Abstract
DNA in human cells is constantly assaulted by endogenous and exogenous DNA damaging agents. It is vital for the cell to respond rapidly and precisely to DNA damage to maintain genome integrity and reduce the risk of mutagenesis. Sophisticated reactions occur in chromatin surrounding the damaged site leading to the activation of DNA damage response (DDR), including transcription reprogramming, cell cycle checkpoint, and DNA repair. Histone proteins around the DNA damage play essential roles in DDR, through extensive post-translational modifications (PTMs) by a variety of modifying enzymes. One PTM on histones, mono-ubiquitylation, has emerged as a key player in cellular response to DNA damage. In this review, we will (1) briefly summarize the history of histone H2A and H2B ubiquitylation (H2Aub and H2Bub, respectively), (2) discuss their roles in transcription, and (3) their functions in DDR.
Keywords: UV damage, RNA Pol II stalling, Deubiquitylation, Chromatin remodeling
1. Prologue
As a part of Mick Smerdon’s “DNA Repair Shop” group, Peng (a former postdoctoral fellow) and I (Rithy, his last graduate student) would like to thank Mick for his guidance and for allowing us to work for him. It has been easy to discuss our projects with Mick, and his breadth of knowledge on DNA repair has allowed us to gain new perspectives on our projects. Our conversations with Mick will not just give us insight on DNA repair but also life. Sometimes a “quick” 5-min conversation in his office may turn into one hour plus tangent on the ETH Zürich University or his makeshift fluorometer, but it will eventually relate back to the original topic of nucleosomes and excision repair. Mick has been a supportive mentor and an invaluable teacher; therefore, we would like to dedicate this review to Mick Smerdon.
2. Introduction
At the primary level of packaging, ~147 base pairs of genomic DNA in eukaryotic cells is wrapped around an octamer of the 4 core histone proteins—H2A, H2B, H3 and H4—to form the nucleosome core particle [1]. Nucleosomes are further packaged into higher-order chromatin structure to compact the genomic DNA into a nucleus. Although DNA packaging is essential for cell survival, chromatin is innately inhibitory to DNA-templated processes, such as transcription, DNA replication, and DNA repair, by restricting the DNA surface area that proteins can bind. Nevertheless, it is essential for DNA-binding proteins to localize to their target DNA sites in response to cellular stimuli. In the case of nucleotide excision repair (NER), which requires at least 30 proteins to repair helix-distorting DNA lesions such as ultraviolet (UV)-induced photolesions [2], repair is greatly inhibited by the presence of nucleosomes, reviewed in Gong et al. [3]. Therefore, cells have created a robust chromatin remodeling system to dynamically alter the chromatin landscape for access of NER factors to lesion sites.
Cells utilize several different strategies to regulate DNA accessibility in chromatin. Various post-translational modifications (PTMs), such as methylation, acetylation, ubiquitylation, and phosphorylation on histones, have emerged as major regulators of chromatin structure, reviewed in Suganuma and Workman, and Zentner and Henikoff [4,5]. Particularly, some histone modifying enzymes are able to respond to DNA damage and mark the damaged sites by inducing specific modifications on histones, creating binding sites for the downstream factors. In this regard, the best characterized histone modification so far is histone H2AX phosphorylation (γ-H2AX) [6]. γ-H2AX can be strongly induced by double-strand breaks (DSBs) and DNA replication stress, and functions as an important signal for initiating the downstream chromatin-associated events, reviewed in Lukas et al. [7].
Histone ubiquitylation, particularly on H2A and H2B, has been documented in previous studies, reviewed in Weake and Workman [8]. H2A and H2B are mainly mono-ubiquitylated, which does not lead to their proteasomal degradation. Instead, attachment of a single ubiquitin moiety significantly changes the histone mass and affects nucleosomal dynamics. The ubiquitin mark on H2B can also stimulate H3 K4 and K79 tri-methylation, thus coordinating ‘cross-talks’ between histone modifications, reviewed in Weake and Workman [8]. We will discuss the functions of H2Aub in the first section and H2Bub in the second section.
3. H2A ubiquitylation and its role in transcription and DDR
Histone H2A is the first protein that was identified to be ubiquitylated, reviewed in Varshavsky [9]. Ubiquitylated H2A was originally detected with 2-dimensional gel electrophoresis and considered a ‘histone-like’ non-histone chromosomal protein [10]. The major ubiquitylation site on H2A was mapped to lysine residue 119 (K119) [11,12]. H2AK119 ubiquitylation occurs in many but not all higher eukaryotes, and ~5–15% of total H2A protein is ubiquitylated in these cells. However, H2Aub has not been detected in the budding yeast Saccharomyces cerevisiae [13].
3.1. H2A ubiquitylation enzymes
Protein ubiquitylation is an enzymatic process requiring the sequential activities of ubiquitin activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3), reviewed in Hochstrasser [14]. Even though H2Aub was discovered in the 1970s, the E3 ligase for this modification was not identified until 2004 [15]. Wang et al. developed an elegant system to fractionate HeLa nuclear proteins and monitor H2A ubiquitylation enzymatic activity in each fraction. With this technique, they found that the human Polycomb repressive complex 1(PRC1)-like is responsible for H2A ubiquitylation at K119 [15]. Three subunits (Ring1, Ring2, and Bmi-1) in this complex contain RING (Really Interesting New Genes) finger domains and can potentially function as E3 ubiquitin ligases. However, only Ring2 (Ring1B/Rnf2) is able to ubiquitylate H2A in vitro, suggesting Ring2 is the E3 ligase for H2AK119. Indeed, in vivo knockdown of Ring2 expression significantly reduces H2Aub in Drosophila [15]. Even though the other two RING domain-containing subunits, Ring1 and Bim1, are unable to ubiquitylate H2AK119 directly, their presence greatly stimulates the ligase activity of Ring2 for H2A [15,16].
In addition to Ring2, Zhou et al. have identified 2A-HUB as another E3 ligase for H2AK119 [17]. 2A-HUB is also a RING finger protein, and is able to ubiquitylate H2AK119 in vitro [17]. Overexpression of 2A-HUB gene in HEK293T cells leads to increased H2Aub levels [17]. However, Zhou et al. did not report the effect of 2A-HUB knockdown on H2Aub, leaving an unaddressed question as to how much Ring2 and 2A-HUB each contributes to H2AK119 ubiquitylation.
K119 is not the only ubiquitylation site on H2A, as recent studies have demonstrated that K13 and K15 of H2A can also be ubiquitylated [18,19]. These two residues are located on the N-terminal tail of H2A protruding out from the nucleosome core and are far from K119 in the nucleosome core particle. Additionally, the ubiquitylation at K13 and K15 is not catalyzed by Ring2 or 2A-HUB, and ubiquitylation of these residues is independent of the canonical ubiquitylation at K119. Instead, the E3 ligase RNF168 (RING finger protein 168) is responsible for ubiquitylating H2A at K13 and 15 in response to DSBs [18,19].
3.2. H2A ubiquitylation at K119 represses transcription
Both E3 ligases for H2AK119, Ring2 and 2A-HUB, are associated with gene repression, suggesting H2AK119 ubiquitylation plays a role in transcription repression. Ring2 has been shown to associate with several important repressive complexes such as PRC1, reviewed in Weake and Workman [8], and 2A-HUB is associated with the histone deacetylase complex N-CoR/HDAC1/3 [17]. Indeed, compelling evidence indicates that H2AK119 ubiquitylation participates in Polycomb silencing [15], X chromosome inactivation [20], and repression of a subset of chemokine genes [17].
In terms of the mechanisms by which H2AK119 ubiquitylation represses gene expression, Zhou et al. have shown the inhibition of RNA Pol II elongation by H2Aub [17]. Specifically, H2Aub is able to prevent the recruitment of the histone chaperone FACT (facilitate chromatin transcription) to the promoters of the repressed genes [17]. Additionally, H2Aub enhances the binding of linker histone H1 to nucleosomes in vitro [21]. The influence of H2Aub on H1 binding is also supported by in vivo studies showing that H2A deubiquitylation by 2A-DUB (H2AK119 deubiquitylase) leads to dissociation of H1 from nucleosomes in human cells [22]. This implies that H2Aub may enhance higher-order chromatin compaction through helping H1 binding. Another mechanism revealed by a recent study indicates that H2Aub represses gene expression via trans-histone crosstalk with H3K27 trimethylation [23], a histone mark linked to gene repression [24]. Specifically, H2Aub in nucleosomes functions as a binding site to recruit Polycomb repressive complex 2 (PRC2), which contains the methyltransferase that tri-methylates H3K27 [23].
3.3. H2AK13/15 ubiquitylation is important for DSB signaling
As mentioned earlier, K13 and K15 on the N-terminal tail of H2A can also be ubiquitylated. The main function of H2AK13/15ub appears to be a signal mark during DDR, particularly to DSBs.
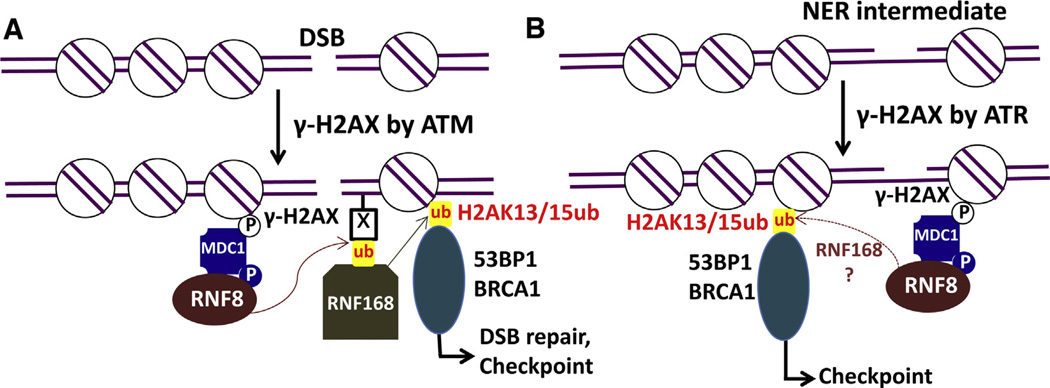
DSBs trigger a cascade of modifications in chromatin surrounding the damaged site, signaling for the recruitment of a variety of downstream factors to activate cell cycle checkpoints and repair. One of the earliest modification events activated by DSBs is H2AX phosphorylation (γ-H2AX) [6], primarily by the kinase ataxia telangiectasia mutated (ATM). Phosphorylated H2AX is directly recognized by MDC1 (mediator of DNA damage checkpoint protein 1) [25], which functions as the scaffold protein at DSBs and is also phosphorylated by ATM. Phospho-MDC1 serves as a binding site for the E3 ligase RNF8 [26,27], which contains a forkhead-associated (FHA) domain and a RING finger domain. The FHA domain of RNF8 is responsible for the recognition of phospho-MDC1, and the RING finger domain is required for its E3 ligase activity [26,27]. Mutation of either the FHA or RING finger domain of RNF8 disrupts the subsequent recruitment of BRCA1 and 53BP1 [26,27], two essential proteins in DSB repair. Although the ubiquitylation activity of RNF8 is important for DSB repair, it remains elusive what proteins are ubiquitylated by RNF8. In vitro studies indicate that both H2A and H2AX can be ubiquitylated by RNF8 [27], and knockdown of RNF8 impairs DSBs-induced H2A and H2AX ubiquitylation in mammalian cells [26,27]. However, a study using nucleosomal H2A as the substrate shows that RNF8 is unable to ubiquitylate H2A in the context of nucleosomes [19]. Instead, the E3 ligase RNF168 is responsible for ubiquitylating H2A in nucleosomes [19], suggesting RNF168 is the direct E3 ligase targeting H2A in chromatin. The involvement of RNF168 in DDR was originally found by Doil et al. via high-throughput RNAi screening for genes required for 53BP1 recruitment to damaged chromosome [28]. The 53BP1 protein is an important regulator in DSB repair that stimulates the end-joining of distal DNA ends. Knockdown of RNF168 strikingly inhibits the accumulation of 53BP1 at DNA breaks [28]. The connection between RNF8 and RNF168 in DDR was further analyzed and it has been shown that the recruitment of RNF168 to DNA damage is dependent on RNF8 [28]. Moreover, RNF168 possesses multiple ubiquitin-binding domains (UBDs), which enable RNF168 to bind to ubiquitylated proteins [29,30]. Collectively, these observations suggest that RNF8 localizes to DSBs prior to RNF168. The ubiquitylation products catalyzed by RNF8 are recognized by RNF168, presumably via its UBDs, leading to the recruitment of RNF168 to DNA breaks (Fig. 1A).
Fig. 1. Model for the induction and function of H2Aub in DDR.
(A) Induction of H2Aub by DSBs. γ-H2AX induced by DSBs recruits MDC1 to the damaged chromatin, where MDC1 is phosphorylated by ATM. Phosphorylated MDC1 is bound by E3 ligase RNF8 to ubiquitylate its substrates. The ubiquitylated products by RNF8 serve as the binding sites for another E3 ligase RNF168, which directly ubiquitylates H2A and H2AX at K13 and K15. RNF168-ubiquitylated H2A is a mark recognized by 53BP1. Through H2Aub and H4K20me2, 53BP1 is recruited to regulate DSB repair and checkpoint. Recruitment of BRCA1 is also facilitated by H2Aub.
(B) Induction of H2Aub by UV photolesions. The ssDNA intermediates accumulated during NER activates γ-H2AX, and γ-H2AX initiates the signaling leading to H2Aub by RNF8. Whether RNF168 is required remains elusive. UV-induced H2Aub facilitates the subsequent recruitment of 53BP1 and BRCA1 to regulate cell cycle arrest.
How does RNF8/ RNF168-mediated H2A ubiquitylation regulate DDR? A recent study demonstrates that H2AK15ub functions as a histone mark that can be directly recognized by 53BP1 [31]. Interestingly, 53BP1 appears to play a unique role in DDR by acting as a histone PTM reader, as it also binds to dimethylation on histone H4K20 (H4K20me2) [32], another DSB-inducible histone mark [33]. Moreover, recognition of H2AK15ub by 53BP1 in nucleosomes requires the presence of H4K20me2, and vice versa [31], suggesting that H2AK15ub functions cooperatively with H4K20me2 to promote 53BP1 recruitment to damaged chromatin.
3.4. H2A ubiquitylation is induced by UV DNA damage
In addition to functioning in DDR to DSBs, H2Aub is also involved in cellular response to UV photolesions. Different from DSBs, DNA damage induced by UV light is mainly helix-distorting pyrimidine dimers. The involvement of H2Aub in UV damage response was first revealed by the finding that UV irradiation induces H2Aub [34]. This induction is dependent on the functional NER pathway and occurs after incision of the damaged strand. In addition, the induced H2A ubiquitylation appears to occur at H2AK119, as the E3 ligase Ring2 is required for this induction [34]. However, another study suggests that H2Aub is first repressed by UV light and subsequently recovered, and its recovery requires the DDB-CUL4 E3 complex [35]. DDB-CUL4 is comprised of UV-damaged DNA binding protein 1 (DDB1), DDB2, and CUL4 [35]. These studies suggest that at least two distinct E3 ubiquitin ligases are able to ubiquitylate H2A in response to UV DNA damage—Ring2 and the DDB-CUL4 complex. But it remains unknown which residue(s) on H2A are ubiquitylated by DDB-CUL4 upon UV irradiation. Intriguingly, histone H3 and H4 are also ubiquitylated by DDB-CUL4 in response to UV damage, and H3 and H4 ubiquitylation destabilizes nucleosomes to allow for the recruitment of NER proteins [36].
A more recent study suggests that H2Aub is important for UV damage signaling by facilitating the recruitment of BRCA1 and 53BP1 to UV lesions [37]. This study demonstrates that UV-induced H2Aub is dependent on the E3 ligase RNF8 and the scaffold protein MDC1 [37]. Whether or not RNF168 is involved in this process remains unknown. A likely mechanism for the induction of H2A ubiquitylation in UV-damaged cells is that the single-strand DNA generated during the NER process acts as the signal to activate γ-H2AX via the kinase ataxia telangiectasia and Rad3-related protein (ATR). The subsequent steps are similar to the DSB pathway, leading to H2A ubiquitylation (Fig. 1B).
Thus, a generic signaling pathway for DNA damage-responsive H2A ubiquitylation appears to exist in mammalian cells. This pathway is initiated by γ-H2AX, catalyzed by ATM (for DSBs) or ATR (for UV lesions). γ-H2AX functions as the essential mark at the DNA damage sites to recruit MDC1, and phosphorylated MDC1 helps the recruitment of RNF8. The ubiquitylation activity of RNF8 is required for the recruitment of RNF168 to directly ubiquitylate H2A and H2AX upon DSBs and possibly other DNA damage such as UV lesions. The ubiquitylated H2A at K15, together with another epigenetic mark, H4K20me2, direct the binding of 53BP1 to damaged chromatin to signal for DNA repair and cell cycle checkpoints.
4. H2B ubiquitylation and its role in transcription and DDR
In 1980, the Bonner’s group found that human and mouse cells ubiquitylate 1–1.5% of their total H2B [38]. For 20 years, it was unknown which enzymes promote the addition of ubiquitin onto H2B. It was not until 2000 that the Osley’s group identified the E2 ubiquitin-conjugating enzyme for H2B (Rad6 in budding yeast) on lysine residue 123 (K123) [13]. Three years later, two different groups identified the E3 ligase Bre1 in S. cerevisiae [39,40]. The identification of the yeast E3 ligase led the Reinberg’s lab to discover the human homologues (RNF20 and RNF40), which perform the same function as Bre1 in ubiquitylating H2B on an analogous site as yeast H2BK123 (K120 in higher eukaryotes) [41]. Their study further showed that RNF20/40 and the E2 conjugating UbcH6 are sufficient for ubiquitylation of nucleosomal H2B in vitro.
H2B lysine residue 123 (120 in humans) is not the only site on H2B that is ubiquitylated. A large-scale protein posttranslational modification study using adult mouse brains identified K34, K46, K108, K116, and K120 as ubiquitylated sites on H2B [42]. Using a mutant His-tagged ubiquitin (G76A) that prevents cleavage by ubiquitin proteases in yeast, the Tansey’s group identified a number of lysine residues on H2B that are ubiquitylated independent of Rad6 and Bre1 [43]. However, since these alternative H2Bub sites have not been well studied, the subsequent paragraphs will focus on H2B ubiquitylation on residue K123 (K120 in humans).
4.1. H2Bub regulates chromatin dynamics
Ubiquitin (~8 kDa) is a large moiety that is over a third the weight of an H2Bub (~22 kDa) molecule. The H2B ubiquitylation site is on the C-terminal α-helix tail on the face of the nucleosome. Since the ubiquitin moiety is a large PTM situated on the nucleosome face, it was suspected that oligo-nucleosome compaction might be inhibited by affecting inter-nucleosomal interactions. To test this, Chatterjee et al. developed a disulfide-directed method to ligate ubiquitin to a specific site on H2B and successfully reconstituted mono- and oligo-nucleosomes containing homogeneously ubiquitylated H2B [44,45]. Using this strategy, H2Bub was shown to disrupt higher-order chromatin compaction [45] but only shows marginal impact on nucleosome stability in vitro [46]. A caveat to these in vitro assays, though, is that both H2B molecules per nucleosome are mono-ubiquitylated, which is not known to occur in vivo. Instead, in vivo studies indicate that H2Bub stabilizes nucleosomes [47] and facilitates nucleosome reassembly in the wake of transcription elongation [48,49]. These observations suggest that H2Bub plays a complex role in modulating nucleosome and chromatin structure. While this modification disrupts internucleosomal interactions, hence inhibiting higher-order chromatin compaction, it improves intra-nucleosomal interactions in the cell, possibly with the help of other cellular components such as the histone chaperone FACT [49].
4.2. H2Bub-H3 methylation crosstalk modulates DNA damage-induced cell cycle arrest
Previous studies have demonstrated that H2Bub promotes diand trimethylation of H3K4 and H3K79 [50–52]. The H2Bub-H3 methylation (H3me) trans-histone crosstalk appears to be conserved in eukaryotes, as H2Bub stimulates H3K4 and H3K79 di- and trimethylation in both budding yeast and human cells [53,54], possibly through an H2Bub-methyltransferase bridge (Set1-COMPASS/Dot1) [55]. However, ubiquitylation does not have to be on H2BK123 (or K120 in humans) or even on H2B itself to propagate methylated H3 in vivo and in vitro [44,56], indicating a striking plasticity in H2Bub-mediated H3 methylation. Furthermore, deletion of the H2B deubiquitylases Ubp8 and Ubp10 was shown to colocalize H2Bub to genomic sites enriched for trimethylated H3K4 (H3 K4me3) and H3K79me3, respectively [57]. This suggests that Ubp8 and Ubp10 may target different cellular pools of H2Bub.
Rad6, the E2-conjugating enzyme for H2Bub, is involved in multiple DNA damage signaling pathways; therefore, it is conceivable that H2Bub may play a role in cellular response to DNA damaging agents. Indeed, when ubiquitylation of H2BK123 is absent in budding yeast, DNA damage checkpoint response is compromised by inhibiting the phosphorylation of Rad9 [58]. Yeast Rad9 (a homolog of human 53BP1) is an adaptor protein that amplifies the initial DNA damage signals from Mec1 and Tel1 (homologs of human ATR and ATM, respectively) [59,60]. Interestingly, a defect in checkpoint signaling was also seen when the H3K79 methyltransferase Dot1 was absent, indicating the importance of the H2Bub-H3K79 crosstalk for proper DNA damage checkpoint signaling in yeast [58]. Epistasis studies using ionizing radiation, which induces DSBs, shows that BRE1 and DOT1 are in the RAD9 epistasis group [61]. Collectively, these data describe the interplay between H2B ubiquitylation and H3 methylation to facilitate DNA damage-induced cell cycle arrest.
4.3. The role of H2Bub in DSB repair and UV lesion repair
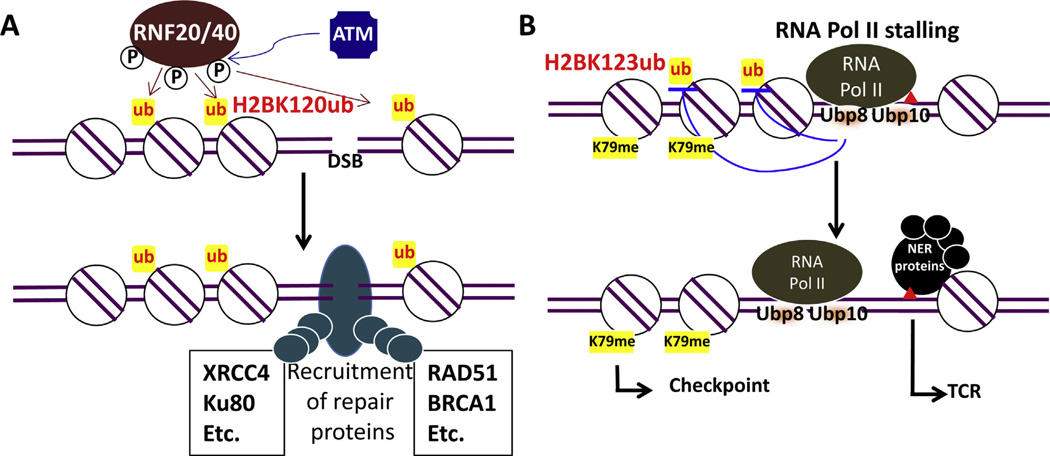
The precise function of H2Bub in DNA repair pathways is not fully understood. However, by analyzing yeast cell sensitivity to ionizing radiation it was found that H2Bub was implicated in DSB repair, and that yeast BRE1 functions in the same epistasis group as RAD51 [61], in addition to RAD9 (see above). Yeast RAD51 is important for homologous recombination (HR) in DSB repair. Similar to yeast, the human RNF20-dependent ubiquitylation of H2B is also required for HR, and H2Bub was shown to recruit BRCA1 and RAD51 to facilitate DNA end resection for strand invasion [62–64]. In addition to HR, H2Bub facilitates the recruitment of non-homologous end joining (NHEJ) repair factors to DSBs [64], suggesting that H2Bub regulates both DSB repair pathways (Fig. 2A). Furthermore, the human H2Bub E3 ligases RNF20 and RNF40 physically interact with ATM, and DSBs stimulate phosphorylation of RNF20 and RNF40 to ubiquitylate H2B [64].
Fig. 2. Functions of H2Bub in DDR. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
(A) A role for H2Bub in DDR to DSBs in mammalian cells. In response to DSBs, ATM phosphorylates E3 ligase complex RNF20/40. Phosphorylated RNF20/40 ubiquitylates H2B at DSBs. H2Bub acts as a signal to recruit downstream DSB repair factors to facilitate repair.
(B) Dual role for H2Bub in DDR to UV photoplesions in budding yeast. RNA Pol II is stalled at a UV-induced lesion (red triangle). H2Bub-dependent H3K79me2 is an important histone mark that can be recognized by yeast DDR protein Rad9, which subsequently promotes Rad53 phosphorylation and cell cycle checkpoint in response to UV irradiation. On the other hand, excessive H2Bub is inhibitory to the rescue of UV-arrested RNA Pol II. In response to UV damage-stalled RNA Pol II, deubiquitylases Ubp8 and Ubp10 are activated and remove ubiquitin from H2B, resulting in nucleosome disassembly and the recruitment of TCR factors.
H2Bub and its dependent H3K79me also play a role in NER. It was found that an H3K79 substitution mutant has a defect in NER at the transcriptionally silent HML locus, which was most likely due to enhanced binding of the silent information regulator (Sir) complex to the HML locus [65]. Analysis on the non-transcribed strand of the constitutively active RPB2 locus showed that repair was deficient in cells that were unable to methylate H3K79, indicating deficiency in the sub-pathway called global genomic repair (GGR) [66]. This study also found a defect in GGR in cells that could not ubiquitylate H2B, indicating H2Bub as well as H2Bub-dependent H3K79me is required for efficient GGR in yeast.
4.4. H2B deubiquitylation responds to RNA polymerase II stalling
In contrast to repair, there have been many studies that focused on the contributions of H2Bub to transcription. During transcription, the Rad6-Bre1 complex is associated with elongating RNA polymerase II (RNA Pol II) to ubiquitylate H2B, reviewed in Weake and Workman [8]. In budding yeast, this is supported by a genome-wide study showing that H2Bub is predominantly located in the coding regions of actively RNA Pol II-transcribed genes [57]. Moreover, using an in vitro reconstituted chromatin transcription system, it was shown that the mammalian histone chaperone FACT recruits the transcription elongation complex PAF, which then recruits ubiquitylation enzymes to ubiquitylate H2B [67]. Thereafter, H2Bub stimulates FACT activity to allow H2A/H2B dimer displacement, permitting RNA Pol II to continue transcription through nucleosomal DNA [67].
The RNA Pol II stalls when there are DNA lesions (e.g., UV photolesions) along the transcribed DNA strand, but cells employ the transcription-coupled repair (TCR) pathway to rescue DNA damage-arrested RNA Pol II [68]. Since H2Bub is tightly associated with transcription elongation, DNA damage-induced RNA Pol II stalling may have an impact on this modification. This hypothesis was tested by our recent study, where we showed that UV radiation caused a rapid and significant decrease in H2Bub in both budding yeast and human fibroblast cells [69]. The decrease in H2Bub was triggered by UV-induced RNA Pol II stalling and was dependent on the H2B-specific deubiquitylases Ubp8 and Ubp10. Moreover, H2B deubiquitylation was important for the rescue of RNA Pol II by TCR, as shown by the impaired TCR of UV-induced lesions in the absence of both Ubp8 and Ubp10 proteins. Furthermore, reduced TCR in a ubp8Δubp10Δ mutant occurred concomitantly with an increase in nucleosome occupancy, indicating that deubiquitylation of H2B is important for destabilizing nucleosomes near stalled RNA Pol II to allow efficient TCR [69]. Therefore, in addition to the role of H2Bub in transcription elongation, deubiquitylation of H2B is associated with transcription stalling and rescue of arrested RNA Pol II (Fig. 2B).
5. Concluding remarks
During the past decade, we have started to understand the important roles of H2Aub and H2Bub in DDR, particularly in response to DSBs and UV photolesions. Even though H2Aub and H2Bub function quite differently in transcription regulation, they both participate in DNA damage signaling by facilitating the recruitment of DDR factors to damaged chromatin. This fits with the general observation that a cascade of protein ubiquitylation occurs in chromatin surrounding DNA damage to recruit downstream DDR proteins.
It is important to note, however, that the repressive effect of H2Aub on transcription is caused by the main ubiquitylation site in its C-terminus, whereas DNA damage-induced ubiquitylation occurs on the N-terminal tail of H2A. Whether or not the N-terminal ubiquitylation of H2A impacts transcription remains unknown. On the other hand, although H2Bub is important for cell cycle checkpoints in response to DNA damage, high level of H2Bub is inhibitory to TCR [69]. This suggests the existence of a precisely controlled mechanism in the cell to fine-tune H2Bub levels at the RNA Pol II stalling sites to coordinate RNA Pol II rescue and cell cycle arrest.
One common theme in histone modifications is the complex ‘crosstalks’ among different PTMs, such as the crosstalk between H2Bub and H3K4me/H3K79me, reviewed in Weake and Workman [8]. Could H2Aub and H2Bub crosstalk with other histone PTMs during DDR? It seems possible that H2AK15ub might interact with H4K20me2 to promote 53BP1 recruitment, since the efficient binding of 53BP1 to nucleosomes require both epigenetic marks [31]. Furthermore, H2B deubiquitylase Ubp8 and H3 acetyltransferase Gcn5 co-exist in the SAGA (Spt-Ada-Gcn5 acetyltransferase) complex [70]. As both Ubp8-dependent H2B deubiquitylation and Gcn5-dependent acetylation are responsive to UV photolesions and promote repair [69,71], it is compelling that these two modifications may interact in NER. More studies will be required to further address the potential interactions between H2Aub, H2Bub and other PTMs for coordinated cellular signaling.
Acknowledgements
We thank John Wyrick and Amelia Hodges for critically reading this manuscript. Work for this review was funded by grants ES002614 (to J. Wyrick and M. Smerdon) and ES004106 (to M. Smerdon) from the National Institute of Environmental Health Sciences (NIEHS).
Footnotes
Conflict of interest
None.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2. 8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 3.Gong F, Kwon Y, Smerdon MJ. Nucleotide excision repair in chromatin and the right of entry. DNA Repair. 2005;4:884–896. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 5.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013;20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 6.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 7.Lukas J, Lukas C, Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 8.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Varshavsky A. The early history of the ubiquitin field. Protein Sci. 2006;15:647–654. doi: 10.1110/ps.052012306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldknopf IL, Taylor CW, Baum RM, Yeoman LC, Olson MO, Prestayko AW, Busch H. Isolation and characterization of protein A24, a histone-like non-histone chromosomal protein. J. Biol. Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- 11.Goldknopf IL, Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc. Natl. Acad. Sci. U. S. A. 1977;74:864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nickel BE, Davie JR. Structure of polyubiquitinated histone H2A. Biochemistry. 1989;28:964–968. doi: 10.1021/bi00429a007. [DOI] [PubMed] [Google Scholar]
- 13.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 14.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 16.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol. Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gatti M, Pinato S, Maspero E, Soffientini P, Polo S, Penengo L. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle. 2012;11:2538–2544. doi: 10.4161/cc.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J. Biol. Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- 21.Jason LJ, Finn RM, Lindsey G, Ausio J. Histone H2A ubiquitination does not preclude histone H1 binding, but it facilitates its association with the nucleosome. J. Biol. Chem. 2005;280:4975–4982. doi: 10.1074/jbc.M410203200. [DOI] [PubMed] [Google Scholar]
- 22.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Muller J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014;21:569–571. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 24.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 26.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 28.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 29.Pinato S, Scandiuzzi C, Arnaudo N, Citterio E, Gaudino G, Penengo L. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol. Biol. 2009;10:55. doi: 10.1186/1471-2199-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinato S, Gatti M, Scandiuzzi C, Confalonieri S, Penengo L. UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol. Cell. Biol. 2011;31:118–126. doi: 10.1128/MCB.00818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, Durocher D. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, Hoeijmakers JH, Vermeulen W, Dantuma NP. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20:1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 37.Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, Vermeulen W. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 2009;186:835–847. doi: 10.1083/jcb.200902150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.West MH, Bonner WM. Histone 2B can be modified by the attachment of ubiquitin. Nucl. Acids Res. 1980;8:4671–4680. doi: 10.1093/nar/8.20.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 40.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 41.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 42.Tweedie-Cullen RY, Reck JM, Mansuy IM. Comprehensive mapping of post-translational modifications on synaptic, nuclear, and histone proteins in the adult mouse brain. J. Proteome Res. 2009;8:4966–4982. doi: 10.1021/pr9003739. [DOI] [PubMed] [Google Scholar]
- 43.Geng F, Tansey WP. Polyubiquitylation of histone H2B. Mol. Biol. Cell. 2008;19:3616–3624. doi: 10.1091/mbc.E08-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 45.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fierz B, Kilic S, Hieb AR, Luger K, Muir TW. Stability of nucleosomes containing homogenously ubiquitylated H2A and H2B prepared using semisynthesis. J. Am. Chem. Soc. 2012;134:19548–19551. doi: 10.1021/ja308908p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 50.Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 51.Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 52.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 53.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi YH, Chandrasekharan MB, Sun ZW, Osley MA, Strahl BD, Jaspersen SL, Shilatifard A. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J. Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chandrasekharan MB, Huang F, Sun ZW. Histone H2B ubiquitination and beyond: regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics. 2010;5:460–468. doi: 10.4161/epi.5.6.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vlaming H, van Welsem T, de Graaf EL, Ontoso D, Altelaar AF, San-Segundo PA, Heck AJ, van Leeuwen F. Flexibility in crosstalk between H2B ubiquitination and H3 methylation in vivo. EMBO Rep. 2014;15:1077–1084. doi: 10.15252/embr.201438793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 2011;25:2242–2247. doi: 10.1101/gad.177220.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 59.Vialard JE, Gilbert CS, Green CM, Lowndes NF. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- 61.Game JC, Williamson MS, Spicakova T, Brown JM. The RAD6/BRE1 histone modification pathway in Saccharomyces confers radiation resistance through a RAD51-dependent process that is independent of RAD18. Genetics. 2006;173:1951–1968. doi: 10.1534/genetics.106.057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, Zou L, Komatsu K. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 63.Kari V, Shchebet A, Neumann H, Johnsen SA. The H2B ubiquitin ligase RNF40 cooperates with SUPT16H to induce dynamic changes in chromatin structure during DNA double-strand break repair. Cell Cycle. 2011;10:3495–3504. doi: 10.4161/cc.10.20.17769. [DOI] [PubMed] [Google Scholar]
- 64.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, Shkedy D, Smorodinsky NI, van Vliet N, Kuster B, Mann M, Ciechanover A, Dahm-Daphi J, Kanaar R, Hu MC, Chen DJ, Oren M, Shiloh Y. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaudhuri S, Wyrick JJ, Smerdon MJ. Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae. Nucl. Acids Res. 2009;37:1690–1700. doi: 10.1093/nar/gkp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tatum D, Li S. Evidence that the histone methyltransferase Dot1 mediates global genomic repair by methylating histone H3 on lysine 79. J. Biol. Chem. 2011;286:17530–17535. doi: 10.1074/jbc.M111.241570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 68.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 69.Mao P, Meas R, Dorgan KM, Smerdon MJ. UV damage-induced RNA polymerase II stalling stimulates H2B deubiquitylation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12811–12816. doi: 10.1073/pnas.1403901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]