Highlights
-
•
Cytosolic Hsp90s are more conserved than those from mitochondrial and ER.
-
•
Cell environment plays a role in the overall physicochemical properties of Hsp90s.
-
•
Serine and tyrosine are favored phosphorylated residues of Hsp90s.
-
•
Mitochondrial and ER Hsp90s have motifs unique to specific organisms.
Abbreviations: GRAVY, grand average of hydropathicity; Hop, Hsp70/90 organizing protein; Hsps, heat shock proteins; M domain, middle domain; ML, Maximum Likelihood; MSA, multiple sequence analysis; NJ, Neighbor Joining
Keywords: Heat shock proteins, Physicochemical properties, Motif analysis, Phosphorylation, Protozoan
Abstract
The treatment of protozoan parasitic diseases is challenging, and thus identification and analysis of new drug targets is important. Parasites survive within host organisms, and some need intermediate hosts to complete their life cycle. Changing host environment puts stress on parasites, and often adaptation is accompanied by the expression of large amounts of heat shock proteins (Hsps). Among Hsps, Hsp90 proteins play an important role in stress environments. Yet, there has been little computational research on Hsp90 proteins to analyze them comparatively as potential parasitic drug targets. Here, an attempt was made to gain detailed insights into the differences between host, vector and parasitic Hsp90 proteins by large-scale bioinformatics analysis. A total of 104 Hsp90 sequences were divided into three groups based on their cellular localizations; namely cytosolic, mitochondrial and endoplasmic reticulum (ER). Further, the parasitic proteins were divided according to the type of parasite (protozoa, helminth and ectoparasite). Primary sequence analysis, phylogenetic tree calculations, motif analysis and physicochemical properties of Hsp90 proteins suggested that despite the overall structural conservation of these proteins, parasitic Hsp90 proteins have unique features which differentiate them from human ones, thus encouraging the idea that protozoan Hsp90 proteins should be further analyzed as potential drug targets.
1. Introduction
Three classes of parasites; protozoans, ectoparasites and helminthes, cause parasitic diseases in humans. Especially protozoan parasitic diseases result in a tremendous health burden worldwide, and the treatment is challenging due to limitations including drug resistance, high cost, low efficacy and poor safety [1], [2].
Parasites survive within host organisms, and some need intermediate hosts to complete their life cycle. Hosts present different environments for parasites; thus for survival, adaptation measures should be taken [3]. Changes in temperature, pH and ionic strength between host environments, as well as the host immune system, might cause enormous stress. Interestingly, some protozoan parasites such as Plasmodium falciparum express large amounts of heat shock proteins (Hsps) to adapt to these changes and to proliferate [4], [5]. This makes Hsps potentially attractive drug targets against parasitic diseases.
Among Hsps, Hsp90 proteins are important in stress environment besides their more complex roles in numerous other physiological processes [6]. Humans have two cytosolic isoforms (Hsp90α and Hsp90β [6]), one mitochondrial isoform (TRAP1 [7]) and one isoform (Grp94 [8]) from endoplasmic reticulum (ER). Hsp90 exists as a homodimer, and each monomer is made up of three domains; the N-terminal ATP binding domain, C-terminal dimerization domain and the middle (M) domain responsible for client protein binding. These domains are highly conserved, and depending on the species Hsp90 contains two highly charged regions [9].
Although Hsp90 became an important anticancer target in the last few decades, limited research has been done in targeting them for parasitic diseases [3], [10], [11]. This is partly due to the view that the high conservation of the protein would make it difficult to develop an inhibitor specific to the Hsp90 of a particular organism. Here, we show that despite the overall structural conservation, parasitic Hsp90 proteins have unique features which separate them from human and vector Hsp90 proteins. The focus of this study is to investigate and compare the differences in sequence composition of human, vector and parasite Hsp90 proteins, and to build a profile of potential sites and features on the protein that could be exploited in selective drug discovery studies. Further, to date, attention in the literature has mainly been given to the ATP binding domain as a drug target. Recently, it was shown that the interface of the M domain of cytosolic Hsp90 and TPR2 domain of Hop (Hsp70/90 organizing protein) have striking differences between human and P. falciparum proteins [12], and it was proposed as potential site for inhibitor design. Thus, this study examines the differences between human and parasitic sequences not only in the N-terminal ATP binding domain as a popular drug targeting site, but in the entire protein sequence with the intention of understanding the possibility of identifying new potential inhibitor sites.
Overall, the study included 104 Hsp90 sequences from parasites, vectors and human. The proteins were divided into three groups according to their subcellular localizations: cytosolic (Group A), ER protein (Group B) and mitochondrial proteins (Group C). Interestingly, the results showed significant differences between proteins of different subcellular localization as well as between the human and protozoan Hsp90 proteins in terms of postulated physicochemical properties, amino acid composition, phosphorylation and motifs. We believe that our findings provide a suitable platform and starting point for further in silico and wet-lab experiments on Hsp90 proteins as potential drug targets for protozoan parasitic diseases.
2. Materials and methods
2.1. Sequence retrieval
Four human Hsp90 (HsHsp90) sequences were retrieved from NCBI. Parasite and vector Hsp90 homologs were searched by NCBI-BLAST using HsHsp90α and HsHsp90β for the cytosolic (Group A); HsGrp94 for the ER (Group B); and HsTRAP1 for the mitochondrial (Group C) proteins. In each case, reverse BLAST was applied to select true orthologs. A total of 100 sequences from 31 parasitic organisms (22 protozoa, 8 helmith and 1 ectoparasite) and 8 vectors were retrieved (S-Data 1). Further, for the phylogenetic tree calculations, six human and P. falciparum Hsp70 sequences, two for each group, were identified to use as outgroup control (S-Data 1).
2.2. Sequence alignment and phylogenetic tree calculations
For all as well as for each group of sequences, multiple sequence analysis (MSA) used MAFFT’s E-INS-i’s protocol [13]. Aligned sequences were analyzed by Jalview [14]. All versus all pairwise sequence alignments were calculated by a Matlab script.
Protein phylogenies were created in MEGA6 [15] using tree-building algorithms; Maximum Likelihood (ML [16], [17]) for all sequences and Neighbor Joining (NJ [18]) for individual groups to measure the distances relative to Hsp70. The overall phylogeny was selected from the top three models with lowest BIC scores, namely Le and Gascuel (LG + G + I and LG + G) [19] and Reverse Transcriptase (rtREV + G + I + F) [16] algorithms. Where the bootstrap values were lower than 60, multifurcation was applied in the main figure (Fig. 1).
Fig. 1.
Phylogenetic tree. Evolutionary history inferred by Maximum Likelihood [17] analysis using the Le and Gascuel model [19]. The discrete Gamma distribution was used to model evolutionary rate differences among sites and rate variation model allowed for some sites to be evolutionarily invariable respectively (5 categories (+G, parameter = 3.6824)) and ([+I], 0.7609% sites) respectively. Color representation: branches; red: Group A, black: Group B and blue: Group C. Sequences; black: vectors (bold italics: hosts), pink: ectoparasite, green: helminthes and maroon: protozoa.
2.3. Physicochemical properties and statistical analysis
For each group, physicochemical properties, i.e. molecular weight (Mr), hydrophobicity, aromaticity, isoelectric point (pI), instability index, aliphatic index, grand average of hydropathicity (GRAVY) and amino acid composition were calculated by Python and Biopython scripts. Calculations were also extended to the N-terminal ATP binding domain analysis. R-scripts were used to calculate boxplots. The Kruskal–Wallis test [20] assessed the statistical significance (p ⩽ 0.05) of the differences between the physicochemical properties measured in the analysis of Hsp90 proteins from different groups.
2.4. Identification of phosphorylation sites
Sequences were submitted to the Netphos 2.0 web-server [21] to calculate the phosphorylation sites for serine, tyrosine and threonine residues. A Python script mapped the results into the alignment file, and colored accordingly in Jalview.
2.5. Homology modeling
Homology models of cytosolic Homo sapiens Hsp90 (HSA_A) and P. falciparum Hsp90 (PFA_A), and the ER bound isoform of Hsp90 for P. falciparum (PFA_B) and Leishmania mexicana (LME_B) were calculated using MODELLER v9.13 [22]. The HHPred web-server [23] was used to identify templates. For the cytosolic proteins (HSA_A and PFA_A) 2CG9 was used as the primary template, while 2O1U was utilized as the main template for both ER bound models (PFA_B and LME_B). HSA_A 3T10, (residues 16–223) and 2CGE (residues 241–677); PFA_B, 3PEH (residues 69–330); LME_B, 2CG9 (residues 119–142 and 149–166) and 1Y6Z (residues 345–357 and 362–371) were incorporated into the modeling alignment to allow for several insert regions. Structural data for some of the model segments (HSA_A: residues 1–17, 230–281 and 695–732; PFA_A: residues 223–240, 307–316, 580–593 and 661–677; PFA_B: residues 1–68 and 808–821; LME_B: residues 1–40 and 731–776) were missing, thus could not be modeled. 100 models were calculated per protein and the model with the lowest DOPE-Z score was selected for unique motif mapping.
2.6. Motif analysis
MEME web-server [24] was used to search for conserved motifs with motif length ranging 6–300 amino acids in each group as well as in the N-terminal domains. A Python script was written to analyze the MEME and MAST log-files, giving as output a heat-map showing conservation. Heat-map results were further analyzed, specifically for cytosolic HSA_A and PFA_A, and ER unique motifs for PFA_B and LME_B, and mapped to their respective homology models by Python scripting and visualized in PyMOL [25].
3. Results and discussion
3.1. Reverse BLAST and phylogenetic tree calculations agreed in grouping the sequences
Since many parasitic sequences used here have not been experimentally annotated, reverse BLAST approach was used to select the true orthologs for each group. Grouping of these sequences according to cellular localization was further confirmed by phylogenetic tree calculations (Fig. 1 and S-Data 2). As shown in Fig. 1, most of the major nodes of the tree were strongly supported by high bootstrap values (>75%). The phylogenetic tree showed clear separations of each group as well as of the Hsp70s control sequences. Taxonomic separation of each Hsp90 group relative to Hsp70 sequences of the same group is presented in S-Data 2A–C. Neighbor Joining distance matrix method was used for this [18]. The controls as expected showed long outgroup branch proving they are distantly related to the Hsp90 proteins. Finer analysis using the distance scale also revealed that the protozoan Hsp90 proteins are located far from human Hsp90 proteins in Groups A, B and C. This shows that as compared to other parasites, they are significantly more distantly related to humans. In the absence of Hsp70s, Hsp90 Group C proteins formed an outgroup with a high bootstrap value (100%), indicating that they are the oldest Hsp90 isoforms (S-Data 2D). Group A proteins shared a more recent common ancestor with Group B proteins than with Group C, as reported previously [26], [27]. As expected, within each group of proteins, ectoparasite and helminthes clustered together with the vector and human sequences, while protozoan sequences formed their own distant clusters.
3.2. Group A proteins are more conserved than Group B and C proteins
To analyze conservation, all versus all pairwise sequence identities were calculated per group (Fig. 2), showing that Group A proteins were the most conserved with most pair identities above 60% (0.6). Group B proteins displayed most pair identities between 0.4 and 0.6, while the least conserved group was Group C. In Groups A and B, human Hsp90 proteins showed high conservation with the helminth, ectoparasite and vector Hsp90 proteins (small rectangles in Fig. 2), while protozoan Hsp90 proteins were relatively distantly related to human Hsp90 proteins and to each other (triangles in Fig. 2). These findings were also supported by phylogenetic tree results presented in Fig. 1. Interestingly, in Group A Hsp90 from Giardia intestinalis (GIN_A), which colonizes and reproduces in the small intestine causing giardiasis, had around 50% sequence identity to all other sequences (green bar in Fig. 2A matrix). This distinct separation was also observed in the phylogenetic tree.
Fig. 2.
Pairwise sequence identity calculations. Top row displays heat maps, where conservation increases from blue to red. Bottom row displays histograms where x-axis represents sequence similarity as a fraction and y-axis represents number of sequence pairs, and right column displays scores as a matrix (similarity scores for every sequence versus every sequence represented as a fraction as per the scale).
Further, within each group alignment, protein sequences were divided into five regions: namely N-terminal ATP-binding domain, linker region 1, M domain, linker region 2 and C-terminal domain (S-Data 3A). HsHsp90 sequences were used to define these regions. For N-terminal, M and C-terminal domains, again, all versus all pairwise sequence identities were calculated (S-Data 3B). Group A proteins showed a higher degree of conservation in their domains than the other groups, with the N-terminal being most conserved, followed by M domain and then C-terminal domain. This order was the same for Group B domains, though the degree of conservation in each domain was much less compared to Group A. In Group C, the N-terminal domain was more conserved than in Group B but less than in Group A; both M and C-terminal domains showed similar conservations.
The N-terminal domain contains the ATP binding site [28], and the high conservation of this domain among the groups can be linked to its functional importance. Yet, it is known that human Hsp90 proteins of organelles have subtle differences in ATPase activity, and the exact mechanism is still under investigation. TRAP1 has higher ATP affinity than the cytosolic Hsp90 [29]. Grp94 has mechanistically important differences in the interaction with ATP [30]. Although overall N terminal domain is structurally conserved, these differences in human Hsp90 proteins and those of other organisms might be attributed to the residue level variations as well as environmental factors discussed later.
The M domain is involved in client protein binding and activating ATP hydrolysis [31]. This domain is also conserved with very few gaps in Group A Hsp90 proteins. Therefore, the high level of conservation might explain why host Hsp90 proteins have been reported to associate with the parasite’s proteins [4]. Conservation was observed in Group B as well, but interestingly, the protozoan Hsp90 M domain has an approximately 30 residue extension (S-Data 3C) suggesting that a different set of substrates might bind to this domain. Furthermore, the region is rich in charged residues indicating that it is exposed at the tertiary structure level. Therefore, this unique extension is a possible site for allosteric inhibitors to selectively bind to protozoan Hsp90 proteins. The M domain of Group C had only very few gaps in the alignment. Uniquely, only the Plasmodium sp. Hsp90 proteins had small insertions.
The C-terminal shows conservation in all groups with few gaps. While the conserved motifs MEEVD (Group A) and KDEL (Group B) at the extreme carboxyl terminus were observed, no conservation was observed in Group C. However, unique patterns were observed in clusters. For instance, host and vector sequences possessed motif L[ED][KRI]H while Leishmania sp. and Trypanosoma sp. P[ST]AD[KQ]. Even though the importance of Group C carboxyl terminus motifs have not been elucidated yet, this level of conservation within the classes of organisms reveals the possibility of functional significance.
The linker regions are the least conserved regions in each group. For instance, Plasmodium sp. cytosolic Hsp90 proteins have a long insertion in the linker region 1. Studies have revealed that the charged linker region plays a crucial role in the sensitivity of ATP [32]. It was shown that PfHsp90 and Hsp90 from Trypanosoma evansi were selectively targeted using low concentrations of the geldanamycin in vivo [10]. Interestingly, T. evansi Hsp90 lacks the extended charged linker but the inhibitor in low concentrations could still bind, which shows that increased sensitivity might be due to the charged linker region in combination with effects from elsewhere in the protein. Interestingly, Group B Babesia and Trypanosoma sp. Hsp90 proteins had a much shorter linker region 1 than other Hsp90 proteins, and lack the variable region.
3.3. Environment plays a role in the overall physicochemical properties of Hsp90 proteins
The environment in cellular compartments is regulated to provide optimal activity to cellular processes. In order to analyze if Hsp90 proteins have environmental specific features, physicochemical properties were calculated for three groups (Fig. 3 and S-Data 4).
Fig. 3.
Distribution of physicochemical properties in each group presented as boxplot. Each box contains three box plots that are ordered to represent Group A, B and C. The middle line shows the central value (median). The first and third quartiles are the edges of the box where the first quartile is the value such that, 25% of the values fall at or below this value and third quartile is the value when 75% of the values fall at or above this value. Whiskers (dashed lines) indicate variability outside the quartiles and this region is calculated by adding 1.5 to the third quartile value and subtracting 1.5 to the first quartile value. The circles represent outliers, points that lie outside the whisker region and are plotted individually.
Isoelectric point calculations indicated that the Hsp90 proteins were acidic in nature (pI < 7.0). Group A proteins showed a very small range of pI (4.81–5.16). The range for Group B proteins was broader (4.45–5.88). In this group, some outliers were observed; Trypanosoma brucei (TBR_B), Plasmodium vivax (PVI_B) and Plasmodium berghei (PBE_B) Blastocystis hominis (BHO_B). Interestingly, Group C proteins had a higher and wider range of pIs (5.38–7.69) than other groups. Only Human TRAP1 (HSA_C) was found to be basic at 7.69.
Instability index measures the stability of a protein in a test tube [33]. Most of the proteins in Groups A and C had instability index higher than 40, thus the mean values of Groups A and C predicted the Hsp90 proteins to have a shorter half-life, while Group B Hsp90 proteins are stable as most of the proteins had an instability index value less than 40. B. hominis (BHO_B) had a uniquely high instability index value.
Since there is a good correlation between aliphatic index and thermostability of globular proteins [34], it can predict thermostability. Overall, Hsp90 proteins had high indices in all groups (73.5–94.0). Yet, ER and mitochondrial proteins had higher values than cytosolic proteins, predicting that Group B and C proteins are more stable at high temperature. Group A had Leishmania infantum (LIN_A) and Leishmania major (LMA_A), and Group B had again B. hominis as outliers, with high aliphatic indices indicating their increased stability at high temperatures relative to HsHsp90.
Grand average hydropathicity (GRAVY) predicts interaction of a protein with water [35], with lower values indicating better interaction. Hsp90 proteins had very low values, indicating good interaction with water. This was supported by hydrophobicity calculations indicating hydrophilic behavior. Interestingly, Group A and B HsHsp90 proteins had lower GRAVY values than parasitic Hsp90 proteins, whereas Group C protozoans were predicted to interact with water better than HsHsp90.
The data produced from the calculation of physicochemical properties in different groups was non-parametric. Thus, whether samples originated from the same distribution was tested using the Kruskal–Wallis test [20], with null hypothesis H0 (same distribution), and H1 as the alternate hypothesis. All the p-values of each property were less than 0.05 (aromaticity: 1.168e−4; instability index: 2.501e−3; hydrophobicity: 3.706e−12; isoelectric point: 9.768e−14; GRAVY: 4.029e−12; aliphatic index: 4.774e−06), therefore there was evidence to reject the null hypothesis that the three groups had the same distribution for each property. The statistical analysis showed that the distributions of the properties in each group are different, implying that the environment plays a major role in the overall properties of a protein.
Further physicochemical analysis was done on N-terminal ATP binding domains only, in each group (Fig. 4). Surprisingly, while isoelectric point, instability index and aromaticity calculations had the same trend as the full length proteins; aliphatic index, GRAVY and hydrophobicity showed almost an opposite tendency to that of the full length proteins. One interesting observation came from isoelectric point calculations. In full length protein analysis, Human TRAP1 (HSA_C) was the only basic (7.69) protein in all groups. N-terminal domain analysis, on the other hand, identified three basic outliers in Group C, namely LIN_C, LMA_C, and Naegleria gruberi (NGR_C). TRAP1 N-terminal domain was acidic (6.11) indicating that the basic feature of this protein is due to other domains.
Fig. 4.
Distribution of physicochemical properties of N-terminal ATP domain in each group presented as boxplot. Each box contains three box plots that are ordered to represent Group A, B and C. Other features are as described in Fig. 3.
3.4. There is more variability in the amino acid distribution across organisms in Groups B and C than in Group A
Fig. 5 and S-Data 5 show the occurrence frequency of each amino acid in an Hsp90 from certain organisms in different groups. Interestingly, the calculations revealed that E, K, L and D are predominant in Group A and B Hsp90 proteins, while D is replaced by S in Group C. The predominance of charged residues (E, K and D) might explain why Hsp90 proteins generally interact well with water as discussed previously. This observation also indicates client specificity of Hsp90 proteins. It was shown that surface electrostatics determine the interaction with the Hsp90 chaperone complex [36].
Fig. 5.
Occurrence frequency of amino acids in Hsp90 proteins. The occurrence frequency of amino acids in each group increases from blue to red. (A) Group A, (B) Group B and (C) Group C. The lines represent the types of organisms as shown in the key; red: protozoa, blue: helminths and black: vectors.
In general, there is an approximately uniform distribution of amino acids across organisms in Group A. On the other hand, the distribution in Groups B and C is more variable. This is particularly true for parasitic Hsp90 proteins. In Group B, the occurrence frequency of E was relatively higher in human Hsp90 than in protozoans. B. hominis was the only protozoan with E composition higher than human. The Leishmania and Plasmodium sp. had higher compositions of V and I respectively than human Hsp90. These protozoans also had relatively higher R composition. In Group C, human Hsp90 has a uniquely high composition of L and R residues which are both positively charged. The predominance of these residues is mainly the reason why the HsHsp90 has a basic nature (Fig. 4). The occurrence frequency of K was higher (around 10%) in P. falciparum, P. vivax and P. yoelii yoelii than in HsHsp90 (6.5%). Interestingly, Plasmodium sp. Hsp90 proteins had also much higher N occurrence (around 10%) than human (2.6%). On the other hand, Plasmodium sp. had very low W, C, M and Y compared to the human Hsp90.
Further, the calculations were repeated for only N-terminal ATP binding domains, and indicated that the domain amino acid compositions had approximately the same trend as for full length sequences (S-Data 5).
3.5. Serine and tyrosine are favored phosphorylation residues
Phosphorylation is an important Hsp90 post-translational modification as it influences client binding, co-chaperone interaction, and inter-domain communications [37], [38]. A number of S, T and Y phosphorylation sites have been discovered in human Hsp90 proteins with important functions [39], [40], [41]. It has also been shown that the function of P. falciparum Hsp90 depends on phosphorylation [4]. Thus better understanding of the phosphorylation site differences between parasitic and human Hsp90 proteins might be critical while targeting the protein.
Here, phosphorylation sites were calculated for each protein and mapped to the sequences in the alignment results (S-Data 6). Group A showed patterns of conservation among S and Y sites but not in T. A similar tendency was observed in Group B and C, although little conservation was detected in the C-terminal domains. Among the predicted phosphorylation sites, some have already been experimentally identified in humans [40], [42]. The experimentally determined sites of HsHsp90α were also shown in the alignment for Group A. To verify the predicted sites, a comparison was made between experimentally elucidated sites [40] and our predictions. The results were mostly in agreement (13/17 Hsp90α and 10/16 Hsp90β) (Table 1). The failed predictions were mostly in the C-terminal domain. Another review showed a strong correlation with our phosphorylation predictions except for Hsp90α t36, Hsp90β t31 and Hsp90β s206 [41]. Additionally, recently, s225 and s254 were identified experimentally as Hsp90β phosphorylation sites [43]. However, this numbering shifts one residue in our predictions (s226 and s255) which correlate with other experimental data [40], [44]. The full peptide sequence, used in Kim et al. [43] experiments, was not provided. This one residue mismatch is most probably due to omitting the first residue (Met) of the protein in their experiments, as there is only one serine residue around those positions (s226 – EKEISDDEAEE and s255 – IEDVGSDEEDDSKGDK).
Table 1.
Comparison of predicted and experimentally established human Hsp90 phosphorylation sites.
| Domain | Experimental residues | Refs. |
|---|---|---|
| N-terminal | Hsp90α – t5, t7, t36⁎, y38, s63, t65, s68, s72, t88⁎, t90, s231 | [40], [49], [37], [50], [51] |
| Hsp90β – t31⁎, y56, y192, s206⁎, s226 | ||
| Middle | Hsp90α – s252, s263, y493⁎ | [52], [53], [54], [55] |
| Hsp90β - s255, t297⁎, y300, s307⁎, s452, y484, s532⁎, y596⁎ | ||
| C-terminal | Hsp90α – y604, t725, s726 | [52], [56] |
| Hsp90β – y619⁎, s718 | ||
Shows the residues that failed to be predicted.
Interestingly, most experimentally studied sites are located in the N-terminal domain while this study predicted a global distribution. There are interesting cases where a site is predicted to be phosphorylated for most organisms, but the residue has not been investigated experimentally, e.g. HsHsp90α 113. There are also residues predicted to be phosphorylated in human and vectors, but not in protozoan parasites, e.g. HsHsp90α 184. These unique sites might be relevant to the function for specific organisms. The percentage of total number of phosphorylated residues of interest over the total number of residues of interest per group was calculated for each case, i.e. S, T and Y (Fig.6A). In Group A and B, of all the S residues, almost 50% were predicted to be phosphorylation sites, and in Group C this ratio was 43%. Y ratios were lower than S ratios but higher than T ones. This is interesting as the amino acid composition results (Fig. 5) showed that T is a more abundant residue in Hsp90 proteins than Y, yet Y was a more favored phosphorylation residue. Y phosphorylation is very important as it affects Hsp90 interactions with distinct client proteins [45], [46]. Overall, S and Y were the favored phosphorylation sites. Fig.6B presents a comparative analysis within each group.
Fig. 6.
Representation of phosphorylation sites as histogram. Frequency is represented as a percentage on the y-axis while the x-axis shows the group or protein names. (A) Ratios of phosphorylated residues as a percentage of total similar residues per group (e.g. [total number of phosphorylated serines in the group/total number of serines in the group] * 100%). The phosphorylation sites are colored accordingly; green: serine, blue: threonine and yellow: tyrosine. (B) Ratios of phosphorylated residues as a percentage of total similar residues per sequence (e.g. [number of phosphorylated serines/total number of serines per sequence] * 100%).
3.6. Group B and C have motifs unique to specific organisms
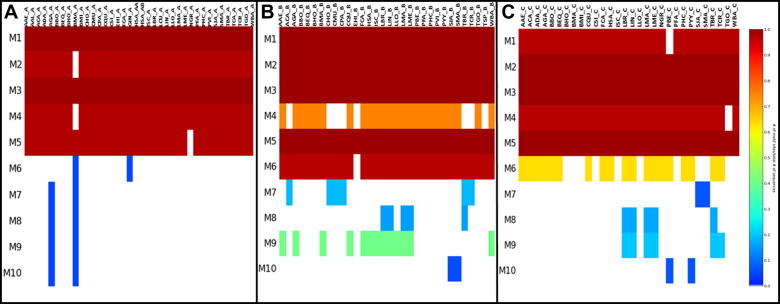
Motif conservations may reflect the functional importance of certain regions in a protein, therefore a detailed motif analysis was conducted (Fig. 7). In Group A, 16 out of 94 motifs found were highly conserved in all proteins. All domains had long conserved regions. Previous studies revealed that host chaperones associate with parasitic proteins in the early stages of infection [4] thereby suggesting human Hsp90 proteins facilitate the trafficking of parasitic proteins. The high level of conservation observed is possibly one reason for chaperone machinery hijacks by parasites. Interestingly, even though motif analysis showed that Group A proteins are highly conserved, at a residue level there are striking differences between human and protozoan parasite proteins, making those residues possible drug target sites. Recent findings [47] support this view, having identified P. falciparum specific residues in the ATP-binding pocket which makes the protein differ from the human one in protein structure and dynamics. To further analyze the residue differences, we looked at the Hsp90–Hop interface from motif conservation perspective. Hsp90 functions as part of a multi-chaperone machine. One of its most important co-chaperones is Hop protein. This co-chaperone is responsible for delivering and transferring client proteins to Hsp90 for folding. As such, besides reserving a binding site for client proteins, Hatherley et al. reported several residues on Hsp90’s surface thought to be responsible for Hop binding [12]. Analyzing the Group A motif data, we found that the 11 Hsp90–Hop interaction residues reported by Hatherley and co-workers were located in motifs 4 and 5. As is indicated in Fig.7A, these motifs were conserved in all organisms, except G. intestinalis (GIN_A). GIN_A was missing motif 5. Interestingly however, a more detailed analysis of motif 4 showed striking differences in key residues. To illustrate these findings, a homology model of human and P. falciparum cytosolic Hsp90 was calculated, and the sequence data was mapped to the structures (Fig. 8). The data shows that while the motif 4 as a whole is conserved, several residues differ between the two organisms, in particular Ala 469 and Thr 482, two of the six Hsp90–Hop interacting residues located in this motif. It thus stands to reason that designing an inhibitor that targets the highly conserved motif 4 would likely have an effect on protein function, with the key residue differences at sequence level, providing the means for the design of an inhibitor with species specificity.
Fig. 7.
Motif analysis output presented as a heat map. The colors represent conservation which increases from blue to red while white represents absence of motifs. (A) Group A; (B) Group B; (C) Group C.
Fig. 8.
Group A Hsp90 and Hop interface analysis for motif 4: pairwise alignment of motif 4 between H. sapiens and P. falciparum (A), mapped to homology models of cytosolic Hsp90 in H. sapiens (B) and P. falciparum (C). In all figures, blue represents conserved residues, green represents residue differences, and red represents Hop–Hsp90 interacting residues. Sequence numbering on pairwise alignment is for H. sapiens (residues 434–513), the corresponding residues for P. falciparum (residues 387–467).
Unlike Group A, both Group B and C proteins showed more variations. Besides conserved motifs, unique patterns of motifs specific to organisms, especially in the protozoans, were also observed (Fig.7B and C). In Group B, Leishmania sp., Babesia sp. and Plasmodium sp. had sets of motifs that were unique to themselves. In Group C, the same trend was observed as Group B. Leishmania sp. and Trypanosoma sp. had unique motifs. Surprisingly, P. falciparum lacked motifs that were conserved in P. berghei (PBE_C) and P. yoelii (PYY_C). Distinct motifs of Leishmania sp., and Plasmodium sp. that were missing in human Hsp90 were mapped to the representative models of PFA_B and LME_B (Fig. 9). Only motif 38 in PFA_B and 26 in LME_B were not included as the structures were not modeled for those regions. While the residue content and length of the motifs differed greatly, the relative correlation of their positions in the two parasitic proteins was interesting. The most notable comparison was observed for motif 18 (PFA_B, position 356–385) and 22 (LME_B, position 427–456), identified as the same length and located at a similar position in the M domain.
Fig. 9.
Unique motifs mapped to the homology models. (A) P. falciparum (top), L. mexicana (bottom). (B) Figure key. The motifs are numbered according to MEME results and colored according to their position for ease of comparison.
While the structural domains of Hsp90 are largely conserved across species, we propose that the unique species specific motif data presented here could provide the required evidence for the identification of novel selective drug target sites; especially in the instances where motifs were found to be unique to a single species. The conservation of the secondary structure of these regions suggests putative functional relevance, which in turn could be selectively targeted using computer-aided drug design methods such as molecular docking and high-throughput screening studies.
3.7. Motif analysis of the ATP binding domain also showed organism specific unique motifs
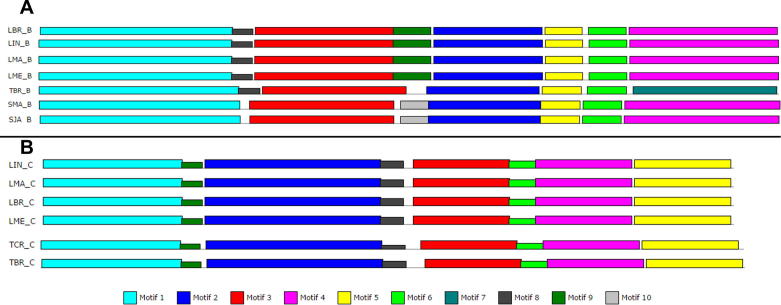
The function of Hsp90 is driven by the binding and hydrolysis of ATP at the N-terminal ATP binding domain. Given its functional importance, the ATP binding domain of Hsp90 has been investigated as a potential drug target site for the treatment of several diseases. However due to the high conservation of the domain, selectivity has been a limiting factor in the clinical trials of inhibitors such as geldanamycin and radicicol. As such a comparative analysis of motif conservation as determined from MEME analysis was carried out (Fig. 10). The ATP binding domain or Bergerat fold was identified in Hsp90’s [28], highlighting three motifs that showed conservation with the type II topoisomerases reported by [48]. These motifs were used as filtering parameters in the analysis of our N-terminal domain motifs. We found that all three motif sequences reported by Prodromou and co-workers [28] were present in our motif results (Group A, motif 1 and 2; Group B motifs 1, 2 and 3; Group C motifs 1, 2 and 3). It was interesting to note that motifs 1, 2, and 3 were completely conserved in Group B, while motifs 1 and 2 were absent in PBE_C (Group C) and Brugia malayi (BMA_A; Group A) respectively. While these motifs show clear conservation of the functionally important regions, several other motifs were identified to be unique to certain organisms. In Group A, BMA_A and Anopheles gambiae (AGA_A) had four unique motifs (7–10) that were found to be located in between the functionally important motifs 1 and 3 along the linear sequence. Motif 6 was found to be unique to BMA_A and GIN_A, located between motifs 2 and 3 in the latter case.
Fig. 10.
Motif analysis of the N-terminal domain of Hsp90 presented as a heat map. The colors represent conservation which increases from blue to red while white represents absence of motifs. (A) Group A; (B) Group B; (C) Group C.
In Group B, interesting differences were observed between human and parasitic organisms. Motif 8 was unique to the Leishmania sp. (LIN_A, LMA_A, LBR_A and LME_A) as well as to T. brucei (TBR_A). Additionally motif 10 was found to be unique to the Schistosoma sp (SMA_A and SJA_A). Here again motifs are located linearly between the functional motifs 1 and 3, and 2 and 3 respectively (Fig. 11). Using the same premise for analysis in Group C, both motif 8 and 9 were found to be specifically unique to the Leishmania species as well as to Trypanosoma cruzi and brucei (TBR_C and TCR_C). Interestingly motif 8 is located between motifs 2 and 3 while motif 9 is located between motifs 1 and 2. This analysis of all three groups’ shows that there exist certain motifs within close vicinity to functionally important subdomains in the N-terminus of Hsp90, and especially in the case of Leishmania and Trypanosoma, targeting design inhibitors to these unique motifs may provide the as yet elusive selective drug for the highly attractive ATP domain. In conclusion, this study aimed to build a profile of potential sites and features on the protein by studying the differences in sequence composition of human, vector and parasite Hsp90 proteins. Doing so, this study provided a number of novel findings. Overall, cytosolic proteins seemed more conserved in many aspects than ER and mitochondrial proteins, yet there were striking differences within each group, especially between human and protozoan parasites. In general, the results supported the view that Hsp90 proteins are interesting potential drug targets especially for protozoan parasitic diseases. We believe that our findings provided a suitable platform and starting point for further in silico and wet-lab experiments.
Fig. 11.
Snap shot of unique motifs to species from MEME motif analysis. (A) Unique motifs in Group B. Motif 8 unique to the Leishmania sp. and Trypanosoma brucei, wedged in between motif 1 and 3. Motif 10 unique to the Schistosoma sp., located between motifs 2 and 3. (B) Unique motifs in Group C. Motifs 8 and 9 were uniquely conserved in the Leishmania species and Trypanosoma cruzi and brucei, being located between motifs 1 and 2 for the former and between 2 and 3 for the latter.
Author contributions
Ö.T.B. conceived and designed the project; N.F. and D.L.P. acquired the data; N.F., D.L.P. and Ö.T.B. analyzed and interpreted the data, and wrote the paper.
Acknowledgements
N.F. and D.L.P. thank Rhodes University for financial support. This work is partially supported by the National Research Foundation of South Africa under Grant No. 93690. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the funder. Authors thank Gregory L. Blatch for his valuable comments.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.fob.2015.11.003.
Appendix A. Supplementary data
List of all the sequences used in this study. A: List of the human Hsp90 proteins with their isoforms. The location of the proteins in the cell is also indicated. B shows the parasitic and host sequences that were retrieved from the NCBI database using the human Hsp90 as query. C: Hsp70 sequences. The location of the proteins in the cell is also indicated. Sequences are colored in tables according to their groups. Blue: protozoa, red: helminths, green: ectoparasite and black: vectors.
Phylogenetic tree calculations. A: Group A, B: Group B, C: Group C, D: All sequences together. The tree building methods used were Neighbor Joining distance matrix for A, B, and C and Maximum Likelihood for D. Using Le_Gascuel_2008 model to calculate trees, the discrete Gamma distribution was used to model evolutionary rate differences among sites. The rate variation model allowed some sites to be evolutionarily invariable specifically A: (5 categories (+G, parameter = 3.6824)) and ([+I], 0.7609% sites); B: (5 categories (+G, parameter = 0.9723)) and ([+I], 0.0000% sites); C: (5 categories (+G, parameter = 1.1790)) and ([+I], 0.0000% sites); D: (5 categories (+G, parameter = 3.6591)) and ([+I], 1.0988% sites) [18]. Color representation SD-2D: Branches; red: Group A, black: Group B and blue: Group C. Sequences; black: vectors (bold italics: hosts), pink: ectoparasite, green: helminthes and maroon: protozoa.
Multiple sequence analysis of Hsp90 proteins. S2-A: Sequence identity calculation of individual domains in Groups A, B and C. S2-B: Multiple sequence alignments of Groups A, B and C. The conserved residues are colored blue in all the groups while the black vertical lines separate domains in the alignment. S2-C: A. Unique region in the middle domain of Group B Hsp90 proteins. The blue highlighting indicates residues conserved throughout the sequences and the red boundary shows the region dissimilar to the non-protozoan sequences. B. A schematic representation of the protozoan Hsp90 indicating its unique middle domain. The arrow shows the insertion. N: N-terminal domain, CL: charged linker, M: middle domain and C: C-terminal domain.
Physicochemical properties calculations using the Biopython module. A: Group A, B: Group B, C: Group C, D: Group A-ATP domain, E: Group B-ATP domain, F: Group C-ATP domain. The background colors represent classification of organisms. Gray: human and host, blue: protozoa, green: helminths and white: ectoparasite.
Amino acid occurrence frequencies of Hsp90 proteins. [Count of a particular residue/sequence length] * 100% A: Group A, B: Group B and C: Group C, D: Group A-ATP domain, E: Group B-ATP domain and F: Group C-ATP domain.
Multiple sequence alignment of all sequences showing positions of predicted phosphorylation sites. A Python script was used to extract information from the Netphos text file and map the residues on the alignment. A: Group A. The numbers above the top row represent the human Hsp90α residues that were experimentally identified. B: Group B, C: Group C. The colors represent, Blue: serine residues, Red: tyrosine residues, Black: threonine residues.
References
- 1.Monzote L., Siddiq A. Drug development to protozoan diseases. Open Med. Chem. J. 2011;5:1–3. doi: 10.2174/1874104501105010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pink R., Hudson A., Mouriès M.-A., Bendig M. Opportunities and challenges in antiparasitic drug discovery. Nat. Rev. Drug Discov. 2005;4:727–740. doi: 10.1038/nrd1824. [DOI] [PubMed] [Google Scholar]
- 3.Roy N., Nageshan R.K., Ranade S., Tatu U. Heat shock protein 90 from neglected protozoan parasites. Biochim. Biophys. Acta. 2012;1823:707–711. doi: 10.1016/j.bbamcr.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Banumathy G., Singh V., Pavithra S.R., Tatu U. Heat shock protein 90 function is essential for Plasmodium falciparum growth in human erythrocytes. J. Biol. Chem. 2003;278:18336–18345. doi: 10.1074/jbc.M211309200. [DOI] [PubMed] [Google Scholar]
- 5.Chiang A.N., Valderramos J., Balachandran R., Chovatiya R.J., Mead B.P., Schneider C. Bioorganic and medicinal chemistry select pyrimidinones inhibit the propagation of the malarial parasite, Plasmodium falciparum. Bioorg. Med. Chem. 2009;17:1527–1533. doi: 10.1016/j.bmc.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prohászka Z., Csermely P., Schnaider T., Csaba S., Nardai G., Amily M.O.C.H.F. The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharm. Ther. 1998;79:129–168. doi: 10.1016/s0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- 7.Felts S.J. The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J. Biol. Chem. 2000;275:3305–3312. doi: 10.1074/jbc.275.5.3305. [DOI] [PubMed] [Google Scholar]
- 8.Dobson C.M., Karplus M. The fundamentals of protein folding: bringing together theory and experiment. Curr. Opin. Struct. Biol. 1999;9:92–101. doi: 10.1016/s0959-440x(99)80012-8. (http://www.ncbi.nlm.nih.gov/pubmed/10047588) [DOI] [PubMed] [Google Scholar]
- 9.Louvion J.F., Warth R., Picard D. Two eukaryote-specific regions of Hsp82 are dispensable for its viability and signal transduction functions in yeast. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13937–13942. doi: 10.1073/pnas.93.24.13937. (http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=19473&tool=pmcentrez&rendertype=abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pallavi R., Roy N., Nageshan R.K., Talukdar P., Pavithra S.R., Reddy R. Heat shock protein 90 as a drug target against protozoan infections. Biochemical characterization of Hsp90 from Plasmodium falciparum and Trypanosoma evansi and evaluation of its inhibitor as a candidate. J. Biol. Chem. 2010;285:37964–37975. doi: 10.1074/jbc.M110.155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shahinas D., Liang M., Datti A., Pillai D.R. A repurposing strategy identifies novel synergistic inhibitors of Plasmodium falciparum heat shock protein 90. J. Med. Chem. 2010;53:3552–3557. doi: 10.1021/jm901796s. [DOI] [PubMed] [Google Scholar]
- 12.Hatherley R., Clitheroe C.-L., Faya N., Tastan Bishop Ö. Plasmodium falciparum Hop: detailed analysis on complex formation with Hsp70 and Hsp90. Biochem. Biophys. Res. Commun. 2015;456:440–445. doi: 10.1016/j.bbrc.2014.11.103. [DOI] [PubMed] [Google Scholar]
- 13.Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimmic M.W., Rest J.S., Mindell D.P., Goldstein R.A. RtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J. Mol. Evol. 2002:65–73. doi: 10.1007/s00239-001-2304-y. [DOI] [PubMed] [Google Scholar]
- 17.Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 18.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. (http://mbe.oxfordjournals.org/content/4/4/406.short) [DOI] [PubMed] [Google Scholar]
- 19.Le S.Q., Gascuel O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 20.Kruskal W.H., Wallis W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 2007;47:583–621. [Google Scholar]
- 21.Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 22.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Biol. Chem. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 23.Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto M., Takahashi Y., Inano K., Horigome T., Sugano H. Characterization of the hydrophobic region of heat shock protein 90. J. Biochem. 1991;110:141–145. doi: 10.1093/oxfordjournals.jbchem.a123532. (http://www.ncbi.nlm.nih.gov/pubmed/1939021) [DOI] [PubMed] [Google Scholar]
- 26.Chen B., Piel W.H., Gui L., Bruford E., Monteiro A. The HSP90 family of genes in the human genome: insights into their divergence and evolution. Genomics. 2005;86:627–637. doi: 10.1016/j.ygeno.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Tastan Bishop Ö., Edkins A.L., Blatch G.L. Sequence and domain conservation of the coelacanth Hsp40 and Hsp90 chaperones suggests conservation of function. J. Exp. Zool. B. Mol. Dev. Evol. 2014;322:359–378. doi: 10.1002/jez.b.22541. [DOI] [PubMed] [Google Scholar]
- 28.Prodromou C., Roe S.M., O’Brien R., Ladbury J.E., Piper P.W., Pearl L.H. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 29.Leskovar A., Wegele H., Werbeck N.D., Buchner J., Reinstein J. The ATPase cycle of the mitochondrial Hsp90 analog Trap1. J. Biol. Chem. 2008;283:11677–11688. doi: 10.1074/jbc.M709516200. [DOI] [PubMed] [Google Scholar]
- 30.Frey S., Leskovar A., Reinstein J., Buchner J. The ATPase cycle of the endoplasmic chaperone Grp94. J. Biol. Chem. 2007;282:35612–35620. doi: 10.1074/jbc.M704647200. [DOI] [PubMed] [Google Scholar]
- 31.Pearl L.H., Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 32.Hainzl O., Lapina M.C., Buchner J., Richter K. The charged linker region is an important regulator of Hsp90 function. J. Biol. Chem. 2009;284:22559–22567. doi: 10.1074/jbc.M109.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guruprasad K., Reddy B.V.B., Pandit M.W. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990;4:155–161. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- 34.Ikai A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980;88:1895–1898. (http://www.ncbi.nlm.nih.gov/pubmed/7462208) [PubMed] [Google Scholar]
- 35.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 36.Citri A., Harari D., Shohat G., Ramakrishnan P., Gan J., Lavi S. Hsp90 recognizes a common surface on client kinases. J. Biol. Chem. 2006;281:14361–14369. doi: 10.1074/jbc.M512613200. [DOI] [PubMed] [Google Scholar]
- 37.Ogiso H., Kagi N., Matsumoto E., Nishimoto M., Arai R., Shirouzu M. Phosphorylation analysis of 90 kDa heat shock protein within the cytosolic arylhydrocarbon receptor complex. Biochemistry. 2004;43:15510–15519. doi: 10.1021/bi048736m. [DOI] [PubMed] [Google Scholar]
- 38.Li J., Richter K., Buchner J. Mixed Hsp90-cochaperone complexes are important for the progression of the reaction cycle. Nat. Struct. Mol. Biol. 2011;18:61–66. doi: 10.1038/nsmb.1965. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Y.G., Gilmore R., Leone G., Coffey M.C., Weber B., Lee P.W. Hsp90 phosphorylation is linked to its chaperoning function. Assembly of the reovirus cell attachment protein. J. Biol. Chem. 2001;276:32822–32827. doi: 10.1074/jbc.M105562200. [DOI] [PubMed] [Google Scholar]
- 40.Mollapour M., Tsutsumi S., Neckers L. Hsp90 phosphorylation, Wee1 and the cell cycle. Cell Cycle. 2010;9:2310–2316. doi: 10.4161/cc.9.12.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mollapour M., Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim. Biophys. Acta. 2012;1823:648–655. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mollapour M., Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. BBA – Mol. Cell Res. 1823;2012:648–655. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S.W., Hasanuzzaman M., Cho M., Heo Y.R., Ryu M.-J., Ha N.-Y. Casein kinase 2 (CK2)-mediated phosphorylation of Hsp90β as a novel mechanism of rifampin-induced MDR1 expression. J. Biol. Chem. 2015;290:17029–17040. doi: 10.1074/jbc.M114.624106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lees-Miller S.P., Anderson C.W. Two human 90-kDa heat shock proteins are phosphorylated in vivo at conserved serines that are phosphorylated in vitro by casein kinase II. J. Biol. Chem. 1989;264:2431–2437. [PubMed] [Google Scholar]
- 45.Adinolfi E., Kim M., Young M.T., Di Virgilio F., Surprenant A. Tyrosine phosphorylation of HSP90 within the P2X7 receptor complex negatively regulates P2X7 receptors. J. Biol. Chem. 2003;278:37344–37351. doi: 10.1074/jbc.M301508200. [DOI] [PubMed] [Google Scholar]
- 46.Brouet A., Sonveaux P., Dessy C., Moniotte S., Balligand J.-L., Feron O. Hsp90 and caveolin are key targets for the proangiogenic nitric oxide-mediated effects of statins. Circ. Res. 2001;89:866–873. doi: 10.1161/hh2201.100319. [DOI] [PubMed] [Google Scholar]
- 47.Wang T., Bisson W.H., Maser P., Scapozza L., Picard D. Differences in conformational dynamics between Plasmodium falciparum and human Hsp90 orthologues enable the structure based discovery of pathogen-selective inhibitors. J. Med. Chem. 2014;57:2524–2535. doi: 10.1021/jm401801t. [DOI] [PubMed] [Google Scholar]
- 48.Bergerat A., de Massy B., Gadelle D., Varoutas P.C., Nicolas A., Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 49.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 50.Rose D.W., Wettenhall R.E., Kudlicki W., Kramer G., Hardesty B. The 90-kilodalton peptide of the heme-regulated eIF-2 αkinase has sequence similarity with the 90-kilodalton heat shock protein. Biochemistry. 1987;26:6583–6587. doi: 10.1021/bi00395a003. [DOI] [PubMed] [Google Scholar]
- 51.Lei H., Venkatakrishnan A., Yu S., Kazlauskas A. Protein kinase A-dependent translocation of Hsp90α impairs endothelial nitric-oxide synthase activity in high glucose and diabetes. J. Biol. Chem. 2007;282:9364–9371. doi: 10.1074/jbc.M608985200. [DOI] [PubMed] [Google Scholar]
- 52.Dephoure N., Zhou C., Villén J., Beausoleil S.A., Bakalarski C.E., Elledge S.J., Gygi S.P. A quantitive atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C., Chen J. Phosphorylation and hsp90 binding mediate heat shock stabilization of p53. J. Biol. Chem. 2003;278:2066–2071. doi: 10.1074/jbc.M206697200. [DOI] [PubMed] [Google Scholar]
- 54.Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., Hurov K.E., Luo J. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 55.Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 56.Albuquerque C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all the sequences used in this study. A: List of the human Hsp90 proteins with their isoforms. The location of the proteins in the cell is also indicated. B shows the parasitic and host sequences that were retrieved from the NCBI database using the human Hsp90 as query. C: Hsp70 sequences. The location of the proteins in the cell is also indicated. Sequences are colored in tables according to their groups. Blue: protozoa, red: helminths, green: ectoparasite and black: vectors.
Phylogenetic tree calculations. A: Group A, B: Group B, C: Group C, D: All sequences together. The tree building methods used were Neighbor Joining distance matrix for A, B, and C and Maximum Likelihood for D. Using Le_Gascuel_2008 model to calculate trees, the discrete Gamma distribution was used to model evolutionary rate differences among sites. The rate variation model allowed some sites to be evolutionarily invariable specifically A: (5 categories (+G, parameter = 3.6824)) and ([+I], 0.7609% sites); B: (5 categories (+G, parameter = 0.9723)) and ([+I], 0.0000% sites); C: (5 categories (+G, parameter = 1.1790)) and ([+I], 0.0000% sites); D: (5 categories (+G, parameter = 3.6591)) and ([+I], 1.0988% sites) [18]. Color representation SD-2D: Branches; red: Group A, black: Group B and blue: Group C. Sequences; black: vectors (bold italics: hosts), pink: ectoparasite, green: helminthes and maroon: protozoa.
Multiple sequence analysis of Hsp90 proteins. S2-A: Sequence identity calculation of individual domains in Groups A, B and C. S2-B: Multiple sequence alignments of Groups A, B and C. The conserved residues are colored blue in all the groups while the black vertical lines separate domains in the alignment. S2-C: A. Unique region in the middle domain of Group B Hsp90 proteins. The blue highlighting indicates residues conserved throughout the sequences and the red boundary shows the region dissimilar to the non-protozoan sequences. B. A schematic representation of the protozoan Hsp90 indicating its unique middle domain. The arrow shows the insertion. N: N-terminal domain, CL: charged linker, M: middle domain and C: C-terminal domain.
Physicochemical properties calculations using the Biopython module. A: Group A, B: Group B, C: Group C, D: Group A-ATP domain, E: Group B-ATP domain, F: Group C-ATP domain. The background colors represent classification of organisms. Gray: human and host, blue: protozoa, green: helminths and white: ectoparasite.
Amino acid occurrence frequencies of Hsp90 proteins. [Count of a particular residue/sequence length] * 100% A: Group A, B: Group B and C: Group C, D: Group A-ATP domain, E: Group B-ATP domain and F: Group C-ATP domain.
Multiple sequence alignment of all sequences showing positions of predicted phosphorylation sites. A Python script was used to extract information from the Netphos text file and map the residues on the alignment. A: Group A. The numbers above the top row represent the human Hsp90α residues that were experimentally identified. B: Group B, C: Group C. The colors represent, Blue: serine residues, Red: tyrosine residues, Black: threonine residues.