Abstract
Structural characterization of new α/γ-peptide foldamers containing the cyclically-constrained γ-amino acid I is described. Crystallographic and 2D NMR analysis shows that γ residue I promotes formation of a 12/10-helical secondary structure in α/γ-peptides. This helix contains two different types of internal H-bond, and the data show that the 12-atom C=O(i)→H-N(i+3) H-bond is more favorable than the 10-atom C=O(i)→H-N(i-1) H-bond. Several foldamer helices featuring topologically distinct H-bonds have been discovered, but our findings are the first to show that such H-bonds may differ in their favorability.
Biology relies on proteins and nucleic acids to carry out diverse functions, most of which require that the biopolymer adopts a complex and specific folding pattern. The relationship between sophisticated function and higher-order structure has inspired many groups to seek non-biological oligomers that favor specific conformations (“foldamers”).1 Extrapolation from the poly-α-amino acid backbone of proteins has led to the study of β-peptides, γ-peptides and higher homologues.2 Secondary structural motifs reminiscent of (but geometrically distinct from) those well-known in proteins, including helices, sheets and reverse turns, have been characterized for these new backbones, and approaches to tertiary structure have been reported.3 The folding rules established for β- and γ-peptides have enabled the development of specific examples that display biomimetic function.4
Expansion beyond the biopolymer prototypes allows deviation from particular structural parameters associated with proteins and nucleic acids. The backbones of these biopolymers, for example, are homogeneous in that each subunit is drawn from a single chemical class (e.g., α-amino acids), but heterogeneous backbones are readily accessed among synthetic oligomers.5 A variety of discrete secondary structures have been identified among peptidic foldamers with mixed backbones, including combinations of α + β residues or α + γ residues. For some applications, such as functional mimicry of a natural α-helix, heterogeneous backbones containing both α and β residues have proven to be superior to the homogeneous β residue backbone.6
Within the regular helices found in proteins, each type of internal non-covalent contact is topologically equivalent across all subunits involved. In an α-helix, for example, all C=O(i)→H-N(i+4) H-bonds should be comparable to one another, excluding terminal effects, since the C=O and H-N groups are all similar. In contrast, different types of internal H-bonds occur within many of the helices that have been documented among peptidic foldamers with unnatural backbones. Such differences are inherent for helices formed by heterogeneous backbones because of subunit diversity. Thus, for example, an α/β-peptide contains H-bond accepting groups (C=O) and donating groups (N-H) from both α and β residues. Different types of H-bonds are found also in foldameric helices in which H-bond directionality alternates along the backbone, whether the backbone is homogeneous (as in the β-peptide 10/12-helix7 and 18/20-helix8) or heterogeneous (as in the α/β-peptide 11/9-helix9 and 18/16-helix10). These systems raise a fundamental question: are the different types of intrahelical H-bonds comparably favorable? Here we describe a new type of α/γ-peptide foldamer and provide the first evidence that distinct types of H-bonds formed within a regular secondary structure can differ in terms of favorability.
The new foldamers contain ring-constrained γ residues of type I (Figure 1). Either enantiomer of the necessary γ-amino acid building block can be efficiently prepared.11 Previous characterization of oligomers containing the stereoisomeric γ residue of type II established that this subunit, alone12 or in alternation with α or with β residues,11, 13 favors helices defined by C=O(i)→H-N(i+3) H-bonds. The present studies of α/γ-peptides containing I were intended to test the generality of the “stereochemical patterning” hypothesis of Martinek, Fülöp et al.14 These workers proposed a correlation between H-bond directionality within foldamer helices and the signs of the torsion angles about the bonds that flank amide linkages (ψ,φ pairs from adjacent residues). If these torsion angles are all of the same sign, then all intrahelical H-bonds should be oriented in the same direction (as observed, for example in the α-helix). If the torsion angles alternate between neighboring amides (i.e., if amides that have positive flanking torsion angles are adjacent to amides that have negative flanking torsion angles and vice versa), then intrahelical H-bonds should alternate in directionality relative to the helix axis.
Figure 1.
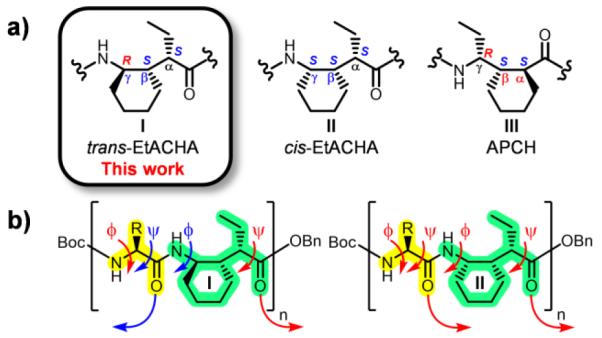
a) Comparison of cyclic γ-amino acids studied by our group. b) Inversion of the Cγ stereocenter of II, forming I, is predicted to invert the adjacent amide H-bond directionality in the full-length H-bonded foldamer.
The Martinek-Fülöp hypothesis led us to predict that altering only the stereocenter adjacent to the φ torsion angle in γ residues (i.e., II → I) would cause a wholesale change in helix geometry in α/γ-peptides, from a conformation with unidirectional H-bonds to a conformation with bidirectional H-bonds. This prediction relies on the fact that α residues can readily adopt conformations in which the backbone torsion angles φ and ψ have the same sign (α-helix) or different signs (β-sheet). γ Residues of type I with absolute configuration of (S,S,R) at the (α,β,γ) carbons must be paired with L-α-amino acid residues, and (R,R,S)-I with D-α residues, to test this prediction.
Our initial efforts were aimed at crystallographic characterization of the new α/γ(I)-peptides. Seven structures were obtained15 for oligomers containing four to six residues, three by racemic crystallization.16 The structures of tetramers 1a-c, collectively, were not wholly consistent with our expectations. All three formed a 12-atom H-bond between the N-terminal Boc C=O and N-H of the α residue closest to the C-terminus (Figure 2). This C=O(i)→H-N(i+3) interaction is characteristic of both the 12/10-helix we expected and the 12-helix previously documented for α/γ-peptides containing γ residue II. Only rac-1a (α residues = Phe), however, displayed the 10-atom C=O(i)→H-N(i-1) H-bond characteristic of the 12/10-helix. In this structure, the 10-atom H-bond was significantly longer than the 12-atom H-bond (N⋯O = 3.00 vs. 2.83 Å). For 1b (α residues = Ala), a second 12-atom H-bond formed, i.e, this α/γ-peptide adopted a 12-helix rather than a 12/10-helix. α/γ-Peptide 1c (α residues = Ala) displayed an interesting variation on the 10-atom H-bond: the carbonyl of Ala-3 was linked to N-H(i-1) via an intermolecular H-bond chain that included a water molecule along with the N-terminal Ala carbonyl of its lattice neighbor.17
Figure 2.
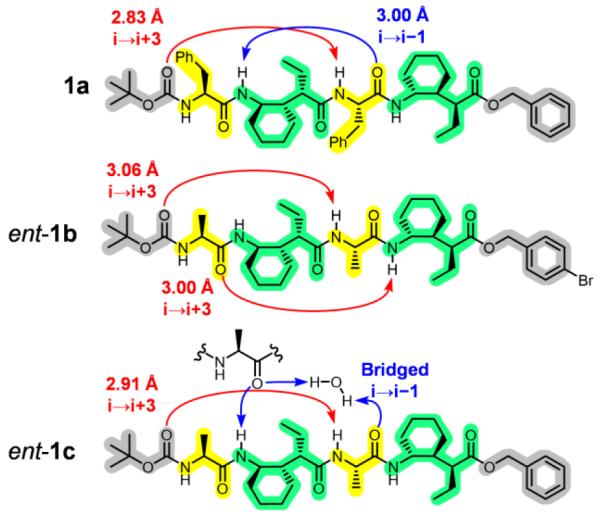
Structures of tetramers 1a-c depicting H-bonds observed in the crystal structures. Enantiomers of 1b and 1c are shown. α Residues are shaded in yellow, γ residues in green, and protecting groups in gray. 12-Atom H-bonds are shown in red, and 10-atom H-bonds in blue. Geometric criteria used for all H-bond assignment are distance N⋯O < 4.0 Å and angle N–H⋯O > 130°.
Longer α/γ-peptides containing γ residue I manifested a stronger preference for the 12/10-helix in the solid state, but the crystal structures offered hints that the 12-atom H-bonds are more favorable than the 10-atom H-bonds. Pentamer 2a (α residues = Val) displayed a 12/10-helical conformation in which the maximum number of amide-to-amide H-bonds was formed, two in 12-atom rings and one in a 10-atom ring (Figure 3). All of the H-bonded N⋯O distances in this structure were relatively long (> 3.00 Å). The two structurally very similar symmetry-independent conformers of pentamer 2b17 (α residues = Ala) adopted a 12/10-helix-like conformation in the crystal, but an ethanol molecule was found to be interpolated between the C=O and H-N groups that would have formed the 10-atom intrahelical H-bond (Figure 3). The two 12-atom H-bonds in this structure featured significantly shorter N⋯O distances than were found in the crystal structure of 2a.17
Figure 3.
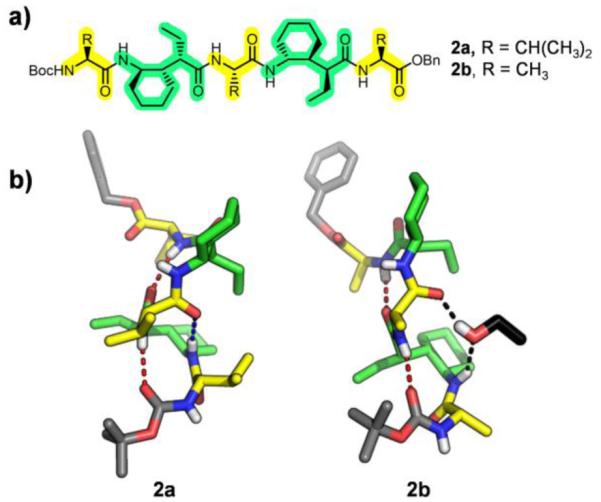
a) Structures of pentamers 2a-b. b) Crystal structures of pentamers 2a and 2b, depicting the inserted EtOH molecule of 2b in black. 12-Atom H-bonds are shown as red dashes, and 10-atom H-bonds as blue dashes.
Hexamer rac-3 (α residues = Phe) provided two crystal forms under slightly different solvent conditions (Figure 4). One crystal contained a 12/10-helix that seemed to unravel toward the C-terminus; this structure contained two short 12-atom H-bonds (2.85/2.86 Å), and a longer 10-atom H-bond (3.02 Å). A second amide-amide 10-atom H-bond would be possible toward the C-terminus, but in this case the poor geometric parameters (N⋯O distance of 4.09 Å and N–H⋯O angle of 135°) suggest at least that this interaction is substantially weaker than all other H-bonds described here.18 The other crystalline form of rac-3 revealed a 12/10-helix-like conformation in which a water molecule was found to be interpolated between the C=O and H-N groups that would have formed the C-terminal 10-atom H-bond. The set of seven crystal structures, collectively, suggests that the new α/γ-peptide backbone has a propensity for 12/10-helix-like conformations, but that H-bonding may be more favorable in the 12-atom rings relative to the 10-atom rings.
Figure 4.
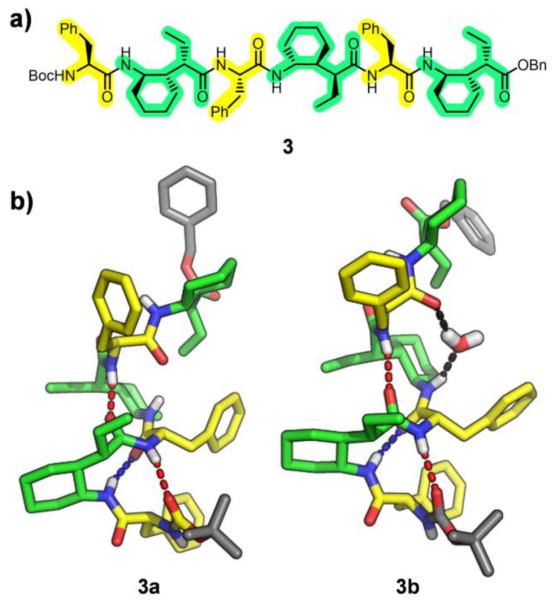
a) Structure of hexamer 3. b) Crystal structures 3a and 3b, depicting the inserted H2O molecule of 3b.
To gain insight on the folding propensity of α/γ(I)-peptides in solution, we employed 2D NMR (COSY, TOCSY, and ROESY) to evaluate the conformations in CDCl3 of five oligomers containing from five to nine residues. Evidence for 12/10-helical folding was obtained in each case in the form of inter-residue NOEs between protons separated by 7 to 12 covalent bonds. Here we focus on nonamer 4. Three non-sequential backbone proton NOE patterns previously reported to be consistent with 12/10-helical secondary structure19 were observed along most or all of 4 (Figure 5). In addition, we observed a set of strong i,i+2 NOEs from CαH of each Val residue to CβH of the next Val residue, which is consistent with the 12/10-helical conformation. NMR-restrained simulated annealing calculations were carried out for each of the five oligomers with the Crystallography and NMR System (CNS) software suite (Figure 6). A total of 24 NOE-derived distance restraints from 4 was used to generate an ensemble of 10 lowest-energy structures. The N-terminal four-residue segment in the average generated from these 10 structures overlays well with the crystal structure of pentamer 2a.
Figure 5.
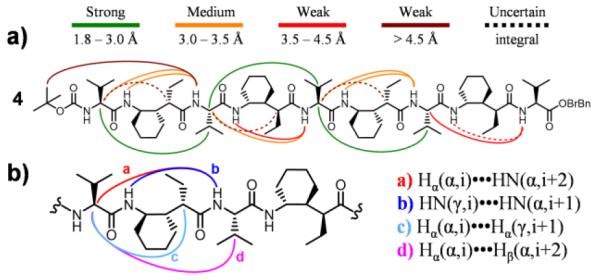
a) Non-sequential ROESY crosspeaks observed in nonamer 4. Uncertain integrals were unambiguously assigned but could not be reliably software-integrated. b) 12/10-Helical crosspeak patterns observed in 4.
Figure 6.
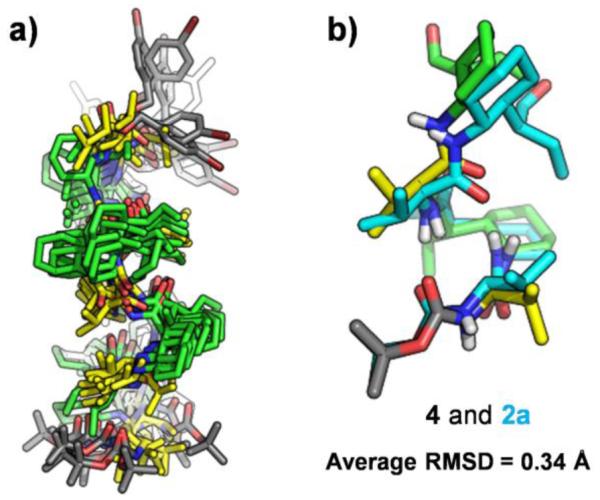
a) NMR ensemble generated by CNS using distance restraints from ROESY data of nonamer 4 in CDCl3. The 10 lowest-energy structures were collected from 1000 trial structures. b) Backbone alignment of the first four residues of the minimized average of the 10 lowest-energy NMR structures with pentamer crystal structure 2a.
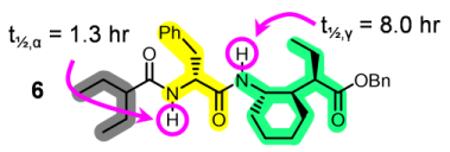
We used H/D exchange (HDX) measurements to test the hypothesis that 12/10-helices formed by the new α/γ(I)-peptide backbone contain H-bond types with different intrinsic favorabilities. These studies focused on octamer 5, Boc-([Phe][γ(I)])4-OBn, for which 2D NMR data provided evidence of 12/10-helical folding in CDCl3.17 Amino acid residue identity exerts a large influence on HDX;20 therefore, results for octamer 5, expressed as half-life (t½) for disappearance of amide 1H-NMR resonances after dissolution in 95:5 CDCl3/CD3OD, were normalized to results for model dipeptide 6. Specifically, t½ values for each α residue NH in 5 were divided by t½ for the α residue NH in 6, and t½ values for each γ residue in 5 were divided by t½ for the γ residue in 6. This normalization should highlight the impact of folding on HDX at each site within the octamer.
The normalized t½ values (Table 1) are generally largest toward the center of 5, which is consistent with the hypothesis that this α/γ-peptide adopts a helical conformation featuring internal H-bonds of alternating directionality. The α residue NH resonances show larger normalized t½ values than do the γ residue NH resonances. This trend is consistent with the hypothesis that the H-bonds formed by the α residue NH groups (12-atom rings) are more favorable than the H-bonds formed by the γ residue NH groups (10-atom rings). Further support for the proposed distinction between 12- and 10-atom internal H-bonds was provided by experiments in which small aliquots of DMSO were added to a CDCl3 solution of octamer 5 (“DMSO titrations”).17
Table 1.
Relative HDX Half-Life Change of Octamer 5.
| Phe-1 | γ(I)-2 | Phe-3 | γ(I)-4 | Phe-5 | γ(I)-6 | Phe-7 | γ(I)-8 | |
|---|---|---|---|---|---|---|---|---|
| t½ (hr) | 2.2 | 102 | 95 | 199 | 121 | – a | 46 | 37 |
| Normalized t½ | 1.7 | 13 | 73 | 25 | 93 | – a | 35 | 4.6 |
|
H-bonded ring
size |
– | 10 | 12 | 10 | 12 | 10 | 12 | – |
Resonance overlapped with phenyl H.
The crystallographic and NMR data reported here show that α/γ-peptide secondary structure can be engineered based on the stereochemical patterning hypothesis of Martinek, Fülöp et al. Altering just one configuration in the dipeptide repeating unit of the backbone, by replacing γ(II) residues with γ(I) residues, leads to a change in conformational preference, from a helix with unidirectional H-bonds to a helix in which H-bond directions alternate. The ability to alter helix type by switching a single configuration within the γ residues represents a significant accomplishment in foldamer design.
Exploration of the new α/γ(I) backbone led to the unexpected discovery that the 12/10-helix formed by these foldamers contains H-bonds that differ in terms of favorability, with the γ residue C=O(i) → α residue H-N(i+3) H-bonds intrinsically superior to the α residue C=O(i) → γ residue H-N(i-1) H-bonds. This possibility was initially suggested by the seven crystal structures described here, in which the α residue C=O(i) → γ residue H-N(i-1) H-bond distances were long, or other molecules were interpolated into these interactions. Such interpolations have occasionally been observed in proteins21 or short conventional peptides,22 and more recently in α/γ-peptides,23 but the frequency of this unusual phenomenon in our structural data set stands out. HDX and DMSO titration results support the conclusion that there is an energetic differentiation between the two types of H-bond in the 12/10-helical conformation formed by the α/γ(I) backbone. We speculate that the γ(I) residue is not ideally preorganized for the 10-atom H-bonding mode.17 Several foldamer secondary structures containing different H-bond types have been discovered, and our results raise the possibility that topologically distinct H-bonds will in general be energetically differentiated.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by NSF grant CHE-1307365. NMR spectrometers used in this work were purchased with support from a generous gift by Paul J. Bender and from NIH (1 S10 RR13866-01). The authors thank Dr. Michael W. Giuliano for helpful discussions regarding CNS calculations, Dr. Young-Hee Shin for assistance in acquiring 2D-NMR, and Dr. Matt Benning for acquiring single-crystal diffraction data of 2b at the Bruker AXS facility in Madison, WI.
Footnotes
Experimental details including synthetic routes, NMR data, details and PDB files of NMR structure calculations of 4 and other foldamers, DMSO titration data, HDX data, helix and H-bond parameters for crystal structures, and crystallographic data, including CIF files. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
REFERENCES
- 1 (a).Gellman SH. Acc. Chem. Res. 1998;31:173. [Google Scholar]; (b) Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. Chem. Rev. 2001;101:3893. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]; (c) Guichard G, Huc I. Chem. Commun. 2011;47:5933. doi: 10.1039/c1cc11137j. [DOI] [PubMed] [Google Scholar]
- 2 (a).Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101:3219. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]; (b) Seebach D, Hook DF, Glättli A. Biopolymers. 2006;84:23. doi: 10.1002/bip.20391. [DOI] [PubMed] [Google Scholar]
- 3.Reinert ZE, Horne WS. Org. Biomol. Chem. 2014;12:8796. doi: 10.1039/c4ob01769b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4 (a).Hamuro Y, Schneider JP, DeGrado WF. J. Am. Chem. Soc. 1999;121:12200. [Google Scholar]; (b) Porter EA, Wang X, Lee HS, Weisblum B, Gellman SH. Nature. 2000;404:565. doi: 10.1038/35007145. [DOI] [PubMed] [Google Scholar]; (c) Stephens OM, Kim S, Welch BD, Hodsdon ME, Kay MS, Schepartz A. J. Am. Chem. Soc. 2005;127:13126. doi: 10.1021/ja053444+. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) English EP, Chumanov RS, Gellman SH, Compton T. J. Biol. Chem. 2006;281:2661. doi: 10.1074/jbc.M508485200. [DOI] [PubMed] [Google Scholar]; (e) Gademann K, Kimmerlin T, Hoyer D, Seebach D. J. Med. Chem. 2001;44:2460. doi: 10.1021/jm010816q. [DOI] [PubMed] [Google Scholar]; (f) Seebach D, Schaeffer L, Brenner M, Hoyer D. Angew. Chem., Int. Ed. 2003;42:776. doi: 10.1002/anie.200390205. [DOI] [PubMed] [Google Scholar]
- 5 (a).Horne WS, Gellman SH. Acc. Chem. Res. 2008;41:1399. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pendem N, Nelli YR, Douat C, Fischer L, Laguerre M, Ennifar E, Kauffmann B, Guichard G. Angew. Chem., Int. Ed. 2013;52:4147. doi: 10.1002/anie.201209838. [DOI] [PubMed] [Google Scholar]; (c) Wu H, Qiao Q, Hu Y, Teng P, Gao W, Zuo X, Wojtas L, Larsen RW, Ma S, Cai J. Chem. Eur. J. 2015;21:2501. doi: 10.1002/chem.201406112. [DOI] [PubMed] [Google Scholar]
- 6 (a).Sadowsky JD, Schmitt MA, Lee HS, Umezawa N, Wang S, Tomita Y, Gellman SH. J. Am. Chem. Soc. 2005;127:11966. doi: 10.1021/ja053678t. [DOI] [PubMed] [Google Scholar]; (b) Sadowsky JD, Fairlie WD, Hadley EB, Lee HS, Umezawa N, Nikolovska-Coleska Z, Wang S, Huang DC, Tomita Y, Gellman SH. J. Am. Chem. Soc. 2007;129:139. doi: 10.1021/ja0662523. [DOI] [PubMed] [Google Scholar]; (c) Johnson LM, Gellman SH. Methods Enzymol. 2013;523:407. doi: 10.1016/B978-0-12-394292-0.00019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seebach D, Gademann K, Schreiber JV, Matthews JL, Hintermann T, Jaun B, Oberer L, Hommel U, Widmer H. Helv. Chim. Acta. 1997;80:2033. [Google Scholar]
- 8.Szolnoki É, Hetényi A, Mándity IM, Fülöp F, Martinek TA. Eur. J. Org. Chem. 2013:3555. [Google Scholar]
- 9.Sharma GV, Nagendar P, Jayaprakash P, Radha Krishna P, Ramakrishna KV, Kunwar AC. Angew. Chem., Int. Ed. 2005;44:5878. doi: 10.1002/anie.200501247. [DOI] [PubMed] [Google Scholar]
- 10 (a).Berlicki L, Pilsl L, Wéber E, Mándity IM, Cabrele C, Martinek TA, Fülöp F, Reiser O. Angew. Chem., Int. Ed. 2012;51:2208. doi: 10.1002/anie.201107702. [DOI] [PubMed] [Google Scholar]; (b) Legrand B, André C, Moulat L, Wenger E, Didierjean C, Aubert E, Averlant-Petit MC, Martinez J, Calmes M, Amblard M. Angew. Chem., Int. Ed. 2014;53:13131. doi: 10.1002/anie.201407329. [DOI] [PubMed] [Google Scholar]
- 11.Guo L, Chi Y, Almeida AM, Guzei IA, Parker BK, Gellman SH. J. Am. Chem. Soc. 2009;131:16018. doi: 10.1021/ja907233q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo L, Zhang W, Reidenbach AG, Giuliano MW, Guzei IA, Spencer LC, Gellman SH. Angew. Chem., Int. Ed. 2011;50:5843. doi: 10.1002/anie.201101301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo L, Almeida AM, Zhang W, Reidenbach AG, Choi SH, Guzei IA, Gellman SH. J. Am. Chem. Soc. 2010;132:7868. doi: 10.1021/ja103233a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mándity IM, Weber E, Martinek TA, Olajos G, Tóth GK, Vass E, Fülöp F. Angew. Chem., Int. Ed. 2009;48:2171. doi: 10.1002/anie.200805095. [DOI] [PubMed] [Google Scholar]
- 15.Crystallographic data has been deposited at the Cambridge Structural Data Centre with accession codes 1056028-1056034 and can be accessed free of charge at www.ccdc.cam.ac.uk/data_request/cif.
- 16 (a).Yeates TO, Kent SB. Annu. Rev. Biophys. 2012;41:41. doi: 10.1146/annurev-biophys-050511-102333. [DOI] [PubMed] [Google Scholar]; (b) Lee M, Shim J, Kang P, Guzei IA, Choi SH. Angew. Chem., Int. Ed. 2013;52:12564. doi: 10.1002/anie.201306404. [DOI] [PubMed] [Google Scholar]
- 17.See Supporting Information.
- 18.Jeffrey GA. An Introduction to Hydrogen Bonding. Oxford University Press; New York: 1997. [Google Scholar]
- 19 (a).Sharma GV, Jadhav VB, Ramakrishna KV, Jayaprakash P, Narsimulu K, Subash V, Kunwar AC. J. Am. Chem. Soc. 2006;128:14657. doi: 10.1021/ja064875a. [DOI] [PubMed] [Google Scholar]; (b) Giuliano MW, Maynard SJ, Almeida AM, Guo L, Guzei IA, Spencer LC, Gellman SH. J. Am. Chem. Soc. 2014;136:15046. doi: 10.1021/ja5076585. [DOI] [PubMed] [Google Scholar]
- 20.Bai Y, Milne JS, Mayne L, Englander SW. Proteins. 1993;17:75. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundaralingam M, Sekharudu YC. Science. 1989;244:1333. doi: 10.1126/science.2734612. [DOI] [PubMed] [Google Scholar]
- 22.Karle IL, Flippen-Anderson J, Uma K, Balaram P. Proc. Natl. Acad. Sci. USA. 1987;85:299. doi: 10.1073/pnas.85.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23 (a).Basuroy K, Dinesh B, Shamala N, Balaram P. Angew. Chem. 2013;125:3218. doi: 10.1002/anie.201209324. [DOI] [PubMed] [Google Scholar]; (b) Basuroy K, Dinesh B, Shamala N, Balaram P. Angew. Chem., Int. Ed. 2013;52:3136. doi: 10.1002/anie.201209324. [DOI] [PubMed] [Google Scholar]; (c) Karle IL, Flippen-Anderson JL, Uma K, Balaram P. Int. J. Pept. Protein Res. 1994;44:491. doi: 10.1111/j.1399-3011.1994.tb00187.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


