ABSTRACT
The ubiquitous bacterial second messenger cyclic di-GMP (c-di-GMP) has recently become prominent as a trigger for biofilm formation in many bacteria. It is generated by diguanylate cyclases (DGCs; with GGDEF domains) and degraded by specific phosphodiesterases (PDEs; containing either EAL or HD-GYP domains). Most bacterial species contain multiples of these proteins with some having specific functions that are based on direct molecular interactions in addition to their enzymatic activities. Escherichia coli K-12 laboratory strains feature 29 genes encoding GGDEF and/or EAL domains, resulting in a set of 12 DGCs, 13 PDEs, and four enzymatically inactive “degenerate” proteins that act by direct macromolecular interactions. We present here a comparative analysis of GGDEF/EAL domain-encoding genes in 61 genomes of pathogenic, commensal, and probiotic E. coli strains (including enteric pathogens such as enteroaggregative, enterohemorrhagic, enteropathogenic, enterotoxigenic, and adherent and invasive Escherichia coli and the 2011 German outbreak O104:H4 strain, as well as extraintestinal pathogenic E. coli, such as uropathogenic and meningitis-associated E. coli). We describe additional genes for two membrane-associated DGCs (DgcX and DgcY) and four PDEs (the membrane-associated PdeT, as well as the EAL domain-only proteins PdeW, PdeX, and PdeY), thus showing the pangenome of E. coli to contain at least 35 GGDEF/EAL domain proteins. A core set of only eight proteins is absolutely conserved in all 61 strains: DgcC (YaiC), DgcI (YliF), PdeB (YlaB), PdeH (YhjH), PdeK (YhjK), PdeN (Rtn), and the degenerate proteins CsrD and CdgI (YeaI). In all other GGDEF/EAL domain genes, diverse point and frameshift mutations, as well as small or large deletions, were discovered in various strains.
IMPORTANCE Our analysis reveals interesting trends in pathogenic Escherichia coli that could reflect different host cell adherence mechanisms. These may either benefit from or be counteracted by the c-di-GMP-stimulated production of amyloid curli fibers and cellulose. Thus, EAEC, which adhere in a “stacked brick” biofilm mode, have a potential for high c-di-GMP accumulation due to DgcX, a strongly expressed additional DGC. In contrast, EHEC and UPEC, which use alternative adherence mechanisms, tend to have extra PDEs, suggesting that low cellular c-di-GMP levels are crucial for these strains under specific conditions. Overall, our study also indicates that GGDEF/EAL domain proteins evolve rapidly and thereby contribute to adaptation to host-specific and environmental niches of various types of E. coli.
INTRODUCTION
Although cyclic di-GMP (c-di-GMP) was first described as an allosteric activator of cellulose synthase already in 1987 (1), it was only in the 21st century that it became clear that this nucleotide second messenger is ubiquitous in the bacterial world and generally promotes biofilm formation, downregulates flagella expression and/or activity, and can modulate virulence, the cell cycle, and development. Furthermore, research on c-di-GMP signaling in a small group of model bacterial species has led to novel general concepts in second messenger signaling (2–6).
c-di-GMP is generated by diguanylate cyclases (DGCs) characterized by GGDEF domains, with this amino acid motif representing the active center (the A-site). Most of these enzymes also contain a secondary and inhibitory binding site for c-di-GMP (the I-site), i.e., their activities are feedback inhibited by their own product. Specific phosphodiesterases (PDEs) that degrade c-di-GMP belong to one of two protein families, featuring either EAL or HD-GYP domains. Structures of DGCs and PDEs have been elucidated and functionally important amino acids have been identified (7). c-di-GMP effector mechanisms operate via many and unexpectedly diverse families of c-di-GMP-binding proteins and RNAs (riboswitches) (8–10). These can target virtually any molecular process in bacterial cells, including transcription, mRNA stability, translation, functional protein-protein interactions, or protein degradation.
One of the most striking features of c-di-GMP signaling is the multiplicity of GGDEF/EAL/HD-GYP domain-encoding genes in most bacterial species. The Escherichia coli K-12 laboratory strain, one of the model species in c-di-GMP-related research, has a complement of 29 of these genes, with 12 and 10 genes encoding GGDEF and EAL domains only, respectively, and 7 genes encoding proteins with both domains. Based on biochemical evidence and knowledge of the functions of specific highly conserved amino acids, 12 of the gene products are DGCs, 13 are PDEs, and 4 are “degenerate” proteins with nonenzymatic functions (11, 12). This is reflected in a novel systematic nomenclature for the genes encoding these enzymes and their products proposed by a group of researchers working on c-di-GMP signaling in E. coli (see the report by Hengge et al. in this issue of the Journal of Bacteriology [13]). For maximal clarity, we use the new designations here but also provide the previous “Y” designations.
For almost half of these GGDEF/EAL domain proteins, the physiological contexts of action and in some cases also molecular functions and interactions have been clarified. For example, several DGCs can contribute to downregulating flagellar rotation via the c-di-GMP-binding protein YcgR (14–16). Another major target for positive control by c-di-GMP is the expression of the biofilm regulator CsgD (15, 17), which activates the biosynthesis of amyloid curli fibers and cellulose, i.e., the components of the extracellular matrix in colony biofilms of E. coli and related bacteria (18, 19). The underlying mechanism provides a paradigm for highly specific “local” c-di-GMP signaling by distinct DGCs and PDEs: the “trigger enzyme” and PDE PdeR (YciR) and the DGC DgcM (YdaM) form a complex with the transcription factor MlrA and thereby act as a core transcriptional switch module that controls transcription of csgD (20). This switch module responds to the cellular level of c-di-GMP, which under standard laboratory conditions increases during entry into stationary phase as a result of decreasing levels of PdeH (YhjH) and increasing levels of DgcE (YegE) (15). DgcC (YaiC) is another DGC that equally specifically activates a cellular target, i.e., cellulose synthase, but here the mechanism is still unclear (21, 22). Highly specific macromolecular interactions are also underlying the functions of the enzymatically inactive “degenerate” GGDEF/EAL domain proteins BluF (YcgF), CsrD (YhdA), and RflP (YdiV) (23–26). However, other GGDEF/EAL domain proteins of E. coli K-12 remain largely uncharacterized at the functional molecular level.
With some rare exceptions (27–30), analyses of GGDEF/EAL domain genes or proteins of E. coli have been performed with laboratory K-12 strains. Therefore, almost nothing is known about how the complement of GGDEF/EAL domain-encoding genes may vary in different E. coli strains, in particular when comparing pathogenic and commensal strains. E. coli is a particularly diverse species with a pangenome several times larger than the core genome conserved in all known strains (31, 32). It colonizes different host-associated niches but also thrives under quite diverse environmental conditions. This diversity suggested that the genomes of E. coli strains may represent a fertile ground for discovering novel GGDEF/EAL domain genes and interesting variations in such genes that are already known, in particular, since different types of pathogenic E. coli differ profoundly in the way they adhere to host tissue (33) and adhesion mechanisms in general are a major target of c-di-GMP signaling. Moreover, our study here was spurred by our analysis of c-di-GMP signaling in the 2011 German outbreak O104:H4 strain, which already led to the discovery of a novel and extremely highly expressed DGC (DgcX) and some other interesting variations in GGDEF/EAL domain genes in enteroaggregative and enterohemorrhagic E. coli (EAEC and EHEC, respectively) (30).
Here we present a more comprehensive genomic comparison of 61 strains—including pathogens of different pathotypes as well as commensals and probiotics—that led to the discovery of additional DGC/PDE-encoding genes and numerous variations with respect to integrity and expression of already known GGDEF/EAL domain proteins. Certain variations correlate with distinct pathotypes and lifestyles of the E. coli strains analyzed and shed light on how rapid evolution of c-di-GMP signaling can contribute to the diversity and specific adaptations of different E. coli strains and pathotypes.
MATERIALS AND METHODS
Bacterial strains and genome sequences.
This study started with a systematic analysis of all of the 39 completed E. coli genomes available on the National Center for Biotechnology Information (NCBI) database in May 2011 (i.e., during the O104:H4 outbreak in Germany). In May 2012 an updated search incorporated an additional nine genomes. In June 2014 an additional 13 genomes were selectively added since there were too many new completed E. coli genomes added to the NCBI database to be included into the present study. The 13 selected genomes were added to complement the previous strains such that the different pathotypes of E. coli are adequately represented. Overall, 61 strains were thus included in this study (see Table S1 in the supplemental material). The strains analyzed included enteric pathogens (enteroaggregative, enterotoxigenic, enterohemorrhagic, enteropathogenic, and adherent and invasive E. coli [EAEC, ETEC, EHEC, EPEC, and AIEC, respectively]), extraintestinal pathogenic E. coli (ExPEC; including uropathogenic E. coli [UPEC] and meningitis-associated E. coli [MNEC]), and commensal E. coli strains (including probiotics such as E. coli Nissle 1917), as well as E. coli strains of nonhuman origin (avian pathogenic E. coli [APEC], porcine ETEC, and environmental isolates). E. coli K-12 W3110 was used as a reference strain here, since it has been extensively used as an experimental model for c-di-GMP signaling in E. coli (20, 24, 34–36).
BLAST analyses.
The Basic Local Alignment Search Tool (BLAST) (37) was used to search for proteins with GGDEF and/or EAL domains encoded in single selected genomes. The 169-amino-acid GGDEF domain of the DgcM (YdaM) protein and the 245-amino-acid EAL domain of the PdeC (YjcC) protein (as identified by the SMART EMBL database) were used as query sequences to detect previously known, as well as unidentified, GGDEF and EAL domain proteins. If a GGDEF/EAL domain gene known from E. coli K-12 was not detected from the genome of a given strain, a second BLAST search was performed using the particular protein from the W3110 strain as the query sequence. This approach showed true absences but also yielded apparent discrepancies in the lengths of the proteins. To verify whether these differences reflected real genomic variations or rather arbitrary differences in the annotation of the respective genome sequence, a follow-up tBLAST search was performed, where the amino acid sequence was searched directly within the nucleotide database by the tBLAST algorithm translating the nucleotide sequence into an amino acid sequence, again using proteins from the W3110 strain as the query sequence. In this way various GGDEF/EAL domain genes that initially seemed to be “missing” were found, whereas others were confirmed to be truly absent from the respective genomes. Apparent discrepancies in protein annotation between strains were further addressed by directly looking at the nucleotide sequences, the annotated start codons and putative Shine-Dalgarno sequences. Many of these cases, in particular when nucleotide sequences were nearly or even complete identical, were found to be due to misannotations of start codons. In other cases, true deleterious disruptions, such as insertions, deletions, or point mutations, were also found at the nucleotide sequence level and were analyzed in detail by alignments (38) with the corresponding nucleotide sequences from strain W3110.
Newly identified GGDEF/EAL domain proteins and their genes, as well as novel sensory input domains, were designated according to the rules for a systematic nomenclature proposed in another publication in this issue (13).
Motif analyses.
All GGDEF/EAL domain-containing proteins were analyzed for potential uncharacterized motifs using the MEME program (39). Default settings were used, except that the condition “any number of repetitions” was selected for the prediction of how single motifs were distributed among the sequences. The locations of the motifs were determined for individual proteins relative to the locations of the putative transmembrane helices using the hydropathy plots generated by the TMHMM program (40).
RESULTS AND DISCUSSION
The E. coli pangenome contains at least 35 genes encoding GGDEF/EAL domain proteins.
Our analysis of GGDEF/EAL domain genes was originally triggered by the outbreak of E. coli O104:H4 infection in May 2011 in Germany (41, 42). At this time the NCBI database genome contained the genome sequences of 39 E. coli strains. Later on, newly completed E. coli genomes were successively added to the analysis (see Materials and Methods for details) such that the final set of 61 genomes included the different pathogroups, as well as commensal strains of E. coli (see Table S1 in the supplemental material). The following pathogroups are represented in our study: enteric pathogens, including EAEC, ETEC, EHEC (also called Shiga toxin [Stx]-producing E. coli [STEC]), EPEC, and AIEC; ExPEC, including UPEC and MNEC; and pathogenic E. coli of nonhuman origin (APEC and porcine ETEC).
Initial BLAST searches (37) using the GGDEF and EAL domain sequences of the DGC DgcM (YdaM) and the PDE PdeC (YjcC), respectively, were followed by a reiterative comparative process that allowed to pinpoint single nucleotide polymorphisms (SNP) in specific genes, as well as the exact extent of smaller or larger deletions (see Materials and Methods). A total of 35 GGDEF/EAL domain-encoding genes (see Table S2 in the supplemental material), as well as numerous small and large variations in the sequences of distinct genes, were identified in the 61 genomes. Previously uncharacterized GGDEF/EAL domain-encoding genes, which, based on the presence of functionally relevant amino acids (see Table S2 in the supplemental material), encode active DGCs or PDEs, were named using a dgc/pde nomenclature, as suggested in the accompanying publication on the nomenclature of c-di-GMP-related enzymes in E. coli (13). The 35 genes include two DGC genes (dgcX and dgcY) and four PDE genes (pdeT, pdeW, pdeX, and pdeY) not found in E. coli K-12 (Table 1).
TABLE 1.
GGDEF/EAL domain proteins not present in E. coli K-12
| Gene designation | Strain | Original gene annotation | GI no. |
|---|---|---|---|
| Genes encoding DGCs not present in E. coli K-12 | |||
| dgcX | LB226692 (EAEC) | HUSEC_04298 | 340741263 |
| 2011C-3493 (EAEC) | O3K_17495 | 407483024 | |
| HUSEC041 (EAEC) | HUSEC41_04052 | 340735556 | |
| 55989 (EAEC) | EC55989_0813 | 218694243 | |
| 2009EL-2071 (EAEC) | O3O_07785 | 407468242 | |
| 2009EL-2050 (EAEC) | O3M_17475 | 410483577 | |
| ETEC H10407 (ETEC) | |||
| E24377A (ETEC) | EcE24377A_0835 | 157155149 | |
| SE11 (commensal E. coli) | ECSE_1457 | 209918648 | |
| dgcY | SMS-3-5 (environmental E. coli) | EcSMS35_1716 | 170517710 |
| O7:K1 strain CE10 (NMEC) | CE10_1648 | 386624007 | |
| Genes encoding PDEs not present in E. coli K-12 | |||
| pdeT (vmpA) | O157:H7 strain TW14359 (EHEC) | ECSP_1197 | 254792284 |
| O157:H7 strain EC4115 (EHEC) | ECH74115_1268 | 209399126 | |
| O157:H7 strain EDL933 (EHEC) | Z1528 | 15801017 | |
| O157:H7 strain Sakai (EHEC) | ECs1272 | 15830526 | |
| O157:H7 strain Xuzhou21 (EHEC) | CDCO157_1207 | 387881786 | |
| O55:H7 strain CB9615 (EPEC) | GI1N-1259 | 291282023 | |
| O55:H7 strain RM12579 (EPEC) | ECO55CA74_06170 | 387506136 | |
| pdeW | E24377A (ETEC) | EcE24377A_E0054 | 157149510 |
| pdeX | 536 (UPEC) | ECP_2965 | 110643119 |
| pdeY (sfaY) | 536 (UPEC) | ECP_0300 | 110342098 |
| IHE3034 (NMEC) | ECOK1_1105 | 386598807 | |
| CFT073 (UPEC) | c1246 | 26247120 | |
| UTI89 (UPEC) | UTI89_C1116 | 91210145 | |
| UM146 (AIEC) | UM146_12325 | 386605046 | |
| ABU 83972 (probiotic E. coli) | ECABU_c12040 | 386638503 |
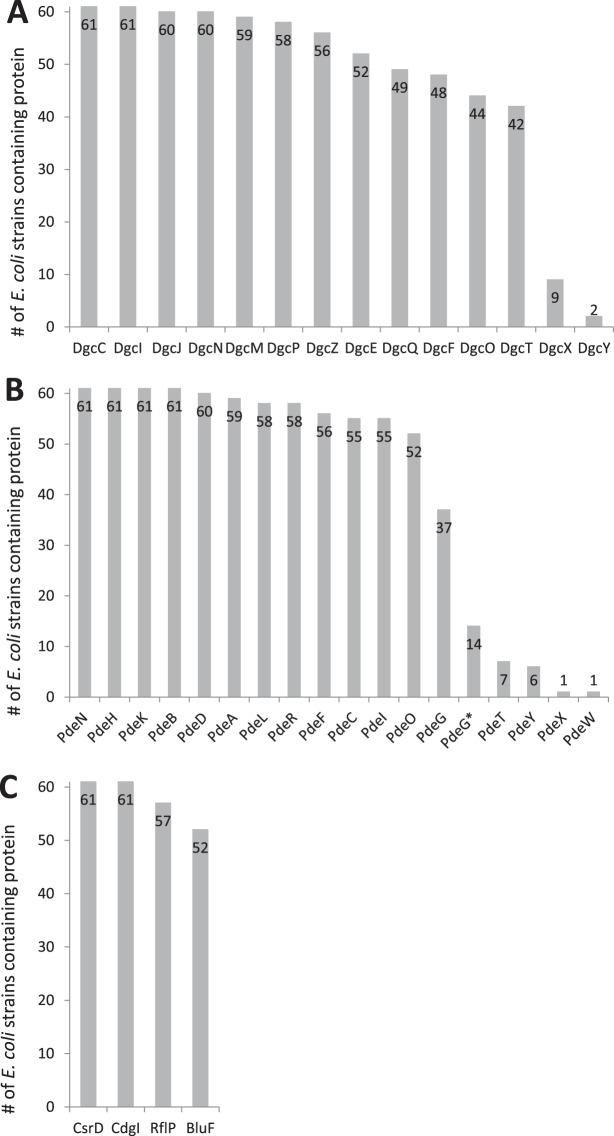
The 35 GGDEF/EAL domain-encoding genes are conserved with various frequencies (Fig. 1). There is a core set of eight genes that are completely conserved among all 61 strains. These encode (i) the two DGCs, DgcC (YaiC) and DgcI (YliF), (ii) the four PDEs PdeB (YlaB), PdeH (YhjH), PdeN (Rtn), and PdeK (YhjK), and (iii) the degenerate GGDEF/EAL proteins CsrD and CdgI (YeaI); these proteins seem to be functionally important independently of all host-related or environmental specialization of different E. coli strains. Furthermore, a large group of 21 GGDEF/EAL domain-encoding genes are found in versions predicted to encode functional proteins in >65% of all strains analyzed, i.e., these genes belong to a complement of ancient and typical E. coli genes but seem dispensable in certain niches or “lifestyles.” Some of these genes also display specific sequence variants that occur frequently in certain groups of strains (for details, see below). Finally, the six GGDEF/EAL domain-encoding genes not present in E. coli K-12 are found in small minorities of strains, indicating that these genes represent recent acquisitions in distinct clades or even single strains that contribute to adaptation to specific host-associated and/or environmental niches.
FIG 1.
Frequency of occurrence of genes encoding intact GGDEF/EAL domain proteins in the genomes of 61 E. coli strains. Mutations in the alleles that do not count as fully intact are described in detail in Table 2. (A) Diguanylate cyclases. (B) c-di-GMP-specific phosphodiesterases. (C) Degenerate GGDEF/EAL domain proteins. An asterisk denotes an allele of pdeG (ycgG) with a 5′ deletion that nevertheless produces an N-terminally truncated yet apparently active PDE (Table 2; see also the text).
Below, the novel DGC and PDE genes and their putative gene products (Table 1), as well as a subset of functionally interesting variations in certain previously known GGDEF/EAL domain genes, are described and discussed in detail. A list of these variations detected in the 61 E. coli strains, including those where a functional consequence is not readily apparent, is given in Table 2 (note that synonymous codons or occasional variations specifying similar amino acids were not included).
TABLE 2.
Genomic variations of the genes encoding GGDEF/EAL domain proteins in 61 E. coli strains
| Gene | Mutation(s)a | Consequence(s) for protein expressionb | Strain(s) (pathotype[s])c |
|---|---|---|---|
| Genes encoding DGCs | |||
| dgcC (yaiC) | |||
| dgcE (yegE) | 1-nt (G) insertion (after nt 413, in codon 138)/frameshift | DgcE (138 + 39 AAs) is C-terminally truncated | 042 (EAEC) |
| 1-nt (A) insertion (after nt 457, in codon 153)/frameshift | DgcE (153 + 24 AAs) is C-terminally truncated | O157:H7 strain EC4115 (EHEC), O157:H7 strain EDL933 (EHEC), O157:H7 strain TW14359 (EHEC), O157:H7 strain Xuzhou21 (EHEC) | |
| 1-nt (A) insertion (after nt 457, in codon 153)/frameshift; 1-nt (G) insertion (after nt 933, in codon 312) | DgcE (153 + 24 AAs) is C-terminally truncated | O157:H7 strain Sakai (EHEC) | |
| 7-nt (GTGATTC) deletion (after nt 1467, in codon 490)/frameshift | DgcE (490 + 16 AAs) is C-terminally truncated | O26:H11 strain 11368 (EHEC) | |
| yegE::ISEC13 (after nt 418, in codon 140) | DgcE (140 + 18 AAs) is C-terminally truncated | O127:H6 strain E2348/69 (EPEC) | |
| In-frame deletion of 12 nt after nt 117 | DgcE is shortened by 4 AAs | B strain REL606 (C; lab strain) | |
| dgcF (yneF) | 1-nt (A) deletion (after nt 348, in codon 117)/frameshift | DgcF (117 + 5 AAs) is C-terminally truncated | H10407 (ETEC) |
| 1-nt (T) deletion (after nt 95, in codon 32) | DgcF (32 + 17 AAs) is C-terminally truncated | O145:H28 strain RM13516 (STEC), O145:H28 strain RM12761 (STEC) | |
| 1-nt (C) deletion (after nt 392, in codon 31) | DgcF (31 + 18 AAs) is C-terminally truncated | O78 (APEC) | |
| 5′ deletion including the first 433 nt | DgcF is not expressed | ATCC 8739 (C; lab strain), BW2952 (C; K-12 derivative; lab strain), DH1 (C; K-12 derivative; lab strain), HS (C) (O9), K-12 substrain DH10B (C; lab strain) K-12 substrain MDS42 (C; lab strain), K-12 substrain MG1655 (C; lab strain), K-12 substrain W3110 (C; lab strain) UMNF18 (porcine ETEC) | |
| dgcI (yliF) | |||
| dgcJ (yeaJ) | 5′ deletion including the first 1,425 nt | DgcJ is not expressed | K-12 substrain MDS42 (C) |
| dgcM (ydaM) | 88-nt deletion (after the nt 561, in codon 187)/frameshift | DgcM (187 + 41 AAs) is C-terminally truncated | O127:H6 strain E2348/69 (EPEC) |
| Whole gene deletion | dgcM is absent | K-12 substrain MDS42 (C; lab strain) | |
| dgcN (yfiN) | 1-nt (G) deletion (after nt 1114, in codon 371)/frameshift | DgcN (371 + 2 AAs) is C-terminally truncated | O55:H7 strain RM12579 (EPEC) |
| dgcO (dosC, yddV) | Whole gene deletion | dgcO is absent | O103:H2 strain 12009 (EHEC) |
| Deletion (−38, +644 and +689, +1219) | DgcO is not expressed | JJ1886 (subclone of ST131), O127:H6 strain E2348/69 (EPEC), IHE3034 (ExPEC, MNEC), PMV-1 (ExPEC), O45:K1:H7 strain S88 (ExPEC), O6:K15:H31 strain 536 (UPEC), CFT073 (UPEC), UTI89 (UPEC), UM146 (AIEC), LF82 (AIEC), O83:H1 strain NRG 857C (AIEC), O1:K1:H7 strain O1 (APEC), ABU 83972 (C), ED1a (C) (O81), Nissle 1917 (C), SE15 (C) (O150:H5) | |
| dgcP (yeaP) | GAA→TAA (stop) (codon 283) | DgcP is shortened by 59 AAs at the C terminus | O157:H7 strain EDL933 (EHEC), O157:H7 strain Sakai (EHEC), O157:H7 strain Xuzhou21 (EHEC) |
| dgcQ (yedQ) | CAG→TAG (stop) (codon 312); codon 314 is ATG | DgcQ is expressed in 2 fragments; the C-terminal GGDEF-containing fragment is inactive | 55989 (EAEC), O104:H4 strain 01-09591 (EAEC), O104:H4 strain LB226692 (EAEC), O104:H4 strain 2009EL-2050 (EAEC), O104:H4 strain 2009EL-2071 (EAEC), O104:H4 strain 2011C-3493 (EAEC) |
| 4-nt (TATC) insertion (after nt 155, in codon 53)/frameshift | DgcQ (53 + 5 AAs) is C-terminally truncated | UMNF18 (porcine ETEC) | |
| 1-nt (A) deletion (after nt 1282, in codon 427)/frameshift | DgcQ (427 + 3 AAs) is C-terminally truncated) | O127:H6 strain E2348/69 (EPEC) | |
| Whole gene deletion | dgcQ is absent | B strain REL606 (C; lab strain), BL21(DE3) (C; lab strain) | |
| TGG/TAG (stop) (codon 114) | DgcQ is shortened by 450 AAs | HS (C) (O9) | |
| Deletion of gene after nt 1233 | DgcQ (411 + 16 AAs) is C-terminally truncated | Nissle 1917 (C) | |
| dgcT (ycdT) | Whole gene deletion | DgcT is absent | 042 (EAEC), UMNK88 (porcine ETEC), O111:H− strain 11128 (EHEC), O26:H11 strain 11368 (EHEC), O145:H28 strain RM13516 (STEC), O145:H28 strain RM13514 (STEC), O145:H28 strain RM12761 (STEC), O145:H28 strain RM12581 (STEC), O127:H6 strain E2348/69 (EPEC), UMN026 (ExPEC) (O7:K1), SMS-3-5 (environmental isolate of unknown pathotype; AR) ED1a (C) (O81), IAI1 (C) (O8), K-12 substrain MDS42 (C; lab strain) |
| 7-nt (TTTGTTT) insertion (after nt 461, in codon 153) | DgcT (152 + 19 AAs) is C- terminally truncated | 55989 (EAEC) | |
| GAG→TAG (stop) (codon 450) | DgcT is shortened by 3 AAs at its C terminus | ETEC H10407 (ETEC) | |
| 1-nt (C) deletion (after nt 483, in codon 162)→frameshift | DgcT is C-terminally truncated (162 + 0 AAs) | O55:H7 strain CB9615 (EPEC), O55:H7 strain RM12579 (EPEC) | |
| ycdT::IS2 (after nt 612, in codon 205) | DgcT (204 + 15 AAs) is C-terminally truncated | K-12 substrain DH10B (C; lab strain) | |
| dgcZ (ydeH) | 5′ deletion including the first 119 nt | DgcZ is not expressed | H10407 (ETEC) |
| GAA→TAA (stop) (codon 259) | DgcZ is shortened by 38 AAs at the C terminus | UMNK88 (porcine ETEC) | |
| ydeH::IS1 (after nt 611, in codon 204) | DgcZ (204 + 6 AAs) is C-terminally truncated | O26:H11 strain 11368 (EHEC) | |
| In-frame deletion of 12 nt in codon 312 | DgcZ is shortened by 4 AAs (probably still active) | O7:K1 strain CE10 (ExPEC, NMEC) | |
| 70-nt deletion (after nt 354, after codon 118)/frameshift | DgcZ (118 + 15 AAs) is C-terminally truncated | ABU 83972 (C) | |
| dgcX | |||
| dgcY | |||
| Genes encoding PDEs | |||
| pdeA (yfeA) | yfeA::IS600 (after nt 405, in codon 135) | PdeA (132 + 0AAs) is truncated | ED1a (C) (O81) |
| Whole gene deletion | pdeA is absent | K-12 substrain MDS42 (C) | |
| pdeB (ylaB) | |||
| pdeC (yjcC) | CAG→TAG (stop) (codon 414) | PdeC is shortened by 114 AAs | H10407 (ETEC) |
| CAA→TAA (stop) (codon 518) | PdeC is shortened by 11 AAs | O127:H6 strain E2348/69 (EPEC), CFT073 (UPEC), ABU 83972 (C), Nissle 1917 (C) | |
| CAG→TAG (stop) (codon 312) | PdeC is shortened by 216 AAs | E. coli B strain REL606 (C; lab strain) | |
| pdeD (yoaD) | 1-nt (C) deletion (after nt 354, in codon 118)/frameshift | PdeD (118 + 17 AAs) is truncated | E24377A (ETEC) |
| pdeF (yfgF) | yfgF::IS1 (after nt 1787, in codon 596) | PdeF (596 + 10 AAs) is C-terminally truncated | UMNK88 (porcine ETEC) |
| TAC→TAA (stop) (codon 445) | PdeF is shortened by 303 AAs | O7:K1 strain CE10 (ExPEC, NMEC), O7:K1 strain IAI39 (ExPEC) | |
| 1-nt (G) insertion (after nt 2121, in codon 708)/frameshift | PdeF (708 + 20 AAs) is C-terminally truncated | O1:K1:H7 strain O1 (APEC) | |
| TAC→TAG (stop) (codon 616) | PdeF is shortened by 541 AAs | HS (C) (O9) | |
| pdeG (ycgG) | 1-nt (G) deletion (after nt 890, in codon 297)/frameshift | PdeG is (297 + 10 AAs) is C-terminally truncated | UMNK88 (porcine ETEC) |
| Whole gene deletion | pdeG is absent | O157:H7 strain EC4115 (EHEC), O157:H7 strain EDL933 (EHEC), O157:H7 strain Sakai (EHEC), O157:H7 strain TW14359 (EHEC), O157:H7 strain Xuzhou21 (EHEC), UTI89 (UPEC), UM146 (AIEC), K-12 substrain MDS42 (C; lab strain) | |
| 5′ deletion including the first 630 nt | PdeG may be expressed without the N terminus | JJ1886 (subclone of ST131), IHE3034 (ExPEC, MNEC), PMV-1 (ExPEC), O45:K1:H7 strain S88 (ExPEC), O6:K15:H31 strain 536 (UPEC), CFT073 (UPEC), LF82 (AIEC), O83:H1 strain NRG 857C (AIEC), O1:K1:H7 strain O1 (APEC), ABU 83972 (C), ED1a (C) (O81), Nissle 1917 (C), SE15 (C) (O150:H5) | |
| 5′ deletion including the first 630 nt; 1-nt (A) deletion (after nt 1506, in codon 502) | PdeG may be expressed without the N terminus; frameshift causes protein extension by 3 AAs | O127:H6 strain E2348/69 (EPEC) | |
| TGG→TAG (stop) (codon 27) | PdeG is shortened by 481 AAs | O7:K1 strain UMN026 (ExPEC) (AR) | |
| pdeH (yhjH) | |||
| pdeI (yliE) | In-frame insertion of 9 nt after nt 404 | PdeI is extended by 3 AAs | O111:H- strain 11128 (EHEC) |
| GAG→TAG (stop) (codon 562) | PdeI is shortened by 221 AAs | O83:H1 strain NRG 857C (AIEC) | |
| CAG→TAG (stop) (codon 51) | PdeI is shortened by 732 AAs | B strain REL606 (C; lab strain), BL21(DE3) (C; lab strain), BL21-Gold(DE3)pLysS AG (C) | |
| TTA→TGA (stop) (codon 446) | YliE is shortened by 337 AAs | SE15 (C) (O150:H5) | |
| pdeK (yhjK) | |||
| pdeL (yahA) | Upstream insertion of a gene encoding an AidA-I adhesin-like protein | 042 (EAEC), 55989 (EAEC), O104:H4 strain 01-09591 (EAEC), O104:H4 strain LB226692 (EAEC), O104:H4 strain 2009EL-2050 (EAEC), O104:H4 strain 2009EL-2071 (EAEC), O104:H4 strain 2009EL-2071 (EAEC), O104:H4 strain 2009EL-2071 (EAEC), O104:H4 strain 2011C-3493 (EAEC), E24377A (ETEC), O103:H2 strain 12009 (EHEC), O111:H− strain 11128 (EHEC), O157:H7 strain EC4115 (EHEC), O157:H7 strain EDL933 (EHEC), O157:H7 strain Sakai (EHEC), O157:H7 strain TW14359 (EHEC), O157:H7 strain Xuzhou21 (EHEC), O26:H11 strain 11368 (EHEC), O145:H28 strain RM13516 (STEC), O145:H28 strain RM13516 (STEC), O145:H28 strain RM13514 (STEC), O145:H28 strain RM12761 (STEC), O145:H28 strain RM12581 (STEC), O55:H7 strain CB9615 (EPEC), O55:H7 strain RM12579 (EPEC), HS (C) (O9), IAI1 (C) (O8), KO11FL (C), W (C; lab strain) | |
| GAA→TAA (stop) (codon 296) | PdeL is shortened by 67 AAs | O78 (APEC) | |
| Whole gene deletion | PdeL is absent | BW2952 (C; K-12 derivative; lab strain) | |
| Whole gene deletion | PdeL is absent | ED1a (C) (O81) | |
| pdeN (rtn) | |||
| pdeO (dosP, yddU) | Whole gene deletion | pdeO is absent | O103:H2 strain 12009 (EHEC) |
| 10-nt (GGTGTATCTC) deletion (after nt 1214, in codon 405)/frameshift | PdeO (405 + 8 AAs) is truncated | O157:H7 strain EC4115 (EHEC), O157:H7 strain EDL933 (EHEC), O157:H7 strain Sakai (EHEC), O157:H7 strain TW14359 (EHEC), O157:H7 strain Xuzhou21 (EHEC) | |
| 1-nt (A) deletion (after nt 1662, in codon 555)/frameshift | PdeO (555 + 1 AAs) is truncated | O7:K1 strain CE10 (ExPEC, NMEC), O7:K1 strain IAI39 (ExPEC) | |
| 1-nt (T) insertion (after nt 1312, in codon 438)/frameshift; 1 nt (G) insertion (after nt 2311, in codon 771) | PdeO is (438 + 0 AAs) is truncated | CFT073 (UPEC) | |
| pdeR (yciR) | TGG→TGA (stop) (codon 432) | PdeR is shortened by 230 AAs | O111:H− strain 11128 (EHEC) |
| 5-nt (GCCCT) deletion (after nt 1573, in codon 524)/frameshift | PdeR (524 + 1 AAs) is C-terminally truncated | O157:H7 strain EC4115 (EHEC), O157:H7 strain TW14359 (EHEC) | |
| pdeT (vmpA) | 5′ deletion including the first 91 nt | PdeT is not expressed | O145:H28 strain RM13516 (STEC), O145:H28 strain RM13514 (STEC), O145:H28 strain RM12761 (STEC), O145:H28 strain RM12581 (STEC) |
| pdeW | |||
| pdeX | |||
| pdeY (sfaY) | |||
| Genes encoding degenerate GGDEF/EAL domain proteins | |||
| bluF (ycgF) | CAA→TAA (stop) (codon 271) | BluF is shortened by 132 AAs | H10407 (ETEC) |
| Whole gene deletion | bluF is absent | O157:H7 strain EC4115 (EHEC), O157:H7 strain EDL933 (EHEC), O157:H7 strain Sakai (EHEC), O157:H7 strain TW14359 (EHEC), O157:H7 strain Xuzhou21 (EHEC) | |
| 4-nt (TTCA) deletion (after nt 716, in codon 238)/frameshift | BluF (238 + 6 AAs) is C-terminally truncated | O55:H7 strain CB9615 (EPEC) | |
| ycgF::IS3411 (after nt 731, in codon 244) | BluF (244 + 15 AAs) is truncated | O7:K1 strain CE10 (ExPEC, NMEC) | |
| Whole gene deletion | bluF is absent | K-12 substrain MDS42 (C; lab strain) | |
| cdgI (yeaI) | |||
| csrD (yhdA) | |||
| rflP (ydiV) | TGG→TAG (stop) (codon 83) | RflP is shortened by 155 AAs | H10407 (ETEC) |
| 1-nt (T) deletion (after nt 218, in codon 73)/frameshift | RflP (73 + 12 AAs) is truncated | B strain REL606 (C; lab strain), BL21(DE3) (C; lab strain), BL21-Gold(DE3)pLysS AG (C) |
nt, nucleotide.
AAs, amino acids.
C, commensal; AR, antibiotic resistant.
DGCs of E. coli: novel genes and variations in previously known dgc genes. (i) DgcX.
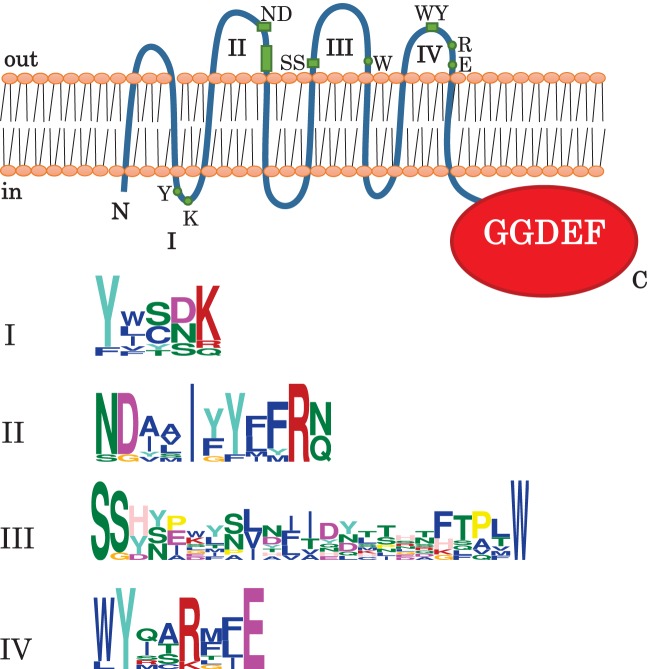
As we have previously described in a study of c-di-GMP signaling in the 2011 outbreak O104:H4 and related strains (30), DgcX is the most highly expressed DGC described thus far in E. coli. It contains a GGDEF domain with intact A- and I-sites linked to an N-terminal domain of unknown function, which is predicted to fold into eight transmembrane helices. A similar putative sensory domain termed MASE4 (membrane-associated sensor) (13) is also present in two other GGDEF domain proteins in E. coli, DgcT (YcdT) and CdgI (YeaI). MASE4-GGDEF proteins do not seem to be widespread (for instance Salmonella does not have any), but we found two and four similar proteins that can be predicted to be active DGCs in Klebsiella pneumoniae and Enterobacter lignolyticus, respectively. An alignment of these with DgcX, DgcT, and CdgI of E. coli (see Fig. S1 in the supplemental material) revealed patches of conserved amino acids in three of the four periplasmic loops, which are rich in aromatic amino acids (Fig. 2). This may represent a binding site for a ligand that itself has a ring structure, e.g., some aromatic compound, a nucleotide or a sugar. Our observation that, in contrast to DgcE and DgcC, DgcX (when cloned on a plasmid without any epitope tagging) was unable to suppress a mutation in dgcE (which results in low curli production), suggests that an unknown signaling molecule may have to activate DgcX via its MASE4 domain (T. Povolotsky and R. Hengge, unpublished data). Among the 61 E. coli strains under study here, the dgcX gene was found in nine strains, with six strains belonging to EAEC of the O104:H4 serotype (Table 1). Its location right next to lambdoid prophages in all of these strains suggests its horizontal spreading by specialized transduction (for further details on DgcX, see reference 30).
FIG 2.
Conserved amino acid motifs in MASE4, a novel sensory input domain present in the DGCs DgcX and DgcT (YcdT) and the degenerate GGDEF domain protein CdgI (YeaI). The most likely transmembrane topology of the eight hydrophobic amino acid stretches of the MASE4 domain and the GGDEF domain are shown schematically. Three highly conserved amino acid motifs in three periplasmic loops were identified by aligning DgcX, DgcT, CdgI, and two and four similar MASE4-GGDEF proteins of K. pneumoniae and E. lignolyticus, respectively (see Fig. S1 in the supplemental material). Their positions in the periplasmic loops and the respective sequence logos are indicated.
(ii) DgcY.
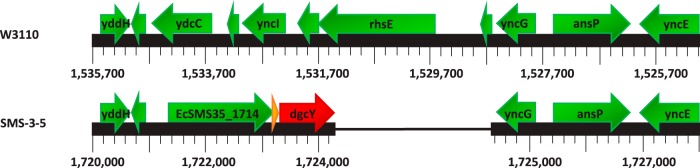
A novel E. coli DGC gene identified here is dgcY, which occurs in two strains only (Table 1). These are E. coli SMS-3-5 (an environmental pathogenic isolate with multiple antibiotic resistances [43]) and the neonatal meningitis E. coli (NMEC) O7:K1 strain CE10 (44). Its gene product is 349 amino acids long and is predicted to be an active DGC, now termed DgcY, since it features a C-terminal GGDEF domain with an A-site (but no inhibitory I-site), which is most closely related to the GGDEF domain of DgcZ (YdeH). Its N-terminal putative sensory domain (termed MASE5 [13]), which is predicted to fold into six transmembrane helices, is unique within E. coli and of unknown function. In both strains, the dgcY gene is preceded by a gene encoding a putative metallo-β-lactamase (EcSMS35_1714) and a small open reading frame (EcSMS35_1715), with the three genes apparently constituting a unique operon not found in any of the other E. coli strains analyzed here (Fig. 3).
FIG 3.
Genomic location of the dgcY gene. The genomic layout of corresponding regions of E. coli K-12 strain W3110 and E. coli SMS-3-5, an antibiotic-resistant environmental isolate, is shown. The location of dgcY (EcSMS35_1716) is indicated by a red arrow. This layout is also representative for the E. coli O7:K1 strain CE10 (dgcY is annotated as CE10_1648).
(iii) DgcE (YegE).
With a length of 1,105 amino acids and its six domains DgcE is by far the largest of all GGDEF/EAL domain proteins of E. coli. It consists of a membrane-inserted MASE1 domain (with eight transmembrane helices), followed by two additional transmembrane segments, three PAS domains, an active GGDEF domain, and a degenerate EAL domain (45) (see Table S2 in the supplemental material). Probably by integrating various signals, YegE-mediated c-di-GMP synthesis plays a key role in initiating the expression of the biofilm regulator CsgD during entry into stationary phase and therefore the production of amyloid curli fibers and cellulose as biofilm matrix components (15, 20). The dgcE gene was found to be corrupted in nine E. coli strains, with all EHEC of the O157:H7 serotype sharing the same disruption (a one-nucleotide insertion after nucleotide 457, which should result in the production of a short N-terminal fragment of DgcE only). Additional mutations were found in another EHEC (O26:H11), as well as in an EPEC (O127:H6) strain, but also in the EAEC strain 042 (Table 2). Thus, many EHEC/EPEC strains have lost DgcE, a key DGC for the synthesis of curli fibers and cellulose, suggesting that the production of a biofilm matrix may be counterproductive for an important and specific activity of EHEC/EPEC strains, possibly their specialized adherence mechanism (see also below).
(iv) DgcF (YneF).
DgcF, a DGC of 472 amino acids, consists of a MASE1 domain connected to a GGDEF domain via two additional transmembrane segments and a HAMP linker domain. No function has been described for DgcF thus far. In E. coli K-12 strains, a deletion that includes the promoter region as well as the first 433 nucleotides of the dgcF coding sequence was originally overlooked, which led to misannotation of an internal codon as a start codon and prediction of a DGC with 315 amino acids only. However, a comparison to other E. coli strains revealed the corruption of dgcF in E. coli K-12, which in fact results in the absence of DgcF (30). Notably, an ETEC strain of porcine origin carries the same large deletion mutation (Table 2). In addition, five other E. coli strains studied here, including two STEC strains, show various one-nucleotide deletion/frameshift mutations in dgcF that should result in the absence of DgcF.
(v) DgcO (DosC, YddV).
DgcO and its cognate PdeO (DosP, YddU), a DGC/PDE pair expressed from an operon, have been found in a complex with RNase E, enolase, and polynucleotide phosphorylase (PNPase), with the latter responding to c-di-GMP, which suggests a regulatory role of DgcO and PdeO in RNA turnover in a specialized degradosome (46). However, no target RNAs and therefore no clear physiological role of this system have been identified thus far. The entire operon was deleted in a specific EHEC strain of the O103:H2 serotype (Table 2). 16 strains display a complex disruption of dgcO consisting of two deletions (the first ranging from nucleotides –38 to +644, followed by 44 nucleotides of the original dgcO sequence and then by a second deletion of 530 nucleotides). Notably, many ExPEC strains, both UPEC and MNEC, carry this corrupted allele and therefore do not possess DgcO. Moreover, the probiotic strain Nissle 1917 also contains this allele.
(vi) DgcQ (YedQ).
DgcQ, a DGC of 564 amino acids, consists of a large periplasmic sensor domain (termed CHASE7 [13]) flanked by two transmembrane helices and followed by the GGDEF domain. DgcQ plays a minor role in reducing flagellar rotation (15), and it has been reported to contribute to cellulose biosynthesis in a particular E. coli strain (E. coli 1094, for which no genome sequence is available) (47). In a variety of E. coli strains, the dgcQ gene is corrupted by various small frameshift and stop codon mutations and, in one case, a complete deletion (Table 2). Notably, in a series of EAEC, all of the O104:H4 serotype, codon 312 is changed to a stop codon, which, however, is almost immediately followed by an ATG (codon 314). Thus, translational coupling could result in a restart, such that DgcQ would be made in two parts, with the second fragment consisting of the second transmembrane helix and the GGDEF domain. This N-terminally truncated DgcQ protein has indeed been observed when this dgcQ allele was cloned onto a plasmid with a C-terminal His6 tag, but this construct did not complement low curli production of a dgcE mutant, suggesting that an intact CHASE7 sensory domain is required for DcgQ activity (Povolotsky and Hengge, unpublished).
(vii) DgcT (YedT).
DgcT (YcdT) is a DGC that has been implicated in the production of poly-GlcNAc (PGA), which is expressed within the host and serves as biofilm matrix component and/or virulence factor in some pathogenic E. coli (48–50). We found that the dgcT gene was entirely deleted in 14 strains and disrupted by small frameshift or stop codon mutations in five additional strains (Table 2). Among the strains with dgcT deletions, there was a high incidence of EHEC/STEC strains, as well as one EPEC strain. In addition, five EHEC and two EPEC strains have an extra PDE gene (pdeT, see below) inserted right after dgcT in an obvious operon, suggesting that this PDE may counteract DgcT activity. This could be an indication that, similar to DgcE-dependent production of a curli and cellulose biofilm matrix (see above), DgcT-driven production of the alternative matrix component PGA may also be detrimental for the specialized adherence mechanism of EHEC/EPEC. Furthermore, two EAEC strains show either a full deletion or an early frameshift mutation in dgcT (Table 2), but in one of these, the EAEC strain 55989, the role of DgcT may be taken over by the strongly expressed DgcX, since these two DGCs show the same type of sensory input domain (see above and reference 30).
c-di-GMP-specific PDEs of E. coli: novel genes and variations in previously known genes.
Our analyses showed the presence, in one or more E. coli strains, of four PDE genes (pdeT, pdeW, pdeX, and pdeY) not found in E. coli K-12 strains, as well as numerous alterations in PDE genes already known from E. coli K-12. However, within four PDE genes—pdeB (ylaB), pdeH (yhjH), pdeK (yhjK), and pdeN (rtn)—not a single mutation was detected in all 61 E. coli strains studied here, even though these genes can be knocked out experimentally (34). This suggests that these PDEs play an important role under some conditions that all E. coli strains experience during their life cycles. For instance, PdeH is crucial for maintaining a low c-di-GMP level in post-exponentially growing flagellum-expressing cells—in pdeH mutants, flagellar rotation is inhibited by the c-di-GMP-binding effector YcgR (see below), which renders these mutants nonmotile despite their expression of flagella (14–16). For the other three strictly conserved PDEs, however, no physiological functions have been reported.
(i) PdeT (VmpA).
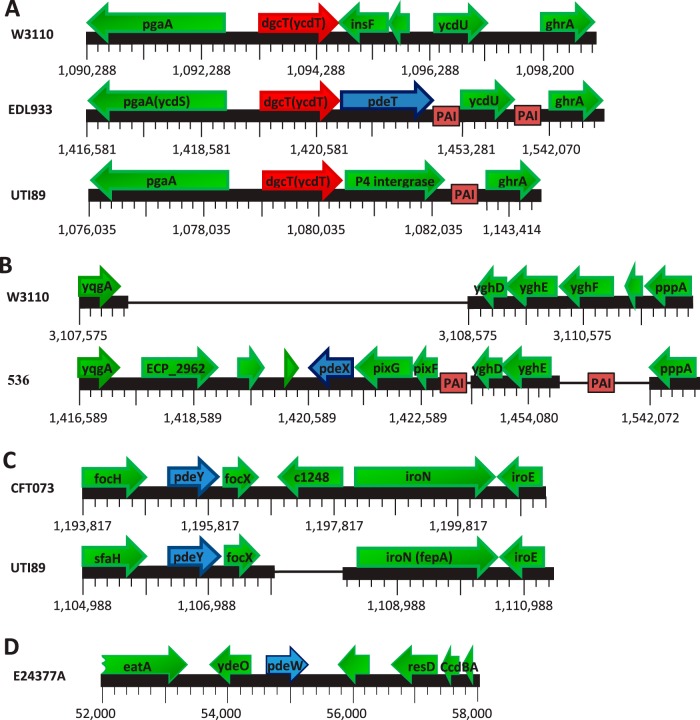
The pdeT gene is inserted downstream of dgcT (ycdT) in five EHEC and two EPEC strains (Table 1, Fig. 4). PdeT features a membrane-integrated periplasmic loop domain, a CSS domain (13), followed by an EAL domain. This functionally uncharacterized CSS domain is also found in a subset of five other PDEs present in all or most other E. coli strains, i.e., PdeB (YlaB), PdeC (YjcC), PdeD (YoaD), PdeG (YcgG), and PdeN (Rtn) (13). PdeT was first described in the classical EHEC strain EDL933, where it was shown to constitute an operon with dgcT and to encode an active PDE (28). Thus, PdeT most likely acts as an antagonist for DgcT, which is believed to be involved in the control of the matrix polymer PGA (48, 49), with the pga operon being located right next to and divergently oriented from dgcT. In functional terms, the insertion of pdeT may thus be equivalent to the corruption of dgcT found in some other EHEC and EPEC strains (see above). In fact, a subset of EHEC strains (all of the O145:H8 serotype) show a 5′-end-truncated pdeT gene (Table 2) but no dgcT, suggesting that this lineage originally possessed a dgcT-pdeT operon but then acquired a large deletion that removed dgcT and the first 91 nucleotides of pdeT. This does not necessarily mean that no PdeT activity is present, since in a similar case of a 5′-truncated gene encoding another CSS-PDE (PdeG), gene product activity was observed (see below).
FIG 4.
Genomic locations of the four PDE genes not found in E. coli K-12, pdeT (vmpA), pdeX, pdeY, and pdeW. (A) An extra PDE gene, pdeT, occurs directly downstream of dgcT in all EHEC O157:H7 strains (which include strain EDL933). In two EPEC strains of the O55:H7 serotype, the overall dgcT-pdeT layout is similar, but ycdT has a one-nucleotide deletion/frameshift mutation that should lead to premature termination of translation, which may be polar on the expression of pdeT (Table 2). In UTI89 and other UPEC strains, dgcT is followed by an integrase gene (termed UTI89_C1089). (B) A large insertion that includes the novel PDE gene pdeX is shown that is present in the UPEC strain 536. (C) Genomic layout of a region containing pdeY in the UPEC strains CFT073 (which is also representative for the commensal E. coli strain ABU83972) and UTI89 (which is also representative for the UPEC strains 536 and UMN146 and the MNEC strain IHE3034). (D) The pdeW gene, which occurs only in the ETEC strain E24377A (annotated as ecE24377A_E0054), is located on an extrachromosomal element, plasmid 2. PAI, insertions of various pathogenicity islands.
(ii) Three novel stand-alone EAL domain PDEs, PdeW, PdeX, and PdeY (SfaY).
The novel genes pdeW and pdeX, which encode PDEs consisting of an EAL domain only, were each detected in only a single E. coli strain (Fig. 4). pdeW (annotated as ecE24377A_E0054) is located on the uncharacterized plasmid 2 of the ETEC O139:H28 strain E24377A. Together with a few other novel genes involved in synthesis of Pix fimbriae, pdeX (annotated as ECP_2965) is inserted in the genome of the classical UPEC strain 536. A third stand-alone EAL domain PDE gene, pdeY, was previously found in the meningitis-associated E. coli strain IHE3034, where it is associated with the sfaX(II) locus involved in the synthesis of S fimbriae and was initially termed sfaY (51). In addition, we find pdeY also in five other E. coli strains, including three widely studied UPEC strains (536, CFT073, and UTI89). Notably, UPEC strain 536 thus has even two of these additional small PDEs, i.e., PdeX and PdeY. The operon layouts in the six pdeY-containing strains are essentially the same, with the exception of the UPEC strain CFT073 and the commensal E. coli ABU 93972 that show an extra gene inserted in this region (c1248 in CFT073) (Fig. 4C).
(iii) PdeG (YcgG).
The 507-amino-acid PdeG belongs to the group of PDEs that combine a membrane-inserted periplasmic loop CSS domain at the N terminus with a C-terminal EAL domain. A number of pathogenic E. coli of various pathotypes are devoid of PdeG due to larger deletions (that include neighboring genes as well), a frameshift-generating one-nucleotide deletion or an early stop codon in pdeG (Table 2). Functional consequences are unclear since knocking out the intact pdeG in a K-12 strain does not produce any phenotype under standard laboratory conditions (34) although PdeG is expressed (G. Klauck and R. Hengge, unpublished data). An interesting allele, in which a larger deletion removes the first 630 nucleotides of pdeG, is found in 14 members of a series of ExPEC, two AIEC, an APEC and several commensal E. coli strains (Table 2). At first glance, such a deletion seems likely to eliminate the expression of the gene. However, the experimental deletion of this shortened allele (c1610), which had not been recognized as a 5-truncated version of an originally longer gene, in the UPEC strain CFT073 resulted in increased biofilm formation (27). This indicates that (i) an N-terminally truncated version of PdeG (denoted as PdeG* in Fig. 1) is in fact expressed from this 5′-incomplete allele, probably from an internal secondary start codon, and (ii) this PdeG* variant, which has an intact EAL domain but no CSS domain, shows PDE activity. Since wild-type PdeG is expressed but inactive under comparable conditions, this suggests an inhibitory role of the CSS domain in the control of PDE activity.
(iv) PdeL (YahA).
The pdeL gene, which encodes a PDE consisting of a putative DNA-binding LuxR-like domain followed by a canonical EAL domain (52), is entirely deleted in two laboratory strains (BW2952 and ED1a) and corrupted by an internal stop codon in an APEC strain (Table 2). In contrast, almost half of the remaining 58 strains with intact pdeL show a large insertion upstream of pdeL (i.e., between betT and pdeL) which contains the gene for an AidA-I-like adhesin (Table 2) (30). The region between this adhesin gene and pdeL does not contain any apparent terminator motifs, suggesting that the two genes constitute an operon. The regulatory regions present upstream of this operon or upstream of pdeL (in the strains that do not contain the AidA-I-like adhesin gene) share important regulatory motifs, e.g., a Cra binding site and the putative promoter (53), with some divergence in between these motifs. The physiological role of the AidA-I-like adhesin and its apparent coregulation with PdeL is not yet clear, mainly because it is widespread among different pathogenic E. coli but also occurs in some commensal strains. Although K-12 strains do not have it, all EHEC strains studied here, as well as all EAEC strains of the O104:H4 serotype (including the 2011 German outbreak strain LB226692), possess this Aid-I-like adhesin gene linked to pdeL.
(v) PdeO (DosP, YddU).
PdeO is a 799-amino-acid protein with two PAS domains and a GAF domain, followed by a degenerate GGDEF and an EAL domain, whose PDE activity is controlled by oxygen via a PAS domain-associated heme (54, 55). It acts as an antagonist to DgcO, with both proteins being part of a specialized degradosome that also contains the c-di-GMP-regulated PNPase (46, 56). Due to a whole dgcO-pdeO operon deletion (in the EHEC O103:H2 strain 12009) or small insertions or deletions that generate frameshifts in pdeO (Table 2), PdeO is absent in several EHEC and ExPEC strains. Since the RNA substrates of this specialized DgcO/PdeO-containing degradosome are unknown, the functional consequences of this absence of PdeO in certain EHEC and ExPEC strains are unclear.
(vi) PdeR (YciR).
PdeR is a 661-amino-acid composite of a PAS domain, a GGDEF domain with hardly detectable activity, and an active EAL domain. It acts as a c-di-GMP-sensing and inhibitory component of the molecular switch mechanism that activates the expression of the biofilm regulator and matrix production activator CsgD in response to rising intracellular c-di-GMP (20). It is thus not surprising that it is highly conserved (Fig. 1). However, there are a few noteworthy exceptions (Table 2). In two EHEC strains of the O157:H7 serotype, a five-nucleotide insertion (in codon 524) produces a frameshift in pdeR and thus should result in an absence of a functional PdeR protein. In the EHEC O111:H− strain 11128 a sense-to-stop codon mutation (in codon 445) produces a similar effect. The consequence of knocking out pdeR is a hyperactivation of CsgD expression and therefore very high production of curli fibers and cellulose. This very high CsgD expression is no longer c-di-GMP regulated but still depends on RpoS-containing RNAP and the transcription factor MlrA (20). In classical O157:H7 EHEC strains, however, a lambdoid stx-carrying phage is inserted within mlrA (57); these strains therefore do not produce CsgD (although derivatives with csgD promoter mutations exist, in which CsgD is expressed again [58]), and the frameshift mutation in pdeR mentioned above should not have any consequences. However, in the equally Stx-producing O111:H− serotype 11128 strain, mlrA is fully intact. Moreover, a distinct aggregative behavior was recently reported for O111 strains that was positively correlated with the production of curli fibers and RpoS function (59). Our finding that the O111:H− strain 11128 is a PdeR-deficient mutant, but wild-type with respect to mlrA, suggests that this strain overproduces CsgD and curli fibers in comparison to most other E. coli strains (i.e., pdeR+ mlrA+ strains). In that respect, it resembles the Stx-producing 2011 outbreak O104:H4 strain, which also combines very high CsgD and curli expression with the production of Stx (30). Curli fibers are highly inflammatory (60, 61) and, if expressed at 37°C, may therefore contribute to systemic absorption of Shiga toxin (30, 62). Notably, curli fibers and cellulose also serve for attachment to surfaces of plants that are of importance for human nutrition and have repeatedly been implicated in EHEC transmission (63–66).
Variations in genes encoding degenerate GGDEF/EAL domain proteins.
Among the four genes for degenerate GGDEF/EAL domain proteins, bluF (ycgF) and rflP (ydiV) show interesting variations described in detail below. The other two genes are absolutely conserved in the 61 E. coli strains studied here, although these genes can be knocked out under laboratory conditions: (i) csrD (yhdA), which encodes a protein involved in the turnover of the regulatory RNAs CsrB and CsrA (23, 67), and (ii) cdgI (yeaI), which shows hardly any expression (34) and encodes an GGDEF domain protein with a degenerate A-site. Purified CdgI is enzymatically inactive but can bind c-di-GMP via its intact I-site (F. Skopp and R. Hengge, unpublished results) and therefore most likely represents a c-di-GMP-binding effector protein acting in an unknown physiological context.
(i) BluF (YcgF).
BluF consists of a blue light-responsive BLUF domain (68), followed by a degenerate EAL domain that neither degrades nor binds c-di-GMP (24). It acts as a blue-light activated direct antagonist to the repressor protein BluR (YcgE) and thereby can induce several small proteins involved in the control of activity of the Rcs phosphorelay system, which, via the sRNA RprA, can downregulate the expression of CsgD (24, 35, 69). Five EHEC strains of the O157:H7 serotype, as well as a commensal and laboratory strain (MDS42), feature a larger deletion that not only eliminates bluF completely but that also extends to pdeG (ycgG) as well (see above). In three additional strains of diverse patho- and serotypes, the bluF gene is affected by a premature stop codon, a frameshifting four-nucleotide deletion, and an IS element insertion (Table 2). Why these bluF mutant strains have lost the environmental modulation (by light) of the globally regulating Rcs/RprA system is currently not apparent.
(ii) RflP (YdiV).
This highly degenerate stand-alone EAL domain protein acts as an inhibitor and proteolytic targeting factor for the flagellar master regulator FlhDC (13, 25, 26). In E. coli K-12, rflP shows very low expression only under standard lab condition (34). However, in other strains or under some unknown conditions, RflP expression might be higher. Then mutants deficient for RflP would have higher levels of FlhDC and increased expression of genes of the flagellar control cascade. This would not only affect flagellar components but would also result in higher expression of two regulatory factors that downregulate CsgD and the biofilm matrix components curli and cellulose: (i) PdeH (YhjH, see above), which keeps c-di-GMP levels low and thereby interferes with the expression of CsgD, and (ii) FliZ, a histone-like protein that downregulates many RpoS-dependent genes (including those involved in activating CsgD expression) (15, 70). It is noteworthy that several widely used laboratory strains (Table 2) carry a one-nucleotide deletion/frameshift mutation in rflP, which may represent a biofilm-reducing laboratory “domestication” that researchers inadvertently have selected for.
Variations in genes encoding c-di-GMP-binding effector proteins.
To date, only four effector proteins that respond to the cellular level of c-di-GMP and directly control the activity of distinct targets have been found in E. coli. These are the two PilZ domain proteins YcgR (71) and BcsA (72), the GIL domain protein BcsE (73), and the “trigger enzyme” and PDE PdeR already described above (20).
(i) YcgR.
By directly interacting with the flagellar basal body, c-di-GMP-bound YcgR slows down flagellar rotation (16, 74, 75). This can be observed during entry into stationary phase in liquid medium (15) and may also occur in macrocolony biofilms where flagella are produced and get entangled in the bottom layer of the colony (76, 77). Notably, several classical EHEC strains of the O157:H7 serotype either carry an IS element within ycgR or exhibit a deletion that eliminates a large 5′ part of ycgR (Table 3), suggesting that these EHEC strains do not shut down flagellar rotation under conditions of high internal c-di-GMP concentration.
TABLE 3.
Genomic variations of the genes encoding c-di-GMP-binding proteins in 61 E. coli strains
| Genea | Mutation(s)b | Consequence for protein expressionc | Strain(s) (pathotype[s]) |
|---|---|---|---|
| ycgR | 1,313-nt insertion of IS element (after nt 594, in codon 199) | YcgR (199 + 35 AAs) is C-terminally truncated | O157:H7 strain EDL933 (EHEC), O157:H7 strain Sakai (EHEC), Xuzhou21 (EHEC) (O157:H7) |
| 5′ deletion including the first 595 nt | YcgR is absent | O157:H7 strain EC4115 (EHEC), O157:H7 strain TW14359 (EHEC) | |
| bcsA | 19-noncontiguous-nt deletion (after nt 83, in codon 28)/frameshift | BcsA (27 + 56 AAs) is C-terminally truncated | IAI39 (ExPEC), CE10 (ExPEC, NMEC) |
| 1-nt (G) deletion (after nt 550, in codon 184)/frameshift | BcsA (184 + 1 AA) is C-terminally truncated | S88 (ExPEC) | |
| 1-nt (A) deletion (after nt 186, in codon 62)/frameshift; 1-nt (G) deletion (after nt 698, in codon 233); 1-nt (A) insertion (after nt 702, in codon 234) | BcsA (62 + 27 AAs) is C-terminally truncated | UMN026 (ExPEC) | |
| Whole gene deletion | BcsA is absent | HS (C) | |
| bcsE | TGT→TGA (stop) (codon 101) | BcsE (100 AAs) is C-terminally truncated | ATCC 8739 (C) |
| 1-nt (C) insertion (after nt 1340, in codon 447) | BcsE (447 + 15 AAs) is C-terminally truncated | O104:H4 strain LB226692 (EAEC), O104:H4 strain 2009EL-2050 (EAEC), O104:H4 strain 2009EL-2071 (EAEC), O104:H4 strain 2011C-3493 (EAEC) | |
| 5′ deletion including the first 779 nt | BcsE is absent | RM13516 (STEC), RM13514 (STEC), RM12761 (STEC), RM12581 (STEC) | |
| Whole gene deletion | BcsE is absent | HS (C) |
All of these genes encoded c-di-GMP-binding proteins.
nt, nucleotide.
AAs, amino acids.
(ii) BcsA.
BcsA is one of two subunits of the membrane-inserted cellulose synthase complex and consists of several domains, including a c-di-GMP-binding PilZ domain which allosterically controls the glucosyltransferase domain (78). Several ExPEC strains show small deletions that result in frameshifts and thus premature termination of BcsA expression. These mutations would not only eliminate BcsA but are also expected to be polar onto the downstream genes bcsB (encoding the other subunit of cellulose synthase), bcsZ and bcsC, i.e., to confer a complete cellulose-negative phenotype. It is conceivable that cellulose production is counterselected for in these strains because it could interfere with adhesion to host tissue via specific fimbriae made by these E. coli strains.
(iii) BcsE.
BcsE binds c-di-GMP via a motif (RxGD) that resembles the I-site of DGCs (73). In E. coli, BcsE, as well as BcsF and BcsG, which are all encoded within a single operon, is required for cellulose biosynthesis (30), whereas in Salmonella it is only required for maximal cellulose production (73), suggesting that BcsE plays a regulatory rather than a structural role in cellulose synthesis. The recently emerged Stx-producing O104:H4 outbreak strains show a C-terminal truncation of bcsE (Table 3) and are cellulose negative, which probably contributes to their virulence because “naked” curli fibers (not in a composite with cellulose) are highly inflammatory (30). In addition, several Stx-producing strains of the O145:H28 serotype show a large deletion that removes most of bcsE and should therefore also be cellulose negative.
Conclusions and perspectives.
In our study we detected a large number of highly diverse mutations (Table 2) in a total of 35 GGDEF/EAL domain-encoding genes in the genomes of 61 E. coli strains which represent the major pathotypes, as well as commensals. Overall, our detailed analysis revealed interesting trends in different types of pathogenic E. coli that seem to reflect different host niches and mechanisms of host cell adherence. Moreover, our findings provide a basis for detailed hypotheses that may guide future experimental analyses of c-di-GMP signaling in these diverse E. coli strains.
In many EHEC strains we observe a tendency to lose DGCs such as DcgE (YegE) and DcgT (YcdT), which are involved in the production of CsgD (and therefore curli and cellulose) and PGA, respectively. Moreover, those EHEC as well as some EPEC that possess an intact dgcT gene, often show an insertion of the extra PDE gene pdeT (vmpA) right downstream of dgcT in a common operon, suggesting that PdeT antagonizes DgcT activity. These strains can therefore be expected to produce low levels of biofilm matrix components, which is often further supported by the absence of MlrA, an activator of csgD transcription, due to insertion of the stx-carrying prophage into the mlrA gene (30, 57). Possibly, matrix production is counterselected because it may interfere with the specialized adherence mechanism of EHEC/EPEC which involves a type III secretion system that induces pedestal formation followed by adhesion via intimin (33). Furthermore, several classical EHEC O157:H7 strains also lack the c-di-GMP binding protein YcgR, suggesting that they continue to be motile under certain conditions of high c-di-GMP levels, which may be those promoting the expression of the type III secretion system effectors EspA and EspB, several types of pili, Tir, and intimin and therefore adhesion to intestinal cells (79). An interesting deviation from this general pattern in classical EHEC/STEC is found in the Stx-producing O111:H− strain 11128. Due to a mutation in pdeR (yciR) and an intact mlrA gene, this strain can be expected to even overproduce CsgD, curli, and cellulose. Indeed, it shows curli-dependent cellular aggregation (59). With respect to Stx and high curli production this strain thus resembles the O104:H4 strains rather than classical STEC (see below).
Also in ExPEC—in both UPEC and MNEC—a trend to reducing c-di-GMP is becoming apparent from our analysis. Many of these strains have lost DgcO (DosC), sometimes together with its cognate PdeO (DosP, encoded in a common operon), which is an oxygen-controlled system that affects an unknown target. Moreover, classical UPEC strains tend to possess additional stand-alone EAL domain PDEs (strain 536 even has two of these, PdeX and PdeY). For UPEC strains, low c-di-GMP levels may be crucial because they depend on motility, which is negatively c-di-GMP regulated, to establish a urinary tract infection. Moreover, they may benefit from downregulating the expression of curli fibers at least during acute infection, since curli can trigger a local immune defense (80).
Finally, EAEC of the O104:H4 serotype, which adhere to intestinal cells in biofilm-like patches with a characteristic stacked brick pattern (81), are characterized by an additional DGC, DgcX. In previous work, we have demonstrated the extremely high expression of the dgcX gene (30). c-di-GMP produced by DgcX may contribute to the very high curli fiber production of these strains (30), as well as to additional adhesion mechanisms. Besides acquiring DgcX, these EAEC strains have lost another DGC, DgcQ (YedQ). In strain 55989, DgcT (YcdT) is also corrupted, and strain 042 has lost both DgcT and DgcE (YegE). Taken together, this indicates a tendency to reduce the diversity of DGCs and to focus c-di-GMP production onto the extremely strongly expressed DgcX. In addition, the membrane-associated DgcX seems to need activation by an unknown molecule binding to a conserved motif on the periplasmic side of its transmembrane MASE4 sensory domain. This unknown ligand could be an intestinal metabolite that may guide EAEC to their optimal sites of adherence.
If high c-di-GMP accumulation by DgcX and strong production of highly inflammatory “naked” curli fibers (due to an absence of cellulose synthesis) occur in combination with Stx production, such as in the recently emerged Stx-producing O104:H4 variants (30), the result may be enhanced virulence, as was observed in the 2011 outbreak (42). It may therefore be useful to complement rapid PCR-based diagnostics as developed during the 2011 outbreak (81) by testing for the presence of dgcX and the status of the mlrA and bcs genes. Overall, our analysis thus indicates that STEC fall into two rather different classes: (i) classical EHEC of several serotypes with reduced c-di-GMP and biofilm matrix production and (ii) nonclassical STEC with high production of c-di-GMP and biofilm matrix, in particular of inflammatory curli fibers, such as the outbreak O104:H4 strain and the Stx-producing O111 strains.
In conclusion, variations in the complement of GGDEF/EAL domain proteins and c-di-GMP-binding effector proteins suggest an intricate interplay of biofilm properties and virulence in pathogenic E. coli. Moreover, these variations within a single bacterial species show that c-di-GMP-related genes and proteins evolve rapidly and thereby contribute to adaptation to host-specific and environmental niches.
Supplementary Material
ACKNOWLEDGMENTS
We thank our coworkers and colleagues Gisela Klauck and Franziska Mika for helpful discussions and Diego Serra for comments on the manuscript.
Financial support was provided by the European Research Council under the European Union's Seventh Framework Programme (ERC-AdG 249780 to R.H.). T.L.P. was supported by a graduate fellowship from the Deutsche Forschungsgemeinschaft (GRK 1673: Functional Molecular Infection Epidemiology).
Individual author contributions were as follows: concept of the study, R.H.; bioinformatic analyses, T.L.P.; interpretation of data, T.L.P. and R.H.; and writing of the paper, R.H. and T.L.P.
We declare that we do not have any conflicts of interest.
Funding Statement
Research reported here has been funded by the European Research Council under the European Union’s Seventh Framework Programme (ERC-AdG 249780 to R.H.). T.L.P. has been partially supported by a fellowship provided by the Graduate Programme GKR 1673 (Functional Molecular Infection Epidemiology) by the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00520-15.
REFERENCES
- 1.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylate. Nature 325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 2.Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 3.Römling U, Gomelsky M, Galperin MY. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 4.Ryan RP, Fouhy Y, Lucey F, Dow JM. 2006. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J Bacteriol 188:8327–8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hengge R. 2009. Principles of cyclic-di-GMP signaling. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 6.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic-di-GMP: the first 25 years of a universal bacterial second messenger. Microb Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signaling. Nat Rev Microbiol 7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 8.Hengge R. 2010. Cyclic-di-GMP reaches out into the bacterial RNA world. Sci Signal 3:pe44. doi: 10.1126/scisignal.3149pe44. [DOI] [PubMed] [Google Scholar]
- 9.Krasteva PV, Giglio KM, Sondermann H. 2012. Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci 21:929–948. doi: 10.1002/pro.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan RP, Tolker-Nielsen T, Dow JM. 2012. When the PilZ don't work: effectors for c-di-GMP action in bacteria. Trends Microbiol 20:235–242. doi: 10.1016/j.tim.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Hengge R. 2010. Role of c-di-GMP in the regulatory networks of Escherichia coli, p 230–252. In Wolfe AJ, Visick KL (ed), The second messenger cyclic-di-GMP. ASM Press, Washington, DC. [Google Scholar]
- 12.Povolotsky TL, Hengge R. 2011. ‘Life-style’ control networks in Escherichia coli: signaling by the second messenger cyclic-di-GMP. J Biotechnol 160:10–16. doi: 10.1016/j.jbiotec.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 13.Hengge R, Galperin MY, Ghigo J-M, Gomelsky M, Green J, Hughes KT, Jenal U, Landini P. 2016. Systematic nomenclature for GGDEF and EAL domain-containing cyclic di-GMP turnover proteins of Escherichia coli. J Bacteriol 198:7–11. doi: 10.1128/JB.00424-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girgis HS, Liu Y, Ryu WS, Tavazoie S. 2007. A comprehensive genetic characterization of bacterial motility. PLoS Genet 3:e154. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev 22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehm A, Kaiser M, Li H, Spangler C, CA K, Ackerman M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 17.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. 2006. Cyclic-di-GMP-mediated signaling within the σS network of Escherichia coli. Mol Microbiol 62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- 18.Römling U, Rohde M, Olsén A, Normark S, Reinköster J. 2000. AgfD, the checkpoint of multicellular and aggregative behavior in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol 36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- 19.Römling U. 2005. Characterization of the rdar morphotype, a multicellular behavior in Enterobacteriaceae. Cell Mol Life Sci 62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain phosphodiesterase YciR acts as a trigger enzyme in a c-di-GMP signaling cascade in Escherichia coli biofilm control. EMBO J 32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 22.Brombacher E, Baratto A, Dorel C, Landini P. 2006. Gene expression regulation by the curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J Bacteriol 188:2027–2037. doi: 10.1128/JB.188.6.2027-2037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki K, Babitzke P, Kushner SR, Romeo T. 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev 20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tschowri N, Busse S, Hengge R. 2009. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue light response of Escherichia coli. Genes Dev 23:522–534. doi: 10.1101/gad.499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. 2011. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J Bacteriol 193:1600–1611. doi: 10.1128/JB.01494-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takaya A, Erhardt M, Karata K, Winterberg K, Yamamoto T, Hughes KT. 2012. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol Microbiol 83:1268–1284. doi: 10.1111/j.1365-2958.2012.08007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spurbeck RR, Tarrien RJ, Mobley HL. 2012. Enzymatically active and inactive phosphodiesterases and diguanylate cyclases are involved in regulation of motility or sessility in Escherichia coli CFT073. mBio 3:e00307-12. doi: 10.1128/mBio.00307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Branchu P, Hindré T, Fang X, Thomas R, Gomelsky M, Claret L, Harel J, Gobert AP, Martin C. 2012. The c-di-GMP phosphodiesterase VmpA absent in Escherichia coli K12 strains affects motility and biofilm formation in the enterohemorrhagic O157:H7 serotype. Vet Immunol Immunopathol 152:132–140. doi: 10.1016/j.vetimm.2012.09.029. [DOI] [PubMed] [Google Scholar]
- 29.Hufnagel DA, DePas WH, Chapman M. 2014. The disulfide bonding system suppresses CsgD-independent cellulose production in Escherichia coli. J Bacteriol 196:3690–3699. doi: 10.1128/JB.02019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richter A, Povolotsky TL, Wieler LH, Hengge R. 2014. C-di-GMP signaling and biofilm-related properties of the Shiga toxin-producing German outbreak Escherichia coli O104:H4. EMBO Mol Med 6:1622–1637. doi: 10.15252/emmm.201404309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. 2011. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci U S A 108:7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaper JB, Nataro JP, Mobley HLT. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 34.Sommerfeldt N, Possling A, Becker G, Pesavento C, Tschowri N, Hengge R. 2009. Gene expression patterns and differential input into curli fimbria regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology 155:1318–1331. doi: 10.1099/mic.0.024257-0. [DOI] [PubMed] [Google Scholar]
- 35.Tschowri N, Lindenberg S, Hengge R. 2012. Molecular function and potential evolution of the biofilm-modulating blue light-signaling pathway of Escherichia coli. Mol Microbiol 85:893–906. doi: 10.1111/j.1365-2958.2012.08147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serra DO, Richter AM, Hengge R. 2013. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol 195:5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 39.Bailey TL, Elkan C. 1995. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3:21–29. [PubMed] [Google Scholar]
- 40.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 41.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karch H, Denamur E, Dobrindt U, Finlay BB, Hengge R, Johannes L, Ron EZ, Tønjum T, Sansonetti PJ, Vicente M. 2012. The enemy within us: lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO Mol Med 4:841–848. doi: 10.1002/emmm.201201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fricke FW, Wright MS, Lindell AH, Harkins DM, Baker-Austin C, Ravel J, Stepanauskas R. 2008. Insights into the environmental resistance gene pool from the genome sequence of the multidrug-resistant environmental isolate Escherichia coli SMS-3-5. J Bacteriol 190:6779–6794. doi: 10.1128/JB.00661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu S, Zhang X, Zhu Y, Kim KS, Yang J, Jin Q. 2011. Complete genome sequence of the neonatal-meningitis-associated Escherichia coli strain CE10. J Bacteriol 193:7005. doi: 10.1128/JB.06284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikolskaya AN, Mulkidjanian AY, Beech IB, Galperin MY. 2003. MASE1 and MASE2: two novel integral membrane sensory domains. J Mol Microbiol Biotechnol 5:11–16. doi: 10.1159/000068720. [DOI] [PubMed] [Google Scholar]
- 46.Tuckerman JR, Gonzalez G, Gilles-Gonzalez M-A. 2011. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J Mol Biol 407:633–639. doi: 10.1016/j.jmb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 47.Da Re S, Ghigo J-M. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J Bacteriol 188:3073–3087. doi: 10.1128/JB.188.8.3073-3087.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackerman M, Schirmer T, Jenal U. 2009. Second messenger signaling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol 72:1500–1516. doi: 10.1111/j.1365-2958.2009.06739.x. [DOI] [PubMed] [Google Scholar]
- 49.Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors O. 2008. The RNA binding protein CsrA controls c-di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol 70:236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al Safadi R, Abu-Ali GS, Sloup RE, Rudrik JT, Waters CM, Eaton KA, Manning SD. 2012. Correlation between in vivo biofilm formation and virulence gene expression in Escherichia coli O104:H4. PLoS One 7:e41628. doi: 10.1371/journal.pone.0041628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sjöström AE, Sondén B, Müller C, Rydström A, Dobrindt U, Wai SN, Uhlin BE. 2009. Analysis of the sfaX(II) locus in the Escherichia coli meningitis isolate IHE3034 reveals two novel regulatory genes within the promoter-distal region of the main S fimbrial operon. Microb Pathog 46:150–158. doi: 10.1016/j.micpath.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Sundriyal A, Massa C, Samoray D, Zehender F, Sharpe T, Jenal U, Schirmer T. 2014. Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger cyclic di-GMP. J Biol Chem 289:6978–6990. doi: 10.1074/jbc.M113.516195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimada T, Yamamoto K, Ishihama A. 2011. Novel members of the Cra regulon involved in carbon metabolism in Escherichia coli. J Bacteriol 193:649–659. doi: 10.1128/JB.01214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wan X, Tuckerman JR, Saito JA, Freitas TAK, Newhouse JS, Denery JR, Galperin MY, Gonzalez G, Gilles-Gonzalez M-A, Alam M. 2009. Globins synthesize the second messenger bis-(3′-5′)-cyclic diguanosine monophosphate in bacteria. J Mol Biol 388:262–270. doi: 10.1016/j.jmb.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 56.Tuckerman JR, Gonzales G, Sousa EH, Wan X, Saito JA, Alam M, Gilles-Gonzalez M-A. 2009. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48:9764–9774. doi: 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- 57.Uhlich GA, Chen CY, Cottrell BJ, Hofmann CS, Dudley EG, Strobaugh TP, Nguyen LH. 2013. Phage insertion in mlrA and variations in rpoS limit curli expression and biofilm formation in Escherichia coli serotype O157:H7. Microbiology 159:1586–1596. doi: 10.1099/mic.0.066118-0. [DOI] [PubMed] [Google Scholar]
- 58.Uhlich GA, Keen JE, Elder RO. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl Environ Microbiol 67:2367–2370. doi: 10.1128/AEM.67.5.2367-2370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diodati ME, Bates AH, Cooley MB, Walker S, Mandrell RE, Brandl MT. 2015. High genotypic and phenotypic similarity among Shiga toxin-producing Escherichia coli O111 environmental and outbreak strains. Foodborne Pathog Dis 12:235–243. doi: 10.1089/fpd.2014.1887. [DOI] [PubMed] [Google Scholar]
- 60.Tükel C, Nishimori JH, Wilson RP, Winter MG, Keestra AM, van Putten JP, Bäumler AJ. 2010. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell Microbiol 12:1495–1505. doi: 10.1111/j.1462-5822.2010.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tükel C, Wilson RP, Nishimori JH, Pezeshki M, Chromy BA, Bäumler AJ. 2009. Responses to amyloids of microbial and host origin are mediated through Toll-like receptor 2. Cell Host Microbe 6:45–53. doi: 10.1016/j.chom.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosser T, Dransfield T, Allison L, Hanson M, Holden N, Evans J, Naylor S, La Ragione R, Low C, Gally DL. 2008. Pathogenic potential of emergent sorbitol-fermenting Escherichia coli O157:NM. Infect Immun 76:5598–5607. doi: 10.1128/IAI.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barak JD, Gorski L, Naraghi-Arani P, Charkowski AO. 2005. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl Environ Microbiol 71:5685–5691. doi: 10.1128/AEM.71.10.5685-5691.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fink RC, Black EP, Hou Z, Sugawara M, Sadowsky MJ, Diez-Gonzalez F. 2012. Transcriptional responses of Escherichia coli K-12 and O157:H7 associated with lettuce leaves. Appl Environ Microbiol 78:1752–1764. doi: 10.1128/AEM.07454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jeter C, Matthysse AG. 2005. Characterization of the binding of diarrheagenic strains of Escherichia coli to plant surfaces and the role of curli in the interaction of the bacteria with alfalfa sprouts. Mol Plant Microbe Interact 18:1235–1242. doi: 10.1094/MPMI-18-1235. [DOI] [PubMed] [Google Scholar]
- 66.Matthysse AG, Deora R, Mishra M, Torres AG. 2008. Polysaccharides cellulose, poly-β-1,6-n-acetyl-d-glucosamine, and colanic acid are required for optimal binding of Escherichia coli O157:H7 strains to alfalfa sprouts and K-12 strains to plastic but not for binding to epithelial cells. Appl Environ Microbiol 74:2384–2390. doi: 10.1128/AEM.01854-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jonas K, Tomenius H, Römling U, Georgellis D, Melefors O. 2006. Identification of YhdA as a regulator of the Escherichia coli carbon storage regulation system. FEMS Microbiol Lett 264:232–237. doi: 10.1111/j.1574-6968.2006.00457.x. [DOI] [PubMed] [Google Scholar]
- 68.Nakasone Y, Ono TA, Ishii A, Masuda S, Terazima M. 2007. Transient dimerization and conformational change of a BLUF protein: YcgF. J Am Chem Soc 129:7028–7035. doi: 10.1021/ja065682q. [DOI] [PubMed] [Google Scholar]
- 69.Mika F, Busse S, Possling A, Berkholz J, Tschowri N, Sommerfeldt N, Pruteanu M, Hengge R. 2012. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol Microbiol 84:51–65. doi: 10.1111/j.1365-2958.2012.08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pesavento C, Hengge R. 2012. The global repressor FliZ antagonizes gene expression by σS-containing RNA polymerase due to overlapping DNA binding specificity. Nucleic Acids Res 40:4783–4793. doi: 10.1093/nar/gks055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ryjenkov DA, Simm R, Römling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 72.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 73.Fang X, Ahmad I, Blanka A, Schottkowski M, Cimdins A, Galperin MY, Römling U, Gomelsky M. 2014. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Microbiol 93:439–452. doi: 10.1111/mmi.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fang X, Gomelsky M. 2010. A posttranslational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol 76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- 75.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Serra DO, Richter AM, Klauck G, Mika F, Hengge R. 2013. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 4:e00103-13. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Serra DO, Hengge R. 2014. Stress responses go three-dimensional: the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ Microbiol 16:1455–1471. doi: 10.1111/1462-2920.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morgan JL, McNamara JT, Zimmer J. 2014. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21:489–496. doi: 10.1038/nsmb.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu J, Wang B, Fang X, MEans WJ, McCormick RJ, Gomelsky M, Zhu MJ. 2013. c-di-GMP signaling regulates Escherichia coli O157:H7 adhesion to colonic epithelium. Vet Microbiol 164:344–351. doi: 10.1016/j.vetmic.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 80.Kai-Larsen Y, Lüthje P, Chromek M, Peters V, Wang X, Holm A, Kádas L, Hedlund KO, Johansson J, Chapman MR, Jacobson SH, Römling U, Agerberth B, Brauner A. 2010. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog 6:e1001010. doi: 10.1371/journal.ppat.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bielaszewska M, Mellmann A, Zhang W, Köck R, Fruth A, Bauwens A, Peters G, Karch H. 2011. Characterization of the Escherichia coli strain associated with an outbreak of hemolytic-uremic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis 11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.