Significance
The ribosome, in analogy with a tree, contains a record of its history, spanning 4 billion years of life on earth. The information contained within ribosomes connects us to the prehistory of biology. Details of ribosomal RNA variation, observed by comparing three-dimensional structures of ribosomes across the tree of life, form the basis of our molecular-level model of the origins and evolution of the translational system. We infer many steps in the evolution of translation, mapping out acquisition of structure and function, revealing much about how modern biology originated from ancestral chemical systems.
Keywords: RNA evolution, translation, origin of life, A-minor interactions
Abstract
We present a molecular-level model for the origin and evolution of the translation system, using a 3D comparative method. In this model, the ribosome evolved by accretion, recursively adding expansion segments, iteratively growing, subsuming, and freezing the rRNA. Functions of expansion segments in the ancestral ribosome are assigned by correspondence with their functions in the extant ribosome. The model explains the evolution of the large ribosomal subunit, the small ribosomal subunit, tRNA, and mRNA. Prokaryotic ribosomes evolved in six phases, sequentially acquiring capabilities for RNA folding, catalysis, subunit association, correlated evolution, decoding, energy-driven translocation, and surface proteinization. Two additional phases exclusive to eukaryotes led to tentacle-like rRNA expansions. In this model, ribosomal proteinization was a driving force for the broad adoption of proteins in other biological processes. The exit tunnel was clearly a central theme of all phases of ribosomal evolution and was continuously extended and rigidified. In the primitive noncoding ribosome, proto-mRNA and the small ribosomal subunit acted as cofactors, positioning the activated ends of tRNAs within the peptidyl transferase center. This association linked the evolution of the large and small ribosomal subunits, proto-mRNA, and tRNA.
The ribosome retains interpretable molecular records of a world of primordial molecules (1) from around 4 billion years ago (2–9). The records are maintained in rRNA secondary and 3D structures, which are fully conserved throughout the tree of life, and in rRNA sequences, which are more variable (SI Appendix, Fig. S1). Here we use information within ribosomes from each major branch of the tree of life to reconstruct much of the emergence of the universal translational machinery.
Large Ribosomal Subunit Evolution
Previously, we reported a 3D comparative method that revealed a molecular level chronology of the evolution of the large ribosomal subunit (LSU) rRNA (10). Insertion fingerprints are evident when comparing 3D structures of LSU rRNAs of various sizes from various species. These insertion fingerprints mark sites where rRNA expands, recording growth steps on a molecular level.
Within the common core of the LSU rRNA, insertion fingerprints were used to identify ancient growth sites. We showed that insertion fingerprints provide a roadmap from the first steps in the formation of the peptidyl transferase center (PTC) (10) located in the ancient heart of the LSU (2–6), culminating in the common core.
Small Ribosomal Subunit, LSU, tRNA, and mRNA Evolution
Here, using the 3D comparative method, we establish a comprehensive and coherent model for the evolution of the entire ribosome. This model covers the LSU rRNA, small ribosomal subunit (SSU) rRNA, tRNA, and mRNA. The evolution of each of these components is reconciled at the molecular level to a common chronology. This evolutionary model, which we call the “accretion model,” is fully grounded in structural data in the form of insertion fingerprints and molecular interactions. The model describes iterative accretion of rRNA fragments in the form of expansion segments. The timeline of the accretion model initiates in ancient proto-biology in the initial building up of the functional centers, proceeds to the establishment of the common core, and continues to the development of large metazoan rRNAs.
Incremental evolution of function is mapped out by stepwise accretion of rRNA. In the extant ribosome, specific segments of rRNA perform specific functions including peptidyl transfer, subunit association, decoding, and energy-driven translocation (11). The model assumes that the correlations of rRNA segments with their functions have been reasonably maintained over the broad course of ribosomal evolution. Therefore the model maps out the time course of acquisition of function. Breadth of function increased as the ribosome grew in size.
rRNA Variation
Expansion segments (“ES” indicates LSU expansion segments, and “es” indicates SSU expansion segments) (7, 8, 12, 13) are small, folding-competent RNA fragments that are inserted into common core rRNA over evolution, increasing rRNA length without substantially perturbing the structure of the common core. Recursive insertion of expansion segments leads to variation in the length of rRNAs. In extant species, rRNA lengths vary according to well-defined rules. (i) rRNA length tends to increase from bacteria/archaea, which approximate the common core, to protists, to metazoa (9, 10). (ii) Size variation is significantly greater in eukaryotes than in prokaryotes and in LSU rRNAs than in SSU rRNAs (8). (iii) Variation is focused at a few specific sites of the common core (7–10, 14, 15). (iv) Variation is excluded from the interior and from functional regions of the rRNA such as the PTC, the decoding center, the core of the subunit interface, and tRNA-binding sites (16). (v) rRNA size generally increases with organismal complexity (10). Here, we consider ancestral expansion segments, which are found within the common core (10) and are denoted as “AES” (LSU) and “aes” (SSU).
Results
Using a 3D comparative method, we establish the order of acquisition of AES and aes. This chronology assimilates serial AES/aes accretion at the sites of expansion marked by insertion fingerprints. Function is acquired commensurate with structural growth.
The SSU from the Common Core to Extant Biology.
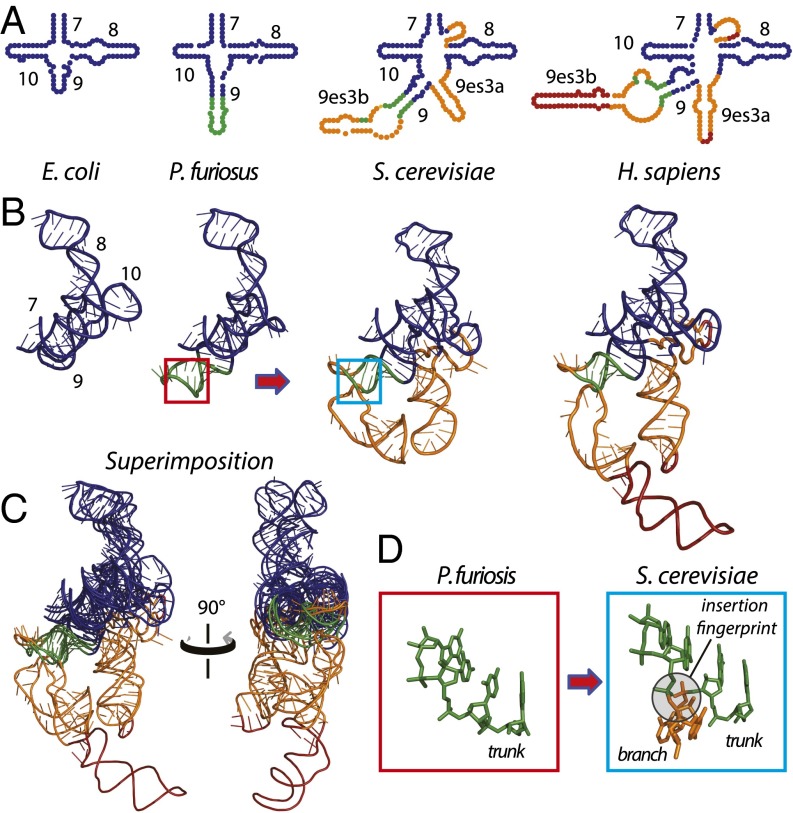
To illustrate SSU expansions that took place after formation of the common core, we use the region of helices 7–10 (Fig. 1 A and B). In the bacterium Escherichia coli, helices 7–10 are 92 nt in length (17). This region of the E. coli SSU is a reasonable approximation of the corresponding region of the common core except for an eight-nucleotide addition to helix 10. In the archaean Pyrococcus furiosus, helix 9 is expanded by 11 nt (18) relative to E. coli (Dataset S1). This rRNA is expanded further in eukaryotes by the addition of three helices constituting expansion segment es3. Helices 7–10 and es3 have a combined length of 174 nt in Saccharomyces cerevisiae (19). This segment of the rRNA is maintained at nearly that size in protists, plants, and invertebrate animals. Helices 7–10 and es3 are expanded further in mammals, reaching a length of 223 nt in Homo sapiens (20). Comparison of rRNAs in three dimensions indicates that from the common core (approximated by E. coli) to mammals this region of the rRNA increases by serial accretion onto an increasing frozen core. Each accretion step adds to previous rRNA but leaves the underlying core unperturbed (Fig. 1C). Sites of eukaryotic expansions are indicated by distinct insertion fingerprints (Fig. 1D). We have used a comparison of S. cerevisiae and E. coli SSU rRNAs to detect insertion fingerprints of eukaryotic expansions on the surface of the common core as shown in SI Appendix, Table S1. Thus, eukaryotic expansions and their insertion fingerprints observed within the SSU rRNA follow patterns previously established within the LSU (10).
Fig. 1.
Accretion of SSU rRNA as illustrated by helices 7–10/es3 from species of increasing complexity. A four-way junction at the surface of the common core, formed by helices 7–10, has expanded by accretion. Accretion adds to the previous rRNA core, leaving insertion fingerprints. (A and B) Secondary (A) and 3D (B) structures are preserved upon the addition of new rRNA. (C) Superimposition of the 3D structures highlights how new rRNA accretes with preservation of ancestral rRNA. (D) A characteristic insertion fingerprint is shown in red and blue boxes. In all panels, the rRNA that approximates the common core is blue. An expansion observed in both archaea and eukaryotes is green. An expansion that is observed only in eukaryotes is gold. An additional expansion in higher eukaryotes (mammals) is red.
The model assumes that the rRNA of an ancestor can be approximated by rRNA components common to its progeny and typically is most similar to the smaller rRNA among progeny (10, 21). Although the growth of rRNA by accretion is a general phenomenon, the progression of increasing size does not hold universally. In rare cases, homologous expansions occur in parallel, leading to small uncertainties in the model. In some instances rRNA does not follow localized monotonic growth in size over evolution. For example, es6 of Drosophila melanogaster (20) contains a helix (21es6b1 as defined in refs. 22 and 23) which is extended further in Trypanosoma brucei (15) but is absent in the mammalian SSU.
The SSU from Its Origins to the Common Core.
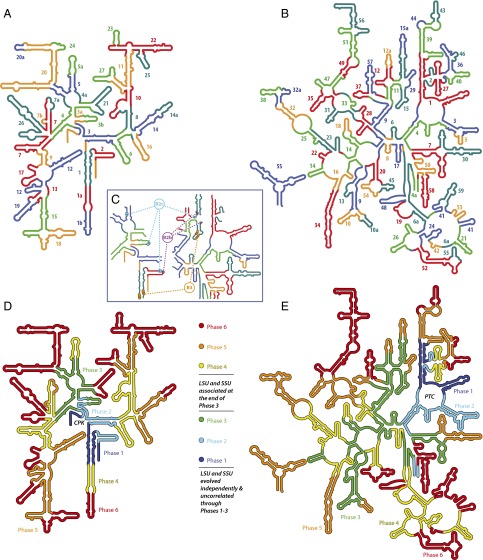
Insertion fingerprints are found throughout the ancient SSU common core and are indistinguishable in form from those at sites of recent eukaryotic expansions. Using these insertion fingerprints, we decompose SSU rRNA into ancestral expansion segments (Fig. 2A and SI Appendix, Fig. S2 and Table S2). In the common core we observe 27 aes in the SSU and 59 AES in the LSU (Fig. 2B). Some of the AES/aes have more tenuous subsequent extensions that are denoted by suffixes “a” and “b.” Our SSU decomposition is generally consistent with Noller’s pattern of continuity of stacking (24).
Fig. 2.
The accretion model mapped onto SSU and LSU secondary structures of E. coli rRNAs. (A and B) Ancestral expansion segments of the SSU (A) and the LSU (B) are numbered by order of acquisition. Insertion fingerprints are located at the seams between the AES or aes. AES/aes colors are arbitrary, chosen to distinguish expansion segments so that no AES or aes is the color of its neighbors. Some ancestral expansion segments appear discontinuous in the secondary structure and so are labeled multiple times. (C) Ancestral bridges B2b, B2c, and B3 mapped onto rRNA secondary structures. (D and E) SSU (D) and LSU (E) common cores are built by the addition of ancestral expansion segments in six phases. Each phase contains ancestral expansion segments that are associated in time and function.
The chronology of AES/aes acquisition is generally indicated by the recursive nature of their accumulation (SI Appendix, Text) and in some cases is based on the directionalities of A-minor interactions. Bokov and Steinberg (2) reasoned that the donating adenosine in an A-minor interaction is dependent on the accepting double helix, whereas the accepting helix is less dependent on the adenosine. If the conformation and structure of the rRNA was maintained throughout its evolution, the donating adenosine is the more recent addition.
Chronological relationships between the subunits (Fig. 2 D and E) are based on the relative ages of the AES/aes that form the intrasubunit bridges and on the directionalities of A-minor interactions within bridges. For the B3 bridge (Fig. 2C) (25) we infer that the SSU bridgehead is younger than the complementary LSU bridgehead because here the SSU donates A-minor interactions and the LSU accepts them. In contrast, the SSU B2a, B4, and B6 bridgeheads are older than the corresponding LSU bridgeheads because in these cases the LSU donates A-minor interactions to the SSU (SI Appendix, Fig. S3). This switching of directionalities of A-minor interactions indicates that the interface grew in size and complexity after initial association. Protein-independent bridges precede protein-assisted bridges, consistent with the appearance of coded proteins at later stages of ribosomal evolution.
The Accretion Model.
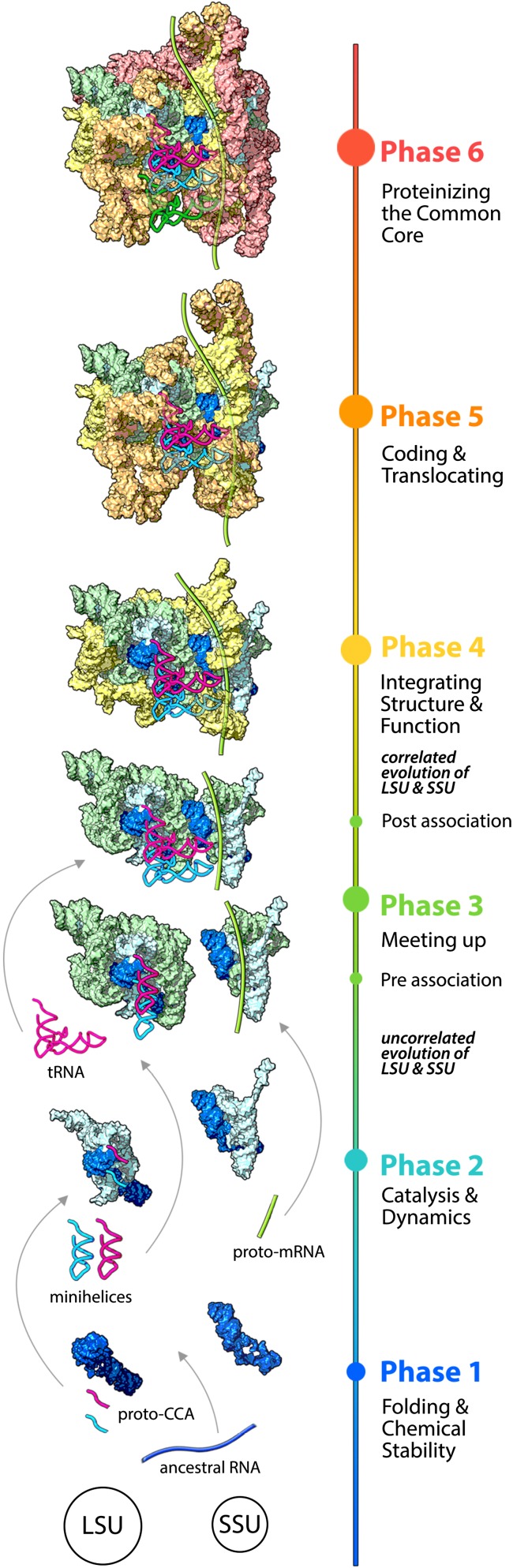
The accretion model describes the origin and evolution of translation in all living systems. Depending on the topology of the insertions, the expansion segments are grouped into four types (X, Y, T, and L). Common core AES/aes have been grouped into six phases (summarized in Fig. 3) with two additional phases exclusive to eukaryotes describing the addition of ES/es to protist and metazoan rRNA. Functions of AES/aes in the ancestral ribosome are known by functions of the corresponding rRNA in the extant ribosome (11). The titles of the phases indicate the major functions acquired during that phase.
Fig. 3.
The first six phases of the accretion model of ribosomal evolution. In Phase 1, ancestral RNAs form stem–loops and minihelices. In Phase 2, the LSU catalyzes the condensation of nonspecific oligomers. The SSU may have a single-stranded RNA-binding function. In Phase 3, the subunits associate, mediated by the expansion of tRNA from a minihelix to the modern L shape. LSU and SSU evolution is independent and uncorrelated during Phase 1–3. In Phase 4, evolution of the subunits is correlated. The ribosome is a noncoding diffusive ribozyme in which proto-mRNA and the SSU act as positioning cofactors. In Phase 5, the ribosome expands to an energy-driven, translocating, decoding machine. Phase 6 marks the completion of the common core with a proteinized surface (the proteins are omitted for clarity). The colors of the phases are the same as in Fig. 2. mRNA is shown in light green. The A-site tRNA is magenta, the P-site tRNA is cyan, and the E-site tRNA is dark green.
Definitions and descriptions of AES/aes, phases, and intersubunit bridges are provided in our previous work (10) for the LSU and in the SI Appendix here for the SSU. Where sequence is specified, as in the context of A-minor interactions, those nucleotides are universally conserved.
Phase 1. RNA Folding.
LSU and SSU structures.
AES 1 is a primordial stem–loop from which the LSU evolved; aes 1 is a primordial stem–loop from which the SSU evolved.
Function.
These RNAs fold into defect-laden stem loops (26), which are stabilized by interactions with metal cations. RNA folding protects against chemical degradation. The evolution of LSU and SSU in Phase 1 is uncorrelated.
Phase 2. Catalysis and Dynamics.
LSU structure.
The PTC (AES 1–5) is a stable and static fold. The P-region is formed by AES 1 and 2. The A-region is AES 3, 4, and 5. The first two insertions are AES 2 and 3 into AES 1. The PTC is completed by insertions of AES 4 and AES 5 onto AES 3. The exit pore is formed by AES 1, 3, and 4 (10).
SSU structure.
The initial expansions of the SSU are accretion of aes 2, 3, and 3a, which produce a quasi-stable fold (aes 1–3a). SSU termini, which are contained within aes 1, reversibly dissociate (SI Appendix, Table S2).
tRNA structure.
Proto-tRNA is composed of a CCA tail (Fig. 3), which acquires the amino acid acceptor stem and then the T-stem and T-loop to form a minihelix (27).
Function.
The LSU is a crude ribozyme, catalyzing nonspecific, noncoded condensation of amino acids and possibly oxyacids (28) by proximity and orientation effects (29). The exit pore/tunnel retains and stabilizes reaction intermediates (10, 30), facilitating the production of oligomers rather than dimers and inhibiting product cyclization. Some of the oligomeric products bind back onto the LSU, conferring advantage. Charged CCA tails and/or minihelices deliver activated substrates to the PTC. SSU function may involve association with single-stranded RNA. The evolution and function of the LSU and SSU are uncorrelated (SI Appendix, Fig. S4).
Phase 3. Meeting up: LSU, SSU, tRNA, and Proto-mRNA.
LSU structure.
The PTC is encased and rigidified, and the exit pore is extended into a short tunnel by AES 6–10. An embryonic subunit interface is formed by AES 11–15.
SSU structure.
The SSU termini dissociate upon accretion of aes 4: The 3′ terminus remains single-stranded when its complement is recruited to participate in the central pseudoknot (CPK). The CPK gains integrity and stability upon accretion of aes 4a, aes 5, and aes 3b (SI Appendix, Fig. S5). The SSU docks onto the LSU forming the B3 bridge (25) between aes 1 and AES 15. The directionality of the A-minor interaction indicates that the SSU B3 bridgehead is younger than the LSU counterpart. Several less-specific magnesium-mediated bridges form (B2b and B2c in Fig. 2C and SI Appendix, Fig. S6). All bridges are protein free.
tRNA and mRNA structure.
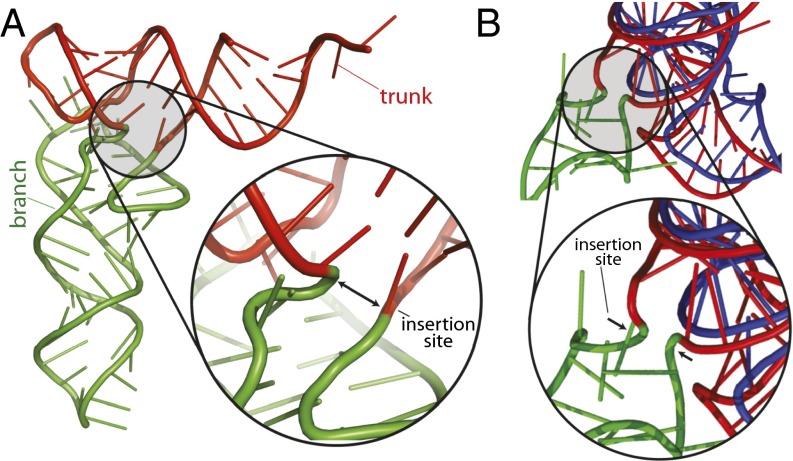
The proto-tRNA minihelix is extended by an insertion (Fig. 4) to form prototypes of the bifunctional L-shaped tRNAs in modern biology. L-shaped tRNAs mediate interactions between the SSU and LSU over a distance of around 70 Å. The LSU, tRNAs, proto-mRNA, and the SSU form a noncovalent quaternary complex. Proto-mRNA is a random population of single-stranded oligomers.
Fig. 4.
Comparison of an ancestral insertion in tRNA with a known insertion in rRNA. (A) The insertion fingerprint in tRNA points to the site of accretion of the D-stem and anti-codon stem–loop (green) onto the ancestral minihelix (red). (B) An insertion fingerprint at the locus of a known expansion (ES 12) in S. cerevisiae. The rRNA for E. coli (which lacks the expansion) is shown in blue and for S. cerevisiae is shown in red, except for the expansion rRNA, which is shown in green.
Function.
Catalytic efficiency and product length are increased. Nascent LSU–SSU association is mediated by proto-mRNAs and tRNAs. The SSU and proto-mRNAs are recruited by tRNA. tRNA on one end associates with the PTC and on the other end with proto-mRNA, which in turn binds to the single-stranded region of the SSU. The SSU and proto-mRNA act as cofactors, positioning and stabilizing tRNAs in the A- and P-regions of the PTC. In the LSU, the tunnel is elongated and rigidified. Correlations in the evolution of the LSU, SSU, tRNA, and mRNA are initiated.
Phase 4. Integrating LSU, SSU, tRNA, and mRNA Structure and Function.
LSU structure.
The LSU gains mass distal to the subunit interface (AES 16–28), assuming a hemispheric shape (SI Appendix, Fig. S7A). The anisotropy of growth is dictated by constraints imposed by association with the SSU (SI Appendix, Fig. S7B). The tunnel is extended and rigidified.
SSU structure.
Newly acquired rRNA creates well-defined binding pockets for tRNAs (SI Appendix, Fig. S8) and a more mature interface with the LSU. The P-site tRNA is stabilized by domain 3′M (the head), which begins with sequential accretion of aes 6, 8, and 10 onto aes 2. The A-site tRNA is stabilized by domain 5′ (the body), which begins with accretion of aes 7 onto aes 4. Domain C (the platform) is elaborated, and intersubunit interactions are enhanced by reinforcement of the bridgehead of B2b as aes 5 is extended by aes 5a. At the end of this phase, primitive binding sites for elongation factors G and Tu (31, 32) are formed by the accretion of aes 9 onto aes 7. Additional bridges with the LSU (B5 and B6) are formed by the elongation of aes 1 (aes 1a).
tRNA and mRNA structure.
tRNAs are optimized to form quasi-stable base-pair triplets with proto-mRNAs, which remain a population of single-stranded oligomers.
Function.
The ribosome is an efficient, noncoding, diffusive ribozyme. The evolutionary pathways of the LSU, the SSU, proto-mRNA, and tRNA are strongly coupled. Proto-mRNAs assist the SSU in positioning A- and P-site tRNAs (SI Appendix, Fig. S8), fixing orientation and proximity within the PTC (33). The subunit interface is well developed, with numerous RNA–RNA interactions. In the LSU, the tunnel is further elongated and rigidified, facilitating the production of longer condensation products by retention and stabilization of reaction intermediates.
Phase 5. Decoding and Translocating.
LSU structure.
The ratcheting system is acquired. The binding sites for elongation factors G and Tu are established (AES 30) along with the L7/L12 stalk (AES 32, 32a, and 38) and the central protuberance (AES 34 and 36) (SI Appendix, Fig. S9). The 5S rRNA associates with and reinforces the central protuberance. The tunnel is extended (AES 31, 33, and 35). The LSU embraces the SSU from each side by A-minor interactions (bridges B2a and B4 are formed by AES 29 and AES 10a). The B6 bridgehead is elaborated by AES 37. Bridges B1a and B1b are formed (25, 34). Most of the bridges (SI Appendix, Fig. S3) established at this phase (including bridges B1a, B1b, B4, B5, and B6) are enhanced by ribosomal proteins.
SSU structure.
Domain 3′ is elaborated, stabilizing the P-site tRNA (aes 11, 14, and 16). Domain 5′ is elaborated, stabilizing the A-site tRNA and enhancing the integrity of the SSU (aes 12, 13, and 15). Short expansions aes 7a and 7b, accreted onto aes 7, interact with each other via base pairing and form the functional pseudoknot (35), which is involved in decoding (36, 37), translocation, and proofreading (38). Intrinsic flexibility incorporated into the SSU (39, 40) allows the domains to move relative to each other (36), providing a mechanical basis for translocation (SI Appendix, Fig. S10).
tRNA and mRNA.
Anticodons in tRNA form specific interactions with mRNA codons. mRNA is polymeric with a defined sequence. The genetic code begins to optimize and expand.
Function.
This transcendental phase of ribosomal evolution marks the transition from a coordinated diffusive catalytic system to an energy-driven, ratcheting, translocating, quasi-decoding machine that initiates the collaboration of functional protein and informational nucleic acids. Ribosomal evolution from this phase on is a tight collaboration between RNA and protein. The ribosome achieves primitive decoding ability. In the LSU, binding sites for elongation factors G and Tu are finalized (41, 42). Dynamics are incorporated into LSU function by the flexibility of the newly acquired L7/L12 stalk and central protuberance. Continued extension of the tunnel facilitates the transition from oligomers to polymers. In the SSU, the head and body are elaborated, enhancing interactions with tRNAs in the P- and A-sites. Decoding and translocation of mRNA is achieved by the acquisition of the functional pseudoknot in the SSU. Relative motions of the subunits involve ratcheting around B3 (25), the oldest intersubunit bridge.
Phase 6. Proteinizing the Ribosomal Surface.
The rRNA common core is finalized (SI Appendix, Fig. S11), and the genetic code is optimized. The ribosomal surface is an integrated patchwork of rRNA and mature ribosomal proteins. Many AES/aes acquired here serve as binding sites for the globular domains of ribosomal proteins. In the LSU, the L1 stalk, responsible for translocation of tRNA from the P-site to the E-site, is formed by AES 43 and 44 (together with AES 39). The central protuberance is stabilized by AES 40 and 46. Development of the exit tunnel continues. In the SSU, domain 5′ is finalized by aes 17, 18, 19, and 26. Domain 3′M and the head and the body are completed by aes 22, 23, and 25. Domain C is completed by aes 20, 20a, 21, and 24. As with the LSU, these rRNA additions to the SSU surface are integrated with ribosomal proteins. The interaction between the two subunits is elaborated by bridges B7a, B7b, and B8. An embellishment of bridge B6 is achieved by a final expansion of aes 1 by aes 1b. All these bridges except B7a involve ribosomal proteins (25). The evolution of Phases 4–6 is summarized in SI Appendix, Fig. S12.
Phase 7. Encasing the Common Core (Eukaryotes).
The ribosome undergoes substantial expansion in eukaryotes. rRNA develops eukaryotic-specific rRNA expansions that are stabilized by eukaryotic-specific proteins (43), forming a secondary rRNA–protein shell.
Phase 8. Elaborating the Surface (Metazoans).
This phase marks the origin of metazoa, in which ribosomes are decorated with tentacle-like rRNA elements that extend well beyond the proteinized subunit surfaces (44). These expansions appear to be docking sites for chaperones and regulatory and auxiliary proteins (9).
Discussion
Using sequences of rRNAs as a telescope in time, Woese and Fox (1) sketched three primary branches in the tree of life on earth. Beyond the root of the Woese’s tree lies the origin of life, which poses some of the most profound and exciting questions in science and tests our understanding of chemical and biological principles. Structures of ribosomes in 3D allow one to peer beyond the root of the tree (3). Here, using structural information, the timeline of life is visualized far back in time, allowing dissection of primordial molecules, reactions, and events at the biochemical origins of life.
The accretion model reconciles the histories of ribosomal components. rRNAs grow by accretion, recursively expanding and adding successive layers, iteratively growing, subsuming, and freezing the rRNA. The ribosomal common core evolves in six phases. The ancient PTC arose from a stem–loop (Phase 1) to begin noncoded synthesis of peptides and esters in Phases 2–4. tRNA extended from a minihelix (Fig. 4A) (27) to deliver a substrate to the primitive LSU while being positioned by the SSU and mRNA (5, 33, 45). Phase 5 marks the transition from a diffusive RNA enzyme to an energy-driven decoding machine that initiates error-prone synthesis via a limited genetic code. During Phase 6, decoding is optimized. The modern cassette of amino acids is used to produce accurately coded proteins, some of which encase the common core. The tunnel is elaborated throughout Phases 1–6 and was central to ribosomal evolution. In a self-reinforcing cycle, tunnel elaboration allows the production of longer oligomers, some of which bind back onto the ribosome, increasing size and stability. Phases 7 and 8 produce tentacle-like rRNA in simple and multicellular eukaryotes. In sum, the ribosome sequentially acquired capabilities for RNA folding, catalysis, subunit association, correlated evolution, decoding, and energy transduction, giving rise to the extant collaboration of functional protein and informational nucleic acids.
Conclusions
The transition from the synthesis of noncoded heterogeneous oligomers to proteins by the ribosome conferred advantages, because some reaction products bound to the ribosome. Proteinization of the ribosome drove a more general proteinization of other processes, giving rise to modern biology as described by the central dogma. The ribosome spawned the existing symbiotic relationship of protein and nucleic acid.
Materials and Methods
Definitions of AES/aes, as well as the criteria used to sort them into temporal order and to partition them into phases, are presented in the SI Appendix for the SSU and in our previous work (10) for the LSU. The margins between the phases are somewhat indistinct, and the original LSU phases (10) were adjusted slightly here to account for data from the SSU. Secondary structures of LSU and SSU rRNAs are taken from our public gallery (apollo.chemistry.gatech.edu/RibosomeGallery/), and data are mapped by the web server RiboVision (23). 3D analysis of ancestral expansion was performed using the 70S ribosome of E. coli (Protein Data Bank ID code 4V9D) (25). Additional information supporting the accretion model is available in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We dedicate this manuscript to the memory of Professor Alexander Rich. We thank Ms. Susann Orth for help in preparing the figures. This work was funded in part by National Aeronautics and Space Agency Astrobiology Institute Grant NNA09DA78A.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509761112/-/DCSupplemental.
References
- 1.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci USA. 1977;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bokov K, Steinberg SV. A hierarchical model for evolution of 23S ribosomal RNA. Nature. 2009;457(7232):977–980. doi: 10.1038/nature07749. [DOI] [PubMed] [Google Scholar]
- 3.Hsiao C, Mohan S, Kalahar BK, Williams LD. Peeling the onion: Ribosomes are ancient molecular fossils. Mol Biol Evol. 2009;26(11):2415–2425. doi: 10.1093/molbev/msp163. [DOI] [PubMed] [Google Scholar]
- 4.Krupkin M, et al. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos Trans R Soc Lond B Biol Sci. 2011;366(1580):2972–2978. doi: 10.1098/rstb.2011.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox GE. Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol. 2010;2(9):a003483. doi: 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith TF, Lee JC, Gutell RR, Hartman H. The origin and evolution of the ribosome. Biol Direct. 2008;3:16. doi: 10.1186/1745-6150-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassouna N, Michot B, Bachellerie JP. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984;12(8):3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerbi SA. Expansion segments: Regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In: Zimmermann RA, Dahlberg AE, editors. Ribosomal RNA—Structure, Evolution, Processing, and Function in Protein Synthesis. CRC; Boca Raton, FL: 1996. pp. 71–87. [Google Scholar]
- 9.Melnikov S, et al. One core, two shells: Bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19(6):560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 10.Petrov AS, et al. Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci USA. 2014;111(28):10251–10256. doi: 10.1073/pnas.1407205111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108(4):557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 12.Ware VC, et al. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 1983;11(22):7795–7817. doi: 10.1093/nar/11.22.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michot B, Bachellerie JP. Comparisons of large subunit rRNAs reveal some eukaryote-specific elements of secondary structure. Biochimie. 1987;69(1):11–23. doi: 10.1016/0300-9084(87)90267-7. [DOI] [PubMed] [Google Scholar]
- 14.Armache JP, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc Natl Acad Sci USA. 2010;107(46):19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashem Y, et al. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature. 2013;494(7437):385–389. doi: 10.1038/nature11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330(6008):1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 17.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armache J-P, et al. Promiscuous behaviour of archaeal ribosomal proteins: Implications for eukaryotic ribosome evolution. Nucleic Acids Res. 2013;41(2):1284–1293. doi: 10.1093/nar/gks1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 20.Anger AM, et al. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497(7447):80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 21.Fitch WM. Toward defining the course of evolution - minimum change for a specific tree topology. Syst Zool. 1971;20(4):406–416. [Google Scholar]
- 22.Petrov AS, et al. Secondary structures of rRNAs from all three domains of life. PLoS One. 2014;9(2):e88222. doi: 10.1371/journal.pone.0088222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernier CR, et al. RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss. 2014;169(1):195–207. doi: 10.1039/c3fd00126a. [DOI] [PubMed] [Google Scholar]
- 24.Noller HF. RNA structure: Reading the ribosome. Science. 2005;309(5740):1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 25.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 26.Moore PB. The RNA Folding Problem. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2nd Ed. Cold Spring Harbor Lab Press; Plainview, NY: 1999. pp. 381–401. [Google Scholar]
- 27.Francklyn C, Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989;337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 28.Rich A. The Possible Participation of Esters as Well as Amides in Prebiotic Polymers. In: Buvet R, Ponnamperuma C, editors. Chemical Evolution and the Origin of Life. North-Holland Publishing Company; Amsterdam: 1971. [Google Scholar]
- 29.Sievers A, Beringer M, Rodnina MV, Wolfenden R. The ribosome as an entropy trap. Proc Natl Acad Sci USA. 2004;101(21):7897–7901. doi: 10.1073/pnas.0402488101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fox GE, Tran Q, Yonath A. An exit cavity was crucial to the polymerase activity of the early ribosome. Astrobiology. 2012;12(1):57–60. doi: 10.1089/ast.2011.0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartman H, Smith TF. GTPases and the origin of the ribosome. Biol Direct. 2010;5:36. doi: 10.1186/1745-6150-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agirrezabala X, et al. Structural insights into cognate versus near-cognate discrimination during decoding. EMBO J. 2011;30(8):1497–1507. doi: 10.1038/emboj.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernhardt HS, Tate WP. The transition from noncoded to coded protein synthesis: Did coding mRNAs arise from stability-enhancing binding partners to tRNA? Biol Direct. 2010;5:16. doi: 10.1186/1745-6150-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gabashvili IS, et al. Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell. 2000;100(5):537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 35.Powers T, Noller HF. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10(8):2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292(5518):897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 37.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111(5):721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 38.Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17(9):1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 39.Mohan S, Donohue JP, Noller HF. Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci USA. 2014;111(37):13325–13330. doi: 10.1073/pnas.1413731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paci M, Fox GE. Major centers of motion in the large ribosomal RNAs. Nucleic Acids Res. 2015;43(9):4640–4649. doi: 10.1093/nar/gkv289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114(1):123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 42.Lancaster L, Lambert NJ, Maklan EJ, Horan LH, Noller HF. The sarcin-ricin loop of 23S rRNA is essential for assembly of the functional core of the 50S ribosomal subunit. RNA. 2008;14(10):1999–2012. doi: 10.1261/rna.1202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 44.Harms J, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107(5):679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 45.Orgel LE. The origin of polynucleotide-directed protein synthesis. J Mol Evol. 1989;29(6):465–474. doi: 10.1007/BF02602917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.