ABSTRACT
Lipids endogenous to skin and mucosal surfaces exhibit potent antimicrobial activity against Porphyromonas gingivalis, an important colonizer of the oral cavity implicated in periodontitis. Our previous work demonstrated the antimicrobial activity of the fatty acid sapienic acid (C16:1Δ6) against P. gingivalis and found that sapienic acid treatment alters both protein and lipid composition from those in controls. In this study, we further examined whole-cell protein differences between sapienic acid-treated bacteria and untreated controls, and we utilized open-source functional association and annotation programs to explore potential mechanisms for the antimicrobial activity of sapienic acid. Our analyses indicated that sapienic acid treatment induces a unique stress response in P. gingivalis resulting in differential expression of proteins involved in a variety of metabolic pathways. This network of differentially regulated proteins was enriched in protein-protein interactions (P = 2.98 × 10−8), including six KEGG pathways (P value ranges, 2.30 × 10−5 to 0.05) and four Gene Ontology (GO) molecular functions (P value ranges, 0.02 to 0.04), with multiple suggestive enriched relationships in KEGG pathways and GO molecular functions. Upregulated metabolic pathways suggest increases in energy production, lipid metabolism, iron acquisition and processing, and respiration. Combined with a suggested preferential metabolism of serine, which is necessary for fatty acid biosynthesis, these data support our previous findings that the site of sapienic acid antimicrobial activity is likely at the bacterial membrane.
IMPORTANCE P. gingivalis is an important opportunistic pathogen implicated in periodontitis. Affecting nearly 50% of the population, periodontitis is treatable, but the resulting damage is irreversible and eventually progresses to tooth loss. There is a great need for natural products that can be used to treat and/or prevent the overgrowth of periodontal pathogens and increase oral health. Sapienic acid is endogenous to the oral cavity and is a potent antimicrobial agent, suggesting a potential therapeutic or prophylactic use for this fatty acid. This study examines the effects of sapienic acid treatment on P. gingivalis and highlights the membrane as the likely site of antimicrobial activity.
INTRODUCTION
Inflammation in the oral cavity ranges from mild and reversible inflammation of the gingiva (gingivitis) to a more chronic inflammation and destruction of tooth-supporting structures (periodontitis). Gingivitis occurs in 50 to 90% of adults worldwide, depending on the widely differing definitions of gingivitis (1). Periodontitis occurs in just over 47% of the U.S. population, with prevalences of 8.7, 30.0, and 8.5% for mild, moderate, and severe periodontitis depending on multiple factors, including oral hygiene, socioeconomic status, educational status, age, and other environmental, genetic, and metabolic risk factors (2). These numbers differ in different populations, and older individuals are at a particularly high risk: 64% of adults 65 years old or older have either moderate or severe periodontitis (2).
Porphyromonas gingivalis is among the influential opportunistic periodontal pathogens contributing to periodontal disease (3). P. gingivalis is present more frequently in patients with periodontitis than in healthy individuals; furthermore, a strong positive relationship is seen between P. gingivalis and multiple diagnostic parameters of periodontitis (3–5). This Gram-negative, black-pigmented anaerobic coccobacillus is recognized as a late colonizer in the development of oral biofilms (3, 6), where multiple virulence factors produced by P. gingivalis contribute to its pathogenicity (7).
A diverse array of nonspecific innate immune factors help maintain periodontal health, including more than 45 antimicrobial proteins in the gingival crevicular fluid and/or saliva (8, 9). Among these innate immune molecules contributing to the control of oral bacteria are several lipids (10, 11). Saliva and mucosal epithelial surfaces contain multiple lipid families, including fatty acids, triglycerides, ceramides, wax esters, cholesterol esters, phospholipids, sterols, sterol esters, and squalene (12–15). Fatty acids of sebaceous origin are particularly potent antimicrobial agents against a wide range of both Gram-negative and Gram-positive bacteria, exhibiting differential and dose-dependent antimicrobial activity that is both bacterium and lipid specific (16–21).
Fatty acids, especially sapienic acid, appear to be more active against oral species of bacteria than against nonoral bacterial species (16, 22). Sapienic acid (C16:1Δ6; 254.4 g/mol) kills P. gingivalis within 6 min (MIC, 58.6 μg/ml; minimal bactericidal concentration [MBC], 62.5 μg/ml), causing disruption of cell membranes that is evident in micrographs (22). Additionally, more than 50% of the sapienic acid used to treat P. gingivalis in our studies stayed associated with the bacterial lipids or was retained intracellularly. Collectively, these findings lead us to speculate that the site of sapienic acid attack on P. gingivalis is likely the bacterial membrane. However, more information is needed for full understanding of the response of P. gingivalis to sapienic acid treatment.
Functional interactions between proteins are the basis for cellular processing, and systematic characterization of these relationships is important for a systems-level understanding of cellular processes (23). In our previous study, we found that sapienic acid treatment of P. gingivalis induces an upregulation of several proteins involved in the biosynthesis of bacterial lipids, metabolism and energy production, degradation of polypeptides, cell adhesion, and virulence (22). Our current study extends those previously reported results, identifies additional proteins in P. gingivalis that are differentially expressed after treatment with sapienic acid, and identifies the primary metabolic pathways affected by sapienic acid treatment.
MATERIALS AND METHODS
Preparation of sapienic acid.
Sapienic acid lipids were obtained from Matreya Inc. (Pleasant Gap, PA) and were dissolved in a chloroform-methanol solution (2:1). Their purity was confirmed by thin-layer chromatography (TLC). Lipids, dried under nitrogen, were then mixed with 0.14 M sterile NaCl to make a 1.0-mg/ml stock solution, which was first sonicated in sequential 5-min increments to suspend the lipid and then diluted to the desired concentration using 0.14 M NaCl.
Porphyromonas gingivalis growth conditions and treatment.
Porphyromonas gingivalis strain 381 was cultured in tryptic soy broth (Difco Laboratories, Detroit, MI) supplemented with vitamin K1 and hemin (Sigma Chemical Co., St. Louis, MO) and was incubated at 37°C in an anaerobic chamber (Coy Laboratory Products Inc., Grass Lake, MI) containing an atmosphere of 85% N2, 10% H2, and 5% CO2 for 3 days.
Six milliliters of the bacterial culture was transferred to 6 ml of fresh medium and was incubated overnight before the optical density at 600 nm was read (Spectronic 20D+ spectrophotometer; Thermo Fisher Scientific, Inc., Waltham, MA) to ensure that the bacteria were in log phase. Half of the culture was then treated with 10 times the MIC of sapienic acid for a final concentration of 625 μg/ml (2.46 mM), and the other half became the negative control and was treated with 0.14 M NaCl. Both samples were incubated anaerobically for 2.5 h. Dilutions of each sample were plated at the beginning and end of the incubation period in order to obtain viable bacterial counts. After the incubation period, bacteria were pelleted and washed with 0.14 M NaCl containing 0.1% sodium azide (Sigma-Aldrich, St. Louis, MO) and the protease inhibitor phenylmethylsulfonylfluoride (PMSF; Sigma-Aldrich, St. Louis, MO). Protein concentrations were determined using a NanoDrop spectrophotometer (ND-1000; Thermo Scientific, Wilmington, DE), and a small amount of each sample was examined via SDS-PAGE before the samples were prepared for the assessment of proteins.
2D-DIGE.
Bacteria treated with either sapienic acid or 0.14 M NaCl (as described above) were pelleted, frozen at −80°C, and shipped to Applied Biomics (Hayward, CA) for 2-dimensional difference in-gel electrophoresis (2D-DIGE). For 2D-DIGE analysis, treated and untreated samples were washed with 10 mM Tris-HCl buffer to remove salts, and cells were lysed to release proteins. Proteins from each sample were then denatured, labeled with Cy3 (treated) or Cy5 (untreated) (GE Healthcare Life Sciences, Pittsburgh, PA), mixed, and then separated first by the isoelectric point and then by molecular weight. The gel was scanned using a Typhoon Trio image scanner (GE Healthcare Life Sciences, Pittsburgh, PA), and images were analyzed using ImageQuant software (version 6.5; GE Healthcare Life Sciences, Pittsburgh, PA). DeCyder in-gel analysis software (GE Healthcare Life Sciences, Pittsburgh, PA) facilitated the in-gel comparative analysis of spots, allowing calculation of the fold change in expression levels by comparing protein expression in sapienic acid-treated samples with that in untreated samples run on separate control gels. All samples with a detectable change in protein levels were then picked from the gel using the Ettan Spot Picker (GE Healthcare Life Sciences, Pittsburgh, PA) and were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectroscopy (TOF/TOF 5800 system; AB Sciex, Framingham, MA). Both peptide mass and the associated fragmentation spectra were submitted to a GPS Explorer workstation equipped with a MASCOT search engine (Matrix Science, Boston, MA) in order to search the P. gingivalis National Center for Biotechnology Information (NCBI) nonredundant (nr) protein database. Searches were performed without constraining the protein molecular weight or isoelectric point, with variable carbamidomethylation of cysteine and oxidation of methionine residues, and with one missed cleavage allowed in the search parameters. Candidates with a >95% confidence interval (CI) for either the protein score or the ion score were considered significant.
Data analysis.
An informatics approach was used to analyze relationships between differentially expressed proteins and the physiologic systems affected. Data analysis was completed using the STRING (Search Tool for the Retrieval of Interacting Proteins) database, version 10 (www.string-db.org), an open-access Web-based analysis program that utilizes information obtained from multiple curated databases to predict physical and functional associations between proteins of interest. Known and predicted associations were derived from evidence of published experimental associations, database associations, gene fusion, neighborhood associations, repeated cooccurrence of proteins in the literature, coexpression of proteins in P. gingivalis or a closely related species, and homology with similar protein sequences in closely related species. STRING then calculated individual confidence scores for each predicted protein association based on the approximate probability that a predicted link exists between two proteins in the same KEGG map (24, 25). Finally, a combined confidence score was computed from individual weighted scores, under the assumption of independence for the various sources, correcting for the probability of random observation of interactions (23, 26). STRING outcome parameter confidence limits for our data analyses were set to “high,” corresponding to a confidence score of 75% or higher.
Statistical enrichment analysis was carried out to detect significant enrichment of differentially expressed proteins within functional systems by utilizing information from functionally annotated databases, such as Gene Ontology (GO), KEGG, Pfam, and InterPro. P values were computed by STRING using a hypergeometric test with Bonferroni multiple-comparison corrections (26). All probabilities were calculated using a 5% level of significance. Several proteins were represented by multiple distinct spots in the 2D-DIGE gel, indicating a different size or charge; therefore, each distinct protein spot was treated as a unique protein in reporting fold changes in expression.
A random background model (Random Graph with Given Degree Sequence [RGGDS]) was utilized by STRING to determine enrichment of STRING interactions within our list of differentially expressed proteins. RGGDS preserves the degree of distribution of proteins within the protein list of interest to create a null model of similar size, independent of known pathway annotations. The probability of a given number of protein associations in this null model is calculated using a hypergeometric test with Bonferroni corrections for multiple comparisons (26). All probabilities were calculated using a 5% level of significance.
RESULTS
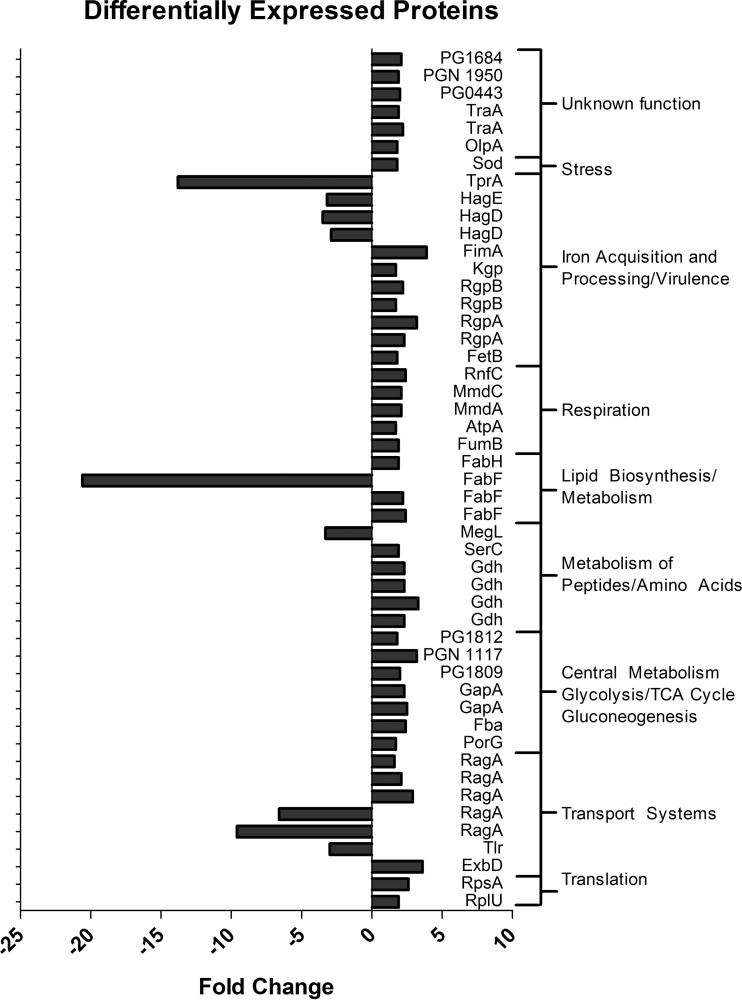
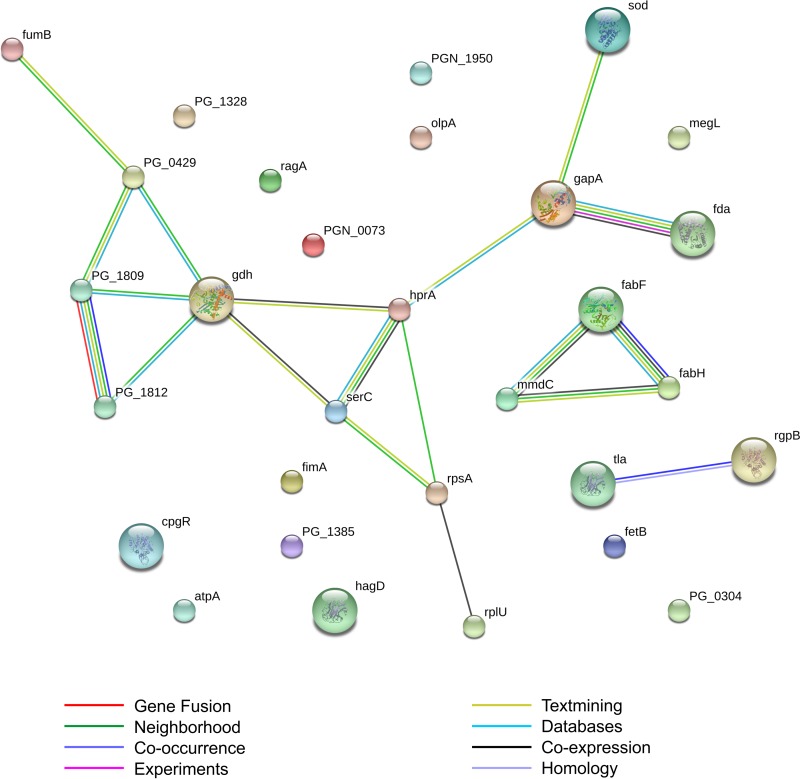
To investigate potential mechanisms involved in the response of P. gingivalis to sapienic acid treatment, we examined the differential expression of proteins in treated versus untreated P. gingivalis samples. A total of 58 differentially expressed protein spots were identified via 2D-DIGE (see Fig. S1 in the supplemental material). Forty-nine of those proteins were identifiable via sequencing and Blast searches (see Table S1 in the supplemental material), of which 29 were unique P. gingivalis proteins recognized by the STRING database software (Fig. 1). Several proteins were represented by multiple distinct spots in the 2D-DIGE gel, indicating a difference in size or charge. Replicate proteins with different sizes or charges were assumed to be modified through posttranslational modifications (e.g., phosphorylation), and each distinct protein spot was treated as a unique protein in reporting fold changes in expression (Fig. 1). However, because specific fold changes in protein expression were not important for the analysis of functional associations between proteins, each protein was included only once in statistical analyses so as to avoid overestimation of protein associations. Functional associations between the 29 unique proteins were significantly enriched; analyses of these data predicted 15 functional associations among these proteins (expected number of interactions, 2.34; P = 2.98 × 10−8). Predicted functional associations between individual proteins are shown in Fig. 2, and specific evidence supporting predictions and confidence scores are shown in Table 1.
FIG 1.
Differentially expressed proteins in P. gingivalis, grouped into functional categories. Fold changes in expression between sapienic acid-treated and untreated samples were calculated from the 2D-DIGE gel using DeCyder in-gel analysis software.
FIG 2.
Predicted functional associations of the 29 P. gingivalis proteins that were differentially expressed with sapienic acid treatment. Functional associations predicted using the STRING database are significantly enriched. The expected number of interactions was 2.34; P = 2.98 × 10−8.
TABLE 1.
Confidence scores for predicted protein-protein associationsa
| Gene 1 | Gene 2 | Source of predicted associations |
Confidence score | |||||
|---|---|---|---|---|---|---|---|---|
| Neighborhood | Cooccurrence | Coexpression | Expts | Databases | Text mining | |||
| fabF | fabH | x | x | x | x | x | 0.997 | |
| fabF | mmdC | x | x | x | x | 0.817 | ||
| fabH | mmdC | x | x | x | 0.740 | |||
| fda | gapA | x | x | x | x | x | 0.996 | |
| ragA | ragB | x | x | x | 0.950 | |||
| fumB | porG | x | x | 0.804 | ||||
| fumB | sod | x | 0.730 | |||||
| PG1809 | porG | x | x | x | x | 0.833 | ||
| gdh | porG | x | x | x | 0.869 | |||
| gdh | PG1809 | x | x | x | 0.837 | |||
| gdh | gapA | x | x | 0.424 | ||||
| PG1812 | PG1809 | x | x | x | x | x | 0.981 | |
| PG1812 | gdh | x | x | x | 0.814 | |||
| PGN_1117 | porG | x | x | 0.701 | ||||
| serC | rpsA | x | x | 0.709 | ||||
| rpsA | rplU | x | x | 0.980 | ||||
Confidence scores were calculated by the STRING database. The combined confidence scores shown here are computed from individual weighted scores, corrected for the probability of random observation of interactions. STRING outcome parameters were set to “high,” corresponding to a confidence score of 75% or higher. Confidence scores for all predicted protein-protein interactions and the sources of those predicted associations are shown here. A high confidence level is ≥0.75; a medium confidence level is ≥0.50.
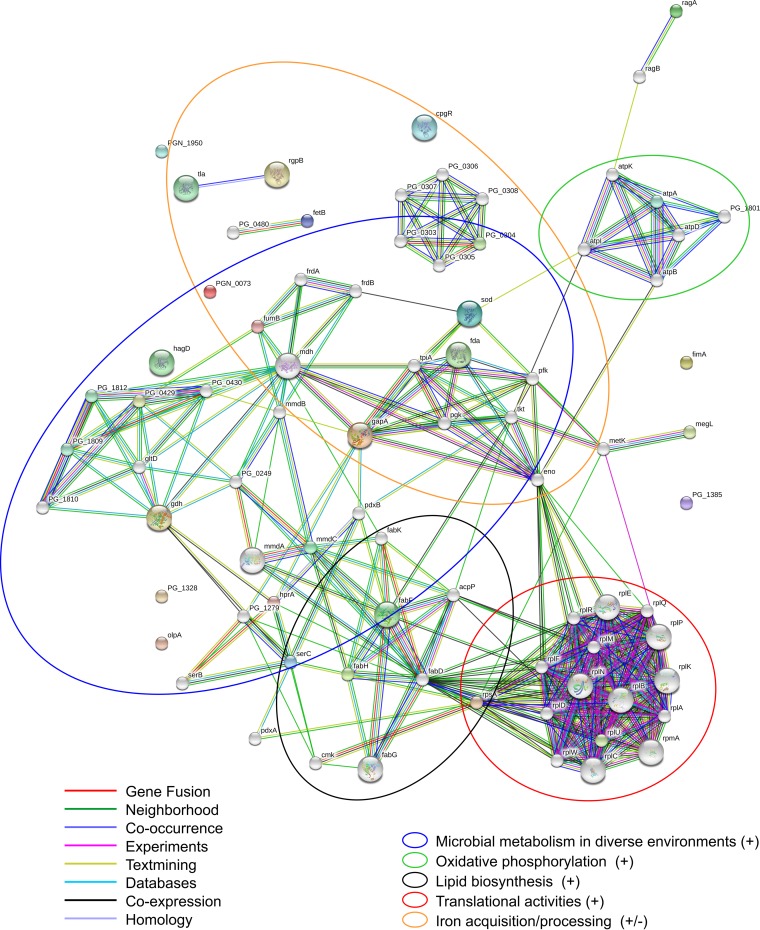
Differentially expressed proteins were associated with several molecular and metabolic systems and pathways, including glycolysis/gluconeogenesis, metabolism, respiration, transport systems, lipid biosynthesis, translation, and virulence (Fig. 1; see also Table S1 in the supplemental material). This suggests that sapienic acid treatment induces changes in many important P. gingivalis metabolic functions (see Fig. S2 in the supplemental material). The identification of the previously mentioned metabolic pathways involved was substantiated when we added 50 additional associated proteins (Fig. 3) to the STRING protein network containing the differentially regulated proteins (Fig. 2). In Fig. 3, the differentially regulated proteins are represented by colored circles, and the additional proteins clustered around these that were detected in our study are represented by white circles. Statistical enrichment analyses showed significant enrichment of differentially expressed proteins in multiple KEGG pathways and GO molecular functions (Table 2), indicating over- or underrepresentation of specific proteins within these pathways. Cellular functions with specific overrepresentations of proteins included respiration, metabolism and energy production, translation, iron acquisition and processing, lipid biosynthesis, and microbial metabolism in diverse environments. The primary pathways with underrepresented proteins were transport systems. Some (though not all) proteins involved in the virulence of P. gingivalis were also underrepresented, but this underrepresentation was not statistically significant.
FIG 3.
Association of differentially regulated P. gingivalis proteins with other P. gingivalis proteins in various metabolic pathways. The predicted functional associations between the 29 unique proteins identifiable by the STRING database (colored circles) are shown with an additional 50 protein associations (white circles), highlighting the roles of our proteins of interest in several important metabolic pathways.
TABLE 2.
Enrichment analysisa
| Pathway or function | GO IDb | Differentially expressed proteins | P valuec | No. of differentially expressed proteins |
|---|---|---|---|---|
| KEGG pathway | ||||
| Carbon metabolism | 1200 | PG2124, PG1278, PG1417, PG1190, PG0548, PG1755, PG1812, PG1809 | 0.0000230* | 8 |
| Microbial metabolism in diverse environments | 1120 | PG1417, PG1278, PG1190, PG2124, PG1755, PG0429, PG1809, PG1812 | 0.0001170* | 8 |
| Citrate cycle (TCA) | 20 | PG1417, PG0429, PG1809, PG1812 | 0.0022100* | 4 |
| Metabolic pathways | 1100 | PG1417, PG1232, PG1278, PG1190, PG1803, PG2124, PG1755, PG1764, PG2141, PG0669, PG0429, PG1809, PG1912 | 0.0062200* | 13 |
| Fatty acid biosynthesis | 61 | PG1764, PG2141 | 0.0466900* | 2 |
| Methane metabolism | 680 | PG1190, PG1278, PG1803 | 0.0507000 | 3 |
| Glycolysis/gluconeogenesis | 10 | PG2124, PG1755 | 0.1990000 | 2 |
| Glycine, serine, and threonine metabolism | 260 | PG1278, PG1190 | 0.2159000 | 2 |
| Biosynthesis of amino acids | 1230 | PG2124, PG1278, PG1755 | 0.2159000 | 3 |
| Fatty acid metabolism | 1212 | PG1764, PG2141 | 0.5110000 | 2 |
| GO molecular functions | ||||
| Cysteine-type endopeptidase activity | 0004197 | PG2024, PG1844, PGN_1970, PG0506 | 0.0202000* | 4 |
| Oxidoreductase activity | 0016491 | PG1545, PG2124, PG1190, PG1232, PG0429, PG1809, PG1812, PG0304 | 0.0395000* | 8 |
| Cysteine-type peptidase activity | 0008234 | PG2024, PG1844, PGN_1970, PG0506 | 0.0395000* | 4 |
| Oxidoreductase activity, acting on the aldehyde or oxo group of donors | 0016903 | PG2124, PG0429, PG1809 | 0.0439900* | 3 |
| Unknown molecular function | 0003674 | PG1545, PG0343, PG1755, PG2124, PG1764, PG2141, PGN_1023, PG0185, PG1190, PG1232, PG1417, PG2024, PG1803, PG1278, PG1297, PGN_1970, PG0506, PG0314, PG0669, PG1844, PG0181, PG0304, PG1328, PG0429, PG1809, PG1812 | 0.0563000 | 26 |
| Catalytic activity | 0003824 | PG1545, PG0343, PG1755, PG2124, PG1764, PG2141, PGN_1023, PG1190, PG1232, PG1417, PG2024, PG1803, PG1278, PG0506, PGN_1970, PG0669, PG1844, PG0304, PG0429, PG1809, PG1812 | 0.2540000 | 21 |
| Endopeptidase activity | 0004175 | PG0506, PGN_1970, PG1844, PG2024 | 0.3521000 | 4 |
| Lyase activity | 0016829 | PG1417, PG0343, PG1755, PG0669 | 0.3549000 | 4 |
Carried out by using the STRING database to detect enrichment of proteins in P. gingivalis metabolic pathways with sapienic acid-induced treatment.
ID, identification number.
P values were computed by STRING using a hypergeometric test with Bonferroni multiple comparisons. All probabilities were calculated using a 5% level of significance. Asterisks indicate a 5% level of statistical significance.
Effects of sapienic acid on central metabolism: glycolysis, gluconeogenesis, and the TCA cycle.
Six metabolic enzymes showed increased expression (1.7- to 3.2-fold) in central metabolic pathways (Fig. 1; see also Fig. S2 and Table S1 in the supplemental material), highlighting the importance of these pathways in the response of P. gingivalis to sapienic acid treatment. Pyruvate ferredoxin/flavodoxin oxidoreductase protein (PorG; PG0548) is predicted to function in the pyruvate-to-acetate pathway of the tricarboxylic acid (TCA) cycle (27) as a thiamine diphosphate-binding domain protein (28). Fructose-1,6-bisphosphate aldolase (Fba; PG1755) is an indispensable enzyme in glycolysis, gluconeogenesis, and the TCA cycle (29, 30) as well as in both anabolic and catabolic amino acid processes (31). Glyceraldehyde-3-phosphate dehydrogenase (GapA; PG2124), involved in the glycolytic pathway, is responsible for the phosphorylation of glyceraldehyde-3-phosphate (29, 32). The last three metabolic enzymes upregulated included 2-oxoglutarate oxidoreductase, subunit gamma (PG1809), acetyl coenzyme A (acetyl-CoA) synthetase (PGN_1117), and 2-ketoisovalerate ferredoxin oxidoreductase (PG1812). These metabolic enzymes are predicted to play roles in KEGG central metabolism pathways (33), but specific functions are thus far limited to the function ascribed to the class of each enzyme and similar functions in related bacterial species.
Sapienic acid modifies amino acid metabolism.
Four enzymes representing different amino acid metabolic pathways were differentially regulated (−3.3- to +3.3-fold) with sapienic acid treatment (Fig. 1; see also Fig. S2 and Table S1 in the supplemental material). P. gingivalis preferentially utilizes aspartate/asparagine, glutamate/glutamine, threonine, serine, leucine, and valine as principal sources of energy acquired by catabolism of amino acids (34). In our samples, the abundance of glutamate dehydrogenase (GDH; PG1232), which is involved in multiple P. gingivalis amino acid metabolic processes, was higher with sapienic acid treatment than in controls. NAD-dependent GDH is involved in the deamination of aspartate to oxaloacetate (35) as well as in the metabolization of glutamate to butyrate, propionate, and ammonia (34). GDH is therefore crucial to the growth of P. gingivalis (36), and a loss of GDH slows P. gingivalis growth, causing a decrease in doubling time and lower cell density yields (36). The NAD dependency of P. gingivalis GDH indicates a primarily catabolic role, contributing to the metabolism of glutamate and aspartate for energy production (35, 36). Although the reverse (anabolic) reaction is also possible under certain conditions (leading to the biosynthesis of glutamate, which can be used to synthesize other amino acids), this is not likely the primary role (35, 36).
Two enzymes involved in the metabolism of glycine, serine, and threonine were upregulated in response to the treatment of P. gingivalis with sapienic acid: glycerate dehydrogenase (HprA; PG1990) and phosphoserine aminotransferase (SerC; PG1278). Serine is particularly important because it also serves as an intermediate for the synthesis of sphingoid bases (31, 33), such as sphingosine and dihydrosphingosine, which are needed for the synthesis of P. gingivalis membrane lipids. P. gingivalis membrane lipids include novel phosphorylated dihydroceramides (modified dihydrosphingosines) containing 17-carbon fully saturated methyl iso-branched fatty acids (37–39). The condensation of serine and palmitoyl-CoA is the rate-limiting step in the biosynthesis of sphingoid bases (40, 41), so the increased abundances of these two enzymes are likely an important component of the adaptive response of P. gingivalis to sapienic acid treatment.
The only downregulated protein (−3.3-fold) involved in amino acid metabolism was methionine gamma-lyase (MegL; PG0343). MegL is a cofactor (e.g., vitamin B6)-binding enzyme (30) that is involved in cysteine, methionine, and selenoamino acid metabolism (33, 42). MegL is unique to microorganisms and plants and catalyzes the committed step of methionine biosynthesis (43).
These data suggest that in response to sapienic acid treatment, P. gingivalis may preferentially metabolize glutamate, aspartate, glycine, serine, and threonine as carbon and energy sources while simultaneously reducing the metabolism of other amino acid sources such as cysteine, methionine, and selenoamino acid. Other bacteria, such as Clostridium sticklandii, which also metabolize multiple amino acids as carbon and energy sources, show preferential amino acid metabolism for energy preservation (44).
Effects of sapienic acid on respiration.
P. gingivalis can metabolize aspartic acid and asparagine to yield succinate through the fumarate electron transport chain (45, 46). In P. gingivalis, fumarate acts as the terminal electron acceptor in a putative respiratory chain composed of transmembrane complexes I to IV. The respiratory chain created by complexes I to IV creates a H+ and/or Na+ gradient, which is used by the fifth complex (ATP synthase type V) to synthesize ATP (46). Similar electron transport phosphorylation (ETP) pathways are responsible for energy conservation in several other bacteria, such as C. sticklandii (44), and may also help conserve energy in P. gingivalis (46). Five proteins from this respiratory chain were more abundant (1.7- to 2.4-fold) following the treatment of P. gingivalis with sapienic acid (Fig. 1; see also Table S1 in the supplemental material), including three complex I components (MmdA, MmdC, and RnfC), one component from complex V (AtpA), and one protein that has not been assigned to a complex yet (FumB).
Complex I of the fumarate respiratory system includes the methylmalonyl-CoA decarboxylase (Mmd) complex (46), two components of which, methymalonyl coenzyme A (MmdA; PG1609) and the biotin carboxyl carrier protein (MmdC; PGN_0503), were upregulated in our studies. The Mmd enzyme complex consists of 10 putative transmembrane segments that help conserve energy (released during decarboxylation reactions), which can be used for ATP synthesis or the regulation of cellular homeostasis via active transport of H+ and Na+ across the membrane to generate a transmembrane Na+ gradient (46).
RnfC (PG0304), also upregulated with sapienic acid treatment, is subunit C of the P. gingivalis RnfABCDGE electron transport complex, which is regulated by iron availability and is predicted to play a role in the transport of electrons to a nitrogenase via ferredoxin (46). Although rnf-like genes have been identified in other bacteria (e.g., Escherichia coli, Clostridium kluyveri, and several Vibrio species), the specific function of these proteins in P. gingivalis remains unclear.
FumB (PG1417) is a class 1 fumarate hydratase (i.e., fumarase) predicted to reversibly catalyze the hydration and dehydration of fumarate to malate during fumarate respiration (30), but the respiration complex to which it belongs is unknown. Although FumB has been identified and classified in P. gingivalis (27), the specific role of FumB in P. gingivalis anaerobic respiration has not been extensively studied yet. However, the FumB fumarase of Escherichia coli, which is oxygen sensitive and is active only under anaerobic conditions, allows this organism to survive under anaerobic conditions through upregulation of the system during anaerobic growth and contributes to the fermentative and noncyclic TCA pathway by converting oxaloacetate and malate to fumarate (47, 48).
AtpA (PG1803) is subunit A of a V-type ATP synthase from complex V, which is dependent on a transmembrane Na+ gradient for ATP synthesis (46). V-type ATP synthases can transport Na+ or H+ ions either into or out of the cell with a few amino acid changes. This reversal of proton transport is thought to prevent toxicity due to excess Na+. For example, V-type ATPases of other microorganisms (e.g., Enterococcus hirae) are not used for general production of ATP but are induced only by high levels of intracellular Na+ or a high pH (49). Given the high energy cost associated with extruding ions from the cell, this is not likely the main function of V-type ATPases under normal P. gingivalis growth conditions, but it may be important under extreme or stressful growth conditions (50). Because multiple components of the fumarate respiratory chain were upregulated in our study, ATP synthesis is likely upregulated to increase the production of energy necessary for increased metabolic events.
Effects of sapienic acid on transport systems and associated surface receptors.
Several components of TonB-linked receptor systems, ExbD, Tlr, and RagA, were differentially regulated (Fig. 1; see also Fig. S2 and Table S1 in the supplemental material). TonB-linked receptors are surface-associated receptors that facilitate the transport of large molecules across the bacterial outer membrane. The energy required by TonB-linked receptors is provided by a complex of Exb and TonB proteins in the cytoplasmic membrane that transport energy from the cytoplasmic membrane to the outer membrane so as to support the movement of large molecules across the outer membrane (51). In P. gingivalis, TonB-linked receptors are known to bind heme and transport it into the cell via the proton motive force of the cytoplasmic membrane (51), but the TonB system may also transport other large molecules across the periplasmic space, like analog systems in other bacteria that involve the transport of specific carbohydrates, vitamins, colicins, or glycoproteins (51–54).
Two TonB-linked receptors, Tlr and RagA, were downregulated with sapienic acid treatment. Tlr was previously named Tla (for TonB-linked adhesion) because the initial sequence mistakenly included a kgp sequence, which codes for an adhesin (55). TonB-linked receptors are essential for growth at low concentrations of hemin (55, 56). RagA is a temperature-regulated TonB-linked receptor that is believed to form a complex on the outer membrane with the hemin-binding lipoprotein RagB. The specific function of RagA is unclear, but because of the hemin-binding ability of RagB, it could also be involved in the transport of heme across the periplasmic space. RagA also has strong similarities to TonB-linked receptors in Bacteroides thetaiotaomicron, which transport macromolecules such as carbohydrates or glycoproteins across the periplasmic space (51, 56). Because P. gingivalis is asaccharolytic, any carbohydrate uptake would likely be linked to anabolic processes (51). RagA also plays a role in the virulence—specifically the soft tissue-destructive abilities—of P. gingivalis. When the rag locus is inactivated, soft tissue destruction in mice is reduced (56).
ExbD is part of the energy-providing complex on the cytoplasmic membrane (51) and is the only component of the TonB system that was upregulated in our samples.
Iron acquisition and processing.
P. gingivalis requires iron for growth; a number of P. gingivalis proteins require iron to function (57). Both iron (57) and hemin (34, 57) levels affect gene expression in multiple biological processes (e.g., metabolism, adherence and invasion, virulence factors, and oxidative stress). Iron is stored in the form of hemin on the outer surface of the cell and is transported into the cell by an energy-dependent process (45). In addition to the TonB-linked receptors discussed in the preceding section, which could be important in the acquisition and storage of iron, FetB is also likely important in iron acquisition and processing. FetB is a heme-binding protein with a GO biological function of anaerobic cobalamin biosynthetic processes (31), but its specific role in P. gingivalis metabolism is unknown. FetB was upregulated with sapienic acid treatment.
Effects of sapienic acid on lipid biosynthesis and metabolism.
Several components of the Gram-negative type 2 fatty acid synthesis (FAS-II) pathway have been described in P. gingivalis but have not been well studied; most of the current knowledge is derived from studies of the FAS-II pathway in E. coli. The FAS-II pathway is characterized by the use of discrete synthases with separate functions, each encoded by unique genes that are essential to fatty acid biosynthesis and thus to bacterial survival. In E. coli, there are three synthases (synthases I, II, and III; FabBFH) located in the cytoplasm (58, 59). Because of differing substrate specificities, each of these isozymes contributes to the regulation of the many products produced by the fatty acid synthase pathway (40).
Two synthases from this pathway, FabH and FabF, were differentially regulated in P. gingivalis with sapienic acid treatment (Fig. 1; see also Table S1 in the supplemental material). FabH was upregulated 1.9-fold. FabF was represented by three distinct spots in the 2D-DIGE gel, indicating proteins of different charges and/or masses. Spots 6 and 7 (see Fig. S1 in the supplemental material) represented 2.4- and 2.2-fold increases in FabF expression with sapienic acid, respectively. The difference in charge between spots 6 and 7 is likely due to posttranslational modifications (e.g., phosphorylation or acetylation), possibly for conversion to active (or inactive) forms of the protein. The exact nature of these modifications is unknown at this time and will require further study. The third FabF spot (spot 12) indicated a 20-fold reduction in expression with sapienic acid treatment; however, because the molecular weight of this spot was lower than that expected for FabF, it is likely an amino-terminal fragment of FabF.
Synthase III (FabH) controls the rate of fatty acid biosynthesis by catalyzing the rate-limiting step (60) and is the only irreversible step in the elongation cycle (40). FabH may also determine whether fatty acids are branched or straight-chain (61). After reduction by a reductase (FabG), synthases I (FabB) and II (FabF) catalyze elongation steps until the fatty acid attains the required length (60). The differences in function between FabB and FabF are not well understood, but studies indicate that synthase II (FabF) is essential for thermal regulation of membrane fatty acid composition (59).
Effects of sapienic acid on virulence proteins.
P. gingivalis produces a number of virulence factors that allow successful colonization, contribute to host inflammation and destruction of tissue, and modulate host immune responses (7, 45). Probably the most versatile of these proteins are the gingipain proteinases RgpA, RgpB, and Kgp, all of which were upregulated (1.7- to 3.2-fold increase) with sapienic acid treatment (Fig. 1; see also Table S1 in the supplemental material). Both the arginine-specific (RgpA and RgpB) and lysine-specific (Kgp) gingipains, in addition to the roles listed above, acquire and degrade heme-containing molecules (51). Although these proteins are well known for their direct involvement in the pathogenesis of P. gingivalis on host tissues, it has been suggested that their primary role may be the acquisition of heme and peptides required for growth (45), given that the acquisition of heme appears to be regulated by the growth phase as much as by heme deficiency (45, 51).
FimA (fimbrillin), which also showed increased (3.9-fold) expression with sapienic acid treatment, is the major component of P. gingivalis type II fimbriae (62). P. gingivalis type II fimbriae mediate adherence to host tissues and salivary molecules (45, 63) and play a role in biofilm formation (64). The expression of fimbriae appears to be upregulated when RgpAB expression is increased and to be downregulated with its decrease (65). Our studies show similar trends for the upregulation of FimA and RgpAB.
P. gingivalis produces at least five hemagglutinins (HagABCDE) that function as adhesins to facilitate the colonization of human cell surface receptors for virulence and nutritional purposes (45). Sapienic acid treatment induced the downregulation of both HagD and HagE (−2.9 to −3.5-fold change), which have high sequence similarities to HagA (73.8% and 93%, respectively) but do not show significant homologies to HagB and HagC (45).
Effects of sapienic acid on stress response proteins.
Superoxide dismutase (Sod) confers tolerance to oxidative stress on P. gingivalis and is known to increase its expression in response to oxidative stress while simultaneously downregulating Fim expression (66). Although both FimA and Sod may be coordinately regulated through the stress protein OxyR (66), both Sod (1.8-fold increase) and FimA were upregulated with sapienic acid treatment. This is atypical of an oxidative response and constitutes part of a possibly unique stress response of P. gingivalis induced by sapienic acid treatment. The previously described universal stress protein of P. gingivalis, UspA (67), was not differentially regulated in our samples.
DISCUSSION
The survival of any successful pathogen requires the ability to adapt quickly to changing environments (68). Mounting a rapid stress response would require additional energy at the lowest possible cost to the cell in order to simultaneously maintain minimal cellular functions for survival. Thus, the rapid increase in metabolic events seen with sapienic acid treatment, including increased levels of energy production, and the preferential use of energy sources and specific metabolic pathways to conserve energy are not unexpected. The downregulation of Hag proteins was also an expected response, since microorganisms typically adapt to various environmental stresses (e.g., pH, temperature, oxygen tension, starvation, and desiccation) by downregulating the expression of virulence genes to allow for optimal growth and survival (69, 70).
The antimicrobial properties of fatty acids, such as sapienic acid, are well documented, and the potential results of fatty acid treatment have been reviewed (71). In our previous study, we proposed that the main site of sapienic acid activity against P. gingivalis is the plasma membrane—either by the creation of pores in the membrane or by the incorporation of sapienic acid into the membrane (22). The present data support our previous findings, suggesting that sapienic acid is likely incorporated into the membrane, either directly by insertion into the membrane or indirectly through the use of sapienic acid as a fatty acid component of P. gingivalis membrane lipids. The endogenous branched fatty acids used in P. gingivalis dihydroceramides are approximately the same length as sapienic acid; therefore, it is possible that sapienic acid competes with the endogenous fatty acids for insertion into P. gingivalis membrane lipids.
Either direct insertion of sapienic acid into the membrane or incorporation of monounsaturated sapienic acid into membrane dihydroceramides in the place of iso-branched, fully saturated fatty acids would have a fluidizing effect on the membrane. With either of these scenarios, it would be advantageous to increase the production of P. gingivalis fatty acids in order to compete with the insertion of toxic sapienic acid into the bacterial membrane and/or to increase the production of membrane lipids that would stabilize the membrane. An increase in fatty acid biosynthesis is supported by the preferential metabolism of serine for use as an intermediate for the sphingoid bases needed for fatty acid metabolism as well as by the upregulation of synthases important for the initiation of fatty acid biosynthesis (FabH) and the extension of the fatty acid chain via FabF. FabF is also essential for regulating membrane fluidity changes due to thermal fluctuations (59), which would be similar to the environment created by sapienic acid fluidization of the membrane.
Identification of the pathways involved in the potent antibacterial activity of sapienic acid against P. gingivalis not only opens the door for the potential use of sapienic acid as a therapeutic agent against P. gingivalis but may also likely identify potential pathway intermediates that could be targeted directly through the use of drugs. In this study, we used an informatics approach to examine the response of P. gingivalis to sapienic acid treatment and the relationships between differentially expressed proteins and affected physiologic systems. However, many of these proteins have not yet been well studied in P. gingivalis, and their functions are inferred from homologs in other bacteria. Further biochemical and metabolic studies are needed to explore the specific functions of many of these proteins in P. gingivalis and to confirm these findings.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge and thank Hongjin Huang of Applied Biomics, Inc. (Hayward, CA, USA), for help with the 2D-DIGE analysis.
This study was conceived by C. L. Fischer and K. A. Brogden. C. L. Fischer prepared the samples for 2D-DIGE and confirmed them by SDS-PAGE. Data were analyzed by C. L. Fischer and were interpreted by C. L. Fischer, D. V. Dawson, D. R. Blanchette, D. R. Drake, P. W. Wertz, and K. A. Brogden. The article was written by C. L. Fischer and K. A. Brogden and was approved by all authors.
We declare no competing financial interests.
Funding Statement
The two training grants were obtained through our Graduate Program director, Christopher Squier. The funders had no role in study design or in the collection and interpretation of data.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00665-15.
REFERENCES
- 1.Pihlstrom BL, Michalowicz BS, Johnson NW. 2005. Periodontal diseases. Lancet 366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 2.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ; CDC Periodontal Disease Surveillance workgroup. 2012. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res 91:914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- 3.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 4.Socransky SS, Haffajee AD. 1992. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol 63(4 Suppl):322–331. [DOI] [PubMed] [Google Scholar]
- 5.Hutter G, Schlagenhauf U, Valenza G, Horn M, Burgemeister S, Claus H, Vogel U. 2003. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 149:67–75. doi: 10.1099/mic.0.25791-0. [DOI] [PubMed] [Google Scholar]
- 6.Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ Jr. 2002. Communication among oral bacteria. Microbiol Mol Biol Rev 66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt SC, Kesavalu L, Walker S, Genco CA. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol 2000 20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 8.Gorr SU. 2012. Antimicrobial peptides in periodontal innate defense. Front Oral Biol 15:84–98. doi: 10.1159/000329673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorr SU. 2009. Antimicrobial peptides of the oral cavity. Periodontol 2000 51:152–180. doi: 10.1111/j.1600-0757.2009.00310.x. [DOI] [PubMed] [Google Scholar]
- 10.Bibel DJ, Aly R, Shah S, Shinefield HR. 1993. Sphingosines: antimicrobial barriers of the skin. Acta Derm Venereol 73:407–411. [DOI] [PubMed] [Google Scholar]
- 11.Drake DR, Brogden KA, Dawson DV, Wertz PW. 2008. Thematic review series: skin lipids. Antimicrobial lipids at the skin surface. J Lipid Res 49:4–11. doi: 10.1194/jlr.R700016-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Brasser A, Barwacz C, Bratt CL, Dawson D, Brogden KA, Drake D, Wertz P. 2011. Free sphingosine in human saliva. J Dent Res 90:3465. [Google Scholar]
- 13.Brasser AJ, Barwacz CA, Dawson DV, Brogden KA, Drake DR, Wertz PW. 2011. Presence of wax esters and squalene in human saliva. Arch Oral Biol 56:588–591. doi: 10.1016/j.archoralbio.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Law S, Wertz PW, Swartzendruber DC, Squier CA. 1995. Regional variation in content, composition and organization of porcine epithelial barrier lipids revealed by thin-layer chromatography and transmission electron microscopy. Arch Oral Biol 40:1085–1091. doi: 10.1016/0003-9969(95)00091-7. [DOI] [PubMed] [Google Scholar]
- 15.Law SL, Squier CA, Wertz PW. 1995. Free sphingosines in oral epithelium. Comp Biochem Physiol B Biochem Mol Biol 110:511–513. doi: 10.1016/0305-0491(94)00194-Y. [DOI] [PubMed] [Google Scholar]
- 16.Fischer CL, Drake DR, Dawson DV, Blanchette DR, Brogden KA, Wertz PW. 2012. Antibacterial activity of sphingoid bases and fatty acids against Gram-positive and Gram-negative bacteria. Antimicrob Agents Chemother 56:1157–1161. doi: 10.1128/AAC.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergsson G, Arnfinnsson J, Steingrimsson O, Thormar H. 2001. Killing of Gram-positive cocci by fatty acids and monoglycerides. APMIS 109:670–678. doi: 10.1034/j.1600-0463.2001.d01-131.x. [DOI] [PubMed] [Google Scholar]
- 18.Bergsson G, Arnfinnsson J, Steingrimsson O, Thormar H. 2001. In vitro killing of Candida albicans by fatty acids and monoglycerides. Antimicrob Agents Chemother 45:3209–3212. doi: 10.1128/AAC.45.11.3209-3212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang CB, Alimova Y, Myers TM, Ebersole JL. 2011. Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch Oral Biol 56:650–654. doi: 10.1016/j.archoralbio.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brogden KA, Drake DR, Dawson DV, Hill JR, Bratt CL, Wertz PW. 2011. Antimicrobial lipids of the skin and oral mucosa, p 75 In Dayan N, Wertz PW (ed), Innate immune system of skin and oral mucosa: properties and impact in pharmaceutics, cosmetics, and personal care products. John Wiley & Sons Inc., Hoboken, NJ. [Google Scholar]
- 21.Kabara JJ, Swieczkowski DM, Conley AJ, Truant JP. 1972. Fatty acids and derivatives as antimicrobial agents. Antimicrob Agents Chemother 2:23–28. doi: 10.1128/AAC.2.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer CL, Walters KS, Drake DR, Dawson DV, Blanchette DR, Brogden KA, Wertz PW. 2013. Oral mucosal lipids are antibacterial against Porphyromonas gingivalis, induce ultrastructural damage, and alter bacterial lipid and protein compositions. Int J Oral Sci 5:130–140. doi: 10.1038/ijos.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. 2007. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res 35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. 2013. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, Daugherty SC, Dodson RJ, Durkin AS, Gwinn M, Haft DH, Kolonay JF, Nelson WC, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin JL, Duncan MJ, Dewhirst FE, Fraser CM. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol 185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huntley RP, Harris MA, Alam-Faruque Y, Blake JA, Carbon S, Dietze H, Dimmer EC, Foulger RE, Hill DP, Khodiyar VK, Lock A, Lomax J, Lovering RC, Mutowo-Meullenet P, Sawford T, Van Auken K, Wood V, Mungall CJ. 2014. A method for increasing expressivity of Gene Ontology annotations using a compositional approach. BMC Bioinformatics 15:155. doi: 10.1186/1471-2105-15-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shams F, Oldfield NJ, Wooldridge KG, Turner DP. 2014. Fructose-1,6-bisphosphate aldolase (FBA)—a conserved glycolytic enzyme with virulence functions in bacteria: ‘ill met by moonlight.’ Biochem Soc Trans 42:1792–1795. doi: 10.1042/BST20140203. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong SY, Bateman A, Punta M, Attwood TK, Sigrist CJ, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeda K, Nagata H, Kuboniwa M, Ojima M, Osaki T, Minamino N, Amano A. 2013. Identification and characterization of Porphyromonas gingivalis client proteins that bind to Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase. Infect Immun 81:753–763. doi: 10.1128/IAI.00875-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dashper SG, Ang CS, Veith PD, Mitchell HL, Lo AW, Seers CA, Walsh KA, Slakeski N, Chen D, Lissel JP, Butler CA, O'Brien-Simpson NM, Barr IG, Reynolds EC. 2009. Response of Porphyromonas gingivalis to heme limitation in continuous culture. J Bacteriol 191:1044–1055. doi: 10.1128/JB.01270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gharbia SE, Shah HN. 1995. Molecular analysis of surface-associated enzymes of Porphyromonas gingivalis. Clin Infect Dis 20(Suppl 2):S160–S166. doi: 10.1093/clinids/20.Supplement_2.S160. [DOI] [PubMed] [Google Scholar]
- 36.Joe A, Murray CS, McBride BC. 1994. Nucleotide sequence of a Porphyromonas gingivalis gene encoding a surface-associated glutamate dehydrogenase and construction of a glutamate dehydrogenase-deficient isogenic mutant. Infect Immun 62:1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nichols FC. 1998. Novel ceramides recovered from Porphyromonas gingivalis: relationship to adult periodontitis. J Lipid Res 39:2360–2372. [PubMed] [Google Scholar]
- 38.Nichols FC, Riep B, Mun J, Morton MD, Bojarski MT, Dewhirst FE, Smith MB. 2004. Structures and biological activity of phosphorylated dihydroceramides of Porphyromonas gingivalis. J Lipid Res 45:2317–2330. doi: 10.1194/jlr.M400278-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Nichols FC, Riep B, Mun J, Morton MD, Kawai T, Dewhirst FE, Smith MB. 2006. Structures and biological activities of novel phosphatidylethanolamine lipids of Porphyromonas gingivalis. J Lipid Res 47:844–853. doi: 10.1194/jlr.M500542-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Magnuson K, Jackowski S, Rock CO, Cronan JE Jr. 1993. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev 57:522–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rütti MF, Richard S, Penno A, von Eckardstein A, Hornemann T. 2009. An improved method to determine serine palmitoyltransferase activity. J Lipid Res 50:1237–1244. doi: 10.1194/jlr.D900001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Sohngen C, Stelzer M, Thiele J, Schomburg D. 2011. BRENDA, the enzyme information system in 2011. Nucleic Acids Res 39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morozova EA, Kulikova VV, Yashin DV, Anufrieva NV, Anisimova NY, Revtovich SV, Kotlov MI, Belyi YF, Pokrovsky VS, Demidkina TV. 2013. Kinetic parameters and cytotoxic activity of recombinant methionine γ-lyase from Clostridium tetani, Clostridium sporogenes, Porphyromonas gingivalis and Citrobacter freundii. Acta Naturae 5:92–98. [PMC free article] [PubMed] [Google Scholar]
- 44.Fonknechten N, Chaussonnerie S, Tricot S, Lajus A, Andreesen JR, Perchat N, Pelletier E, Gouyvenoux M, Barbe V, Salanoubat M, Le Paslier D, Weissenbach J, Cohen GN, Kreimeyer A. 2010. Clostridium sticklandii, a specialist in amino acid degradation: revisiting its metabolism through its genome sequence. BMC Genomics 11:555. doi: 10.1186/1471-2164-11-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamont RJ, Jenkinson HF. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev 62:1244–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meuric V, Rouillon A, Chandad F, Bonnaure-Mallet M. 2010. Putative respiratory chain of Porphyromonas gingivalis. Future Microbiol 5:717–734. doi: 10.2217/fmb.10.32. [DOI] [PubMed] [Google Scholar]
- 47.Tseng CP. 1997. Regulation of fumarase (fumB) gene expression in Escherichia coli in response to oxygen, iron and heme availability: role of the arcA, fur, and hemA gene products. FEMS Microbiol Lett 157:67–72. doi: 10.1111/j.1574-6968.1997.tb12754.x. [DOI] [PubMed] [Google Scholar]
- 48.Herzberg M, Kaye IK, Peti W, Wood TK. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J Bacteriol 188:587–598. doi: 10.1128/JB.188.2.587-598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikegami M, Kawano M, Takase K, Yamato I, Igarashi K, Kakinuma Y. 1999. Enterococcus hirae vacuolar ATPase is expressed in response to pH as well as sodium. FEBS Lett 454:67–70. doi: 10.1016/S0014-5793(99)00776-0. [DOI] [PubMed] [Google Scholar]
- 50.Häse CC, Fedorova ND, Galperin MY, Dibrov PA. 2001. Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol Mol Biol Rev 65:353–370. doi: 10.1128/MMBR.65.3.353-370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olczak T, Simpson W, Liu X, Genco CA. 2005. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol Rev 29:119–144. doi: 10.1016/j.femsre.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Genco CA. 1995. Regulation of hemin and iron transport in Porphyromonas gingivalis. Adv Dent Res 9:41–47. doi: 10.1177/08959374950090010801. [DOI] [PubMed] [Google Scholar]
- 53.Murakami Y, Masuda T, Imai M, Iwami J, Nakamura H, Noguchi T, Yoshimura F. 2004. Analysis of major virulence factors in Porphyromonas gingivalis under various culture temperatures using specific antibodies. Microbiol Immunol 48:561–569. doi: 10.1111/j.1348-0421.2004.tb03552.x. [DOI] [PubMed] [Google Scholar]
- 54.Nagano K, Murakami Y, Nishikawa K, Sakakibara J, Shimozato K, Yoshimura F. 2007. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J Med Microbiol 56:1536–1548. doi: 10.1099/jmm.0.47289-0. [DOI] [PubMed] [Google Scholar]
- 55.Slakeski N, Dashper SG, Cook P, Poon C, Moore C, Reynolds EC. 2000. A Porphyromonas gingivalis genetic locus encoding a heme transport system. Oral Microbiol Immunol 15:388–392. doi: 10.1034/j.1399-302x.2000.150609.x. [DOI] [PubMed] [Google Scholar]
- 56.Shi L, Bielawski J, Mu J, Dong H, Teng C, Zhang J, Yang X, Tomishige N, Hanada K, Hannun YA, Zuo J. 2007. Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res 17:1030–1040. doi: 10.1038/cr.2007.100. [DOI] [PubMed] [Google Scholar]
- 57.Anaya-Bergman C, Rosato A, Lewis JP. 2015. Iron- and hemin-dependent gene expression of Porphyromonas gingivalis. Mol Oral Microbiol 30:39–61. doi: 10.1111/omi.12066. [DOI] [PubMed] [Google Scholar]
- 58.Zheng CJ, Yoo JS, Lee TG, Cho HY, Kim YH, Kim WG. 2005. Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS Lett 579:5157–5162. doi: 10.1016/j.febslet.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 59.Cronan JE Jr, Rock CO. 1987. Biosynthesis of membrane lipids, p 474–497. In Neidhardt FC, Ingraham JL, Magasanik B, Low BL, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol 1 American Society for Microbiology, Washington, DC. [Google Scholar]
- 60.Heath RJ, Rock CO. 1996. Regulation of fatty acid elongation and initiation by acyl-acyl carrier protein in Escherichia coli. J Biol Chem 271:1833–1836. doi: 10.1074/jbc.271.4.1833. [DOI] [PubMed] [Google Scholar]
- 61.Choi KH, Kremer L, Besra GS, Rock CO. 2000. Identification and substrate specificity of beta-ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis. J Biol Chem 275:28201–28207. doi: 10.1074/jbc.M003241200. [DOI] [PubMed] [Google Scholar]
- 62.Nagano K, Hasegawa Y, Abiko Y, Yoshida Y, Murakami Y, Yoshimura F. 2012. Porphyromonas gingivalis FimA fimbriae: fimbrial assembly by fimA alone in the fim gene cluster and differential antigenicity among fimA genotypes. PLoS One 7:e43722. doi: 10.1371/journal.pone.0043722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakagawa I, Amano A, Kuboniwa M, Nakamura T, Kawabata S, Hamada S. 2002. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect Immun 70:277–285. doi: 10.1128/IAI.70.1.277-285.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bagaitkar J, Demuth DR, Daep CA, Renaud DE, Pierce DL, Scott DA. 2010. Tobacco upregulates P. gingivalis fimbrial proteins which induce TLR2 hyposensitivity. PLoS One 5:e9323. doi: 10.1371/journal.pone.0009323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuramitsu HK. 1998. Proteases of Porphyromonas gingivalis: what don't they do? Oral Microbiol Immunol 13:263–270. doi: 10.1111/j.1399-302X.1998.tb00706.x. [DOI] [PubMed] [Google Scholar]
- 66.Wu J, Lin X, Xie H. 2008. OxyR is involved in coordinate regulation of expression of fimA and sod genes in Porphyromonas gingivalis. FEMS Microbiol Lett 282:188–195. doi: 10.1111/j.1574-6968.2008.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen W, Honma K, Sharma A, Kuramitsu HK. 2006. A universal stress protein of Porphyromonas gingivalis is involved in stress responses and biofilm formation. FEMS Microbiol Lett 264:15–21. doi: 10.1111/j.1574-6968.2006.00426.x. [DOI] [PubMed] [Google Scholar]
- 68.McKenzie RM, Johnson NA, Aruni W, Dou Y, Masinde G, Fletcher HM. 2012. Differential response of Porphyromonas gingivalis to varying levels and duration of hydrogen peroxide-induced oxidative stress. Microbiology 158:2465–2479. doi: 10.1099/mic.0.056416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scherber CM, Schottel JL, Aksan A. 2009. Membrane phase behavior of Escherichia coli during desiccation, rehydration, and growth recovery. Biochim Biophys Acta 1788:2427–2435. doi: 10.1016/j.bbamem.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 70.Forng RY, Champagne C, Simpson W, Genco CA. 2000. Environmental cues and gene expression in Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans. Oral Dis 6:351–365. [DOI] [PubMed] [Google Scholar]
- 71.Desbois AP, Smith VJ. 2010. Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl Microbiol Biotechnol 85:1629–1642. doi: 10.1007/s00253-009-2355-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.