SUMMARY
Complex genomic rearrangements (CGRs) are a hallmark of many human diseases. Recently, CGRs were suggested to result from microhomology-mediated break-induced replication (MMBIR), a replicative mechanism involving template switching at positions of microhomology. Currently, the cause of MMBIR and the proteins mediating this process remains unknown. Here, we demonstrate in yeast, that a collapse of homology-driven break-induced replication (BIR) caused by defective repair DNA synthesis in the absence of Pif1 helicase leads to template-switches involving 0–6 nucleotides of homology, followed by resolution of recombination intermediates into chromosomal rearrangements. Importantly, we show that these microhomology-mediated template-switches, indicative of MMBIR, are driven by translesion synthesis (TLS) polymerases Polζ and Rev1. Thus, an interruption of BIR involving fully homologous chromosomes in yeast triggers a switch to MMBIR catalyzed by TLS polymerases. Overall, our study provides important mechanistic insights into the initiation of MMBIR associated with genomic rearrangements, similar to those promoting diseases in humans.
INTRODUCTION
Genomic rearrangements underlie a host of human diseases. In many cases, rearrangements can be so complex that currently proposed models cannot explain their formation. In particular, recent studies have discovered cases of complex genomic rearrangements (CGRs) in cancer and other diseases known as chromothripsis, which is characterized by a massive number of chromosomal rearrangements that are typically localized to a single chromosome (Berger et al., 2011; Stephens et al., 2011; Kloosterman et al., 2011b; Kloosterman et al., 2012; Malhotra et al., 2013; Molenaar et al., 2012; Zack et al., 2013). Typically, the copy number of chromosomal areas involved in chromothripsis is either not changed, or is reduced due to deletions. It is unknown how chromothripsis occurs, but it was proposed to result from the shattering of a chromosome and its subsequent reconstitution via non-homologous end joining (NHEJ).
Another class of CGRs has been found in patients with various congenital disorders and was named chromoanasynthesis (Liu et al., 2011a; Carvalho et al., 2011; Carvalho et al., 2013; Beck et al., 2015; Carvalho et al., 2015). The distinguishing feature of these events is the combination of chromosomal rearrangements with copy number gains. It was proposed that these CGRs form through a replicative process where DNA synthesis undergoes frequent template switches leading to chromosomal rearrangements. The most direct support for a replication-based process has recently come from studies of the neurological disorders Pelizaeus-Merzbacher disease (PMD) and MECP2 duplication syndrome, which result from CNVs at different loci on the X chromosome (Lee et al., 2007; Carvalho et al., 2013; Carvalho et al., 2011; Beck et al., 2015). Specifically, it was observed that the regions with CNVs in these diseases were not comprised of simple tandem duplications that could be explained by non-allelic homologous recombination or by NHEJ; rather, the CNVs consisted of DNA regions containing interspersed segments that were duplicated, triplicated, and quadruplicated, with many containing microhomologies at their junctions. This pattern was explained by an unusual type of DNA synthesis called microhomology-mediated break-induced replication (MMBIR) (Hastings et al., 2009a; Payen et al., 2008). According to current models (Hastings et al., 2009a; Hastings et al., 2009b), MMBIR is initiated by DNA breakage generating a single DNA end, and proceeds with multiple template switches at positions of microhomologies that could be as short as 1-3bp, leading to varying levels of amplification and rearrangements (Lee et al., 2007; Liu et al., 2011a; Carvalho et al., 2013; Carvalho et al., 2011; Beck et al., 2015). The MMBIR model has since been used to explain telomere healing (Lowden, et al., 2011; Yatsenko, et al., 2012) and CGRs in a number of diseases including cancer (Lawson et al., 2011; Vissers et al., 2009; Wang et al., 2015). In addition, MMBIR-like events have been described in various model systems including bacteria (Slack et al., 2006; Lin et al., 2011), yeast (Payen et al., 2008), Arabidopsis (Kwon et al., 2010; Marechal et al., 2009), Caenorhabditis elegans (Meier et al., 2014), and mouse embryonic stem cells (Arlt et al., 2012). However, despite the broad occurrence of MMBIR and its important role in CGR formation, what triggers initiation of MMBIR remains unknown.
An important mechanistic insight into MMBIR has come from sequence analyses of various CGRs in humans. The limitation of such analyses however, is that the enzymatic requirements for MMBIR cannot be established, nor can the mechanisms regulating its usage be determined. Nevertheless, based on the initial sequence analyses of CGRs found in patients with PMD and MECP2 duplication syndrome (Carvalho et al., 2011; Carvalho et al., 2013; Beck et al., 2015) it was proposed that MMBIR is coupled to homology-mediated break induced replication (BIR). In particular, it was suggested that initiation of MMBIR observed in these patients involved the following two steps. First, a breakage of a replication fork led to homology-driven BIR involving two highly homologous inverted repeats which generated an inverted segment and led to a copy number gain (Carvalho et al., 2011; Beck et al., 2015). For the second step, a switch from homology-mediated BIR to MMBIR was postulated. The involvement of MMBIR was supported by a series of template switching events mediated by microhomologies that resulted in genomic changes of various sizes from large-scale copy number variations (CNVs) to small templated insertions (Carvalho et al., 2013; Beck et al., 2015). However, frequent template switching events that were previously described in association with BIR in model organisms always occured at regions of significant homology or homeology (Smith et al., 2007; Anand et al., 2014; Stafa et al., 2014) and therefore do not represent MMBIR events. Thus, the hypothesis that BIR switches to MMBIR remains unconfirmed, and the mechanisms underlying this process are unknown.
Here, using an experimental system in yeast where BIR can be efficiently induced and tracked, we provide two major insights into the mechanism of switching from homology-to microhomology-driven BIR. First, we demonstrate that re-routing of BIR into MMBIR results from an interruption of BIR due to a deficiency of repair-specific DNA synthesis that was modeled in our study by the lack of Pif1 helicase. Second, we show that polymerases ζ and Rev1 are responsible for DNA synthesis initiated at 0–6 nucleotide microhomologies during MMBIR. Together, our data provide the first demonstration of a transition from BIR to MMBIR and reveal the identity of the proteins that mediate this transition and drive MMBIR. Overall, results presented here lay the foundation for further studies on the mechanism of MMBIR in other systems including humans.
RESULTS
Experimental system to study switching from homology- to microhomology-mediated BIR
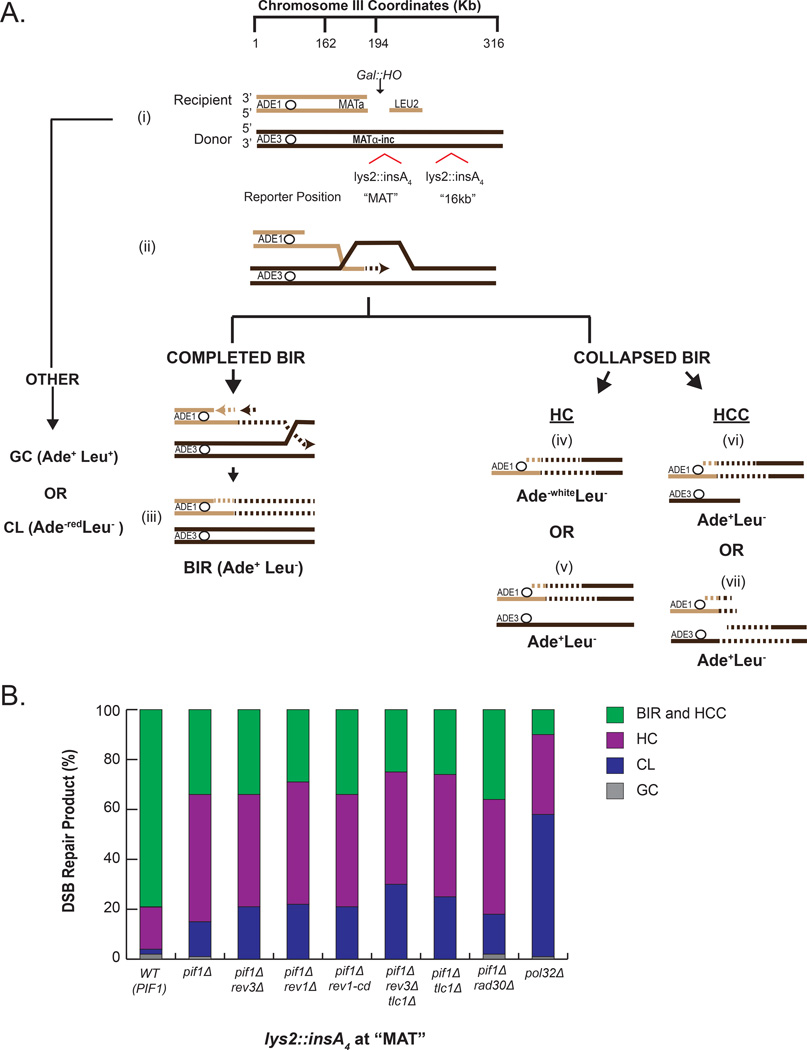
In this study, using a yeast model, we asked whether an interruption of BIR resulting from a deficiency in repair DNA synthesis could trigger a switch from BIR to MMBIR. Towards this goal, we used pif1Δ mutant strains disomic for chromosome III (Figure 1A, Table S1), where BIR is successfully initiated but is always interrupted (Wilson et al., 2013; Saini et al., 2013). In this assay, BIR is induced by an HO endonuclease-generated double strand break (DSB) at a truncated (MATa-containing) chromosome III by the addition of galactose (Figure 1A). Repair by BIR is initiated by strand invasion into a full-length (MATα-inc-containing) copy of chromosome III that is refractory to cutting followed by DNA synthesis that in wild-type (PIF1) strains can proceed for over 100kb to the end of the chromosome. However, in the absence of Pif1, which is needed for extensive synthesis, BIR interrupts within the first 20kb, and is often terminated by the resolution of recombination intermediates into half-crossovers (HC), where the unfinished BIR product fuses to a broken template chromosome (Wilson et al., 2013; Saini et al., 2013). In this study, by using auxotrophic markers positioned at the ends of chromosome III (donor and recipient), we confirmed that DSBs induced in pif1Δ strains frequently (in more than 50% o cases) resulted in HCs represented by Ade−white Leu− and portion of Ade+ Leu− events (Figure 1A, 1B, S2A, S2C, and see also Experimental Procedures for details). Also, a significant portion of Ade+ Leu− outcomes resulted from half-crossover initiated cascades, a type of chromosomal rearrangement initiated by half-crossovers (HCC; Figure 1 A(vi and vii), 1B, Figure S2B, S2C, and see Experimental Procedures). Finally, the frequency of chromosome loss (CL; Ade−red Leu−) was also increased in pif1Δ strains as compared to PIF1 (Figure 1B, S2A, S2C). In contrast, in PIF1 strains (WT) more than 70% of repair outcomes likely resulted from BIR that proceeded to the end of the donor chromosome (Figure 1A, 1B, S2A, S2B, S2C), consistent with previous observations (Sakofsky et al., 2014; Vasan et al., 2014). Overall, interruption of BIR that in yeast, frequently occurs in pif1Δ strains within a defined localized genomic region (20kb from break-site), allowed us to investigate molecular events that are specifically linked to BIR interruption.
Figure 1. Experimental system to model BIR collapse.
(A) DSB repair in a yeast strain disomic for chromosome III that has genetic markers at the ends of both copies of chromosome III used to identify DSB repair outcomes. (i) DSB is induced in a truncated copy of chromosome III (recipient chromosome) at MATa by a galactose inducible HO endonuclease. A full-length copy of chromosome III (donor) where MATα-inc is refractory to cutting by HO and contains a lys2::insA4 frameshift reporter inserted at one of two positions: MAT or 16kb. (ii) Broken (recipient) chromosome invades intact homologous donor chromosome and initiates DNA repair synthesis that proceeds via a migrating bubble. (iii) DNA synthesis proceeds to the end of the chromosome resulting in a completed BIR product. (iv-vii) BIR synthesis is interrupted, which leads to the resolution of BIR intermediates and the formation of one of the following half-crossover events: (iv) Ade−white Leu− half-crossovers (HC), (v) half-crossover that segregated with an intact sister chromatid of the broken donor similar to (Deem et al., 2008) (HC), (vi and vii) half-crossover initiated cascades (HCC) where the broken chromosome is stabilized by ectopic recombination or de novo telomere formation as represented by purple lines (see also Figure S1). Dotted lines indicate nascent DNA. Abbreviations: gene conversion (GC); chromosome loss (CL). (B) Distribution of DSB repair events in strains containing lys2::insA4 reporter inserted at MAT. Classification of repair events was based on phenotypes described in (A). See Figure S2A and Experimental Procedures for details of phenotypic analysis.
Microhomology-mediated template switches associated with BIR interruption
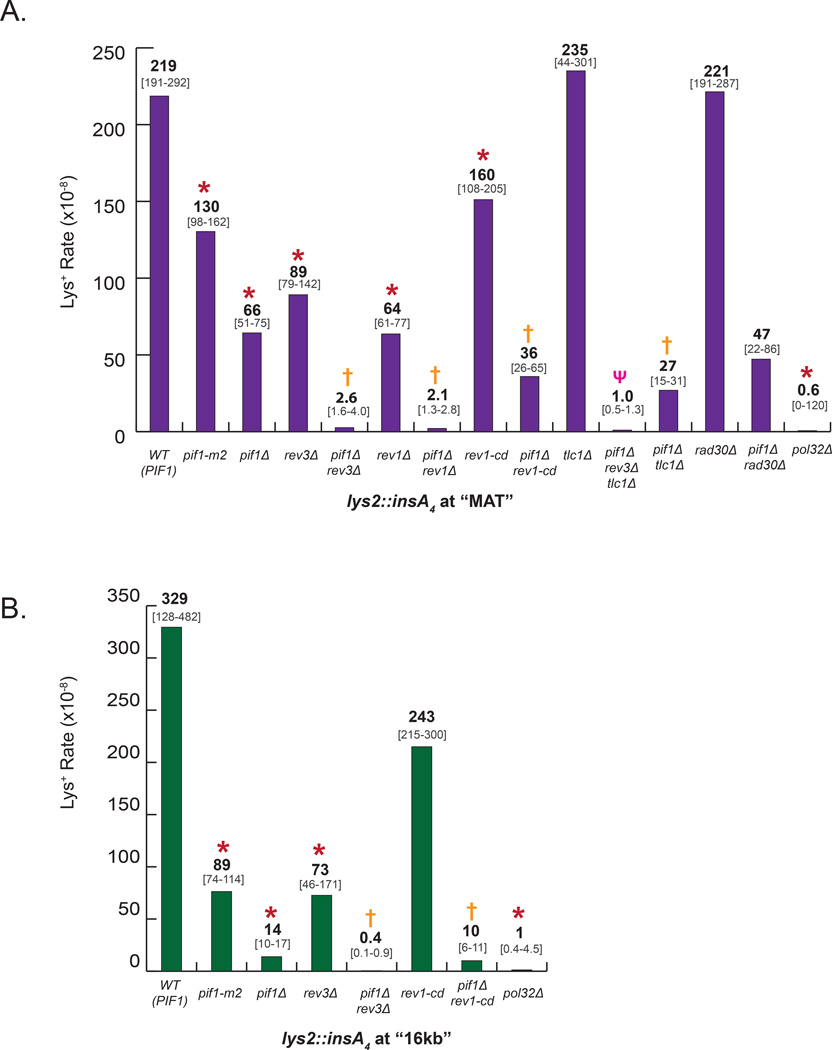
Upon DSB induction, we followed the fidelity of DNA synthesis within the first 20kb by using lys2::insA4 reporters located either at MAT locus (close to the position of strand invasion) or 16kb centromere-distal to MAT on the donor chromosome (Figure 1A). We compared the frequency (Figure 2A, 2B) and the spectra (Figure 3A, 3B) of Lys+ mutations in wild-type cells where BIR was usually completed, and in Pif1 deficient cells where BIR frequently collapsed. The frequency of Lys+ in strains containing lys2::insA4 reporters at 16kb position was reduced 24-fold in pif1Δ as compared to PIF1, (Figure 2B, Table S2), similar to (Saini et al., 2013), and was explained by the fact that synthesis infrequently reaches this position in pif1Δ mutants (Saini et al., 2013). Conversely, we observed that the decrease of mutagenesis in pif1Δ with the reporter at MAT (referred to herein as pif1Δ-MAT) was much less dramatic (only three-fold as compared to (Figure 2A, Table S2)) suggesting that even in the absence of Pif1 the invading strand is often extended, but eventually DNA synthesis interrupts in the MAT vicinity, leading to the formation of HC events. Thus, the pif1Δ strains with the frameshift reporter at MAT represents an optimal model system for studying mutations associated with collapsed BIR.
Figure 2. Formation of Lys+ frameshift mutations in pif1Δ require Polζ and Rev1.
The rate of Lys+ mutations for WT (PIF1) and derivative mutant strains was measured among Ade+ DSB repair outcomes 7hrs ater the addition of galactose for strains with lys2::insA4 reporter at MAT (A) or at 16kb position (B). Median mutation rates are shown; bracketed numbers below show 95% CI for strains with ≥6 experiments or the median range for strains with 4 or 5 experiments. Data for WT and pif1Δ at 16kb was similar to Saini et al. (Saini et al., 2013). Asterisks (*), crosses (†) and psi (ψ) symbols indicate statistically significant differences from WT (PIF1), pif1Δ, and pif1Δ rev3Δ strains, respectively. See Table S2 for P-values and for the description of statistical analysis.
Figure 3. Collapse of BIR leads to MMBIR mutations.
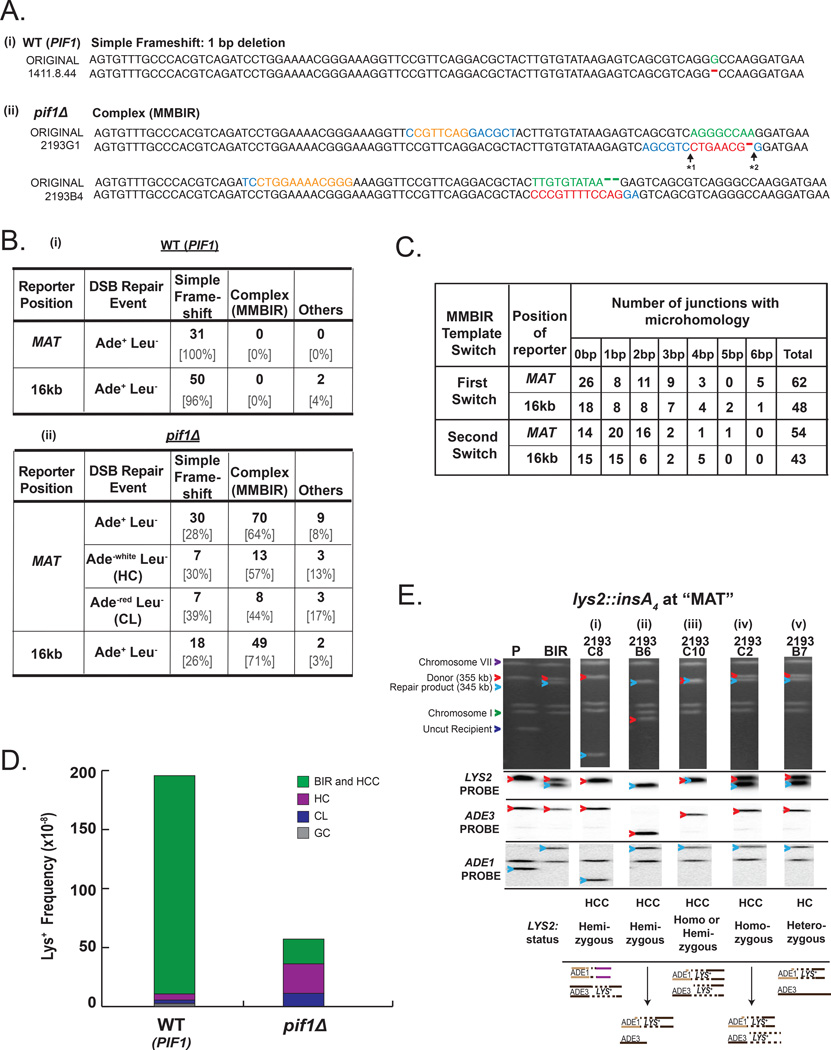
(A) Representative Lys+ mutant sequences obtained in WT (PIF1) and pif1Δ strains aligned to the original Lys− sequence. (i) 1bp deletion mutation, the main class of mutations associated with completed BIR in PIF1. (ii) MMBIR mutations in pif1Δ . Red letters indicate MMBIR mutations; orange letters indicate DNA sequence that served as a template for MMBIR; green letters indicate DNA sequences that were replaced by MMBIR, blue letters indicate microhomology. “* 1” and “* 2” indicate the junctions of the first and second template switch during the formation of MMBIR mutations, respectively. (B) Lys+ mutation spectra associated with completed (PIF1) (i) and interrupted BIR (pif1Δ) (ii). Simple frameshifts include 1bp deletions and 2bp insertions that are sometimes also associated with 1bp or 2bp substitutions. (C) Microhomologies at the junctions of the first and second MMBIR template switching events that occurred in Ade+ Leu− pif1Δ strains with reporters at MAT and 16kb positions. (D) Distribution of DSB repair events among Lys+ outcomes in WT (PIF1) and pif1Δ strains containing lys2:insA4 reporters at MAT. See Figure S2D and Experimental Procedures for details. (E) Chromosome III structure of representative MMBIR Ade+ Lys+ mutants obtained in pif1Δ-MAT strain. Upper panel: PFGE gel stained with ethidium bromide. Subsequent panels below show Southern blot analysis using LYS2-specific, ADE3-specific, and ADE1-specific probes as indicated. Red arrowheads indicate ADE3-containing chromosomes, while blue arrowheads indicate ADE1-containing chromosomes. The structure of chromosome III for each representative Lys+ mutant is shown below the gel, and only Lys+ copies of LYS2 are depicted. “P” denotes AM1003 derivative strain prior to induction of HO break. See Table S4, S5, S6 for full list of MMBIR mutations.
Using Sanger sequencing, Lys+ mutations from pif1Δ-MAT and WT-MAT were characterized, and a difference in the mutation spectra was detected between the two strains (Figure 3A, 3B). Specifically, while mutations from WT-MAT were entirely (31 out of 31) 1bp deletions (Figure 3A(i), 3B(i), Table S4), only 28% (30 out of 109) of Lys + events in pif1Δ-MAT were simple frameshifts (1bp deletions or 2bp insertions), while the majority of mutations (70 out of 109) were more complex (Figure 3A(ii), 3B(ii), Table S4). These complex mutations resulted from the replacement of 4 to 29 adjacent nucleotides by another sequence ranging from 4 to 34 nucleotides in length. For example, in the case depicted in Figure 3A(ii), the nucleotides shown in green were replaced with the sequence shown in red. Our analysis suggested that the majority of these insertions (similar to those shown in red in Figure 3A(ii)) were copied from nearby sequences since we identified templates corresponding to these complex mutations within the lys2::insA4 reporter. In particular, for 70 analyzed complex insertion mutations, we identified 64 templates (50 full templates and 14 partial templates; Table S4). We propose that the formation of each of these insertions included at least 2 consecutive template switching events (Figure 4) that occurred through the following steps: (i) in the absence of Pif1, DNA synthesis associated with BIR was interrupted, leading to the dissociation of the 3’ end of the newly synthesized DNA from its template; (ii) the 3’ end annealed to a ssDNA region located behind the BIR bubble or in the D-loop (not shown), which then copied a short stretch of DNA, and (iii) a second template switching event then occurred creating an insertion at the position where BIR was initially interrupted, followed by further synthesis to create a functional LYS2.
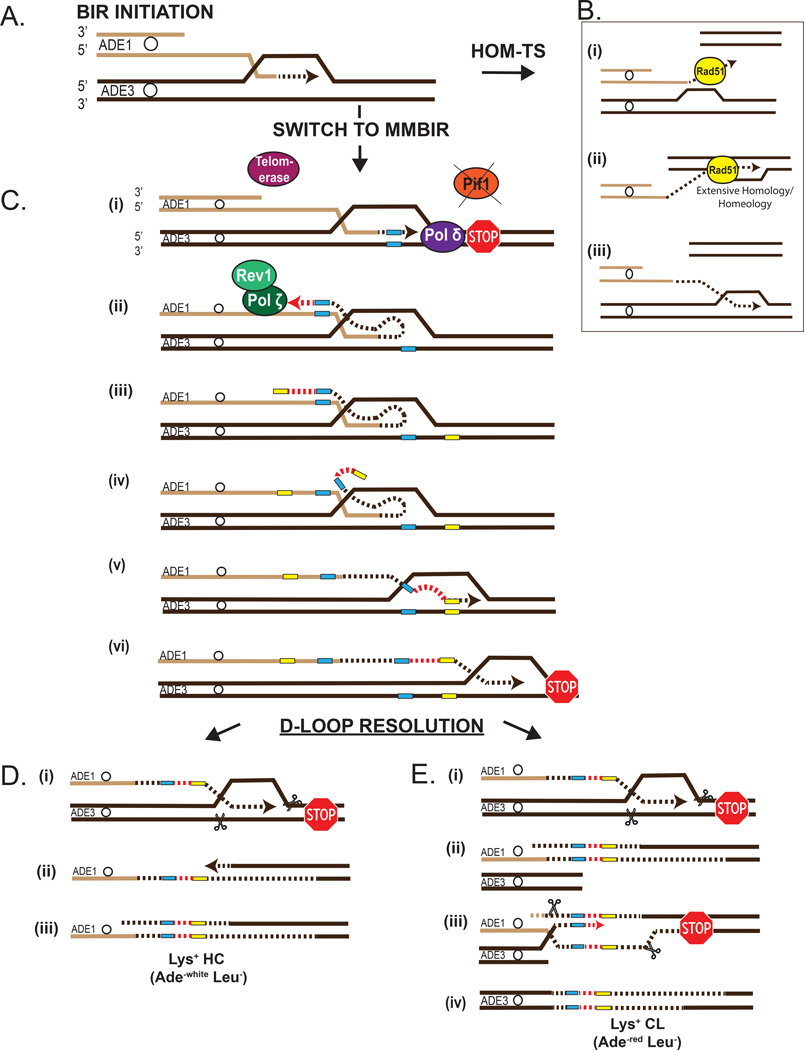
Figure 4. Proposed mechanism of switching from homology-driven BIR to MMBIR.
(A) Initiation of BIR. (B) Processive BIR associated with homology- (homeology-) mediated template switching (HOM-TS) (Anand et al., 2014; Smith et al., 2007; Stafa et al., 2014). (i) dissociation of the 3’ end. (ii) Rad51-mediated invasion of the 3’ end into an area with significant homology or homeology. (iii) Second template switch back to original homologous chromosome. (C) Defective BIR synthesis leads to MMBIR that is followed by resolution resulting in HC events. (i) interruption of BIR induced by the absence of Pif1, (ii) and (iii) annealing at microhomology (blue rectangles) located in ssDNA behind the BIR bubble followed by DNA synthesis (red dotted lines) driven by Polζ/Rev1. In the absence of Polζ/Rev1 telomerase creates GT-rich mutations (as illustrated in Figure 5B). (iv) strand dissociation, and (v) annealing of 3’ end at microhomology (yellow rectangles) in original track of BIR. Resolution of interrupted BIR intermediates leads to HC events with Lys+ mutations. Two events shown here are HC (Ade−white Leu−) (D) or CL event (Ade−red Leu−) (E).
Importantly, the switching between templates observed within lys2::insA4 reporter at MAT in pif1Δ cells occurred at positions of 0–6 bp long microhomologies. In particular, we found microhomologies from 1-6 bp at 58% (36 out of 62) (Figure 3C) of the junctions where the position of the first template switching (details in Figure 3A) was determined and at 74% (40 out of 54) of the junctions where the position of the second template switching was determined (Figure 3C, Table S4). In addition, we also observed that in multiple cases no microhomology was found immediately at the junctions of either the first (in 26 out of 62) or the second (in 14 out of 54) template switching event (Figure 3C, Table S4), and therefore they were categorized as “0bp microhomology”. However, in some of these cases, microhomologies could be identified at ≥1 nucleotide away from the junction (imperfect microhomologies; see Figure S2E, S2F for examples). We propose that microhomologies located at the junction and possibly even imperfect microhomologies could facilitate template switching (Figure 4). Importantly, the presence of only short or no microhomology at the junctions suggests that the template switches found in pif1Δ greatly differ from homologous/homeologous template switching (HOM-TS; Figure 4B) events previously described in association with BIR (Smith et al., 2007; Anand et al., 2014; Stafa et al., 2014). This new class of complex mutations formed by microhomology-mediated template switching is indicative of the MMBIR pathway and referred to herein as MMBIR mutations. We also found that MMBIR mutations were frequent in pif1Δ with lys2::insA4 reporter at 16kb (pif1Δ -16kb), with 71% (49 of 69) Lys+ mutations resulting from templated MMBIR (Figure 3B(ii), Table S5). These events were also frequently mediated by microhomologies that were found at 63% (30 out of 48) of the first template switch junctions and at 65% (28 out of 43) of the second template switch junctions (Figure 3C, Table S5). We also observed a significant number of MMBIR mutations where no microhomologies were found at either the first (18 out of 48) or second (15 out of 43) template switch junctions (Figure 3C, Table S5). Overall, these results obtained in a yeast model system represent the first documentation of a transition from BIR to MMBIR. Importantly, the frequency of MMBIR events reported here is likely to be underestimated since our assay revealed only those events that formed a functional LYS2.
Pulse field gel electrophoresis (PFGE) analysis demonstrated that 39% (20 out of 51) and 71% (12 out of 17) of outcomes containing MMBIR mutations formed in pif1Δ-MAT and in pif1Δ-16kb respectively, contained a rearrangement in one of the two copies of chromosome III (Table S4, S5). The analysis of these rearranged Lys + outcomes combined from MAT and 16kb reporter positions showed that the majority of them (24 out of 32) possessed only one copy of the LYS2 gene that was located in the non-rearranged copy of chromosome III (Table S4 and S5). These events likely resulted from half crossover-initiated cascades (HCC) (Vasan et al., 2014), where HC led to the breakage of the donor chromosome, which was repaired by ectopic recombination forming a rearranged chromosome (Figure 3E(i) and (ii), and described in Figure S1). An additional 16% of these chromosomal rearrangement events (5 out of 32) also likely resulted from HCC events where a broken donor chromosome recombined with an HC product, which caused the donor and recipient chromosomes to be the same size (Figure 3E(iii), Table S4, S5 (marked by asterisks)).
Since BIR in pif1Δ is always interrupted (Saini et al., 2013; Wilson et al., 2013), the formation of MMBIR events that did not contain visible chromosomal rearrangements was likely to also represent HC events, which could have resulted from two scenarios. First, they could be formed by HCC, which is supported by our finding that 5 out of 36 non-rearranged MMBIR events were homozygous for LYS2 (Figure 3E(iv), Table S4, S5). Alternatively, the formation of non-rearranged MMBIR events could also result from HC that segregated with an intact copy of a donor chromosome (Figure 3E(v)) (Deem et al., 2008).
We also asked whether MMBIR mutations were frequent among Ade−white Leu− HC outcomes in pif1Δ. Since these events could not be directly selected for, we first selected for Lys+ mutants among all (unselected) DSB repair events in pif1Δ, and then determined the fraction of Ade −white Leu− HC events among them by replicaplating on various media. We found that approximately 22% of all Lys+ repair outcomes pif1-MAT were Ade−white Leu− HCs (Figure 3D, Figure S2D). In addition, we found that over half (57%) of Lys+ HCs were MMBIR events in pif1Δ-MAT (Figure 3B(ii), Table S6), thus confirming that HCs were frequently associated with MMBIR events. We also analyzed Lys+ HCs isolated from WT-MAT, but failed to detect MMBIR events among them (Table S6). This is not surprising since the resolution of BIR intermediates leading to HC events in PIF1 is unlikely to occur in the vicinity of any given reporter, thus making it difficult to detect MMBIR events.
In addition, we observed that a signcant fraction (19%) o Lys mutations in pif1Δ -MAT were chromosome loss events (CLs, Ade −red Leu−; Figure 3D, S2D). Moreover, about half (44%) of these Lys + CLs were MMBIR events (Figure 3B(ii), Table S6). These data suggested that Lys+ CLs pif1Δ are frequently formed via resolution of a BIR intermediate allowing the Lys + mutation to be inherited on the donor chromosome (as shown in Figure 4E) rather than from the failure to initiate BIR as previously assumed (Deem et al., 2008).
Surprisingly, the frequency and spectrum of Lys+ mutations that we observed in pif1-m2 mutants were very different from those in pif1Δ. The pif1-m2 mutation is known to result in mostly mitochondrial localization of Pif1, and therefore we expected that the phenotype of pif1-m2 would be similar to pif1Δ (Schulz and Zakian, 1994). Instead, we observed that the level of mutagenesis in pif1-m2-MAT was only 1.7x lower (Figure 2A, Table S2) than WT (PIF1) as compared to a 3x reduction in pif1Δ, while mutagenesis in pif1-m2-16kb was only reduced 4x (Figure 2B, Table S2) versus 24x in pif1Δ. Moreover, practically all Lys+ mutations at both positions were 1bp deletions similar to WT, with only one MMBIR mutation found at 16kb (Table S4, S5). Thus as previously observed, pif1-m2 is a hypomorphic mutant that likely retains partial Pif1 activity in the nucleus (Ribeyre et al., 2009), which allows BIR synthesis to proceed beyond the areas of reporters thus precluding MMBIR detection.
Polζ drives MMBIR
It was previously demonstrated that during S-phase replication, Polζ promotes the formation of complex mutations (Northam et al., 2014). Therefore, we hypothesized that complex mutations associated with the collapse of BIR were also promoted by Polζ. To test this hypothesis, we investigated mutagenesis in a pif1Δ rev3Δ double mutant (REV3 encodes the catalytic subunit of the Polζ ). We found that in the absence of Polζ in pif1Δ background, the level of DSB-induced Lys+ mutations was reduced 25-fold at MAT and 34-fold at 16kb positions as compared to the mutation level in pif1Δ when Polζ is present (Figure 2, Table S2), which demonstrated that the formation of the majority of mutations in pif1Δ requires Polζ. Moreover, for both reporter positions, no single MMBIR event was found in pif1Δ rev3Δ (Figure 5A, 5B, Table S4, S5). Therefore, the formation of all MMBIR mutations is mediated by Polζ, consistent with our hypothesis. Importantly, the level of mutagenesis in pif1Δ rev3Δ at MAT was still 16x higher as compared to the level of spontaneous Lys+ mutations measured in isogenic strains lacking an HO cut site (No-DSB control) (Table S2, S3), suggesting the existence of another, Rev3-independent pathway of mutagenesis. In contrast, the level of mutagenesis in pif1Δ rev3Δ at 16kb was similar to the level of spontaneous mutagenesis (Table S2, S3). One possible explanation for this difference in mutation frequencies at MAT and 16kb could be that Rev3 helps to extend DNA repair synthesis in the absence of Pif1 to the 16kb position.
Figure 5. Pol ζ/Rev1 complex mediates formation of MMBIR mutations.
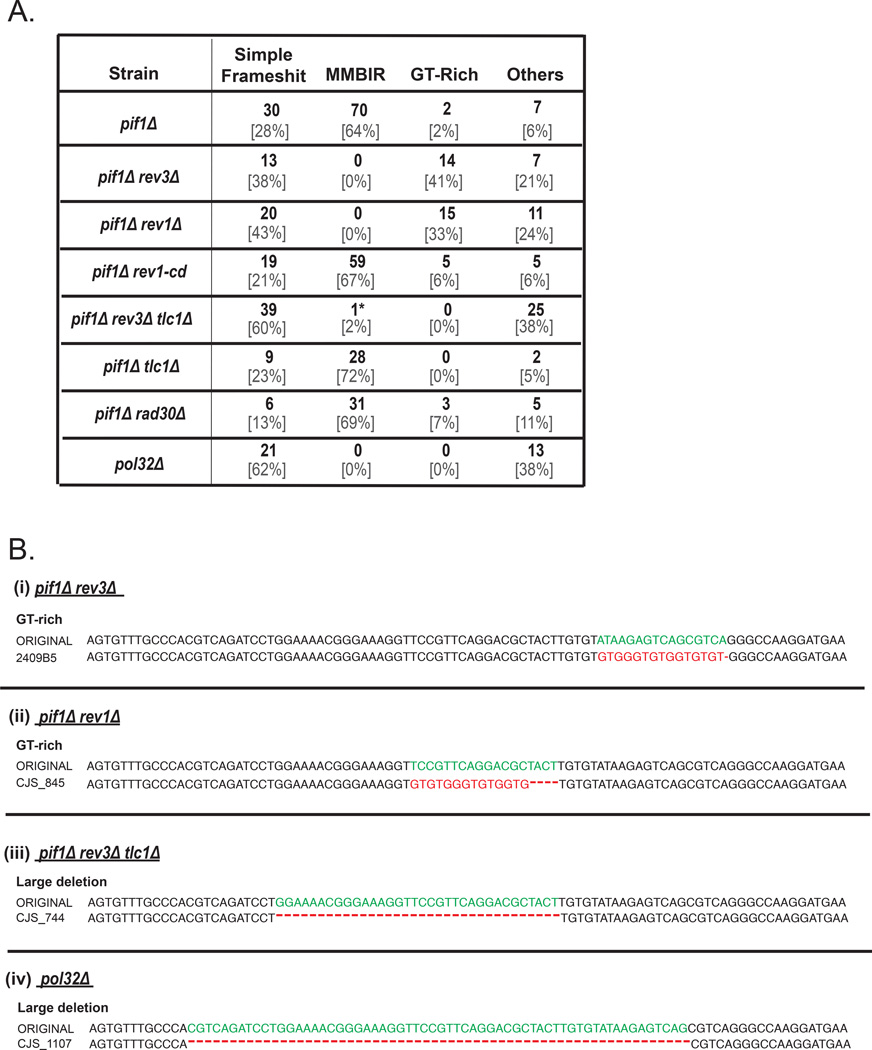
(A) Lys+ mutation spectra for Ade+ Leu− outcomes formed in various pif1Δ-MAT derivative strains. GT-rich class represents complex mutations where a telomere-like GT-rich sequence shown in red letters in B (i and ii) replace a stretch of DNA shown in green letters. Asterisk (*) denotes one rare outcome of ectopic MMBIR similar to rare ectopic template switching events observed in the absence of Polζ in (Northam et al., 2014). (B) Representative Lys+ mutant sequences aligned to the original Lys− sequence. (i and ii) GT-rich mutations common in pif1Δ rev3Δ and pif1Δ rev1Δ . (iii and iv) large deletions common in pif1Δ rev3Δ tlc1Δ and in pol32Δ. See legend to Figure 3B and Table S4.
In addition, we tested whether MMBIR occurs in the absence of Pol32, another known source of BIR collapse (Deem et al., 2008; Smith et al., 2009). We found that in the pol32Δ mutant, BIR mutagenesis was greatly reduced to a level below that of the double pif1Δ rev3Δ mutant, and no MMBIR was found (Figure 2, 5A, Table S2, S4). The majority of residual mutations were large deletions (Figure 5B(iv)). This is consistent with Pol32 being a subunit of both Polδ, and Polζ (Makarova et al., 2012), and therefore required for the formation of MMBIR mutations.
Similar experiments were also performed in pif1Δ rev1Δ mutants. REV1 encodes another translesion polymerase that plays two distinct roles in Polζ -mediated translesion bypass during S-phase replication. In particular, Rev1 inserts cytosines (catalytic function) and also plays a non-catalytic (structural) role in repair via its interaction with Polζ (as reviewed in (Pavlov et al., 2006)). We observed that the frequency of Lys+ mutations in pif1Δ rev1Δ was drastically (32-fold) reduced as compared to pif1Δ (Figure 2A, Table S2) and all MMBIR mutations were eliminated (Figure 5, Table S4), consistent with the idea that Rev1 assists Polζ in generating MMBIR events. We further asked whether the catalytic activity of Rev1 was important by using a rev1-cd mutation that disrupts the catalytic activity of Rev1, but does not affect the ability of Rev1 to interact with Polζ (Northam et al., 2014). We observed that the frequency of Lys+ mutations was mildly, but significantly decreased (2x at MAT and 1.4x at 16kb) in pif1Δ rev1-cd as compared to pif1Δ (Figure 2A, 2B, Table S2). Importantly, this reduction in mutagenesis proportionately lowered all types of mutations (Figure 5A), suggesting that the catalytic activity of Rev1 is likely to be required not for the formation of MMBIR mutations per se, but rather for the extension of repair-specific synthesis. Indeed, we observed that rev1-cd, as well as rev1Δ and rev3Δ decreased the level of frameshift mutagenesis even in PIF1 background (Figure 2, Table S2), suggesting that in these mutant backgrounds repair-specific DNA synthesis extend less frequently to the reporter positions. Therefore, we propose that Rev1 (including its catalytic activity) as well as of Polζ are important for making repair DNA synthesis more processive. We also tested the role of TLS polymerase Polη (encoded by RAD30) in MMBIR mutagenesis. We found that the mutation frequency in pif1Δ rad30Δ was not significantly different from pif1Δ and that 69% of Lys+ mutations (31 out of 45) were MMBIR (Figure 2A, 5A, Table S2, S4), suggesting that Polη does not play an important role in MMBIR mutagenesis.
Telomerase controls an alternative pathway of MMBIR-associated mutagenesis
While analyzing residual Lys+ mutations formed in pif1Δ rev3Δ at MAT, we found that many of these mutations were complex with a characteristic guanine- and thymidine-(GT-) rich pattern (Figure 5, Table S4) suggesting that they could have formed by telom erase. In addition, many of these mutations were adjacent to a ‘GT-seeding sequence’, which further supported the idea that telomerase contributed to the formation of these mutations. Indeed, deleting TLC1 (encoding RNA component of telomerase; Singer and Gottschling, 1994) in both pif1Δ rev3Δ-MAT and in pif1Δ-MAT backgrounds reduced the frequency of Lys+ and eliminated all GT-rich mutations (Figure 2A, Figure 5, Table S2, S4). This indicates that telomerase is responsible for the observed GT-rich insertions and therefore contributes to the formation of complex mutations. In addition, the reduction of mutation frequency indicates that telomerase can extend interrupted repair synthesis, both in the absence or presence of Rev3. Additionally, no GT-rich mutations were found at the 16kb position in pif1Δ rev3Δ (Table S5), consistent with a lack of Rev3-independent mutagenesis in pif1Δ-16kb (see earlier).
DISSCUSION
Our study using yeast as a model provides two major insights into the mechanism of MMBIR, that has been implicated in non-recurrent genomic rearrangements in humans (Liu et al., 2011a, Carvalho et al., 2011; Carvalho et al., 2013; Beck et al., 2015; Carvalho et al., 2015). First, we define the conditions when MMBIR is initiated, and second, we identify the enzymes driving this pathway.
MMBIR is triggered by the collapse of classical BIR
By using a controlled system in yeast we were able to demonstrate that MMBIR initiates due to an impediment of DNA synthesis during BIR, which was modeled in our experiments by the absence of Pif1, an enzyme required for extensive DNA repair synthesis. According to our model (Figure 4), an impediment of DNA synthesis resulting in a collapse of BIR triggers two important events that collectively initiate MMBIR. The first event is annealing of the dissociated 3’ DNA end at regions of microhomology located in ssDNA accumulated behind or inside of the BIR bubble. Importantly, since ssDNA is highly persistent during BIR, it is likely to promote a higher frequency and variety of microhomology-mediated template switching events. The second event involves the exchange of DNA polymerase Polδ that is driving normal BIR, with the translesion polymerase Polζ. Polζ initiates MMBIR by extending short and imperfect primers formed by the 3’ end annealed at microhomology or by extending a DNA end without any microhomology at all, as it was frequently observed in our study as well as in studies of template switching initiated by stalling of S-phase replication (Northam et al., 2014). It remains unclear how Polζ extended DNA ends in those cases where no microhomology was found at the junctions. One possibility is that imperfect microhomology located near the junctions of template switching could facilitate the formation of an annealing intermediate containing mispaired DNA at the 3’ end that could be extended by Polζ.
In addition to the initial template switching and Polζ-mediated synthesis, the formation of Lys+ mutations described in this study also required a second template switch that occurred at microhomology (or without any homology at all), which brought DNA synthesis back to its original position along the track of BIR. This second template switch could have been caused by the low processivity of Polζ (reviewed in (Pavlov et al., 2006)). Alternatively, if the first template switch resulted in annealing to ssDNA in the D-loop, it is possible that the instability of the D-loop, or its migration could also promote a second template switching event. In either case, the second switch allowed synthesis to continue enough to generate Lys+ mutants, but it was eventually terminated, followed by resolution into HCs, or more complex rearrangements. In addition, in the absence of Polζ, the formation of complex mutations could be alternatively mediated by telomerase that is especially potent in the absence of Pif1, a known telomerase suppressor (Dewar and Lydall, 2010; Schulz and Zakian, 1994).
In this study we specifically looked at yeast MMBIR events caused by an interruption of BIR resulting from a deficiency in Pif1, however, we predict that MMBIR may also initiate in other circumstances when repair-specific synthesis is interrupted. For example, interruption can occur due to (i) various defects in BIR replication machinery (e.g., Polδ defects), (ii) DNA damage accumulation in the template for BIR leading-strand synthesis, or (iii) formation of secondary structures that impede progression of BIR leading-strand synthesis. These sources of interruption can lead to a collapse of BIR that initiates microhomology-mediated template switching occurring at any position along the track of BIR. Therefore, to detect these non-localized events, the development of new high-throughput screening will likely be needed since the probability of detecting them with reporter-based assays is low.
Overall, we propose that MMBIR results from interruption of BIR progression and involves an exchange of DNA polymerases. Importantly, MMBIR cannot be explained by homology-driven template switching commonly associated with processive BIR (HOM-TS) (Smith et al., 2007; Anand et al., 2014; Stafa et al., 2014) (Figure 4B), since HOM-TS only occur between significantly homologous or homeologous DNA regions, and therefore cannot mediate template switching between 0–6 bps occurring in MMBIR. Significantly, MMBIR demonstrated in this study operates in the presence of a fully homologous template and with functional strand exchange proteins, and therefore it is not the absence of these cellular components that initiates MMBIR, but rather it is a consequence of interrupted DNA synthesis.
MMBIR model: implications for understanding complex genomic rearrangements in humans
The MMBIR model was proposed by Lupski and colleagues to explain the formation of complex genomic rearrangements associated with neurological diseases in humans (Lee et al., 2007; Liu et al., 2011a; Carvalho et al., 2011; Carvalho et al., 2013; Beck et al., 2015; Carvalho et al., 2015). While these studies provided initial insights for how chromosomal rearrangements could occur, it was insufficient to prove MMBIR. Testing this model became even more important since the MMBIR mechanism has been readily embraced to explain the formation of a variety of CGRs in humans (Carvalho et al., 2011; Carvalho et al., 2013; Beck et al., 2015; Carvalho 2015; Zhang et al., 2015; Wang et al., 2015; Kidd et al., 2010; Conrad et al., 2010; Arlt et al., 2012), see Table S8 for details. For example, two recent studies of CNVs in humans identified a number of MMBIR-like insertions at breakpoints of rearrangements (Conrad et al., 2010; Wang et al., 2015). Interestingly, MMBIR-like events were also found at the borders of AOH (absence of heterozygosity) regions in humans that could result from half-crossover events (Carvalho et al., 2015) – rearrangements that were linked to MMBIR in our study. Finally, a recent study of chromothripsis that was experimentally induced by a micro-nuclear formation, revealed multiple templated insertions that likely resulted from MMBIR (Zhang et al., 2015), thus linking MMBIR to chromothripsis. Overall, the broad implication of MMBIR in the formation of CGRs in humans made it important to directly test this model including the hypothesis proposed by Lupski and colleagues that the initiation of MMBIR results from interruption of classical BIR (Carvalho et al., 2013; Beck et al., 2015; Carvalho et al., 2015).
Here, using an experimental system in yeast, we directly demonstrated that the collapse of classical BIR can lead to initiation of MMBIR. Moreover, we discovered that in yeast TLS polymerases Rev1 and Polζ drive MMBIR. Based on this finding, we propose that MMBIR associated with neurological or other disorders in humans may also involve Polζ, or one of several other known human TLS polymerases. Another important outcome of our study is the refinement of an MMBIR signature in yeast: small templated insertions that represent inverted copies of their template and that are associated with chromosomal rearrangements. This will allow for a direct search for analogous MMBIR events in humans using whole-genome databases. This search however, will likely require the development of new bioinformatics tools such as the one we recently described (Segar et al., 2014) that can allow for robust detection of MMBIR events often overlooked by existing algorithms due to the intrinsic complexity of the MMBIR pattern. We believe that in the future this approach will lead to the characterization of patterns and frequencies of MMBIR events across various human diseases. Specifically, we propose to search for an MMBIR signature in cancer cells that underwent NRT (non-reciprocal translocation) cycles (Sabatier et al., 2005), since they are likely formed by an HC mechanism. The search for MMBIR mutations could also be applied to cell lines where the overexpression of oncogenes leading to replication collapse initiates BIR followed by CGRs (Costantino et al., 2014). While the role of Pif1 in BIR in humans remains to be determined, Pif1 helicase has been recently implicated in the recovery from oncogene-induced replication stress (Gagou et al., 2014), a process that requires PolD3 (Pol32)-mediated BIR (Costantino et al., 2014). Thus Pif1 levels may impact the frequency of BIR/MMBIR events in humans.
Finally, we propose that some CGRs that were previously explained by NHEJ – based on the lack a copy number increase and/or on the absence of homology at their junctions (see for example in (Carvalho et al., 2011; Arlt et al., 2012, Kloosterman et al., 2011b, Kloosterman et al., 2011a)), should be revisited. We propose that they alternatively can result from MMBIR since our findings indicate that MMBIR in yeast often occurs with no microhomology, and also are linked to HCs, which do not result in copy number increase.
EXPERIMENTAL PROCEDURES
Yeast Strains and growth conditions
All yeast strains are isogenic to AM1003 strain that is disomic for chromosome III (Deem et al., 2008). In these strains, HO-induced DSBs were introduced into a truncated copy of chromosome III (recipient) by the addition of galactose. The DSB created a broken end that initiated BIR through recombination with a full-length copy of chromosome III (donor) that is refractory to cutting due to a point mutation in its HO recognition sequence at MATα-inc (Figure 1A) (Deem et al., 2008). In addition, lys2::insA4 reporters were placed in the donor chromosome at different positions (MAT or 16kb) (Figure 1A) to assess frameshift mutations associated with DSB repair (Deem et al., 2011). Mutagenic copying of the lys2::insA4 reporters restored the reading frame and produced a Lys+ phenotype, which usually results from 1bp deletions in WT (PIF1) strains. Rich medium (yeast extract-peptone-dextrose (YEPD)) and synthetic complete medium were made as described in (Guthrie and Fink, 1991). YEP-lactate and YEP-galactose used for DSB induction were similar to (Deem et al., 2008), and 5-fluoroorotic acid (5-FOA) was added to synthetic complete medium with trace amounts of uracil (30mg/mL). See Supplemental Experimental Procedures for further details.
Determining Lys+ mutation rates and spectra
The rate of Lys+ mutagenesis was determined among all DSB repair outcomes similar to (Deem et al., 2011) or among Ade+ outcomes that preserved both chromosome III copies as in (Saini et al., 2013). Lys+ mutation spectra were determined similar to (Deem et al., 2011) using the Codon Code Aligner DNA Sequence Analysis Program: http://www.codoncode.com/aligner/. See Supplemental Experimental Procedures for further details.
Characterization of DSB repair isolates
DSB repair isolates were characterized by phenotype, similar to (Deem et al., 2008). HCs were represented by Ade−white Leu−, as well as by a portion of Ade+ Leu− outcomes where HC segregated with an intact donor during mitosis (see Supplemental Experimental Procedures for details). The Ade+ Leu− outcomes were referred to as BIR/HCC because a significant portion of these events in pif1Δ contained chromosomal rearrangements, indicative of HCC. The presence of chromosomal rearrangements was determined using PFGE, similar to (Deem et al., 2008) (see Supplemental Experimental Procedures for details). In addition, even non-rearranged Ade+ Leu− outcomes in pif1Δ were more likely to result from multiple rounds of HCs rather than from completed BIR based on our previous observations (Saini et al., 2013).
Supplementary Material
ACKNOWLEGEMENTS
We thank Drs. James Lupski, Phil Hastings, and Claudia Carvalho for their helpful comments on the manuscript. We thank Polina Shcherbakova for generously providing pR306-rev1-cd plasmid. We thank David Cooper and Liping Liu for their help in analysis of Lys+ mutants. This work was unded by the US National Institutes of Health grants R01GM084242 to A.M., and GM080600 and the CPRIT RP140456 to G.I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
C.J.S., S.A., and A.M. designed the experiments. C.J.S., S.A., A.K.D., and W.C. constructed strains. S.A., C.J.S., and A.K.D. performed the experiments. C.J.S., A.M., S.A., and G.I., analyzed the data. C.J.S., A.M., A.K.D., and G.I. wrote the manuscript. C.J.S. and S.A. contributed equally to this work.
REFERENCES
- Anand RP, Tsaponina O, Greenwell PW, Lee CS, Du W, Petes TD, Haber JE. Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev. 2014;28:2394–2406. doi: 10.1101/gad.250258.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet. 2012;8:e1002981. doi: 10.1371/journal.pgen.1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Carvalho CM, Banser L, Gambin T, Stubbolo D, Yuan B, Sperle K, McCahan SM, Henneke M, Seeman P, et al. Complex Genomic Rearrangements at the PLP1 Locus Include Triplication and Quadruplication. PLoS Genet. 2015;11:e1005050. doi: 10.1371/journal.pgen.1005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Pehlivan D, Ramocki MB, Fang P, Alleva B, Franco LM, Belmont JW, Hastings PJ, Lupski JR. Replicative mechanisms for CNV formation are error prone. Nat Genet. 2013;45:1319–1326. doi: 10.1038/ng.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Pfundt R, King DA, Lindsay SJ, Zuccherato LW, Macville MV, Liu P, Johnson D, Stankiewicz P, Brown CW, et al. Absence of Heterozygosity due to Template Switching during Replicative Rearrangements. Am J Hum Genet. 2015;96:1–10. doi: 10.1016/j.ajhg.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, Hines-Dowell S, Seaver LH, et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. 2011;43:1074–1081. doi: 10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Barker K, Vanhulle K, Downing B, Vayl A, Malkova A. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics. 2008;179:1845–1860. doi: 10.1534/genetics.108.087940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar JM, Lydall D. Pif1- and Exo1-dependent nucleases coordinate checkpoint activation following telomere uncapping. The EMBO journal. 2010;29:4020–4034. doi: 10.1038/emboj.2010.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagou ME, Ganesh A, Phear G, Robinson D, Petermann E, Cox A, Meuth M. Human PIF1 helicase supports DNA replication and cell growth under oncogenic-stress. Oncotarget. 2014;5:11381–11398. doi: 10.18632/oncotarget.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink G. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009a;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009b;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, Kallicki J, Kaul R, Wilson RK, Eichler EE. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B, Hochstenbach R, Poot M, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011a;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Hoogstraat M, Paling O, Tavakoli-Yaraki M, Renkens I, Vermaat JS, van Roosmalen MJ, van Lieshout S, Nijman IJ, Roessingh W, et al. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol. 2011b;12:R103. doi: 10.1186/gb-2011-12-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Tavakoli-Yaraki M, van Roosmalen MJ, van Binsbergen E, Renkens I, Duran K, Ballarati L, Vergult S, Giardino D, Hansson K, et al. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 2012;1:648–655. doi: 10.1016/j.celrep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Kwon T, Huq E, Herrin DL. Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc Natl Acad Sci. 2010;107:13954–13959. doi: 10.1073/pnas.1004326107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson AR, Hindley GF, Forshew T, Tatevossian RG, Jamie GA, Kelly GP, Neale GA, Ma J, Jones TA, Ellison DW, et al. RAF gene fusion breakpoints in pediatric brain tumors are characterized by significant enrichment of sequence microhomology. Genome Res. 2011;21:505–514. doi: 10.1101/gr.115782.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Lin D, Gibson IB, Moore JM, Thornton PC, Leal SM, Hastings PJ. Global chromosomal structural instability in a subpopulation of starving Escherichia coli cells. PLoS Genet. 2011;7:e1002223. doi: 10.1371/journal.pgen.1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011a;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowden MR, Flibotte S, Moerman DG, Ahmed S. DNA synthesis generates terminal duplications that seal end-to-end chromosome fusions. Science. 2011;332:468–471. doi: 10.1126/science.1199022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Lindberg M, Faust GG, Leibowitz ML, Clark RA, Layer RM, Quinlan AR, Hall IM. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 2013;23:762–776. doi: 10.1101/gr.143677.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A, Parent JS, Veronneau-Lafortune F, Joyeux A, Lang BF, Brisson N. Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci. 2009;106:14693–14698. doi: 10.1073/pnas.0901710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier B, Cooke SL, Weiss J, Bailly AP, Alexandrov LB, Marshall J, Raine K, Maddison M, Anderson E, Stratton MR, et al. C. elegans whole-genome sequencing reveals mutational signatures related to carcinogens and DNA repair deficiency. Genome Res. 2014;24:1624–1636. doi: 10.1101/gr.175547.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- Northam MR, Moore EA, Mertz TM, Binz SK, Stith CM, Stepchenkova EI, Wendt KL, Burgers PM, Shcherbakova PV. DNA polymerases zeta and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res. 2014;42:290–306. doi: 10.1093/nar/gkt830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV, Rogozin IB. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int Rev Cytol. 2006;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatier L, Ricoul M, Pottier G, Murnane JP. The loss of a single telomere can result in instability of multiple chromosomes in a human tumor cell line. Mol Cancer Res. 2005;3:139–150. doi: 10.1158/1541-7786.MCR-04-0194. [DOI] [PubMed] [Google Scholar]
- Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–392. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA, Malkova A. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–1648. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- Segar MW, Sakofsky CJ, Malkova A, Liu Y. MMBIRFinder: a tool to detect microhomology-mediated break-induced replication. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2014 doi: 10.1109/TCBB.2014.2359450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Lam AF, Symington LS. Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase delta complex. Mol Cell Biol. 2009;29:1432–1441. doi: 10.1128/MCB.01469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Stafa A, Donnianni RA, Timashev LA, Lam AF, Symington LS. Template switching during break-induced replication is promoted by the Mph1 helicase in Saccharomyces cerevisiae. Genetics. 2014;196:1017–1028. doi: 10.1534/genetics.114.162297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan S, Deem A, Ramakrishnan S, Argueso JL, Malkova A. Cascades of genetic instability resulting from compromised break-induced replication. PLoS Genet. 2014;10:e1004119. doi: 10.1371/journal.pgen.1004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, de Vries BB, Gilissen C, Hoischen A, et al. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum Mol Genet. 2009;18:3579–3593. doi: 10.1093/hmg/ddp306. [DOI] [PubMed] [Google Scholar]
- Wang Y, Su P, Hu B, Zhu W, Li Q, Yuan P, Li J, Guan X, Li F, Jing X, et al. Characterization of 26 deletion CNVs reveals the frequent occurrence of micro-mutations within the breakpoint-flanking regions and frequent repair of double-strand breaks by templated insertions derived from remote genomic regions. Hum Genet. 2015;134:589–609. doi: 10.1007/s00439-015-1539-4. [DOI] [PubMed] [Google Scholar]
- Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, et al. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko SA, Hixson P, Roney EK, Scott DA, Schaaf CP, Ng YT, Palmer R, Fisher RB, Patel A, Cheung SW, et al. Human subtelomeric copy number gains suggest a DNA replication mechanism for formation: beyond breakage-fusion-bridge for telomere stabilization. Hum Genet. 2012;131:1895–1910. doi: 10.1007/s00439-012-1216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.