ABSTRACT
Over the course of the last 3 decades the role of the second messenger cyclic di-GMP (c-di-GMP) as a master regulator of bacterial physiology was determined. Although the control over c-di-GMP levels via synthesis and breakdown and the allosteric regulation of c-di-GMP over receptor proteins (effectors) and riboswitches have been extensively studied, relatively few effectors have been identified and most are of unknown functions. The obligate predatory bacterium Bdellovibrio bacteriovorus has a peculiar dimorphic life cycle, in which a phenotypic transition from a free-living attack phase (AP) to a sessile, intracellular predatory growth phase (GP) is tightly regulated by specific c-di-GMP diguanylate cyclases. B. bacteriovorus also bears one of the largest complement of defined effectors, almost none of known functions, suggesting that additional proteins may be involved in c-di-GMP signaling. In order to uncover novel c-di-GMP effectors, a c-di-GMP capture-compound mass-spectroscopy experiment was performed on wild-type AP and host-independent (HI) mutant cultures, the latter serving as a proxy for wild-type GP cells. Eighty-four proteins were identified as candidate c-di-GMP binders. Of these proteins, 65 did not include any recognized c-di-GMP binding site, and 3 carried known unorthodox binding sites. Putative functions could be assigned to 59 proteins. These proteins are included in metabolic pathways, regulatory circuits, cell transport, and motility, thereby creating a potentially large c-di-GMP network. False candidate effectors may include members of protein complexes, as well as proteins binding nucleotides or other cofactors that were, respectively, carried over or unspecifically interacted with the capture compound during the pulldown. Of the 84 candidates, 62 were found to specifically bind the c-di-GMP capture compound in AP or in HI cultures, suggesting c-di-GMP control over the whole-cell cycle of the bacterium. High affinity and specificity to c-di-GMP binding were confirmed using microscale thermophoresis with a hypothetical protein bearing a PilZ domain, an acyl coenzyme A dehydrogenase, and a two-component system response regulator, indicating that additional c-di-GMP binding candidates may be bona fide novel effectors.
IMPORTANCE In this study, 84 putative c-di-GMP binding proteins were identified in B. bacteriovorus, an obligate predatory bacterium whose lifestyle and reproduction are dependent on c-di-GMP signaling, using a c-di-GMP capture compound precipitation approach. This predicted complement covers metabolic, energy, transport, motility and regulatory pathways, and most of it is phase specific, i.e., 62 candidates bind the capture compound at defined modes of B. bacteriovorus lifestyle. Three of the putative binders further demonstrated specificity and high affinity to c-di-GMP via microscale thermophoresis, lending support for the presence of additional bona fide c-di-GMP effectors among the pulled-down protein repertoire.
INTRODUCTION
The second messenger cyclic di-GMP (c-di-GMP) is a main modulator of bacterial behavior. It affects the transition of individual motile cells between a free-living planktonic state to an immotile state or to multicellular entities such as biofilms or flocs and can also regulate virulence in pathogenic bacteria (1). This transition is accompanied by numerous physiological, morphological, and metabolic changes brought about by signals transduced through the allosteric binding of c-di-GMP to effectors (receptor proteins) and to riboswitch aptamers; binding directly affects catalysis, protein interactions, and gene expression (1). In Caulobacter crescentus, a bacterium with a dimorphic life cycle, in which the transition from the planktonic to the sessile state is coupled to cell division, c-di-GMP has strong effects on the dividing cell phenotype (2). Strikingly, while c-di-GMP is not necessary for C. crescentus reproduction (2), oscillating levels of the messenger regulate the progression of cell division (3). By impacting on the physiology and morphology of the bacterium, c-di-GMP is thus involved in ecological adaptation. c-di-GMP levels are tightly regulated via synthesis and breakdown by diguanylate cyclases (DGCs) and phosphodiesterases (PDEs), respectively, both globally at the cell level and locally at the subcellular level (4, 5). DGCs contain the signature domain GGDEF, while PDEs can be identified through their EAL or HD-GYP domains (1); effector proteins bind c-di-GMP through PilZ domains or through catalytically nonfunctional GGDEF or EAL domains that retain high c-di-GMP affinity (4, 6). Experimental evidence suggests that c-di-GMP can bind to other “unorthodox” domains (7–11), bringing a larger fraction of proteins and the functions they perform into the c-di-GMP network.
B. bacteriovorus is an obligate predatory bacterium that subsists on Gram-negative bacteria. Cell cycle progress and reproduction in B. bacteriovorus have been shown to depend upon c-di-GMP signaling (4). B. bacteriovorus maintains a biphasic life cycle that includes a free-living, invasive and motile attack phase (AP) and an intracellular, sessile growth phase within the invaded prey cell (called a bdelloplast [12]). Strikingly, the transition to the sessile, replicative state involves prey sensing and confinement of the predator within the prey cell (12). The predator consumes the prey cytoplasm during its growth phase (GP), forming a filamentous, multinucleoid cell that finally splits, generating progeny AP cells (Fig. 1). AP and GP cells not only differ in morphology, but they also express totally different gene sets in an almost exclusive pattern (13). Importantly, spontaneous mutations in bd0108 and in rhlB (bd3461) or pcnB (bd3464) enable B. bacteriovorus to grow axenically (i.e., in the absence of prey) in a rich medium as host-independent (HI) mutants (14). Bd0108 interacts with the type IVa pilus machinery (51), while RhlB and PcnB are components of the RNA degradosome. HI mutants exhibit deregulated transitions between the AP to the GP and alterations in gene and protein expression patterns and yet maintain characteristic GP features, in particular filamentous, polynucleoid growth, and multiple parallel septation events during division (15).
FIG 1.
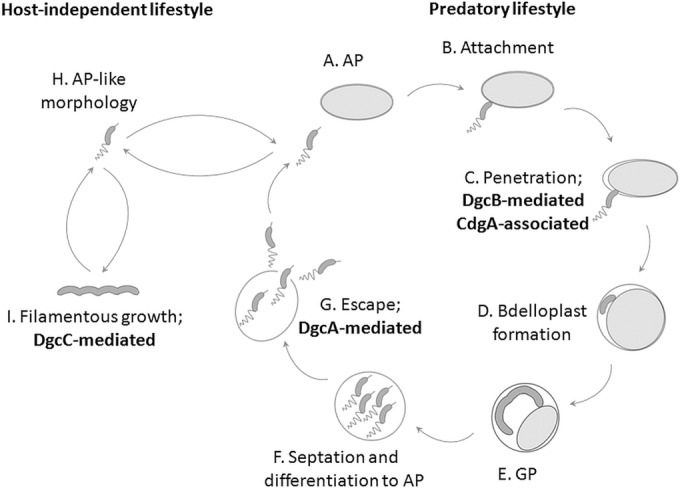
Cyclic di-GMP control over the progress of B. bacteriovorus cell cycle: wild-type B. bacteriovorus maintains an obligate predatory biphasic life cycle. (A) Nonreplicating free-living and fast-swimming vibrioid AP cells search for prey. (B and C) Encountering of the prey cell is followed by irreversible attachment (B) and then by DgcB-dependent and CdgA-associated invasion into the prey periplasm (C). CdgA is a cyclic di-GMP effector that interacts with the invasion-essential type IVa pilus regulatory hub. (D) During entry and settlement, the bdelloplast is formed. (E) Within the bdelloplast, the predator enters GP, consumes the prey and grows as a multinucleoid filament. (F and G) Upon nutrient depletion, the filament divides to progeny AP cells that lyse and escape from the bdelloplast (F), the latter necessitating DgcA, which is essential for development of both flagellar and gliding motility (G). The nascent AP cells are ready for another predation cycle. (H and I) HI mutants are not restricted to the predatory lifestyle and, while remaining predatory, they can grow saprophytically via the DgcC pathway, retaining the dimorphic cell cycle.
Thus far, the c-di-GMP network of B. bacteriovorus is known to include five DGCs, one EAL PDE, six HD-GYP PDEs, one putative c-di-GMP-I riboswitch, and an unusually large number (see Results) of PilZ effectors (16, 17). Three of the five DGCs exert remarkable phenotypic effects: DgcA (Bd0367) and DgcB (Bd0742) are essential for the predatory lifestyle and, in contrast, DgcC (Bd1434) is essential for axenic growth. CdgA (Bd3125), an inactive DGC, is required for rapid prey penetration (4). It is probably recruited to a regulatory protein hub situated at the prey-invasion pole (16). The putative c-di-GMP-I riboswitch is a standalone c-di-GMP aptamer massively expressed in AP and downregulated upon entry to the GP. It was proposed to act as a c-di-GMP sink during the AP (13). These data suggest that B. bacteriovorus has an extensive c-di-GMP machinery implicated in the regulation of its complex and unique lifestyle. We further proposed that novel classes of c-di-GMP effectors controlled functions hitherto not known to be under c-di-GMP regulation through uncharacterized binding domains.
In an effort to define the c-di-GMP signaling network of B. bacteriovorus in finer details, proteins from AP cells and axenic HI mutant cells, mimicking the GP, were pulled down using a c-di-GMP capturing compound (cdG-CC) as bait (18). Eighty-four c-di-GMP binding proteins were identified by mass spectroscopy, including 65 candidate c-di-GMP binding proteins missing any known c-di-GMP binding site and three c-di-GMP binders bearing recently described unorthodox binding sites. The c-di-GMP binding specificity of three c-di-GMP binding proteins, a hypothetical protein bearing a PilZ domain, an acyl coenzyme A (acyl-CoA) dehydrogenase, and a two-component system response regulator, was further demonstrated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. bacteriovorus type strain HD100 (ATCC 15356) was grown in two-membered suspension cultures with ∼2 × 109 PFU ml−1 of Escherichia coli ML35 (ATCC 43827) in HEPES buffer (25 mM HEPES, 2 mM CaCl2·2H2O, 3 mM MgCl2·6H2O [pH 7.4 to 7.6]) at 28°C and 180 rpm. Axenic B. bacteriovorus HD100 mutant M1.1 (kindly provided by Eckhard Strauch [14]) was cultured in PYE (10.0 g liter−1 peptone, 3.0 g liter−1 yeast extract, 2 mM CaCl2·2H2O, 3 mM MgCl2·6H2O [pH 7.4 to 7.6]) to an optical density at 600 nm (OD600) of 0.2. Attack-phase (AP) cells and HI M1.1 cultures were obtained from four replicates of 1-liter cultures. The cultures were pelleted at 4°C (17,000 × g, 10 min) and kept frozen at −20°C until protein extraction.
Protein extraction.
Pellets were resuspended in lysis buffer (6.7 mM morpholineethanesulfonic acid, 6.7 mM HEPES, 200 mM NaCl, 6.7 mM potassium acetate [pH 7.5]) supplemented with protease inhibitor (Complete Mini; Roche) and Benzonase nuclease (Sigma-Aldrich), and lysed using a French press three times at 20,000 lb/in2. After ultracentrifugation (100,000 × g, 50 min), the supernatants were passed through a PD10 (GE Healthcare) to remove nucleotides. Protein concentration was determined by the Bradford protein assay (Pierce Biotechnology). These supernatants (i.e., soluble proteins) were used for the capture-compound mass spectrometry (CCMS) experiments. The pellets, containing the membrane fractions, were washed once with lysis buffer containing 400 mM NaCl, resuspended in the same buffer with 0.5% n-dodecyl-β-d-maltopyranoside (DDM; Anatrace), and incubated for 2 h at 4°C with gentle agitation on a rotary wheel. Preparations were later ultracentrifuged (100,000 × g, 1 h). The protein concentration within the supernatant was determined with the BCA protein assay (Pierce Biotechnology). This fraction, used in CCMS experiments represents integral and membrane-associated proteins.
Capture of c-di-GMP binding proteins.
Capture experiments were carried out as described earlier (18, 19). All experiments were performed in 200-μl 12-tube PCR strips (Thermo Scientific). Soluble and membrane proteins (350 μg of protein per sample) were mixed with 5 and 10 μM cdG-CC (Caprotec Bioanalytics GmbH, Berlin) and with 20 μl of 5× capture buffer (100 mM HEPES, 250 mM potassium acetate, 50 mM magnesium acetate, 50% glycerol [pH 7.5]). The volume was adjusted with H2O to 100 μl, followed by incubation overnight at 4°C in the dark on a rotary wheel. Three control experiments were run in parallel. In one control treatment, cdG-CC was excluded. In the other two experiments, 1 mM c-di-GMP was added to compete with cdG-CC present at 5 and 10 μM. All reaction mixtures were incubated 30 min at 4°C on a rotary wheel prior to addition of cdG-CC. Reactions were UV irradiated for 4 min at 310 nm (Caprobox; Caprotec Bioanalytics), resulting in the proteins covalently binding cdG-CC. Then, 50 μl of magnetic streptavidin beads (Dynabeads MyOne Streptavidin C1; Invitrogen) and 25 μl of 5× wash buffer (250 mM Tris [pH 7.5], 5 M NaCl), with 0.1% n-octyl-β-glucopyranoside (β-OG) supplement added only to the soluble fraction samples, were added, and the mixtures were incubated for an additional 1 h at 4°C on a rotary wheel. The beads were then collected with a magnet (caproMag; Caprotec Bioanalytics GmbH, Berlin). Beads from the soluble fraction samples were washed 6 times with wash buffer, including 0.1% β-OG. Beads from the membrane fraction samples were washed six times with 200 μl of wash buffer amended with 0.1% DDM, twice with 200 μl of wash buffer (including 0.05% DDM), and once with 200 μl of wash buffer added (including 0.025% DDM).
Tryptic digestion for MS analysis.
Beads from the soluble fraction samples were washed once with 200 μl of H2O, six times with 200 μl of 80% acetonitrile, and twice with 200 μl of H2O before being resuspended in 20 μl 100 mM ammonium bicarbonate. Beads from the membrane fraction samples were washed three times with 200 μl of 100 mM ammonium bicarbonate with 2 M urea before being resuspended in 20 μl of 100 mM ammonium bicarbonate with 8 M urea. Then, 0.5 μl of 200 mM sulfhydryl reductant Tris[2-carboxyethyl]-phosphine (TCEP) was added to each sample, and the beads were shaken at 500 rpm for 1 h at 60°C. Samples were then cooled to 25°C, and 0.5 μl of 400 mM iodoacetamide was added before the beads were incubated in the dark for 30 min at 25°C. Next, 0.5 μl of 500 mM N-acetylcysteine was added, and the beads were further shaken 500 rpm for 10 min at room temperature. Only membrane fraction beads were further incubated overnight with 1 μg of endoproteinase Lys-C (Wako) at 37°C, after the addition of 100 μl of 100 mM ammonium bicarbonate. Next, 1 μg of porcine trypsin (Promega) was added to all of the samples. Soluble fraction samples were incubated overnight, and membrane fraction samples were incubated 6 h at 37°C and 500 rpm. Beads were collected with a magnet. Supernatants of soluble fraction samples were mixed with 3 μl of 5% trifluoroacetic acid (TFA) and 1 μl of 2 M hydrochloric acid (HCl). Supernatants of membrane fraction samples were mixed with 15 trifluoroacetic acid and 5 μl of 2 M HCl. Peptides were purified on C18 Microspin columns (Harvard Apparatus) and dried in a SpeedVac.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Analyses were performed on a dual pressure LTQ-Orbitrap Velos mass spectrometer connected to an electrospray ion source (Thermo Scientific). Peptide separation was carried out using an Easy Nano-LC system (Thermo Scientific) equipped with a RP-HPLC column (75 μm by 15 cm) packed with C18 resin (Magic C18 AQ, 3 μm; Michrom BioResources) using a linear gradient from 96% solvent A (0.15% formic acid, 2% acetonitrile) and 4% solvent B (98% acetonitrile, 0.15% formic acid) to 35% solvent B over 40 min at a flow rate of 0.3 μl min−1. The data acquisition mode was set to obtain one high-resolution MS scan in the FT part of the mass spectrometer at a resolution of 60,000 (full width at half maximum [FWHM]), followed by tandem mass spectrometry (MS/MS) scans in the linear ion trap of the 20 most intense ions. In order to increase the efficiency of the MS/MS attempts, the charged state screening modus was enabled to exclude unassigned and singly charges ions. Collision induced dissociation was triggered when the precursor exceeded 100 ion counts. The dynamic exclusion duration was set to 15 s. The ion accumulation times were set to 300 ms (MS) and 50 ms (MS/MS).
Database search.
Mass spectrometry raw spectra were converted into MASCOT generic files (mgf) and searched with MASCOT version 2.3. B. bacteriovorus HD100 Uniprot/Tremble databases were downloaded via the European Bioinformatics Institute (http://www.ebi.ac.uk/). In silico trypsin digestion was performed after the amino acids lysine and arginine, unless they were followed by proline, tolerating two missed cleavages in fully tryptic peptides. Database search parameters were set to allow oxidized methionine (+15.99491 Da) as variable modifications and carboxyamidomethylation (+57.021464 Da) of cysteine residues as fixed modification. For MASCOT searches using high-resolution scans the precursor mass tolerance was set to 15 ppm, and the fragment mass tolerance was set to 0.6 Da. The protein false discovery rate was set to 1%. Mascot search of CCMS experiments were imported into Scaffold (Proteomesoftware, version 3), which was used to extract spectral count.
Data analysis.
Proteins interacting unspecifically with unloaded beads were taken off the analysis. Of the protein subset interacting with cdG-CC (see Table S1 in the supplemental material) proteins enriched by >2-fold in at least half of the experiments per fraction type (designated AP soluble, AP membrane, HI soluble, or HI membrane), compared to competition controls, were regarded as candidate binders. Of four independent experiments carried out in duplicate for the four AP soluble, four AP membrane, four HI soluble, and four HI membrane protein fractions (32 experiments in total), five, six, six, and eight experiments were analyzed, respectively, due to insufficient signal for the other samples in the MS analysis (see Table S1 in the supplemental material).
Protein expression and purification.
Genes bd2717, bd2402, and bd2924 were cloned into pET14b according to the restriction-free cloning procedure for multicomponents assembly, using the primers listed in Table 1. Plasmids were transformed into E. coli BL21(DE3). Transformants were subsequently cultured in 1 liter of Luria-Bertani broth with 100 μg ml−1 ampicillin at 37°C at 180 rpm, supplemented with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) in mid-log phase (OD600 = 0.4) to induce the production of His-tagged Bd2717, Bd2402, and Bd2924. After additional agitation for 2 h at 37°C and 180 rpm, the cells were pelleted (10,000 × g, 10 min), resuspended in 20 ml of phosphate-buffered saline (PBS; 20 mM Na2HPO4, 300 mM NaCl [pH 7.4]), and lysed by sonication. Soluble proteins were collected after two rounds of centrifugation (17,000 × g, 30 min, 4°C) and the His-tagged proteins were purified next by affinity chromatography with HisPur Ni-NTA resin (Thermo) according to the manufacturer's instructions. After protein elution, imidazole was removed by overnight dialysis with PBS and 10% glycerol. The batch was divided into aliquots and kept in −20°C.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| 5pET_bd2717 | AGCAGCGGCCTGGTGCCGCGCGGCAGCCATATGGCCACACCGCTGTC |
| 3pET_bd2717 | TCAGCTTCCTTTCGGGCTTTGTTAGCAGCCCTATGAATTTATTTCTTGGGGTACT |
| 5pET_bd2402 | AGCAGCGGCCTGGTGCCGCGCGGCAGCCATGTGGAATCAGGCAAATCTAAG |
| 3pET_bd2402 | TCAGCTTCCTTTCGGGCTTTGTTAGCAGCCTTATTTCTTTTTAAAGAAGCCCG |
| 5pET_bd2924 | AGCAGCGGCCTGGTGCCGCGCGGCAGCCATATGAAAAACTTTTACCAAGACGG |
| 3pET_bd2924 | TCAGCTTCCTTTCGGGCTTTGTTAGCAGCCCTATAAAGAATACTTCTTCATAGCGATA |
Microscale thermophoresis (MST).
Proteins were labeled with a Monolith NT RED protein labeling kit according to the supplied labeling protocol (NanoTemper Technologies). The PBS nuffer was exchanged after labeling with c-di-GMP binding buffer (10 mM Tris, 5 mM MgCl2·6H2O, 250 mM NaCl, 10% glycerol, 1 mM dithiothreitol [pH 8.0]) to prevent unwanted Tris labeling. Labeled proteins were used at a final concentration of 50 nM. Cyclic di-GMP (Biolog) or cyclic di-AMP (Biolog) were titrated in 1:1 dilutions starting with 5 mM. The experiments were performed in c-di-GMP binding buffer supplemented with 0.05% Tween, and measurement took place in standard treated capillaries (NanoTemper Technologies). All binding reaction mixtures were incubated for 10 min at room temperature before they were loaded into the capillaries. The measurements were done with a NanoTemper Monolith NT.115 instrument. All measurements were performed with 50% light-emitting diode (LED) and 40% infrared (IR) laser using a laser-on time of 30 s and a laser-off time of 5 s.
RESULTS
High-throughput identification of potential cyclic di-GMP binding proteins in B. bacteriovorus.
Putative c-di-GMP binding proteins were separated from the total soluble and membrane protein fractions of nonreplicative AP cells of the wild-type strain HD100 and of replicating M1.1 axenic HI mutant cells, mutated in bd0108 and pcnB (bd3464 [14]), using a c-di-GMP-specific capture compound (cdG-CC [18]), and identified by LC-MS/MS (CCMS). Replicating M1.1 HI cultures, mostly containing growing filaments, were chosen over wild-type GP cells obtained from bdelloplasts, since the latter are severely contaminated with prey content, hindering precise protein identification to be compared to AP cells (see below). In total, 777 proteins were detected by MS; 128 also interacted unspecifically with the beads. Of 649 proteins that interacted with cdG-CC (see Table S1 in the supplemental material) 84 (12.9%) were enriched by >2-fold in at least half of the experiments, compared to controls in which an excess of soluble c-di-GMP was present, competing for binding of the putative effectors with the cdG-CC-loaded beads (Fig. 2 and see Table S2 in the supplemental material). Thirty-two of them (38.1%) were captured in the soluble fraction and 52 (61.9%) were pulled from the membrane fraction. The proportions of putative effectors in the soluble and membrane fractions were similar within each of the phases, since 21 and 25% were specific to the AP, 41 and 49% were specific to the HI cultures (i.e., the GP), and 22 and 26%, respectively, were identified in both phases (see Table S2 in the supplemental material). The putative subcellular location of the identified proteins is included in Table S2 in the supplemental material (column I).
FIG 2.
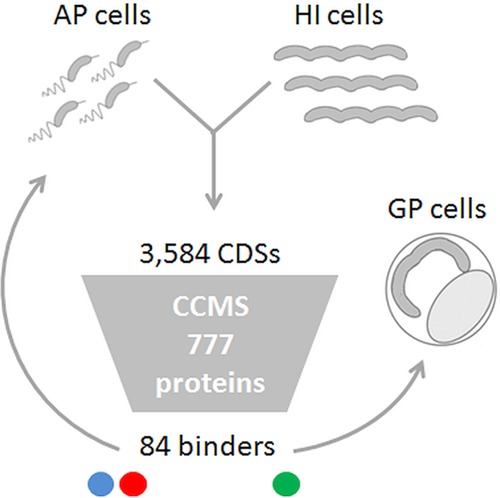
Experimental scheme. Total protein harvested from fresh B. bacteriovorus HD100 AP cells and axenically replicating B. bacteriovorus HD100 M1.1 HI mutant cells was investigated by CCMS. Of 3,584 encoded proteins (23), 777 were detected by LC-MS/MS, but only 84 proteins presented competitive cyclic di-GMP specific binding. These binders demonstrated c-di-GMP binding exclusive to the AP (blue circle), unique to the HI mutants (green circle), or in both (red circle). AP-specific and common binders represent cyclic di-GMP effectors functional in the AP. HI-specific binders represent cyclic di-GMP effectors functional in the GP. The functionality of shared binders in the GP cannot be determined due to possible deregulated protein production in HI mutants (15).
Cyclic di-GMP binding proteins with known binding domains.
To date, three functional and two degenerate GGDEF proteins, six HD-GYP PDEs and one EAL PDE, and fifteen PilZ c-di-GMP effectors have been recognized in B. bacteriovorus (4, 17). A reanalysis of B. bacteriovorus proteome using the NCBI and Pfam databases revealed three additional PilZ proteins (Bd0760, Bd1616, and Bd2147). One of them (Bd0760) was identified as a c-di-GMP binder in the CCMS experiment, along with 10 other known PilZ proteins (see Table S2 in the supplemental material). Noteworthy, conservation of the PilZ consensus sequence, RXXXR-X20–30-(D/N)X(S/A)XXG, did not correlate with c-di-GMP binding. Indeed, nonbinding PilZ proteins are common, including the eponymous PilZ from Pseudomonas aeruginosa (17). Only three of the five GGDEF proteins were identified as c-di-GMP binders in this assay, along with the EAL PDE (Bd1971) and one HD-GYP PDE (Bd2421). The other five HD-GYP PDE carry mutations in the consensus sequence HD-HHEXXDGXGYP (summarized in Table S2 in the supplemental material) (20, 21) that might result in the loss of binding ability, explaining their absence from the set of retrieved proteins. In addition, proteins carrying domains recently shown to bind c-di-GMP were identified as binders: Bd0156, a NifA-like transcription factor holds an AAA+ domain with an intact Walker A motif (10). Bd2590, a CRP (cyclic AMP receptor protein), retains three of the four positions involved in c-di-GMP binding in Clp, a CRP derivate that binds c-di-GMP instead of cAMP in Xanthomonas campestris (see Fig. S1 in the supplemental material) (11). Bd1551, is a polyribonucleotide nucleotidyltransferase (PNPase), a member of the RNA degradosome. In E. coli, PNPase activity, and thus RNA turnover, is regulated by c-di-GMP binding (9). Although the c-di-GMP binding site is still unknown, the orthologues exhibit high relatedness with 51% sequence identity, 68% sequence similarity, and the sharing of all five PNPase domains. We conclude that the CCMS experiment was efficient, revealing 19 of the 33 predicted complement of c-di-GMP effectors in B. bacteriovorus. These 19 known or predicted (KOP) binders, a class we define as all binders bearing characterized c-di-GMP-binding domains, include 12 unclassified proteins, as well as 3 proteins involved in c-di-GMP turnover, 3 proteins involved in transcription and RNA metabolism, and 1 protein involved acting as a transporter (Fig. 3A; see also Table S3 in the supplemental material).
FIG 3.
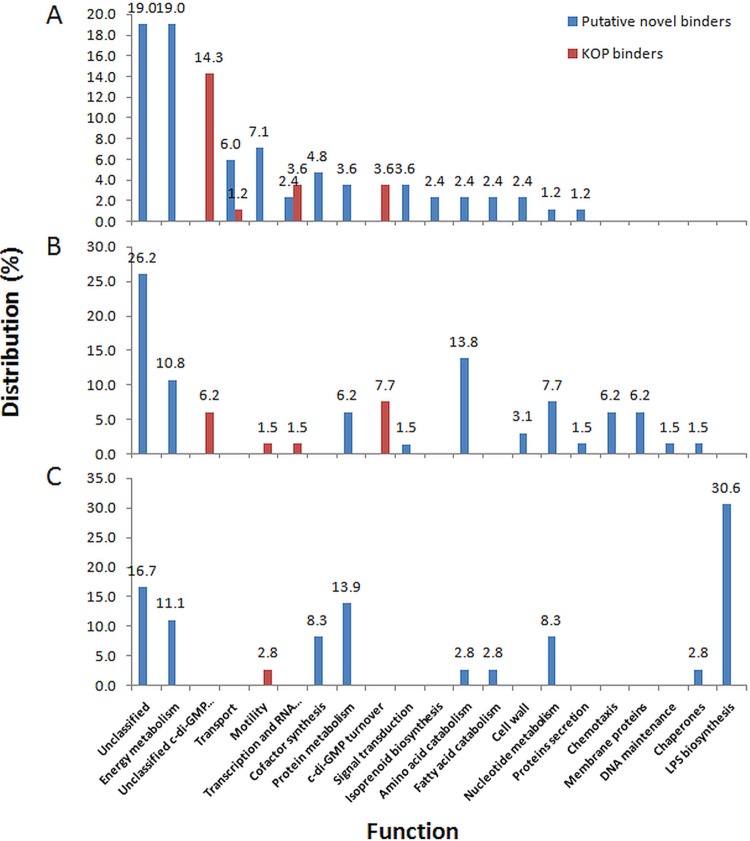
Functional classification of the CCMS-identified cyclic di-GMP binders in B. bacteriovorus HD100 (84 binders) (A), P. aeruginosa (65 binders) (B), and S. Typhimurium SL1344 (36 binders) (C). Functional data for P. aeruginosa and S. Typhimurium were obtained from Nesper et al. (18). Only 5 of 20 functions (energy metabolism, motility, protein metabolism, amino acid catabolism, and nucleotide metabolism) were shared between the three bacteria. Of these categories, “energy metabolism” was enriched in all three. Other functions were differentially represented, possibly reflecting the adaptation of the c-di-GMP network to the different lifestyles of the three bacteria.
Putative cyclic di-GMP binding proteins with uncharacterized binding domains.
Of the 84 proteins identified in the CCMS experiments, 65 were hitherto not known to bind c-di-GMP, i.e., they are putative novel c-di-GMP effectors. No function could be assigned to 16 (19.0% in total) of the proteins; the other binders were classified according to 13 functional classes (Fig. 3A; see also Table S3 in the supplemental material) as follows (number of proteins, percent total): energy metabolism (16, 19%), i.e., respiration and the TCA cycle; transcription and RNA metabolism (2, 2.4%); protein metabolism (3, 3.6%); signal transduction (3, 3.6%); metabolism of nucleic (1, 1.2%), amino (2, 2.4%) and fatty (2, 2.4%) acids; cofactor synthesis (2, 2.4% for coenzyme A; 1, 1.2% for porphyrin; and 1, 1.2% for adenosylcobalamin); isoprenoid synthesis (2, 2.4%); cell wall maintenance (2, 2.4%); membrane transport (5, 6.0%); protein secretion (1, 1.2%); and motility (3, 3.6% for flagellar motility; 3, 3.6% % for gliding). Although c-di-GMP was earlier shown to potentially regulate proteins in 12 of these functional classes (17, 18, 22), the specific enzymatic functions within each of the classes were almost all different from those of the proteins identified in the present study (e.g., the prediction of c-di-GMP control over the synthesis of porphyrin and adenosylcobalamin); these data also suggest that additional pathways, such as the isoprenoid pathway, may be regulated by c-di-GMP (Fig. 3A; see also Table S3 in the supplemental material).
Potential false-positive c-di-GMP binders.
Fifteen of the putative novel binders are found in operons in which at least another protein was also identified as a binder in the pulldown. More specifically, they form six pairs and a triplet of genes (see Table S2 in the supplemental material) (13). The genes bd1833 and bd1834 are annotated as two consecutive malate dehydrogenases (23), but they probably encode the C and N termini, respectively, of a single, wrongly annotated CDS, as a pairwise alignment with the homolog bdt1809 from the closely related B. bacteriovorus strain Tiberius (accession number YP_007022765) suggests (data not shown). The operon-clustered genes bd0179-bd0182 (along with bd0836), bd2728-bd2729, bd3412-bd3413, and bd3897-bd3898-bd3900 and the dispersed genes bd0286-bd2611 and bd0604-bd3395-bd3403 encode proteins that are known to form complexes in other bacteria (24–29). Therefore, it may be that only one of the complex members actually bind c-di-GMP, the other(s) being carried over during the experiment. Also, 20 candidate binders hold allosteric binding sites for other cofactors, most of which nucleotides (see Table S2 in the supplemental material), and could, putatively, unspecifically bind the capture compound. Furthermore, of the 84 candidates, 55 (65.5%) cytosolic and 15 (17.9%) membrane may directly interact with c-di-GMP, the latter via cytosolic segments, since the membrane cell cycle kinase CckA in C. crescentus (3). However, nine (10.7%), one (1.2%), two (2.4%), and an additional two proteins are, respectively, assigned to the periplasm, the outer membrane, the extracellular milieu, and an unspecified noncytoplasmic location (see Table S2 in the supplemental material). Since c-di-GMP has hitherto been shown to only be present in the cytosol, these proteins may not bind it directly. Hence, the interaction of those with c-di-GMP is either indirect (e.g., the extracellular flagellar proteins FlgM and FliC were probably copurified with FliF [see Discussion]), results from unspecific binding, or takes place prior to secretion (see discussion).
Validation of novel cyclic di-GMP effectors.
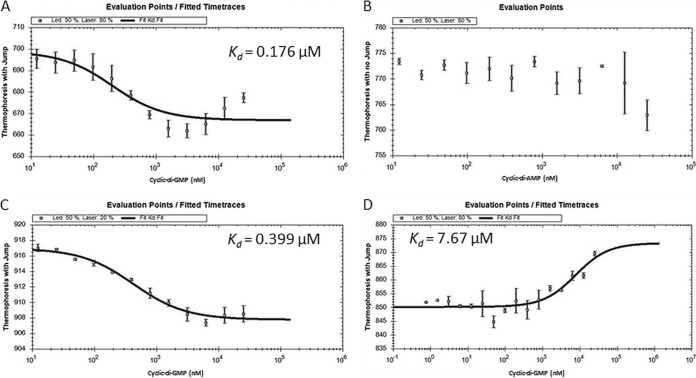
Bd2402, a two-component response regulator, and Bd2924, one of the various uncharacterized c-di-GMP-dependent dehydrogenases (EC 1.3.99-) that are largely represented in the c-di-GMP complement of effectors, represent two very different functional classes. Both exhibited reproducible c-di-GMP binding and were selected to further validate c-di-GMP binding and test its specificity using microscale thermophoresis (MST). The PilZ domain effector Bd2717 of unknown function was chosen to serve as a positive control. MST quantifies molecular interactions by monitoring the motion of (bio)molecules across a microscopic temperature gradient (30). It was successfully used to measure c-di-GMP binding by EAL and PilZ domain proteins (31, 32). All three proteins were fused with a His tag and purified by nickel affinity chromatography (see Fig. S2 in the supplemental material). The MST assay clearly showed the high affinity of Bd2717 for c-di-GMP, exhibiting a Kd = 0.176 μM, also revealing the binding of two c-di-GMP molecules per effector protein (Fig. 4; see also Fig. S3 in the supplemental material). This was indicated by the conservation of R111 at the position preceding the PilZ consensus sequence—RXXXR-X20–30-(D/N)X(S/A)XXG—that was previously shown to promote binding of two intercalated c-di-GMP molecules per binding site (33). The PilZ effector did not exhibit any change in thermophoretic motion when mixed with c-di-AMP, thus providing experimental support for c-di-GMP binding specificity. The MST binding assay also demonstrated the high affinity of the response regulator Bd2402 for c-di-GMP with a Kd = 0.399 μM (Fig. 4). Specificity was shown as this effector did not bind c-di-AMP. Careful examination of the Bd2402 sequence detected a PilZ domain-like consensus sequence (R151QKRKVIESIEEVSGFDLPFLYSLLVETKSGGYLNIYNADGSVSG195, where boldface indicates conserved residues) within a DUF4388 domain, suggesting that this region may contain the binding site. Finally, the acyl-CoA dehydrogenase Bd2924 also selectively bound c-di-GMP with a Kd = 7.67 μM and did not bind c-di-AMP (Fig. 4; see also Fig. S4 in the supplemental material). This relatively low affinity to c-di-GMP indicates that Bd2924 responds to relatively high cellular c-di-GMP concentrations. It may thus represent a class of low-affinity receptors in contrast to Bd2717 and Bd2402. Based on these results, we feel confident to infer that the pulldown assay truly selected for proteins with high affinity and selectivity for c-di-GMP.
FIG 4.
Validation of cyclic di-GMP binding by microscale thermophoresis (MST). The PilZ effector Bd2717, the two-component response regulator Bd2402, and the acyl-CoA dehydrogenase Bd2924 were His tagged, produced, purified, and fluorescently labeled before subjected to MST analysis with increasing concentrations of cyclic di-GMP or cyclic di-AMP. The thermophoretic movement of the three proteins was affected by c-di-GMP but not by c-di-AMP. (A) Bd2717 bound c-di-GMP with Kd = 0.176 μM. (B) The thermophoretic movement of Bd2717 unaffected by c-di-AMP, provided as an example. (C) Bd2402 bound c-di-GMP with Kd = 0.399 μM. (C) Bd2924 bound c-di-GMP with Kd = 7.67 μM. Values are averages of three triplicates. Error bars indicate the standard deviations. MST plots of Bd2402 and Bd2924 with c-di-AMP are presented in Fig. S4 in the supplemental material.
Cell cycle phase specificity of cyclic di-GMP effectors.
In B. bacteriovorus, gene expression in the AP and in the GP is highly specific, i.e., the sets of genes expressed in the AP and in the GP are almost completely segregated (13). We therefore hypothesized that c-di-GMP binders may also exhibit such phase specificity. Replicating HI mutants, like wild-type GP cells, exhibit the filamentous phenotype of a polynucleoid elongating cell culminating in coordinated division into progeny cells; the proteome of growing HI culture is in concordance with the phenotype (15), and the expression of many genes is similar in HI and GP cells (13, 34). Therefore, we considered replicating HI mutants to be an adequate substitute for wild-type GP cells that, as mentioned above, impose severe contamination originating from the prey. Accordingly, both AP cells and axenically replicating M1.1 HI cultures were analyzed. The ratio of c-di-GMP binders expressed in the AP is high (34 of 84 genes [40.5%]) compared to the general transcriptome (467 uniquely or significantly enhanced AP genes of 2,090 expressed genes [22.3%]). Classification of the pulled-down complement based on the phase specificity of the actual c-di-GMP binding revealed that 21 (25%) of the proteins bind c-di-GMP exclusively in the AP cells; 22 (26%) bind it in both the AP and the HI cultures, and 41 (49%) bind c-di-GMP uniquely in the HI culture (Fig. 5). Hence, 51% are predicted to be functional during AP. However, the cyclic-di-GMP binding profile and the gene expression profile (13) do not fully overlap: 10 of 21 and 25 of 41 genes expressed in the AP or the GP, respectively, according to the transcriptome analysis, specifically bound c-di-GMP in accordance to their gene expression profile, the remaining binding in the other phase (see Table S2 in the supplemental material). In addition, 15 (18%) binders were clearly detected by MS in both the AP and the axenic growth phases but exhibited specific and competitive c-di-GMP binding only in one of the two phases (see Table S2 in the supplemental material). These behaviors suggest that phase-specific posttranslational activation of some sort may occur for these proteins.
FIG 5.
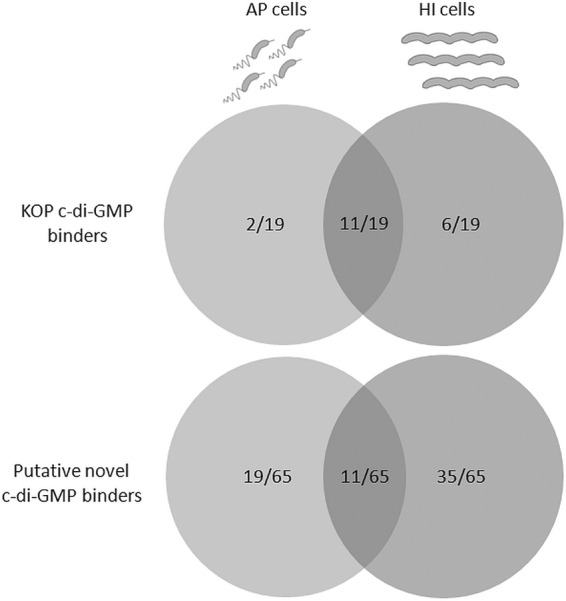
Phase-dependent cyclic di-GMP binding. A total of 84 c-di-GMP binders in B. bacteriovorus HD100, divided into 19 KOP and 65 putative novel, demonstrated three c-di-GMP binding patterns: AP specific (21/84), HI specific (41/84), and common AP and HI binding (22/84). It was thus concluded that at least 41 of 84 binders were GP related. The common binders are either AP-specific binders, abnormally produced and functional in the HI mutant (15), or binders shared between the AP and the GP. Since 8 of these were encoded by AP genes and 14 are GP genes, this group might be a composite group.
DISCUSSION
Turnover of cyclic di-GMP deeply affects the bacterial cell. This is achieved through local and global changes in the concentration of this secondary messenger (2–5), mediated by synthesis and degradation performed by cyclic di-GMP metabolic enzymes with characterized binding domains (GGDEF, HD-GYP, and EAL [1]). A most striking example of these effects is found in the obligate bacterial predator B. bacteriovorus, in which particular diguanylate cyclases control major transitions in the bacterium life cycle (4). However, only few c-di-GMP binding domains are known in effector proteins, which have mostly unknown functions (17). Nevertheless, the profound effects of cyclic di-GMP upon bacterial cells versus the dearth of known c-di-GMP effectors suggest that many more c-di-GMP binders have yet to be recognized and that these effectors transduce alterations in c-di-GMP concentration into output at the biochemical and signaling levels.
Recently, two pulldown approaches for high-throughput identification of c-di-GMP binding proteins were developed, each based on a unique synthetic ligand. Both approaches were used to identify c-di-GMP binding proteins in P. aeruginosa, producing data sets with a very limited overlap; the CCMS technique yielded 54 putative novel binders (18), whereas the approach of Duvel et al. (22) provided 140. Markedly, only five proteins were identified in both. CCMS showed robustness, generating reproducible results and reliably enriching the c-di-GMP complement in C. crescentus and P. aeruginosa. It also appeared to be less noisy than its alternative (18, 22). CCMS was thus selected for the current study.
B. bacteriovorus possesses a large complement of putative c-di-GMP effectors.
Eighty-four putative c-di-GMP binding proteins were discovered, including 65 hitherto-unknown binders. Noteworthy, only three novel proteins were shared between B. bacteriovorus and P. aeruginosa and none with S. Typhimurium, also analyzed by CCMS (see Table S2 in the supplemental material) (18), despite the fact that seven and six functional groups predicted as c-di-GMP-regulated are shared between B. bacteriovorus and P. aeruginosa and between B. bacteriovorus and Salmonella enterica serovar Typhimurium, respectively (Fig. 3). The remarkable adaptation of the c-di-GMP network to the unique cell differentiation process in C. crescentus (3) further suggests that the c-di-GMP network is locally adapted and independently shaped in different species.
Validation of three c-di-GMP binding proteins.
Bd2717, used to validate the CCMS experiment exhibited high affinity toward c-di-GMP in MST experiments (Kd = 0.176 μM), as expected from a PilZ domain-containing protein, which have Kd values at the submicromolar range (17, 35). This modified PilZ domain contains a longer than usual 34-amino-acid long linker and an additional arginine (R111) positioned aside the motif RXXXR, attributes that explain the binding of c-di-GMP dimers (33). The response regulator Bd2402 also bound c-di-GMP with high affinity (Kd = 0.399 μM), possibly through a novel variant of the PilZ domain (see Results). In contrast, Bd2924 probably contains a novel binding domain, since no motif related to a c-di-GMP domain was detected. Although its dissociation constant is higher by up to an order of magnitude (Kd = 7.67 μM) over the ones measured for the other effectors, it is still within the known range of c-di-GMP binding affinities, shown to distribute from as low as <50 nM up to a few μM, enabling a diversity of concentration-dependent responses (36). Finally, high specificity for c-di-GMP was demonstrated since none of the three effectors tested bound to c-di-AMP, a molecule structurally highly resembling c-di-GMP (37). In summary, the high affinity and specificity measured for these effectors and the presence of many KOP binders in the pulldown set (see Results) suggest that additional candidate binders isolated by the CCMS experiment might be actually bona fide c-di-GMP effectors. However, some proteins identified in the set of pulled-down candidates may nonspecifically bind to c-di-GMP, possibly through allosteric binding sites for other cofactors (see Table S2 in the supplemental material). Others have been shown to form protein complexes in other bacteria, like the flagellum and the gliding apparatuses, transporters, and complexes with various metabolic functions. Several of those candidates are also encoded in operons in B. bacteriovorus. It is plausible that only one complex member is a genuine c-di-GMP effector (see Table S2 and see below). Moreover, 14 of the 84 candidates are inaccessible to the cytoplasm (see Results), where c-di-GMP is present. Hence, these candidates are either indirect or unspecific c-di-GMP binders.
Cyclic-di-GMP signaling might bridge between cell cycle phases.
Of the 84 (74%) putative c-di-GMP binding proteins, 62 were specific to the AP or to the HI samples (see Results; see also Table S2 in the supplemental material). Although in HI mutants the transition from the AP to the GP is deregulated, similarities between replicating HI and GP cells are high, as described above. In addition, the M1.1 HI strain used here exhibits a predatory phenotype and shows a very high degree of genome conservation to the wild-type strain, with only four missense mutations in addition to the nonsense mutations in genes bd0108 and pcnB, which underlie the axenic phenotype (14). We therefore interpret the results from the HI samples as relevant to the GP. Accordingly, the 22 candidates, common to the AP and to the HI culture, may include genuine AP-specific binders, as well as effectors truly common to both the AP and the GP, implying that the apparent c-di-GMP phase specificity may be even larger than that identified in the present study. The set of putative c-di-GMP effectors is mainly populated by proteins containing uncharacterized c-di-GMP binding domains (65/84), and yet putative functions could be attributed to most (49/65), thereby enabling functional mapping of the putative c-di-GMP network (Fig. 3A). Some appeared to be phase specific: proteins related to peptidoglycan (Bd0160, Bd0967) and to cofactor biosynthesis (Bd1196, Bd1602, and Bd3560) were active in the replicating HI culture, whereas flagellum-related proteins (Bd0604, Bd3395, and Bd3404) were active in the AP (see Table S2 in the supplemental material).
The distribution of c-di-GMP binders between the AP and the GP was not congruent with the expression of their encoding genes (13). Only 35 of 62 proteins (56.4%) showed congruence, i.e., c-di-GMP binding by the pulled-down proteins and expression of their encoding genes cooccurred during the same cell cycle phase (see Table S2 in the supplemental material). The remaining 27 noncongruent entries included all seven putative c-di-GMP effectors (Bd0025, Bd0407, Bd1608, Bd1833, Bd2174, Bd2611, and Bd2729) specifically binding in AP and assigned to metabolism (thereby encoded by GP expressed genes) and the three (Bd0156, Bd1080, and Bd1590) GP-specific c-di-GMP-binding transcriptional regulators (thereby encoded in AP-expressed genes) (see Table S2 in the supplemental material). Other noncongruent entries include response regulators, singletons, or are not functionally annotated. These discrepancies may indicate that the cyclic-di-GMP network adds regulatory plasticity by enabling proteins to be differentially active between phases, even though the transcriptome is sharply phase specific (13). Posttranslational regulation was previously proposed to play a major role in segregating activity of coexpressed c-di-GMP network proteins (38). It may thus regulate protein activity in a cell cycle-dependent manner.
B. bacteriovorus flagellar components might bind c-di-GMP.
The suggested network encompasses numerous metabolic and signaling processes, as well as gliding and flagellar proteins. Thus far, c-di-GMP is known to negatively affect flagellar motility through various effectors. For example, in P. aeruginosa through c-di-GMP binding to the transcription factor FleQ (39) or the PDE Pch (PA5017), which in response, interacts with CheA and potentially with FliF (40), and in E. coli and Salmonella, through the interaction of the c-di-GMP effector YcgR with the flagellar motor components FliG and FliM (41). In B. bacteriovorus, Hobley et al. (4) showed that mutations in DGCs affect flagellar biosynthesis. Thus far, a direct interaction between c-di-GMP and flagellar apparatus proteins was not detected, as the identification of some of its constituents—the flagellin FliC1 (42), the hook protein FlgE, and the MS ring component FliF—in the CCMS experiment suggests for the first time. Since these polypeptides are not known to directly interact one with the other, additional “bridging” components of this larger protein complex were probably missed by the pulldown. Indeed, FlgC, a flagellar basal-body rod protein, was detected at low levels in samples in which FliF, FlgE and FliC1 were found (see Table S1 in the supplemental material; see the “AP membrane” tab). Since only FliF, of these three proteins, may directly interact with the cytosol through its C terminus (28), it may thus be the c-di-GMP binding component of a copurified complex containing the other flagellar proteins found here. It is noteworthy that the pulled PDE Bd1971 and P. aeruginosa Pch (that possibly interacts with P. aeruginosa FliF) share very low homology (15.8% global identity and 25.5% global similarity) except for the common EAL domain (33% local identity and 57% local similarity).
Flagellar activity is essential for prey penetration, and yet soon after prey invasion motility is inactivated (42). It had earlier been proposed that the rapid breakdown of the putative c-di-GMP riboswitch upon entry into GP may release c-di-GMP (13). If this is true, c-di-GMP may bind to and inactivate the flagellar apparatus at the early stages of predation.
Central metabolism may be regulated by c-di-GMP.
The confirmation of Bd2924, an acyl-CoA dehydrogenase, as a genuine c-di-GMP effector provides for the first time experimental support that c-di-GMP may affect central metabolism. Additional candidate binders are embedded in the metabolic pathways of the cofactors adenosylcobalamine, porphyrin and CoA, isoprenoids, amino and fatty acids, and the cell wall (Fig. 3A and see Table S3 in the supplemental material). Potential c-di-GMP binding effectors may affect key reactions of the bacterium energy metabolism. This includes the rate-limiting oxidative decarboxylation from α-ketoglutarate (2-oxoglutarate) to succinyl-CoA (Bd2828 and Bd2829 [25]), which produces NADH; additional reductant-producing reactions such as the oxidation of succinate to fumarate (Bd0025), producing FADH2 (43); the action of malate dehydrogenase (oxaloacetate-decarboxylating), yielding pyruvate and NADPH (Bd1833 and Bd1834); acetyl-CoA (and reductants) production from pyruvate (Bd0394) through the last steps in fatty acid metabolism; and amino acid degradation (Bd1835, Bd2070, Bd2728, and Bd2729). In addition, 2-methylcitrate dehydratase (Bd2500) provides a modulation point in the propanoyl-CoA/succinyl-CoA pathway. Candidate c-di-GMP binding proteins were found in complexes I, II, and IV of the respiratory pathway and in the ATP synthase complex V (see Table S3 in the supplemental material). It is not yet known whether these interactions result from specific c-di-GMP binding or from unspecific nucleotide binding. If c-di-GMP binding is confirmed, this would be the first experimental evidence that the secondary messenger directly regulates ATP production. Three candidate binders are implicated in nucleic acid metabolism: Bd0081 and Bd1196 catalyze the formation of 3′,5′ cyclic AMP/GMP and of dAMP/dGMP, respectively. Bd0049 is a transaldolase that links between the pentose phosphate pathway and glycolysis (KEGG bba00030 [http://www.genome.jp/kegg-bin/show_pathway?bba00030]). B. bacteriovorus is unable to import most sugars and mostly depends on noncarbohydrate metabolism for carbon and energy (44). Accordingly, this enzyme may as well participate in the utilization of prey-derived ribose for energy production and for the biosynthesis of non-nucleic-acid material, as shown by Hespell and Odelson (45).
The data presented here suggest that an overlying, coordinated c-di-GMP regulatory network directly interacts with the catabolism of energy-rich substrates and the production of reducing equivalents. Although yet to be confirmed, c-di-GMP may also affect the electrochemical gradient and energy production. These features may at least in part explain the extraordinary energy efficiency of B. bacteriovorus, which was shown to produce more than twice as much biomass per mole of ATP than E. coli (46).
c-di-GMP may interact with the secretion machinery.
The secretin GspD (Bd1597, a PulD homolog) was identified in the CCMS experiment. This outer membrane protein is the pore protein through which folded polypeptides are secreted to the extracellular space through a type II secretion pathway (47). B. bacteriovorus has no known syringe-like secretion systems; most probably, the majority of the very large complement of its secreted proteins is exported through the Sec pathway (48). At this stage, we can only hypothesize that c-di-GMP modulation affects GspD while in the cytoplasm, perhaps by controlling its own secretion through interactions with other components of the secretion machinery.
Interactions between regulatory networks.
Three candidate binders possibly bridge between the c-di-GMP and the cAMP networks in B. bacteriovorus. bd2590, coding for a Clp-like c-di-GMP binder is cotranscribed with bd2591, coding for a standard CRP (13); the PDE Bd1971 carries a CRP domain; Bd0081 is an adenylate cyclase that putatively binds to c-di-GMP. c-di-GMP/cAMP cross-regulation has been described in Vibrio cholerae, where cAMP represses biofilm formation by suppressing DGC gene expression (49), and in P. aeruginosa, where a type IV pilus functions as a surface sensor, promoting the coordinated and hierarchical production of cAMP, followed by c-di-GMP, as part of the regulation of the early steps of biofilm formation (50). The interconnection observed here between the c-di-GMP and cAMP networks includes alleged “bilingual” proteins and coregulated effectors that suggest direct cross-regulation between these two secondary messengers. The suggested c-di-GMP network also interacts with gene regulation through a NifA-like transcription factor (Bd0156) and a LysR-type transcription factor (Bd1080), as well as possibly with environmental sensing through two-component system proteins (Bd0367, Bd2402, and Bd3393). Although these data suggest that c-di-GMP interacts with other regulatory networks at the sensing and at the gene expression levels, the interconnections appear to be infrequent, being limited to one of six adenylate cyclases, three of >60 response regulators and histidine kinases, and one of 16 LysR regulators. Finally, the polyribonucleotide nucleotidyltransferase (Pnp, Bd1151), a component of the RNA degradosome, may interact directly with c-di-GMP, like its orthologue in E. coli (9). Noteworthy, RhlB and PcnB, other components of the RNA degradosome, are directly implicated in the formation of axenic HI mutants (14). Taken together, these observations let us suppose that c-di-GMP regulation over RNA turnover affect B. bacteriovorus cell cycle progress.
Supplementary Material
ACKNOWLEDGMENTS
This study was support by the Israel Science Foundation (grant 1583/12) and the German Israel Foundation (grant I-1217-342.13/2012).
This article is dedicated to Felix Frolow.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00422-15.
REFERENCES
- 1.Hengge R. 2010. Cyclic-di-GMP reaches out into the bacterial RNA world. Sci Signal 3:pe44. doi: 10.1126/scisignal.3149pe44. [DOI] [PubMed] [Google Scholar]
- 2.Abel S, Bucher T, Nicollier M, Hug I, Kaever V, Abel zur Wiesch P, Jenal U. 2013. Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the Caulobacter cell cycle. PLoS Genet 9:e1003744. doi: 10.1371/journal.pgen.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lori C, Ozaki S, Steiner S, Bohm R, Abel S, Dubey BN, Schirmer T, Hiller S, Jenal U. 2015. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523:236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 4.Hobley L, Fung RKY, Lambert C, Harris MATS, Dabhi JM, King SS, Basford SM, Uchida K, Till R, Ahmad R, Aizawa S-I, Gomelsky M, Sockett RE. 2012. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog 8:e1002493. doi: 10.1371/journal.ppat.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signaling cascade in Escherichia coli biofilm control. EMBO J 32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 7.Fang X, Ahmad I, Blanka A, Schottkowski M, Cimdins A, Galperin MY, Römling U, Gomelsky M. 2014. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Micro 93:439–452. doi: 10.1111/mmi.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An S-Q, Caly DL, McCarthy Y, Murdoch SL, Ward J, Febrer M, Dow JM, Ryan RP. 2014. Novel cyclic di-GMP effectors of the YajQ protein family control bacterial virulence. PLoS Pathog 10:e1004429. doi: 10.1371/journal.ppat.1004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuckerman JR, Gonzalez G, Gilles-Gonzalez M-A. 2011. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J Mol Biol 407:633–639. doi: 10.1016/j.jmb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Baraquet C, Harwood CS. 2013. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci U S A 110:18478–18483. doi: 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin KH, Lee YC, Tu ZL, Chen CH, Tseng YH, Yang JM, Ryan RP, McCarthy Y, Dow JM, Wang AH, Chou SH. 2010. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol 396:646–662. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 12.Rotem O, Pasternak Z, Jurkevitch E. 2014. Bdellovibrio and like organisms, p 3–17. In Rosenberg E, DeLong EF, Stackebrandt E Lory S, Thompson F (ed), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 13.Karunker I, Rotem O, Dori-Bachash M, Jurkevitch E, Sorek R. 2013. A global transcriptional switch between the attack and growth forms of Bdellovibrio bacteriovorus. PLoS One 8:e61850. doi: 10.1371/journal.pone.0061850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roschanski N, Klages S, Reinhardt R, Linscheid M, Strauch E. 2011. Identification of genes essential for prey-independent growth of Bdellovibrio bacteriovorus HD100. J Bacteriol 193:1745–1756. doi: 10.1128/JB.01343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dori-Bachash M, Dassa B, Pietrokovski S, Jurkevitch E. 2008. Proteome-based comparative analyses of growth stages reveal new cell cycle-dependent functions in the predatory bacterium Bdellovibrio bacteriovorus. Appl Environ Microbiol 74:7152–7162. doi: 10.1128/AEM.01736-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milner DS, Till R, Cadby I, Lovering AL, Basford SM, Saxon EB, Liddell S, Williams LE, Sockett RE. 2014. Ras GTPase-like protein MglA, a controller of bacterial social motility in myxobacteria, has evolved to control bacterial predation by Bdellovibrio. PLoS Genet 10:e1004253. doi: 10.1371/journal.pgen.1004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nesper J, Reinders A, Glatter T, Schmidt A, Jenal U. 2012. A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J Proteomics 75:4874–4878. doi: 10.1016/j.jprot.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 19.Laventie BJ, Nesper J, Ahrne E, Glatter T, Schmidt A, Jenal U. 2015. Capture compound mass spectrometry: a powerful tool to identify novel c-di-GMP effector proteins. J Vis Exp doi: 10.3791/51404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lovering AL, Capeness MJ, Lambert C, Hobley L, Sockett RE. 2011. The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of cyclic-di-GMP phosphodiesterases. mBio 2:e00163-11. doi: 10.1128/mBio.00163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey TL, Johnson J, Grant CE, Noble WS. 2015. The MEME suite. Nucleic Acids Res 43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duvel J, Bertinetti D, Moller S, Schwede F, Morr M, Wissing J, Radamm L, Zimmermann B, Genieser HG, Jansch L, Herberg FW, Haussler S. 2012. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods 88:229–236. doi: 10.1016/j.mimet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett RE, Schuster SC. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 24.Lazzaroni JC, Dubuisson JF, Vianney A. 2002. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie 84:391–397. doi: 10.1016/S0300-9084(02)01419-0. [DOI] [PubMed] [Google Scholar]
- 25.Frank RAW, Price AJ, Northrop FD, Perham RN, Luisi BF. 2007. Crystal structure of the E1 component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J Mol Biol 368:639–651. doi: 10.1016/j.jmb.2007.01.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollenstein K, Frei DC, Locher KP. 2007. Structure of an ABC transporter in complex with its binding protein. Nature 446:213–216. doi: 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- 27.Kagawa Y. 1999. Biophysical studies on ATP synthase. Adv Biophys 36:1–25. doi: 10.1016/S0065-227X(99)80003-3. [DOI] [PubMed] [Google Scholar]
- 28.Ogawa R, Abe-Yoshizumi R, Kishi T, Homma M, Kojima S. 2015. Interaction of the C-terminal tail of FliF with FliG from the Na+-driven flagellar motor of Vibrio alginolyticus. J Bacteriol 197:63–72. doi: 10.1128/JB.02271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwata S. 1998. Structure and function of bacterial cytochrome c oxidase. J Biochem 123:369–375. doi: 10.1093/oxfordjournals.jbchem.a021946. [DOI] [PubMed] [Google Scholar]
- 30.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. 2010. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 31.Sundriyal A, Massa C, Samoray D, Zehender F, Sharpe T, Jenal U, Schirmer T. 2014. Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger cyclic di-GMP. J Biol Chem 289:6978–6990. doi: 10.1074/jbc.M113.516195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fei N, Haussinger D, Blumli S, Laventie BJ, Bizzini LD, Zimmermann K, Jenal U, Gillingham D. 2014. Catalytic carbene transfer allows the direct customization of cyclic purine dinucleotides. Chem Commun 50:8499–8502. doi: 10.1039/C4CC01919A. [DOI] [PubMed] [Google Scholar]
- 33.Ko J, Ryu KS, Kim H, Shin JS, Lee JO, Cheong C, Choi BS. 2010. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by PilZ domain proteins. J Mol Biol 398:97–110. doi: 10.1016/j.jmb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Lambert C, Chang CY, Capeness MJ, Sockett RE. 2010. The first bite–profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One 5:e8599. doi: 10.1371/journal.pone.0008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. 2007. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J 26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hengge R. 2009. Principles of c-di-GMP signaling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 37.Sureka K, Choi PH, Precit M, Delince M, Pensinger DA, Huynh TN, Jurado AR, Goo YA, Sadilek M, Iavarone AT, Sauer JD, Tong L, Woodward JJ. 2014. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell 158:1389–1401. doi: 10.1016/j.cell.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seshasayee AS, Fraser GM, Luscombe NM. 2010. Comparative genomics of cyclic-di-GMP signaling in bacteria: posttranslational regulation and catalytic activity. Nucleic Acids Res 38:5970–5981. doi: 10.1093/nar/gkq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Micro 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulasekara BR, Kamischke C, Kulasekara HD, Christen M, Wiggins PA, Miller SI. 2013. c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. ELife 2:e01402. doi: 10.7554/eLife.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambert C, Evans KJ, Till R, Hobley L, Capeness M, Rendulic S, Schuster SC, Aizawa S-I, Sockett RE. 2006. Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol Micro 60:274–286. doi: 10.1111/j.1365-2958.2006.05081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berg JM, Tymoczko JL, Stryer L. 2002. Biochemistry. W.H. Freeman, New York, NY. [Google Scholar]
- 44.Hespell RB, Rosson RA, Thomashow MF, Rittenberg SC. 1973. Respiration of Bdellovibrio bacteriovorus strain 109J and its energy substrates for intraperiplasmic growth. J Bacteriol 113:1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hespell RB, Odelson DA. 1978. Metabolism of RNA-ribose by Bdellovibrio bacteriovorus during intraperiplasmic growth on Escherichia coli. J Bacteriol 136:936–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rittenberg SC, Hespell RB. 1975. Energy efficiency of intraperiplasmic growth of Bdellovibrio bacteriovorus. J Bacteriol 121:1158–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korotkov KV, Gonen T, Hol WGJ. 2011. Secretins: dynamic channels for protein transport across membranes. Trends Biochem Sci 36:433–443. doi: 10.1016/j.tibs.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barabote RD, Rendulic S, Schuster SC, Saier MH Jr. 2007. Comprehensive analysis of transport proteins encoded within the genome of Bdellovibrio bacteriovorus. Genomics 90:424–446. doi: 10.1016/j.ygeno.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fong JC, Yildiz FH. 2008. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J Bacteriol 190:6646–6659. doi: 10.1128/JB.00466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GC, O'Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6:e02456-14. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Capeness MJ, Lambert C, Lovering AL, Till R, Uchida K, Chaudhuri R, Alderwick LJ, Lee DJ, Swarbreck D, Liddell S, Aizawa S, Sockett RE. 2013. Activity of Bdellovibrio hit locus proteins, Bd0108 and Bd0109, links type IVa pilus extrusion/retraction status to prey-independent growth signaling. PLoS One 8:e79759. doi: 10.1371/journal.pone.0079759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.