Abstract
Background
Chronic psychological stress is associated with enhanced abdominal pain and altered intestinal barrier function that may result from a perturbation in the hypothalamic–pituitary–adrenal (HPA) axis. The glucocorticoid receptor (GR) exploits diverse mechanisms to activate or suppress congeneric gene expression, with regulatory variation associated with stress-related disorders in psychiatry and gastroenterology.
Purpose
During acute and chronic stress, corticotropin-releasing hormone (CRH) drives secretion of adrenocorticotropic hormone (ACTH) from the pituitary, ultimately leading to the release of cortisol (human) and corticosterone (rodent) from the adrenal glands. Cortisol binds with the GR in the cytosol, translocates to the nucleus, and activates the NR3C1 (nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)) gene. This review focuses on the rapidly developing observations that cortisol is responsible for driving circadian and ultradian bursts of transcriptional activity in the CLOCK (clock circadian regulator) and PER (period circadian clock 1) gene families, and this rhythm is disrupted in major depressive disorder, bipolar disorder, and stress-related gastrointestinal and immune disorders. GR regulates different sets of transcripts in a tissue-specific manner, through pulsatile waves of gene expression that includes occupancy of glucocorticoid response elements located within constitutively open spatial domains in chromatin. Emerging evidence supports a potentially pivotal role for epigenetic regulation of how GR interacts with other chromatin regulators to control the expression of its target genes. Dysregulation of the central and peripheral GR regulome has potentially significant consequences for stress-related disorders affecting the brain–gut axis.
Keywords: chronic stress, CLOCK genes, epigenomics, intestinal barrier function, irritable bowel syndrome, mental health disorders
Background
The human glucocorticoid receptor α isoform, herein referred to as GR, is responsible for the maintenance of physiological homeostasis. GR has been implicated in the etiology of a wide range of stress-related disorders. Molecular mechanisms underlying GR gene expression are widespread and diverse in the human genome. They include opportunistic nucleosome occupancy, preprogramming of chromatin to permit indiscriminate binding of steroid receptors, and distal enhancer-promoter looping to transcriptionally poised genes (1, 2, 3, 4, 5, 6, 7, 8, 9). The GR controls target genes through long-distance looping to promoters within the three-dimensional milieu of euchromatin (10, 11, 12). A range of cooperative interactions with trophic factors, coactivators, corepressors, and other nuclear receptors, including brain-derived neurotrophic factor (BDNF), are important in GR regulation (13). Although greater than 1% of all transcripts in the human genome are regulated by the GR, circumscribed cell- and tissue-type expression is a prominent characteristic of glucocorticoid regulation.
Allele-specific bias is not uncommon among genes that are glucocorticoid responsive, including asymmetric expression of targets resulting from allele-specific methylation and parent-of-origin of monoallelic effects (14). In brain, studies in humans and in rodent models show that chronic stress and early life adversity disrupts normal ultradian GR pulsatile chromatin interactions during critical periods of development in an allele-specific manner. This stress-induced disruption has been associated with psychiatric disorders which persists and may be transmitted through maternal or paternal alleles across generations (15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26). Similarly, chronic stress and altered regulation of GR expression and function have been implicated in irritable bowel syndrome, intestinal barrier dysfunction, inflammatory bowel diseases, and alterations in the gut microbiome (27, 28, 29, 30).
Epigenetic modifications are known to play an important role in chronic stress–associated altered gene transcription (31). Epigenetics refers to external modifications to DNA and histones affecting gene transcription that are independent of DNA sequence, but are often driven by DNA sequence variation. One example of an epigenetic change is DNA methylation, which involves the addition of a methyl group to specific DNA sites, thereby preventing the expression of specific genes. Another example is histone modification. DNA wraps around histone proteins to form compact DNA–histone complexes. Modifications that relax the DNA–histone compaction state, such as acetylation at specific histone sites, allow accessibility to proteins that “read” genes and, thereby, promote gene transcription (32).
Insights from the central nervous system and psychiatry
Expression quantitative trait loci (eQTLs) are genomic loci that regulate gene expression. eQTLs may act in cis (locally) or trans (at a distance) to a gene. Cis-eQTLs, methylation-quantitative trait loci (meQTLs), and QTLs associated with the histone mark H3K27ac, which indicates enhancer and promoter sites of active gene regulation, are significantly enriched in domains containing the GR response element (GRE) (33). These domains are targeted by cis-regulatory, transcription factor–enriched sites within the human NR3C1 gene that contain individual SNPs, and elsewhere, depending on cell type (3, 34). Chromatin remodeling, which can be observed near GR-regulated genes using live cell imaging immediately after treatment with the synthetic glucocorticoid dexamethasone, occurs with a well-defined ultradian rhythm (6, 9, 35). Numerous studies have shown that loss of data about the chromatin environment of the human NR3C1 gene limits understanding of the clinical consequences of genetic variation (36, 37, 38). In the context of genetic association studies, one confounding problem has been the influence of population structure, as the allele frequency of a given variant, but also haplotype structure, varies widely depending on ethnicity (39).
Most studies show that abuse, neglect, and/or lack of parenting during early childhood are associated with blunted cortisol cycling in adolescence (21, 40, 41); although the directionality of effect is still controversial as a consequence of variability in study designs (42). Blunted cortisol and ACTH responses are associated with major depressive disorder, especially in females (43). In humans, chronic stress of the mother while the infant is in utero, and early life stress between birth and 5 years of age has been shown to affect DNA methylation of specific sites within the NR3C1 gene (25, 26, 44). It has been proposed that there is a critical period during child development when abuse and trauma can exert an irreversible lifelong and transgenerational impact (24). Hyper-suppression of plasma cortisol is a feature of adult patients with major depressive disorder and/or posttraumatic stress disorder who have been abused as children. The flattening of the normal circadian cortisol curve in women who were abused as children suggests a mechanism that involves genomic variants in critical genes that govern NR3C1 gene expression interacting with epigenetic modifications to disrupt regulated GR periodicity. For example, data suggest that early childhood trauma interacts with a risk allele in the FKBP5 (FK506 binding protein 5) gene that encodes a molecular chaperone of the GR. Early childhood stress caused by sexual or physical abuse or neglect, especially between the ages of 0–5 years, results in demethylation of CpG sites within intron 7 of the FKBP5 gene, leading to overexpression of FKBP5, GR resistance, and dysregulation of the central GR regulome (14). This may lead to flattening of the diurnal plasma cortisol curve in such individuals and put them at greater risk of developing major depressive disorder, posttraumatic stress, and functional bowel disorders later in life. Further research is needed to understand the molecular substrate of these phenomena.
Rhythmic patterning of gene expression
The GR is the major central nervous system determinant of metabolic rhythmicity in humans. Through the hypothalamic–pituitary–adrenal (HPA) axis, the GR regulates peripheral glucocorticoids, such as cortisol (45, 46). Mouse and human studies show that about 10% of all genes exhibit diurnal rhythmicity. Neurons of the suprachiasmatic nucleus of the hypothalamus are the only cell type known to exhibit endogenous circadian rhythmicity in the absence of all inputs (47), although when these neurons are dissociated in mutant Cry1 −/− knockout mice, their endogenous rhythms become disrupted and asynchronous. In peripheral tissues, inputs such as light, temperature, and metabolic signaling reset various rhythms (48, 49, 50, 51, 52, 53). Peripheral tissue clocks regulate a variety of metabolic processes, including lipogenesis, insulin secretion, glucose clearance, lipid accumulation, and food absorption (49). Circadian clocks are based on feedback loops that involve a heterodimer of the transcription factors CLOCK (circadian locomotor output cycles kaput) and brain and muscle ARNTL (aryl hydrocarbon receptor nuclear translocator–like; formally known as brain and muscle ARNT-like 1 [BMAL1]), which program transcriptional dynamics of rhythmicity, including those of the period (PER1, PER2, and PER3) and cryptochrome (CRY1 and CRY2) gene families. Maintenance of the circadian and ultradian rhythmicity of cortisol release appears to be regulated through acetylation and deacetylation of the GR by the CLOCK gene (46, 54, 55), and through CRY1 and CRY2 rhythmic repression of GR expression in tissues, such as the liver (56). Disruption of the periodicity of GR expression is associated with disease states characterized by glucocorticoid resistance or sensitivity (57, 58). The periodicity of GR function is important based on two observations: 1. During acute physiological states, such as the stress response, the need for rapid transactivation or transrepression of GR-regulated gene expression, mediated by the HPA axis, may explain the presence of preassembled, spatial chromatin domains where GR can bind in seconds (6, 9, 12, 33, 37, 38, 57, 59, 60, 61), and 2. Cortisol release exhibits both ultradian and circadian rhythmicity, and the temporal dynamics of GR binding to nuclear sites follows the circadian cycle as visualized using microscopy in living cells (6, 9, 61). GR chromatin immunoprecipitation sequencing (ChIP-Seq) studies confirm that the GR occupies existing DNase I hypersensitive sites, allowing for rapid binding to GREs and inverted repeat negative GREs (62). DNase I hypersensitivity indicates open chromatin, and is a primary indicator of sites of transcriptional regulation. Although the GR initiates the opening of closed chromatin, along with the recruitment of other factors that can assist in “tethered” binding, similar to the androgen receptor (63), it also interacts with specific transcripts that are transcriptionally poised, acting through long-distance “looping” (11, 12).
Recent studies show that chronic exposure to cortisol in human cell lines or corticosterone in mouse mammary epithelial cells unmasks GR-binding sites not evident after pulsatile steroid exposure (64). This may help explain why chronic stress in humans, which is accompanied by continuous cortisol secretion, acts to blunt healthy cortisol-mediated GR responses later in life, compared to pulsatile increases in hormone levels in blood that are a normal component of the acute stress response (65, 66, 67). Thus, in a murine cell model involving a 60-min exposure to corticosterone, ChIP-Seq demonstrated the presence of thousands of unique GR-binding sites, not found after 60-min pulses of hormone (64). Additionally, the Per1 transcription factor was more significantly associated with chronic exposure than Hes1 (Hes family bHLH transcription factor 1), which was associated with pulsatile exposure (64). In humans, this may be related to the disruption of circadian, ultradian, and seasonal sleep habits associated with a variety of diseases.
The complexity of NR3C1 gene transcriptional regulation
The NR3C1 promoter lacks a consensus TATA box and CCAAT motif, although it contains binding sites for transcription factors, such as AP1 (activator protein 1), SP1 (specificity protein 1), nuclear factor-κ (NFKB1), CREB (cAMP response element–binding protein), EGR1/NGF1A (early growth response 1/nerve growth factor 1), and others. The CLOCK gene has inherent histone acetyltransferase activity, and acetylates lysine moieties located within the hinge region of the GR, a lysine cluster containing a KXKK motif, and represses NR3C1 transcriptional activity with a diurnal rhythm (46, 54, 55). Neurotrophins, such as NGF (nerve growth factor) and BDNF (brain-derived neurotrophic factor), also regulate NR3C1 transcriptional activity, at least in animal models. For example, rat cortical neurons treated with both BDNF and dexamethasone produced a unique set of GR-regulated genes associated with neuronal growth and differentiation (13). The BDNF receptor NTRK2 (neurotrophic tyrosine kinase, receptor, type 2) increased GR induction of these genes, whereas BDNF has been shown to phosphorylate serine within the N-terminus of the GR, increasing nucleosome occupancy and cofactor recruitment at glucocorticoid binding sites (13). Methylation of the exon 1F promoter of the NR3C1 gene is variable, but hyper-methylation is associated with: 1. blunted cortisol response in adults who have experienced childhood loss (68), 2. increased salivary cortisol response in the children of mothers who were depressed during pregnancy and, 3. late gestational age if this methylation occurs in the fetus (69). Hyper-methylation of NR3C1 has also been found in postmortem brain tissue of suicide victims with a history of child abuse (70, 71). Ultra-rapid negative feedback of NR3C1 expression is mediated by FKBP5, which can lead to sustained repression of NR3C1 expression through hyper-methylation of the 1F promoter, concomitant with abnormal disease states associated with glucocorticoid resistance (72, 73). A functional nGRE in exon 6 of the NR3C1 gene and a long-range interaction occurring between this intragenic response element and the transcription start site are instrumental in this repression. This auto-regulatory mechanism of repression shows that the GR concentration can coordinate repression with excess ligand, regardless of the combinatorial associations of tissue-specific transcription factors. Consequently, the chronic nature of inflammatory conditions involving long-term glucocorticoid administration may lead to constitutive repression of NR3C1 gene transcription, and thus to glucocorticoid resistance. This is one of several long-distance, epigenetic mechanisms used to stop expression of NR3C1 and its target genes (10, 11, 12).
Chromatin dynamics
Upon entry into the nucleus, the GR diffuses throughout chromatin and opportunistically finds poised spatial domains of open chromatin, as defined by DNase I hypersensitivity and chromatin conformation capture (5, 8, 12). The GR also modifies chromatin in these domains, rapidly cycling in and out of GREs, and recruiting other cofactors (59, 60). Trans-repression can be exerted by three mechanisms: 1. through “negative” palindromic GREs characterized by inverted repeats (IR nGREs), which act by direct binding of glucocorticoid agonist–liganded GR, producing a silencing repressor complex through the association of NCOR2 (nuclear receptor corepressor 2) complex corepressors and histone deacetylases (7), 2. the GR forms complexes with molecules, such as histone H3 lysine 9 methyltransferase G9a, which apparently can act as a molecular scaffold in modulating the periodicity of a subset of GR-regulated genes (74), and 3. indirect suppression using corepressors, such as GRIP1 (75). Euchromatin is characterized by DNase 1 hypersensitivity and specific combinations of histone marks that define active genomic regulatory elements, such as promoters H3K4me3 and H3K27ac, and enhancers H3K4me1 and H3K27ac. An enhancer can either increase or decrease transcription. Recent research demonstrates that, in brain, the DNA sequence CAC is a common site of methylation, in contrast to other tissues, where CpG is most often methylated (76). Following activation by endogenous cortisol in humans or by exogenous glucocorticoids, such as dexamethasone, GR protein is rapidly expressed and binds to molecular co-chaperones, such as heat shock protein (hsp90), p23 (prostaglandin E synthase 3-cytosolic), and the immunophilin FKBP5, which are important for its translocation to the nucleus. GR protein also participates in a process called assisted loading at some sites in chromatin (61), where it transiently increases accessibility to closed nucleosomes, enabling other nuclear receptors, such as the estrogen receptor, to bind at the same GRE, in what has been called a hit-and-run cycle (5, 9). Research has shown that GR binding to open chromatin, as defined by DNase I hypersensitivity, can be defined as: 1. opportunistic binding to spatial clusters of constitutively open chromatin that can also be used by other transcription factors, 2. GR binding to transcriptionally poised genes and transcripts located within already DNase I–hypersensitive sites, and 3. reprogramming of closed chromatin to open chromatin by activated GR bound to glucocorticoid, which probably occurs at the most conserved GRE motifs (62, 77). The transcription factor AP1 is present at all of these GRE-binding sites (77).
Long-range interactions that control GR-regulated genes by three-dimensional looping
The architecture of the three-dimensional genome provides the basis for the regulation of gene expression, bringing enhancers and promoters into close proximity, although they may be far apart in linear sequence (78). Thousands of significant long-range looping interactions have been found to link gene promoters and distal loci, reinforcing the notion that many, if not all, gene promoters engage with distal elements using a three-dimensional loop of chromatin that confers spatial proximity, as detected by chromatin conformation capture (10). A common class of long-range interactions mediated by the GR involves the looping of promoters to sites bound by the insulator protein CTCF, the CCCTC-binding factor (zinc finger protein) whose gene (CTCF) is not only highly regulated by glucocorticoids (11, 12) but exhibits significant allele-specific methylation in humans (79, 80, 81, 82, 83, 84, 85, 86). One example of glucocorticoid-mediated looping is trans-repression of the NR3C1 gene through intragenic auto-regulation (4). In some examples of tissue-specific transcription induced by glucocorticoids, there is evidence that looping between GREs and their target genes takes place in a hormone-independent manner. For example, using associated chromatin trapping, lipocalin 2 (LCN2) was found to be looped to a GRE upstream of the Cip1-interacting zinc finger protein (CIZ1) gene prior to treatment with dexamethasone, although both genes are regulated by the GR (12). The loop structure is absent in pituitary (AtT-20) cells, in which glucocorticoid induction of these genes does not occur (12).
Cell and tissue-specific regulation of target genes by the GR
There is no distinct set of GR-regulated genes in the human genome; instead, results from many studies show that these genes tend to be tissue-specific, at least in human and animal cell lines (87, 88). Although some studies have attempted to define all of the GR-regulated or glucocorticoid-sensitive genes or transcripts in the whole genome, detailed comparison of GR-regulated genes from different tissues, species, and cell types shows negligible overlap. However, there is considerable evidence from both rodent and human studies that CLOCK, PER, and CRY gene families are highly regulated by nuclear receptors and constitute part of a master transcriptional system of peripheral circadian rhythmicity in humans.
The GR regulates the expression of restricted sets of tissue-specific transcripts in mouse and rat hippocampus, dorsal root ganglion (DRG) neurons, human brain, human liver, and various cell lines, as well as differences following maternal care in the mouse Nr3c1 gene in different tissues and cell types, as determined by assessing DNA methylation by sequencing. For example, in liver, Grøntved et al (88) showed that for both mouse and human, C/EBP (CCAAT/enhancer–binding protein) maintains chromatin accessibility and enables GR recruitment to GREs and IR GREs in most GR-binding sites. C/EBP is highly regulated by glucocorticoids in liver but not brain (87, 88). The ubiquitous CTCF insulator, sometimes acting in concert with cohesion, broadly defines the boundaries of specific gene bodies, as well as topologically associated domains (TADs), which may be regulated by an interaction between the GR and transcriptionally poised genes, similar to enhancer-regulated genes (11). In addition, certain genes, such as MMTV (mouse mammary tumor virus) and FKBP5 are transcriptionally poised for specific GR control, fitting into a model of tissue-specific gene expression (11, 89, 90, 91, 92).
In the mammalian central nervous system, responsiveness to glucocorticoids appears to be mediated by both the mineralocorticoid receptor and the GR in some cells of the hippocampus (93). However, although the mineralocorticoid receptor is expressed in many tissues, such as the kidney, colon, heart, hippocampal formation, brown adipose tissue, and sweat glands, its expression is often protected from glucocorticoid activation through co-localization of HSD11K (11-beta-dehydrogenase isozyme 2), a GR-regulated gene, whose product inactivates cortisol to cortisone (94, 95, 96, 97, 98). In the context of inflammatory disease, GR acts in a reciprocal manner with NFKB1, a phenomenon that has been well-characterized; although a genome-wide coactivation analysis shows that GRE and NFKB1 bind only in a fraction of GR- and p65-(NF-κβ protein 65) binding sites (78). Studies of neuronal PC-12 cells suggest that greater than 40% of GR-binding sites are not GREs, as defined by the canonical motif, as well as related motifs, consisting of A/G-G—ACA---T/A-GT-CTC (87). Of the 1031 sites that bound the GR, only 25 were shared with 3,857 GR-binding sites in A549 cells derived from human lung epithelium, and 73 shared with the 8,265 GR-binding sites found in the mouse adipocyte 3T3-L1 cell line (8, 87).
Cooperativity, cross talk, pioneer molecules, and transcription factors
As described above, pleiotropy and promiscuity are prominent characteristics of GR interactions with chromatin, and the subsequent regulation of gene expression. Other steroid receptors, such as the estrogen receptor, benefit from GR reprogramming of chromatin in a cell type–specific manner. For example, in a mouse mammary tumor cell line designed to exclude progesterone receptor cross talk, ChiP-Seq analysis shows that priming with dexamethasone treatment significantly increases the binding of estrogen receptor to sites within DNase-I–accessible chromatin compared to treatment with estradiol alone, suggesting that glucocorticoids provide accessibility to estrogen receptor–binding sites that are only accessible after GR programming (38). Specific cross talk between NFKB1 and GR involves significant reprogramming of gene expression under conditions when both tumor necrosis factor (TNF) and glucocorticoids are present, lending support to the notion that GR and p65 are recruited by each other, and probably by other factors, such as AP1, to binding sites gained on coactivation, in a mutually dependent manner (78). At many genomic sites where GR binding causes increased chromatin accessibility, concurrent steady-state binding levels for another receptor are actually increased (61). Thus, although GR occupancy of a GRE may be short-lived, it acts cooperatively through assisted loading to enable GRE binding for the estrogen receptor at the same site (61). The mechanisms by which the GR enables other transcription factors to bind, resulting in either a positive or negative transcriptional outcome at composite response elements, has been extensively studied using the MMTV promoter as a model (99, 100, 101). In the central nervous system, CRH expressed by neurons of the paraventricular nucleus of the hypothalamus is critical for the release of ACTH from the anterior pituitary, and the subsequent release of cortisol from the adrenal glands. At the CRH promoter in these neurons, glucocorticoid-dependent negative feedback regulation, which is critical for limitation of the HPA-mediated stress response, is controlled by a composite element encompassing high-affinity binding sites for AP1 and GR at adjacent elements within the nGRE. CRH trans-repression by glucocorticoids involves formation of a repressor complex consisting of GR, MECP2 (methyl CpG binding protein 2), and HDAC1 (histone deacetylase 1), as well as recruitment of DNMT3b and associated changes in proximal promoter methylation (102). GR trans-repression is known to act through intermediary transcription factors that impact the nervous and immune systems, including NFKB1 (nuclear factor NF-kappa-B p105 subunit), AP1, CREB1, NFATC1 (nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1), STAT6 (signal transducer and activator of transcription 6, interleukin-4 induced), IRF3 (interferon regulatory factor 3), STAT3 (signal transducer and activator of transcription 3), GATA3 (GATA binding protein 3), and TBX21 (T-box 21) (103). Transcription factors are also involved in extensive remodeling of nucleosome structure by the GR.
GR rhythmicity, dysregulation, and the brain–gut axis
The CLOCK gene, whose translational product has a helix–loop–helix structure, has a primary self-oscillatory component that drives circadian rhythm by acting by acetylating the hinge region of the GR (54, 55). This mechanism normally represses GR-regulated transcriptional activity, acting in a 180-degree phase shift relative to the HPA-mediated oscillatory cycle (46). Changes in this inverse relationship between HPA-mediated and CLOCK cycling have been implicated in shift work disorders (63, 104), cardiovascular diseases such as hypertension (104), obesity associated with sleep deprivation (105), and a range of inflammatory disorders (106), including irritable bowel syndrome, gastroesophageal reflux disease, and peptic ulcer disease (107).
Dysregulation of the HPA-mediated stress response is critical to the decoupling of the phasing of GR-regulated and CLOCK-regulated gene expression in many psychiatric disorders (108, 109, 110, 111, 112, 113, 114, 115), although the molecular mechanisms that synchronize the periodicity of this relationship are not completely understood. Studies of the regulation of PER2, a clock-controlled gene, show a unique overlapping of GRE and E-box motifs in its proximal promoter. PER2 responds rapidly to glucocorticoid-induction through a complex including ARNTL (aryl hydrocarbon receptor nuclear translocator–like protein), also known as brain and muscle ARNTL (BMAL1), and CLOCK, and is involved in delaying circadian rhythmicity mediated by the GR (116). However, many GR-regulated genes are expressed in a variety of different phases relative to the circadian cycle in the mouse (117). The circadian clock is regulated at the cellular level by a transcriptional feedback loop. CLOCK:ARNTL heterodimers activate the expression of the PER and CRY genes, acting as transcription factors directed to the PER and CRY promoters via E-box elements. PER and CRY proteins form heterodimers and suppress the activity of CLOCK:ARNTL, completing the feedback loop. In addition, ARNTL has been shown to directly mediate circadian control of protein translation (118).
It is largely unknown how chronic stress directly or indirectly affects GR expression and function in the GI tract. Chronic, intermittent water-avoidance stress (WAS), 1 h per day for 10 days, significantly reduces the levels of the GR and epithelial tight junction protein levels in the colon but not the jejunum (119). These changes correlate with increased permeability to low molecular weight molecules, and a significant increase in visceral pain in response to colorectal distension. GR appears to play a pivotal role in these responses based on the observation that the GR antagonist RU-486 significantly reversed chronic stress–associated decrease in epithelial tight junction protein levels, increased paracellular permeability, and visceral hyperalgesia. It is probably relevant that the baseline level of GR expression in healthy rats is markedly higher in the mucosa of the colon compared to the jejunum (119).
Ontogenetic maturation of a circadian clock from the fetal stage until weaning has been demonstrated in the rat colon (120). This study suggests that the molecular mechanism for entraining the circadian clock in the colon appears to involve maternal breast-feeding, but subsequently, a suprachiasmatic nucleus–independent developmental signal switches the colonic clock from a maternal-dependent to maternal-independent stage. Consistent with reports that cortisol is responsible for driving circadian surges of transcriptional activity in the CLOCK and PER gene families in the central nervous system, expression of the epithelial tight junction proteins occludin and claudin-1 are under circadian control in the mouse large intestine (121), which supports an earlier report that the circadian clock regulated intestinal permeability (122). The expression of occludin and claudin-1 in the large intestine epithelium demonstrated daily oscillations, which require normal Per2 activity. In addition, the temporal changes of the expression levels were inversely associated with colonic permeability and differential susceptibility to dextran sodium sulfate (DSS)–induced colitis between wild-type mice and mice with a mutation of the clock gene Period 2 (Per2; mPer2(m/m)). Occludin and claudin-1 mRNAs showed daily oscillations; Clock and Arntl (Bmal1) bonded directly to the E-box elements of occludin and claudin-1 promoters; and overexpression of Clock and Arntl activated occludin and claudin-1 promoters containing E-box elements. These observations suggest that occludin and claudin-1 are CGs with E-box elements in the promoter region directly regulated by Clock:Arntl heterodimers. The study did not exclude the possibility that posttranscriptional mechanisms may also underlie the circadian regulation of occludin and claudin-1 expression. Colonic permeability was also under circadian control, which was inversely associated with occludin and claudin-1 expression levels. Of interest, IL-1β and TNF-α mRNA levels did not show circadian oscillations in wild-type mice, and were comparable between wild-type and mPer2m/m mice. Therefore, Clock:Arntl heterodimers probably regulate colonic permeability in a circadian manner by timing the expression levels of claudin-1 and occludin, thereby affecting tight junction function. It remains to be determined whether the changes in occludin and claudin-1 expression are primarily responsible for daily changes in tight junction permeability and susceptibility to DSS-induced colitis or, alternatively, the reduced susceptibility to DSS-induced colitis in mPer2m/m mice may be due to some type of immune cell dysfunction. Nevertheless, the chimeric mouse experiments suggest that the Per2 mutation in non-hematopoietic cells is responsible for the protection against DSS-induced colitis.
Role of the GR in chronic stress-induced visceral hyperalgesia
Chronic stress is associated with down-regulation of the anti-nociceptive endocannabinoid CB1 receptor (CNR1 gene) and upregulation of the pro-nociceptive endovanilloid TRPV1 receptor in the subpopulation of nociceptive DRG neurons innervating the pelvic organs, including the colon. The effect of chronic stress on both CB1 and TRPV1 receptor expression and function are prevented by treatment with RU-486, supporting an important role for GR in this pathway (123, 124). Subsequent observations support a potentially pivotal role for epigenetic regulatory pathways in chronic stress–induced visceral hyperalgesia (125). Male rats were subjected to WAS or subcutaneous injections of corticosterone that reproduced chronic stress levels. Lumbar 4–5 and lumbar 6–sacral 2 DRGs were collected and compared between stressed and sham-stressed control rats. L4–L5 DRG neurons contribute to the somatosensory innervation of the lower extremities by way of the sciatic nerve, whereas L6–S2 DRG neurons innervate pelvic organs, including the colon. DNA methylation was measured at genes encoding NR3C1 and CNR1. Protein levels of CNR1 and DNA (cytosine-5-)-methyltransferase 1 (DNMT1) were knocked down in DRG neurons in situ with small interfering RNAs. Visceral pain was measured in response to colorectal distention. Chronic stress was associated with upregulation of DNMT1-mediated methylation at CpG sites involving both NR3C1 and CNR1 promoter regions, resulting in downregulation of NR3C1-induced expression of CNR1 in L6–S2, but not L4–L5 DRGs. Gene silencing of DNMT1 in L6–S2 DRG neurons of rats reduced DNA methylation and prevented chronic stress–induced increases in visceral pain. These studies indicate that the NR3C1 receptor acts as a positive transcription factor on CNR1 expression. Chronic stress increases DNA methylation at upstream CpG promoter sites for NR3C1 expression, resulting in decreased levels of the anti-nociceptive CNR1 receptor, culminating in enhanced visceral pain.
Chronic, intermittent stress also alters the level of NR3C1 in regions of the central nervous system involved in visceral pain registration. In the WAS rodent model, exposing the central nucleus of the amygdala (CeA) to elevated glucocorticoid levels was associated with significant reduction in NR3C1 receptor expression but no changes in mineralocorticoid receptor expression (126), which is consistent with findings from experiments in the hippocampus and studies using cell cultures (127). Epigenetic mechanisms appear to play a role in chronic stress–induced changes in NR3C1 receptor levels in the amygdala (128). Trichostatin A, a potent histone deacetylase inhibitor, significantly attenuated chronic stress–associated visceral hyperalgesia. Methylation of NR3C1 was increased following WAS, which was associated with decreased NR3C1 expression. In contrast, methylation of the corticotropin-releasing factor (CRF) promoter was decreased after WAS with a concomitant increase in CRF expression – an anticipated result. In a subsequent study, the authors examined the hypothesis that histone deacetylation contributes to the maintenance of chronic anxiety and pain induced by prolonged exposure of the CeA to corticosterone (129). Bilateral infusions of a histone deacetylase inhibitor into the CeA attenuated anxiety-like behavior, as well as somatic and visceral hypersensitivity resulting from elevated corticosterone exposure. Deacetylation of histone 3 lysine 9 (H3K9), through the coordinated action of the NAD+-dependent protein deacetylase sirtuin-6 (SIRT6) and NFKB1-sequestered NR3C1 expression, leading to disinhibition of CRF. These observations suggest that epigenetic programming in the amygdala, involving both methylation and histone modification, are important in the maintenance of chronic anxiety and pain.
Limitations of animal models
Much of what we know about how the GR regulates gene expression and behavior has been obtained from studies in rodents, with extension to human mechanisms of disease. However, recent studies show that although the complexity of GR interactions with the three-dimensional structure of programmed and un-programmed chromatin exhibits some degree of similarity between species, population differences in allele-specific and tissue-specific expression appear to be limited to humans (82). Programmed chromatin has been prepared by factors such as AP1, ER-alpha, or remodeling factors for subsequent transient occupation by the GR. Unprogrammed chromatin has not undergone this preparation for GR occupancy (88). In addition, rodents provide poor models of the human inflammatory response (130), and do not adequately represent higher cognitive contributions to stress-related psychiatric disease and ameliorative drug therapy (131, 132, 133). Studies from human cell lines underscore the inadequacy of the rodent to mirror the long-distance, enhancer-based regulation of GR transcription that is a defining characteristic of glucocorticoid regulation of gene expression in humans (1, 134, 135, 136, 137, 138, 139).
Summary
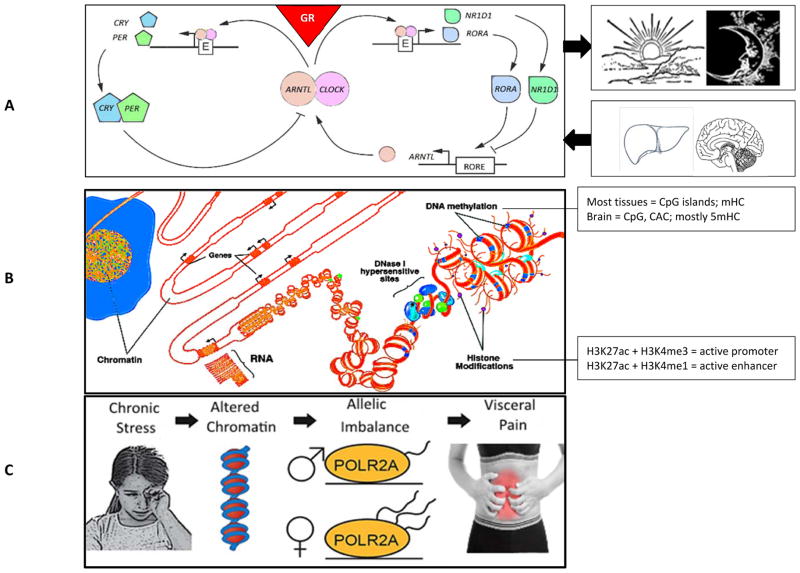
Diurnal rhythms in the HPA axis and chronic intermittent stress have differential, region-specific, and cell-specific effects involving the brain–gut axis, including intestinal barrier function, peripheral pain signaling mechanisms, and central nervous system modulation of pain perception. It appears that epigenetic regulation of NR3C1 expression plays a potentially key role in these actions. Epidemiological studies suggest that chronic stress–associated functional gastrointestinal disorders, such as irritable bowel syndrome, are more prevalent in females and individuals who have experienced significant stress early in life (140). Future studies will need to address the mechanistic basis for the distinct effects of gender and early life stress as predisposing risk factors in functional bowel disorders, and the potential role of stress-induced alterations in intestinal barrier function on visceral hyperalgesia. It is probable that alterations in rhythmic gene transcription, epigenetic regulation of gene transcription, and allele-specific gene expression play a significant role in the pathophysiology of functional gastrointestinal disorders that are characteristically inducible and reversible. These concepts are summarized in Figure 1.
Figure 1.
The glucocorticoid receptor, regulation of gene expression, and its clinical consequences. (A) The glucocorticoid receptor (GR) and regulation of the CLOCK gene family. This pathway contributes to regulation of circadian rhythmicity, and is driven by the hypothalamic-pituitary-adrenal axis and metabolic inputs from liver. Of all the thousands of genes whose expression is regulated by the GR, the most highly regulated include members of the CLOCK gene family such as PER1 (33). Products of ARNTL and CLOCK form a heterodimer that acts a transcription factor and binds to the E-box (E) in the promoter of other GR-regulated CLOCK and related genes, including CRY, PER, NR1D1, and RORA. In general, both CRY and PER genes are expressed in a circadian manner, acting either by repressing the CLOCK:ARNTL heterodimer, or blocking GR from binding to the GR response element (GRE) sequence. This latter mechanism is described in the section: GR rhythmicity, dysregulation, and the brain–gut axis. NR1D1 and RORA form a heterodimer that acts as a transcription factor that binds to the RORE sequence that is located in genes such as ARNTL. The ARNTL protein also acts independently of its role as a transcription factor to mediate translational control of specific proteins in a circadian manner (118). (B) Overview of results from the NIH Roadmap Epigenomics Mapping Consortium. Chromosomes are located in chromatin-bound territories in the nucleus. Euchromatin is characterized by DNase 1 hypersensitivity and specific combinations of histone marks that define active genomic regulatory elements, such as promoters H3K4me3 and H3K27ac, and enhancers H3K4me1 and H3K27ac. An enhancer can either increase or decrease transcription. Recent research demonstrates that, in brain, the DNA sequence CAC is a common site of methylation, in contrast to other tissues where CpG is most often methylated. Also, in brain, 5-hydroxymethylcytosine (5hmC), a reactive species, is common. In contrast, in the periphery, methylcytosine (hmC) is common (76). (C) Chronic and/or early life stress appear to lead to alteration of chromatin and allele-biased gene expression, concomitant with psychiatric disorders and comorbid conditions including irritable bowel syndrome. In such cases, allele skewing of transcription (21, 22) is most likely a consequence of epigenetic alterations arising from environmental adversity that contributes to a variety of stress-related disorders distinct from parental imprinting (29, 30). POLR2A polymerase (RNA) II (DNA directed) polypeptide A, 220kDa is a highly conserved, ubiquitously expressed gene that encodes the largest subunit of RNA polymerase II, the polymerase responsible for synthesizing messenger RNA in eukaryotes and, therefore, plays an important role in gene transcription. Allelic imbalance refers to differential expression of the maternal vs paternal allele. POLR2A, DNA-dependent RNA polymerase II; H3K4me3, histone H3 trimethyl Lys4; H3K27ac, histone H3 Lys27 acetylation; H3Kme1, histone H3 monomethyl Lys4. of methylation, in contrast to other tissues where CpG is most often methylated. Also, in brain, 5-hydroxymethylcytosine (5hmC), a reactive species, is common. In contrast, in the periphery, methylcytosine (hmC) is common (76). (C) Chronic and/or early life stress appear to lead to alteration of chromatin and allele-biased gene expression, concomitant with psychiatric disorders and comorbid conditions including irritable bowel syndrome. In such cases, allele skewing of transcription (21, 22) is most likely a consequence of epigenetic alterations arising from environmental adversity that contributes to a variety of stress-related disorders distinct from parental imprinting (29, 30). POLR2A polymerase (RNA) II (DNA directed) polypeptide A, 220kDa is a highly conserved, ubiquitously expressed gene that encodes the largest subunit of RNA polymerase II, the polymerase responsible for synthesizing messenger RNA in eukaryotes and, therefore, plays an important role in gene transcription. Allelic imbalance refers to differential expression of the maternal vs paternal allele. POLR2A, DNA-dependent RNA polymerase II; H3K4me3, histone H3 trimethyl Lys4; H3K27ac, histone H3 Lys27 acetylation; H3Kme1, histone H3 monomethyl Lys4.
Key Messages.
Chronic stress is associated with the exacerbation of common mental health and gastrointestinal disorders, including depression and irritable bowel syndrome.
Emerging evidence indicates that chronic stress alters physiological cortisol-mediated pulsatile (circadian and ultradian) gene transcription in a tissue-, cell- and allele-specific manner.
Epigenetic studies indicate that chronic stress selectively alters both the DNA methylome and chromatin compaction state affecting gene transcription in central and peripheral pathways involved in a variety of important functions including mood and pain perception.
Chronic and early life stress–associated disruption in glucocorticoid receptor–regulated gene transcription has a potentially significant impact on the brain–gut axis throughout the lifespan and across generations.
Acknowledgments
The authors wish to thank Barbara Boughen for editorial support.
Funding
JWW receives grant support from NIH 1R01DK098205 and Takeda Pharmaceuticals.
Footnotes
Conflicts of Interest
None
References
- 1.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG, et al. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. 1985;318:635–41. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oakley RH, Cidlowski JA. Cellular processing of the glucocorticoid receptor gene and protein: New mechanisms for generating tissue-specific actions of glucocorticoids. J Biol Chem. 2011;286:3177–84. doi: 10.1074/jbc.R110.179325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramamoorthy S, Cidlowski JA. Ligand-induced repression of the glucocorticoid receptor gene is mediated by an NCoR1 repression complex formed by long-range chromatin interactions with intragenic glucocorticoid response elements. Mol Cell Biol. 2013;33:1711–22. doi: 10.1128/MCB.01151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burd CJ. Analysis of chromatin dynamics during glucocorticoid receptor activation. Mol Cell Biol. 2012;32:1805–17. doi: 10.1128/MCB.06206-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miranda TB, Morris SA, Hager GL. Complex genomic interactions in the dynamic regulation of transcription by the glucocorticoid receptor. Mol Cell Endrocrinol. 2013;380:16–24. doi: 10.1016/j.mce.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–41. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 8.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–8. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stavreva DA, Varticovski L, Hager GL. Complex dynamics of transcription regulation. Biochim Biophys Acta. 2012;1819:657–66. doi: 10.1016/j.bbagrm.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–44. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paakinaho V, Makkonen H, Jääskeläinen T, Palvimo JJ. Glucocorticoid receptor activates poised FKBP51 locus through long-distance interactions. Mol Endocrinol. 2010;24:511–25. doi: 10.1210/me.2009-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakim O, John S, Ling JQ, Biddie SC, Hoffman AR, Hager GL. Glucocorticoid receptor activation of the Ciz1-Lcn2 locus by long range interactions. J Biol Chem. 2009;284:6048–52. doi: 10.1074/jbc.C800212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert WM, Xu CF, Neubert TA, Chao MV, Garabedian MJ, Jeanneteau FD. Brain-derived neurotrophic factor signaling rewrites the glucocorticoid transcriptome via glucocorticoid receptor phosphorylation. Mol Cell Biol. 2013;33:3700–14. doi: 10.1128/MCB.00150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TW, Mercer KB, et al. Allele-specific FKBP5 DNA demethylation mediates gene–childhood trauma interactions. Nat Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heim C, Newport DJ, Mletzko T, Miller AH, Nemeroff CB. The link between childhood trauma and depression: Insights from HPA axis studies in humans. Psychoneuroendocrinology. 2008;33:693–710. doi: 10.1016/j.psyneuen.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 16.van Zuiden M, Geuze E, Willemen HL, Vermetten E, Maas M, Amarouchi K, Kavelaars A, Heijnen CJ. Glucocorticoid receptor pathway components predict posttraumatic stress disorder symptom development: A prospective study. Biol Psychiatry. 2012;71:309–16. doi: 10.1016/j.biopsych.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 17.van Wingen GA, Geuze E, Vermetten E, Fernández G. Perceived threat predicts the neural sequeale of combat stress. Mol Psychiatry. 2011;16:664–71. doi: 10.1038/mp.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maniglio R. Prevalence of child sexual abuse among adults and youths with bipolar disorder: A systematic review. Clin Psychol Rev. 2013;33:561–73. doi: 10.1016/j.cpr.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, Tang Y, Gillespie CF, et al. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299:1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delahanty DL, Raimonde AJ, Spoonster E. Initial posttraumatic urinary cortisol levels predict subsequent PTSD symptoms in motor vehicle accident victims. Biol Psychiatry. 2000;48:940–7. doi: 10.1016/s0006-3223(00)00896-9. [DOI] [PubMed] [Google Scholar]
- 21.Cicchetti D, Rogosch FA, Gunnar MR, Toth SL. The differential impacts of early physical and sexual abuse and internalizing problems on daytime cortisol rhythm in school-aged children. Child Dev. 2010;81:252–69. doi: 10.1111/j.1467-8624.2009.01393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee ER, Alisch RS. Early-life disruption of epigenetic marks may contribute to the origins of mental illness. Epigenomics. 2012;4:355–7. doi: 10.2217/epi.12.35. [DOI] [PubMed] [Google Scholar]
- 23.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–9. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crews D, Gillette R, Scarpino SV, Manikkam M, Savenkova MI, Skinner MK. Epigenetic transgenerational inheritance of altered stress responses. Proc Natl Acad Sci U S A. 2012;109:9143–8. doi: 10.1073/pnas.1118514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heim C, Binder EB. Current research trends in early life stress and depression: Review of human studies on sensitive periods, gene–environment interactions, and epigenetics. Exp Neurol. 2012;233:102–11. doi: 10.1016/j.expneurol.2011.10.032. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman A, Spengler D. The lasting legacy of social stress on the epigenome of the hypothalamic–pituitary–adrenal axis. Epigenomics. 2012;4:431–44. doi: 10.2217/epi.12.34. [DOI] [PubMed] [Google Scholar]
- 27.Kim YS, Lee MY, Choi CS, Sohn YW, Park BR, Choi MG, Nah YH, Choi SC. The effect of chronic variable stress on bowel habit and adrenal function in rats. J Gastroenterol Hepatol. 2008;23:1840–6. doi: 10.1111/j.1440-1746.2008.05524.x. [DOI] [PubMed] [Google Scholar]
- 28.Larauche M, Mulak A, Taché Y. Stress and visceral pain: From animal models to clinical therapies. Exp Neurol. 2012;233:49–67. doi: 10.1016/j.expneurol.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larauche M, Mulak A, Taché Y. Stress-related alterations of visceral sensation: Animal models for irritable bowel syndrome study. J Neurogastroenterol Motil. 2012;17:213–34. doi: 10.5056/jnm.2011.17.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradford K, Shih W, Videlock EJ, Presson AP, Naliboff BD, Mayer EA, Chang L. Association between early adverse life events and irritable bowel syndrome. Clin Gastroenterol Hepatol. 2012;10:385–90. doi: 10.1016/j.cgh.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peña CJ, Bagot RC, Labonté B, Nestler EJ. Epigenetic signaling in psychiatric disorders. J Mol Biol. 2014;426:3389–412. doi: 10.1016/j.jmb.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ordog T, Syed SA, Hayashi Y, Asuzu DT. Epigenetics and chromatin dynamics: A review and a paradigm for functional disorders. Neurogastroenterol Motil. 2012;24:1054–68. doi: 10.1111/nmo.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–71. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talens RP, Boomsma DI, Tobi EW, Kremer D, Jukema JW, Willemsen G, Putter H, Slagboom PE, et al. Variation, patterns, and temporal stability of DNA methylation: Considerations for epigenetic epidemiology. FASEB J. 2010;24:3135–44. doi: 10.1096/fj.09-150490. [DOI] [PubMed] [Google Scholar]
- 36.Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, Bryois J, Giger T, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife. 2013;2:e00523. doi: 10.7554/eLife.00523. Erratum in eLife 2013; 2: e01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008;29:611–24. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 38.Miranda TB, Voss TC, Sung MH, Baek S, John S, Hawkins M, Grøntved L, Schiltz RL, et al. Reprogramming the chromatin landscape: Interplay of the estrogen and glucocorticoid receptors at the genomic level. Cancer Res. 2013;73:5130–40. doi: 10.1158/0008-5472.CAN-13-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornejo Castro EM, Carr DF, Jorgensen AL, Alfirevic A, Pirmohamed M. HLA-allelotype associations with nevirapine-induced hypersensitivity reactions and hepatotoxicity: a systematic review of the literature and meta-analysis. Pharmacogenet Genomics. 2015;25:186–98. doi: 10.1097/FPC.0000000000000124. [DOI] [PubMed] [Google Scholar]
- 40.Tarullo AR, Gunnar MR. Child maltreatment and the developing HPA axis. Horm Behav. 2006;50:632–39. doi: 10.1016/j.yhbeh.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Lupien SJ, McEwen BS, Gunnar MR, Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci. 2009;10:434–45. doi: 10.1038/nrn2639. [DOI] [PubMed] [Google Scholar]
- 42.Evans BE, Greaves-Lord K, Euser AS, Tulen JH, Franken IH, Huizink AC. Determinants of physiological and perceived physiological stress reactivity in children and adolescents. PLoS One. 2013;8:e61724. doi: 10.1371/journal.pone.0061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dinan TG. Glucocorticoids and the genesis of depressive illness. A psychobiological model. Br J Psychiatry. 1994;164:365–71. doi: 10.1192/bjp.164.3.365. [DOI] [PubMed] [Google Scholar]
- 44.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 45.Charmandari E, Chrousos GP, Lambrou GI, Pavlaki A, Koide H, Ng SS, Kino T. Peripheral CLOCK regulates target-tissue glucocorticoid receptor transcriptional activity in a circadian fashion in man. PLoS One. 2010;6:e25612. doi: 10.1371/journal.pone.0025612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nader N, Chrousos GP, Kino T. Interactions of the circadian CLOCK system and the HPA axis. Trends Endocrinol Metab. 2010;21:277–86. doi: 10.1016/j.tem.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohawk JA, Takahashi JS. Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends neurosci. 2011;34:349–58. doi: 10.1016/j.tins.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–10. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 49.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–42. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lonard DM, O’Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–14. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 51.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–61. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–40. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 54.Nader N, Chrousos GP, Kino T. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: Potential physiological implications. FASEB J. 2009;23:1572–83. doi: 10.1096/fj.08-117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grimaldi B, Nakahata Y, Sahar S, Kaluzova M, Gauthier D, Pham K, Patel N, Hirayama J, et al. Chromatin remodeling and circadian control: master regulator CLOCK is an enzyme. Cold Spring Harb Symp Quant Biol. 2007;72:105–12. doi: 10.1101/sqb.2007.72.049. [DOI] [PubMed] [Google Scholar]
- 56.Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–6. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Charmandari E. Primary generalized glucocorticoid resistance and hypersensitivity. Horm Res Paediatr. 2011;76:145–55. doi: 10.1159/000330759. [DOI] [PubMed] [Google Scholar]
- 58.Ramamoorthy S, Cidlowski JA. Exploring the molecular mechanisms of glucocorticoid receptor action from sensitivity to resistance. Endocr Dev. 2013;24:41–56. doi: 10.1159/000342502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagaich AK, Walker DA, Wolford R, Hager GL. Rapid Periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell. 2004;14:163–74. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- 60.George AA, Schiltz RL, Hager GL. Dynamic access of the glucocorticoid receptor to response elements in chromatin. Int J Biochem Cell Biol. 2009;41:214–24. doi: 10.1016/j.biocel.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–54. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–6. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Slagsvold T, Kraus I, Frønsdal K, Saatcioglu F. DNA binding-independent transcriptional activation by the androgen receptor through triggering of coactivators. J Biol Chem. 2001;276:31030–6. doi: 10.1074/jbc.M104310200. [DOI] [PubMed] [Google Scholar]
- 64.Stavreva DA, Coulon A, Baek S, Sung MH, John S, Stixova L, Tesikova M, Hakim O, et al. Dynamics of chromatin accessibility and long-range interactions in response to glucocorticoid pulsing. Genome Res. 2015;25:845–57. doi: 10.1101/gr.184168.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ouellet-Morin I, Odgers CL, Danese A, Bowes L, Shakoor S, Papadopoulos AS, Caspi A, Moffitt TE, et al. Blunted cortisol responses to stress signal social and behavioral problems among maltreated/bullied 12-year-old children. Biol Psychiatry. 2011;70:1016–23. doi: 10.1016/j.biopsych.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carpenter LL, Shattuck TT, Tyrka AR, Geracioti TD, Price LH. Effect of childhood physical abuse on cortisol stress response. Psychopharmacology (Berl) 2011;214:367–75. doi: 10.1007/s00213-010-2007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cohen S, Janicki-Deverts D, Doyle WJ, Miller GE, Frank E, Rabin BS, Turner RB. Chronic stress, glucocorticoid receptor resistance, inflammation, and disease risk. Proc Natl Acad Sci U S A. 2012;109:5995–9. doi: 10.1073/pnas.1118355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tyrka AR, Price LH, Marsit C, Walters OC, Carpenter LL. Childhood adversity and epigenetic modulation of the leukocyte glucocorticoid receptor: Preliminary findings in healthy adults. PLoS ONE. 2012;7:e30148. doi: 10.1371/journal.pone.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Filiberto AC, Maccani MA, Koestler D, Wilhelm-Benartzi C, Avissar-Whiting M, Banister CE, Gagne LA, Marsit CJ. Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. Epigenetics. 2011;6:566–72. doi: 10.4161/epi.6.5.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Labonte B, Yerko V, Gross J, Mechawar N, Meaney MJ, Szyf M, Turecki G. Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biol Psychiatry. 2012;72:41–8. doi: 10.1016/j.biopsych.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 71.McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;3:342–48. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen HL, Li LR. Glucocorticoid receptor gene polymorphisms and glucorticoid resistance in inflammatory bowel disease: A meta-analysis. Dig Dis Sci. 2012;57:3065–74. doi: 10.1007/s10620-012-2293-2. [DOI] [PubMed] [Google Scholar]
- 73.Maltese P, Palma L, Sfara C, de Rocco P, Latiano A, Palmieri O, Corritore G, Annese V, et al. Glucocorticoid resistance in Crohn’s disease and ulcerative colitis: an association study investigating GR and FKBP5 gene polymorphisms. Pharmacogeneomics J. 2012;12:432–8. doi: 10.1038/tpj.2011.26. [DOI] [PubMed] [Google Scholar]
- 74.Bittencourt D, Wu DY, Jeong KW, Gerke DS, Herviou L, Ianculescu I, Chodankar R, Siegmund KD, et al. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc Natl Acad Sci U S A. 2012;109:19673–8. doi: 10.1073/pnas.1211803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uhlenhaut NH, Barish GD, Yu RT, Downes M, Karunasiri M, Liddle C, Schwalie P, Hübner N, et al. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol Cell. 2013;49:158–71. doi: 10.1016/j.molcel.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luo C, Ecker JR. Epigenetics. Exceptional epigenetics in the brain. Science. 2015;348:1094–5. doi: 10.1126/science.aac5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He X, Chatterjee R, John S, Bravo H, Sathyanarayana BK, Biddie SC, FitzGerald PC, Stamatoyannopoulos JA, et al. Contribution of nucleosome binding preferences and co-occurring DNA sequences to transcription factor binding. BMC Genomics. 2013;14:428. doi: 10.1186/1471-2164-14-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rao NA, McCalman MT, Moulos P, Francoijs KJ, Chatziioannou A, Kolisis FN, Alexis MN, Mitsiou DJ, et al. Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res. 2011;21:1404–16. doi: 10.1101/gr.118042.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Cell Curr Biol. 2000;10:853–6. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 80.Rosa AL, Wu YQ, Kwabi-Addo B, Coveler KJ, Reid Sutton V, Shaffer LG. Allele-specific methylation of a functional CTCF binding site upstream of MEG3 in the human imprinted domain of 14q32. Chromosome Res. 2005;13:809–19. doi: 10.1007/s10577-005-1015-4. [DOI] [PubMed] [Google Scholar]
- 81.Prickett AR, Barkas N, McCole RB, Hughes S, Amante SM, Schulz R, Oakey RJ. Genome-wide and parental allele-specific analysis of CTCF and cohesin DNA binding in mouse brain reveals a tissue-specific binding pattern and an association with imprinted differentially methylated regions. Genome Res. 2013;23:1624–35. doi: 10.1101/gr.150136.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tycko B. Allele-specific DNA methylation: Beyond imprinting. Hum Mol Genet. 2012;19:R210–20. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holwerda SJ, de Laat W. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120369. doi: 10.1098/rstb.2012.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biddie SC, Conway-Campbell BL, Lightman SL. Dynamic regulation of glucocorticoid signaling in health and disease. Rheumotology (Oxford) 2012;51:403–12. doi: 10.1093/rheumatology/ker215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gicquel C, El-Osta A, Le Bouc Y. Epigenetic regulation and fetal programming. Best Pract Res Clin Endocrinol Metab. 2008;22:1–16. doi: 10.1016/j.beem.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 86.Essien K, Vigneau S, Apreleva S, Singh LN, Bartolomei MS, Hannenhalli S. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol. 2009;10:R131. doi: 10.1186/gb-2009-10-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Polman JA, Welten JE, Bosch DS, de Jonge RT, Balog J, van der Maarel SM, de Kloet ER, Datson NA. A genome-wide signature of glucocorticoid receptor binding in neuronal PC12 cells. BMC Neurosci. 2012;13:118. doi: 10.1186/1471-2202-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grøntved L, John S, Baek S, Liu Y, Buckley JR, Vinson C, Aguilera G, Hager GL. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J. 2013;32:1568–83. doi: 10.1038/emboj.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, et al. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 2010;5:e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Echeverría PC, Mazaira G, Erlejman A, Gomez-Sanchez C, Piwien Pilipuk G, Galigniana MD. Nuclear import of the glucocorticoid receptor-hsp90 complex through the nuclear pore complex is mediated by its interaction with Nup62 and importin beta. Mol Cell Biol. 2009;29:4788–97. doi: 10.1128/MCB.00649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Makkonen H, Kauhanen M, Paakinaho V, Jääskeläinen T, Palvimo JJ. Long-range activation of FKBP51 transcription by the androgen receptor via distal intronic enhancers. Nucleic Acids Res. 2009;37:4135–48. doi: 10.1093/nar/gkp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee RS, Tamashiro KL, Yang X, Purcell RH, Harvey A, Willour VL, Huo Y, Rongione M, et al. Chronic corticosterone exposure increases expression and decreases deoxyribonucleic acid methylation of Fkbp5 in mice. Endocrinology. 2010;151:4332–43. doi: 10.1210/en.2010-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Medina A, Seasholtz AF, Sharma V, Burke S, Bunney W, Jr, Myers RM, Schatzberg A, Akil H, et al. Glucocorticoid and mineralocorticoid receptor expression in the human hippocampus in major depressive disorder. J Psychiatric Res. 2013;47:307–14. doi: 10.1016/j.jpsychires.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cottrell EC, Secki JR. Prenatal stress, glucocorticoids and the programming of adult disease. Front Behav Neurosci. 2009;3:19. doi: 10.3389/neuro.08.019.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mune T, White PC. Apparent mineralocorticoid excess: Genotype is correlated with biochemical phenotype. Hypertension. 1996;27:1193–9. doi: 10.1161/01.hyp.27.6.1193. [DOI] [PubMed] [Google Scholar]
- 96.Kato K, Sasano H, Ohara S, Sekine H, Mochizuki S, Mune T, Yasuda K, Nagura H, et al. Coexpression of mineralocorticoid receptors and 11beta-hydroxysteroid dehydrogenase 2 in human gastric mucosa. J Clin Endocrinol Metab. 1999;84:2568–73. doi: 10.1210/jcem.84.7.5845. [DOI] [PubMed] [Google Scholar]
- 97.Driver PM, Kilby MD, Bujalska I, Walker EA, Hewison M, Stewart PM. Expression of 11 beta-hydroxysteroid dehydrogenase isozymes and corticosteroid hormone receptors in primary cultures of human trophoblast and placental bed biopsies. Mol Hum Reprod. 2001;7:357–63. doi: 10.1093/molehr/7.4.357. [DOI] [PubMed] [Google Scholar]
- 98.Samarasinghe RA, Witchell SF, DeFranco DB. Cooperativity and complementarity: Synergies in non-classical and classical glucocorticoid signaling. Cell Cycle. 2012;11:2819–27. doi: 10.4161/cc.21018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kassel O, Herrlich P. Crosstalk between the glucocorticoid receptor and other transcription factors: molecular aspects. Mol Cell Endocrinol. 2007;275:13–29. doi: 10.1016/j.mce.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 100.Wu J, Bresnick EH. Glucocorticoid and growth factor synergism requirement for Notch4 chromatin domain activation. Mol Cell Biol. 2007;27:2411–22. doi: 10.1128/MCB.02152-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Latchoumanin O, Mynard V, Devin-Leclerc J, Dugué MA, Bertagna X, Catelli MG. Reversal of glucocorticoids-dependent proopiomelanocortin gene inhibition by leukemia inhibitory factor. Endocrinology. 2007;148:422–32. doi: 10.1210/en.2006-0460. [DOI] [PubMed] [Google Scholar]
- 102.Sharma D, Bhave S, Gregg E, Uht R. Dexamethasone induces a putative repressor complex and chromatin modifications in the CRH promoter. Mol Endocrinol. 2013;27:1142–52. doi: 10.1210/me.2013-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ratman D, Vanden Berghe W, Dejager L, Libert C, Tavernier J, Beck IM, De Bosscher K. How glucocorticoid receptors modulate the activity of other transcription factors: A scope beyond tethering. Mol Cell Endocrinol. 2013;380:41–54. doi: 10.1016/j.mce.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 104.Kino T, Chrousos GP. Circadian CLOCK-mediated regulation of target-tissue sensitivity to glucocorticoids: Implications for cardiometabolic diseases. Endocr Dev. 2011;20:116–26. doi: 10.1159/000321232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goel N, Stunkard AJ, Rogers NL, Van Dongen HP, Allison KC, O’Reardon JP, Ahima RS, Cummings DE, et al. Circadian rhythm profiles in women with night eating syndrome. J Biol Rhythms. 2009;24:85–94. doi: 10.1177/0748730408328914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chrousus GP. The hypothalamic–pituitary–adrenal axis and immune-mediated inflammation. N Engl J Med. 1995;332:1351–62. doi: 10.1056/NEJM199505183322008. [DOI] [PubMed] [Google Scholar]
- 107.Konturek PC, Brzozowski T, Konturek SJ. Gut clock: Implications of circadian rhythms in the gastrointestinal tract. J Physiol Pharmacol. 2011;62:139–50. [PubMed] [Google Scholar]
- 108.Mansour HA, Monk TH, Nimgaonkar VL. Circadian genes and bipolar disorder. Ann Med. 2005;37:196–205. doi: 10.1080/07853890510007377. [DOI] [PubMed] [Google Scholar]
- 109.Gillin JC, Buchsbaum M, Wu J, Clark C, Bunney W., Jr Sleep deprivation as a model experimental antidepressant treatment: Findings from functional brain imaging. Depress Anxiety. 2001;14:37–49. doi: 10.1002/da.1045. [DOI] [PubMed] [Google Scholar]
- 110.Ehlert U, Gaab J, Heinrichs M. Psychoneuroendocrinological contributions to the etiology of depression, posttraumatic stress disorder, and stress-related bodily disorders: the role of the hypothalamus–pituitary–adrenal axis. Biol Psychol. 2001;57:141–52. doi: 10.1016/s0301-0511(01)00092-8. [DOI] [PubMed] [Google Scholar]
- 111.Nestler EJ, Barrot M, DiLeone RJ, Eisch AJ, Gold SJ, Monteggia LM. Neurobiology of depression. Neuron. 2002;34:13–25. doi: 10.1016/s0896-6273(02)00653-0. [DOI] [PubMed] [Google Scholar]
- 112.Serretti A, Cusin C, Benedetti F, Mandelli L, Pirovano A, Zanardi R, Colombo C, Smeraldi E. Insomnia improvement during antidepressant treatment and CLOCK gene polymorphism. Am J Med Genet B Neuropsychiatr Genet. 2005;137B(1):36–9. doi: 10.1002/ajmg.b.30130. [DOI] [PubMed] [Google Scholar]
- 113.Schulz P. Biological clocks and the practice of psychiatry. Dialogues Clin Neurosci. 2007;9:237–55. doi: 10.31887/DCNS.2007.9.3/pschulz. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wirz-Justice A. Biological rhythm disturbances in mood disorders. Int Clin Psychopharmacol. 2006;21:S11–5. doi: 10.1097/01.yic.0000195660.37267.cf. [DOI] [PubMed] [Google Scholar]
- 115.Keller J, Flores B, Gomez RG, Solvason HB, Kenna H, Williams GH, Schatzberg AF. Cortisol circadian rhythm alterations in psychotic major depression. Biol Psychiatry. 2006;60:275–81. doi: 10.1016/j.biopsych.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 116.Cheon S, Park N, Cho S, Kim K. Glucocorticoid-mediated Period2 induction delays the phase of circadian rhythm. Nucleic Acids Res. 2013;41:6161–74. doi: 10.1093/nar/gkt307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol Cell Biol. 2012;32:3756–67. doi: 10.1128/MCB.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Güttler T, Davis F, et al. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell. 2015;161:1138–51. doi: 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zheng G, Wu SP, Hu Y, Smith DE, Wiley JW, Hong S. Corticosterone mediates stress-related increased intestinal permeability in a region-specific manner. Neurogastroenterol Motil. 2013;25:e127–39. doi: 10.1111/nmo.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Polidarová L, Olejníková L, Paušlyová L, Sládek M, Soták M, Pácha J, Sumová A. Development and entrainment of the colonic circadian clock during ontogenesis. Am J Physiol Gastrointest Liver Physiol. 2014;306:G346–56. doi: 10.1152/ajpgi.00340.2013. [DOI] [PubMed] [Google Scholar]
- 121.Kyoko OO, Kono H, Ishimaru K, Miyake K, Kubota T, Ogawa H, Okumura K, Shibata S, Nakao A. Expressions of tight junction proteins Occludin and Claudin-1 are under the circadian control in the mouse large intestine: implications in intestinal permeability and susceptibility to colitis. PLoS One. 2014;9:e98016. doi: 10.1371/journal.pone.0098016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Summa KC, Voigt RM, Forsyth CB, Shaikh M, Cavanaugh K, Tang Y, Vitaterna MH, Song S, et al. Disruption of the circadian clock in mice increases intestinal permeability and promotes alcohol-induced hepatic pathology and inflammation. PLoS One. 2013;8:e67102. doi: 10.1371/journal.pone.0067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hong S, Fan J, Kemmerer ES, Evans S, Li Y, Wiley JW. Reciprocal changes in vanilloid (TRPV1) and endocannabinoid (CB1) receptors contribute to visceral hyperalgesia in the water avoidance stressed rat. Gut. 2009;58:202–10. doi: 10.1136/gut.2008.157594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hong S, Zheng G, Wu X, Snider NT, Owyang C, Wiley JW. Corticosterone mediates reciprocal changes in CB 1 and TRPV1 receptors in primary sensory neurons in the chronically stressed rat. Gastroenterology. 2011;140:627–37. doi: 10.1053/j.gastro.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hong S, Zheng G, Wiley JW. Epigenetic regulation of genes that modulate chronic stress-induced visceral pain in the peripheral nervous system. Gastroenterology. 2015;148:148–57. doi: 10.1053/j.gastro.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tran L, Greenwood-Van Meerveld B. Altered expression of glucocorticoid receptor and corticotropin-releasing factor in the central amygdala in response to elevated corticosterone. Behav Brain Res. 2012;234:380–5. doi: 10.1016/j.bbr.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 127.Erdeljan P, Andrews MH, MacDonald JF, Matthews SG. Glucocorticoids and serotonin alter glucocorticoid receptor mRNA levels in fetal guinea-pig hippocampal neurons, in vitro. Reprod Fertil Dev. 2005;17:743–9. doi: 10.1071/rd05043. [DOI] [PubMed] [Google Scholar]
- 128.Tran L, Chaloner A, Sawalha AH, Greenwood Van-Meerveld B. Importance of epigenetic mechanisms in visceral pain induced by chronic water avoidance stress. Psychoneuroendocrinology. 2013;38:898–906. doi: 10.1016/j.psyneuen.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 129.Tran L, Schulkin J, Ligon CO, Greenwood-Van Meerveld B. Epigenetic modulation of chronic anxiety and pain by histone deacetylation. Mol Psychiatry. 2014 Oct 7; doi: 10.1038/mp.2014.122. [DOI] [PubMed] [Google Scholar]
- 130.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–12. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hyman SE. How far can mice carry autism research? Cell. 2014;158:13–14. doi: 10.1016/j.cell.2014.06.032. [DOI] [PubMed] [Google Scholar]
- 132.Nestler EJ, Hyman SJ. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13:1161–9. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Markou A, Chiamulera C, Geyer MA, Tricklebank M, Steckler T. Removing obstacles in neuroscience drug discovery: the future path for animal models. Neuropsychopharmacology. 2009;34:74–89. doi: 10.1038/npp.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kundaje A, Kyriazopoulou-Panagiotopoulou S, Libbrecht M, Smith CL, Raha D, Winters EE, Johnson SM, Snyder M, et al. Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Res. 2012;22:1735–47. doi: 10.1101/gr.136366.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang Z, Willard HF. Evidence for sequence biases associated with patterns of histone methylation. BMC Genomics. 2012;13:367–72. doi: 10.1186/1471-2164-13-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–8. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, et al. Landscape of transcription in human cells. Nature. 2012;469:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cheng C, Alexander R, Min R, Leng J, Yip KY, Rozowsky J, Yan KK, Dong X, et al. Understanding transcriptional regulation by integrative analysis of transcription factor binding data. Genome Res. 2012;22:1658–67. doi: 10.1101/gr.136838.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chey WD, Kurlander J, Eswaran S. Irritable bowel syndrome: a clinical review. JAMA. 2015;313:949–58. doi: 10.1001/jama.2015.0954. [DOI] [PubMed] [Google Scholar]