Abstract
We establish the genotype-phenotype correlations for the complete spectrum of POLG syndromes, by refining our previously described protocol for mapping pathogenic mutations in the human POLG gene to functional clusters in the catalytic core of the mitochondrial replicase, Pol γ (1). We assigned 136 mutations to five clusters and identify segments of primary sequence that can be used to delimit the boundaries of each cluster. We report that compound heterozygotes with two mutations from different clusters manifested more severe, earlier onset POLG syndromes, whereas two mutations from the same cluster are less common and generally are associated with less severe, later onset POLG syndromes. We also show that specific cluster combinations are more severe than others, and have a higher likelihood to manifest at an earlier age. Our clustering method provides a powerful tool to predict the pathogenic potential and predicted disease phenotype of novel variants and mutations in POLG, the most common nuclear gene underlying mitochondrial disorders. We propose that such a prediction tool would be useful for routine diagnostics for mitochondrial disorders.
Keywords: mitochondria, POLG, mitochondrial DNA, mitochondrial DNA replication, mitochondrial DNA polymerase
Introduction
Mitochondrial dysfunction due to impaired energy production via oxidative phosphorylation (OXPHOS) causes a variety of diseases, known collectively as mitochondrial disorders (2). Base substitutions, deletions or depletion of mitochondrial DNA (mtDNA) resulting in production of dysfunctional and/ or depletion of OXPHOS proteins is an important cause of mitochondrial dysfunction (2). In animal mitochondria, Pol γ is the only known DNA polymerase, and therefore it is responsible for maintenance of mtDNA integrity associated with mtDNA replication and repair (3). Human Pol γ is a heterotrimer consisting of a catalytic subunit, Pol γA, and a dimer of an accessory subunit, Pol γB (4). Encoded by the POLG gene, Pol γA, known as the catalytic core, is a 140 kDa polypeptide that contains the 5'-3' DNA polymerase (pol), 3'-5' exonuclease (exo), and 5'-dRP lyase activities (3). Mutations in the POLG gene lead to accumulation of mtDNA deletions as well as mtDNA depletion, manifesting mitochondrial disorders termed POLG syndromes (5). The most severe form of POLG syndrome, known as Alpers syndrome, is caused by compound heterozygosity of two recessive POLG mutations, and leads to hepatocerebral mtDNA depletion syndrome during infancy and death at an early age (6). Late onset POLG syndromes, such as progressive external ophthalmoplegia (PEO) can be caused by dominant or recessive compound heterozygous mutations, and are associated with varying degrees of tissue specific mtDNA depletion and/ or single or multiple mtDNA deletions (7).
The crystal structure of Pol γ was determined in its apoenzyme form (PDB code 3IKM, (8)). Its catalytic core has three major domains: an N-terminal Exo domain that contains the exo active site; a C-terminal Pol domain that contains the pol active site; and a spacer domain that separates the Exo and Pol domains in primary sequence. Three subdomains are defined in the Pol domain: the palm subdomain represents the most highly conserved structural module between non-homologous DNA polymerases, and contains the pol catalytic site coordinating two Mg2+ ions; the fingers subdomain is involved in binding the incoming dNTP substrate, and the thumb subdomain forms a major surface of the DNA binding channel. The spacer domain comprises two subdomains: the accessory interacting domain (AID), which forms the major hydrophobic contact with the proximal accessory subunit, and the intrinsic processivity (IP) domain, which forms a region of the upstream DNA binding channel and does not contact the accessory subunits. Both subdomains form distinct regions of the upstream DNA binding channel, but the flexible AID contributes to DNA binding affinity only when the accessory subunit stabilizes it in the correct position (9). The extreme N-terminal residues of the catalytic core do not show sequence conservation with other Exo domains in the family A polymerase group and are thus designated as a separate domain N-terminal domain (NTD), which presumably contains the mitochondrial leader sequence.
Pol γ is a member of the family A polymerase group and shares the highest sequence similarity with bacteriophage T7 DNA polymerase (T7 Pol) (8). We generated a structural model of the Pol γ ternary complex by superimposition of the apo-holoenzyme Pol γ structure (PDB code 3IKM) (1) with the structure of T7 DNA polymerase (PDB code 1T8E) with bound primer template DNA, incoming ddNTP and Mg2+ ions. We then mapped 58 Alpers mutations onto this structural model and reported that they cluster into distinct regions, which we termed clusters 1-5 (1). Because Pol γ is a family A polymerase, functional insight can be extrapolated from the extensive studies of conserved elements in bacterial DNA polymerase I (Pol I) and T7 Pol (10-15). This and evaluation of the unique features of Pol γ by biochemical analysis of yeast, fly, and human Pol γ variants (16-18), revealed that each cluster defines a unique functional region, which exhibits a distinct biochemical defect when affected by a pathogenic mutation (1). Cluster 1 mutations are predicted to cause a primary defect in pol activity, and affect residues involved directly in catalysis, or indirectly by affecting architectural residues that may disrupt the position of catalytic residues. Catalytic residues include those binding the two Mg2+ ions, those that make contact with the incoming dNTP, and those that make contact with the first, second and third nucleotide pairs in the nascent dsDNA, which are critical for correct positioning of the substrate in the pol active site. For example, the human pathogenic mutation R943H affects an amino acid residue that contacts the γ phosphate of the incoming dNTP, and has been shown in vitro to have a 30-fold increase in Km for dNTPs and a 5-fold decrease in kcat, which together reduce pol activity 150-fold without affecting DNA binding affinity (19). Cluster 2 mutations are predicted to cause a primary defect in DNA binding affinity, and affect residues of the IP and AID that form the upstream DNA binding channel, which enhance DNA binding affinity via contacts with nucleotides upstream of the third base pair of primer template. Biochemical analysis of recombinant fly Pol γ with a triple alanine substitution in amino acid residues K768/D769/F770 are located in the DNA binding channel wall of the IP domain, and cause a small decrease in binding affinity (17). Cluster 3 mutations are predicted to affect the pol: exo activity ratio and may have variable defects in DNA binding affinity. Many of the cluster 3 mutations map to a helix-coil-helix module (residues 295-312) located in the Exo domain that has been termed the “orienter” module (18). L304R of the “orienter” module has been studied biochemically in recombinant yeast Pol γ, and exhibits 3-fold increased exo activity, 20-fold decreased pol activity and 10-fold decreased DNA binding affinity (18). In addition, cluster 3 mutations map to the partitioning loop, which is a novel module conserved in Pol γ (residues 1050-1095) that is located between the fingers and palm subdomains and is not present in any other known polymerase (1). To date, no biochemical data is available for residues of the partitioning loop, although it has been shown that yeast strains homozygous for a mutation that is equivalent to G1051R in human Pol γ cause a 10-fold increase in point mutational frequency in vivo (20). Cluster 4 mutations map to the Exo domain along the distal accessory subunit interface, and are predicted to cause a biochemical defect similar to the R232G variant, which was shown in vitro to have reduced pol rate, increased exo activity and wild type (WT) DNA binding affinity (21). Cluster 5 mutations are located in the periphery of the IP subdomain and have not been shown to cause a biochemical defect. For example, human recombinant Pol γ harboring R627Q or R627W mutations exhibited no defects in vitro when analyzed for DNA binding affinity, pol activity and stimulation by the accessory subunit (22).
In applying our clustering model to the reported Alpers mutations, we found that Alpers syndrome was found only in compound heterozygous patients bearing two mutations from different clusters in Pol γ (1). Since publication, several new reports have been published that have added dozens of new mutations and mutation combinations to the POLG syndrome library (23-26). Here, we examine potential genotype-phenotype correlations by analyzing all reported POLG mutations available to date, not restricted to a phenotype or age-of-onset, utilizing our clustering model. In total, we assign 136 mutations to the five clusters and identify segments of primary sequence that can be used to delimit the boundaries of each cluster. We demonstrate the validity of our clustering model as the first to establish genotype-phenotype correlations for POLG syndromes, and show that it can be used to predict the pathogenic potential and biochemical defects of novel mutations, and to provide information about the likely severity of the POLG syndrome for compound heterozygotes with novel combinations of mutations.
Methods
Computational analysis
We docked primer-template DNA (ptDNA) into the putative DNA-binding channel of the Pol γ apo-holoenzyme crystal structure (PDB code 3IKM, (8)) by superposition of the closed ternary complex of T7 Pol bound to ptDNA and dNTP (PDB code 1T8E, (27)) using PyMOL (http://www.pymol.org/). As we described previously, we found that the best overall alignment was obtained by aligning the palm subdomain of Pol γA (residues 815-910, 1095-1239) to the palm subdomain of T7 Pol (residues 409-487, 611-704) (1). This model was used to map and evaluate all reported POLG disease mutations.
Statistical analysis
We used Pearson's chi-square analysis to calculate the significance of each age of onset (infantile, childhood, juvenile, adult), and the mutation combination affecting the same (=0) or different cluster (=1) of Pol γ. Logistic regression analysis was used to calculate odds ratios for having an infantile-onset disease. We conducted all statistical analyses with Stata (version 11.0). Two-sided p values were used with a significance level of 0·05. We thank Dr. Kirsi Pietiläinen for help in statistical analysis.
Results
Cluster assignment of all reported POLG disease mutations
The Alpers mutations represent only a subset of the total library of reported disease-causing mutations in POLG. To investigate further the utility of the five functional clusters in Pol γ that we defined earlier to evaluate Alpers mutations (1), we compiled a list of all pathogenic point mutations reported to date by combining the POLG database entries (http://tools.niehs.nih.gov/polg/) with new data from recent reports (23-26, 28, 29). The list comprises a total of 136 pathogenic point mutations, including both dominant and recessive mutations. We included in our analysis POLG mutations that represented the only disease-causing defect identified in the patient, exclusive of mutations in other genes associated with mitochondrial dysfunction, and excluded consideration of null mutations. By excluding patients with mutations in multiple genes associated with mitochondrial dysfunction, a clear link can be established between functional defects in Pol γ and the severity of disease manifestations. Furthermore, we considered a pathogenic mutation to be dominant only if the family history demonstrated a dominant mode of inheritance, leaving out potential de novo mutations.
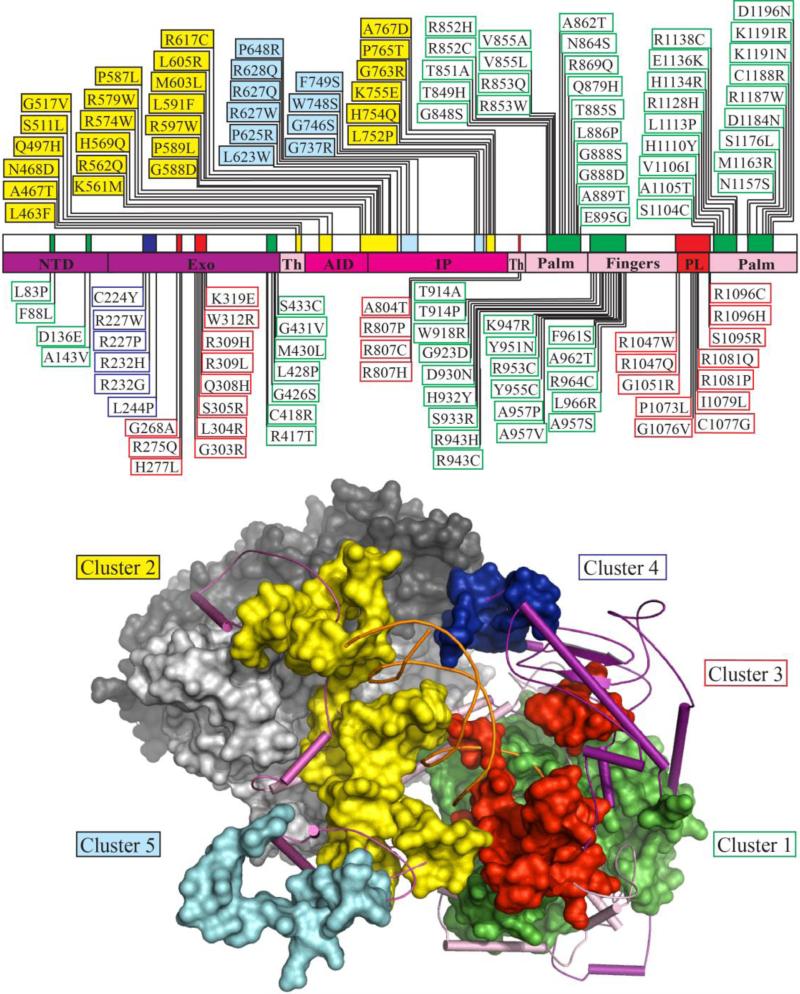
We mapped each of the pathogenic mutations onto our structural model and assigned them to a cluster by evaluating them individually. Although a cluster may contain residues that are distant in primary sequence, assignment was straightforward in most cases because each cluster occupies a distinct structural region in the Pol γ tertiary structure. Some mutations mapped to areas equidistant from two different clusters, and because we did not define explicitly the boundaries of the structural regions for each cluster in our previous analysis, we made use of additional criteria for cluster assignment. To do so, we analyzed each point mutation based on reported biochemical data to predict the functional defect it would cause. In this way, cluster assignment is based not only on structural location but also upon functional insight. In total, we assigned 136 pathogenic mutations to the five functional clusters we defined earlier (1) as shown in Figure 1. Due to the large library of reported mutations, we concluded that a sufficient number of mutations were assigned to each cluster to use their map positions as the sole means of defining the structural region and primary sequence occupied by each cluster. Mutations from each cluster are divided into several subclusters of 10-100 amino acid residues, and are distributed across the length of the POLG gene (Figure 1, upper schematic). Despite the large separation of subclusters in primary sequence, the elements of a cluster fold into compact structural regions as illustrated in Figure 1 (lower panel).
Figure 1. Clustering of 136 pathogenic mutations within five functional modules in the catalytic subunit of human Pol γ.
Upper panel, schematic diagram of the human POLG gene illustrating the clustering of 136 pathogenic mutations into discrete blocks of amino acid residues, which we term subclusters. Mutant alleles and subclusters are colored according to the cluster to which they belong: cluster 1, green; cluster 2, yellow; cluster 3, red; cluster 4, blue; cluster 5, cyan (see the text for details). The palm (residues 815-910 and 1096-1239), fingers (residues 911-1048), and thumb (Th, residues 440-475 and 785-814) subdomains of the Pol domain are colored pink, and the partitioning loop (PL, residues 1049-1095) is red. The accessory (subunit) interacting domain (AID, residues 476-570) and the intrinsic processivity (IP, residues 571-784) subdomains of the spacer domain are colored in magenta. The N-terminal domain (NTD, residues 1-170) and the Exo domain (residues 171-439) are colored in purple. Lower panel, structural model of the human Pol γ apo-holoenzyme (PDB code 3IKM) with docked primer template DNA shown as orange ribbon (see Computational methods for details). The catalytic subunit of Pol γ is shown as a cartoon representation of the secondary structural elements (SSEs), with regions defined by clusters illustrated as space-filled modules, colored according to the schematic. The proximal and distal accessory subunits are shown as surface representations in light and dark gray, respectively.
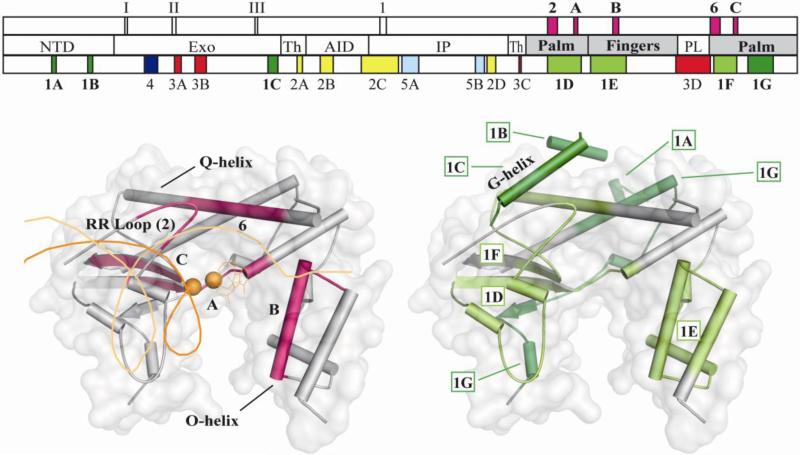
68 mutations were assigned to cluster 1, defining seven subclusters located in the NTD, Exo and Pol domains. The primary biochemical defect caused by a cluster 1 mutation is predicted to be reduced pol activity. As expected, most mutations affect residues of the Pol domain, including the five conserved motifs within the Pol domain of family A polymerases that are essential for 5'-3' DNA polymerase activity (Figure 2, upper schematic, (12)). Subcluster 1e (residues 914-966) spans the O-helix and its environs (Figure 2, lower panel), the function of which is to bind the correct dNTP substrate by transitioning between an open and closed complex (30). The conserved amino acid residues on the O-helix are known as the Pol B motif (residues 943-958), and function to establish specific contacts with correctly base paired dNTP in the closed conformation (12). POLG mutations that disrupt the specific contacts with the incoming dNTP (H932Y, R943H, K947R, Y951N, and Y955C) will reduce fidelity and increase Km (dNTP), without affecting DNA binding affinity (19, 31). Thus, mutations affecting this site are most likely dominant because they are capable of competing for dNTP binding with wild type (WT) Pol γ but are unable to polymerize nucleotides effectively, and are predicted to cause mtDNA damage that is associated with enzyme stalling (32).
Figure 2. Architectural and functional subclusters of the Pol domain.
Upper panel, schematic diagram of the POLG gene as shown in Figure 1, with an additional upper section indicating the location of the three Exo motifs, labeled as I, II, and III, and the six Pol motifs, labeled as 1 (motif 1), 2 (motif 2), A (Pol A motif), B (Pol B motif), 6 (motif 6) and C (Pol C motif), which are conserved throughout family A polymerases (see text). Motifs and subclusters that are illustrated in the bottom panels are in shown bold. Bottom panels, SSEs and transparent surface representation of the palm and fingers subdomains of Pol γ (PDB code 3IKM) are shown (subdomains are colored in gray in the middle section of the schematic). Bottom-left panel, the five motifs of the Pol domain, which are colored in pink in the upper section of the schematic, are shown as pink SSEs and are labeled accordingly. Docked primer template DNA is shown as orange ribbon, Mg2+ ions are shown as orange spheres, and the incoming dNTP is shown as orange sticks (see Computational methods for details). Bottom-right panel, subclusters 1D, 1E and 1F encompass one or more of the five conserved motifs of the Pol domain and are colored in light green. Subclusters 1A, 1B, 1C and 1G, colored in dark green, are located further structurally from the pol active site, and are considered to play an architectural role.
Subcluster 1d (residues 848-895) comprises the RR loop (residues 845-863) (1) and the conserved Pol A motif (residues 887-896) of the palm subdomain (Figure 2) (3). The RR loop is equivalent to motif 2 in Pol I, and is critical for binding correctly base-paired ptDNA in the minor groove and the template DNA backbone (12). The Pol A motif in Pol I contacts the primer strand, binds Mg2+ via amino acid residue D705 (D890 in Pol γ) and discriminates against ribonucleotide incorporation via residue E710 (E895 in Pol γ) (33). Mutations in the Pol A motif at either D890 or E895 in Pol γ would likely manifest a dominant lethal phenotype, and the only reported patient to date with the E895G mutation died immediately after birth (34). Motif 2 mutations should always cause reduced pol activity but may also cause a DNA binding defect depending on the residue, such as the 5-fold defect reported for G848S and R852C (35). The DNA binding affinity of T851A, R853Q, Q879H, T885S may decrease slightly, but the reduction in pol activity is much greater (35). Overall, we feel it is unlikely for dominant mutations to reside in motif 2 because these mutant forms of Pol γ would not compete with WT Pol γ for DNA binding.
Subcluster 1f (residues 1104-1138) comprises motif 6 (residues 1097-1110) and the Pol C motif (residues 1134-1141) in the palm subdomain (Figure 2) (according to the nomenclature defined for Pol I (12)). Motif 6 is located on the Q-helix and binds correctly base paired template strand and the ptDNA minor groove in Pol I via the residues N845 and Q849 ((13), equivalent to N1098 and Q1102 in Pol γ, respectively). The Pol C motif in Pol γ binds Mg2+ via D1135, whereas H1134 and E1136 contact the primer strand. Subcluster 1g (residues 1157-1196) maps to a C-terminal region of the palm subdomain and forms an anti-parallel beta strand adjacent to the Pol A motif. Only K1191 is predicted to contact the primer terminus, whereas the rest of subcluster 1g serves an architectural role (1). Mutations affecting motif 6 have been reported to cause DNA binding defects along with pol defects in Pol I Klenow (13). However, an alanine substitution at the equivalent residue to Pol γ H1134 in Klenow (H881A) caused a decrease in kcat from 6- to 66- fold (13) but retained DNA binding affinity, suggesting that a mutation in H1134 might be dominant.
We assigned mutations to subclusters 1d-g with high confidence because there is extensive biochemical data available for the Pol domain, in addition to the presence of the highly conserved amino acid sequence motifs. In contrast, a subcluster of seven mutations (R417T, C418R, G426S, L428P, M430L, G431V, S433C) mapped to the G-helix (Figure 2, lower right panel), a structural element that had not been studied previously. According to the current Exo domain assignment of residues 170-440 by Lee et al. (8), this subcluster would reside in the Exo domain, although earlier T7 Pol (15) and Pol I structures (36) label the G-helix as part of the palm subdomain within the Pol domain. Regardless of the domain assignment, this subcluster is adjacent to motif 2 and motif 6, and we propose it serves an architectural role that contributes to pol activity indirectly, and assign it as subcluster 1c. Four mutations from the NTD were also observed to be closer structurally to functional Pol motifs as compared to Exo motifs; we thus assigned D136E and A143V as subcluster 1b (residues 136-143), and both L83P and F88L were assigned as subcluster 1a (residues 83-88).
25 mutations from cluster 2, which we predict to cause primary defects in DNA binding affinity, outline the putative DNA binding channel, and map to four subclusters within the thumb subdomain of the Pol domain (2a, residues 463-468), and the AID (2b, residues 497-517) and IP (2c, residues 561-617 and 2d, 752-767) subdomains of the spacer domain. Subcluster 2c contains motif 1, which in Pol I was shown to fold into a loop and binds DNA in the channel (12). Motif 1, together with subcluster 2d, form the major face of the DNA binding channel. Subcluster 2a maps to a region of the thumb subdomain at the accessory subunit interface where A467T, N468D, and L463F are positioned. Our group has characterized residue A467T biochemically, and it was shown to retain 70% of WT DNA binding affinity in a reconstituted holoenzyme form (22). Extensive biochemical analysis of the spacer domain in general has demonstrated that the hydrophobic core of the IP subdomain is critical for shaping the DNA binding channel wall (17, 22). Mutations that alter the IP subdomain, such as A467T, are predicted to perturb the channel to block DNA from entering, which is then observed as reduced DNA binding affinity.
8 mutations from cluster 3 map to the orienter module (18, 52) of the Exo domain, and another 12 mutations from the same cluster map to the partitioning loop (1) of the Pol domain, which are defined by two subclusters (3b, residues 303-319 and 3d, 1047-1096, respectively). Additionally, a recent report from the Foury group (37) has shown that variants of yeast Pol γ bearing substitutions in residues of the thumb subdomain M602I (R802 in Hs) and the Exo II motif of the Exo domain A228V, R231K, R233W (S272, R275, H277 in Hs) also produce the altered pol:exo activity ratio that characterizes cluster 3 mutations. Therefore, we assign pathogenic residues R807P, R807C, R807H, and R804T as subcluster 3c (residues 804-807), and the Exo II motif mutations G268A, R275Q, and H277L as subcluster 3a (residues 268-277). In support of the proposed subcluster 3c, we note that the SYW triple alanine substitution in recombinant fly Pol γ, which was found to exhibit increased exo activity with both decreased pol activity and DNA binding affinity (17), maps adjacent to subcluster 3c in human Pol γ (residues S799/F800/W801). Similarly, the Exo II motif has been demonstrated to alter the pol: exo activity ratio in biochemical variants of Pol I by decreasing the affinity of the primer strand for the exo active site, consistent with the decrease in exo activity and WT pol activity observed for the R233W variant in yeast Pol γ (H277 in human Pol γ) (37).
Six mutations in cluster 4 (comprising residues 224-244), C224Y, R227P, R227W, R232G, R232H and L244P, map to a single region on the subunit interface of the Exo domain. Biochemical studies of a human R232G variant in reconstituted holoenzyme form showed a decrease in pol rate and an increase in exo activity, with unchanged DNA binding affinity that derive from loss of a direct stimulation of pol activity by the distal accessory subunit (21). This, and an earlier study by Lee et al., support a distinct role for the distal accessory subunit in enhancing processivity (53). We propose that the other residues of cluster 4 have similar biochemical characteristics.
Ten mutations from cluster 5 are also located in the IP domain, but they map to the distal region far removed from the DNA binding channel, and define subcluster 5a (residues 623-648) and subcluster 5b (residues 737-749).
Cluster assignment can be used to predict the pathogenicity of a novel mutation
Our clustering model provides an annotated guide to assign function to the entire mutational spectrum in the POLG gene, and we propose that it can be used to evaluate the potential pathogenicity of novel POLG mutations. In particular, when a novel mutation maps to a specific subcluster within the five functional clusters we have defined, we predict that it will likely be pathogenic and result in the biochemical defect associated with that subcluster. At the same time, we would argue that mutations that do not map to these specific amino acid sequence blocks are more likely to be neutral polymorphisms. In support of this model, we found that only 12 out of 87 reported single nucleotide polymorphisms (SNPs) from the human dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/) map within subclusters (Figure S1). Three of these are conservative substitutions of highly similar amino acids (F1092L, F1164L and F1164I), and likely have no functional consequences. These examples would suggest that although a mutation may affect a residue within a functional subcluster, conservative substitutions with amino acids with highly similar chemical properties may not be pathogenic, and this possibility should be taken into consideration when evaluating novel mutations. Additionally, missense mutations reported as SNPs in the SNP database are not necessarily non-pathogenic, as they could be uncommon recessive mutations that are masked when paired with a wild type allele.
Although the mutation library is large, the POLG mutational map is likely not yet saturated, so routine sequencing of patients with POLG syndrome will continue to fill out the spectrum. We predict that most novel mutations will map within our current clusters because they are by definition “mutational hotspots” and importantly, we observe from their positions within the crystal structure that they encompass the known functional motifs required for the biochemical activities of Pol γ. For example, based upon the high sequence conservation with family A polymerases, we note that the essential elements of the pol active site such as the Pol A-C motifs, motif 2, and motif 6 are each populated by human pathogenic mutations that map within our current cluster 1 (Figures 1 and 2).
Cluster combinations of compound heterozygotes correlate with age of onset of POLG syndrome
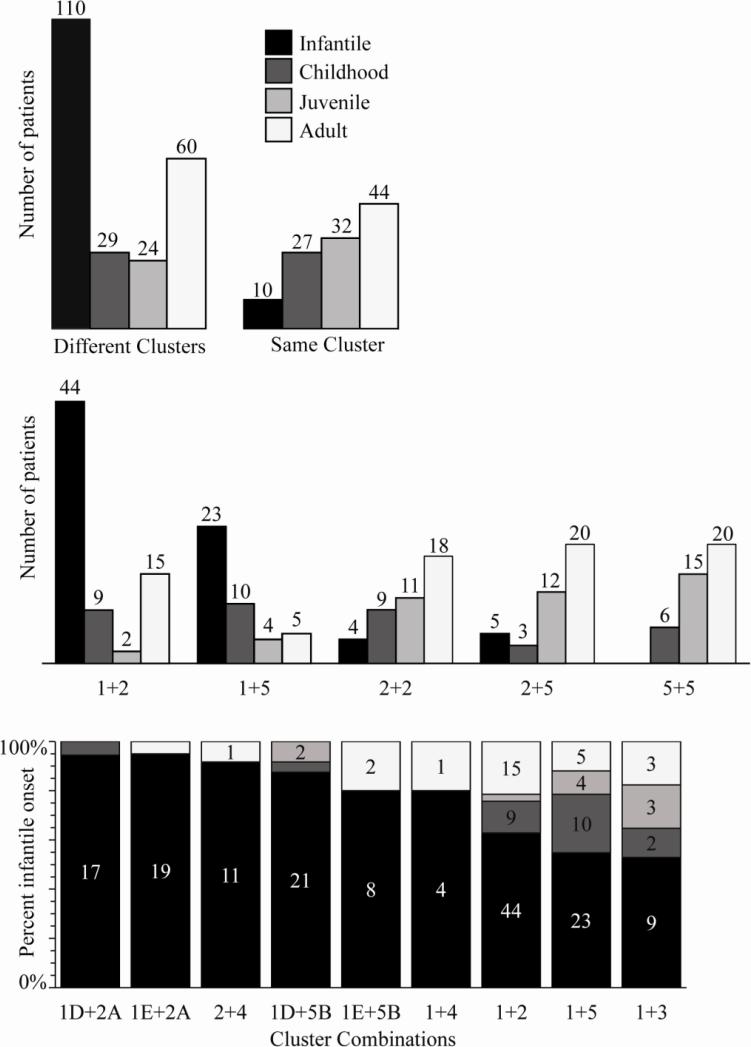
Previously, we evaluated the combination of mutations in compound heterozygotes that cause Alpers syndrome using our original clustering model, and found that Alpers was triggered exclusively by the combination of two mutations from different clusters (1). Here, we evaluate all mutations causing POLG syndromes (caused by compound heterozygosity) using the model refined by the designation of subclusters within the original five functional clusters. Whereas clinical presentation and age of onset are extremely variable for POLG syndromes, in general the severity of symptoms decreases with increasing age of onset (38). We were able to catalogue a total of 340 compound heterozygous patients harboring two pathogenic mutations and we classified each patient by the age of onset reported. Each combination was assigned to one of four age groups: infantile, < 3 years of age; childhood, ages 3-12; juvenile, 13-20 years; and adult, > 20 years. The data are compiled and evaluated in Figure 3. Compound heterozygous patients with two mutations from the same cluster were less common (113/340) and were generally associated with late onset POLG syndromes (Figure 3, upper panel). Compound heterozygous patients with two mutations from different clusters were much more common (227/340), and comprised 110/120 of the infantile onset combinations. These results argue that the combination of mutations from two different clusters is not just a specific trigger for Alpers syndrome; rather, in 123 unique mutation combinations in 227 compound heterozygous patients, it serves to represent a universal genotype-phenotype correlation for all POLG syndromes.
Figure 3. Analysis of mutation combinations as cluster combinations reveals predictive genotype-phenotype correlations.
The data used to compile the information in this figure was derived from the references included as Supplemental data. The number of mutation combinations manifesting POLG syndrome at each of the four age groups is shown in each panel for specific cluster combinations. Age of onset trends can be used to predict the severity of POLG syndrome for an individual with compound heterozygous mutations in POLG. Upper panel, POLG syndrome age of onset trends for mutation combinations of two mutations from different clusters versus the same cluster. Middle panel, patient data show trends in which severe cluster combinations have an earlier age of onset, whereas less severe or uncommon cluster combinations have a later age of onset. Lower panel, earlier age of onset trends for more severe cluster/ subcluster combinations.
Critical functions: clusters 1, 3, and 4
We predict that mutations affecting cluster 1 will reduce primarily pol activity, and these have been associated with increased replication fork stalling in vivo (32,52,53). The stalling phenotype is characterized by slow progression of or suspended replication forks, and causes an accumulation of replication intermediates that can lead to double-stranded DNA breaks, mtDNA deletions and base substitution mutations (39-41). It is thus unlikely that a compound heterozygote with two cluster 1 mutations would be viable unless the defects are very mild. Indeed, only 11 such combinations of 14 different mutations have been reported in 18 patients (Figure S2), and involve mutations such as G923D and A957S that retain 25% WT pol (19), which is a relatively mild pol defect compared to other cluster 1 mutations, such as the 100-fold reduction of pol activity engendered by the Y955C mutation (32). Although the partitioning loop has not been studied biochemically, cluster 3 mutations may produce a biochemical phenotype similar to that of the L260R variant in yeast Pol γ (L304R in Hs Pol γ), which is characterized by an increase in exo activity, and decreases in pol activity and DNA binding affinity (18). Only four combinations have been reported in 12 patients for compound heterozygotes with two cluster 3 mutations (Figure S2), which may indicate that most of these combinations are lethal. Cluster 4 mutations are predicted to increase exo activity and decrease pol rate, while retaining WT DNA binding affinity (21). Interestingly, cluster 4 mutations have not been reported in combination with other cluster 4 mutations, or together with cluster 3 mutations. When Pol γ is stalled, the primer strand can translocate between the exo active site the pol active site, a mode described as idling by Atanassova et al. (32). We speculate that cluster 3 and 4 mutants may manifest an idling phenotype because of their enhanced exo: pol activity ratios, which is likely to increase non-productive turnover by mutant Pol γ, via exonucleolytic hydrolysis of correctly incorporated nucleotides. In compound heterozygotes, two idling Pol γs may be lethal, explaining the absence of cluster 3 + 4 combinations, as well as cluster 4 + 4 combinations. Taken together, the lack of complementation observed for clusters 1, 3, and 4 suggests that they serve essential functions in mtDNA metabolism.
Moderately severe dysfunction: clusters 2 and 5
Mutations affecting cluster 2 will most likely cause a primary defect in DNA binding affinity. Processivity, defined as the number of nucleotides incorporated in a single DNA binding event, varies directly with pol rate and DNA binding affinity and as a consequence, cluster 2 mutations reduce processivity. Individuals carrying homozygous A467T mutations present as juveniles with POLG syndrome (26, 42, 43). As we showed that Pol γ carrying the A467T substitution shows a DNA binding affinity 70% of that of WT (22), this suggests that moderate reductions in DNA binding and pol activity may be tolerated throughout childhood. It thus seems likely that most same cluster combinations of cluster 2 mutations will not be lethal, and instead will lead to development of later-onset POLG syndrome. Consistent with this hypothesis, 42 such combinations have been reported, and the majority show juvenile or adult onset (Figure 3, middle panel).
We have studied three cluster 5 mutations, W748S, R627Q and R627W in the reconstituted human Pol γ holoenzyme, but all three revealed no biochemical phenotype in our hands (22, 44). These mutations map to the distal surface of the IP domain, far removed from the DNA binding channel. Despite the lack of a documented biochemical phenotype, cluster 5 mutations have been reported in 139 compound heterozygous patients with POLG syndrome manifesting at all ages (Figures 3 and S2). Therefore, this region clearly carries a biologically-relevant function in mtDNA metabolism because cluster 5 mutations cause mtDNA instability (44, 45). As for cluster 2 mutations, cluster 5 mutations are found together in compound heterozygote individuals with juvenile or adult onset POLG syndrome. Compound heterozygotes with a cluster 2 mutation and a cluster 5 mutation have been reported in 40 patients, and a majority of the combinations resulted in juvenile or adult onset POLG syndrome (Figure 3, middle panel). Notably, this cluster combination is the only one that did not typically cause infantile or childhood onset POLG syndromes. To this point, 39/39 patients that were homozygous for W748S only developed a mild adult-onset POLG syndrome (46-48). In sum, cluster 2 and cluster 5 mutations, in contrast to those in clusters 1, 3 and 4, are observed to complement each other, perhaps suggesting that individual amino acid residues within these clusters serve less critical functions in mtDNA metabolism.
Severe combination: cluster 1 + 2
Compound heterozygotes with cluster 1 and cluster 2 mutations have been reported in 70 patients, and are the most abundant of all cluster combinations (Figure 3, middle and lower panels). The majority (44/70) of these combinations caused infantile onset POLG syndrome. In fact, the most severe combinations observed in this study are the subcluster combinations 1D + 2A and 1E + 2A. These combinations cause infantile onset POLG syndrome in 17/18 and 19/20 cases, respectively. Cluster 1 mutants have reduced pol activity but will compete for DNA binding and cause enzyme stalling when bound. Cluster 2 mutations show reduced DNA binding affinity and would be unable to compete for DNA binding with a cluster 1 mutant. We predict this would result in more stalled enzyme complexes than for bound cluster 2 complexes alone. Because cluster 2 mutants show only modest decreases in pol activity, mtDNA depletion will progress gradually until a threshold level is reached to give rise to symptoms of POLG syndrome.
The two most common cluster 2 mutations are A467T and P587L, and the latter is always reported in cis with T251I in patients. The occurrence of two mutations together suggests either a genetic founder effect (as in Hakonen et al. (46)), or synergic or compensatory effect of the variants. T251I maps to the Exo domain, but we consider it to be a SNP for the following reasons: T251 is not a conserved amino acid, is located on the protein surface, and is not in a reasonable proximity to any functional region. In contrast, P587L is conserved in mammals and fly, is positioned in the putative DNA binding channel, and two pathogenic mutations, G588D and P589L, are located directly adjacent to P587 on the same hairpin. By considering T251I as a SNP and excluding it from the functional analysis, P587L can be compared directly to A467T for its relative pathogenicity. Although no mechanistic data is available for POLG carrying the P587L allele, we suggest it will reduce DNA binding affinity to a much lesser extent than A467T. Direct comparison of the two mutations is possible in two different cluster 1 mutations. One patient was reported as compound heterozygous for G848S/A467T with infantile onset (43), whereas another was reported as compound heterozygous for G848S/P587L with adult onset (26). Homozygosity for P587L+T251I results in a late-onset myopathy phenotype (7).The same pattern is observed with K1191N (7, 26).
Severe combination: cluster 1+5
Compound heterozygotes with cluster 1 and cluster 5 mutations were reported showing that 23/42 mutation combinations had infantile onset POLG syndrome (Figure 3, middle panel). Such a high occurrence of the most severe form of POLG syndrome, together with the relatively large number of combinations documented, indicates that the combined defects from cluster 1 and cluster 5 are highly prone to cause mtDNA depletion at a young age. In particular, the most common cluster 5 mutation, W748S, manifested infantile onset POLG syndrome in 28/31 patients when found in combination with cluster 1 mutations. Two particularly severe combinations are the subcluster combinations 1D + 5B and 1E + 5B; both result in infantile onset POLG syndrome in >80% of affected patients (Figure 3, lower panel).
Other combinations
Cluster 4 mutations were reported in a total of 17 combinations with cluster 1 and cluster 2 (Figure 3, lower panel). The incidence of infantile onset POLG syndrome was high (15/17), and may indicate that biochemical defects of cluster 4 mutations are uniquely severe in vivo. Infantile onset was less common for compound heterozygotes with cluster 3 mutations in combination with cluster 1 (9/17), cluster 2 (10/22) and cluster 5 (3/12) (Figure S2). These results suggest that cluster 4 mutations are more pathogenic than cluster 3 mutations.
Expected incidence of specific symptoms caused by POLG syndrome can be predicted by the age of onset
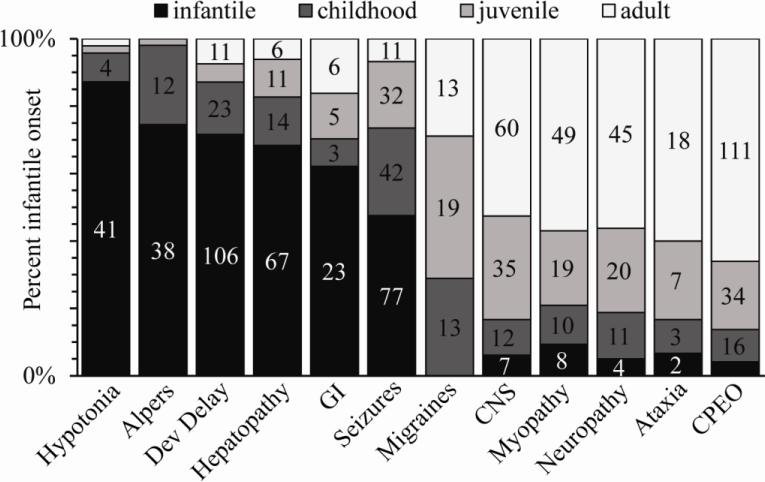
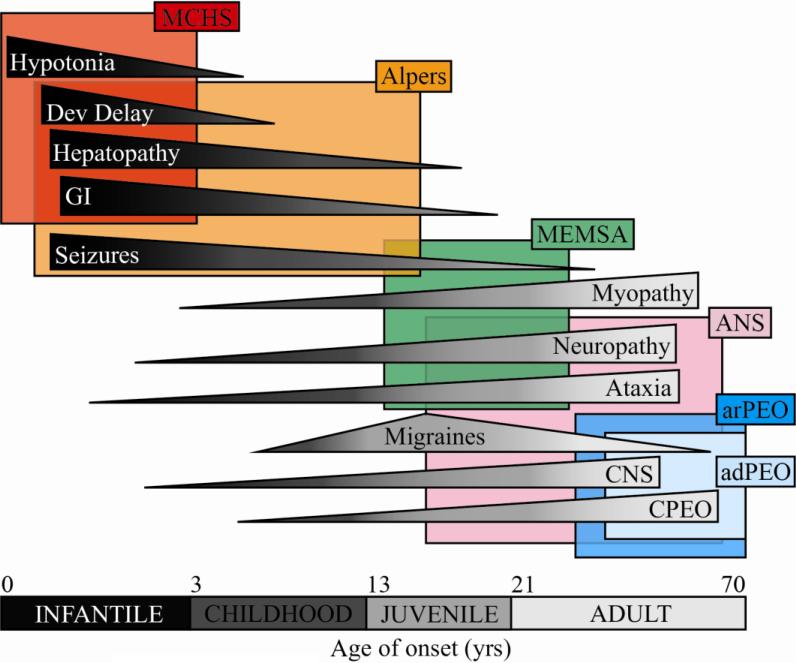
The clinical manifestations reported for each compound heterozygous patient were compiled in effort to study the incidence and timeline of symptoms caused by POLG syndrome. To analyze the clinical descriptions in terms of individual symptoms, it was necessary to group similar and synonymous symptoms into symptom groups (Table S1). Very common symptoms, such as ataxia, seizures, hypotonia, and migraines, were left as individual entries because their sample size was sufficiently large, and these symptoms have been established as hallmarks for specific POLG syndromes. For other symptom groups, it was necessary to combine different symptoms into a group of related symptoms based on the affected tissue type resulting in the observed symptom. For example, the CPEO group includes all synonyms for ophthalmoplegia as well as diplopia and ptosis, two symptoms that both indicate impaired function of the extraocular muscles. Neurological disorders are divided into two subcategories; the neuropathy group includes symptoms caused by damage to peripheral neurons, and the CNS group includes symptoms caused by damage to neurons of the central nervous system. The developmental delay group includes symptoms related to an encephalopathy that involve an altered mental state. The myopathy, hepatopathy, and GI groups involve symptoms that affect or are an indicator of dysfunction in muscle tissue, the liver, or the gastrointestinal tract, respectively. Figure 4 shows the incidence of each symptom group in patients as a function of age of onset. The more severe symptoms, such as hepatopathy, hypotonia, and developmental delay have a high incidence among patients with infantile onset POLG syndrome, whereas less severe symptoms such as CPEO manifest mainly in patients with adult onset POLG syndrome. These data demonstrate that the symptoms caused by POLG syndromes represent a continuum of manifestations that become less severe as the age of onset increases. These syndromes have been characterized in the past according to specific groups of co-occurring symptoms that present in patients (Figure 5). For example, Alpers syndrome is characterized by seizures, developmental delay, and liver failure. Figure 5 displays the timeline of POLG syndromes, and outlines the co-occurring symptoms that define each of the specific syndromes that have been used to characterize patients with pathogenic POLG mutations.
Figure 4. Age of onset correlation for reported symptoms.
Symptoms associated with POLG syndromes manifest typically at different ages. The symptoms reported in the patients have been grouped together based on affected tissue types, and ordered according to their ages of onset. The more severe symptoms and forms of POLG syndromes are at the left end of the figure, while the less severe symptoms are at the right end. Detailed information of the symptom grouping is shown in Table 1.
Figure 5. Symptoms associated with different POLG syndromes.
Depending on the specific mutations and cluster combinations of the POLG gene mutations in individual patients, POLG syndromes can become symptomatic at different ages and manifest as pathogenic conditions in different tissue types. The figure shows a continuing spectrum of decreasing symptom severity from top to bottom, as well as a delayed age of onset from left to right. MCHS, Childhood myocerebrohepatopathy spectrum; MEMSA, Myoclonic epilepsy myopathy sensory ataxia; ANS, Autonomic Nervous System Dysfunction; arPEO/ adPEO, Autosomal Recessive/ Dominant Progressive External Opthalmoplegia.
Discussion
Due to the mutational diversity in the POLG gene and the variable clinical features of POLG syndromes, establishment of genotype-phenotype correlations has remained elusive (49). Furthermore, a number of de novo variants emerge in routine diagnosis laboratories, and interpretation of their pathogenic role is challenging. We show here that detailed protein structural and functional studies, with the aid of a crystal structure, combined with knowledge from experimental and disease mutations can be combined to predict clinical consequences of an identified variant, and to distinguish likely pathogenic mutations from polymorphic variants. In this report, we demonstrate clear genotype-phenotype correlations for predicting the severity and hence age of onset of compound heterozygous POLG syndromes. We refine our clustering model of Pol γ (1), proposing five functional clusters, each of which comprise subclusters that define structural regions of the catalytic core of Pol γ. Within these clusters, we can predict the likely biochemical defects and intrinsic pathogenicity of novel point mutations. In applying this clustering protocol to nearly 140 combinations of POLG compound heterozygous mutations, we showed that each cluster combination can be used to give information of the potential severity of disease outcome. Strikingly, we were able to predict correctly the age of onset in 90% of infantile-onset POLG syndromes via the presence of two mutations from different clusters in compound heterozygotes of POLG. Furthermore, we show that age of onset can be used to predict the symptoms that a patient will manifest, and provide a timeline of the disease progression that may be expected.
Individuals carrying mutations in POLG can manifest a disease at any age, depending on the mutation combination as well as the severity of the Pol γ functional defects (5, 26, 50). Presumably, the most severe Pol γ mutations would be embryonic lethal in compound heterozygous form with any other, even slightly deleterious mutation, and we speculate that they may also have the potential to be dominant. This is likely the reason for example, that nucleotide changes that yield amino acid substitutions in the Mg2+ binding residues D890 and D1135 have never been reported. We found a similar trend in our clustering analysis showing a lower occurrence of compound heterozygotes from the same cluster for clusters 1, 3 and 4. The low occurrence of same-cluster compound heterozygotes may indicate that the majority of potential combinations of this type are lethal, and the lack of complementation supports our assignment of a common biochemical defect. This does not, however, apply to IP-domain mutations, which as homozygous are a common cause of severe progressive disorder that has a later disease-onset in juvenile age or early adulthood. Therefore, combinations of mutations that cause the most severe form of the POLG syndrome allow embryonic development, but lead to early infantile death.
We believe that our clustering model of POLG mutations can be used as a tool to aid in the diagnosis of patients with mitochondrial disorders. The POLG gene shows considerable variation, and novel polymorphic changes are common. A valuable existing tool, the polymerase gamma database (http://tools.niehs.nih.gov/polg/), lists reported mutations, but their pathogenic role sometimes remains unclear, especially for rare changes. Our prediction tool adds considerable power via structural analysis for interpretation of the consequences of identified rare variants, and can provide information for novel changes. Furthermore, it predicts the combinatorial effect of compound heterozygous changes for a patient, and provides information of typical clinical manifestations and ages of onset. We note that these predictions may not always be accurate, both because of the variability in disease manifestation introduced by genetic background, and the possibility that a number of the patients reported with POLG syndrome may also have other inherited mutations in their mitochondrial or nuclear DNAs in addition to the reported mutations in the POLG gene; although we excluded known cases of digenic mitochondrial diseases in this study, the latter possibility remains problematic and would be addressed by whole exome sequencing. Despite these potential drawbacks, we believe that our clustering protocol provides additional information that will prove important not only for diagnosis and genetic counselling, but also for treatment: e.g., the epilepsy drug valproate is toxic to the liver of patients with POLG mutations (51), and as epilepsy is a common initial symptom in children with POLG diseases, many patients have died before receiving a liver transplant. The ability to predict pathogenic likelihood for new POLG variants will be important when considering valproate as a treatment in severe epilepsies. Finally, because the predictions are based on reported patient data, the ever- increasing number of documented POLG mutations will serve to enhance the diagnostic power of the clustering protocol.
Supplementary Material
Highlights.
Five functional modules in the POLG catalytic core can be assigned.
Human POLG disease mutations map into functional clusters.
Mapping POLG mutations defines genotype-phenotype relationships.
Onset and severity of POLG syndromes depends on functional cluster combinations.
Pathogenicity of new genetic variants of POLG can be assessed.
Acknowledgments
The authors wish to acknowledge Drs. Anu Suomalainen and Liliya Euro for their contributions to the conceptual framework of this study that was developed in our earlier joint report (1).
Funding
This work was supported by National Institutes of Health Grant GM45295 to L.S.K. A.N. was supported by the Academy of Finland.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
None
References
- 1.Euro L, Farnum GA, Palin E, Suomalainen A, Kaguni LS. Clustering of Alpers disease mutations and catalytic defects in biochemical variants reveal new features of molecular mechanism of the human mitochondrial replicase, Pol {gamma}. Nucleic Acids Res. 2011;39:9072–9084. doi: 10.1093/nar/gkr618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ylikallio E, Suomalainen A. Mechanisms of mitochondrial diseases. Ann Med. 2011;44:41–59. doi: 10.3109/07853890.2011.598547. [DOI] [PubMed] [Google Scholar]
- 3.Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 4.Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J Biol Chem. 2006;281:374–82. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- 5.Saneto RP, Naviaux RK. Polymerase gamma disease through the ages. Dev Disabil Res Rev. 2010;16:163–74. doi: 10.1002/ddrr.105. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen KV, Sharief FS, Chan SS, Copeland WC, Naviaux RK. Molecular diagnosis of Alpers syndrome. J Hepatol. 2006;45:108–16. doi: 10.1016/j.jhep.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 7.Horvath R, Hudson G, Ferrari G, Futterer N, Ahola S, Lamantea E, Prokisch H, Lochmuller H, McFarland R, Ramesh V, et al. Phenotypic spectrum associated with mutations of the mitochondrial polymerase gamma gene. Brain. 2006;129:1674–84. doi: 10.1093/brain/awl088. [DOI] [PubMed] [Google Scholar]
- 8.Lee YS, Kennedy WD, Yin YW. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009;139:312–24. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YS, Lee S, Demeler B, Molineux IJ, Johnson KA, Yin YW. Each monomer of the dimeric accessory protein for human mitochondrial DNA polymerase has a distinct role in conferring processivity. J Biol Chem. 2009;285:1490–9. doi: 10.1074/jbc.M109.062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh K, Modak MJ. Contribution of polar residues of the J-helix in the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I (Klenow fragment): Q677 regulates the removal of terminal mismatch. Biochemistry. 2005;44:8101–10. doi: 10.1021/bi050140r. [DOI] [PubMed] [Google Scholar]
- 11.McCain MD, Meyer AS, Schultz SS, Glekas A, Spratt TE. Fidelity of mispair formation and mispair extension is dependent on the interaction between the minor groove of the primer terminus and Arg668 of DNA polymerase I of Escherichia coli. Biochemistry. 2005;44:5647–59. doi: 10.1021/bi047460f. [DOI] [PubMed] [Google Scholar]
- 12.Loh E, Loeb LA. Mutability of DNA polymerase I: implications for the creation of mutant DNA polymerases. DNA Repair (Amst) 2005;4:1390–8. doi: 10.1016/j.dnarep.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Singh K, Modak MJ. Presence of 18-A long hydrogen bond track in the active site of Escherichia coli DNA polymerase I (Klenow fragment). Its requirement in the stabilization of enzyme-template-primer complex. J Biol Chem. 2003;278:11289–302. doi: 10.1074/jbc.M211496200. [DOI] [PubMed] [Google Scholar]
- 14.Patel PH, Suzuki M, Adman E, Shinkai A, Loeb LA. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J Mol Biol. 2001;308:823–37. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 15.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–8. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 16.Johnson AA, Tsai Y, Graves SW, Johnson KA. Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochemistry. 2000;39:1702–8. doi: 10.1021/bi992104w. [DOI] [PubMed] [Google Scholar]
- 17.Luo N, Kaguni LS. Mutations in the spacer region of Drosophila mitochondrial DNA polymerase affect DNA binding, processivity, and the balance between Pol and Exo function. J Biol Chem. 2005;280:2491–7. doi: 10.1074/jbc.M411447200. [DOI] [PubMed] [Google Scholar]
- 18.Szczepanowska K, Foury F. A cluster of pathogenic mutations in the 3'-5' exonuclease domain of DNA polymerase gamma defines a novel module coupling DNA synthesis and degradation. Hum Mol Genet. 2010;19:3516–29. doi: 10.1093/hmg/ddq267. [DOI] [PubMed] [Google Scholar]
- 19.Graziewicz MA, Longley MJ, Bienstock RJ, Zeviani M, Copeland WC. Structure-function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nat Struct Mol Biol. 2004;11:770–6. doi: 10.1038/nsmb805. [DOI] [PubMed] [Google Scholar]
- 20.Baruffini E, Ferrero I, Foury F. Mitochondrial DNA defects in Saccharomyces cerevisiae caused by functional interactions between DNA polymerase gamma mutations associated with disease in human. Biochim Biophys Acta. 2007;1772:1225–35. doi: 10.1016/j.bbadis.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Lee YS, Johnson KA, Molineux IJ, Yin YW. A single mutation in human mitochondrial DNA polymerase Pol gammaA affects both polymerization and proofreading activities of only the holoenzyme. J Biol Chem. 2010;285:28105–16. doi: 10.1074/jbc.M110.122283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luoma PT, Luo N, Loscher WN, Farr CL, Horvath R, Wanschitz J, Kiechl S, Kaguni LS, Suomalainen A. Functional defects due to spacer-region mutations of human mitochondrial DNA polymerase in a family with an ataxia-myopathy syndrome. Hum Mol Genet. 2005;14:1907–20. doi: 10.1093/hmg/ddi196. [DOI] [PubMed] [Google Scholar]
- 23.Ferreira M, Evangelista T, Almeida LS, Martins J, Macario MC, Martins E, Moleirinho A, Azevedo L, Vilarinho L, Santorelli FM. Relative frequency of known causes of multiple mtDNA deletions: two novel POLG mutations. Neuromuscul Disord. 2011;21:483–8. doi: 10.1016/j.nmd.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 24.Sato K, Yabe I, Yaguchi H, Nakano F, Kunieda Y, Saitoh S, Sasaki H. Genetic analysis of two Japanese families with progressive external ophthalmoplegia and parkinsonism. J Neurol. 2011;258:1327–32. doi: 10.1007/s00415-011-5936-x. [DOI] [PubMed] [Google Scholar]
- 25.Tang S, Dimberg EL, Milone M, Wong LJ. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE)-like phenotype: an expanded clinical spectrum of POLG1mutations. J Neurol. 2011;259:862–8. doi: 10.1007/s00415-011-6268-6. [DOI] [PubMed] [Google Scholar]
- 26.Tang S, Wang J, Lee NC, Milone M, Halberg MC, Schmitt ES, Craigen WJ, Zhang W, Wong LJ. Mitochondrial DNA polymerase gamma mutations: an ever expanding molecular and clinical spectrum. J Med Genet. 2011;48:669–81. doi: 10.1136/jmedgenet-2011-100222. [DOI] [PubMed] [Google Scholar]
- 27.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–61. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baruffini E, Horvath R, Dallabona C, Czermin B, Lamantea E, Bindoff L, Invernizzi F, Ferrero I, Zeviani M, Lodi T. Predicting the contribution of novel POLG mutations to human disease through analysis in yeast model. Mitochondrion. 2010;11:182–90. doi: 10.1016/j.mito.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Khan A, Trevenen C, Wei XC, Sarnat HB, Payne E, Kirton A. Alpers Syndrome: The Natural History of a Case Highlighting Neuroimaging, Neuropathology, and Fat Metabolism. J Child Neurol. 2011;27:636–40. doi: 10.1177/0883073811423973. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–25. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batabyal D, McKenzie JL, Johnson KA. Role of histidine 932 of the human mitochondrial DNA polymerase in nucleotide discrimination and inherited disease. J Biol Chem. 2010;285:34191–201. doi: 10.1074/jbc.M110.156182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atanassova N, Fuste JM, Wanrooij S, Macao B, Goffart S, Backstrom S, Farge G, Khvorostov I, Larsson NG, Spelbrink JN, et al. Sequence-specific stalling of DNA polymerase gamma and the effects of mutations causing progressive ophthalmoplegia. Hum Mol Genet. 2011;20:1212–23. doi: 10.1093/hmg/ddq565. [DOI] [PubMed] [Google Scholar]
- 33.Astatke M, Ng K, Grindley ND, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc Natl Acad Sci U S A. 1998;95:3402–7. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spinazzola A, Invernizzi F, Carrara F, Lamantea E, Donati A, Dirocco M, Giordano I, Meznaric-Petrusa M, Baruffini E, Ferrero I, et al. Clinical and molecular features of mitochondrial DNA depletion syndromes. J Inherit Metab Dis. 2009;32:143–58. doi: 10.1007/s10545-008-1038-z. [DOI] [PubMed] [Google Scholar]
- 35.Kasiviswanathan R, Longley MJ, Chan SS, Copeland WC. Disease mutations in the human mitochondrial DNA polymerase thumb subdomain impart severe defects in mitochondrial DNA replication. J Biol Chem. 2009;284:19501–10. doi: 10.1074/jbc.M109.011940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Kong Y, Korolev S, Waksman G. Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates. Protein Sci. 1998;7:1116–23. doi: 10.1002/pro.5560070505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foury F, Szczepanowska K. Antimutator Alleles of Yeast DNA Polymerase Gamma Modulate the Balance between DNA Synthesis and Excision. PLoS One. 2011;6:e27847. doi: 10.1371/journal.pone.0027847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suomalainen A, Isohanni P. Mitochondrial DNA depletion syndromes--many genes, common mechanisms. Neuromuscul Disord. 2010;20:429–37. doi: 10.1016/j.nmd.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 39.Srivastava S, Moraes CT. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum Mol Genet. 2005;14:893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet. 2009;18:1028–36. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–51. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong LJ, Naviaux RK, Brunetti-Pierri N, Zhang Q, Schmitt ES, Truong C, Milone M, Cohen BH, Wical B, Ganesh J, et al. Molecular and clinical genetics of mitochondrial diseases due to POLG mutations. Hum Mutat. 2008;29:E150–72. doi: 10.1002/humu.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart JD, Tennant S, Powell H, Pyle A, Blakely EL, He L, Hudson G, Roberts M, du Plessis D, Gow D, et al. Novel POLG mutations associated with neuromuscular and liver phenotypes in adults and children. J Med Genet. 2009;46:209–14. doi: 10.1136/jmg.2008.058180. [DOI] [PubMed] [Google Scholar]
- 44.Palin EJ, Lesonen A, Farr CL, Euro L, Suomalainen A, Kaguni LS. Functional analysis of H. sapiens DNA polymerase gamma spacer mutation W748S with and without common variant E1143G. Biochim Biophys Acta. 2010;1802:545–51. doi: 10.1016/j.bbadis.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uusimaa J, Hinttala R, Rantala H, Paivarinta M, Herva R, Roytta M, Soini H, Moilanen JS, Remes AM, Hassinen IE, et al. Homozygous W748S mutation in the POLG1 gene in patients with juvenile-onset Alpers syndrome and status epilepticus. Epilepsia. 2008;49:1038–45. doi: 10.1111/j.1528-1167.2008.01544.x. [DOI] [PubMed] [Google Scholar]
- 46.Hakonen AH, Heiskanen S, Juvonen V, Lappalainen I, Luoma PT, Rantamaki M, Goethem GV, Lofgren A, Hackman P, Paetau A, et al. Mitochondrial DNA polymerase W748S mutation: a common cause of autosomal recessive ataxia with ancient European origin. Am J Hum Genet. 2005;77:430–41. doi: 10.1086/444548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winterthun S, Ferrari G, He L, Taylor RW, Zeviani M, Turnbull DM, Engelsen BA, Moen G, Bindoff LA. Autosomal recessive mitochondrial ataxic syndrome due to mitochondrial polymerase gamma mutations. Neurology. 2005;64:1204–8. doi: 10.1212/01.WNL.0000156516.77696.5A. [DOI] [PubMed] [Google Scholar]
- 48.Van Goethem G, Luoma P, Rantamaki M, Al Memar A, Kaakkola S, Hackman P, Krahe R, Lofgren A, Martin JJ, De Jonghe P, et al. POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology. 2004;63:1251–7. doi: 10.1212/01.wnl.0000140494.58732.83. [DOI] [PubMed] [Google Scholar]
- 49.Milone M, Benarroch EE, Wong LJ. POLG-related disorders: Defects of the nuclear and mitochondrial genome interaction. Neurology. 2011;77:1847–52. doi: 10.1212/WNL.0b013e318238863a. [DOI] [PubMed] [Google Scholar]
- 50.Blok MJ, van den Bosch BJ, Jongen E, Hendrickx A, de Die-Smulders CE, Hoogendijk JE, Brusse E, de Visser M, Poll-The BT, Bierau J, et al. The unfolding clinical spectrum of POLG mutations. J Med Genet. 2009;46:776–85. doi: 10.1136/jmg.2009.067686. [DOI] [PubMed] [Google Scholar]
- 51.Stewart JD, Horvath R, Baruffini E, Ferrero I, Bulst S, Watkins PB, Fontana RJ, Day CP, Chinnery PF. Polymerase gamma gene POLG determines the risk of sodium valproate-induced liver toxicity. Hepatology. 2010;52:1791–6. doi: 10.1002/hep.23891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stumpf JD, Bailey CM, Spell D, Stillwagon M, Anderson KS, Copeland W,C. mip1 containing mutations associated with mitochondrial disease causes mutagenesis and depletion of mtDNA in Saccharomyces cerevisiae. Hum Mol Genet. 2010;11:2123–33. doi: 10.1093/hmg/ddq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee YS, Demeler B, Molineux IJ, Johnson KA, Yin YW. Each monomer of the dimeric accessory protein for human mitochondrial DNA polymerase has a distinct role in conferring processivity. J Biol Chem. 2010;285:1490–9. doi: 10.1074/jbc.M109.062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.