Abstract
Whole genome duplications (WGD) have had strong impacts on species diversification by triggering evolutionary novelties, however, relatively little is known about the balance between gene loss and forces involved in the retention of duplicated genes originating from a WGD. We analyzed putative Salicoid duplicates in willows, originating from the Salicoid WGD, which took place more than 45 Mya. Contigs were constructed by de novo assembly of RNA-seq data derived from leaves and roots from two genotypes. Among the 48,508 contigs, 3,778 pairs were, based on fourfold synonymous third-codon transversion rates and syntenic positions, predicted to be Salicoid duplicates. Both copies were in most cases expressed in both tissues and 74% were significantly differentially expressed. Mean Ka/Ks was 0.23, suggesting that the Salicoid duplicates are evolving by purifying selection. Gene Ontology enrichment analyses showed that functions related to DNA- and nucleic acid binding were over-represented among the non-differentially expressed Salicoid duplicates, while functions related to biosynthesis and metabolism were over-represented among the differentially expressed Salicoid duplicates. We propose that the differentially expressed Salicoid duplicates are regulatory neo- and/or subfunctionalized, while the non-differentially expressed are dose sensitive, hence, functionally conserved. Multiple evolutionary processes, thus drive the retention of Salicoid duplicates in willows.
Whole genome duplication (WGD or polyploidy) has long been recognized as an important evolutionary force that create biological novelty and complexity1,2 and polyploidization is regarded as the trigger for the rapid diversification of angiosperms3,4,5. In fact, most flowering plant lineages have undergone one or more rounds of ancient polyploidization events4,5. For example, the Arabidopsis thaliana genome shows signs of two recent WGDs and one triplication event that is likely shared by all eudicots6,7,8 and the poplar, Populus trichocarpa genome shows signs of this ancient triplication event, as well as a more recent WGD9. A WGD introduces massive genetic redundancy, as immediately after the WGD, every gene will have a copy, and the two copies are often referred to as paralogs or duplicates. Over evolutionary times, polyploid genomes can undergo diploidization10,11 (also known as fractionation12), making the genomes more diploid-like. Since the duplicates have redundant functions immediately after they are formed, one of the duplicate copies might become nonfunctional by accumulating deleterious mutations (nonfunctionalization or pseudogenization), without any effects on fitness of the individual. During diploidization, therefore, massive gene loss and genome rearrangements usually take place.
Many duplicates, however, escape nonfunctionalization and are retained even long after the WGD, which is for example seen in the cotton (Gossypium raimondii)13 and poplar (P. trichocarpa) genomes9 that contain approximately 2,000 and 8,000 pairs of retained paralogous genes respectively. The genes retained in duplicates are assumed to follow distinctive modes of evolution, resulting in functional diversification or conservation of redundant function. According to the neofunctionalization model1,14, the degenerating copy escapes nonfunctionalization by acquiring a substitution that lead to a new gene function, which is expected to be fixed by drift15, meanwhile, the other copy retains the ancestral function. Following neofunctionalization, the functions of the copies are expected to be maintained by purifying selection15. The subfunctionalization, or more specifically the duplication-degeneration-complementation (DDC) model14, proposes that both copies acquire degenerate substitutions that damage their functions, however both will eventually be fixed in the population by drift15. As neither of the copies can perform its original function, that is, they are subfunctionalized, both will be maintained by purifying selection. Both neo- and subfunctionalization are thus expected to lead to functional diversification of the retained duplicates, which can be changes in regulatory regions, leading to differential expression or to changes in the coding sequences, resulting in differences in protein functions.
Subfunctionalization can also be manifested as the partitioning of expression of the duplicates between different developmental stages or between different tissues. Examples of rapid subfunctionalization by tissue-specific reciprocal silencing have previously been demonstrated16,17,18. A third possible outcome for retained duplicates is that their functions are conserved, throughout evolutionary times, meaning that the duplicates escape functional diversification. Some classes of genes, for example transcription factors and genes involved in signal transduction are overrepresented among duplicates with conserved redundant functions19. It is hypothesised that these genes are sensitive to changes in expression levels and therefore must be present in the same number of genomic copies as the genes with whose product they interact (summarized in the gene balance model20,21).
The Salicoid WGD is the most recent WGD that has been detected in poplars (Genus: Populus)9,22. Interestingly, this WGD is shared with the willows (genus: Salix)9,23, suggesting that it should have taken place before the divergence of the two lineages, more than 45 million years ago (Mya)24,25. As sequence divergence was estimated to be around three times higher between willow and poplar duplicates (with a predicted origin from the Salicoid WGD) compared to orthologs23, it suggests that the WGD happened long before the speciation event (assuming a constant molecular clock). In a more recent study based on whole-genome sequence data, the divergence time was estimated to 52 Mya and the WGD to six million years prior to the lineage split26. These estimates, therefore suggest that the Salicoid WGD is relatively ancient and happened sometime between 45 and 58 Mya. Both willow and poplar genomes are organized in nineteen chromosomes (n = 19) and they display high levels of macrosynteny, although several major rearrangements have been detected23. Willow genomes are generally smaller than poplar genomes and they also contain fewer protein coding genes. For example, the predicted genome size of P. trichocarpa is 485 million base pairs (Mbp) and the genome contains 41,335 protein coding genes9, while the predicted genome size of Salix purpurea is 450 Mbp and contains 37,865 putative protein coding genes (Salix purpurea v1.0, DOE-JGI, http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Spurpurea). The recently sequenced Salix suchowensis genome is possibly even smaller as it was estimated to be 425 Mbp with only 25,599 putative protein coding genes26. Based solely on these observations, we therefore expect fewer Salicoid duplicates in willows than in poplars. In order to study mechanisms operating on retained paralogous genes after a WGD, we first aimed to identify retained Salicoid duplicates among expressed genes in willows and secondly to investigate sequence and gene expression divergence between the Salicoid duplicates. This gave us important information on evolutionary processes involved in the retention of duplicated genes after a WGD.
Results
The construction and filtering of the de novo transcriptome assembly
As we generated a total of 769 × 106 100 bp reads from all the samples, the total output of the reads was 76.9 × 109 bp, representing about 180-fold of the predicted willow genome size (Table 1). After trimming adapter sequences and removing low-quality bases, 682 million sequencing reads from all samples were combined and a de novo assembly was built with Trans-ABySS27,28. The total number of generated contigs was 392,355 with lengths ranging from 61 to 15,602 bp (Fig. 1). To remove lowly expressed and erroneous contigs, the assembly was filtered on expression levels (at least one fragment per kilobase of contig per million mapped fragments (FPKM)) and on contig lengths (at least 500 bp in length). This resulted in 52,215 significantly expressed contigs, with a minimum length of 500 bp (Fig. 1). These were finally translated to peptide sequences and only contigs with open reading frames were retained, resulting in 48,508 contigs in the final assembly. This filtered assembly was used for further analysis. When the contigs were mapped to the putative protein coding transcripts in the draft genome of S. purpurea, 32,563 matches were retrieved. As the S. purpurea draft genome contains 37,865 protein coding transcripts, the contigs, thus represent as many as 86% of all protein coding transcripts present in the current version of the S. purpurea genome. As a quality control of the assembly, the proportion of sequencing reads that were integrated in the assembly was estimated. The trimmed sequencing reads were mapped back to the unfiltered assembly with 392,355 contigs, resulting in successful mapping of 80.1% of the sequencing reads, of which 73.8% aligned uniquely. This demonstrates that the reads were well represented in the assembly. On the other hand, when the sequencing reads were mapped to the filtered assembly with 48,508 contigs, on average 26.2% of all the trimmed reads mapped (Table 1). Of these, on average 73.2% mapped to unique positions and 58.9% aligned with no mismatches (Table 1). This shows that a large proportion of sequencing reads in the unfiltered assembly were present in short and/or lowly expressed contigs and/or contigs that had no detectable open reading frames, which were thus not present in the filtered assembly.
Table 1. Summary of Illumina sequencing, assembly and mapping.
| 520 leaves | % | 520 roots | % | 592 leaves | % | 592 roots | % | Sum | % | |
|---|---|---|---|---|---|---|---|---|---|---|
| Total no of reads | 207,824,532 | 168,557,233 | 187,381,473 | 205,123,179 | 768,886,417 | |||||
| Total no of bases (Gbp) | 20.8 | 16.9 | 18,7 | 20.5 | 76.9 | |||||
| No of reads after trimming | 174,156,958 | 148,667,480 | 168,291,008 | 190,426,984 | 681,542,430 | |||||
| Average read length after trimming | 98.4 | 98.5 | 97.9 | 98.7 | ||||||
| Total mapped readsa | 64,492,170 | 37.0b | 35,511,470 | 23.9b | 38,630,740 | 23.0b | 39,889,518 | 20.9b | 178,523,898 | 26.2b |
| Total unmapped readsc | 109,664,788 | 63.0b | 113,156,010 | 76.1b | 129,660,268 | 77.0b | 150,537,466 | 79.1b | 503,018,532 | 73.8b |
| Multi-position matchese | 19,530,798 | 30.3d | 8,648,426 | 24.4d | 9,284,534 | 24.0d | 10,379,248 | 26.1d | 47,843,006 | 26.8d |
| Unique matchesf | 44,961,372 | 69.7d | 26,863,044 | 75.6d | 29,346,206 | 76.0d | 29,510,270 | 74.0d | 130,680,892 | 73.2d |
aNumber of filtered sequencing reads that were aligned to the contigs in the filtered assembly.
bRelative to the number of reads after trimming.
cSequencing reads that could not be aligned to the contigs in the filtered assembly.
dRelative to the number of mapped reads.
eSequencing reads that aligned to two or more positions in the contigs in the filtered assembly.
fSequencing reads aligned to only one position in the contigs in the filtered assembly.
Figure 1. Length distribution of the contigs generated by the Trans-ABySS assembler.
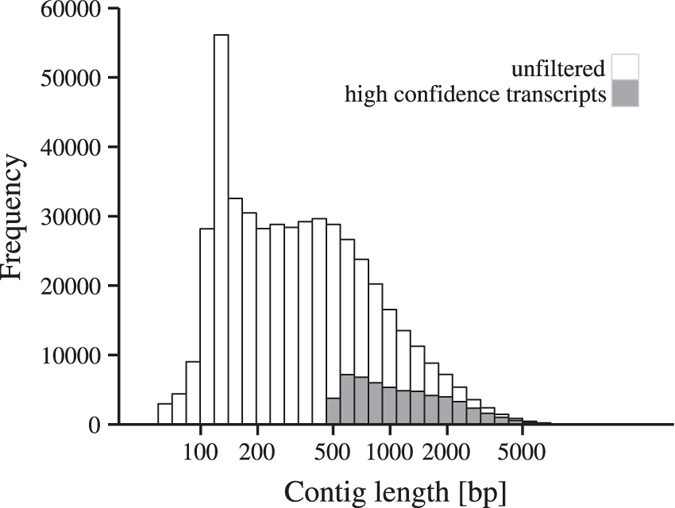
The white distribution shows the contig lengths in the unfiltered assembly and the light grey distribution shows the contig lengths in the filtered assembly.
Identification and verification of Salicoid duplicates
Putative duplicate pairs were identified by reciprocal BLAST using BLASTP, assuming that for every contig, the second best hit was its duplicate copy (the best BLAST hit was assumed to be to itself). This analysis resulted in 11,279 predicted duplicate pairs. To identify Salicoid duplicate pairs, we estimated sequence divergence, or more specifically, we determined the fourfold synonymous third-codon transversion rate (4DTV) between each pair. Previous studies in P. trichocarpa9 and S. suchowensis26 demonstrated that Salicoid duplicate pairs have 4DTV values approximately between 0.1 and 0.2. Guided by the distribution of 4DTV values in Fig. 1, we identified 3,981 pairs in the peak, with 4DTV values between 0.04 and 0.2 (Fig. 2, Supplementary Table S1 online). 203 pairs that were located within 100 kb of each other (using the S. purpurea genome as reference) were filtered away, as they, possibly, were tandem duplicates. 3,778 duplicate pairs were therefore predicted to be Salicoid duplicates, which means that 23% of all expressed genes in the leaves and the roots of these willows are retained Salicoid duplicates.
Figure 2. 4DTV rates between all predicted duplicates.
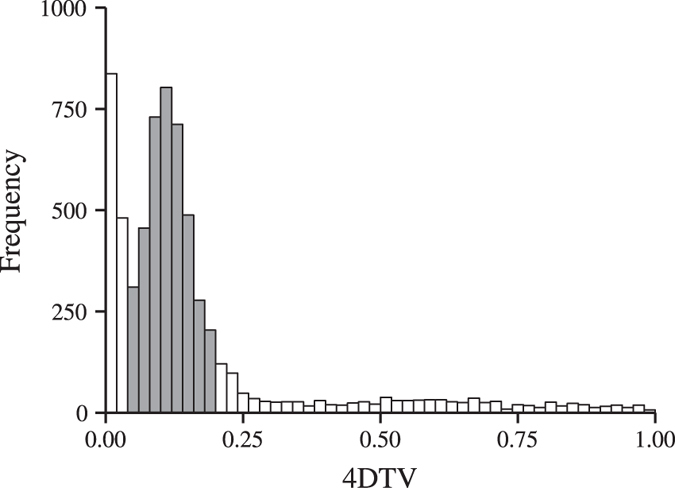
The predicted Salicoid duplicates are highlighted in light grey.
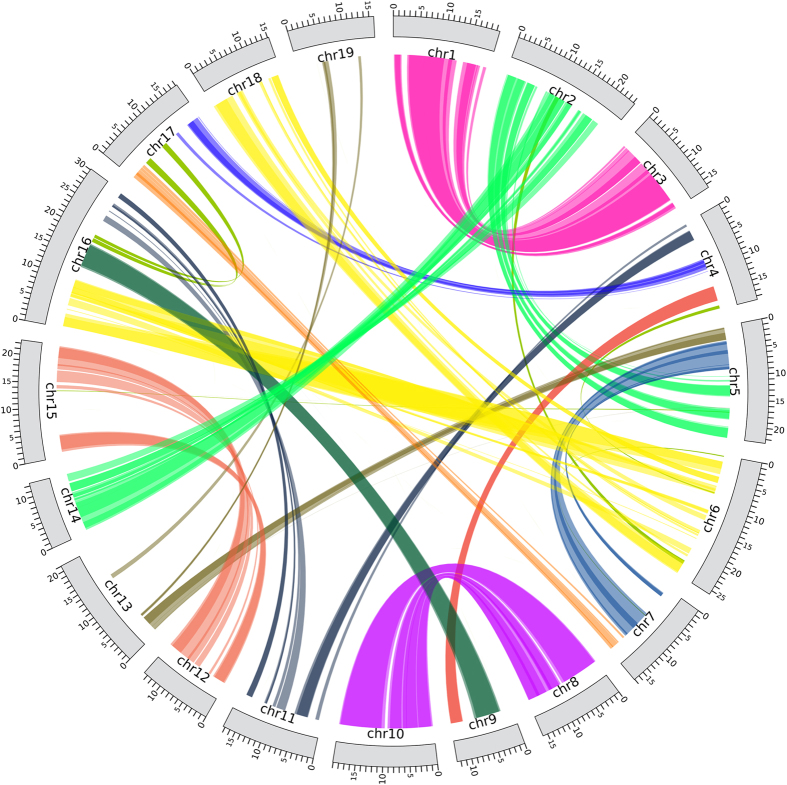
Homeologous genomic segments originating from the Salicoid WGD have been identified in the P. trichocarpa genome9. For example, several chromosomes are more or less completely homeologous, e.g. chromosome 8 and 10, 12 and 15, while others contain large homeologous segments, e.g. 5 and 7, and 2, 5 and 149. It is thus expected that Salicoid duplicate copies in a pair map to homeologous chromosomes. In order to test this, we mapped each copy to the S. purpurea genome, assuming conserved synteny between the willow species as well as between willows and poplars. A total of 6,004 duplicate copies (3,002 pairs) mapped to one of the nineteen chromosomes, while the rest either did not map or mapped to scaffolds. For 2,789 pairs (93%), the two copies were located on different chromosomes and in most cases they were located on chromosomes previously described as homeologous9 (Fig. 3). This observation, based on synteny, thus strongly supports that the predicted duplicates indeed are Salicoid duplicates.
Figure 3. Positions of Salicoid duplicate copies in the S. purpurea genome.
The lines connect the two copies in every pair. Most Salicoid duplicates are located on homeologous chromosomes originating from the Salicoid WGD, for example on chromosome 8 and 10, 12 and 15, 5 and 7 and on 2, 5 and 14. Homeologous chromosomes were defined in the P. trichocarpa genome9. The image was created with Circos46.
Gene expression divergence of Salicoid duplicates
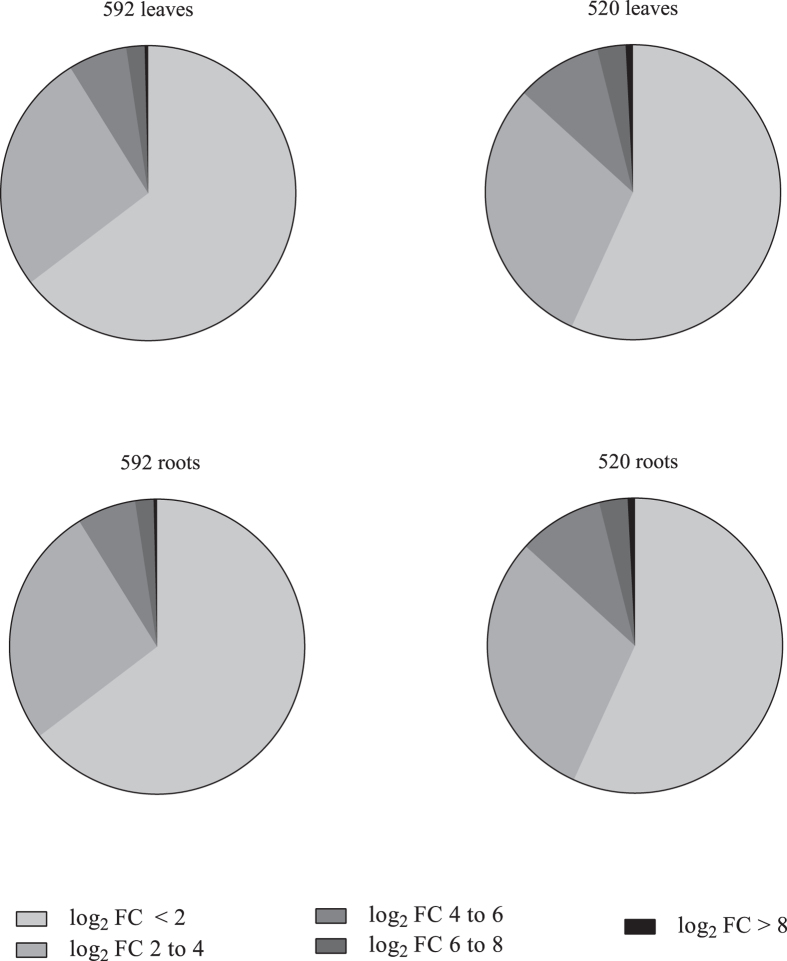
To study expression differences i.e. gene expression divergence between the Salicoid duplicate pairs, gene expression was quantified for every Salicoid duplicate copy in the two tissues and genotypes separately (Supplementary Table S2 online). For 3,704 pairs, both copies were expressed in at least one of the tissues and genotypes and for 70 pairs, both copies were expressed in only the leaves and 127 in only the roots, which means that for most Salicoid duplicate pairs both copies were expressed in both tissues. For three pairs, the two copies were expressed in different tissues, thus their expression were partitioned among the tissues. Differential expression between the Salicoid duplicate copies was investigated in the two tissues and genotypes separately (Supplementary Table S3 online). On average, 2,872 (76%) pairs displayed significant levels of differential expression (False discovery rate (FDR) ≤ 0.05) across the tissues and genotypes (520 leaves: 3,079, 520 roots: 2,784, 592 leaves: 3,029 and 592 roots: 2,595). Most copies displayed low levels of differentiation (log2 fold change (FC) between 0.4 and 2). Some copies were, however highly differentially expressed (Fig. 4A–D).
Figure 4. Levels of differential expression between Salicoid duplicates in the two genotypes and tissues.
FC = fold change.
Sequence divergence of Salicoid duplicates
We examined the level of sequence divergence by estimating the Ka/Ks ratio (non-synonymous substitutions per non-synonymous sites/synonymous substitutions per synonymous sites) for every Salicoid duplicate pair. The ratios centred around a peak at 0.23 (Fig. 5), and only 21 of the pairs had Ka/Ks ratios > 1. This indicates overall slow rates of protein evolution at these genes, suggesting that the retained Salicoid duplicates are evolving under purifying selection. To investigate the association between coding-sequence divergence (i.e. Ka/Ks, Ka, Ks and 4DTV) and gene expression divergence we used Spearman Rank correlations. We found a weak, albeit significant positive correlation between Ka/Ks and differential expression, while all other comparisons were non-significant (Table 2). To further investigate the association between coding-sequence and gene expression divergence, we also compared Ka/Ks ratios between different classes of Salicoid duplicates after classification based on level of differential expression and found that Ka/Ks was lowest for the pairs with log2 FC < 2 and highest for the Salicoid duplicates with log2 FC > 8 (Supplementary Fig. S1 online). These differences were however not statistically significant.
Figure 5. Distribution of Ka/Ks values between the Salicoid duplicates.
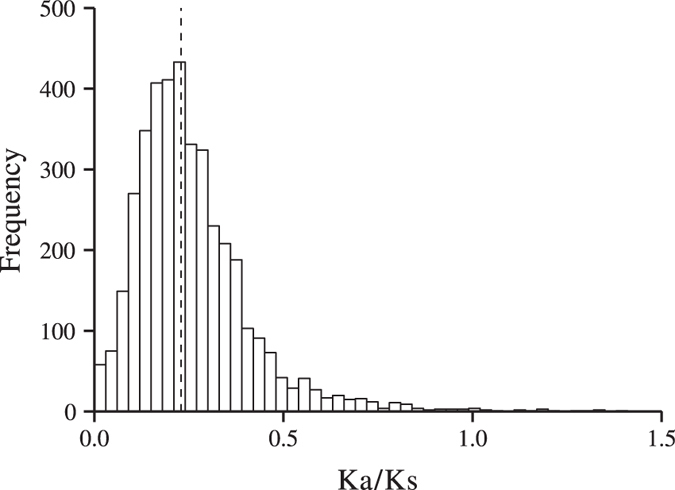
The median is indicated by the dashed line.
Table 2. Spearman rank correlation coefficients (r) and the level of significance (P-value) between differential expression and Ka/Ks, Ka, Ks and 4DTV.
| Ka/Ks | Ka | Ks | 4DTV | |
|---|---|---|---|---|
| 520 leaves | 0.051. P < 0.01 | 0.008, ns. | 0.001, ns. | 0.013, ns. |
| 520 roots | 0.089. P < 0.0001 | −0.009, ns. | 0.0014, ns. | −0.001, ns. |
| 592 leaves | 0.039. P < 0.05 | 0.013, ns. | 0.016, ns. | 0.015, ns. |
| 592 roots | 0.087. P < 0.0001 | −0.013, ns. | −0.001, ns. | −0.003, ns. |
ns. = not significant.
Functional annotation and GO enrichment analyses
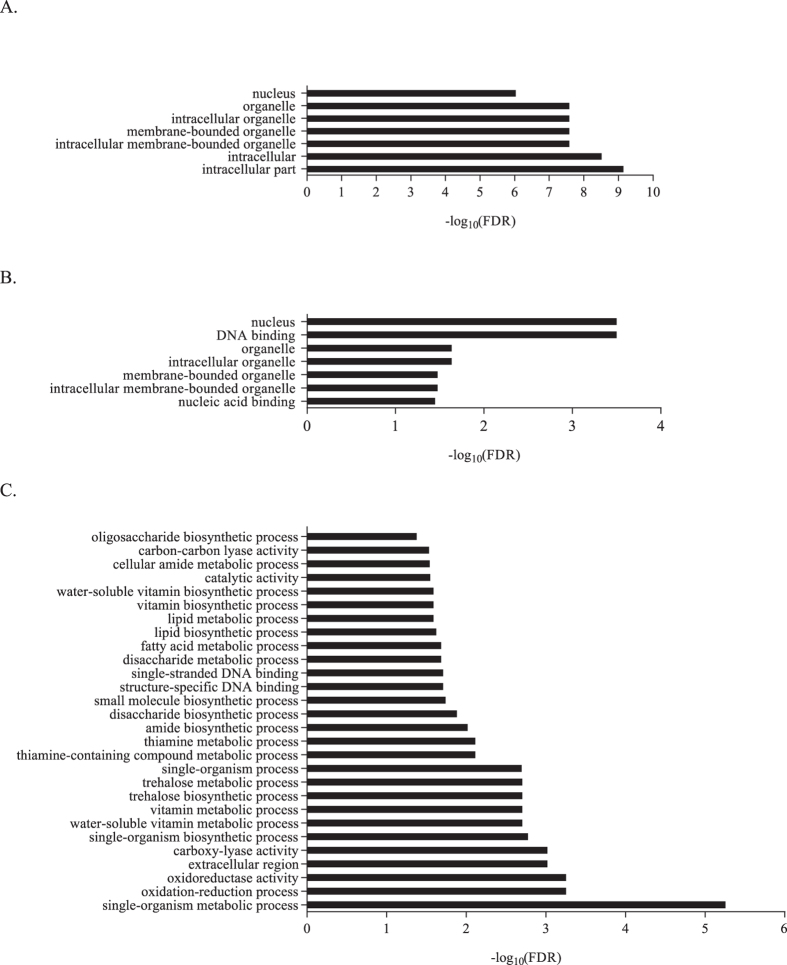
To functionally classify the contigs, we used the Blast2GO software for annotation and 33% of all contigs and 88% of the Salicoid duplicates were annotated with gene ontology (GO) terms (Supplementary Table S4 online). We found that the two copies in a pair were always annotated with the same terms, which was expected given the high degree of sequence similarities. To investigate if any functional categories were overrepresented among the Salicoid duplicates, we performed GO enrichment analyses using all contigs as reference. First we tested for enrichment in all Salicoid duplicates and found that several GO terms related to intracellular and nucleus components in the cellular component category were overrepresented (Fig. 6A). We then tested for enrichment among the non-differentially expressed Salicoid duplicates and found overrepresentation of for example DNA- and nuclear binding terms (Fig. 6B). Lastly, we tested for enrichment among the differentially expressed Salicoid duplicates and found overrepresentation of several terms related to metabolism and biosynthesis (Fig. 6C).
Figure 6. Enriched GO terms.
(A) All Salicoid duplicates vs. all contigs. (B) The non-differentially expressed Salicoid duplicates vs. all contigs. (C) The differentially expressed Salicoid duplicates vs. all contigs.
Discussion
In this study we present results from analyses of retained duplicates in willows that originate from a relatively ancient genome duplication that occurred more than 45 Mya, prior to the divergence of poplars and willows. For the analyses, we used 48,508 contigs that were generated by de novo assembly of RNA-seq data derived from leaf and root tissues of two willow genotypes. The assembly was produced by assembling trimmed sequencing reads using the Trans-ABySS assembler and by filtering on gene expression levels, contig lengths and the presence of open reading frames. The filtered contigs mapped to 32,563 of the 37,865 predicted protein coding transcripts in the S. purpurea genome, thus representing as much as 86% of the transcribed part of the S. purpurea genome, showing that extensive gene expression is taking place in these willow leaf and root tissues. Similarly, in leaves of P. trichocarpa, as many as 33,044 genes were expressed29. This demonstrates that RNA-seq data from a few tissues gives a good representation of the transcribed part of willow and poplar genomes.
Among the contigs, 11,279 putative duplicate pairs were identified, of which 3,778 pairs were predicted to be Salicoid duplicates. Our strategy to use RNA-seq data from two tissues to obtain expressed genes and then relying on sequence divergence for the identification of the Salicoid duplicates, is conservative and we therefore do not expect to have sampled all Salicoid duplicates present in the genomes of these willows. The observed number is however surprisingly low (given that we have sampled approximately 86% of all protein coding genes (based on the S. purpurea genome)), compared to the nearly 8,000 pairs that were detected in the P. trichocarpa genome9. It is hence likely that willow genomes contain fewer retained Salicoid duplicates than poplar genomes, which could simply be because they overall have smaller genomes. It is however also possible that willows have lost relatively more Salicoid duplicate copies than poplars, which was suggested by Dai et al.26. An interesting hypothesis is that the Salicoid WGD actually led to the split of the two lineages, which follows the hypothesis that if gene losses occur independently in different populations this can lead to population differentiation30. According to this hypothesis, different duplicate copies might have been lost in the willow and poplar lineages respectively, leading to the retention of different Salicoid duplicate pairs.
For most Salicoid duplicates, both copies were expressed in both tissues, indicating that subfunctionalization was not manifested by the partitioning of expression between the two tissues. When testing for differential expression between the copies, we found that on average 76% of the Salicoid duplicates were significantly differentially expressed and more than 40% were highly differentially expressed (log2 FC > 2). Expression divergence manifested as significant differential expression was therefore found for a large fraction of the retained Salicoid duplicates. The high prevalence of differentially expressed Salicoid duplicates in our dataset as well as in poplar31, suggests that expression divergence is a key process in retention of Salicoid duplicates across the two lineages. If we hypothesise that the observed expression differences between the copies are functionally meaningful, implying that the differentially expressed Salicoid duplicates are functionally diverged. Following that argument, it is possible that the differentially expressed Salicoid duplicates are neofunctionalized1 and/or subfunctionalized, following the predictions of the DDC model14. Unfortunately, we cannot distinguish between the two processes of neo- and subfunctionalization, as we do not know the ancestral gene function, before the Salicoid WGD. Interestingly, expression divergence as a result of subfunctionalization and/or neofunctionalization has previously been demonstrated in a diverse range of plant species, for example in cotton (Gossypium raimondii)13, soybean (Glycine max)32, maize (Zea mays)33 and Arabidopsis34, thus manifesting the importance of these process in the maintenance of retained duplicates in plants.
In contrast, a substantial fraction of the Salicoid duplicates were not differentially expressed. One possibility is that these duplicates did not have time to become neo- and/or subfunctionalized, however, as expression divergence is expected to evolve rapidly13, there should have been enough time for neo- and/or subfunctionalization to have evolved since the Salicoid WGD. A more likely explanation is therefore that the expression of these duplicates is evolutionary conserved, following the predictions of the gene balance model20. This is further supported by the observation that the functional categories nucleic acid binding and DNA binding are overrepresented among the non-differentially expressed Salicoid duplicates, which include transcription factors and other regulatory elements that are expected to be dose sensitive20. In addition, our analyses of Ka/Ks ratios indicate extensive purifying selection on the non-differentially expressed Salicoid duplicates (Ka/Ks = 0.23) as well as on the differentially expressed (Ka/Ks = 0.24) Salicoid duplicates, which is expected15. Similar low values were obtained for Salicoid duplicates in poplars31.
We found no strong correlations between differential expression and any of the measures of coding-sequence divergence (Ks, Ka/Ks, Ks och 4DTV) for the Salicoid duplicates, however we observed a weak, but significant, positive correlation between DE and Ka/Ks. Given the low correlation coefficients, it is doubtful that these correlations have any biological meaning, therefore our results, favours the uncoupling of the mechanisms of gene expression and sequence divergence in the Salicoid duplicates. This indicates that substitutions in coding sequences have little impact on expression divergence, suggesting that regulatory substitutions play a more significant role. Interestingly, this fits well with the hypothesis that the observed expression differences are the result of neo- and/or subfunctionalization of regulatory regions.
To conclude, in this study, we have identified retained duplicates with a presumed origin from the Salicoid WGD. We estimated the expression and coding sequence divergence between the Salicoid duplicates and found two classes of duplicates; the differentially expressed and the non-differentially expressed. We hypothesise that the differentially expressed Salicoid duplicates are regulatory neo- and/or subfunctionalized, while the non-differentially expressed are dose sensitive and therefore functionally conserved. This shows that similar evolutionary processes are operating on retained Salicoid duplicates in both willows and poplars31. Our analyses, furthermore, suggest that willows have much fewer retained Salicoid duplicates than the poplars. This indicates that neo- and/or subfunctionalization occurred in the period between the WGD and the divergence of the two lineages. However, gene loss following the Salicoid duplication continued independently in two lineages. Whether or not this process played a role in the diversification of the two lineages needs to be investigated further.
Methods
Sample collection, RNA extractions and sequencing
Fifteen biological replicates for each of two willow genotypes (520 and 592) were cultivated in a walk-in growth chamber with 20 °C constant temperature, 70% relative humidity and 1 h photoperiod (300 μmol photosynthetically active radiation (PAR) m−2 s−1) with regular watering. The two genotypes are siblings from a cross between S. viminalis x (S. viminalis x S. schwerinii)23. After 69 days, two fully developed young leaves and about one centimetre of several root tips were sampled from each plant and immediately snap frozen in liquid nitrogen and stored at −80 °C while awaiting RNA extractions. Approximately 100 mg of leaves and 30 mg of root tips were used for the RNA extractions, which were performed using a Spectrum Plant Total RNA Kit from Sigma-Aldrich with a On-Column DNase I digestion set, also from Sigma-Aldrich. Eighteen samples were selected for sequencing, representing: i) five biological replicates of 520 leaves, ii) five biological replicates of 592 leaves, iii) four biological replicates of 520 roots, and iv) four biological replicates of 592 roots. The RNA samples were first treated with DNase, after which one library per sample was prepared using Illumina’s TruSeq RNA Sample Prep Kit v1, where polyA-fragments were selected, followed by cDNA synthesis and ligation of amplification and sequencing adapters. Sequencing libraries were individually barcoded and then pooled with nine libraries per lane and sequenced on an Illumina HiSeq 2000 instrument. All samples were sequenced as paired-end with 100 bp read length. Library preparations and sequencing were performed by the SNP&SEQ Technology Platform in Uppsala, Sweden.
Sequencing read processing and de novo assembly
The raw sequencing reads were first pre-processed by trimming the adapters and low quality bases with the software Trimmomatics (Version 0.32)35. An average quality of 20 was maintained across the sliding window of four bases (SLIDINGWINDOW:4:20) and a minimum length of 75 bp of the reads after trimming (MINLEN:75) was required. The trimmed sequencing reads from all eighteen samples (i.e. libraries) were combined and assembled de novo using the paired end read assembler Trans-ABySS27,28 with k-mer size of 61, k-cov of 5and otherwise with default settings. The assembly was filtered based on transcript lengths ( ≥500 bp) and gene expression levels, which was achieved by mapping the trimmed reads for all samples to the contigs using the program RNA-Seq by Expectation-Maximization (RSEM)36 with default settings. A contig was considered to be expressed if it contained at least one mapped fragment per kilobase of contig per million fragments mapped in total (FPKM). Coding and peptide sequences were generated using the TransDecoder tool from the Trinity package37. To investigate the proportion of sequencing reads that were integrated in the unfiltered and the filtered assembly, the sequencing reads from each of the different samples were separately mapped using Bowtie238 with a maximum of two mismatches. Different quality metrics measurements i.e. i) total number of reads, ii) total mapped reads, iii) uniquely mapped reads and iv) reads that mapped to more than one position were calculated.
Identification of Salicoid duplicates
In order to identify duplicates among the filtered contigs, reciprocal BLAST was performed using BLASTP. Two genes were predicted to be duplicates if the reciprocal second best BLAST hit had an e-value lower than 1 × 10−10. The best BLAST hit was always excluded as it was assumed to be to itself. Fourfold synonymous third-codon transversion rates (4DTV) were estimated between the pairs and the peak with 4DTV values between 0.04 and 0.2 were predicted to contain the Salicoid duplicates9. The reason for using 4DTV rates and not the total number of synonymous substitutions is that transversions occur much more seldom than transitions, therefore back mutations are less likely to have happened at these positions. To estimate 4DTV rates, the peptide sequences of the predicted duplicates were aligned using the ClustalW algorithm39 (Version 2.1, gapopen penalty: 10, gapextension penalty: 1) and the alignments were converted to a codon alignment using PAL2NAL40. 4-fold degenerate sites were located using an in-house python script and the ratio of transversion per 4-fold degenerate site was calculated. This raw value was corrected for multiple substitutions according to Hellsten et al. (2007)41. The genomic positions of the Salicoid duplicates in the S. purpurea genome were determined by BLAST and the Salicoid duplicates that occurred within 100 kb of each other were considered to be tandem duplicates and were filtered out.
Ka/Ks calculations
For each Salicoid duplicate pair, the number of non-synonymous substitutions per non-synonymous sites (Ka), the number of synonymous substitutions per synonymous sites (Ks) and the Ka/Ks ratio were calculated in the codeml program in the PAML package using runmode -2 (Version 4.7a)42,43.
Gene expression divergence and its correlation with sequence divergence
Gene expression was quantified by mapping the trimmed reads against every Salicoid duplicate copy using Bowtie38 allowing a maximum of two mismatches and applying a seed length of 25 bp. Normalised gene expression measured as transcripts per million (TPM) and expected read counts were calculated separately for each tissue and genotype in RSEM36 with default settings. A copy was considered expressed in the tissue and genotype if the average TPM was above 1. We used two different measures of expression divergence. First, we examined the partitioning of gene expression between the duplicates across the two tissues, by counting pairs where both copies were expressed (TPM > 1) in at least one of the tissues and pairs where one copy was expressed in one tissue and the other was expressed in the other tissue. Secondly, in every tissue and genotype, differential expression was tested for between the Salicoid duplicates using the R/Bioconductor package edgeR44 with the expected read counts calculated in RSEM36 as input. Duplicates were defined as differentially expressed when the false discovery rate (FDR) was ≤ 0.05. In addition, for all predicted Salicoid duplicates, correlations between differential expression and various estimates of sequence divergence (Ka/Ks, Ka, Ks and 4DTV) were analysed by Spearman rank correlations.
Gene annotations
The protein primary transcripts and Gene Ontology (GO) annotation terms for the Arabidopsis genome were downloaded from The Arabidopsis Information Resource (TAIR) database v. 10 (https://www.arabidopsis.org). A BLAST search was performed with the filtered assembly against the primary transcripts. The best hits were retrieved based on e-values lower than 1 × 10−10 and bitscore above 40. The locus identifiers and their respective gene IDs were extracted from the best hit results. An in-house python script was used to assign GO terms to the willow transcripts based on the BLAST results.
GO enrichment analysis
GO enrichment analyses were performed for different sets using the Blast2GO software45. Blast2GO uses Fischer’s exact test to represent relationships between reference and test data sets. The GO terms with corrected p-values ≤ 0.05 were considered to be overrepresented. The set of all annotated contigs was used as the reference set and analyses were done with three different test sets: i) all annotated Salicoid duplicates, ii) all annotated differentially expressed Salicoid duplicates, and iii) all annotated non-differentially expressed Salicoid duplicates.
Additional Information
Data availability: The raw sequencing reads are available in the European Nucleotide Archive (ENA; www.ebi. ac.uk/ena) with the reference number PRJEB10883.
How to cite this article: Harikrishnan, S. L. et al. Sequence and gene expression evolution of paralogous genes in willows. Sci. Rep. 5, 18662; doi: 10.1038/srep18662 (2015).
Supplementary Material
Acknowledgments
Sequencing was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. This research was financed by a Project grant for junior researchers to S.B. from the Swedish Research Council (Project grant 2011-3544) and sequencing was financed by the Swedish Energy Agency.
Footnotes
Author Contributions S.L.H. and P.P. carried out the analyses and S.B. acquired funding, designed the study and drafted the manuscript. All authors read and approved the final manuscript.
References
- Ohno S. Evolution by genome duplication. (George Allen and Unwin, 1970). [Google Scholar]
- Zhang J. Z. Evolution by gene duplication: an update. Trends Ecol Evol 18, 292–298 (2003). [Google Scholar]
- Van de Peer Y., Fawcett J. A., Proost S., Sterck L. & Vandepoele K. The flowering world: a tale of duplications. Trends Plant Sci 14, 680–688 (2009). [DOI] [PubMed] [Google Scholar]
- De Bodt S., Maere S. & Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol 20, 591–597 (2005). [DOI] [PubMed] [Google Scholar]
- Jiao Y. et al. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100 (2011). [DOI] [PubMed] [Google Scholar]
- Vision T. J., Brown D. G. & Tanksley S. D. The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117 (2000). [DOI] [PubMed] [Google Scholar]
- Jaillon O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007). [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000). [DOI] [PubMed] [Google Scholar]
- Tuskan G. A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 (2006). [DOI] [PubMed] [Google Scholar]
- Wolfe K. H. Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet 2, 333–341 (2001). [DOI] [PubMed] [Google Scholar]
- Doyle J. J. et al. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42, 443–461 (2008). [DOI] [PubMed] [Google Scholar]
- Thomas B. C., Pedersen B. & Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res 16, 934–946 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renny-Byfield S. et al. Ancient gene duplicates in Gossypium (cotton) exhibit near-complete expression divergence. Genome Biol Evol 6, 559–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A. et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H. & Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11, 97–108 (2010). [DOI] [PubMed] [Google Scholar]
- Adams K. L., Cronn R., Percifield R. & Wendel J. F. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100, 4649–4654 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K. L., Percifield R. & Wendel J. F. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168, 2217–2226 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggs R. J. et al. Tissue-specific silencing of homoeologs in natural populations of the recent allopolyploid Tragopogon mirus. New Phytol 186, 175–183 (2010). [DOI] [PubMed] [Google Scholar]
- Blanc G. & Wolfe K. H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16, 1679–1691 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., Riddle N. C., Auger D. L. & Veitia R. A. Dosage balance in gene regulation: biological implications. Trends genet 21, 219–226 (2005). [DOI] [PubMed] [Google Scholar]
- Birchler J. A. & Veitia R. A. The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol 186, 54–62 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterck L. et al. EST data suggest that poplar is an ancient polyploid. New Phytol 167, 165–170 (2005). [DOI] [PubMed] [Google Scholar]
- Berlin S., Lagercrantz U., von Arnold S., Öst T. & Rönnberg-Wästljung A. C. High-density linkage mapping and evolution of paralogs and orthologs in Salix and Populus. BMC Genomics 11, 129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchester S. R., Judd W. S. & Handley B. Foliage and fruits of early poplars (Salicaceae: Populus) from the eocene of Utah, Colorado, and Wyoming. Int J Plant Sci 167, 897–908 (2006). [Google Scholar]
- Boucher L. D., Manchester S. R. & Judd W. S. An extinct genus of Salicaceae based on twigs with attached flowers fruits, and foliage from the Eocene Green River Formation of Utah and Colorado, USA. Am J Bot 90, 1389–1399 (2003). [DOI] [PubMed] [Google Scholar]
- Dai X. et al. The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res 24, 1274–1277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J. T. et al. ABySS: A parallel assembler for short read sequence data. Genome Res 19, 1117–1123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G. et al. De novo assembly and analysis of RNA-seq data. Nat Methods 7, 909–912 (2010). [DOI] [PubMed] [Google Scholar]
- Tang S. et al. Analysis of the drought stress-responsive trancriptome of Black Cottonwood (Populus trichocarpa) using deep RNA sequencing. Plant Mol Biol Rep 33, 423–438 (2015). [Google Scholar]
- Sémon M. & Wolfe K. H. Consequences of genome duplication. Curr Opin Genet Dev 17, 505–512 (2007). [DOI] [PubMed] [Google Scholar]
- Rodgers-Melnick E. et al. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res 22, 95–105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin A. et al. The fate of duplicated genes in a polyploid plant genome. Plant J 73, 143–153 (2013). [DOI] [PubMed] [Google Scholar]
- Hughes T. E., Langdale J. A. & Kelly S. The impact of widespread regulatory neofunctionalization on homeolog gene evolution following whole-genome duplication in maize. Genome Res 24, 1348–1355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte J. M. et al. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol 23, 469–478 (2006). [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M. & Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. & Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29, 644–U130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. & Salzberg S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Suyama M., Torrents D. & Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34, W609–612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U. et al. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biology 5, 31 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13, 555–556 (1997). [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz S. et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36, 3420–3435 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.