Abstract
Single-nucleotide polymorphisms within intron 1 of the FTO (fat mass and obesity-associated) gene are associated with enhanced FTO expression, increased body weight, obesity and type 2 diabetes mellitus (T2DM). The N6-methyladenosine (m6A) demethylase FTO plays a pivotal regulatory role for postnatal growth and energy expenditure. The purpose of this review is to provide translational evidence that links milk signaling with FTO-activated transcription of the milk recipient. FTO-dependent demethylation of m6A regulates mRNA splicing required for adipogenesis, increases the stability of mRNAs, and affects microRNA (miRNA) expression and miRNA biosynthesis. FTO senses branched-chain amino acids (BCAAs) and activates the nutrient sensitive kinase mechanistic target of rapamycin complex 1 (mTORC1), which plays a key role in translation. Milk provides abundant BCAAs and glutamine, critical components increasing FTO expression. CpG hypomethylation in the first intron of FTO has recently been associated with T2DM. CpG methylation is generally associated with gene silencing. In contrast, CpG demethylation generally increases transcription. DNA de novo methylation of CpG sites is facilitated by DNA methyltransferases (DNMT) 3A and 3B, whereas DNA maintenance methylation is controlled by DNMT1. MiRNA-29s target all DNMTs and thus reduce DNA CpG methylation. Cow´s milk provides substantial amounts of exosomal miRNA-29s that reach the systemic circulation and target mRNAs of the milk recipient. Via DNMT suppression, milk exosomal miRNA-29s may reduce the magnitude of FTO methylation, thereby epigenetically increasing FTO expression in the milk consumer. High lactation performance with increased milk yield has recently been associated with excessive miRNA-29 expression of dairy cow mammary epithelial cells (DCMECs). Notably, the galactopoietic hormone prolactin upregulates the transcription factor STAT3, which induces miRNA-29 expression. In a retrovirus-like manner milk exosomes may transfer DCMEC-derived miRNA-29s and bovine FTO mRNA to the milk consumer amplifying FTO expression. There is compelling evidence that obesity, T2DM, prostate and breast cancer, and neurodegenerative diseases are all associated with increased FTO expression. Maximization of lactation performance by veterinary medicine with enhanced miRNA-29s and FTO expression associated with increased exosomal miRNA-29 and FTO mRNA transfer to the milk consumer may represent key epigenetic mechanisms promoting FTO/mTORC1-mediated diseases of civilization.
Keywords: Cancer, Diabetes, DNMT, Epigenetics, FTO, Milk, miRNA-29, mTORC1, N6-Methyladenosine, Obesity, Transcriptome
Background
FTO, fat mass- and obesity-associated gene (MIM 612938) maps to chromosome 16q12.2 and is widely expressed in a variety of human tissues with highest levels detected in the brain, pancreatic islets, and the liver [1, 2]. The 505 amino acid-long human FTO and its orthologs are present in vertebrate evolution for at least 450 million years [3]. FTO mRNA is most abundant in hypothalamic nuclei governing energy balance [4, 5]. FTO was identified as an obesity susceptibility gene by several large-scale genome association studies [1, 6, 7]. Single nucleotide polymorphisms (SNPs) in the first intron of FTO are highly associated with obesity and obesity-related traits [1, 6, 7]. In various populations FTO has been confirmed to be a major risk gene promoting obesity [8–19]. Obesity is a well-known risk factor for the development of type 2 diabetes mellitus (T2DM). Indeed, FTO has been identified as a critical T2DM susceptibility locus [20–28]. Obesity and T2DM-associated genetic variations of FTO are associated with increased primary transcript levels of FTO mRNA [14, 29, 30].
Not only genetic polymorphisms of FTO, but also the methylation status of FTO, especially CpG hypomethylation of intron 1 has been linked to increased T2DM prevalence [31]. It is not known whether demethylated CpG loci in intron 1 map directly to regulatory regions and SNPs. Notably, FTO methylation in human pancreatic islets of T2DM patients is significantly reduced compared to healthy controls [32]. Thus, not only genetic but also epigenetic modifications of FTO appear to modify FTO expression. It is well appreciated that dietary factors induce epigenetic alterations, which have pivotal long-term biological consequences [33].
This paper highlights the potential role of milk as an epigenetic modifier of the human genome paying special attention to cow milk-mediated overactivation of FTO and its impact on the transcriptome of the human milk consumer.
Review
FTO regulates fetal and postnatal growth
The FTO gene is widely expressed in both fetal and adult tissues [1]. The mouse mutant Fused toes (Ft) is a dominant trait characterized by partial syndactyly of the forelimbs and massive thymic hyperplasia in heterozygotes [34]. Homozygous Ft/Ft embryos die at midgestation and exhibit absent Fto expression in fibroblasts [35]. Fto-null mice exhibit postnatal growth retardation and a significant reduction in adipose tissue and lean body mass [36]. Mice lacking Fto display postnatal growth retardation with shorter body length, lower body weight, lower bone mineral density, and reduced serum levels of insulin-like growth factor 1 (IGF-1) [37]. Remarkably, specific Fto deletion in the central nervous system (CNS) results in a similar phenotype as whole body Fto deletion pointing to a crucial role of Fto in the CNS to promote postnatal growth [37]. Studies of human cultured skin fibroblasts from subjects with an R316Q mutation that inactivates FTO enzymatic activity showed impaired proliferation and accelerated senescence [2].
Milk is the exclusive nutrient environment provided by mammals promoting postnatal growth during the lactation period [38]. Milk activates the nutrient-sensitive kinase mechanistic target of rapamycin complex 1 (mTORC1), which induces mTORC1-dependent translation [39]. FTO plays a crucial role in mRNA transcription [40], a requirement for mTORC1-dependent translation. Thus, from a mechanistic point of view milk has to interact with both FTO and mTORC1 of the milk recipient.
FTO controls energy homeostasis and protein intake
In mice, overexpression of Fto leads to a dose-dependent increase in body and fat mass, irrespective of whether mice are fed a standard or a high-fat diet [41]. However, mice with increased Fto expression on a high-fat diet develop glucose intolerance [41]. FTO plays a critical role in controlling feeding behavior and energy expenditure [42]. SNPs of FTO have been linked to higher energy intake and increased appetite [40, 43–49]. FTO mRNA is present mainly in sites related to hunger/satiation control [50]. Changes in hypothalamic FTO expression are associated with cues related to energy intake [50]. Fasting induced cytoplasmic Fto expression in some neurons of rat hypothalamus [51], whereas under conditions of nutrient availability Fto is concentrated in nuclear speckles [30]. Interestingly, FTO has been found to mediate circadian rhythms and inhibits CLOCK-BMAL1-induced transcription [52]. The BMI-increasing allele of FTO showed a significant association with higher dietary protein intake [53].
Epigenetic regulation of FTO
The FTO and RPGRIP1L genes are located on the long arm of chromosome 16 and share a CpG island with 51 CpG dinucleotides. The obesity-associated SNPs of FTO are located in intron 1 [30]. FTO methylation has been linked to environmental influences such as dietary factors [29]. It is generally accepted that CpG hypomethylation increases transcriptional activity, whereas DNA CpG methylation is associated with gene silencing [54]. Recent evidence indicates that CpG hypomethylation is generally associated with gene silencing at CpG-island promotors of genes, whereas high levels of intragenic methylation are associated with increased transcription [55, 56]. There is compelling evidence that FTO SNPs of intron 1 are associated with increased levels of FTO expression [14, 29, 30]. Toperoff et al. [57] demonstrated that a CpG site in the first intron of the FTO gene showed small but significant hypomethylation in T2DM patients relative to controls. The same group later confirmed that CpG sites in the first intron of FTO of peripheral blood leukocytes exhibited significant hypomethylation in T2DM cases relative to controls [31]. Furthermore, significant FTO hypomethylation has been confirmed in human pancreatic islets of T2DM patients compared to healthy controls [32]. Importantly, decreased methylation of FTO CpG11 sites have been associated with increased FTO mRNA expression [58]. Thus, preliminary indirect evidence supports the view that hypomethylation of specific CpG sites of the FTO gene enhance FTO expression. Thus, a critical question arises: Do we have evidence that milk, the exclusive and sufficient nutrient environment of a newborn mammal, shape the epigenome and transcriptome of its recipient by increasing FTO expression?
Milk miRNA-29s: potential suppressors of DNA methyltransferases
Secreted exosomal miRNAs represent an important layer of gene regulation and intercellular communication [59–64]. MiRNAs bind through partial sequence homology to the 3′-untranslated region (UTR) of their target mRNAs and cause either translational block or mRNA degradation [64]. Melnik et al. [38] have suggested that milk functions like an “epigenetic transfection system” via transfer of milk-derived exosomes. Of all human body fluids, milk contains the highest amount of RNAs [65]. In fact, mRNA- and miRNA-containing exosomes have been discovered in human milk [66–69], bovine milk [70–74], and milk of other mammals [75–78]. An accumulating body of evidence supports the view that milk miRNAs are bioactive compounds in foods [79]. Whereas a recent study in a mouse model screening for selected mouse milk miRNAs (miRNA-375, miRNA-200c/141) revealed rapid milk miRNA degradation in the intestinal fluid [80], Wolf et al. [81] demonstrated that bovine milk exosomes including miRNA-29b and miRNA-200c are taken up by human intestinal cells via endocytosis depending on exosome and cell surface glycoproteins. It has been confirmed in the suckling wallaby that highly expressed milk miRNAs are detected at signifcant higher levels in the neonate blood serum, confirming that milk miRNAs are absorbed in the gut of the young [78]. Milk is obviously a unique mammalian transfer system of exosomal miRNAs and mRNAs from the mother to her infant [38, 82]. MiRNAs of commercial cow´s milk survive processing such as pasteurization, homogenization and refrigeration for at least 2 weeks [83]. Remarkably, bovine milk exosomes are highly resistant against harsh degrading conditions [72, 77]. Bovine milk exosomes containing miRNAs and mRNAs are taken up by human macrophages [72]. Baier et al. [84] provided evidence that cow’s milk miRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow’s milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. Importantly, bovine milk miRNA-29b, which is identical with human miRNA-29b (http://www.microrna.org), is taken up by blood mononuclear cells of the milk consumer in a dose-dependent manner and modifies mRNA expression such as RUNX2 [84]. These observations imply that milk-derived miRNA-29b may also affect other miRNA-29b targets such as the mRNAs of the DNA demethylase (DNMT) 3A and 3B [85]. Enforced expression of miRNA-29b results in marked reduction of the expression of DNMT1, DNMT3A, and DNMT3B at both mRNA and protein levels.
It has been demonstrated that miRNA-29b indirectly downregulates DNMT1 by targeting Sp1, a transactivator of DNMT1 [86–90]. MiRNA-21, another abundant miRNA type of bovine milk [70, 74] which shares sequence homology with human miRNA-21 (http://www.microrna.org), indirectly downregulates DNMT1 by targeting RASGRP1 [91]. It is noteworthy to mention that murine mammary miRNA-21 expression is controlled by prolactin-induced upregulation of STAT5 [92].
Thus, milk appears to modify the infant’s epigenome via suppressing DNA de novo methylation (DNMT3A and DNMT3B) as well as DNA maintenance methylation (DNMT1) [54]. Physiologically, milk consumption is an early life environmental exposure that may modify epigenetic signatures [93]. DNA methylation plays a critical role in genomic imprinting during preimplantation development [94–100] as well as during early differentiation [101]. Remarkably, early nutrition during the postnatal period apparently influences the adult phenotype via DNA methylation [102–104]. It is conceivable that milk via miRNA-29b/miRNA-21/DNMT signaling promotes CpG demethylation at intron 1 of FTO resulting in increased expression of FTO, which functions as a critical amplifier of the transcriptional machinery for postnatal growth.
Human breast milk also contains and transfers exosomal miRNA-29b and miRNA-21 to the suckling infant [66–69, 105]. At present, no quantitative studies are available which allow a comparison of the amounts of miRNA-29b and miRNA-21 of commercial cow´s milk and human breast milk, respectively.
Essential amino acids increase FTO expression
Milk proteins are a rich nutrient source of branched-chain essential amino acids (BCAAs) and glutamine [106, 107]. In mouse hypothalamic N46 cells, mouse embryonic fibroblasts (MEFs) and in human HEK293 cells, FTO mRNA and protein levels are significantly downregulated by total amino acid deprivation [108]. Remarkably, FTO functions as an amino acid sensor [109] and couples BCAA availability to mTORC1 signaling [110], which plays a crucial role in translation [39]. In contrast, FTO knockdown results in the upregulation of genes involved in cellular response to starvation [30] such as ATG5 and BECN1, which initiate autophagy by inhibiting mTORC1 and mTORC1-dependent translation [111, 112].
The availability of BCAAs plays a fundamental role in mTORC1 activation [113–120]. The rate-controlling and irreversible step of BCAA catabolism is catalyzed by the multienzyme mitochondrial branched-chain α-keto acid dehydrogenase (BCKD) [121]. The dihydrolipoyl transacylase (E2) forms the core of the BCKD complex and is of critical importance for BCKD activity [122]. Notably, miRNA-29b targets the mRNA of dihydrolipoyl transacylase (E2) and thereby inhibits BCKD activity and mitochondrial BCAA catabolism [123]. Thus, milk miRNA-29b enhances the availability of BCAAs, an important requirement for FTO expression [108]. It is conceivable that milk-mediated transfer of BCAAs and miRNA-29b-mediated preservation of BCAAs may enhance the recipient´s FTO- and mTORC1 activity.
It is important to note that human breast milk contains 1.2 g/100 mL milk protein in comparison to cow´s milk that contains 3.5 g protein/100 mL. In comparison to human breast milk, equivalent volumes of cow´s milk transfer three times more BCAAs to the milk consumer [38, 39] and thus may overstimulate BCAA-driven FTO activation.
FTO controls the RNA methylome
FTO belongs to the superfamily of Fe(II) 2-oxoglutarate (2-OG)-dependent dioxygenases [4]. FTO catalyzes the demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA to thymine and uracil [124]. Jia et al. [125–127] recently detected that N6-methyladenosine (m6A) in nuclear RNA is a major substrate of the demethylase FTO. The m6A mark is the most prevalent internal (non-cap) modification present in mRNAs of all higher eukaryotes [128, 129]. The discovery that FTO is an m6A demethylase indicates that this modification is reversible and dynamically regulated, suggesting that reversible RNA methylation may affect gene expression and cell fate decisions by modulating multiple RNA-related cellular pathways, which potentially provide rapid responses to environmental signals such as nutrient and especially milk availability in mammals [129] (Table 1).
Table 1.
Biological impacts of FTO from fetal to adult life
| FTO expression | Biological alterations | References |
|---|---|---|
| Increased placental FTO expression | Increased fetal and birth weight | [226–228] |
| Absent FTO expression (fused toes mutant mice) | Murine embryos die at midgestation | [34, 35] |
| Fto-null mice | Postnatal growth retardation, reduced adipose tissue and lean body mass, shorter body length, lower bone mineral density, lower serum IGF-1 | [36, 37] |
| Fto deletion in murine CNS | Postnatal growth retardation | [37] |
| SNPs with higher FTO expression | Higher energy intake, increased appetite, higher dietary protein intake | [40, 43–49, 53] |
| Hypothalamic FTO expression | Regulation of hunger/satiation, energy intake and circadian rhythm | [50–52] |
| Early hypothalamic FTO over-expression in the rat | Postweaning hyperphagia | [155] |
| Fto over-expression in mice | Increase in body and fat mass, glucose intolerance during high-fat diet, metabolic syndrome | [41, 213] |
| FTO over-expression | Increased gluconeogenesis | [266] |
| FTO over-expression | Increased adipogenesis, reduced thermogenesis of preadipocytes, insulin resistance | [146, 249, 251, 268] |
| FTO deficiency in adipocytes | Increased expression of UCP-1 (thermogenin), induction of BAT phenotype with increased thermogenesis | [252] |
| Obesigenic FTO variants | Increased BMI in children at 8 yrs, higher risk of early menarche at 12 yrs | [234, 244] |
| Obesigenic FTO variants | Increased BMI and obesity in adults | [8–19] |
| Obesigenic FTO variants | Increased risk of T2DM in adults | [20–28] |
| FTO rs9939609 A-allele | Increased risk of coronary heart disease | [214, 215] |
| Obesigenic FTO variants | Increased risk of cancer, especially PCa and BCa | [276, 277, 291–298] |
| Obesigenic FTO SNPs | Reduction in frontal lobe volume of the brain, impaired verbal fluency, increased risk of AD | [319–323] |
| FTO rs9939609 SNP | Reduced leucocyte telomere length, accelerated aging | [342] |
Berulava et al. [30] demonstrated that FTO is highly enriched in nuclear speckles, which serve as for storage and modification of pre-mRNA splicing factors. Proposed functions for m6A modification include mRNA splicing, export, stability, and immune tolerance [130]. FTO overexpression greatly reduced the levels of m6A in cellular RNA underlining that m6A is a major physiologic target of FTO [125–127].
Wang et al. [131] recently demonstrated that m6A is selectively recognized by the human YTH domain family 2 (YTHDF2) protein to promote mRNA degradation. Liu et al. [132] showed that m6A-dependent RNA structural switches regulate RNA–protein interactions. m6A alters the local structure in RNA and long non-coding RNA to facilitate binding of heterogeneous nuclear ribonucleoprotein C (HNRNPC), an abundant nuclear RNA-binding protein responsible for pre-mRNA processing [133–137]. These m6A-switch-regulated HNRNPC-binding activities affect the abundance as well as alternative splicing of target mRNAs, underlining the pivotal regulatory role of m6A-switches on gene expression and RNA maturation [132]. Thus, dynamic m6A modifications are recognized by selective binding proteins, which affect the translation status and lifetime of mRNAs. m6A methylation not only occurs in mRNA but also in ribosomal RNA (rRNA), transfer RNA (tRNA), small nucleolar RNA (snoRNA), long non-coding RNA (lncRNA), and miRNAs [138–142]. Overexpression of FTO in HeLa cells reduced the level of m6A in purified poly (A) RNA by 18 %, whereas FTO knockdown increased m6A levels by 23 % in poly (A) mRNA [138, 143]. Adenosine methyltransferases (‘writers’), m6A demethylating enzymes (‘erasers’) such as FTO and ALKBH5 and m6A-binding proteins (‘readers’) thus define cellular pathways for the post-transcriptional regulation of mRNAs. The m6A methylation pattern thus constitutes the mRNA ‘epitranscriptome’ [138, 143].
Furthermore, the m6A status modifies the interaction of miRNAs with their target mRNAs. A significant portion of m6A occurs in close vicinity to the 3´UTRs of mRNA transcripts, which correlate with their miRNA binding sites [138]. Knockdown of FTO affects the steady state levels of several miRNAs pointing to a further layer of FTO-dependent posttranscriptional regulation of gene expression [142]. As miRNAs inhibit their target mRNAs, decreased expression of FTO suppresses global mRNA transcription, whereas overexpression of FTO enhances transcriptional activity. Moreover, m6A marks act as a key post-transcriptional modification initiating miRNA biogenesis [144].
Milk, lactation’s nutrient system operating during the postnatal growth period between mother and infant may shape the infants epitranscriptome via RNA m6A demethylation to increase transcription, required for postnatal growth, body mass, protein and fat mass accretion.
FTO regulates adipogenesis-related RUNX1T1 mRNA splicing
Recent evidence links FTO overexpression to enhanced expression of the pro-adipogenic short isoform of the transcription factor RUNX1T1. FTO controls mRNA splicing by regulating the ability of the splicing factor SRSF2 to bind to mRNA in an m6A-dependent manner [145]. The pro-adipogenic short isoform of RUNX1T1 stimulates mitotic clonal expansion (MCE) of MEFs and thus enhances adipocyte numbers [146]. In contrast, mRNA m6A methylation downregulates adipogenesis in porcine adipocytes [147]. Thus, FTO negatively regulates m6A levels and positively regulates adipogenesis, while methyltransferase-like 3 (METTL3), which catalyzes the formation of m6A in RNA [148], positively correlates with m6A levels and suppresses adipogenesis [147]. In accordance, long-term feeding of commercial pasteurized cow´s milk to young mice increased body weight, caloric intake, and epididymal fat mass as compared to milk-free controls [149].
FTO controls appetite via ghrelin mRNA demethylation
The “hunger hormone” ghrelin functions as a neuropeptide in the CNS and regulates energy homeostasis [150, 151]. Ghrelin increases appetite by triggering receptors in the arcuate nucleus [152]. The ghrelin receptor is expressed in many brain areas important for feeding control, including hypothalamic nuclei involved in energy balance regulation and reward-linked areas such as the ventral tegmental area (VTA) [153]. It has recently been recognized that ghrelin signaling at the level of the mesolimbic system is one of the key molecular substrates that provides a physiological signal connecting gut and reward pathways [153]. In some way, milk intake should trigger ghrelin signaling enhancing appetite and reward pathways to secure postnatal food intake for mammalian development.
Karra et al. [154] found that subjects homozygous for the FTO “obesity-risk” rs9939609 A allele have dysregulated circulating levels of acyl-ghrelin and attenuated postprandial appetite reduction. FTO overexpression reduced ghrelin mRNA m6A methylation, concomitantly increasing ghrelin mRNA and peptide levels. Peripheral blood cells from homozygous (AA) subjects for the FTO “obesity-risk” rs9939609 variant exhibited increased FTO mRNA, reduced ghrelin m6A mRNA, and increased ghrelin mRNA abundance [154].
At weaning, hypothalamic FTO mRNA expression was increased significantly in the offspring of obese female Sprague–Dawley rats and FTO was correlated with both visceral and epididymal fat mass [155]. In these rats early hypothalamic FTO overexpression contributes to postweaning hyperphagia [155]. Among carriers of the risk allele of the FTO SNP rs9939609 an association was found between BMI growth and the duration of exclusive breastfeeding (EXBF). In girls, EXBF interacts with the SNP at baseline and can reverse the increase in BMI. In boys, EXBF reduces BMI both in carriers and non-carriers of the risk allele with an association found after 10 years of age. Six months of EXBF will put the boys’ BMI growth curves back to the normal range [156]. Thus, human breast milk appears to control the appropriate postnatal magnitude of FTO expression and activity.
FTO couples leucyl-tRNA synthase to mTORC1
FTO expression is of pivotal importance for mTORC1 signaling and represents the critical mechanistic link between transcription and translation. MEFs of FTO-deficient mice (Fto−/− mice) exhibit slower growth rates and reduced mRNA translation compared with wild-type MEFs [110]. Postnatal growth retardation and significant reduction in adipose tissue and lean body mass has been observed in Fto−/− mice [36]. Severe growth retardation has also been observed in humans with homozygous FTO loss-of-function mutations [2]. During tRNA charging with amino acids amino-acyl-tRNA synthetases (AARS) work together as part of the multi-synthetase complex (MSC), which is essential for mRNA translation [157–159]. In higher eukaryotic systems, several different AARSs including leucyl-tRNA synthetase form a macromolecular protein complex with three nonenzymatic cofactors AIMP1/p43, AIMP2/p38, and AIMP3/p18 [159]. Notably, Fto−/− MEFs exhibit reduced protein levels of MSC components [110]. A key role for mTORC1 activation plays leucyl-tRNA synthase (LRS) [160, 161]. LRS has been proposed to function as an intracellular amino acid sensor [161]. In yeast, LRS Cdc60 interacts with the Rag GTPase Gtr1 of the EGOC in a leucine-dependent manner [160]. This interaction is necessary and sufficient to mediate leucine signaling to TORC1 and is disrupted by the engagement of Cdc60 in editing mischarged tRNA(Leu) [160]. Han et al. [161] demonstrated that LRS directly binds to Rag GTPase, the mediator of amino acid signaling to mTORC1, in an amino acid-dependent manner and functions as a GTPase-activating protein (GAP) for Rag GTPase to activate mTORC1. Importantly, FTO-deficient MEFs exhibit reduced LRS protein expression [110].
It is tempting to speculate that FTO-mediated demethylation of m6A in leucyl-tRNA may modify LRS regulatory functions. Furthermore, Lo et al. [162] detected splice variants of LRS, which differ in catalytic activity. It is possible that LRS mRNA splice variants modify the affinity for leucine binding, thereby change the functional interaction of LRS with Rag GTPase activating mTORC1. This mRNA-splicing dependent mechanism may resemble the generation RUNX1T1 short splice variants observed in the regulation of adipogenesis [145, 146] (Table 2).
Table 2.
Biological functions of the m6A demethylase FTO
| Epigenetic FTO functions | Transcriptional effects | References |
|---|---|---|
| FTO-catalyzed demethylation of m6A in mRNAs |
Increased transcription and mRNA stability, modification of m6A-dependent alternative splicing and miRNA binding to target mRNAs | [125–127, 138–143] |
| FTO over-expression | Generation of the pro-adipogenic short isoform of RUNX1T1 promoting MCE increasing adipocyte numbers | [145, 146] |
| Increased FTO mRNA | Reduction of ghrelin mRNA m6A methylation resulting in increased ghrelin mRNA abundance | [154] |
| FTO-deficient MEFs | Reduced protein expression of the mTORC1 activator leucyl-tRNA synthetase | [110] |
| Increased FTO expression | Association of FTO CpG hypomethylation in T2DM with recuded m6A levels in mRNA of T2DM patients | [31, 32, 265] |
| FTO over-expression | Increased expression of C/EBPβ mRNA, the key transcription factor of gluconeogenesis and adipogenesis | [266, 267] |
| FTO over-expression | Loss of stem cell self-renewal capability | [207] |
| FTO over-expression | Increased expression of PRL mRNA enhancing PIP expression involved in PC and BC progression | [185–187] |
| FTO over-expression | Interaction with APO ϵ4, increasing the risk of AD | [321] |
Dairy cattle FTO mutations increase milk yield
mRNA abundance of FTO in mammary epithelial cells has been shown during lactation of the rabbit [163]. Recently, genetic variations of bovine FTO have been identified that increase milk fat and protein yield in German Holstein dairy cattle [164]. Five SNPs and two haplotype blocks in a 725 kb region covering FTO and the neighboring genes RPGRIP1L, U6ATAC, and 5 S rRNA were associated with milk fat and protein yield [164]. Furthermore, Sorbolini et al. [165] confirmed that FTO belongs to the selection signatures in Piemontese and Marchigiana cattle known to affect productive traits.
Physiologically cattle digest grass, whereas high performance dairy cows are fed with soybean and grain concentrates that provide higher amounts of BCAAs promoting FTO expression and FTO-driven transcription increasing lactation performance. Thus, increased expression of FTO in DCMECs increases milk yield.
Is bovine FTO mRNA retroconverted into the human genome?
The human FTO gene shares 86.8 % amino acid sequence homology with bovine FTO [3]. It is thus conceivable that upregulated bovine FTO mRNA of high performance DCMECs may reach the human milk consumer via intake of mRNA containing milk exosomes. RNA-dependent DNA polymerase activity (reverse transcriptase, RT) has been detected in the milk of several mammalian species including humans [166–173]. Importantly, milk exosomes contain both mRNA and RT [174]. Around 42 % of the human genome is made up of retrotransposons, which operate via RNA intermediates [175]. Recently, Irmak et al. [176] provided translational evidence for the hypothesis that milk exosomes function as an RNA-based gene delivery system between mother and infant. This implies that bovine milk exosomes via transfer of RT and mRNA—such as bovine FTO mRNA—may affect the composition and function of retrotransposons of the human milk consumer. The integration of milk-derived bovine FTO mRNA into the human genome via retrotransposition may be a further mechanism amplifying FTO gene expression [177] (Fig. 1). Most transposable elements are silenced by CpG methylation. Potential milk-miRNA-29/DNMT-mediated changes of the epigenetic state of transposable elements may affect regions encompassing neighboring genes [102]. Evidence has been presented that transposable elements are targets for early nutritional effects on epigenetic gene regulation [102].
Fig. 1.
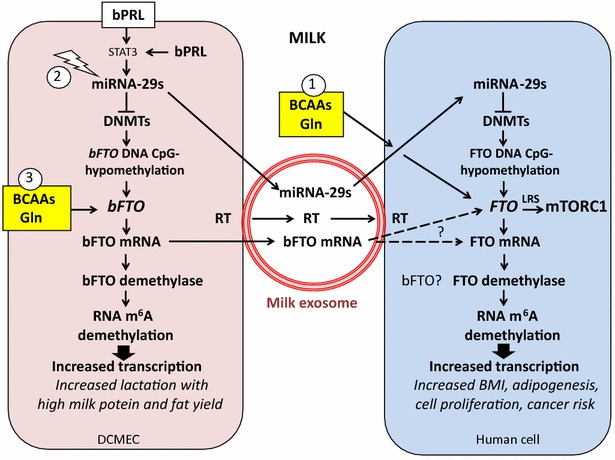
Working model presenting milk-mediated epigenetic activation of FTO-driven transcription. Milk functions in analogy to a retroviral infection via exosome transfer to the cells of the milk recipient. Milk exosomes transport bioactive miRNA-29s that reduce the expression of DNA methyltransferases (DNMTs) resulting in epigenetic activation of human FTO via DNA hypomethylation. Bovine milk exosomes apparently transfer bovine FTO mRNA and reverse transcriptase (RT) that may result in bFTO mRNA retroconversion into the human genome. (1) Milk provides abundant amounts of branched-chain amino acids (BCAAs) and glutamine (Gln) that activate FTO and mTORC1. (2) Bovine milk production is enhanced by increased expression of miRNA-29s and certain bFTO gene polymorphisms. (3) Dairy cow feeding with BCAA-enriched concentrated feedingstuffs upregulates bFTO mRNA and protein expression. Efforts of veterinary medicine intended to maximize milk yield associated with increased dairy cow mammary epithelial cell (DCMEC) transcription have an epigenetic impact on FTO expression of the human milk consumer. FTO, the critical m6A demethylase, upregulates transcription, adipogenesis, and gluconeogenesis promoting postnatal growth
Bovine miRNA-29 increases lactation performance
MiRNA-29a and miRNA-29b have been identified as important miRNAs involved in the regulation of lactation [178, 179]. Bian et al. [180] investigated the roles of miRNA-29s in epigenetic regulation of DCMECs. They showed that miRNA-29s regulate the DNA methylation level by inversely targeting both DNMT3A and DNMT3B in DCMECs. MiRNA-29 s stimulate lactation via decreasing the promoter methylation of CSN1S1 (casein-α s1), EIF5 (E74-like factor 5), PPARγ (peroxisome proliferator-activated receptor-γ), SREBP1 (sterol regulatory element binding protein-1) and GLUT1 (glucose transporter 1) and increase triglyceride biosynthesis [180]. In contrast, inhibition of miRNA-29s causes global DNA hypermethylation and increases the methylation levels of the promoters of important lactation-related genes CSN1S1, ElF5, PPARγ, SREBP1 and GLUT1 and reduces the secretion of lactoprotein, triglycerides and lactose by DCMECs [180]. It is thus conceivable that high expression of bovine miRNA-29b via inhibition of DNMT3A and DNMT3B in high performance dairy cows also decreases bovine FTO methylation thereby increasing FTO-dependent transcription. Furthermore, selecting cows with high miRNA-29 expression may increase the miRNA-29 content of commercial milk exosomes. Thus, milk enriched in miRNA-29b and bovine FTO mRNA may epigenetically enhance the FTO expression of the milk consumer (Fig. 1).
FTO catalyzes prolactin mRNA demethylation
Prolactin (PRL) is the central hormone in female mammals to produce milk [181, 182].
PRL is essential for maintaining lactation and is galactopoietic in dairy cows [183, 184]. m6A residues in an intron-specific region of bovine PRL pre-mRNA have been identified [185–187]. Treatment of CHO cells with the methylation inhibitor neplanocin resulted in a 4–6fold increase in nuclear bPRL precursor compared to control cells [186]. FTO-mediated upregulation of bovine PRL expression will enhance the transcription factors STAT3 and STAT5 in DCMECs [188]. Notably, STAT3 promotes the expression of miRNA-29 [189, 190]. In a vicious cycle, miRNA-29-mediated suppression of DNMTs may epigenetically further enhance bovine FTO expression via FTO CpG demethylation augmenting lactation. Remarkably, PRL induces the expression of prolactin-inducible protein (PIP), a component of milk that is widely expressed in breast cancer (BC) and prostate cancer (PC) promoting cancer cell growth and proliferation [191–194]. Bovine milk miRNA-29b has been demonstrated to enhance the expression of the osteogenic transcription factor RUNX2 [84], which interacts synergistically with androgen receptor to enhance PIP expression [195]. Thus, FTO-mediated activation of PRL expression may stimulate oncogenic signaling via increased PIP expression.
Bacterial and viral infections upregulate miRNA-29
Mastitis, the inflammation of mammary glands resulting from bacterial infection, is a common problem in milk production of dairy cattle. Lipopolysaccharide (LPS) injection into murine lactating mammary glands increased the expression of STAT3 [196], promoting miRNA-29 expression [189, 190].
Viral infections such as papilloma virus infections as well enhance miRNA-29 expression via STAT3 signaling [189, 190, 196–198]. It is of most critical concern that oncogenic bovine viruses (e.g. polyoma-, papilloma- or single-stranded DNA viruses) contaminate milk and dairy products [199–201]. Thus, unnoticed infection of dairy cattle may promote epigenetic activation of human FTO via miRNA-29-mediated suppression of DNMTs. Vice versa, overactivated FTO-mediated demethylation of single-stranded DNA and RNA may modify oncogenic viral transcription and mRNA splicing [4, 124, 202, 203]. Furthermore, the transfer of milk exsomes, which resembles a retrovirus-like infection, may promote the spreading of oncogenic bovine viruses.
FTO-driven diseases of civilization
m6A has been identified as a conserved epitranscriptomic modification of eukaryotic mRNAs. Deficiency of m6A formation has been proven to affect circadian rhythm, cell meiosis, embryonic stem cell proliferation, and thus is implicated in obesity, cancer and other human diseases [130, 138, 141, 203–213]. FTO is a relevant factor for the development of obesity and the metabolic syndrome in mice [203]. As observed in Swedish men and women, FTO rs9939609 A-allele carriers have an increased risk of coronary heart disease (CHD) [214, 215]. There is evidence in humans that milk consumption correlates with the risk of CHD [216, 217]. It is thus conceivable that increased FTO expression from fetal to adult life promotes diseases of civilization.
Fetal macrosomia and increased birth weight
Fetal overgrowth and increased birth weight are risk factors for the development of diseases of civilization such as obesity [218–226]. Sébert et al. [227] identified FTO as a critical factor for fetal programming of obesity-related disorders. The authors found a significant relation between placental FTO gene expression and fetal weight at 110 days gestation [227]. In humans, FTO is highly expressed in the placenta and is associated with increased fetal weight and length [228]. Bassols et al. [228] suggested that FTO controls important genes related to fetal growth. Liu et al. [58] investigated placental FTO expression in relation to the promoter methylation of FTO in a Chinese population. Intriguingly, the methylation rates of CpG11 sites were significantly decreased in high birth weight newborns [58]. The investigators concluded that high placental FTO expression is associated with increased birth weight [58]. Milk exosome-mediated transfer of bovine miRNA-29 and miRNA-21 may via downregulation of DNMTs reduce the methylation of critical CpG sites of the FTO promoter explaining milk’s function as an epigenetic enhancer of FTO expression. The Generation R study observed the association of increased fetal and infant growth with an increased risk of obesity during early childhood [229]. A higher peak weight velocity, which generally occurs in the first month after birth, was associated with an increased risk of overweight and obesity at 4 years of age [229]. Accumulating evidence underlines that milk consumption increases placental, fetal and birth weight [230–232].
Increased growth trajectories in childhood
The A allele of the FTO rs9939609 SNP is associated with a high BMI from 5.5 years onwards [233]. Established obesity loci including FTO affect the level and the rate of change in BMI at 8 years in children [234]. The National Health and Nutrition Examination Survey (NHANES) provided evidence that milk consumption in children increases BMI [235]. Among children of 5–10 years of age, those in the highest quartile for milk intake had higher BMI [236]. Notably, milk had more consistent positive associations with BMI than any other dairy product, and these are strongest among children of 2–4 years of age [236].
Increased linear growth
FTO is highly expressed in the hypothalamus and pituitary gland [1, 2, 236]. FTO controls the somatotropic (growth hormone-IGF-1) axis [37, 236]. Fto-deleted mice exhibit reduced serum levels of IGF-1 [37]. Adult obesity susceptibility variants including FTO conferred a faster tempo of height growth that was evident before puberty [237]. Remarkably, milk consumption is known to shift the prepubertal somatotropic axis [238] and increases growth hormone (GH) and total serum IGF-1 levels [239, 240]. It is widely accepted that cow´s milk consumption accelerates linear growth in children [241]. Thus, milk consumption may increase the somatotropic axis in an FTO-dependent manner.
Early menarche and diabetes risk
By using a BMI-increasing-allele-score including FTO it has been demonstrated that each 1 kg/m2 increase in childhood BMI was predicted to result in a 6.5 % higher absolute risk of early menarche before age 12 years [242]. These findings support a causal effect of BMI on early menarche [242]. It is of considerable concern that early menarche is associated with an increased risk of T2DM and obesity [243, 244]. In fact, NHANES [245] provided evidence that higher milk intake of children is associated with an increased risk of early menarche. However, milk consumption after the age of 9 years did not predict the age at menarche [246], which points to a sensitive window affecting BMI-menarche interactions during early childhood.
Obesity
There is compelling evidence that FTO is one of the world´s major risk genes increasing BMI and promoting obesity [8–19]. Notably, carriers of the FTO risk allele rs8050136 have an increased risk of CHD mediated by BMI [247]. FTO deficiency in mice led to reduction in adipose tissue [213]. The association between FTO and fat mass in humans develops by the postnatal age of 2 years [248]. Striking evidence underlines that FTO-mediated mRNA demethylation regulates mRNA alternative splicing in the control of mitotic clonal expansion (MCE) resulting in adipogenesis [146, 249]. Zhang et al. [250] confirmed that the demethylase activity of FTO is required for 3T3 L1 preadipocyte differentiation. The level of m6A is decreased in cells overexpressing FTO [250].
Furthermore, the obesity-associated FTO allele represses mitochondrial thermogenesis in adipocyte precursor cells in a tissue-autonomous manner associated with a shift of energy-dissipating beige adipocytes to energy-storing white adipocytes [251]. FTO-deficient adipocytes exhibit a reduced de novo lipogenesis and fourfold higher expression of uncoupling protein 1 (UCP1, thermogenin) mRNA and protein compared with control cells. The upregulation of UCP1 in FTO-deficient adipocytes enhances mitochondrial uncoupling of the respiratory chain, allowing for fast substrate oxidation with a low rate of ATP production [252]. Thus, FTO is not only involved in adipocyte differentiation but also inhibits browning, the development of brown adipose tissue (BAT). BAT was believed to show rapid involution in early childhood, leaving only vestigial amounts in adults. However, recent evidence suggests that its expression in adults is far more common than previously appreciated [253]. Rockstroh et al. [254] detected BAT-like and UCP1-positive adipocytes in 10.3 % of 87 lean children (aged 0.3–10.7 years) and in one overweight infant, whereas they did not find brown adipocytes in obese children or adults. It is conceivable, that during the period of lactation milk-derived FTO signaling reduces UCP1 expression in BAT and thus reduces BAT-driven thermogenesis, a requirement for early postnatal survival. Milk (FTO)-mediated suppression of UCP1 would allow the generation of higher levels of ATP required for biosynthetic pathways during postnatal growth.
Milk provides abundant amounts of BCCAs and glutamine required for FTO expression [108]. According to a recent epidemiological study with 177,330 individuals a positive association between the BMI-increasing allele of FTO variant and higher dietary protein intake has been observed supporting the link between FTO and adiposity and dietary protein intake [53].
Milk via transfer of abundant BCAAs and exosomal miRNA-29 may thus activate FTO expression promoting adipogenic transcription (Fig. 1). In fact, feeding commercial pasteurized cow’s milk increased BMI and fat mass in mice [149]. There is further evidence that milk consumption in children, adolescents and adults increases BMI and obesity [235, 255–258].
Type 2 diabetes mellitus
FTO has been confirmed as an important diabetes susceptibility locus [20–28] associated with increased primary transcript levels of FTO mRNA [14, 30]. Patients with the FTO risk allele showed significantly higher serum insulin concentrations and HOMA-IR compared with subjects without the risk allele [259]. Bell et al. [260] were the first who identified variant-CpG restricted haplotype-specific methylation within the FTO T2DM and obesity susceptibility locus tagged by SNP rs8050136. Several studies identified increased expression of the miRNA-29 family as a biomarker for T1DM and T2DM [261–264]. MiRNA-29s via DNMT suppression may decrease the CpG methylation level of FTO in pancreatic β-cells. In fact, recent evidence underlines that increased FTO expression is related to CpG hypomethylation of FTO [31, 32]. Toperoff et al. [31] found that DNA methylation of a specified regulatory site in peripheral blood leukocytes (PBLs) is associated with impaired glucose metabolism and T2DM. Dayeh et al. [32] identified 1649 CpG sites and 853 genes, including FTO with differential DNA methylation in T2DM islets. FTO methylation in human pancreatic β-cells of non-diabetic patients differed significantly from FTO methylation of T2DM patients, respectively [32]. Modest differences in DNA methylation of individual CpG sites may exert big effects on gene expression over long periods of time [31, 32]. As DNA hypomethylation commonly increases gene activity [54], it is conceivable that CpG hypomethylation of FTO in T2DM pancreatic β-cells may decrease m6A methylation of mRNAs. Indeed, Shen et al. [265] demonstrated that the m6A contents in the RNA from T2DM patients and diabetic rats were significantly lower compared with controls. The mRNA expression level of FTO was significantly higher in T2DM patients than that of the controls and was associated with the risk of T2DM. Moreover, the m6A contents were inversely correlated with FTO mRNA expression [265].
There is recent evidence that FTO overexpression is also involved gluconeogenesis associated with upregulation of mRNA expression of the rate-limiting gluconeogenic enzymes glucose-6-phosphatase (G6P) and mitochondrial phosphoenolpyruvate carboxykinase (PEPCK) [266]. FTO overexpression resulted in increased mRNA and protein expression of CCAAT/enhancer-binding protein-β (C/EBPβ), which promotes G6P and PEPCK expression [266]. Notably, C/EBPβ activates expression of C/EBPα and PPAR-γ, which further drives the adipocyte phenotype [267] after FTO-driven mitotic clonal expansion (MCE) of adipocytes [146, 249, 250]. Thus, FTO counteracts the activity of metformin, which inhibits gluconeogenesis and adipogenesis, whereas FTO overexpression upregulates gluconeogenesis and adipogenesis.
Interestingly, FTO mRNA levels were increased in subcutaneous adipose tissue (SAT) of T2DM patients, and treatment with Rosiglitazone improved insulin sensitivity and reduced SAT FTO mRNA levels [268]. SAT FTO mRNA and protein levels were increased in insulin resistant women (high HOMA) compared to insulin sensitive women (low HOMA) [268]. Furthermore, FTO expression was transiently increased in early 3T3-L1 adipocyte differentiation, which coincides with the induction of PPARγ2 expression [268]. There is compelling evidence that omental adipose tissue (OAT) FTO expression is associated with adiposity, whereas SAT FTO expression is associated with insulin resistance [268].
Remarkably, the EPIC-InterAct Study (n = 340,234) provided evidence for an increased T2DM risk in association with milk consumption in contrast to other dairy products [269]. Obviously, it is a fundamental function of milk to increase FTO-dependent β-cell mRNA transcription and insulin production, which drive mTORC1-dependent translation [39]. However, persistent milk-mediated activation of β-cell transcription and translation will induce premature aging and β-cell endoplasmic reticulum (ER) stress promoting early onset of T2DM [264]. Notably, high intakes of milk, but not meat, increase serum insulin levels and insulin resistance in 8-year-old boys [270]. Early insulin resistance is commonly treated with the anti-diabetic drug metformin, which is a multifunctional inhibitor of mTORC1 [271]. Remarkably, metformin has recently been shown to induce DNA methylation [272], thus may directly counteract milk-mediated epigenetic upregulation of FTO expression.
Cancer
FTO polymorphisms can regulate the expression of genes at large kilobases of distance as well as the expression of FTO itself, and regions for transcription factor binding. To date it has been observed that variants rs9939609, rs17817449, rs8050136, rs1477196, rs6499640, rs16953002, rs11075995 and rs1121980 are associated with the risk of developing cancer [273].
There is recent interest in the role of m6A mRNA methylation in stem cell and cancer stem cell homeostasis [204–207, 212]. m6A RNA methylation is required to maintain mouse embryonic stem cells in their ground state [207]. Loss of m6A methylation is associated with a loss of self-renewal capability [207]. m6A RNA methylation is regulated by miRNAs and promotes reprogramming to pluripotency [209].
Long interspersed element 1 (L1) retrotransposon mobilization occurs exclusively in cancers of epithelial origin [274]. According to a recent hypothesis L1 promoter hypomethylation is correlated with epithelial to mesenchymal transition (EMT) promoting metastasis. Furthermore, human L1 retrotransposition, which is dependent on RNA intermediates, is associated with genetic instability [275]. Milk-derived miRNA-29s via DNMT suppression may be involved in L1 hypomethylation and L1 activation promoting cancer initiation and progression.
Prostate cancer
Prostate cancer (PC) is the most common cancer of men in Western societies. The rs9939609 A allele, which was associated with higher BMI, was positively associated with high-grade PC [276]. Machiela et al. [277] identified 10 T2DM markers including FTO that were associated with increased risk for PC. This points to a common genetic or epigenetic basis of T2DM and PC. In fact, an association of T2DM and PC has recently been confirmed in American Indians [278]. During a mean follow-up of 8.5 years, 2446 men of 129,502 participants of the EPIC study developed PC. Waist circumference and waist-hip ratio were positively associated with risk of advanced disease [279].
There is compelling evidence that total dairy protein intake is related to PC risk [280]. According to the EPIC (n = 142,251) daily intake of 35 g dairy protein increased PC risk by 32 % [280]. The Physicians’ Health study provided evidence that only whole milk was consistently associated with higher incidence of fatal PC in the entire cohort and higher PC-specific mortality among cases [281]. In fact, feeding reconstituted milk protein powder but not whole milk in two mouse models of benign and neoplastic lesions did not promote PC progression [282]. Notably, daily milk consumption in adolescence (vs. less than daily), but not in midlife or currently, was associated with a 3.2-fold risk of advanced PC in the population-based Iceland cohort of 8894 men [283]. Thus, there is good reason to suggest that milk-mediated upregulation of FTO increases transcriptional activity driving an mRNA landscape for the development and progression of PC during childhood and adolescence. Moreover, milk-mediated epigenetic activation of FTO may be linked to FTO-driven activation of mTORC1 [110], which is closely involved in the pathogenesis of PC [284, 285]. Indeed, cow’s milk stimulated the growth of LNCaP cells producing an average increase in cancer cell growth rate of over 30 % [286]. Furthermore, FTO-mediated upregulation of PRL may enhance oncogenic PIP expression [192, 193]. There is accumulating evidence for the beneficial effect of metformin in the treatment and prevention of PC, especially in patients with T2DM [287–290]. Metformin´s anti-cancer effects may be traced back to its capability to methylate DNA [272], which may attenuate FTO-PRL-PIP well as FTO-mTORC1 signaling.
Breast cancer
Breast cancer (BC) is the most common cancer of women in Western societies and is often associated with obesity [291]. A recent epigenome-wide association study reveals decreased average methylation levels years before BC diagnosis [292].
Genome-wide SNP studies strongly suggest that the FTO locus is associated with estrogen receptor (ER)-negative BC [293]. This signal is tagged by rs11075995, located in a 40 kb LD block in intron 1 of FTO, within an enhancer region that appears to be active in both normal and triple-negative BC cells. da Cunha et al. [294] observed a 4.59-fold increased risk for women who have the allele combination FTO rs1121980/FTO rs9939609/MC4R rs17782313 indicating an interaction between FTO and MC4R polymorphisms in BC development. FTO rs16953002 AA genotype conferred significant increased BC risk compared to GG genotype in a Chinese population [295]. In another Chinese study 5 susceptibility loci including FTO correlated with BC [296]. Of 41 recently discovered BC susceptibility variants, associations were found between rs1432679 (EBF1), rs17817449 (MIR1972-2: FTO), rs12710696 (2p24.1), and rs3757318 (ESR1) and adjusted absolute and percent mammographic dense areas, respectively [297]. Singh et al. presented a BC cell model featuring an embryo-like gene expression with amplification of FTO [298]. Thus, substantial evidence links FTO to pathogenesis of BC or subtypes of BC.
From 1916 to 1975, BC risk increased 2.7-times in Norway and has been associated with changes of life style factors after World War II including milk intake [299]. In fact, women consuming 0.75 L or more of full-fat milk daily had a relative risk of 2.91 compared with those who consumed 0.15 L or less [300]. Remarkably, consumption of commercial whole and non-fat milk increased the incidence of 7,12-dimethylbenz(a)anthracene (DMBA)-induced mammary tumors in rats [301]. Tumor numbers, volume and incidence doubled after 20 weeks of milk consumption in these DMBA-induced mammary tumors [301]. Notably, PRL binding to DMBA-induced mammary tumors was three times higher than that observed in lactating mammary glands of the rat [302, 303]. Further administration of PRL enhanced tumor growth [302, 303], whereas pharmacological suppression of PRL secretion inhibited DMBA-induced mammary carcinogenesis in the rat [304]. It is of critical concern that large prospective epidemiological studies show correlations between circulating levels of PRL with an increased risk of ER-positive invasive BC [305].
Breastfeeding apparently meets the appropriate species-specific signaling axis of FTO- and mTORC1-mediated postnatal programming. Women, who had been breast-fed have a reduced lifetime risk of developing BC [306]. Breastfeeding reduces the risk of developing BC [307], specifically of triple-negative BC [308].
A large Swedish cohort study demonstrated that people with lactose intolerance and low consumption of milk and other dairy products had a decreased risk of BC, lung, and ovarian cancers [309]. These data imply that cow´s milk consumption increases transcription, translation and cell proliferation in BC. Notably, there is preliminary evidence that metformin treatment reduces BC recurrence in T2DM patients [310, 311]. Metformin-induced DNA methylation [272] may reduce FTO-PRL signaling that increases PIP expression in BC [192, 193]. Remarkably, PIP regulates proliferation of luminal A type BC cells in an ER-independent manner [312], supporting the role of FTO in ER-negative BC [293].
Neurodegenerative diseases
Environmental factors such as diet contribute significantly to risk of Alzheimer’s disease (AD) and Parkinson’s disease (PD) [313]. Accumulating evidence points to the important role of dietary epigenetic regulation in the pathophysiology of AD [314–316]. AD and PD are both tauopathies. mTORC1 induces abnormally hyperphosphorylated tau proteins, which aggregate resulting in compromised microtubule stability [317]. mTORC1 is involved in regulating tau distribution in subcellular organelles and in the initiation of tau secretion from cells to extracellular space [318]. Notably, FTO via LRS plays a crucial role for mTORC1 activation [110].
Carriers of common FTO polymorphisms rs9939609 A allele exhibit a reduction in frontal lobe volume of the brain and an impaired verbal fluency performance [319, 320]. A population-based study from Sweden found that carriers of the FTO rs9939609 A allele have an increased risk for incident AD [320]. Furthermore, an interaction between FTO and APOE was found, with increased risk for dementia for those carrying both FTO AA and APOE ϵ4 [321]. Genetic variation in introns 1 and 2 of the FTO gene may contribute to AD risk [322]. Remarkably, impaired satiation and increased feeding behavior has been reported in the triple-transgenic AD mouse model [323], which may point to increased FTO expression.
A dietary pattern associated with a lower AD risk was characterized by higher intakes of salads, nuts, fish, tomatoes, poultry, cruciferous vegetables, fruits, and dark and green leafy vegetables and a lower intake of high-fat dairy products, red meat, organ meat, and butter [324].
Overweight is more prevalent in PD patients [325, 326]. In early PD, weight gain was revealed over 3 years accompanied by an increase in fat mass and waist circumference [327]. Whereas FTO gene polymorphisms have not yet been studied in PD patients, several studies show an association between milk consumption and the risk of PD [328–333]. DNA methylation plays a pivotal role in the pathogenesis of age-related neurodegeneration and cognitive defects [334]. Milk-miRNA-29-mediated suppression of DNA methylation and FTO-enhanced mRNA m6A demethylation via persistent milk consumption may thus play an important role in the pathogenesis of neurodegenerative diseases.
Future prospects
The continuous interaction between the individual´s genetic makeup and environmental factors such as early nutrition result in a spectrum of states ranging from healthy aging to age-related diseases of civilization [102–104, 335]. Milk is a highly specialized nutrient and signaling system of mammalian evolution that apparently shapes the epitranscriptome of the milk recipient via FTO-mediated modifications of RNA nucleotides. Future research should unravel milk´s biological impact on the recently recognized dual axis of coordinated regulation between the genome and the epigenome and the transcriptome and the epitranscriptome, respectively [336]. More scientific insights into milk’s epigenetic signature, which links the maternal lactation genome to the infant’s epitranscriptome as well as the impact of pasteurized cow’s milk on the epitranscriptome of the milk consumer will allow a deeper understanding of milk-mediated postnatal programming as well as the pathogenesis of Western diseases of civilization.
Conclusions
Early-life experiences play a critical role for lifelong metabolic programming. Current research focuses on the role of epigenetic causes of excess adult weight gain and early onset of age-related diseases [337]. Importantly, the FTO gene has been recognized to play a crucial role in the early-life determination of body weight, body composition and energy balance [337]. Milk is the early nutritional environment and life experience of all mammals. There is increasing evidence that milk is not “just food” but represents a sophisticated signaling system of mammals to promote anabolism for postnatal mTORC1-mediated growth [38]. It has recently been demonstrated that milk stimulates mTORC1-dependent translation [39], a pivotal requirement for cell growth and proliferation. There is overwhelming evidence that the well-preserved gene FTO plays a predominant role in DNA demethylation and m6A-dependent mRNA demethylation [1, 4, 125, 129–132]. Thus, FTO promotes transcription and increases genomic transcriptional activity, a requirement for postnatal growth. Milk functions apparently as an “epigenetic interface” generated by the lactation genome that controls the FTO-dependent transcriptome of the milk recipient (Fig. 1). FTO is thus an evolutionary checkpoint coordinating the program of the lactation driving transcription, translation and anabolism of the newborn mammal. FTO-mediated demethylation of mRNAs increases transcriptional activity and generates mRNA splice variants that are critically involved in adipogenesis [144–146], ghrelin-regulated appetite control [150], and LRS-mediated mTORC1 activation [110, 157–162] (Fig. 2).
Fig. 2.
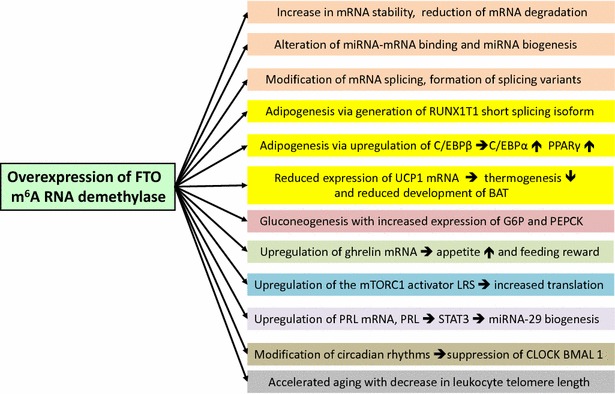
Synopsis of m6A mRNA modifications induced by increased FTO expression. The RNA m6A eraser FTO drives crucial epigenetic mechanisms fundamentally involved in the pathogenesis of diseases of civilization
Remarkably, FTO plays a critical role in milk production. The mRNA of PRL, the most important hormone promoting lactation, is regulated via m6A methylation [180, 186] and thus depends on FTO activity. In this context it is not surprising that bovine FTO variants enhance lactation performance [164]. Moreover, enhanced expression of miRNA-29 in DCMECs increases milk yield [180]. Abundance of miRNA-29s in DCMECs of high performance dairy cows may downregulate DNMT-mediated methylation of bovine FTO, thereby increasing bovine FTO mRNA and protein levels of DCMECs. Bovine miRNA-29s, which are identical with human miRNA-29s, and bovine FTO mRNA, which is highly homologous to human FTO mRNA, may reach the consumer of pasteurized fresh milk via uptake of milk exosomes [84]. Viral and bacterial infections of dairy cows may further increase miRNA-29 levels [189, 190, 196–201] (Fig. 1). Thus, the efforts of veterinary medicine intensifying lactation performance and milk yield apparently overstimulate FTO signaling of the human milk consumer, an overlooked interaction in the pathogenesis of Western diseases. The presented working model allows a new understanding of milk as mammal´s temporal amplifier of the epitranscriptome of the milk recipient. m6A is a common modification of mRNA with potential roles in fine-tuning the RNA life cycle and mRNA expression control [143, 338]. Milk-activated FTO operates as a methylation “eraser” to promote transcription and translation for adequate growth during the lactation period [129, 143]. It is conceivable that individuals carrying obesigenic genetic FTO variants with increased FTO expression may be even more susceptible to milk-mediated epigenetic activation of FTO. FTO should be regarded as the driver of the transcriptome, whereas mTORC1 drives the translational machinery interconnected via an FTO-mTORC1 crosstalk.
Persistent milk-mediated epigenetic FTO signaling may explain the epidemic of age-related diseases of civilization. It is thus not surprising that an increased mortality in relation to high milk intake has recently been observed in a Swedish cohort of men and women [339]. The antagonistic pleiotropy theory of aging postulates that genes beneficial early in life operate at the cost of aging when persistently activated later in life [340]. Persistent overactivation of evolutionary developmental genes, such as FTO and MTOR, which are most important for perinatal programming, appear to be the major health hazard promoting aging and early onset of age-related diseases. In fact, obesity-related risk allele carriers of FTO gene show dose-dependent increments in BMI during aging. Moreover, the obesity-related risk allele is associated with reduced medial prefrontal cortical function during aging [341]. In addition, presence of the FTO rs9939609 polymorphism risk allele in a Korean population was inversely associated with leukocyte telomere length [342].
Future research should characterize the epigenetic FTO-activating potential of milk versus other fermented dairy products. The relative contribution of milk-derived regulatory mechanisms that activate FTO expression such as BCAAs and exosomal miRNAs, have to be determined in detail. It is of critical interest to provide experimental evidence showing that milk consumption modifies the FTO methylation status resulting in increased FTO expression and activity. Furthermore, it is important to study differences in epigenetic FTO activation levels between human breast milk, bovine milk, and commercial milk of high performance dairy cows. Special attention should be paid to the effects of bovine milk exosomal miRNAs and mRNAs in the epigenetic control of FTO expression during sensitive periods of pre- and postnatal FTO-mediated metabolic programming.
Acknowledgements
The author thanks Gerd Schmitz, University of Regensburg, for discussions of milk and lipidomics, and Harald zur Hausen, Deutsches Krebsforschungszentrum Heidelberg, for valuable insights into bovine oncogenic viruses in commercial milk, and Helena Ord, student of Princeton University, for language improvements of the manuscript.
Funding
There are no sources of funding.
Competing interests
The author declares that he has no competing interests.
Abbreviations
- AD
Alzheimer’s disease
- AARS
amino-acyl-tRNA synthetase
- APOE
apolipoprotein E
- ATG5
autophagy 5, S. cerevisiae, homolog of
- BAT
brown adipose tissue
- BC
breast cancer
- BCAA
branched-chain amino acid
- BCGN1
beclin 1
- BMAL1
brain and muscle ARNT-like protein 1
- BMI
body mass index
- C/EBPβ
CCAAT/enhancer-binding protein-β
- CHD
coronary heart disease
- CLOCK
circadian locomotor output cycles kaput
- CNS
central nervous system
- CpG
cytosine-phosphate-guanine
- CSN1S1
casein alpha s1 gene
- DCMEC
dairy cow mammary epithelial cell
- DMBA
7,12-dimethylbenz(a)anthracene
- DNMT
DNA methyltransferase
- EIF5
E74-like factor 5 gene
- EMT
epithelial to mesenchymal transition
- EPIC
European Prospective Investigation into Cancer and Nutrition
- ER
estrogen receptor
- EXBF
exclusive breast feeding
- FTO
fat mass- and obesity-associated gene
- GLUT1
glucose transporter 1 gene
- HNRNPC
heterogeneous nuclear ribonucleoprotein C
- HOMA
homeostasis model assessment
- IGF-1
insulin-like growth factor 1
- LPS
lipopolysaccaride
- LRS
leucyl-tRNA synthase
- MCE
mitotic clonal expansion
- MEF
mouse embryonic fibroblast
- m6A
N6-methyl-adenosine
- METTL3
methyltransferase-like 3
- miRNA
microRNA
- MSC
multi-synthetase complex
- mTORC1
mechanistic target of rapamycin complex 1
- NHANES
National Health and Nutrition Examination Survey
- PD
Parkinson´s disease
- PC
Prostate cancer
- PIP
prolactin-inducible protein
- PRL
prolactin
- PPARγ
peroxisome proliferator-activated receptor-γ
- rRNA
ribosomal RNA
- RT
reverse transcriptase
- RUNX2
runt-related transcription factor 2
- SAT
subcutaneous adipose tissue
- SNP
single nucleotide polymorphism
- snRNA
small nuclear RNA
- snoRNA
small nucleolar RNA
- STAT
signal transducer and activator of transcription
- SREBP1
sterol regulatory element binding protein-1
- tRNA
transfer RNA
- T2DM
type 2 diabetes mellitus
- UCP-1
uncoupling protein 1
- UTR
untranslated region
- VTA
ventral tegmental area
References
- 1.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS, et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet. 2009;85:106–111. doi: 10.1016/j.ajhg.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fredriksson R, Hägglund M, Olszewski PK, Stephansson O, Jacobsson JA, Olszewska AM, et al. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology. 2008;149:2062–2071. doi: 10.1210/en.2007-1457. [DOI] [PubMed] [Google Scholar]
- 4.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larder R, Cheung MK, Tung YC, Yeo GS, Coll AP. Where to go with FTO? Trends Endocrinol Metab. 2011;22:53–59. doi: 10.1016/j.tem.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 7.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinney A, Nguyen TT, Scherag A, Friedel S, Brönner G, Müller TD, et al. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS One. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha SW, Choi SM, Kim KS, Park BL, Kim JR, Kim JY, et al. Replication of genetic effects of FTO polymorphisms on BMI in a Korean population. Obesity (Silver Spring) 2008;16:2187–2189. doi: 10.1038/oby.2008.314. [DOI] [PubMed] [Google Scholar]
- 10.Chang YC, Liu PH, Lee WJ, Chang TJ, Jiang YD, Li HY, et al. Common variation in the fat mass and obesity-associated (FTO) gene confers risk of obesity and modulates BMI in the Chinese population. Diabetes. 2008;57:2245–2252. doi: 10.2337/db08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hotta K, Nakata Y, Matsuo T, Kamohara S, Kotani K, Komatsu R, et al. Variations in the FTO gene are associated with severe obesity in the Japanese. J Hum Genet. 2008;53:546–553. doi: 10.1007/s10038-008-0283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubacek JA, Bohuslavova R, Kuthanova L, Kubinova R, Peasey A, Pikhart H, et al. The FTO gene and obesity in a large Eastern European population sample: the HAPIEE study. Obesity (Silver Spring) 2008;16:2764–2766. doi: 10.1038/oby.2008.421. [DOI] [PubMed] [Google Scholar]
- 13.Peeters A, Beckers S, Verrijken A, Roevens P, Peeters P, Van Gaal L, et al. Variants in the FTO gene are associated with common obesity in the Belgian population. Mol Genet Metab. 2008;93:481–484. doi: 10.1016/j.ymgme.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Villalobos-Comparán M, Teresa Flores-Dorantes M, Teresa Villarreal-Molina M, Rodríguez-Cruz M, García-Ulloa AC, Robles L, et al. The FTO gene is associated with adulthood obesity in the Mexican population. Obesity (Silver Spring). 2008;16:2296–2301. doi: 10.1038/oby.2008.367. [DOI] [PubMed] [Google Scholar]
- 15.Zhang F, Xu L, Jin L, Wang XF. A common variant in the FTO gene is associated with obesity in the Uyghur population. J Endocrinol Invest. 2008;31:1043. doi: 10.1007/BF03345646. [DOI] [PubMed] [Google Scholar]
- 16.Cornes BK, Lind PA, Medland SE, Montgomery GW, Nyholt DR, Martin NG. Replication of the association of common rs9939609 variant of FTO with increased BMI in an Australian adult twin population but no evidence for gene by environment (G × E) interaction. Int J Obes (Lond) 2009;33:75–79. doi: 10.1038/ijo.2008.223. [DOI] [PubMed] [Google Scholar]
- 17.González-Sánchez JL, Zabena C, Martínez-Larrad MT, Martínez-Calatrava MJ, Pérez-Barba M, Serrano-Ríos M. Variant rs9939609 in the FTO gene is associated with obesity in an adult population from Spain. Clin Endocrinol (Oxf) 2009;70:390–393. doi: 10.1111/j.1365-2265.2008.03335.x. [DOI] [PubMed] [Google Scholar]
- 18.Zabena C, González-Sánchez JL, Martínez-Larrad MT, Torres-García A, Alvarez-Fernández-Represa J, Corbatón-Anchuelo A, et al. The FTO obesity gene. Genotyping and gene expression analysis in morbidly obese patients. Obes Surg. 2009;19:87–95. doi: 10.1007/s11695-008-9727-0. [DOI] [PubMed] [Google Scholar]
- 19.Meisel SF, Beeken RJ, van Jaarsveld CH, Wardle J. The association of FTO SNP rs9939609 with weight gain at university. Obes Facts. 2015;8:243–251. doi: 10.1159/000434733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, et al. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet. 2008;9:59. doi: 10.1186/1471-2350-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legry V, Cottel D, Ferrières J, Arveiler D, Andrieux N, Bingham A, et al. Effect of an FTO polymorphism on fat mass, obesity, and type 2 diabetes mellitus in the French MONICA Study. Metabolism. 2009;58:971–975. doi: 10.1016/j.metabol.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 24.Abbas S, Raza ST, Ahmed F, Ahmad A, Rizvi S, Mahdi F. Association of genetic polymorphism of PPARγ-2, ACE, MTHFR, FABP-2 and FTO genes in risk prediction of type 2 diabetes mellitus. J Biomed Sci. 2013;20:80. doi: 10.1186/1423-0127-20-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raza S, Abbas S, Ahmad A, Ahmed F, Zaidi Zh, Mahdi F. Association of glutathione-S-transferase (GSTM1 and GSTT1) and FTO gene polymorphisms with type 2 diabetes mellitus cases in Northern India. Balkan J Med Genet. 2014;17:47–54. doi: 10.2478/bjmg-2014-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fawwad A, Siddiqui IA, Zeeshan NF, Shahid SM, Basit A. Association of SNP rs9939609 in FTO gene with metabolic syndrome in type 2 diabetic subjects, rectruited from a tertiary care unit of Karachi, Pakistan. Pak J Med Sci. 2015;31:140–145. doi: 10.12669/pjms.311.6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keaton JM, Cooke Bailey JN, Palmer ND, Freedman BI, Langefeld CD, Ng MC, et al. A comparison of type 2 diabetes risk allele load between African Americans and European Americans. Hum Genet. 2014;133:1487–1495. doi: 10.1007/s00439-014-1486-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Razquin C, Marti A, Martinez JA. Evidences on three relevant obesogenes: MC4R, FTO and PPARγ. Approaches for personalized nutrition. Mol Nutr Food Res. 2011;55:136–149. doi: 10.1002/mnfr.201000445. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Loos RJ, Powell JE, Medland SE, Speliotes EK, Chasman DI, et al. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490:267–272. doi: 10.1038/nature11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berulava T, Horsthemke B. The obesity-associated SNPs in intron 1 of the FTO gene affect primary transcript levels. Eur J Hum Genet. 2010;18:1054–1056. doi: 10.1038/ejhg.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toperoff G, Kark JD, Aran D, Nassar H, Ahmad WA, Sinnreich R, et al. Premature aging of leukocyte DNA methylation is associated with type 2 diabetes prevalence. Clin Epigenetics. 2015;7:35. doi: 10.1186/s13148-015-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dayeh T, Volkov P, Salö S, Hall E, Nilsson E, Olsson AH, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10:e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathers JC, Strathdee G, Relton CL. Induction of epigenetic alterations by dietary and other environmental factors. Adv Genet. 2010;71:3–39. doi: 10.1016/B978-0-12-380864-6.00001-8. [DOI] [PubMed] [Google Scholar]
- 34.van der Hoeven F, Schimmang T, Volkmann A, Mattei MG, Kyewski B, Rüther U. Programmed cell death is affected in the novel mouse mutant Fused toes (Ft) Development. 1994;120:2601–2607. doi: 10.1242/dev.120.9.2601. [DOI] [PubMed] [Google Scholar]
- 35.Peters T, Ausmeier K, Rüther U. Cloning of Fatso (Fto), a novel gene deleted by the Fused toes (Ft) mouse mutation. Mamm Genome. 1999;10:983–986. doi: 10.1007/s003359901121. [DOI] [PubMed] [Google Scholar]
- 36.Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Brüning JC, et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 37.Gao X, Shin YH, Li M, Wang F, Tong Q, Zhang P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One. 2010;5:e14005. doi: 10.1371/journal.pone.0014005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melnik BC, John SM, Schmitz G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr J. 2013;12:103. doi: 10.1186/1475-2891-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melnik BC. Milk- a nutrient system of mammalian evolution promoting mTORC1-dependent translation. Int J Mol Sci. 2015;16:17048–17087. doi: 10.3390/ijms160817048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Q, Saunders RA, Szkudlarek-Mikho M, Serna Ide L, Chin KV. The obesity- associated Fto gene is a transcriptional coactivator. Biochem Biophys Res Commun. 2010;401:390–395. doi: 10.1016/j.bbrc.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L, et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fawcett KA, Barroso I. The genetics of obesity: FTO leads the way. Trends Genet. 2010;26:266–274. doi: 10.1016/j.tig.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wardle J, Carnell S, Haworth CM, Farooqi IS, O’Rahilly S, Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. J Clin Endocrinol Metab. 2008;93:3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- 44.Wardle J, Llewellyn C, Sanderson S, Plomin R. The FTO gene and measured food intake in children. Int J Obes (Lond). 2009;33:42–45. doi: 10.1038/ijo.2008.174. [DOI] [PubMed] [Google Scholar]
- 45.Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity- associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359:2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- 46.Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring). 2008;16:1961–1965. doi: 10.1038/oby.2008.318. [DOI] [PubMed] [Google Scholar]
- 47.Haupt A, Thamer C, Staiger H, Tschritter O, Kirchhoff K, Machicao F, et al. Variation in the FTO gene influences food intake but not energy expenditure. Exp Clin Endocrinol Diabetes. 2009;117:194–197. doi: 10.1055/s-0028-1087176. [DOI] [PubMed] [Google Scholar]
- 48.Timpson NJ, Emmett PM, Frayling TM, Rogers I, Hattersley AT, McCarthy MI, et al. The fat mass- and obesity-associated locus and dietary intake in children. Am J Clin Nutr. 2008;88:971–978. doi: 10.1093/ajcn/88.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Velders FP, De Wit JE, Jansen PW, Jaddoe VW, Hofman A, Verhulst FC, et al. FTO at rs9939609, food responsiveness, emotional control and symptoms of ADHD in preschool children. PLoS One. 2012;7:e49131. doi: 10.1371/journal.pone.0049131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olszewski PK, Fredriksson R, Olszewska AM, Stephansson O, Alsiö J, Radomska KJ, et al. Hypothalamic FTO is associated with the regulation of energy intake not feeding reward. BMC Neurosci. 2009;10:129. doi: 10.1186/1471-2202-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vujovic P, Stamenkovic S, Jasnic N, Lakic I, Djurasevic SF, Cvijic G, et al. Fasting induced cytoplasmic Fto expression in some neurons of rat hypothalamus. PLoS One. 2013;8:e63694. doi: 10.1371/journal.pone.0063694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang CY, Shie SS, Hsieh IC, Tsai ML, Wen MS. FTO modulates circadian rhythms and inhibits the CLOCK-BMAL1-induced transcription. Biochem Biophys Res Commun. 2015;464:826–832. doi: 10.1016/j.bbrc.2015.07.046. [DOI] [PubMed] [Google Scholar]
- 53.Qi Q, Kilpeläinen TO, Downer MK, Tanaka T, Smith CE, Sluijs I, et al. FTO genetic variants, dietary intake and body mass index: insights from 177,330 individuals. Hum Mol Genet. 2014;23:6961–6972. doi: 10.1093/hmg/ddu411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harb Perspect Biol. 2014;6:a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet. 2011;20:670–680. doi: 10.1093/hmg/ddq513. [DOI] [PubMed] [Google Scholar]
- 56.Shenker NS, Flower KJ, Wilhelm-Benartzi CS, Dai W, Bell E, Gore E, et al. Transcriptional implications of intragenic DNA methylation in the oestrogen receptor alpha gene in breast cancer cells and tissues. BMC Cancer. 2015;15:337. doi: 10.1186/s12885-015-1335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toperoff G, Aran D, Kark JD, Rosenberg M, Dubnikov T, Nissan B, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21:371–383. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu ZW, Zhang JT, Cai QY, Zhang HX, Wang YH, Yan HT, et al. Birth weight is associated with placental fat mass- and obesity-associated gene expression and promoter methylation in a Chinese population. J Matern Fetal Neonatal Med. 2014;10:1–6. doi: 10.3109/14767058.2014.987749. [DOI] [PubMed] [Google Scholar]
- 59.Valadi H, Ekstöm K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome- mediated transfer of mRNA and microRNAs is novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 60.Liang H, Huang L, Cao J, Zen K, Chen X, Zhang CY. Regulation of mammalian gene expression by exogeneous microRNAs. Wiley Interdiscip Rev RNA. 2012;3:733–742. doi: 10.1002/wrna.1127. [DOI] [PubMed] [Google Scholar]
- 61.Chen X, Liang H, Zhang J, Zen K, Thang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 62.Ludwig AK, Giebel B. Exosomes: small vesicles participating in intercellular communication. Int J Biochem Cell Biol. 2012;44:11–15. doi: 10.1016/j.biocel.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Horizontal transfer of microRNAs: molecular mechanism and clinical applications. Protein Cell. 2012;3:28–37. doi: 10.1007/s13238-012-2003-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 65.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune- regulatory agent in breast milk. Silence. 2010;1:7. doi: 10.1186/1758-907X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, et al. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. 2012;8:118–123. doi: 10.7150/ijbs.8.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Munch EM, Harris RA, Mohammad M, Benham AL, Pejerrey SM, Showalter L, et al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS One. 2013;8:e50564. doi: 10.1371/journal.pone.0050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alsaweed M, Hepworth AR, Lefèvre C, Hartmann PE, Geddes DT, Hassiotou F. Human milk microRNA and total RNA differ depending on milk fractionation. J Cell Biochem. 2015;116:2397–2407. doi: 10.1002/jcb.25207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X, Gao C, Li H, Huang L, Sun Q, Dong Y, et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010;20:1128–1137. doi: 10.1038/cr.2010.80. [DOI] [PubMed] [Google Scholar]
- 71.Hata T, Murakami K, Nakatani H, Yamamoto Y, Matsuda T, Aoki N. Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biochem Biophys Res Commun. 2010;396:528–533. doi: 10.1016/j.bbrc.2010.04.135. [DOI] [PubMed] [Google Scholar]
- 72.Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci. 2012;95:4831–4841. doi: 10.3168/jds.2012-5489. [DOI] [PubMed] [Google Scholar]
- 73.Sun Q, Chen X, Yu J, Zen K, Zhang CY, Li L. Immune modulatory function of abundant immune-related microRNAs in microvesicles from bovine colostrum . Protein Cell. 2013;4:197–210. doi: 10.1007/s13238-013-2119-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pieters BC, Arntz OJ, Bennink MB, Broeren MG, van Caam AP, Koenders MI, et al. Commercial cow milk contains physically stable extracellular vesicles expressing immunoregulatory TGF-β. PLoS One. 2015;10:e0121123. doi: 10.1371/journal.pone.0121123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen T, Xi QY, Ye RS, Cheng X, Qi QE, Wang SB, et al. Exploration of microRNAs in porcine milk exosomes. BMC Genom. 2014;15:100. doi: 10.1186/1471-2164-15-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gu Y, Li M, Wang T, Liang Y, Zhong Z, Wang X, et al. Lactation-related microRNA expression profiles of porcine breast milk exosomes. PLoS One. 2012;7:e43691. doi: 10.1371/journal.pone.0043691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Time- dependent expression profiles of microRNAs and mRNAs in rat milk whey. PLoS One. 2014;9:e88843. doi: 10.1371/journal.pone.0088843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Modepalli V, Kumar A, Hinds LA, Sharp JA, Nicholas KR, Lefevre C. Differential temporal expression of milk miRNA during the lactation cycle of the marsupial tammar wallaby (Macropus eugenii) BMC Genom. 2014;15:1012. doi: 10.1186/1471-2164-15-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zempleni J, Baier SR, Howard KM, Cui J. Gene regulation by dietary microRNAs. Can J Physiol Pharmacol. 2015;93:1097–1102. doi: 10.1139/cjpp-2014-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Title AC, Denzler R, Stoffel M. Uptake and function studies of maternal milk- derived microRNAs. J Biol Chem. 2015;290:23680–23691. doi: 10.1074/jbc.M115.676734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolf T, Baier SR, Zempleni J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma Caco-2 cells and rat small intestinal IEC-6 cells. J Nutr. 2015;145:2201–2206. doi: 10.3945/jn.115.218586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Howard KM, Jati Kusuma R, Baier SR, Friemel T, Markham L, Vanamala J, et al. Loss of miRNAs during processing and storage of cow’s (Bos taurus) milk. J Agric Food Chem. 2015;63:588–592. doi: 10.1021/jf505526w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK- 293 kidney cell cultures, and mouse livers. J Nutr. 2014;144:1495–1500. doi: 10.3945/jn.114.196436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, et al. MicroRNA- 29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jia LF, Huang YP, Zheng YF, Lyu MY, Wei SB, Meng Z, et al. miR-29b suppresses proliferation, migration, and invasion of tongue squamous cell carcinoma through PTEN-AKT signaling pathway by targeting Sp1. Oral Oncol. 2014;50:1062–1071. doi: 10.1016/j.oraloncology.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 88.Zhao S, Wu J, Zheng F, Tang Q, Yang L, Li L, et al. β-Elemene inhibited expression of DNA methyltransferase 1 through activation of ERK1/2 and AMPKα signalling pathways in human lung cancer cells: the role of Sp1. J Cell Mol Med. 2015;19:630–641. doi: 10.1111/jcmm.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meunier L, Siddeek B, Vega A, Lakhdari N, Inoubli L, Bellon RP, et al. Perinatal programming of adult rat germ cell death after exposure to xenoestrogens: role of microRNA miR-29 family in the down-regulation of DNA methyltransferases and Mcl-1. Endocrinology. 2012;153:1936–1947. doi: 10.1210/en.2011-1109. [DOI] [PubMed] [Google Scholar]
- 90.Cicchini C, de Nonno V, Battistelli C, Cozzolino AM, De Santis Puzzonia M, Ciafrè SA, et al. Epigenetic control of EMT/MET dynamics: HNF4α impacts DNMT3s through miRs-29. Biochim Biophys Acta. 2015;1849:919–929. doi: 10.1016/j.bbagrm.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 92.Feuermann Y, Kang K, Shamay A, Robinson GW, Hennighausen L. MiR-21 is under control of STAT5 but is dispensable for mammary development and lactation. PLoS One. 2014;9:e85123. doi: 10.1371/journal.pone.0085123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tammen SA, Friso S, Choi SW. Epigenetics: the link between nature and nurture. Mol Aspects Med. 2013;34:753–764. doi: 10.1016/j.mam.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 95.Feil R, Walter J, Allen ND, Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development. 1994;120:2933–2943. doi: 10.1242/dev.120.10.2933. [DOI] [PubMed] [Google Scholar]
- 96.Ubeda F, Wilkins JF. Imprinted genes and human disease: an evolutionary perspective. Adv Exp Med Biol. 2008;626:101–115. doi: 10.1007/978-0-387-77576-0_8. [DOI] [PubMed] [Google Scholar]
- 97.Radford EJ, Ferrón SR, Ferguson-Smith AC. Genomic imprinting as an adaptative model of developmental plasticity. FEBS Lett. 2011;585:2059–2066. doi: 10.1016/j.febslet.2011.05.063. [DOI] [PubMed] [Google Scholar]
- 98.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 100.Li Y, Sasaki H. Genomic imprinting in mammals: its life cycle, molecular mechanisms and reprogramming. Cell Res. 2011;21:466–473. doi: 10.1038/cr.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. 2013;5:553–568. doi: 10.2217/epi.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 104.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet. 2006;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- 105.Alsaweed M, Hartmann PE, Geddes DT, Kakulas F. MicroRNAs in breastmilk and the lactating breast: potential immunoprotectors and developmental regulators for the infant and the mother. Int J Environ Res Public Health. 2015;12:13981–14020. doi: 10.3390/ijerph121113981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Millward DJ, Layman DK, Tomé D, Schaafsma G. Protein quality assessment: impact of expanding understanding of protein and amino acid needs for optimal health. Am J Clin Nutr. 2008;87:1576S–1581S. doi: 10.1093/ajcn/87.5.1576S. [DOI] [PubMed] [Google Scholar]
- 107.Lenders CM, Liu S, Wilmore DW, Sampson L, Dougherty LW, Spiegelman D, et al. Evaluation of a novel food composition database that includes glutamine and other amino acids derived from gene sequencing data. Eur J Clin Nutr. 2009;63:1433–1439. doi: 10.1038/ejcn.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cheung MK, Gulati P, O’Rahilly S, Yeo GS. FTO expression is regulated by availability of essential amino acids. Int J Obes (Lond). 2013;37:744–747. doi: 10.1038/ijo.2012.77. [DOI] [PubMed] [Google Scholar]
- 109.Gulati P, Yeo GS. The biology of FTO: from nucleic acid demethylase to amino acid sensor. Diabetologia. 2013;56:2113–2121. doi: 10.1007/s00125-013-2999-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gulati P, Cheung MK, Antrobus R, Church CD, Harding HP, Tung YC, et al. Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proc Natl Acad Sci USA. 2013;110:2557–2562. doi: 10.1073/pnas.1222796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jegga AG, Schneider L, Ouyang X, Zhang J. Systems biology of the autophagy-lysosomal pathway. Autophagy. 2011;7:477–489. doi: 10.4161/auto.7.5.14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 113.Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296:E592–E602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sengupta S, Peterson T, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Laplante M, Sabatini DM. mTOR signaling. Cold Spring Harb Perspect Biol. 2012;4 (pii:a011593). [DOI] [PMC free article] [PubMed]
- 116.Kim J, Guan KL. Amino acid signaling in TOR activation. Ann Rev Biochem. 2011;80:1001–1032. doi: 10.1146/annurev-biochem-062209-094414. [DOI] [PubMed] [Google Scholar]
- 117.Kim S, Buel GR, Blenis J. Nutrient regulation of the mTOR complex 1 signaling pathway. Mol Cells. 2013;35:463–473. doi: 10.1007/s10059-013-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jewell JL, Guan KL. Nutrient signaling to mTOR and cell growth. Trends Biochem Sci. 2013;38:233–242. doi: 10.1016/j.tibs.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Efeyan A, Sabatini DM. Nutrients and growth factors in mTORC1 activation. Biochem Soc Trans. 2013;41:902–905. doi: 10.1042/BST20130063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oshiro N, Rapley J, Avruch J. Amino acids activate mammalian target of rapamycin (mTOR) complex 1 without changing Rag GTPase guanyl nucleotide charging. J Biol Chem. 2014;289:2658–2674. doi: 10.1074/jbc.M113.528505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brosnan JT, Brosnan ME. Branched-chain amino acids: enzyme and substrate regulation. J Nutr. 2006;136:207S–211S. doi: 10.1093/jn/136.1.207S. [DOI] [PubMed] [Google Scholar]
- 122.AEvarsson A, Chuang JL, Wynn RM, Turley S, Chuang DT, Hol WG. Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease. Structure. 2000;8:277–91. [DOI] [PubMed]
- 123.Mersey BD, Jin P, Danner DJ. Human microRNA (miR29b) expression controls the amount of branched chain alpha-ketoacid dehydrogenase complex in a cell. Hum Mol Genet. 2005;14:3371–3377. doi: 10.1093/hmg/ddi368. [DOI] [PubMed] [Google Scholar]
- 124.Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM. The AlkB family of Fe(II)/α-ketoglutarate dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J Biol Chem. 2015;290:20734–20742. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tuck MT. The formation of internal 6-methyladenine residues in eucaryotic messenger RNA. Int J Biochem. 1992;24:379–386. doi: 10.1016/0020-711X(92)90028-Y. [DOI] [PubMed] [Google Scholar]
- 129.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29:108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19:1848–1856. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine- dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine- dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McCloskey A, Taniguchi I, Shinmyozu K, Ohno M. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science. 2012;335:1643–1646. doi: 10.1126/science.1218469. [DOI] [PubMed] [Google Scholar]
- 135.Rajagopalan LE, Westmark CJ, Jarzembowski JA, Malter JS. hnRNP C increases amyloid precursor protein (APP) production by stabilizing APP mRNA. Nucleic Acids Res. 1998;26:3418–3423. doi: 10.1093/nar/26.14.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, Stévant I, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cieniková Z, Damberger FF, Hall J, Allain FH, Maris C. Structural and mechanistic insights into poly(uridine) tract recognition by the hnRNP C RNA recognition motif. J Am Chem Soc. 2014;136:14536–14544. doi: 10.1021/ja507690d. [DOI] [PubMed] [Google Scholar]
- 138.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39(Database issue):D195–201. [DOI] [PMC free article] [PubMed]
- 140.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, et al. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37(Database issue):D118–21. [DOI] [PMC free article] [PubMed]
- 141.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 142.Berulava T, Rahmann S, Rademacher K, Klein-Hitpass L, Horsthemke B. N6-adenosine methylation in MiRNAs. PLoS One. 2015;10:e0118438. doi: 10.1371/journal.pone.0118438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Merkestein M, Laber S, McMurray F, Andrew D, Sachse G, Sanderson J, et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat Commun. 2015;6:6792. doi: 10.1038/ncomms7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang X, Zhu L, Chen J, Wang Y. mRNA m6A methylation downregulates adipogenesis in porcine adipocytes. Biochem Biophys Res Commun. 2015;459:201–207. doi: 10.1016/j.bbrc.2015.02.048. [DOI] [PubMed] [Google Scholar]
- 148.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- 149.Bar Yamin H, Barnea M, Genzer Y, Chapnik N, Froy O. Long-term commercial cow’s milk consumption and its effects on metabolic parameters associated with obesity in young mice. Mol Nutr Food Res. 2014;58:1061–1068. doi: 10.1002/mnfr.201300650. [DOI] [PubMed] [Google Scholar]
- 150.Dickson SL, Egecioglu E, Landgren S, Skibicka KP, Engel JA, Jerlhag E. The role of the central ghrelin system in reward from food and chemical drugs. Mol Cell Endocrinol. 2011;340:80–87. doi: 10.1016/j.mce.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 151.Burger KS, Berner LA. A functional neuroimaging review of obesity, appetitive hormones and ingestive behavior. Physiol Behav. 2014;136:121–127. doi: 10.1016/j.physbeh.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hewson AK, Dickson SL. Systemic administration of ghrelin induces Fos and Egr-1 proteins in the hypothalamic arcuate nucleus of fasted and fed rats. J Neuroendocrinol. 2000;12:1047–1049. doi: 10.1046/j.1365-2826.2000.00584.x. [DOI] [PubMed] [Google Scholar]
- 153.Perello M, Dickson SL. Ghrelin signalling on food reward: a salient link between the gut and the mesolimbic system. J Neuroendocrinol. 2015;27:424–434. doi: 10.1111/jne.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Karra E, O’Daly OG, Choudhury AI, Yousseif A, Millership S, Neary MT, et al. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J Clin Invest. 2013;123:3539–3551. doi: 10.1172/JCI44403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Caruso V, Chen H, Morris MJ. Early hypothalamic FTO overexpression in response to maternal obesity—potential contribution to postweaning hyperphagia. PLoS One. 2011;6:e25261. doi: 10.1371/journal.pone.0025261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Abarin T, Yan Wu Y, Warrington N, Lye S, Pennell C, Briollais L. The impact of breastfeeding on FTO-related BMI growth trajectories: an application to the Raine pregnancy cohort study. Int J Epidemiol. 2012;41:1650–1660. doi: 10.1093/ije/dys171. [DOI] [PubMed] [Google Scholar]
- 157.Quevillon S, Robinson JC, Berthonneau E, Siatecka M, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J Mol Biol. 1999;285:183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- 158.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 159.Park SG, Choi EC, Kim S. Aminoacyl-tRNA synthetase-interacting multifunctional proteins (AIMPs): a triad for cellular homeostasis. IUBMB Life. 2010;62:296–302. doi: 10.1002/iub.324. [DOI] [PubMed] [Google Scholar]
- 160.Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 161.Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 162.Lo WS, Gardiner E, Xu Z, Lau CF, Wang F, Zhou JJ, et al. Human tRNA synthetase catalytic nulls with diverse functions. Science. 2014;345:328–332. doi: 10.1126/science.1252943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xing J, Jing W, Jiang Y. Molecular characterization and expression analysis of fat mass and obesity-associated gene in rabbit. J Genet. 2013;92:481–488. doi: 10.1007/s12041-013-0298-z. [DOI] [PubMed] [Google Scholar]
- 164.Zielke LG, Bortfeldt RH, Reissmann M, Tetens J, Thaller G, Brockmann GA. Impact of variation at the FTO locus on milk fat yield in Holstein dairy cattle. PLoS One. 2013;8:e63406. doi: 10.1371/journal.pone.0063406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sorbolini S, Marras G, Gaspa G, Dimauro C, Cellesi M, Valentini A, et al. Detection of selection signatures in Piemontese and Marchigiana cattle, two breeds with similar production aptitudes but different selection histories. Genet Sel Evol. 2015;47:52. doi: 10.1186/s12711-015-0128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schlom J, Spiegelman S, Moore D. RNA-dependent DNA polymerase activity in virus-like particles isolated from human milk. Nature. 1971;231:97–100. doi: 10.1038/231097a0. [DOI] [PubMed] [Google Scholar]
- 167.Schlom J, Spiegelman S, Moore DH. Reverse transcriptase and high molecular weight RNA in particles from mouse and human milk. J Natl Cancer Inst. 1972;48:1197–1203. [PubMed] [Google Scholar]
- 168.McCormick JJ, Larson LJ, Rich MA. RNase inhibition of reverse transcriptase activity in human milk. RNase inhibition of reverse transcriptase activity in human milk. Nature. 1974;251:737–740. doi: 10.1038/251737a0. [DOI] [PubMed] [Google Scholar]
- 169.McFarlane ES, Ryan SM, Mann E. RNA-dependent DNA-polymerase activity in human milk. J Med Microbiol. 1975;8:447–449. doi: 10.1099/00222615-8-3-447. [DOI] [PubMed] [Google Scholar]
- 170.Sanner T. Removal of inhibitors against RNA-directed DNA polymerase activity in human milk. Cancer Res. 1976;36:405–408. [PubMed] [Google Scholar]
- 171.Dumaswala RU, Talageri VR, Karande KA, Joshi BJ, Ranadive KJ. Characterization of RNA-directed DNA polymerase in the milk of strain ICRC mice. Indian J Biochem Biophys. 1978;15:493–496. [PubMed] [Google Scholar]
- 172.Kantor JA, Lee YH, Chirikjian JG, Feller WF. DNA polymerase with characteristics of reverse transcriptase purified from human milk. Science. 1979;204:511–513. doi: 10.1126/science.86209. [DOI] [PubMed] [Google Scholar]
- 173.Littlewood TD, Tomley FM, Owen LN. Preliminary report on the presence of RNA-dependent DNA polymerase in canine milk. Res Vet Sci. 1984;37:97–100. [PubMed] [Google Scholar]
- 174.Das MR, Padhy LC, Koshy R, Sirsat SM, Rich MA. Human milk samples from different ethnic groups contain RNase that inhibits, and plasma membrane that stimulates, reverse transcription. Nature. 1976;262:802–805. doi: 10.1038/262802a0. [DOI] [PubMed] [Google Scholar]
- 175.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 176.Irmak MK, Oztas Y, Oztas E. Integration of maternal genome into the neonate genome through breast milk mRNA transcripts and reverse transcriptase. Theor Biol Med Model. 2012;9:20. doi: 10.1186/1742-4682-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol Spectr. 2015;3:MDNA3-0061-2014. [DOI] [PMC free article] [PubMed]
- 178.Li Z, Liu H, Jin X, Lo L, Liu J. Expression profiles of microRNAs from lactating and non-lactating bovine mammary glands and identification of miRNA related to lactation. BMC Genom. 2012;13:731. doi: 10.1186/1471-2164-13-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wicik Z, Gajewska M, Majewska A, Walkiewicz D, Osińska E, Motyl T. Characterization of microRNA profile in mammary tissue of dairy and beef breed heifers. J Anim Breed Genet. 2015 [Epub ahead of print]. [DOI] [PubMed]
- 180.Bian Y, Lei Y, Wang C, Wang J, Wang L, Liu L, et al. Epigenetic regulation of miR-29s affects the lactation activity of dairy cow mammary epithelial cells. J Cell Physiol. 2015;230:2152–2163. doi: 10.1002/jcp.24944. [DOI] [PubMed] [Google Scholar]
- 181.Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr Rev. 1998;19:225–268. doi: 10.1210/edrv.19.3.0334. [DOI] [PubMed] [Google Scholar]
- 182.Grattan DR. 60 years of neuroendocrinology: the hypothalamo- prolactin axis. J Endocrinol. 2015;226:T101–T122. doi: 10.1530/JOE-15-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lacasse P, Lollivier V, Dessauge F, Bruckmaier RM, Ollier S, Boutinaud M. New developments on the galactopoietic role of prolactin in dairy ruminants. Domest Anim Endocrinol. 2012;43:154–160. doi: 10.1016/j.domaniend.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 184.Lacasse P, Ollier S. The dopamine antagonist domperidone increases prolactin concentration and enhances milk production in dairy cows. J Dairy Sci. 2015;98:7856–7864. doi: 10.3168/jds.2015-9865. [DOI] [PubMed] [Google Scholar]
- 185.Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. Mapping of N6- methyladenosine residues in bovine prolactin mRNA. Proc Natl Acad Sci USA. 1984;81:5667–5671. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Carroll SM, Narayan P, Rottman FM. N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol Cell Biol. 1990;10:4456–4465. doi: 10.1128/MCB.10.9.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Narayan P, Ludwiczak RL, Goodwin EC, Rottman FM. Context effects on N6-adenosine methylation sites in prolactin mRNA. Nucleic Acids Res. 1994;22:419–426. doi: 10.1093/nar/22.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Clarkson RW, Boland MP, Kritikou EA, Lee JM, Freeman TC, Tiffen PG, et al. The genes induced by signal transducer and activators of transcription (STAT)3 and STAT5 in mammary epithelial cells define the roles of these STATs in mammary development. Mol Endocrinol. 2006;20:675–685. doi: 10.1210/me.2005-0392. [DOI] [PubMed] [Google Scholar]
- 189.Adoro S, Cubillos-Ruiz JR, Chen X, Deruaz M, Vrbanac VD, Song M, et al. IL- 21 induces antiviral microRNA-29 in CD4 T cells to limit HIV-1 infection. Nat Commun. 2015;6:7562. doi: 10.1038/ncomms8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rozovski U, Calin GA, Setoyama T, D’Abundo L, Harris DM, Li P, et al. Signal transducer and activator of transcription (STAT)-3 regulates microRNA gene expression in chronic lymphocytic leukemia cells. Mol Cancer. 2013;12:50. doi: 10.1186/1476-4598-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Clark JW, Snell L, Shiu RP, Orr FW, Maitre N, Vary CP, et al. The potential role for prolactin-inducible protein (PIP) as a marker of human breast cancer micrometastasis. Br J Cancer. 1999;81:1002–1008. doi: 10.1038/sj.bjc.6690799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Naderi A. Prolactin-induced protein in breast cancer. Adv Exp Med Biol. 2015;846:189–200. doi: 10.1007/978-3-319-12114-7_8. [DOI] [PubMed] [Google Scholar]
- 193.Naderi A, Vanneste M. Prolactin-induced protein is required for cell cycle progression in breast cancer. Neoplasia. 2014;16:329–342. doi: 10.1016/j.neo.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tian W, Osawa M, Horiuchi H, Tomita Y. Expression of the prolactin-inducible protein (PIP/GCDFP15) gene in benign epithelium and adenocarcinoma of the prostate. Cancer Sci. 2004;95:491–495. doi: 10.1111/j.1349-7006.2004.tb03238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Baniwal SK, Little GH, Chimge NO, Frenkel B. Runx2 controls a feed-forward loop between androgen and prolactin-induced protein (PIP) in stimulating T47D cell proliferation. J Cell Physiol. 2012;227:2276–2282. doi: 10.1002/jcp.22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kobayashi K, Oyama S, Uejyo T, Kuki C, Rahman MM, Kumura H. Underlying mechanisms involved in the decrease of milk secretion during Escherichia coli endotoxin induced mastitis in lactating mice. Vet Res. 2013;44:119. doi: 10.1186/1297-9716-44-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fang J, Hao Q, Liu L, Li Y, Wu J, Huo X, et al. Epigenetic changes mediated by microRNA miR29 activate cyclooxygenase 2 and lambda-1 interferon production during viral infection. J Virol. 2012;86:1010–1020. doi: 10.1128/JVI.06169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J, et al. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV-related target genes for miR-29. J Pathol. 2011;224:484–495. doi: 10.1002/path.2873. [DOI] [PubMed] [Google Scholar]
- 199.Zur Hausen H, de Villiers EM. Dairy cattle serum and milk factors contributing to the risk of colon and breast cancers. Int J Cancer. 2015;137:959–967. doi: 10.1002/ijc.29466. [DOI] [PubMed] [Google Scholar]
- 200.Whitley C, Gunst K, Müller H, Funk M, Zur Hausen H, de Villiers EM. Novel replication-competent circular DNA molecules from healthy cattle serum and milk and multiple sclerosis-affected human brain tissue. Genome Announc. 2014;2:4. doi: 10.1128/genomeA.00849-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Funk M, Gunst K, Lucansky V, Müller H, ZurHausen H, deVilliers EM. Isolation of protein-associated circular DNA from healthy cattle serum. Genome Announc. 2014;2:4. doi: 10.1128/genomeA.00846-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. 1985;5:2298–2306. doi: 10.1128/MCB.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinform. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon- Divon M, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 206.Lin S, Gregory RI. Methyltransferases modulate RNA stability in embryonic stem cells. Nat Cell Biol. 2014;16:129–131. doi: 10.1038/ncb2914. [DOI] [PubMed] [Google Scholar]
- 207.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 209.Liu J, Jia G. Methylation modifications in eukaryotic messenger RNA. J Genet Genomics. 2014;41:21–33. doi: 10.1016/j.jgg.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 210.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013;41(Database issue):D262–7. [DOI] [PMC free article] [PubMed]
- 211.Sibbritt T, Patel HR, Preiss T. Mapping and significance of the mRNA methylome. Wiley Interdiscip Rev RNA. 2013;4:397–422. doi: 10.1002/wrna.1166. [DOI] [PubMed] [Google Scholar]
- 212.Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 213.Ikels K, Kuschel S, Fischer J, Kaisers W, Eberhard D, Rüther U. FTO is a relevant factor for the development of the metabolic syndrome in mice. PLoS One. 2014;9:e105349. doi: 10.1371/journal.pone.0105349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gustavsson J, Mehlig K, Leander K, Lissner L, Björck L, Rosengren A, et al. FTO genotype, physical activity, and coronary heart disease risk in Swedish men and women. Circ Cardiovasc Genet. 2014;7:171–177. doi: 10.1161/CIRCGENETICS.111.000007. [DOI] [PubMed] [Google Scholar]
- 215.Gustavsson J, Mehlig K, Leander K, Berg C, Tognon G, Strandhagen E, et al. FTO gene variation, macronutrient intake and coronary heart disease risk: a gene-diet interaction analysis. Eur J Nutr. 2015 [Epub ahead of print]. [DOI] [PubMed]
- 216.Armstrong BK, Mann JI, Adelstein AM, Eskin F. Commodity consumption and ischemic heart disease mortality, with special reference to dietary practices. J Chronic Dis. 1975;28:455–469. doi: 10.1016/0021-9681(75)90056-9. [DOI] [PubMed] [Google Scholar]
- 217.Grant WB. Milk and other dietary influences on coronary heart disease. Altern Med Rev. 1998;3:281–294. [PubMed] [Google Scholar]
- 218.Boney CM, Verma A, Tucker R, Vohr BR. Metabolic syndrome in childhood: association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics. 2005;115:e290–e296. doi: 10.1542/peds.2004-1808. [DOI] [PubMed] [Google Scholar]
- 219.Sørensen HT, Sabroe S, Rothman KJ, Gillman M, Fischer P, Sørensen TI. Relation between weight and length at birth and body mass index in young adulthood: cohort study. BMJ. 1997;315:1137. doi: 10.1136/bmj.315.7116.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Leunissen RW, Stijnen T, Hokken-Koelega AC. Influence of birth size on body composition in early adulthood: the programming factors for growth and metabolism (PROGRAM)-study. Clin Endocrinol (Oxf) 2009;70:245–251. doi: 10.1111/j.1365-2265.2008.03320.x. [DOI] [PubMed] [Google Scholar]
- 221.Brüske I, Flexeder C, Heinrich J. Body mass index and incidence of asthma in children. Curr Opin Allergy Clin Immunol. 2014;14:155–160. doi: 10.1097/ACI.0000000000000035. [DOI] [PubMed] [Google Scholar]
- 222.Skilton MR, Siitonen N, Würtz P, Viikari JS, Juonala M, Seppälä I, et al. High birth weight is associated with obesity and increased carotid wall thickness in young adults: the cardiovascular risk in young Finns study. Arterioscler Thromb Vasc Biol. 2014;34:1064–1068. doi: 10.1161/ATVBAHA.113.302934. [DOI] [PubMed] [Google Scholar]
- 223.Bukowski R, Chlebowski RT, Thune I, Furberg AS, Hankins GD, Malone FD, et al. Birth weight, breast cancer and the potential mediating hormonal environment. PLoS One. 2012;7:e40199. doi: 10.1371/journal.pone.0040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Spracklen CN, Wallace RB, Sealy-Jefferson S, Robinson JG, Freudenheim JL, Wellons MF, et al. Birth weight and subsequent risk of cancer. Cancer Epidemiol. 2014;38:538–543. doi: 10.1016/j.canep.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lewis RM, Demmelmair H, Gaillard R, Godfrey KM, Hauguel-de Mouzon S, Huppertz B, et al. The placental exposome: placental determinants of fetal adiposity and postnatal body composition. Ann Nutr Metab. 2013;63:208–215. doi: 10.1159/000355222. [DOI] [PubMed] [Google Scholar]
- 226.Symonds ME, Mendez MA, Meltzer HM, Koletzko B, Godfrey K, Forsyth S, et al. Early life nutritional programming of obesity: mother-child cohort studies. Ann Nutr Metab. 2013;62:137–145. doi: 10.1159/000345598. [DOI] [PubMed] [Google Scholar]
- 227.Sébert SP, Hyatt MA, Chan LL, Yiallourides M, Fainberg HP, Patel N, et al. Influence of prenatal nutrition and obesity on tissue specific fat mass and obesity-associated (FTO) gene expression. Reproduction. 2010;139:265–274. doi: 10.1530/REP-09-0173. [DOI] [PubMed] [Google Scholar]
- 228.Bassols J, Prats-Puig A, Vázquez-Ruíz M, García-González MM, Martínez- Pascual M, Avellí P, et al. Placental FTO expression relates to fetal growth. Int J Obes (Lond) 2010;34:1365–1370. doi: 10.1038/ijo.2010.62. [DOI] [PubMed] [Google Scholar]
- 229.Mook-Kanamori DO, Durmuş B, Sovio U, Hofman A, Raat H, Steegers EA, et al. Fetal and infant growth and the risk of obesity during early childhood: the Generation R Study. Eur J Endocrinol. 2011;165:623–630. doi: 10.1530/EJE-11-0067. [DOI] [PubMed] [Google Scholar]
- 230.Olsen SF, Halldorsson TI, Willett WC, Knudsen VK, Gillman MW, Mikkelsen TB, et al. Milk consumption during pregnancy is associated with increased infant size at birth: prospective cohort study. Am J Clin Nutr. 2007;86:1104–1110. doi: 10.1093/ajcn/86.4.1104. [DOI] [PubMed] [Google Scholar]
- 231.Heppe DH, van Dam RM, Willemsen SP, den Breeijen H, Raat H, Hofman A, et al. Maternal milk consumption, fetal growth, and the risks of neonatal complications: the Generation R Study. Am J Clin Nutr. 2011;94:501–509. doi: 10.3945/ajcn.111.013854. [DOI] [PubMed] [Google Scholar]
- 232.Melnik BC, John SM, Schmitz G. Milk consumption during pregnancy increases birth weight, a risk factor for the development of diseases of civilization. J Transl Med. 2015;13:13. doi: 10.1186/s12967-014-0377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sovio U, Mook-Kanamori DO, Warrington NM, Lawrence R, Briollais L, Palmer CN, et al. Association between common variation at the FTO locus and changes in body mass index from infancy to late childhood: the complex nature of genetic association through growth and development. PLoS Genet. 2011;7:e1001307. doi: 10.1371/journal.pgen.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Warrington NM, Howe LD, Paternoster L, Kaakinen M, Herrala S, Huikari V, et al. A genome-wide association study of body mass index across early life and childhood. Int J Epidemiol. 2015;44:700–712. doi: 10.1093/ije/dyv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wiley AS. Dairy and milk consumption and child growth: is BMI involved? An analysis of NHANES 1999-2004. Am J Hum Biol. 2010;22:517–525. doi: 10.1002/ajhb.21042. [DOI] [PubMed] [Google Scholar]
- 236.Rosskopf D, Schwahn C, Neumann F, Bornhorst A, Rimmbach C, Mischke M, et al. The growth hormone–IGF-I axis as a mediator for the association between FTO variants and body mass index: results of the Study of Health in Pomerania. Int J Obes (Lond) 2011;35:364–372. doi: 10.1038/ijo.2010.158. [DOI] [PubMed] [Google Scholar]
- 237.Elks CE, Loos RJ, Hardy R, Wills AK, Wong A, Wareham NJ, et al. Adult obesity susceptibility variants are associated with greater childhood weight gain and a faster tempo of growth: the 1946 British Birth Cohort Study. Am J Clin Nutr. 2012;95:1150–1156. doi: 10.3945/ajcn.111.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rich-Edwards JW, Ganmaa D, Pollak MN, Nakamoto EK, Kleinman K, Tserendolgor U, et al. Milk consumption and the prepubertal somatotropic axis. Nutr J. 2007;6:28. doi: 10.1186/1475-2891-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Qin LQ, He K, Xu JY. Milk consumption and circulating insulin-like growth factor-I level: a systematic literature review. Int J Food Sci Nutr. 2009;60(Suppl 7):330–340. doi: 10.1080/09637480903150114. [DOI] [PubMed] [Google Scholar]
- 240.Hoppe C, Udam TR, Lauritzen L, Mølgaard C, Juul A, Michaelsen KF. Animal protein intake, serum insulin-like growth factor I, and growth in healthy 2.5-year-old Danish children. Am J Clin Nutr. 2004;80:447–452. doi: 10.1093/ajcn/80.2.447. [DOI] [PubMed] [Google Scholar]
- 241.Hoppe C, Mølgaard C, Michaelsen KF. Cow’s milk and linear growth in industrialized and developing countries. Annu Rev Nutr. 2006;26:131–173. doi: 10.1146/annurev.nutr.26.010506.103757. [DOI] [PubMed] [Google Scholar]
- 242.Mumby HS, Elks CE, Li S, Sharp SJ, Khaw KT, Luben RN, et al. Mendelian Randomisation Study of Childhood BMI and Early Menarche. J Obes. 2011;2011:180729. doi: 10.1155/2011/180729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Janghorbani M, Mansourian M, Hosseini E. Systematic review and meta- analysis of age at menarche and risk of type 2 diabetes. Acta Diabetol. 2014;51:519–528. doi: 10.1007/s00592-014-0579-x. [DOI] [PubMed] [Google Scholar]
- 244.Wilson DA, Derraik JG, Rowe DL, Hofman PL, Cutfield WS. Earlier menarche is associated with lower insulin sensitivity and increased adiposity in young adult women. PLoS One. 2015;10:e0128427. doi: 10.1371/journal.pone.0128427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wiley AS. Milk intake and total dairy consumption: associations with early menarche in NHANES 1999-2004. PLoS One. 2011;6:e14685. doi: 10.1371/journal.pone.0014685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Carwile JL, Willett WC, Wang M, Rich-Edwards J, Frazier AL, Michels KB. Milk consumption after age 9 years does not predict age at menarche. J Nutr. 2015;145:1900–1908. doi: 10.3945/jn.115.214270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ahmad T, Chasman DI, Mora S, Paré G, Cook NR, Buring JE, et al. The fat- mass and obesity-associated (FTO) gene, physical activity, and risk of incident cardiovascular events in white women. Am Heart J. 2010;160:1163–1169. doi: 10.1016/j.ahj.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.López-Bermejo A, Petry CJ, Díaz M, Sebastiani G, de Zegher F, Dunger DB, et al. The association between the FTO gene and fat mass in humans develops by the postnatal age of two weeks. J Clin Endocrinol Metab. 2008;93:1501–1515. doi: 10.1210/jc.2007-2343. [DOI] [PubMed] [Google Scholar]
- 249.Ben-Haim MS, Moshitch-Moshkovitz S, Rechavi G. FTO: linking m6A demethylation to adipogenesis. Cell Res. 2015;25:3–4. doi: 10.1038/cr.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhang M, Zhang Y, Ma J, Guo F, Cao Q, Zhang Y, et al. The demethylase a activity of FTO (Fat Mass and Obesity Associated Protein) is required for preadipocyte differentiation. PLoS One. 2015;10:e0133788. doi: 10.1371/journal.pone.0133788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tews D, Fischer-Posovszky P, Fromme T, Klingenspor M, Fischer J, Rüther U, et al. FTO deficiency induces UCP-1 expression and mitochondrial uncoupling in adipocytes. Endocrinology. 2013;154:3141–3151. doi: 10.1210/en.2012-1873. [DOI] [PubMed] [Google Scholar]
- 253.Stephens M, Ludgate M, Rees DA. Brown fat and obesity: the next big thing? Clin Endocrinol (Oxf). 2011;74:661–670. doi: 10.1111/j.1365-2265.2011.04018.x. [DOI] [PubMed] [Google Scholar]
- 254.Rockstroh D, Landgraf K, Wagner IV, Gesing J, Tauscher R, Lakowa N, et al. Direct evidence of brown adipocytes in different fat depots in children. PLoS One. 2015;10:e0117841. doi: 10.1371/journal.pone.0117841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Berkey CS, Rocket HR, Willet WC, Colditz GA. Milk, dairy fat, dietary calcium, and weight gain. Arch Pediatr Adolesc Med. 2005;159:543–550. doi: 10.1001/archpedi.159.6.543. [DOI] [PubMed] [Google Scholar]
- 256.Matthews VL, Wien M, Sabaté J. The risk of child and adolescent overweight is related to types of food consumed. Nutr J. 2011;10:71. doi: 10.1186/1475-2891-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Arnberg K, Mølgaard C, Michaelsen KF, Jensen SM, Trolle E, Larnkjær A. Skim milk, whey, and casein increase body weight and whey and casein increase plasma C-peptide concentration in overweight adolescents. J Nutr. 2012;142:2083–2090. doi: 10.3945/jn.112.161208. [DOI] [PubMed] [Google Scholar]
- 258.Barr SI, McCarron DA, Heaney RP, Dawson-Hughes B, Berga SL, Stern JS, et al. Effects of increased consumption of fluid milk on energy and nutrient intake, body weight, and cardiovascular risk factors in healthy older adults. Am J Diet Assoc. 2000;100:810–817. doi: 10.1016/S0002-8223(00)00236-4. [DOI] [PubMed] [Google Scholar]
- 259.Serralde-Zúñiga AE, Guevara-Cruz M, Tovar AR, Herrera-Hernández MF, Noriega LG, Granados O, et al. Omental adipose tissue gene expression, gene variants, branched-chain amino acids, and their relationship with metabolic syndrome and insulin resistance in humans. Genes Nutr. 2014;9:431. doi: 10.1007/s12263-014-0431-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Bell CG, Finer S, Lindgren CM, Wilson GA, Rakyan VK, Teschendorff AE, et al. Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS One. 2010;5:e14040. doi: 10.1371/journal.pone.0014040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Arnold N, Koppula PR, Gul R, Luck C, Pulakat L. Regulation of cardiac expression of the diabetic marker microRNA miR-29. PLoS One. 2014;9:e103284. doi: 10.1371/journal.pone.0103284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ślusarz A, Pulakat L. The two faces of miR-29. Diabetogenic miRs. J Cardiovasc Med (Hagerstown). 2015;16:480–90. [DOI] [PMC free article] [PubMed]
- 263.Kurtz CL, Peck BC, Fannin EE, Beysen C, Miao J, Landstreet SR, et al. MicroRNA-29 fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes. Diabetes. 2014;63:3141–3148. doi: 10.2337/db13-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Melnik BC. The pathogenic role of persistent milk signaling in mTORC1- and milk-microRNA-driven type 2 diabetes mellitus. Curr Diabetes Rev. 2015;11:46–62. doi: 10.2174/1573399811666150114100653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Shen F, Huang W, Huang JT, Xiong J, Yang Y, Wu K, et al. Decreased N(6)- methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J Clin Endocrinol Metab. 2015;100:E148–E154. doi: 10.1210/jc.2014-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Guo F, Zhang Y, Zhang C, Wang S, Ni Y, Zhao R. Fatmass and obesity associated (FTO) gene regulates gluconeogenesis in chicken embryo fibroblast cells. Comp Biochem Physiol A: Mol Integr Physiol. 2015;179:149–156. doi: 10.1016/j.cbpa.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 267.Zhang JW, Tang QQ, Vinson C, Lane MD. Dominant-negative C/EBP disrupts mitotic clonal expansion and differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci USA. 2004;101:43–47. doi: 10.1073/pnas.0307229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Bravard A, Veilleux A, Disse E, Laville M, Vidal H, Tchernof A, et al. The expression of FTO in human adipose tissue is influenced by fat depot, adiposity, and insulin sensitivity. Obesity (Silver Spring) 2013;21:1165–1173. doi: 10.1002/oby.20110. [DOI] [PubMed] [Google Scholar]
- 269.Sluijs I, Forouhi NG, Beulens JW, van der Schouw YT, Agnoli C, Arriola L, et al. The amount and type of dairy product intake and incident type 2 diabetes: results from the EPIC-InterAct Study. Am J Clin Nutr. 2012;96:382–390. doi: 10.3945/ajcn.111.021907. [DOI] [PubMed] [Google Scholar]
- 270.Hoppe C, Mølgaard C, Vaag A, Barkholt V, Michaelsen KF. High intakes of milk, but not meat, increase s-insulin and insulin resistance in 8-year-old boys. Eur J Clin Nutr. 2005;59:393–398. doi: 10.1038/sj.ejcn.1602086. [DOI] [PubMed] [Google Scholar]
- 271.Melnik BC, Schmitz G. Metformin: an inhibotor of mTORC1 singaling. J Endocrinol Diabetes Obes. 2014;2:1029. [Google Scholar]
- 272.Yan L, Zhou J, Gao Y, Ghazal S, Lu L, Bellone S, et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin- induced DNA methylation. Oncogene. 2015;34:3076–3084. doi: 10.1038/onc.2014.236. [DOI] [PubMed] [Google Scholar]
- 273.Hernández-Caballero ME, Sierra-Ramírez JA. Single nucleotide polymorphisms of the FTO gene and cancer risk: an overview. Mol Biol Rep. 2015;42:699–704. doi: 10.1007/s11033-014-3817-y. [DOI] [PubMed] [Google Scholar]
- 274.Carreira PE, Richardson SR, Faulkner GJ. L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J. 2014;281:63–73. doi: 10.1111/febs.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/S0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 276.Lewis SJ, Murad A, Chen L, Davey Smith G, Donovan J, Palmer T, et al. Associations between an obesity related genetic variant (FTO rs9939609) and prostate cancer risk. PLoS One. 2010;5:e13485. doi: 10.1371/journal.pone.0013485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Machiela MJ, Lindström S, Allen NE, Haiman CA, Albanes D, Barricarte A, et al. Association of type 2 diabetes susceptibility variants with advanced prostate cancer risk in the Breast and Prostate Cancer Cohort Consortium. Am J Epidemiol. 2012;176:1121–1129. doi: 10.1093/aje/kws191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Best LG, García-Esquinas E, Yeh JL, Yeh F, Zhang Y, Lee ET, et al. Association of diabetes and cancer mortality in American Indians: the Strong Heart Study. Cancer Causes Control. 2015 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 279.Pischon T, Boeing H, Weikert S, Allen N, Key T, Johnsen NF, et al. Body size and risk of prostate cancer in the European prospective investigation into cancer and nutrition. Cancer Epidemiol Biomarkers Prev. 2008;17:3252–3261. doi: 10.1158/1055-9965.EPI-08-0609. [DOI] [PubMed] [Google Scholar]
- 280.Allen NE, Key TJ, Appleby PN, Travis RC, Roddam AW, Tjønneland A, et al. Animal foods, protein, calcium and prostate cancer risk: the European Prospective Investigation into Cancer and Nutrition. Br J Cancer. 2008;98:1574–1581. doi: 10.1038/sj.bjc.6604331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Song Y, Chavarro JE, Cao Y, Qiu W, Mucci L, Sesso HD, et al. Whole milk intake is associated with prostate cancer-specific mortality among US male physicians. J Nutr. 2013;143:189–196. doi: 10.3945/jn.112.168484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Bernichtein S, Pigat N, Capiod T, Boutillon F, Verkarre V, Camparo P, et al. High milk consumption does not affect prostate tumor progression in two mouse models of benign and neoplastic lesions. PLoS One. 2015;10:e0125423. doi: 10.1371/journal.pone.0125423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Torfadottir JE, Steingrimsdottir L, Mucci L, Aspelund T, Kasperzyk JL, Olafsson O, et al. Milk intake in early life and risk of advanced prostate cancer. Am J Epidemiol. 2012;175:144–153. doi: 10.1093/aje/kwr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Melnik BC, John SM, Carrera-Bastos P, Cordain L. The impact of cow’s milk- mediated mTORC1-signaling in the initiation and progression of prostate cancer. Nutr Metab (Lond). 2012;9:74. doi: 10.1186/1743-7075-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Tate PL, Bibb R, Larcom LL. Milk stimulates growth of prostate cancer cells in culture. Nutr Cancer. 2011;63:1361–1366. doi: 10.1080/01635581.2011.609306. [DOI] [PubMed] [Google Scholar]
- 287.Hwang IC, Park SM, Shin D, Ahn HY, Rieken M, Shariat SF. Metformin association with lower prostate cancer recurrence in type 2 diabetes: a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2015;16:595–600. doi: 10.7314/APJCP.2015.16.2.595. [DOI] [PubMed] [Google Scholar]
- 288.Raval AD, Thakker D, Vyas A, Salkini M, Madhavan S, Sambamoorthi U. Impact of metformin on clinical outcomes among men with prostate cancer: a systematic review and meta-analysis. Prostate Cancer Prostatic Dis. 2015;18:110–121. doi: 10.1038/pcan.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Tseng CH. Metformin significantly reduces incident prostate cancer risk in Taiwanese men with type 2 diabetes mellitus. Eur J Cancer. 2014;50:2831–2837. doi: 10.1016/j.ejca.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 290.Mayer MJ, Klotz LH, Venkateswaran V. Metformin and prostate cancer stem cells: a novel therapeutic target. Prostate Cancer Prostatic Dis. 2015;18:303–309. doi: 10.1038/pcan.2015.35. [DOI] [PubMed] [Google Scholar]
- 291.Jain R, Strickler HD, Fine E, Sparano JA. Clinical studies examining the impact of obesity on breast cancer risk and prognosis. J Mammary Gland Biol Neoplasia. 2013;18:257–266. doi: 10.1007/s10911-013-9307-3. [DOI] [PubMed] [Google Scholar]
- 292.van Veldhoven K, Polidoro S, Baglietto L, Severi G, Sacerdote C, Panico S, et al. Epigenome-wide association study reveals decreased average methylation levels years before breast cancer diagnosis. Clin Epigenetics. 2015;7:67. doi: 10.1186/s13148-015-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–398. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.da Cunha PA, de Carlos Back LK, Sereia AF, Kubelka C, Ribeiro MC, Fernandes BL, et al. Interaction between obesity-related genes, FTO and MC4R, associated to an increase of breast cancer risk. Mol Biol Rep. 2013;40:6657–6664. doi: 10.1007/s11033-013-2780-3. [DOI] [PubMed] [Google Scholar]
- 295.Zeng X, Ban Z, Cao J, Zhang W, Chu T, Lei D, et al. Association of FTO mutations with risk and survival of breast cancer in a Chinese population. Dis Markers. 2015;2015:101032. doi: 10.1155/2015/101032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Zhang B, Li Y, Li L, Chen M, Zhang C, Zuo XB, et al. Association study of susceptibility loci with specific breast cancer subtypes in Chinese women. Breast Cancer Res Treat. 2014;146:503–514. doi: 10.1007/s10549-014-3041-4. [DOI] [PubMed] [Google Scholar]
- 297.Stone J, Thompson DJ, Dos Santos Silva I, Scott C, Tamimi RM, Lindstrom S, et al. Novel associations between common breast cancer susceptibility variants and risk-predicting mammographic density measures. Cancer Res. 2015;75:2457–2467. doi: 10.1158/0008-5472.CAN-14-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Singh B, Shamsnia A, Raythatha MR, Milligan RD, Cady AM, Madan S, et al. Highly adaptable triple-negative breast cancer cells as a functional model for testing anticancer agents. PLoS One. 2014;9:e109487. doi: 10.1371/journal.pone.0109487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Tretli S, Gaard M. Lifestyle changes during adolescence and risk of breast cancer: an ecologic study of the effect of World War II in Norway. Cancer Causes Control. 1996;7:507–512. doi: 10.1007/BF00051882. [DOI] [PubMed] [Google Scholar]
- 300.Gaard M, Tretli S, Løken EB. Dietary fat and the risk of breast cancer: a prospective study of 25,892 Norwegian women. Int J Cancer. 1995;63:13–17. doi: 10.1002/ijc.2910630104. [DOI] [PubMed] [Google Scholar]
- 301.Qin LQ, Xu JY, Tezuka H, Li J, Arita J, Hoshi K, et al. Consumption of commercial whole and non-fat milk increases the incidence of 7,12- dimethylbenz(a)anthracene-induced mammary tumors in rats. Cancer Detect Prev. 2007;31:339–343. doi: 10.1016/j.cdp.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 302.Smith RD, Hilf R, Senior AE. Prolactin binding to mammary gland, 7,12- dimethylbenz(a)-anthracene-induced mammary tumors, and liver in rats. Cancer Res. 1976;36:3726–3731. [PubMed] [Google Scholar]
- 303.Meites J. Relation of prolactin and estrogen to mammary tumorigenesis in the rat. J Natl Cancer Inst. 1972;48:1217–1224. [PubMed] [Google Scholar]
- 304.Heuson JC, Waelbroeck C, Legros N, Gallez G, Robyn C, L’Hermite M. Inhibition of DMBA-induced mammary carcinogenesis in the rat by 2-br- ergocryptine (CB 154), an inhibitor of prolactin secretion, and by nafoxidine (U- 11,100 A), an estrogen antagonist. Gynecol Invest. 1971;2:130–137. doi: 10.1159/000301858. [DOI] [PubMed] [Google Scholar]
- 305.Tworoger SS, Hankinson SE. Prolactin and breast cancer etiology: an epidemiologic perspective. J Mammary Gland Biol Neoplasia. 2008;13:41–53. doi: 10.1007/s10911-008-9063-y. [DOI] [PubMed] [Google Scholar]
- 306.Martin RM, Middleton N, Gunnell D, Owen CG, Smith GD. Breast-feeding and cancer: the Boyd Orr cohort and a systematic review with meta-analysis. J Natl Cancer Inst. 2005;97:1446–1457. doi: 10.1093/jnci/dji291. [DOI] [PubMed] [Google Scholar]
- 307.Collaborative Group on Hormonal Factors in Breast Cancer Breast cancer and breastfeeding: collaborative reanalysis of individual data from 47 epidemiological studies in 30 countries, including 50302 women with breast cancer and 96973 women without the disease. Lancet. 2002;360:187–195. doi: 10.1016/S0140-6736(02)09454-0. [DOI] [PubMed] [Google Scholar]
- 308.Islami F, Liu Y, Jemal A, Zhou J, Weiderpass E, Colditz G, et al. Breastfeeding and breast cancer risk by receptor status-a systematic review and meta- analysis. Ann Oncol. 2015. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 309.Ji J, Sundquist J, Sundquist K. Lactose intolerance and risk of lung, breast and ovarian cancers: aetiological clues from a population-based study in Sweden. Br J Cancer. 2015;112:149–152. doi: 10.1038/bjc.2014.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Tseng CH. Metformin may reduce breast cancer risk in Taiwanese women with type 2 diabetes. Breast Cancer Res Treat. 2014;145:785–790. doi: 10.1007/s10549-014-2985-8. [DOI] [PubMed] [Google Scholar]
- 311.Hatoum D, McGowan EM. Recent advances in the use of metformin: can treating diabetes prevent breast cancer? Biomed Res Int. 2015;2015:548436. doi: 10.1155/2015/548436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Baniwal SK, Chimge NO, Jordan VC, Tripathy D, Frenkel B. Prolactin-induced protein (PIP) regulates proliferation of luminal A type breast cancer cells in an estrogen-independent manner. PLoS One. 2013;8:e62361. doi: 10.1371/journal.pone.0062361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Campdelacreu J. Parkinson disease and Alzheimer disease: environmental risk factors. Neurologia. 2014;29:541–549. doi: 10.1016/j.nrl.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 314.Chouliaras L, Rutten BP, Kenis G, Peerbooms O, Visser PJ, Verhey F, et al. Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog Neurobiol. 2010;90:498–510. doi: 10.1016/j.pneurobio.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 315.Otaegui-Arrazola A, Amiano P, Elbusto A, Urdaneta E, Martínez-Lage P. Diet, cognition, and Alzheimer’s disease: food for thought. Eur J Nutr. 2014;53:1–23. doi: 10.1007/s00394-013-0561-3. [DOI] [PubMed] [Google Scholar]
- 316.Davinelli S, Calabrese V, Zella D, Scapagnini G. Epigenetic nutraceutical diets in Alzheimer’s disease. J Nutr Health Aging. 2014;18:800–805. doi: 10.1007/s12603-014-0552-y. [DOI] [PubMed] [Google Scholar]
- 317.Tang Z, Bereczki E, Zhang H, Wang S, Li C, Ji X, et al. Mammalian target of rapamycin (mTor) mediates tau protein dyshomeostasis: implication for Alzheimer disease. J Biol Chem. 2013;288:15556–15570. doi: 10.1074/jbc.M112.435123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Tang Z, Ioja E, Bereczki E, Hultenby K, Li C, Guan Z, et al. mTor mediates tau localization and secretion: implication for Alzheimer’s disease. Biochim Biophys Acta. 2015;1853:1646–1657. doi: 10.1016/j.bbamcr.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 319.Ho AJ, Stein JL, Hua X, Lee S, Hibar DP, Leow AD, et al. A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc Natl Acad Sci USA. 2010;107:8404–8409. doi: 10.1073/pnas.0910878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Benedict C, Jacobsson JA, Rönnemaa E, Sällman-Almén M, Brooks S, Schultes B, et al. The fat mass and obesity gene is linked to reduced verbal fluency in overweight and obese elderly men. Neurobiol Aging. 2011; 32:1159.e1–5. [DOI] [PubMed]
- 321.Keller L, Xu W, Wang HX, Winblad B, Fratiglioni L, Graff C. The obesity related gene, FTO, interacts with APOE, and is associated with Alzheimer’s disease risk: a prospective cohort study. J Alzheimers Dis. 2011;23:461–469. doi: 10.3233/JAD-2010-101068. [DOI] [PubMed] [Google Scholar]
- 322.Reitz C, Tosto G, Mayeux R. Luchsinger JA; NIA-LOAD/NCRAD Family Study Group; Alzheimer’s Disease Neuroimaging Initiative. Genetic variants in the Fat and Obesity Associated (FTO) gene and risk of Alzheimer’s disease. PLoS One. 2012;7:e50354. doi: 10.1371/journal.pone.0050354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Adebakin A, Bradley J, Gümüsgöz S, Waters EJ, Lawrence CB. Impaired satiation and increased feeding behaviour in the triple-transgenic Alzheimer’s disease mouse model. PLoS One. 2012;7:e45179. doi: 10.1371/journal.pone.0045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Gu Y, Nieves JW, Stern Y, Luchsinger JA, Scarmeas N. Food combination and Alzheimer disease risk: a protective diet. Arch Neurol. 2010;67:699–706. doi: 10.1001/archneurol.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Morales-Briceño H, Cervantes-Arriaga A, Rodríguez-Violante M, Calleja- Castillo J, Corona T. Overweight is more prevalent in patients with Parkinson’s disease. Arq Neuropsiquiatr. 2012;70:843–846. doi: 10.1590/S0004-282X2012001100004. [DOI] [PubMed] [Google Scholar]
- 326.Sääksjärvi K, Knekt P, Männistö S, Lyytinen J, Heliövaara M. Prospective study on the components of metabolic syndrome and the incidence of Parkinson’s disease. Parkinsonism Relat Disord. 2015;21:1148–1155. doi: 10.1016/j.parkreldis.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 327.Vikdahl M, Carlsson M, Linder J, Forsgren L, Håglin L. Weight gain and increased central obesity in the early phase of Parkinson’s disease. Clin Nutr. 2014;33:1132–1139. doi: 10.1016/j.clnu.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 328.Chen H, Zhang SM, Hernán MA, Willett WC, Ascherio A. Diet and Parkinson’s disease: a potential role of dairy products in men. Ann Neurol. 2002;52:793–801. doi: 10.1002/ana.10381. [DOI] [PubMed] [Google Scholar]
- 329.Park M, Ross GW, Petrovitch H, White LR, Masaki KH, Nelson JS, et al. Consumption of milk and calcium in midlife and the future risk of Parkinson disease. Neurology. 2005;64:1047–1051. doi: 10.1212/01.WNL.0000154532.98495.BF. [DOI] [PubMed] [Google Scholar]
- 330.Chen H, O’Reilly E, McCullough ML, Rodriguez C, Schwarzschild MA, Calle EE, et al. Consumption of dairy products and risk of Parkinson’s disease. Am J Epidemiol. 2007;165:998–1006. doi: 10.1093/aje/kwk089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Kyrozis A, Ghika A, Stathopoulos P, Vassilopoulos D, Trichopoulos D, Trichopoulou A. Dietary and lifestyle variables in relation to incidence of Parkinson’s disease in Greece. Eur J Epidemiol. 2013;28:67–77. doi: 10.1007/s10654-012-9760-0. [DOI] [PubMed] [Google Scholar]
- 332.Sääksjärvi K, Knekt P, Lundqvist A, Männistö S, Heliövaara M, Rissanen H, et al. A cohort study on diet and the risk of Parkinson’s disease: the role of food groups and diet quality. Br J Nutr. 2013;109:329–337. doi: 10.1017/S0007114512000955. [DOI] [PubMed] [Google Scholar]
- 333.Jiang W, Ju C, Jiang H, Zhang D. Dairy foods intake and risk of Parkinson’s disease: a dose-response meta-analysis of prospective cohort studies. Eur J Epidemiol. 2014;29:613–619. doi: 10.1007/s10654-014-9921-4. [DOI] [PubMed] [Google Scholar]
- 334.Xu X. DNA methylation and cognitive aging. Oncotarget. 2015;6:13925–13934. doi: 10.18632/oncotarget.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Prattichizzo F, Giuliani A, Ceka A, Rippo MR, Bonfigli AR, Testa R, et al. Epigenetic mechanisms of endothelial dysfunction in type 2 diabetes. Clin Epigenetics. 2015;7:56. doi: 10.1186/s13148-015-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Li S, Mason CE. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet. 2014;15:127–150. doi: 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- 337.Sebert S, Salonurmi T, Keinänen-Kiukaanniemi S, Savolainen M, Herzig KH, Symonds ME, et al. Programming effects of FTO in the development of obesity. Acta Physiol (Oxf) 2014;210:58–69. doi: 10.1111/apha.12196. [DOI] [PubMed] [Google Scholar]
- 338.Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Michaëlsson K, Wolk A, Langenskiöld S, Basu S, Warensjö Lemming E, Melhus H, et al. Milk intake and risk of mortality and fractures in women and men: cohort studies. BMJ. 2014;349:g6015. doi: 10.1136/bmj.g6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Blagosklonny MV. Revisiting the antagonistic pleiotropy theory of aging: TOR- driven program and quasi-program. Cell Cycle. 2010;9:3151–3156. doi: 10.4161/cc.9.16.12814. [DOI] [PubMed] [Google Scholar]
- 341.Chuang YF, Tanaka T, Beason-Held LL, An Y, Terracciano A, Sutin AR, et al. FTO genotype and aging: pleiotropic longitudinal effects on adiposity, brain function, impulsivity and diet. Mol Psychiatry. 2015;20:133–139. doi: 10.1038/mp.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Shin C, Kim NH, Baik I. Sex-specific association between longitudinal changes in adiposity, FTO rs9939609 polymorphism, and leukocyte telomere length. J Am Coll Nutr. 2015;11:1–10. doi: 10.1080/07315724.2015.1005197. [DOI] [PubMed] [Google Scholar]


