap values, all having very long branches (electronic supplementary material, figure S1). In fact, even though the reference species used were the same in both trees, the bootstrap values obtained for all the families decrease when the sequences of S. roscoffensis are incorporated (compared with the X. bocki's tree).
Figure 4.
Xenoturbella bocki Wnt ligands. Phylogenetic analysis of X. bocki Wnt relatives was performed as explained previously [29], using a maximum-likelihood approach with the program RAxML and 100 bootstrap replications. The alignment of the reference species was provided by Dr J. C. Croce [80] and included the sequences from the species Pl, Lg, Hs and Xt (see below for abbreviations); the other sequences are used in Kusserow et al. [81] and were downloaded from the NCBI database, the accession numbers are indicated after the name. Protein families are highlighted in different colours. Xenoturbella bocki Wnts are marked with a rectangle. Sr, Symsagittifera roscoffensis; Xb, Xenoturbella bocki; Lg, Lottia gigantea; Hs, Homo sapiens; Xt, Xenopus tropicalis; Nv, Nematostella vectensis; Pl, Paracentrotus lividus; Bf, Branchiostoma floridae; Pd, Platynereis dumerilii; Ag, Anopheles gambiae.
Table 2.
Complement of each Wnt subtype in the indicated species. Bold font indicates the species analysed in this study.
| total | Wnt1 | Wnt2 | Wnt3 | Wnt4 | Wnt5 | Wnt6 | Wnt7(a-b) | Wnt8(a-b) | Wnt9(a-b) | Wnt10(a-b) | Wnt11 | Wnt16 | WntA | Wnts unclassified | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Symsagittifera roscoffensis | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| Xenoturbella bocki | 11 (12?) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 (2?) | 0 | 1 | 1 | 1 | 0 | 0 |
| Homo sapiens [80] | 19 | 1 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 1 | 1 | 0 | 0 |
| Nematostella vectensis [81] | 12 | 1 | 1 | 0 | 1 | 1 | 1 | 2 | 2b | 0 | 1 | 1 | 0 | 1 | 0 |
| Platynereis dumerilii [81] | 7 | 1 | 1a | 0 | 1 | 0 | 0 | 1a | 0 | 1a | 1a | 0 | 0 | 1 | 0 |
| Lottia gigantea [80] | 11 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| Anopheles gambiae [81] | 7 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 |
| Xenopus tropicalis [80] | 21 | 1 | 2 | 2 | 1 | 2 | 1 | 3 | 2 | 2 | 2 | 2 | 1 | 0 | 0 |
| Branchiostoma floridae [81] | 13 | 1 | 1a | 1 | 1 | 1 | 1a | 2b | 1 | 2a | 1a | 1 | 0 | 0 | 0 |
| Paracentrotus lividus [80] | 11 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 |
aSequences not included in our analysis.
bOne of the sequences not included in our analysis.
Previous studies carried out by Srivastava et al. [72] in H. miamia found five Wnt homologues which were branching together with one Wnt relative of the cnidarian Hydra vulgaris. We decided to try including those Wnt protein sequences in our analysis and see how they were placed in relationship to the Wnt relatives in our acoel species. Our results show that all acoel sequences, classifiable or not, present extremely low bootstrap values (electronic supplementary material, figure S2). Consequently, after this analysis, complemented with BLAST searches, we decided that it was most appropriate to name our acoel Wnts as UC (unclassified).
The difficulty in classifying these acoel genes could be due either to those being homologous to ancestral bilaterian genes lost in other lineages (unlikely) or to these sequences differing enormously from all others known in bilaterians, because of a high rate of sequence evolution (a more plausible scenario). Most probably, acoel Wnts (and members of the other gene families) are highly divergent [29], something that can be seen clearly in different known phylogenetic analysis, where acoel branches tend to be very long. However, and independently of the high evolutionary rate at which the sequences of members of different families change, we see that in the acoel there are fewer genes from the Wnt family than in the xenoturbellid, again showing in this case an inverse relationship between this gene family complexity and the organizational/structural complexity of the NS.
Needless to say, further studies of other gene families that are important for NS development are needed, as is a better understanding of the expression patterns of the genes identified, before we can start to unravel this apparent paradox of inverse correlation between molecular complexity and the degree of centralization of the NS. We are aware that the apparent simplicity of the NS structure does not indicate functional simplicity. For instance, it has been demonstrated that in hydra (having another ‘simple’ NS) different subsets of neurons in the nerve net express different neuropeptides [82]; hence although the nerve net might look structurally simple, in fact it is a structure that is highly (from a functional point of view) regionalized. The X. bocki nerve net may be a similar case.
7. Transformation from nerve nets to compact brains in the Xenacoelomorpha: an evolutionary scenario
Within Xenacoelomorpha, we consider the epidermal nerve net of xenoturbellids as the ancestral condition (because of the relative position of xenoturbellids within this group and because Xenoturbella has the shortest branch in all molecular phylogenies). This assumption would be consistent with either of the two hypotheses regarding the placement of this clade in the metazoan tree: as basal bilaterians or as the sister taxa of Ambulacraria. Let us now revisit these two alternatives.
If xenacoelomorphs did in fact branch at the base of bilaterian animals, their sister taxa would be all the remaining bilaterian lineages. Trying to infer the structure of the NS in the last common ancestor of protostomes and deuterostomes is a very complex matter, and a subject of acrimonious debates. The reason is that as the centralized NSs of the representatives of these two superphyla are generally organized in different positions and are diverse in their morphology, it is highly contentious to establish possible homologies. Consequently, with the available morphological and molecular data, two main hypotheses exist (that will be further discussed in the next section of the paper): their common ancestor had either a centralized, complex, NS [83] or a diffuse nerve-net [84–86]. As we cannot deduce the structure of the deuterostome and protostome ancestor, we can compare the Xenacoelomorph NS, representing the first offshoot of the Bilateria, with that of the animal taxa that would have diverged prior to it: the cnidarians. The last comparison should allow us to polarize the NS character evolution for all bilaterians.
Cnidarians are mostly marine animals that usually present radial symmetry. Their body is formed of two layers, the ectoderm and the endoderm, which are separated by an extracellular matrix called the mesoglea [87]. The cnidarian NS consists of a diffuse nerve net that extends throughout the whole body. However, the neuron distribution is not uniform: the nerve net density is higher in certain anatomical areas, for instance in the head of Hydra as opposed to its body column [88]. In addition to this nerve net, we can also find annular-like structures around the oral opening in Nematostella, or at the base of tentacles in some Hydra sp. [88–90]; but in none of these cases has any large concentration of neurons (ganglia) been identified [82]. Cnidarians often possess both endodermal and ectodermal NSs [91]. As highlighted by Koizumi et al. [82], it has been observed that nerve cells residing either in ectoderm or the endoderm express different types of peptides. These differences are also seen in different nerve cells populations. The lesson to be learned here is that even if nerve net organization superficially looks simple, it actually contains many subsets of different neurons, expressing different peptides, with very specific body distributions [82]. Whether this is also the case for the nerve net of the xenacoelomorphs, as stated above, remains to be seen. In any case, if xenacoelomorphs were placed at a basal position within bilaterian animals, the cnidarian nerve-net structure of the NS would be an arrangement comparable to that we find in Xenoturbella. Moreover, it is important to note that Xenoturbella gives a very short branch in all molecular phylogenies, which has prompted us (and others) to suggest that they may represent good proxies for the ancestor of the Xenacoelomorpha.
The alternative position suggested for the Xenacoelomorpha is a sister group of Ambulacraria (echinoderms plus hemichordates). Both groups have NSs arranged as ectodermal nerve nets and nerve condensations/cords. The NS of echinoderms consists of an ectodermal nerve net and a nerve ring around the mouth. However, as they present pentameral symmetry, an obviously derived character, it is not possible for us to make any clear comparison between this arrangement and those present in other bilateral animals (though this is also a matter of debate).
Hemichordates, in contrast, have a body divided into three morphological regions: proboscis, collar and trunk. The taxon Hemichordata includes two classes: enteropneusts and pterobranchs. Enteropneusts have a diffuse basiepithelial nerve net present in all three body regions and show structural dorsoventral and anteroposterior polarities [92–94]. Moreover, they also possess areas of nerve plexus thickenings, the so-called dorsal and ventral nerve cords. The dorsal cord runs from the proboscis and extends along the whole length of the animal, whereas the ventral one is located in the trunk and is thicker. Both these cords are described as mostly axon tracks, in general showing no capacity as processing centres [94,95], except in one study where some integrative function of the ventral cord was described [96]. Pterobranchs, however, are sessile and have a concentration of neurons at the base of their anterior tentacles, a structure that has been called a brain [83]. They also have neurite concentrations and associated neurons in the tentacles, the stalk and in between the gill slits. Although they were traditionally considered a basal group inside hemichordates, their relationship with enteropneusts is not clear. Some phylogenetic analysis using molecular data suggests that are derived and placed within (and not as a sister group of) enteropneusts [97], while others reject this hypothesis completely [98] or are unable to resolve the relationships [99]. In any case, here again in the Hemichordata, we see that the ancestral state for the NS organization is most probably that of a diffuse nerve net (though with extensive internal structure; see, for instance, [86]). This arrangement would also be shared with the echinoderms. All in all, regardless of whether pterobranchs or enteropneusts are more similar to ancestral hemichordates, the common ancestor of Ambulacraria is expected to have a diffuse nerve net [100]. In this new context, a nerve net as seen in Xenoturbella (and in the acoels and nemertodermatids) would be again consistent with the placement of Xenacoelomorpha as a sister group of the Ambulacraria.
Taking into consideration both possible phylogenetic positions of Xenacoelomorpha and the fact, also stated above, that Xenoturbella has the shortest branch in all molecular phylogenies obtained for this clade, it seems reasonable to assume that the nerve net-like organization present in Xenoturbella most probably represents the basal condition for Xenacoelomorpha. This has a profound implication for the evolution of NSs within the clade, and the observed changes of NS organization within it, with more compact brains developing in more derived acoel lineages (probably the result of specific adaptive changes happening over time).
We are fully aware that even if these evolutionary scenarios seem feasible, we cannot exclude the possibility that the Xenoturbella NS has been secondarily reduced from that of a more complex ancestor. It would be interesting, though, to have access to more species within this clade (xenoturbellids) since, right now, our inferences are based on morphological information derived from only two different species [31,32].
8. Independent events in nervous system centralization across metazoan phyla
There are currently two different hypotheses regarding the emergence of centralized NSs in animals [84]: the single origin and the multiple origins hypotheses. In short, the single origin hypothesis argues that the last common ancestor of all the bilaterian animals (or Urbilateria) already had a centralized NS. This would imply that it was secondarily simplified several times in the metazoan lineages. This hypothesis relies on the identification of multiple genes involved in NS patterning that are well conserved across bilaterians (such as bone morphogenic protein/decapentaplegic genes and their antagonists, plus the many regulators of anteroposterior patterning). However, we have to recognize that those genes could also have been co-opted several times in evolution (in toto or a fraction of them; see, for instance, [83]) or used for the general patterning of the neuroepithelia, instead of being devoted to the specification of concrete neural areas/structures [84–86].
The alternative hypothesis is the multiple origin of centralized NSs, which states that they have evolved, independently, more than once [84,85,101,102]. The hypothesis relies on the observation that there is a ‘patchy’ distribution of clades with complex brains within the metazoan tree [85]. In fact, it is known that in each of the two bilaterian major clades (Deuterostomia and Protostomia), animals having compact brains and animals with diffuse nerve nets (and different degrees in between) coexist, even within the same taxon. Some authors such as Moroz [84] go a step further and argue that even neuronal cells might have originated more than once in the course of evolution. In this hypothesis, the urbilaterian ancestor might have had a non-centralized NS with a nerve net-like organization (with some general patterning mechanisms already in place). Centralization of the nervous structures may have derived from such a net (or net-like) arrangement between five and nine times in metazoans according to Moroz [84,103], or at least four according to Northcutt [85].
Among the well-described cases, one paradigmatic example of the possibility of independent brain origins is described in the molluscs. Here, we just provide the essential facts as a good illustration of the evolutionary process, without delving into the details. In a review published by Moroz [84], he arrives at the conclusion that three or four independent events of centralization of the NS have occurred in Mollusca over evolutionary time. The most basal molluscan lineages present a NS organization called tetraneury (fig. 2 in [84]) which consists of two pairs of longitudinal nerve cords, ventral and lateral (with the second pair more dorsally situated), which are connected through commissures along the whole body axis [104]. This configuration does not present any obvious signs of centralization [105]. However, in more recent lineages of Mollusca, such as Cephalopoda and Gastropoda, the presence of areas with centralized nervous tissue is clearly observable. In this way, we see in basal groups of these lineages similar nervous patterns to the tetraneury arrangement, whereas in more derived clades we find different patterns and degrees of aggregation in individual ganglia (described in detail in [106] for Cephalopoda and [107] for Gastropoda). This description of events happening in the Mollusca is just an illustrative example; but Northcutt [85] also mentions similar processes occurring in other metazoan groups: arthropods, annelids and, of course, chordates (fig. 2 in [85]).
Incorporating into this discussion what is now known from the NS in Xenacoelomorpha, we postulate that this process of independent centralization has also taken place within this clade. Moreover, we hypothesize that this evolutionary process seen in several metazoan lineages from simple NSs to more complex brains might have been facilitated by the modular construction mechanisms that early neurogenesis affords to most animals, always modulated by the functional and regulatory versatility of the gene networks involved [108]. Most of the molecular machinery of these modules was most probably already present in the Urbilateria (or before). Many genes, such as those involved in anteroposterior and dorsoventral patterning, might have modulated the position of neurons and circuits along the body axis. Downstream of those general patterning mechanisms, the modulation of those gene batteries that regulate early neurogenesis would have allowed certain plasticity in the construction of the NSs and thus resulted in the huge variability of architectures that we now observe in the different metazoan phyla. How this was achieved, at the molecular regulatory levels and over evolutionary time, still remains to be understood through the comparative analysis of many taxa.
9. A speculative note: why having a brain?
In the previous paragraphs, we have described an interesting case of a NS ‘centralization’ observed in some members of Xenacoelomorpha. We have also shown how this event is not circumscribed to this specific clade, most probably occurring independently in many other phyla. This parallel evolution of centralized NSs merits some closer attention, specifically to those factors that drive it and the physiological advantages that having a centralized (compact) NS would provide to its carriers [28].
The driving force behind ‘centralization’ was most probably the accumulation of sensory structures in one particular pole of the body (though there are also ganglia in many bilaterians that are not centred on sensory receptors). The proximity of a central processing unit may have given some ‘computational’ advantages to animals that maintained these neuronal aggregations (brains). However, not only computational advantages but also energy saving may have been involved since signals into and out of such a central processing unit would have had to travel shorter distances. It is important to point out that any increase in brain size or wiring density would be selected only if the costs of production or maintenance were outweighed by performance. We speculate that sensory organs (or cells) and brains would have co-evolved in different lineages and over evolutionary time, always sustained by mutual, reinforcing, positive feedback: sensory organs driving centralization of nervous structures in their vicinity, which at the same time would be instrumental in bringing other sensory structures (cells) to the same area. Moreover, the evolution of these structures would need the parallel rewiring of the gene regulatory networks involved in their patterning [109]. The final size, shape and (network) functionality of the brain would depend on many internal and external causal factors. It is well known that variability in brain size or architecture correlates with several environmental, life-history or behavioural traits (though most experimental data have been gathered from vertebrate studies; for a review, see [110]). Along these lines, we have speculated [6] that richer environments or the presence of sexual conflict might have driven the appearance of more centralized NSs in the acoel class Crucimusculata. More basal groups of acoels are interstitial and feed on dissolved organic matter, whereas in the Crucimusculata, the diets of many taxa are based on the capture of diatoms, crustaceans and other worms, or even on cannibalism [6]. Thus Crucimusculata are active predators requiring a more sophisticated repertoire of behaviour.
What are the advantages of having a brain or a centralized NS? Many authors have speculated with the idea that brain, ganglion or neural cells are arranged in any animal in accordance with a major goal: to minimize the cost of connection between the components (the ‘save wire’ principle [111,112]). This principle that is derived from component network optimization theory assumes that a network is selected if it minimizes the total nerve connection length (taken as a proxy for total connection costs). This condition constrains the geometry of the network, provided that energy consumption is efficient. The principle has been thoroughly analysed in the context of the arrangement of the Caenorhabditis elegans NS [111]; the rat olfactory cortex and the amygdala [113]; and the cat sensory and macaque visual cortex [114]. In fact, Cherniak [111] has shown that in C. elegans (or in H. sapiens), the brain position (at the front of the body) minimizes total sensory/motor connection costs and this is due to the fact that in those brains there are more connections to and from the front than there are to the rear of the animal (accommodating all sensory information flow). The results can be extrapolated to other systems, where input/output connections to neighbouring sensory structures are most prevalent. An extreme version of this optimization model assumes that brains (or complex neural layouts) can be self-assembled based only on physical processes, for free, without the intervention of genomic mechanisms [113]. This, according to the proponents will ‘lower the information load on the genome, and also in the process yield optimality’ [113, p. 52]. Interestingly, this proposal might not be so far-fetched in view of some new human embryonic stem cell-derived three-dimensional culture studies that show the high potential for self-assembly that some tissues have [115].
Another set of arguments in favour of a more compact arrangement of neural components relies on the energetics of maintaining any working neural network. It is well known that NSs consume metabolic energy continuously and at a high rate. A good fraction of it is dedicated to moving signals along the axons and across synapses. Brains have countered these metabolic constrains by adopting energy-efficient designs. An efficient layout with high component density is critical for energy efficiency. This suggests a need for miniature, highly connected components in the networks (brains). Miniature designs have some intrinsic problems (such as noise; not to be discussed here) but they can be compensated by other properties (see, for instance, a discussion in [116]). Also, neural function varies with temperature, ionic concentrations, etc. so that if a neuronal network is built as a large distributed system, its properties may vary across space, making uniform coordination very difficult. Thus, again it is better to have the components clustered in a reduced space.
Other factors that come to mind and might be involved include good developmental reasons for having neurons linked in close proximity. If a neuron A needs to connect to a neuron B, it is easier if they are born close together, so that the instructions for growing a connection and finding the target are simple.
All in all, it is probably a combination of all these factors that contributes to the fitness of a structure such as the brain, and thus it has been selected for, over evolutionary time, in many independent lineages.
As with many other adaptive scenarios proposed in the bibliography, this needs to be checked experimentally. We propose that testing the hypothesis would be specifically interesting (and feasible) in the context of a coherent group of animals (a monophyletic clade) with members showing different degrees of NS centralization and, perhaps most importantly, a relatively reduced number of participating neurons. Xenacoelomorphs should provide here a useful system to analyse the evolution of neural centralization and also should provide an experimentally amenable subject to test the functional advantages (and constraints) underlying the origin of brains.
Supplementary Material
Acknowledgements
We have to acknowledge Prof. Gilles Laurent, from the Max Planck Institute for Brain Research, Frankfurt am Main, for discussing (and clarifying) with us some of the major centralization/cephalization issues. Christopher Evans (Universitat Barcelona) is acknowledged for helping us to improve the English quality of the text. Dr Josep Abril's (UB) help with bioinformatic procedures is most appreciated. We acknowledge all the members of the ‘Xenacoelomorpha Sequencing Consortium’ for sharing with us their enthusiasm for these enigmatic critters. Thanks to the referees for their constructive criticisms, as well as their suggestions, because they have helped us to improve this manuscript.
Data accessibility
All the sequences included in our study keep the original reference number assigned in the preliminary genome/transcriptome analyses. The whole genome/transcriptome original sequences will be made publicly available along with the forthcoming publication of the Xenacoelomorpha Genome Project consortium paper.
Authors' contributions
B.G. and E.P.-A. carried out the experiments, were responsible for data acquisition and performed the data analysis. All authors drafted and critically revised the manuscript. P.M. acquired the funding for the project.
Competing interests
The authors declare that they have no competing interests.
Funding
We acknowledge the generous support obtained from the EU Programme ‘ASSEMBLE’, which covered some of the costs associated to our acoel collection trips. A grant from the Spanish Ministry of Economy (BFU2012–32806) supports the work in the laboratory of P.M. Funds from the Generalitat de Catalunya (2009-SGR-001018) were also used in this project. B.G. and E.P.-A. have PhD (APIF-UB) fellowships from the Universitat de Barcelona.
References
- 1.Ehlers U. 1985. Das phylogenetische system der Plathelmintes. Stuttgart, Germany: Gustav Fischer Verlag. [Google Scholar]
- 2.Sterrer W. 1998. New and known Nemertodermatida (Platyhelminthes–Acoelomorpha): a revision. Belg. J. Zool. 128, 55–92. [Google Scholar]
- 3.Tyler S, Rieger RM. 1977. Ultrastructural evidence for the systematic position of the Nemertodermatida (Turbellaria). Acta Zool. Fenn. 154, 193–207. [Google Scholar]
- 4.Smith JPS, Tyler S. 1985. The acoel turbellarians: kingpins of metazoan evolution or a specialized offshoot? In The origins and relationships of lower invertebrates (eds Conway-Morris C, George JD, Gibson R, Platt HM), pp. 123–142. Oxford, UK: Oxford University Press. [Google Scholar]
- 5.Rieger RM, Tyler S, Smith JPS III, Rieger GE. 1991. Platyhelminthes: Turbellaria. In Microscopic anatomy of invertebrates, vol. 3 (eds Harrison FW, Bogitsh BJ), pp. 7–140. New York, NY: Wiley-Liss Inc. [Google Scholar]
- 6.Achatz JG, Martinez P. 2012. The nervous system of Isodiametra pulchra (Acoela) with a discussion on the neuroanatomy of the Xenacoelomorpha and its evolutionary implications. Front. Zool. 9, 27 ( 10.1186/1742-9994-9-27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Achatz JG, Chiodin M, Salvenmoser W, Tyler S, Martinez P. 2013. The Acoela: on their kind and kinships, especially with nemertodermatids and xenoturbellids (Bilateria incertae sedis). Org. Divers. Evol. 13, 267–286. ( 10.1007/s13127-012-0112-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakano H. 2015. What is Xenoturbella? Zool. Lett. 1, 22 ( 10.1186/s40851-015-0018-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philippe H, Brinkmann H, Copley RR, Moroz LL, Nakano H, Poustka AJ, Wallberg A, Peterson KJ, Telford MJ. 2011. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature 470, 255–258. ( 10.1038/nature09676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hejnol A, et al. 2009. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. R. Soc. B 276, 4261–4270. ( 10.1098/rspb.2009.0896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hejnol A. 2015. Acoelomorpha and Xenoturbellida. In Evolutionary developmental biology of invertebrates 1 (ed. Wanninger A.), pp. 203–214. Vienna, Austria: Springer. [Google Scholar]
- 12.Tyler S, Rieger RM. 1975. Uniflagellate spermatozoa in Nemertoderma (Turbellaria) and their phylogenetic significance. Science 188, 730–732. ( 10.1126/science.1124394) [DOI] [PubMed] [Google Scholar]
- 13.Smith JPS, Teyler S, Rieger RM. 1986. Is the Turbellaria polyphyletic? Hydrobiologia 132, 71–78. ( 10.1007/BF00046223) [DOI] [Google Scholar]
- 14.Lundin K, Hendelberg J. 1996. Degenerating epidermal bodies (‘pulsatile bodies’) in Meara stichopi (Plathelminthes, Nemertodermatida). Zoomorphology 116, 1–5. ( 10.1007/BF02526924) [DOI] [Google Scholar]
- 15.Lundin K. 1997. Comparative ultrastructure of the epidermal ciliary rootlets and associated structures in species of the Nemertodermatida and Acoela (Plathelminthes). Zoomorphology 117, 81–92. ( 10.1007/s004350050033) [DOI] [Google Scholar]
- 16.Lundin K. 2000. Phylogeny of the Nemertodermatida (Acoelomorpha, Platyhelminthes). Zool. Scr. 29, 65–74. ( 10.1046/j.1463-6409.2000.00028.x) [DOI] [Google Scholar]
- 17.Lundin K, Sterrer W. 2001. The Nemertodermatida. In Interrelationships of the Platyhelminthes (Systematics Association special volume series), vol. 60 (eds Littlewood DTJ, Bray RA), pp. 24–27. London, UK: Taylor and Francis. [Google Scholar]
- 18.Pedersen KJ, Pedersen LR. 1986. Fine structural observation of the extracellular matrix (ECM) of Xenoturbella bocki, Westblad 1949. Acta Zool. 67, 103–113. ( 10.1111/j.1463-6395.1986.tb00854.x) [DOI] [Google Scholar]
- 19.Hyman LH. 1951. The invertebrates, Platyhelminthes and Rhynchocoela, vol. II New York, NY: McGraw-Hill Book Co. [Google Scholar]
- 20.Carranza S, Baguñà J, Riutort M. 1997. Are the Platyhelminthes a monophyletic primitive group? An assessment using 18S rDNA sequences. Mol. Biol. Evol. 14, 485–497. ( 10.1093/oxfordjournals.molbev.a025785) [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Trillo I, Riutort M, Littlewood DTJ, Herniou EA, Baguñà J. 1999. Acoel flatworms : earliest extant bilaterian metazoans, not members of Platyhelminthes. Science 283, 1919–1923. ( 10.1126/science.283.5409.1919) [DOI] [PubMed] [Google Scholar]
- 22.Baguñà J, Riutort M. 2004. The dawn of bilaterian animals: the case of acoelomorph flatworms. BioEssays 26, 1046–1057. ( 10.1002/bies.20113) [DOI] [PubMed] [Google Scholar]
- 23.Richter S, et al. 2010. Invertebrate neurophylogeny: suggested terms and definitions for a neuroanatomical glossary. Front. Zool. 7, 29 ( 10.1186/1742-9994-7-29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raikova OI. 2004. Neuroanatomy of basal bilaterians (Xenoturbellida, Nemertodermatida, Acoela) and its phylogenetic implications. PhD thesis, Department of Biology, Abo Akademi University, Turku, Finland.
- 25.Haszprunar G. 1996. Platyhelminthes and Platyhelminthomorpha: paraphyletic taxa. J. Zool. Syst. Evol. Res. 34, 41–48. ( 10.1111/j.1439-0469.1996.tb00808.x) [DOI] [Google Scholar]
- 26.Harzsch S, Müller CHG. 2007. A new look at the ventral nerve centre of Sagitta: implications for the phylogenetic position of Chaetognatha (arrow worms) and the evolution of the bilaterian nervous system. Front. Zool. 4, 14 ( 10.1186/1742-9994-4-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunn CW, et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749. ( 10.1038/nature06614) [DOI] [PubMed] [Google Scholar]
- 28.Stach T. 2015. Xenoturbella. In Structure and evolution of invertebrate nervous systems (eds Schmidt-Rhaesa A, Harzsch S, Purschke G), pp. 62–66. Oxford, UK: Oxford University Press. [Google Scholar]
- 29.Perea-Atienza E, Gavilan B, Chiodin M, Abril JF, Hoff KJ, Poustka AJ, Martinez P. 2015. The nervous system of Xenacoelomorpha: a genomic perspective. J. Exp. Biol. 218, 618–628. ( 10.1242/jeb.110379) [DOI] [PubMed] [Google Scholar]
- 30.Nielsen C. 2010. After all: Xenoturbella is an acoelomorph! Evol. Dev. 12, 241–243. ( 10.1111/j.1525-142X.2010.00408.x) [DOI] [PubMed] [Google Scholar]
- 31.Westblad E. 1949. Xenoturbella bocki n.g, n.sp, a peculiar, primitive turbellarian type. Ark. Zool. 1, 3–29. [Google Scholar]
- 32.Israelsson O. 1999. New light on the enigmatic Xenoturbella (phylum uncertain): ontogeny and phylogeny. Proc. R. Soc. Lond. B 266, 835–841. ( 10.1098/rspb.1999.0713) [DOI] [Google Scholar]
- 33.Rouse GW, Wilson NG, Vrijenhoek RC. 2013. First Xenoturbella spp. (Xenoturbellida) from the Pacific. In Society for Integrative and Comparative Biology 2013 Annual Meeting, 3–7 January 2013, San Francisco, CA. See http://www.sicb.org/meetings/2013/schedule/abstractdetails.php?id=334.
- 34.Raikova OI, Reuter M, Jondelius U, Gustafsson MKS. 2000. An immunocytochemical and ultrastructural study of the nervous and muscular systems of Xenoturbella westbladi (Bilateria inc. sed.). Zoomorphology 120, 107–118. ( 10.1007/s004350000028) [DOI] [Google Scholar]
- 35.Israelsson O. 2007. Ultrastructural aspects of the ‘statocyst’ of Xenoturbella (Deuterostomia) cast doubt on its function as a georeceptor. Tissue Cell 39, 171–177. ( 10.1016/j.tice.2007.03.002) [DOI] [PubMed] [Google Scholar]
- 36.Ehlers U. 1991. Comparative morphology of statocysts in the Plathelminthes and the Xenoturbellida. Hydrobiologia 227, 263–271. ( 10.1007/BF00027611) [DOI] [Google Scholar]
- 37.Westblad E. 1937. Die Turbellarien-Gattung Nemertoderma Steinböck. Acta Soc. Fauna Flora Fenn. 60, 45–89. [Google Scholar]
- 38.Meixer J. 1938. Turbellaria (Strudelwürmer). I. Allgemeiner Teil. In Die Tierwelt der Nord- und Ostsee, 33/IVb (eds Grimpe G, Wagler E, Remane A), pp. 1–146. Leipzig, Germany: Akademische Verlagsgesellschaft. [Google Scholar]
- 39.Tyler S. 2001. The early worm: origins and relationships of the lower flatworms. In Interrelationships of the Platyhelminthes (eds Littlewood DTJ, Bray RA), pp. 3–12. London, UK: Taylor and Francis. [Google Scholar]
- 40.Raikova OI, Reuter M, Jondelius U, Gustafsson MKS. 2000. The brain of the Nemertodermatida (Platyhelminthes) as revealed by anti-5HT and anti-FMRFamide immunostainings. Tissue Cell 32, 358–365. ( 10.1054/tice.2000.0121) [DOI] [PubMed] [Google Scholar]
- 41.Raikova OI, Reuter M, Gustafsson MKS, Maule AG, Halton DW, Jondelius U. 2004. Basiepidermal nervous system in Nemertoderma westbladi (Nemertodermatida): GYIRFamide immunoreactivity. Zoology 107, 75–86. ( 10.1016/j.zool.2003.12.002) [DOI] [PubMed] [Google Scholar]
- 42.Børve A, Hejnol A. 2014. Development and juvenile anatomy of the nemertodermatid Meara stichopi (Bock) Westblad 1949 (Acoelomorpha). Front. Zool. 11, 50 ( 10.1186/1742-9994-11-50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westblad E. 1949. On Meara stichopi (Bock) Westblad, a new representative of Turbellaria Archoophora. Ark. Zool. 1, 43–57. [Google Scholar]
- 44.Reisinger E. 1960. Was it Xenoturbella? Z. wiss. Zool. 164, 188–198. [Google Scholar]
- 45.Ax P. 1963. Relationships and phylogeny of the Turbellaria. In The lower Metazoa (ed. Dougherty EC.), pp. 191–224. Berkeley, CA: University California Press. [Google Scholar]
- 46.Ferrero EA. 1973. A fine structure analysis of the statocyst in Turbellaria Acoela. Zool. Scr. 2, 5–16. ( 10.1111/j.1463-6409.1973.tb00793.x) [DOI] [Google Scholar]
- 47.Enrico AF, Bedini C. 1991. Ultrastructural aspects of nervous system and statocyst morphogenesis during embryonic development of Convoluta psammophila Bekl. (Turbellaria, Acoela). Hydrobiol. 227/Dev. Hydrobiol. 69, 131–137. [Google Scholar]
- 48.Jondelius U, Wallberg A, Hooge M, Raikova OI. 2011. How the worm got its pharynx: phylogeny, classification and Bayesian assessment of character evolution in Acoela. Syst. Biol. 60, 845–871. ( 10.1093/sysbio/syr073) [DOI] [PubMed] [Google Scholar]
- 49.Smith JPS, Tyler S. 1985. Fine structure and evolutionary implications of the frontal organ in Turbellaria Acoela. I. Diopisthoporus gymnopharyngeus sp.n. Zool. Scr. 14, 91–102. ( 10.1111/j.1463-6409.1985.tb00180.x) [DOI] [Google Scholar]
- 50.Bery A, Cardona A, Martinez P, Hartenstein V. 2010. Structure of the central nervous system of a juvenile acoel, Symsagittifera roscoffensis. Dev. Genes Evol. 220, 61–76. ( 10.1007/s00427-010-0328-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Semmler H, Chiodin M, Bailly X, Martinez P, Wanninger A. 2010. Steps towards a centralized nervous system in basal bilaterians: insights from neurogenesis of the acoel Symsagittifera roscoffensis. Dev. Growth Differ. 52, 701–713. ( 10.1111/j.1440-169X.2010.01207.x) [DOI] [PubMed] [Google Scholar]
- 52.Crezée M. 1978. Paratomella rubra Rieger and Ott, an amphiatlantic acoel turbellarian. Cah. Biol. Mar. 19, 1–9. [Google Scholar]
- 53.Ehlers U. 1992. On the fine structure of Paratomella rubra Rieger and Ott (Acoela) and the position of the taxon Paratomella Dörjes in a phylogenetic system of the Acoelomorpha (Plathelminthes). Microfauna Mar. 7, 265–293. [Google Scholar]
- 54.Steinböck VO. 1966. Die Hofsteniiden (Turbellaria Acoela). Grundsätzliches zur Evolution der Turbellarien. J. Zool. Syst. Evol. Res. 4, 58–195. ( 10.1111/j.1439-0469.1966.tb00494.x) [DOI] [Google Scholar]
- 55.Corrêa DD. 1960. Two new marine Turbellaria from Florida. Bull. Mar. Sci. Gulf Caribb. 10, 208–216. [Google Scholar]
- 56.Bock S. 1923. Eine neue Turbellariengattung aus Japan. Uppsal Univ. Arsskr. Mat. Och Nat. 1, 1–52. [Google Scholar]
- 57.Crezée M. 1975. Monograph of the Solenofilomorphidae (Turbellaria: Acoela). Int. Rev. Gesamten Hydrobiol. 60, 769–845. ( 10.1002/iroh.19750600604) [DOI] [Google Scholar]
- 58.Brose N, Petrenko AG, Südhof TC, Jahn R. 1992. Synaptotagmin: a calcium sensor on the synaptic vesicle surface. Science 256, 1021–1025. ( 10.1126/science.1589771) [DOI] [PubMed] [Google Scholar]
- 59.Graff L. 1891. Die Organisation der Turbellaria Acoela. Leipzig, Germany: W. Engelmann. [Google Scholar]
- 60.Delage Y. 1886. Etudes histologiques sur les planaires rhabdocoeles acoeles (Convoluta schultzii O. Sch.). Arch. Zool. Exp. Gén. 4, 109–144. [Google Scholar]
- 61.Reisinger E. 1925. Ein landbewohnender Archiannelide. (Zugleich ein Beitrag zur Systematik der Archianneliden). Z. Morphol. Tiere 3, 197–254. ( 10.1007/BF00408145) [DOI] [Google Scholar]
- 62.Ramachandra NB, Gates RD, Ladurner P, Jacobs DK, Hartenstein V. 2002. Embryonic development in the primitive bilaterian Neochildia fusca: normal morphogenesis and isolation of POU genes Brn-1 and Brn-3. Dev. Genes Evol. 212, 55–69. ( 10.1007/s00427-001-0207-y) [DOI] [PubMed] [Google Scholar]
- 63.Henry JQ, Martindale MQ, Boyer BC. 2000. The unique developmental program of the acoel flatworm, Neochildia fusca. Dev. Biol. 220, 285–295. ( 10.1006/dbio.2000.9628) [DOI] [PubMed] [Google Scholar]
- 64.Magie CR, Pang K, Martindale MQ. 2005. Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis. Dev. Genes Evol. 215, 618–630. ( 10.1007/s00427-005-0022-y) [DOI] [PubMed] [Google Scholar]
- 65.Hejnol A, Martindale MQ. 2009. Coordinated spatial and temporal expression of Hox genes during embryogenesis in the acoel Convolutriloba longifissura. BMC Biol. 7, 65 ( 10.1186/1741-7007-7-65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno E, Nadal M, Baguñà J, Martínez P. 2009. Tracking the origins of the bilaterian Hox patterning system: insights from the acoel flatworm Symsagittifera roscoffensis. Evol. Dev. 11, 574–581. ( 10.1111/j.1525-142X.2009.00363.x) [DOI] [PubMed] [Google Scholar]
- 67.Sikes JM, Bely AE. 2010. Making heads from tails: development of a reversed anterior–posterior axis during budding in an acoel. Dev. Biol. 338, 86–97. ( 10.1016/j.ydbio.2009.10.033) [DOI] [PubMed] [Google Scholar]
- 68.Sinigaglia C, Busengdal H, Leclère L, Technau U, Rentzsch F. 2013. The bilaterian head patterning gene six3/6 controls aboral domain development in a cnidarian. PLoS Biol. 11, e1001488 ( 10.1371/journal.pbio.1001488) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moreno E, De Mulder K, Salvenmoser W, Ladurner P, Martínez P. 2010. Inferring the ancestral function of the posterior Hox gene within the bilateria: controlling the maintenance of reproductive structures, the musculature and the nervous system in the acoel flatworm Isodiametra pulchra. Evol. Dev. 12, 258–266. ( 10.1111/j.1525-142X.2010.00411.x) [DOI] [PubMed] [Google Scholar]
- 70.Young T, Deschamps J. 2009. Hox, Cdx, and anteroposterior patterning in the mouse embryo. Curr. Top. Dev. Biol. 88, 235–255. ( 10.1016/S0070-2153(09)88008-3) [DOI] [PubMed] [Google Scholar]
- 71.Hejnol A, Martindale MQ. 2008. Acoel development indicates the independent evolution of the bilaterian mouth and anus. Nature 456, 382–386. ( 10.1038/nature07309) [DOI] [PubMed] [Google Scholar]
- 72.Srivastava M, Mazza-Curll KL, van Wolfswinkel JC, Reddien PW. 2014. Whole-body acoel regeneration is controlled by Wnt and Bmp-Admp signaling. Curr. Biol. 24, 1107–1113. ( 10.1016/j.cub.2014.03.042) [DOI] [PubMed] [Google Scholar]
- 73.Bertrand N, Castro DS, Guillemot F. 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530. ( 10.1038/nrn874) [DOI] [PubMed] [Google Scholar]
- 74.Guillemot F. 2007. Spatial and temporal specification of neural fates by transcription factor codes. Development 134, 3771–3780. ( 10.1242/dev.006379) [DOI] [PubMed] [Google Scholar]
- 75.Strotmann R, Schröck K, Böselt I, Stäubert C, Russ A, Schöneberg T. 2011. Evolution of GPCR: change and continuity. Mol. Cell. Endocrinol. 331, 170–178. ( 10.1016/j.mce.2010.07.012) [DOI] [PubMed] [Google Scholar]
- 76.Krishnan A, Almén MS, Fredriksson R, Schiöth HB. 2013. Remarkable similarities between the hemichordate (Saccoglossus kowalevskii) and vertebrate GPCR repertoire. Gene 526, 122–133. ( 10.1016/j.gene.2013.05.005) [DOI] [PubMed] [Google Scholar]
- 77.Fritzsch G, Böhme MU, Thorndyke M, Nakano H, Israelsson O, Stach T, Schlegel M, Hankeln T, Stadler PF. 2008. PCR survey of Xenoturbella bocki Hox genes. J. Exp. Zool. B Mol. Dev. Evol. 310B, 278–284. ( 10.1002/jez.b.21208) [DOI] [PubMed] [Google Scholar]
- 78.Komiya Y, Habas R. 2014. Wnt signal transduction pathways. Organogenesis 4, 68–75. ( 10.4161/org.4.2.5851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petersen CP, Reddien PW. 2009. Wnt signaling and the polarity of the primary body axis. Cell 139, 1056–1068. ( 10.1016/j.cell.2009.11.035) [DOI] [PubMed] [Google Scholar]
- 80.Robert N, Lhomond G, Schubert M, Croce JC. 2014. A comprehensive survey of wnt and frizzled expression in the sea urchin Paracentrotus lividus. Genesis 52, 235–250. ( 10.1002/dvg.22754) [DOI] [PubMed] [Google Scholar]
- 81.Kusserow A, et al. 2005. Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433, 156–160. ( 10.1038/nature03158) [DOI] [PubMed] [Google Scholar]
- 82.Koizumi O, Sato N, Goto C. 2004. Chemical anatomy of Hydra nervous system using antibodies against Hydra neuropeptides: A review. Hydrobiologia 530–531, 41–47. ( 10.1007/s10750-004-2636-x) [DOI] [Google Scholar]
- 83.Holland LZ, Carvalho JE, Escriva H, Laudet V, Schubert M, Shimeld SM, Yu J-K. 2013. Evolution of bilaterian central nervous systems: a single origin? Evodevo 4, 27 ( 10.1186/2041-9139-4-27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moroz LL. 2009. On the independent origins of complex brains and neurons. Brain. Behav. Evol. 74, 177–190. ( 10.1159/000258665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Northcutt RG. 2012. Evolution of centralized nervous systems: two schools of evolutionary thought. Proc. Natl Acad. Sci. USA 109, 10 626–10 633. ( 10.1073/pnas.1201889109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pani AM, Mullarkey EE, Aronowicz J, Assimacopoulos S, Grove EA, Lowe CJ. 2012. Ancient deuterostome origins of vertebrate brain signalling centres. Nature 483, 289–294. ( 10.1038/nature10838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bouillon J. 1994. Embranchement des cnidaires (Cnidaria). In Traité de Zoologie. Cnidaires, Cténaires, III. Fascicule II (ed. Grassé PP.), pp. 1–28. Paris, France: Masson. [Google Scholar]
- 88.Galliot B, Quiquand M, Ghila L, de Rosa R, Miljkovic-Licina M, Chera S. 2009. Origins of neurogenesis, a cnidarian view. Dev. Biol. 332, 2–24. ( 10.1016/j.ydbio.2009.05.563) [DOI] [PubMed] [Google Scholar]
- 89.Koizumi O, Itazawa M, Mizumoto H, Minobe S, Javois LC, Grimmelikhuijzen CJP, Bode HR. 1992. Nerve ring of the hypostome in Hydra. I. Its structure, development, and maintenance. J. Comp. Neurol. 326, 7–21. ( 10.1002/cne.903260103) [DOI] [PubMed] [Google Scholar]
- 90.Marlow HQ, Srivastava M, Matus DQ, Rokhsar D, Martindale MQ. 2009. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev. Neurobiol. 69, 235–254. ( 10.1002/dneu.20698) [DOI] [PubMed] [Google Scholar]
- 91.Nakanishi N, Renfer E, Technau U, Rentzsch F. 2012. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development 139, 347–357. ( 10.1242/dev.071902) [DOI] [PubMed] [Google Scholar]
- 92.Bullock TH. 1945. The anatomical organization of the nervous system of Enteropneusta. Q. J. Microsc. Sci. 86, 55–112. [PubMed] [Google Scholar]
- 93.Knight-Jones EW. 1952. On the nervous system of Saccoglossus cambrensis (Enteropneusta). Phil. Trans. R. Soc. Lond. B 236, 315–354. ( 10.1098/rstb.1952.0004) [DOI] [Google Scholar]
- 94.Lowe CJ. 2008. Molecular genetic insights into deuterostome evolution from the direct-developing hemichordate Saccoglossus kowalevskii. Phil. Trans. R. Soc. B 363, 1569–1578. ( 10.1098/rstb.2007.2247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruppert EE. 2005. Key characters uniting hemichordates and chordates: homologies or homoplasies? Can. J. Zool. 83, 8–23. ( 10.1139/z04-158) [DOI] [Google Scholar]
- 96.Pickens PE. 1970. Conduction along the ventral nerve cord of a hemichordate worm. J. Exp. Biol. 53, 515–528. [Google Scholar]
- 97.Cameron CB, Garey JR, Swalla BJ. 2000. Evolution of the chordate body plan: new insights from phylogenetic analyses of deuterostome phyla. Proc. Natl Acad. Sci. USA 97, 4469–4474. ( 10.1073/pnas.97.9.4469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cannon JT, Kocot KM, Waits DS, Weese DA, Swalla BJ, Santos SR, Halanych KM. 2014. Report phylogenomic resolution of the hemichordate and echinoderm clade. Curr. Biol. 24, 1–6. ( 10.1016/j.cub.2014.10.016) [DOI] [PubMed] [Google Scholar]
- 99.Osborn KJ, Kuhnz LA, Priede IG, Urata M, Gebruk AV, Holland ND. 2012. Diversification of acorn worms (Hemichordata, Enteropneusta) revealed in the deep sea. Proc. R Soc. B 279, 1646–1654. ( 10.1098/rspb.2011.1916) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Swalla BJ, Smith AB. 2008. Deciphering deuterostome phylogeny: molecular, morphological and palaeontological perspectives. Phil. Trans. R Soc. B 363, 1557–1568. ( 10.1098/rstb.2007.2246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jager M, Chiori R, Alié A, Dayraud C, Quéinnec E, Manuel M. 2011. New insights on ctenophore neural anatomy: immunofluorescence study in Pleurobrachia pileus (Müller, 1776). J. Exp. Zool. B Mol. Dev. Evol. 316B, 171–187. ( 10.1002/jez.b.21386) [DOI] [PubMed] [Google Scholar]
- 102.Moroz LL, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114. ( 10.1038/nature13400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moroz LL. 2015. Convergent evolution of neural systems in ctenophores. J. Exp. Biol. 218, 598–611. ( 10.1242/jeb.110692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wanninger A. 2009. Shaping the things to come: ontogeny of lophotrochozoan neuromuscular systems and the tetraneuralia concept. Biol. Bull. 216, 293–306. ( 10.2307/25548161) [DOI] [PubMed] [Google Scholar]
- 105.Bullock TH, Horridge GA. 1965. Structure and function in the nervous systems of invertebrates. San Francisco, CA: Freeman. [Google Scholar]
- 106.Budelmann BU. 1995. The cephalopod nervous system: what evolution has made of the molluscan design. In The nervous systems of invertebrates: an evolutionary and comparative approach (eds Breidbach O, Kutsh W), pp. 115–138. Basel, Switzerland: Birkhäuser Verlag. [Google Scholar]
- 107.Bulloch AGM, Ridgway RL. 1995. Comparative aspects of gastropod neurobiology. In The nervous systems of invertebrates: an evolutionary and comparative approach (eds Breidbach O, Kutsh W), pp. 88–113. Basel, Switzerland: Birkhäuser Verlag. [Google Scholar]
- 108.Hartenstein V, Stollewerk A. 2015. The evolution of early neurogenesis. Dev. Cell 32, 390–407. ( 10.1016/j.devcel.2015.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Davidson EH, Peter IS. 2015. Genomic control process: development and evolution. San Diego, CA: Elsevier Science Publishing Co Inc. [Google Scholar]
- 110.Gonda A, Herczeg G, Merilä J. 2013. Evolutionary ecology of intraspecific brain size variation: a review. Ecol. Evol. 3, 2751–2764. ( 10.1002/ece3.627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cherniak C. 1994. Component placement optimization in the brain. J. Neurosci. 14, 2418–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chklovskii DB. 2004. Synaptic connectivity and neuronal morphology: two sides of the same coin. Neuron 43, 609–617. ( 10.1016/j.neuron.2004.08.012) [DOI] [PubMed] [Google Scholar]
- 113.Cherniak C, Rodriguez-Esteban R. 2010. Information processing limits on generating neuroanatomy: global optimization of rat olfactory cortex and amygdala. J. Biol. Phys. 36, 45–52. ( 10.1007/s10867-009-9162-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cherniak C, Mokhtarzada Z, Rodriguez-Esteban R, Changizi K. 2004. Global optimization of cerebral cortex layout. Proc. Natl Acad. Sci. USA 101, 1081–1086. ( 10.1073/pnas.0305212101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y. 2013. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl Acad. Sci. USA 110, 20 284–20 289. ( 10.1073/pnas.1315710110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Laughlin SB, Sejnowski TJ. 2003. Communication in neuronal networks. Science 301, 1870–1874. ( 10.1126/science.1089662) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the sequences included in our study keep the original reference number assigned in the preliminary genome/transcriptome analyses. The whole genome/transcriptome original sequences will be made publicly available along with the forthcoming publication of the Xenacoelomorpha Genome Project consortium paper.