Abstract
Osteoblasts, the bone forming cells, affect self-renewal and expansion of hematopoietic stem cells (HSCs), as well as homing of healthy hematopoietic cells and tumor cells into the bone marrow. Constitutive activation of β-catenin in osteoblasts is sufficient to alter the differentiation potential of myeloid and lymphoid progenitors and to initiate the development of acute myeloid leukemia (AML) in mice. We show here that Notch1 is the receptor mediating the leukemogenic properties of osteoblast-activated β-catenin in HSCs. Moreover, using cell-specific gene inactivation mouse models, we show that FoxO1 expression in osteoblasts is required for and mediates the leukemogenic properties of β-catenin. At the molecular level, FoxO1 interacts with β-catenin in osteoblasts to induce expression of the Notch ligand, Jagged-1. Subsequent activation of Notch signaling in long-term repopulating HSC progenitors induces the leukemogenic transformation of HSCs and ultimately leads to the development of AML. These findings identify FoxO1 expressed in osteoblasts as a factor affecting hematopoiesis and provide a molecular mechanism whereby the FoxO1/activated β-catenin interaction results in AML. These observations support the notion that the bone marrow niche is an instigator of leukemia and raise the prospect that FoxO1 oncogenic properties may occur in other tissues.
Introduction
Over the last few years it has become increasingly apparent that stromal cells within the bone marrow microenvironment influence the fate of hematopoietic stem cells (HSC) 1–8. In particular, osteoblasts, the bone-forming cells, influence hematopoietic stem cell (HSC) fate 9–12. Osteoblastic cells support and expand HSCs in vitro and increase engraftment in vivo 13. They can both stimulate 11, 12 and limit HSC expansion 14, promote quiescence 3, 15, 16, initiate HSC mobilization 17, 18, and regulate B lymphopoiesis 19, 20 and myeloproliferation 21. They also integrate sympathetic nervous system signaling and HSC regulation 22. In addition, mesenchymal stem cells (MSCs) of the osteoblast lineage affect HSC function in mouse models of myeloproliferative neoplasms (MPN) 23, 24 resulting in the development of myelodysplasia (MDS) and acute myeloid leukemia (AML) 25. Osteoblastic cells also differentially regulate the progression of chronic versus acute myeloid leukemia 26.
Recent findings have extended the regulatory role of osteoblasts beyond hematopoiesis and into the initiation of hematological disease by showing that genetic alterations in osteoblasts can induce AML in mice and are associated with AML development in humans 27. Indeed, constitutive activation of β-catenin signaling in mouse osteoblasts disrupts hematopoiesis by shifting the differentiation potential of HSC progenitors to the myeloid lineage, compromising B-lymphopoiesis and inducing anemia. As a result, granulocyte/monocyte progenitors accumulate and AML develops which, is characterized by infiltration of blood, bone marrow, spleen and liver with immature myeloid and dysplastic cells. This type of osteoblast-induced AML phenotype can be transferred to healthy mice and is associated with clonal evolution at the cytogenetic level. The pathway transmitting the leukemogenic signal of activated β-catenin in osteoblasts involves induction of Notch signaling in HSC progenitors. These observations are relevant to human disease, because β-catenin nuclear accumulation in osteoblasts and activation of Notch signaling in hematopoietic cells was detected in a population of patients with MDS, AML, and AML that arose from a prior MDS.
The molecular mechanisms underlying the oncogenic function of β-catenin in osteoblasts are incompletely understood. With the goal to improve this knowledge we sought transcription factors that could interact with β-catenin and affect its activity in osteoblasts. Among the many candidates we focused on FoxO1, a member of the FoxO family of Forkhead transcription factors, because it interacts with β-catenin in osteoblasts, as well as in other cells types, where it acts to oppose β-catenin function 28–31. At the same time, FoxO1 is required to maintain the function and immature state of leukemia-initiating cells 32 raising the possibility that, at least part of the AML-potentiating properties of FoxO1 might be due to its expression in osteoblasts in addition to its expression in hematopoietic cells. These observations led us to hypothesize that, that FoxO1 may also influence the leukemogenic function of constitutive active β-catenin in osteoblasts.
In testing this working hypothesis through genetic and molecular means, we observed that FoxO1 in osteoblasts does interact with β-catenin, to upregulate Notch ligand expression on osteoblasts and to subsequently induce Notch signaling in long term repopulating HSC progenitors. This interaction leads to anemia, myeloid expansion and AML development.
Materials and Methods
Animals
FoxO1fl/fl, α1(I)Collagen-Cre [α1(I)Col-Cre], and Catnb+/lox(ex3) mice have been reported 33–36. Specific deletion of Notch1 and Notch2 in hematopoietic cells was obtained by breeding Notch1fl/fl 42 (Purchased from Jax, Stock# 007181) or Notch2fl/fl mice 37 (purchased from the Jackson Laboratory, Stock# 010525) with Vav-cre transgenic mice 38 (purchased from the Jackson Laboratory Stock# 008610). The comparative analysis of all histological and flow cytometry measurements was performed at 1 month of age because Ctnnb1CAosb and Ctnnb1CAosb;FoxO1osb−/− mice die between 4 and 6 weeks of age. Additional details are provided in the supplementary Information. All the protocols and experiments were conducted according to the guidelines of the Institute of Comparative Medicine, Columbia University.
Microarray
Total RNA was extracted from primary osteoblasts isolated from mouse calvaria using Trizol reagent (Invitrogen). Microarray analysis was performed using the GeneChip 3′ IVT Express kit and mouse genome 430 2.0 array gene chips (Affymetrix). Detailed protocol is provided in Supplementary Information.
Hematological measurements and peripheral blood morphology
Blood was collected by cardiac puncture and cell counts were performed on a FORCYTE Hematology Analyzer (Oxford Science Inc.). Further details are included in Supplementary Information.
Reporter constructs and luciferase assays
Mouse FoxO1, FoxO3, FoxO4 and β-catenin expression constructs were transfected in HEK293T, OB-6 or primary osteoblasts. Further details about the preparation of reporter constructs and luciferase assays are given in Supplementary Information.
Antibodies and Flow Cytometry analysis
Freshly isolated bone marrow cells and spleen cells were resuspended in flow-staining buffer (PBS plus 2% FBS) and primary conjugated antibodies were added. After 30 minutes of incubation at 4°C, cells were washed twice before flow cytometry analysis. Detailed staining protocol and listing of antibodies are given in Supplementary Information.
Histological analysis of murine bone, spleen and liver
Murine long bones, spleen and liver were collected from one month old mice, fixed overnight in 10% neutral formalin solution, embedded in paraffin, sectioned at 5 μm, and stained with haematoxylin and eosin (H&E). Murine bones were decalcified prior to paraffin embedding. Immunohistochemistry details are provided in Supplementary Information.
Bone marrow transplantation
Ctnnb1CAosb mice, Notch1fl/fl;vav-cre, Notch2fl/fl;vav-cre and their WT control littermates were all CD45.2 congenic mice. Therefore, for transplantation experiments, donor derived bone marrow cells were labeled with CellTrace Far Red DDAO-SE fluorescent dye (Invitrogen) according to the manufacturer’s instructions. Further details are given in the Supplementary Information.
Assessment of chimerism
Engraftment efficiency in recipients was monitored by donor contribution of cells with red fluorescence in the blood, bone marrow, spleen and thymus of recipients using FACS analysis. Additional details are provided in Supplementary Information.
Statistical analysis
All data are represented as mean ± standard deviation. Statistical analyses were performed using a one-way ANOVA followed by Student-Newman-Keuls test and a p value less than 0.05 was considered significant.
Results
FoxO1 promotes β-catenin signaling in osteoblasts
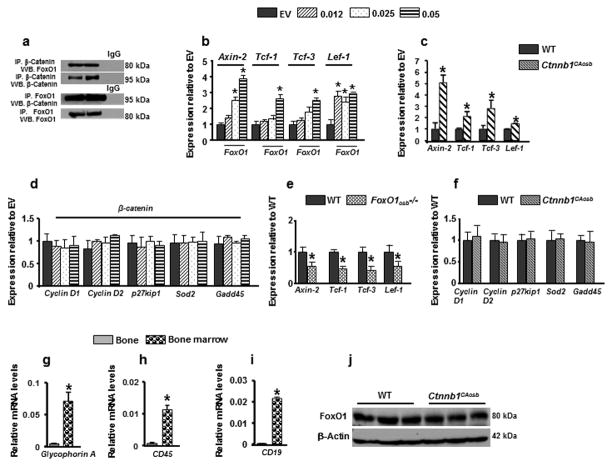
To determine whether FoxO1 affects β-catenin signaling in osteoblasts, we examined if the two endogenous proteins interact. FoxO1 physically associated with β-catenin in osteoblasts (Figure 1a). Consistent with this observation, expression of the β-catenin transcriptional targets, Axin2, Tcf1, Tcf3 and Lef1 was increased following forced expression of FoxO1 in osteoblasts (Figure 1b). Expression of the same β-catenin target genes was also upregulated in the bone of Ctnnb1CAosb mice (Figure 1c). In contrast, expression of the FoxO1 targets cyclin D1, D2, p27Kip1, Superoxide Dismutase 2 (Sod2) and Gadd45 were not affected by forced expression of β-catenin in osteoblasts (Figure 1d). In vivo, expression of Axin2, Tcf1, Tcf3 and Lef1 was decreased in bones from mice harboring an osteoblast-specific inactivation of FoxO1 (FoxO1osb−/−) as compared to wild type (WT) littermates (Figure 1e). In contrast, expression of the FoxO1 targets was not altered in the bone of Ctnnb1CAosb mice expressing the constitutively active β-catenin allele in osteoblasts (Figure 1f). Confirming the purity of the material used to assess gene expression in bone, expression of the blood-specific genes GlycophorinA, CD19 and CD45 was barely detectable in bone (Figures 1g–i). FoxO1 protein levels were not altered in Ctnnb1CAosb mice (Figure 1j). Taken together, these observations suggest that FoxO1 and β-catenin could form a functional complex in which FoxO1 acts as a coactivator of β–catenin required for β-catenin activity.
Figure 1. FoxO1 interacts with β-catenin in osteoblasts.
(a) Coimmunoprecipitation (IP) of β-Catenin and FoxO1 in cell lysates from osteoblast cells followed by immunoblot analysis. Blots were representatives of n=3. (b–f) Real-Time PCR analysis of (b) Axin-2, Tcf-1, Tcf-3 and Lef-1 expression in osteoblasts transfected with increasing concentrations of FoxO1 construct. EV denotes Empty vector. Data are representative of 3 independent experiments. *p < 0.05 versus EV-transfected cells. (c) Real-Time PCR analysis of Axin-2, Tcf1, Tcf-3 and Lef-1 in the bones of wild type and Ctnnb1CAosb mice (Identical data are shown in Figure 2b–e). (d) Real-Time PCR analysis of Cyclin D1, Cyclin D2, p27kip1, Sod2 and Gadd45 expression in osteoblasts transfected with increasing concentrations of β-catenin. EV denotes Empty vector. Data are representative of three independent experiments. *p < 0.05 versus EV-transfected cells. (e) Quantitative Real-Time PCR analysis of Axin-2, Tcf1, Tcf-3 and Lef-1 in the bones of wild type and FoxO1osb−/− mice. Total RNA was isolated from flushed long bones. n=4. *p < 0.05 versus wild type. (f) Quantitative Real-Time PCR analysis of Cyclin D1, Cyclin D2, p27kip1, Sod2 and Gadd45 gene expression in bones of wild type and Ctnnb1CAosb mice. Total RNA was isolated from flushed long bones. (g–i) Real-Time PCR analysis of (g) glycophorin A (h) CD45 (i) CD19 in the flushed long bones and bone marrow. n=4. *p < 0.05 versus WT. (j) Immunoblot analysis showing FoxO1 protein levels in bones lysates of wild type and Ctnnb1CAosb mice. Blot is representative of n=3. Results are mean ± SD. All mice were 1 month of age.
FoxO1 induces anemia and potentiates myeloid expansion in response to activated β-catenin
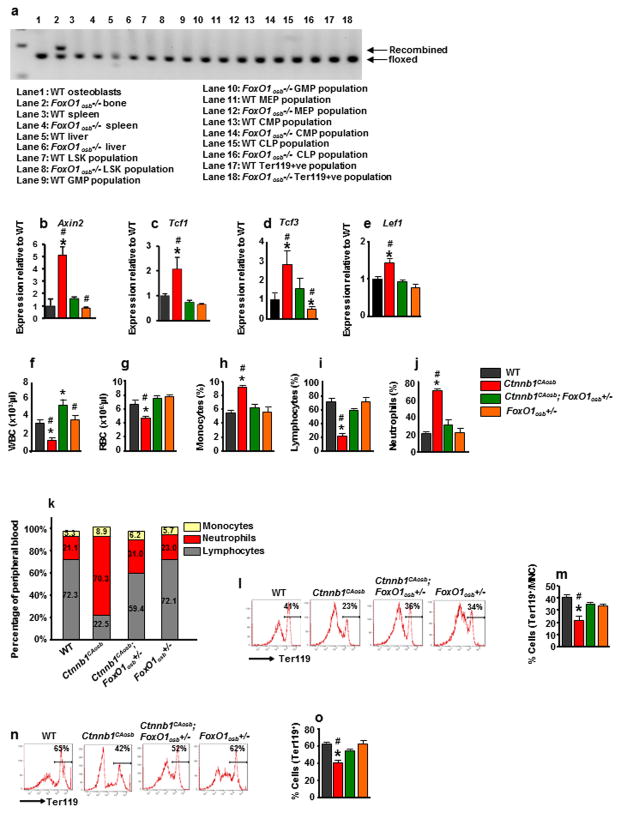
To investigate whether FoxO1 synergizes with β-catenin in osteoblasts to deregulate hematopoiesis in Ctnnb1CAosb mice, we generated mice lacking FoxO1 in osteoblasts by crossing mice harboring a floxed allele of FoxO1 with mice expressing Cre under the control of 2.3 kb of the proximal promoter of the mouse pro- α1(I)Collagen gene 39. Prior to analyzing these mutant mice we verified we had deleted FoxO1 in osteoblasts but not in HSC or other cell types (Figure 2a). The expression of Axin2, Tcf1, Tcf3 and Lef1 was normalized in the compound mutant mice confirming that removing one allele of FoxO1 had normalized β-catenin signaling in osteoblasts (Figures 2 b–e).
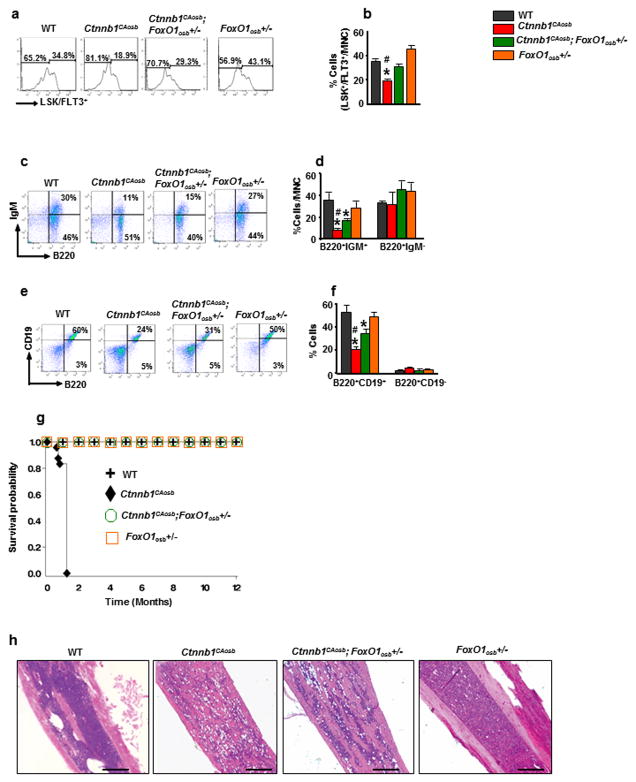
Figure 2. FoxO1 haploinsufficiency in osteoblasts rescues anemia and peripheral blood leukocytosis and monocytosis of Ctnnb1CAosb mice.
(a) FoxO1 recombination in the femur, spleen, liver and indicated hematopoietic populations from wild-type and FoxO1osb−/− mice. Bone marrow cells were flushed from the femur prior to DNA extraction. (b–e) Real-Time PCR analysis of (b) Axin2 (c) Tcf1 (d) Tcf3 and (e) Lef1 gene expression in wild type, Ctnnb1CAosb, Ctnnb1CAosb;FoxO1osb+/− and FoxO1osb+/− mice. (f–j) Total count of (f) WBCs, (g) RBCs, (h) Monocytes (i) Lymphocytes and (j) Neutrophils in the blood. (k) Relative percentage of white blood cell populations in the peripheral blood of wild type and Ctnnb1CAosb mice. (l) Flow cytometry analysis showing erythroid cells and (m) Percentage of Ter119+ cells in the bone marrow. (n) Representative images of Flow cytometry analysis showing erythroid cells and (o) Percentage of Ter119+ population in the spleen. In (b–e) N- 4 mice per group. In (f–o) N=6 mice per group. *p < 0.05 versus WT. and # p < 0.05 versus Ctnnb1CAosb;FoxO1osb+/−. Results are mean ± SD. All mice were 1 month of age.
We reasoned that if the activating mutation of β-catenin in osteoblasts causes leukemia in a FoxO1-dependent manner then removing one FoxO1 allele should normalize hematopoiesis in Ctnnb1CAosb mice. Consistent with this hypothesis and cell culture results presented in Figure 1, anemia, peripheral monocytosis, neutrophilia and lymphocytopenia, features observed in Ctnnb1CAosb mice 27, were all corrected in Ctnnb1CAosb;FoxO1osb+/− mice (Figures 2f–k and supplementary Table 1). Likewise, the decrease in erythroid cells observed in both the bone marrow and spleen of Ctnnb1CAosb mice were rescued in Ctnnb1CAosb;FoxO1osb+/− animals (Figures 2l–o). Total bone marrow cellularity was decreased in Ctnnb1CAosb mice but was partially rescued in Ctnnb1CAosb;FoxO1osb+/− animals (Supplementary Table 1).
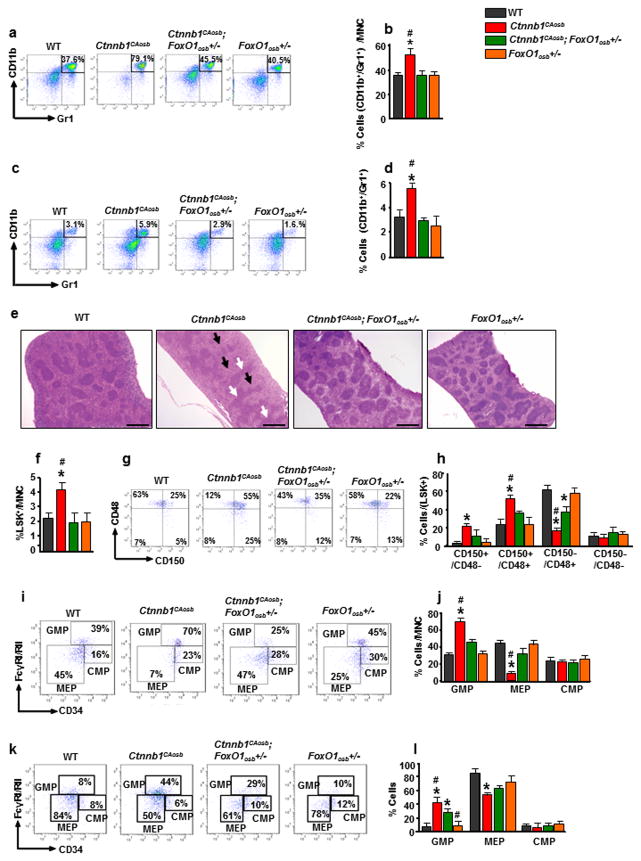
Ctnnb1CAosb mice are characterized by a shift in the differentiation of HSCs to the myeloid lineage at the expense of B-lymphopoiesis 27. Therefore, we examined whether haploinsufficiency at the FoxO1 locus in osteoblasts would correct deregulation of these two lineages in the bone marrow and spleen. We observed that the increase in the relative percentage of myeloid (CD11b+/Gr1+) cells seen in the Ctnnb1CAosb bone marrow was normalized in Ctnnb1CAosb;FoxO1osb+/− mice (Figures 3a and b). Likewise, the percentage of myeloid cells in the spleen of Ctnnb1CAosb;FoxO1osb+/− mice was similar to that of WT animals (Figures 3c and d). The loss of typical splenic architecture, expansion of red pulp and coalescence of white pulp, observed in Ctnnb1CAosb mice was inhibited in Ctnnb1CAosb;FoxO1osb+/− mice (Figure 3e).
Figure 3. FoxO1 haploinsufficiency rescues myeloid lineage expansion in Ctnnb1CAosb mice.
(a) Representative flow cytometry image and (b) Flow cytometry analysis showing percentage of CD11b+/Gr1+ cells in the bone marrow. (c) Representative FACS image and (d) Percentage of CD11b+/Gr1+ population in the spleen. (e) Normal splenic architecture in Ctnnb1CAosb;FoxO1osb+/− mice. White arrows indicate coalescence of white pulp and black arrows indicate expansion of red pulp. Scale bars, 500μm. (f) Percentage of LSK cells in the bone marrow (g) Representative FACS image of LSK+ subpopulations/CD150/CD48 population in the bone marrow and (h) Flow cytometry analysis showing percentage of LT-HSCs, ST-HSCs and MPPs in the bone marrow. (i and j) Flow cytometry analysis showing (i) Representative FACS images and (j) Percentage of myeloid progenitor populations in the bone marrow. (k) Representative images of flow cytometry analysis showing myeloid progenitor populations and (l) Percentage of myeloid progenitor populations in the spleen. N=6 mice per group. *p < 0.05 versus WT and # p < 0.05 versus Ctnnb1CAosb;FoxO1osb+/−. MNC: mononuclear cells. Results are mean ± SD. All mice were 1 month of age.
Because monocytes and granulocytes originate from the same progenitors, we examined the hematopoietic stem and progenitor cell (HSPC) populations in the bone marrow. The HSPC pool size, defined by the percentage of Lin-Sca+c-Kit+ (LSK) cells, is 2- fold greater than in WT littermates (Figure 3f). FoxO1 haploinsufficiency in osteoblasts of Ctnnb1CAosb;FoxO1osb+/− mice reversed the defect in LSK cells. Similarly, it prevented the increase in the LSK+/CD150+/CD48− subset of long term repopulating HSC progenitors (LT-HSCs) (Figures 3g and h). In the bone marrow, within the myeloid progenitor population (Lin-Kit+Sca1−), alterations in the granulocyte/monocyte progenitor subset (CD34/FcgRII/III, GMP population), observed in Ctnnb1CAosb mice were rescued in Ctnnb1CAosb; FoxO1osb+/− mice (Figures 3i and j). Likewise, Ctnnb1CAosb;FoxO1osb+/− mice showed normal GMP population percentage in the spleen (Figures 3k and l).
Taken together these findings indicate that defects in bone marrow, peripheral and spleen hematopoiesis caused by constitutive active β-catenin signaling in osteoblasts are fully rescued by removing one allele of FoxO1 from osteoblasts.
Constitutively active β-catenin acts through FoxO1 in osteoblasts to induce AML development
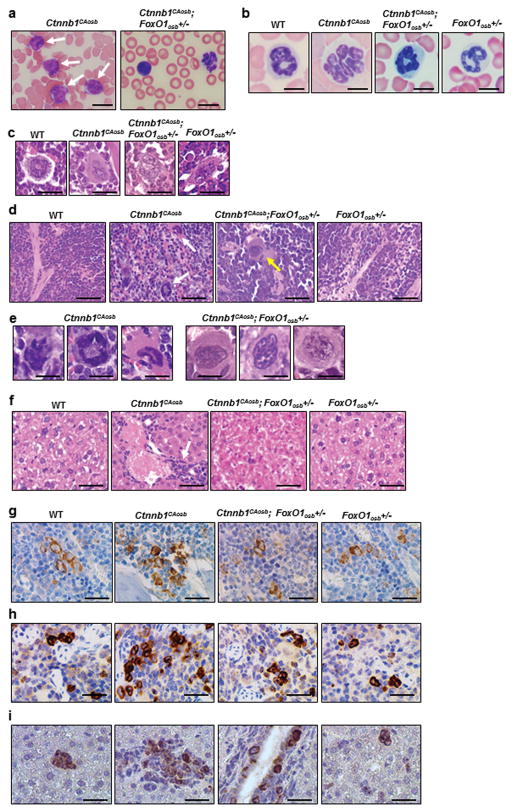
We next asked whether FoxO1, in addition to mediating deregulation of hematopoiesis in Ctnnb1CAosb mice, is also involved in the development of myeloid malignancy. Histological analysis showed that the cellular alterations observed in Ctnnb1CAosb mice associated with their AML phenotype were rescued in Ctnnb1CAosb;FoxO1osb+/− animals. Specifically, monocytic and myeloid cells of the long bones, spleen and liver were similar in Ctnnb1CAosb;FoxO1osb+/− and wild type mice (Figures 4a–f). The appearance of blasts and neutrophils with nuclear hypersegmentation in the blood (Figures 4a and b), infiltration of myeloid and monocytic cells, and increased atypical micro-megakaryocytes in the bone marrow (Figure 4c) and spleen (Figure 4d and e), observed in Ctnnb1CAosb animals were all eliminated in Ctnnb1CAosb;FoxO1osb+/− mice. The liver of Ctnnb1CAosb mice is characterized by focal blast infiltration at periportal sites rather than extensive infiltration across the entire tissue. This phenotype is similar to other mouse AML models which present with variable extent of leukemia blast infiltration 40 such as the AML model of human AML1/MDS1/EVI1 fusion protein 41. In the Ctnnb1CAosb model limited infiltration in the liver may be due to the dependency of AML on the abnormal marrow niche. Inactivation of one allele of FoxO1 in osteoblasts abrogated\monocyte infiltration or clusters of atypical cells in the liver of Ctnnb1CAosb;FoxO1osb+/− mice (Figure 4f). Myeloperoxidase staining of immature myeloid cells in bone marrow, spleen and liver of Ctnnb1CAosb;FoxO1osb+/− mice, was similar to that of WT animals, establishing that leukemogenesis did not occur in these mice (Figures 4g–i).
Figure 4. FoxO1 haploinsufficiency in osteoblasts prevents AML in Ctnnb1CAosb mice.
(a and b) Lack of (a) blasts (white arrows) and (b) hypersegmented neutrophils in the blood of Ctnnb1CAosb;FoxO1osb+/− mice. Scale bars, 10 μm. (c) Normal megakaryocytes in bone marrow and (d) Normal spleen histology in Ctnnb1CAosb;FoxO1osb+/− mice. Solid arrows in Ctnnb1CAosb spleen indicate atypical megakaryocytes. Dotted arrow in Ctnnb1CAosb;FoxO1osb+/− spleen indicates normal megakaryocyte. (e) Normal megakaryocytes in the spleen of Ctnnb1CAosb;FoxO1osb+/− mice. (f) Normal liver histology in Ctnnb1CAosb;FoxO1osb+/− mice. Arrow indicates a cluster of immature cells in the liver of Ctnnb1CAosb mice. (g and h) MPO staining of (g) bone marrow and (h) spleen showing invasion of myeloid cells and in (i) Liver showing focal aggregation of immature myeloid cells in Ctnnb1CAosb but not in Ctnnb1CAosb;FoxO1osb+/− littermates. In figures c–i, scale bars, 20 μm. All mice were 1 month of age.
The reduction of the lymphoid-biased multipotential LSK+/FLT3+ progenitors in the bone marrow (Figures 5a and b) was fully reversed by FoxO1 haploinsufficiency in osteoblasts of Ctnnb1CAosb;FoxO1osb+/− mice. B-lymphopoiesis was partially reversed in Ctnnb1CAosb;FoxO1osb+/− mice (Figures 5c–f). Lastly, FoxO1 haploinsufficiency in osteoblasts prevented the early lethality phenotype of Ctnnb1CAosb mice since Ctnnb1CAosb;FoxO1osb+/− mice lived and were healthy for at least one year, the entire time that they were observed (Figure 5g). Importantly, reduction of FoxO1 levels in osteoblasts of Ctnnb1CAosb;FoxO1osb+/− mice did not rescue the osteopetrotic phenotype that is observed in Ctnnb1CAosb mice, suggesting that AML development occurs independent of osteopetrosis (Figure 5h). Collectively, these results indicate that the development of AML by constitutive activation of β-catenin in osteoblasts depends on FoxO1. In addition they demonstrate an oncogenic role of FoxO1 in osteoblasts that is triggered by constitutive activation of β-catenin in the same cells and potentiates leukemogenesis.
Figure 5. FoxO1 haploinsufficiency in osteoblasts attenuates altered B-lymphopoiesis and prevents lethality in Ctnnb1CAosb mice.
(a and b) Flow cytometry analysis showing (a) Representative images of LSK+/FLT3+ population and (b) Percentage of LSK+/FLT3+ cells in the bone marrow. (c) Flow cytometry analysis showing representative images for B-cell populations in the bone marrow. (d) Proportion of B-cell populations in the bone marrow. (e) Flow cytometry analysis showing representative image for B-cell populations in the spleen. (f) Proportion of B-cell populations in the spleen. N=6 mice per group. *p < 0.05 versus WT and # p < 0.05 versus Ctnnb1CAosb;FoxO1osb+/−. (g) Kaplan-Meier survival curve showing life span in indicated groups. Wild type (n=10), Ctnnb1CAosb (n=8), Ctnnb1CAosb;FoxO1osb+/− (n=10) and FoxO1osb+/− (n=10). (h) H&E staining of paraffin embedded long bone sections. Scale bars, 250 μm. Results are mean ± SD. MNC: mononuclear cells. In a–f and h, mice were 1 month of age.
A β-catenin/FoxO1 interaction in osteoblasts regulates HSC function through Notch signaling
Next, we sought to investigate the molecular mechanisms through which FoxO1 synergizes with β-catenin to induce AML development. We have previously shown that constitutive activation of β-catenin in osteoblasts induces AML, in part by promoting the expression of Jagged-1 in the same cells. As a result Notch signaling is activated in long term HSC progenitors (LT-LSK cells)27. To obtain genetic proof of this mechanism, we inactivated Notch signaling specifically in hematopoietic cells of Ctnnb1CAosb mice by inactivating either the Notch1 or the Notch2 receptor. Bone marrow cells isolated from C57BL/6J (CD45.2) mice lacking Notch1 or Notch2 in hematopoietic cells (Notch1fl/fl;Vav-Cre and Notch2fl/fl;Vav-Cre mice), or their WT littermates (Notch1fl/fl and Notch2fl/fl mice), were labelled with far red florescent dye and transplanted into the liver of 2 day-old, lethally-irradiated CD45.2 Ctnnb1CAosb mice or WT littermates as previously described. We verified impaired Notch signaling in Notch1fl/fl;Vav-Cre knockout bone marrow cells (Supplementary Figure 1a). Overall chimerism, 2 weeks after transplantation, reached 50%–70% in all examined tissues (Supplementary Figure 1b and c). Our analysis indicated 40%–50% and 67%–80% chimerism within the LSK and LT-LSK populations, the Leukemia Initiating Population from Ctnnb1CAosb mice, respectively (Supplementary Figure 1d and e). Far red-labeled donor cells were able to differentiate into the myeloid (CD11b+), B-lymphoid (CD19+), and erythroid (Terr 119+) lineages (Supplementary Figure 1f–k).
Notch signaling was also assessed in LSK cells following 10 weeks of transplantation. In the transplanted Ctnnb1CAosb mice Notch signaling was decreased as tested by the expression of Notch transcriptional targets (Figures 6a and b). At this time Notch1 and Notch2 recombination in the marrow and spleen of transplanted Ctnnb1CAosb mice reached 70%–90% (Figures 6c and d). This degree of recombination along with our observation that host hematopoiesis is ablated in lethally irradiated neonate Ctnnb1CAosb recipients suggest that the LSK cells used to assess Notch signaling are mainly derived from donor hematopoiesis.
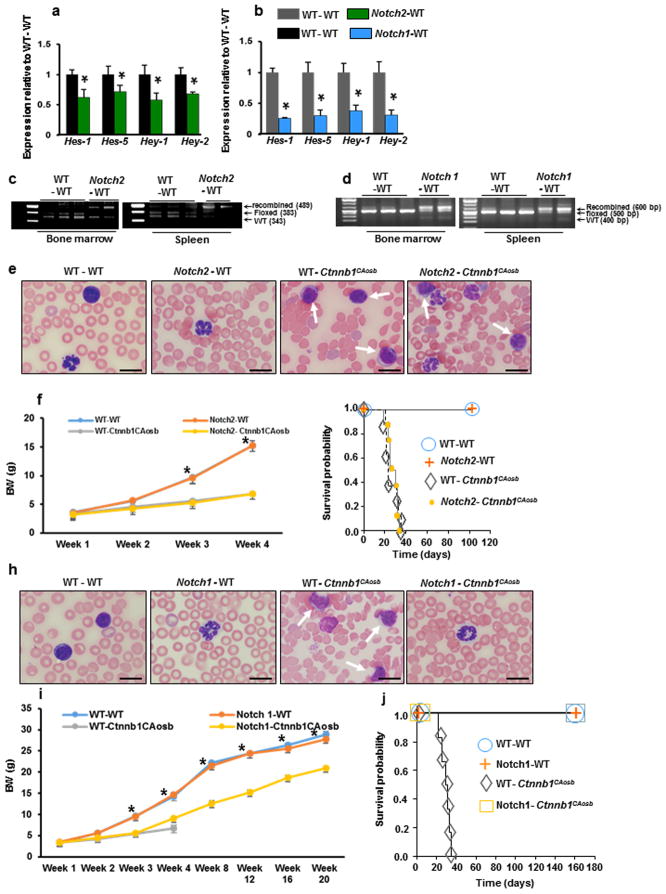
Figure 6. Notch1 inactivation in hematopoietic cells inhibits AML development in Ctnnb1CAosb mice.
Bone marrow cells were obtained from mice with a deletion of Notch1 or Notch2 in the hematopoietic population, (Notch1fl/fl;Vav-Cre and Notch2fl/fl;Vav-Cre mice), or control littermates, (Notch1fl/fl and Notch2fl/fl mice). From each respective donor mouse, 1×105 bone marrow cells were transplanted into the liver of each lethally-irradiated (600 rads, split dose) neonate Ctnnb1CAosb or wild type recipient. (a and b) Quantitative Real-Time PCR analysis of Notch target genes Hes 1, Hes 5, Hey 1 and Hey 2 in the LSK population of wild type mice transplanted with (a) Wild type and Notch2-deficient bone marrow cells and (b) Wild type mice transplanted with bone marrow cells from wild type and Notch1-deficient bone marrow cells, 10 weeks following transplantation. (c and d) PCR analysis of (c) Notch2 and (d) Notch1 allele excision in transplanted recipients. (e) Blasts in the blood of Ctnnb1CAosb mice transplanted with Notch2 deficient bone marrow cells. Scale bars, 10 μm. (f) Body weight of mice transplanted with Notch2-deficient or Notch2 wild type control (Notch2fl/fl) bone marrow cells. (g) Survival curve in mice transplanted with Notch2 deficient bone marrow cells. (h) Lack of blasts in the blood of Ctnnb1CAosb mice transplanted with Notch1 deficient bone marrow cells. Scale bars, 10 μm. (i) Body weight of mice transplanted with Notch1-deficient or Notch1 wild type control (Notch1fl/fl) bone marrow cells. (j) Survival curve in mice transplanted with Notch1 deficient bone marrow cells. In A–B, n=3 mice per group. In g and j, n= 6 mice per group. In f, n=8 mice in WT-WT, Notch2 - WT transplanted groups at all indicated time points. n= 8 mice in WT- Ctnnb1CAosb transplanted group at week 1, 2, 3 and n=3 mice at week 4. n =8 mice in Notch2- Ctnnb1CAosb transplanted group at week 1, 2, 3 and n=4 mice at week 4. *p < 0.05 versus Notch1- Ctnnb1CAosb transplanted group. In i, n=6 mice per group in WT-WT, Notch1-WT and Notch1- Ctnnb1CAosb transplanted groups at all indicated time points. n=6 mice in WT- Ctnnb1CAosb transplanted group at week 1, 2, 3 and n=3 mice at week 4. *p < 0.05 versus Notch 1- Ctnnb1CAosb transplanted group. In A and B *p < 0.05 versus WT-WT transplanted mice. Data are pooled from two independent experiments. Results are mean ± SD. Solid arrows indicate blasts and dotted arrows indicated hypersegmented neutrorphils.
Recipients of Notch2-deficient hematopoietic cells showed heavy infiltration of blood with blasts and dysplastic neutrophils (Figure 6e); their numbers ranged from 18–75% and 20–70%, respectively, within 20 days following transplantation. Transplanted mice maintained low body weight (Figure 6f), and similar to the Ctnnb1CAosb mice transplanted with wild type bone marrow cells, died within 5 weeks following transplantation (Figure 6g). In contrast, transplantation of Ctnnb1CAosb neonates with Notch1-deficient hematopoietic cells prevented AML development (Figure 6h), progressively increased body weight (Figure 6i) and rescued lethality as transplanted Ctnnb1CAosb mice survived for at least 23 weeks (Figure 6j), the entire time they were observed. These results indicate that activation of Notch signaling in HSCs of Ctnnb1CAosb mice mediates the leukemogenic signal of osteoblasts and that this event is facilitated by Notch1.
In view of this observation, we asked whether β-catenin and FoxO1 act synergistically to activate Jagged-1/Notch signaling and/or whether FoxO1, in response to activated β-catenin induces a distinct pathway that deregulates HSC fate and is also required for AML development.
To distinguish between these two possibilities we performed an unbiased approach, microarray analysis. To identify potential targets we looked for complementary changes in the expression of secreted molecules in microarrays of Ctnnb1CAosb and FoxO1osb−/− osteoblasts. The aim of this experiment was to identify FoxO1 targets and to generate compound mutant mice to examine whether β-catenin and FoxO1 synergize in vivo to regulate expression of the identified target. For these genetic epistasis experiments we needed to know the genes that are deregulated in the FoxO1−/− osteoblasts so that we could verify that they were not deregulated in the FoxO1+/− osteoblasts. Microarray analysis of 12,000 genes from Ctnnb1CAosb and 34,000 genes from FoxO1osb−/− osteoblasts identified only one gene with opposite changes in its expression in both cell types, Jagged -1, a Notch ligand (Supplementary Table 2).
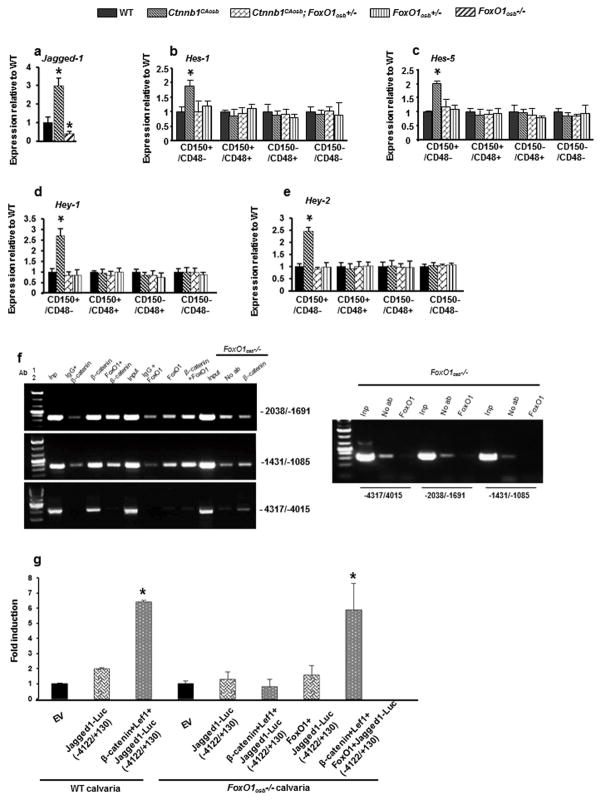
Further experiments confirmed the results of the microarray analysis since, Jagged-1 was upregulated in Ctnnb1CAosb and downregulated in FoxO1osb−/− compared to WT osteoblasts (Figure 7a). We subsequently examined whether FoxO1 mediates the effects of constitutive activation of β-catenin in osteoblasts to increase Notch signaling in LT-LSK cells, the leukemia initiating cells (LICs) which can transfer the AML of Ctnnb1CAosb mice to healthy animals. Similar to the reversal of LSK expansion, removal of one FoxO1 allele from osteoblasts of Ctnnb1CAosb mice reversed the increased expression of the Notch targets Hes1, Hes5, Hey1 and Hey2 specifically in the leukemia-initiating LSK+/CD150+/CD48− subpopulation (LT-HSCs) (Figures 7b–e). These results demonstrate that Notch signaling in hematopoietic cells mediates the leukemogenic signal of activated β-catenin in osteoblasts. They also show that FoxO1 is required for activation of the Jagged-1/Notch pathway by β-catenin in osteoblasts and for AML development.
Figure 7. β-catenin/FoxO1 interaction in osteoblasts regulates HSC function through Notch signaling.
(a) Expression levels of Jagged-1 in WT, Ctnnb1CAosb and FoxO1osb−/− bone. (b–e) Expression of (b) Hes1 and (c) Hes5 (d) Hey1 and (e) Hey2 in LSK+ subpopulations. In (a–e) n=4 mice per group and *p < 0.05 versus WT. (f) Re-Chip analysis of β-catenin and FoxO1. Complexes immunoprecipitated with the indicated first antibodies were eluted form the protein G-sepharose beads and after dilution were immunoprecipitated with the indicated second antibodies. Left panel: FoxO1-deficient osteoblasts were subjected to a single chromatin immunoprecipitation in the absence (no ab) or the presence of the anti-βcatenin antibody. Non immunoprecipitated chromatin was included (input). Promoter occupancy was assessed by PCR amplification using primers corresponding to the (−2039/−1691), (−1431/−1085), or the (−4317/−4015) region of Jagged1 promoter. Right panel: Lack of FoxO1 antibody binding to extracts from FoxO1-deficient osteoblasts. (g) Luciferase activity in primary osteoblasts isolated from WT and FoxO1osb−/− mice co-transfected with β-catenin, Lef1, FoxO1 and Jagged1-Luc (−4112/+130) reporter constructs. Results show fold induction over empty vector. *p< 0.05 versus respective Jagged1-Luc. n=3 replicates. Results are mean ± SD. All mice were 1 month of age.
Transcriptional control of Jagged-1 by β-catenin/FoxO1 signaling in osteoblasts
The regulation of Jagged-1 expression in osteoblasts by canonical Wnt signaling and FoxO1 suggested that it might be a direct target of TCF-1 and/or or FoxO1 in osteoblasts. A close inspection of the DNA sequence of the Jagged-1 promoter region revealed the presence of multiple potential TCF-1 (C/TCTTTG) and FoxO1 (TGTTTT) elements located up to nucleotide −4075 (TCF-1:−4075, −3072, −2626, −2578, −2343, −1993, −1957, −1566, −1232, −1221, −782, FoxO1: −3875, −3861, −3270, −2805, −2442, −2048, −1847, −1835, −1430, −1294). Chromatin immunoprecipitation assays in osteoblasts has shown recruitment of β–catenin to the proximal 2kb promoter region of Jagged-1 (27). An antibody against β–catenin could efficiently immunoprecipitate the −2038/−1691 and −1431/−1085 region of the Jagged-1 promoter spanning elements −1993/−1988, −1957/−1952, −1232/−1227 and −1221/−1216 while weaker binding was observed using primers amplifying the distal −4317/−4015 and −3089/−2776 region of Jagged-1 promoter (Figure 7f). No binding was detectable using primers to amplify the −2675/−2306 or the most proximal −864/−610 region of the promoter. Similarly, using an antibody against FoxO1, recruitment was shown to the −2038/−1691 and −1431/−1085 region of the Jagged-1 promoter while binding to the −4317/−4015 and −3089/−2776 region was not significant. Upon FoxO1 deficiency binding of β–catenin to the −2038/−1691 and −1431/−1085 region of the Jagged-1 promoter was markedly reduced. As a control, FoxO1 antibody did not bind to FoxO1-deficient osteoblasts.
In order to confirm the existence of β-catenin/FoxO1 complex in the same DNA region, rechIP experiments were performed. In these assays, chromatin was first precipitated with antibodies against β–catenin, then was eluted from the beads and subjected to a second immunopurification with antibodies against FoxO1 and vice versa. As shown in Figure 7f, specific signals were observed with both combinations of sequential immunoprecipitations using primers amplifying the −2038/−1691 and −1431/−1085 region of the Jagged-1 promoter. There is no significant rechIP signal even if β-catenin alone is present at the −4317/−4015 region of the promoter showing the simultaneous presence of the two proteins at the elements located within the proximal 2kb region of the Jagged-1 promoter (Figure 7f). The biological importance of these sites was examined by transient transfection assays in FoxO1-deficient or wild type primary osteoblasts using a reporter construct containing the 4.1 Kb of the Jagged-1 promoter fused to the luciferase gene (Jag1-4.1-luc). Overexpression of β-catenin/TCF-1 alone in FoxO1-deficient osteoblasts could not transactivate the Jagged-1 promoter; while re-introduction of FoxO1 induced a 5-fold increase (Figure 7g). Thus, β-catenin is able to form a complex with FoxO1 that enhances β-catenin binding and co-operatively contribute to Jagged-1 upregulation.
Taken together, these observations suggest that β-catenin activation in osteoblasts in conjunction with FoxO1 induces expression of the Notch ligand, Jagged-1, which in turn triggers downstream activation of Notch signaling in adjacent HSCs.
FoxO3 and FoxO4 compensate FoxO1 homozygous deficiency in Ctnnb1CAosb;FoxO1−/− osteoblasts and lead to AML development
In addition to FoxO1, osteoblasts express FoxO3 and FoxO4 and all 3 transcription factors are involved in the regulation of bone mass 31, 42, 43. Based on this expression pattern and the fact that the three isoforms have redundant functions when expressed in hematopoietic cells 44, we examined whether FoxO3 and FoxO4 can compensate for the leukemogenic function of FoxO1 in its absence. For this purpose we generated mice expressing constitutive active β-catenin but lacking both FoxO1 alleles in osteoblasts (Ctnnb1CAosb;FoxO1osb−/− mice). Ctnnb1CAosb;FoxO1osb−/− mice showed deregulated hematopoiesis and presence of blasts and dysplastic cells in the blood, marrow, spleen and liver, indicating that they had developed AML (Supplementary Figures 2a–h). Ctnnb1CAosb;FoxO1osb−/− mice have a compound bone phenotype: they are osteopetrotic due to activation of β-catenin but also have low osteoblast numbers due to FoxO1 deficiency in osteoblasts (Supplementary Figures 3a–c). Bone volume was similar to that of Ctnnb1CAosb mice, probably because the decrease in osteoclast numbers is pronounced enough to compensate for the decrease in osteoblast numbers.
To understand the mechanism of AML development we measured Jagged-1 expression in their osteoblasts and found that it was increased in the osteoblasts of Ctnnb1CAosb;FoxO1osb−/− mice, to the same level as in osteoblasts of Ctnnb1CAosb mice (Supplementary Figure 4a). Dose response assays comparing the ability of FoxO1, FoxO3 and FoxO4 to activate a Jagged-1 reporter construct show that FoxO1 is a more potent activator than FoxO3 and FoxO4 with 3.5- to 10-fold activity at concentrations of 0.01 – 0.5 μM. (Supplementary Figure 4b). Moreover, combination of FoxO3 and FoxO4 at equal doses of 0.06 or 0.2 μM increased Jagged-1 promoter activity to similar levels as 0.2 μM of FoxO1 Taken together, these results indicate that decreased levels of FoxO1 in Ctnnb1CAosb;FoxO1osb+/− osteoblasts prevent FoxO3 and FoxO4 from binding to the Jagged-1 promoter and upregulate Jagged-1 expression and therefore prevent AML development. Homozygous deletion of FoxO1 in Ctnnb1CAosb;FoxO1osb−/− osteoblasts allows FoxO3 and FoxO4 to compensate in upregulating Jagged-1 expression by β-catenin and the AML phenotype to develop.
Discussion
In this study we identified a novel function of FoxO1 in osteoblasts as a leukemia potentiator and deciphered the molecular bases of this function. Specifically, we show that FoxO1 synergizes with activated β-catenin in osteoblasts to initiate a program of gene expression that disrupts hematopoiesis by altering HSC lineage determination, ultimately leading to AML. This entire signaling cascade, which mediates the crosstalk between osteoblasts and HSCs, is initiated by activation of canonical Wnt signaling in osteoblasts. In turn, stabilized β-catenin acts through FoxO1 to promote expression of Notch ligands in osteoblasts, leading to activation of Notch signaling in HSCs. These findings demonstrate that osteoblasts are able to produce signals that affect the fate of HSCs and simultaneously induce an oncogenic process. They also show that FoxO1, a known tumor suppressor, assumes the opposite role in osteoblasts, where it acts as a tumor inducer capable of triggering AML in response to activated β-catenin signaling.
Similar to our finding that FoxO1 is pro-leukemogenic when expressed in osteoblasts, and in spite of the fact that FoxOs are primarily known as tumor suppressors, indications of a tumorigenic role for them have been previously reported in chronic and acute myeloid leukemia. High expression of FOXO3 has been associated with adverse prognosis in AMLs exhibiting normal cytogenetics 45. Furthermore, genetic ablation of FoxO3 reduced disease burden in a mouse model of chronic myeloid leukemia 46. Lastly, activation of both FoxO1 and FoxO3 has been implicated in human AML as means of inducing myeloid block 32. Therefore, whether it acts through osteoblasts or through leukemia blasts FoxO1 is an inducer or potent modulator of myeloid leukemia.
Our observations, together with current evidence indicate that FoxO1 has different functions in activating β-catenin signaling in osteoblasts depending on the stage of osteoblast differentiation, and these distinct functions may differentially regulate normal and malignant hematopoiesis. Indeed, FoxO1 inhibits β-catenin signaling by tethering it away from its transcriptional complex(31) in osteoblast progenitors which are implicated in normal hematopoiesis by affecting HSC lineage determination survival and proliferation 1, 2, 20, 47–49, HSC expansion 11, 12, 20 and quiescence 14, 50, erythroid lineage 11, 12 and B lymphopoiesis 19, 20. In contrast, we show that FoxO1 synergizes with β-catenin in osteoblasts to induce malignant hematopoiesis and the development of AML. These observations suggest that cell differentiation along the osteoblast lineage is a complex process and that manipulating FoxO1/β-catenin signaling in the osteoblast or its progenitor can have different repercussions on hematopoiesis.
An “osteoblastic niche” has been proposed as a specialized microenvironment which provides a quiescent state for the maintenance of hematopoietic cell stemness 9. Changes in it may precede and promote the initiation of genetic events by creating a pre-malignant state characterized by disruption of quiescence-inducing signals or increases in proliferating signaling 51. Our study directly implicates osteoblasts in the development of leukemogenesis. Similarly, deletion of Dicer1 in Osterix-expressing osteoblast progenitors has been shown to induce MDS with pancytopenia and emergence of secondary leukemia 25. These two studies address two different pathways: one that involves microRNA regulation of gene expression, and one that affects expression of canonical Wnt signaling targets. Currently it is not known whether the two pathways intersect, since specific microRNAs expressed in osteoblasts and affecting hematopoiesis have not been identified. This important question needs to be addressed in the future.
The fact that Ctnnb1CAosb mice have an osteopetrotic phenotype raised the possibility that part of the hematopoietic dysfunction may be due to narrowing of the bone marrow cavity. However, our observations demonstrate that leukemogenesis occurs independent of osteopetrosis in Ctnnb1CAosb mice. Inactivation of one FoxO1 allele as well as pharmacological inhibition of Notch signaling 27 rescued dysfunctional hematopoiesis and leukemia in Ctnnb1CAosb mice without affecting osteopetrosis. In support of these observations, HSC progenitors are maintained in osteopetrotic mice and osteoclasts do not affect HSC maintenance or mobilization 52. Additionally, in contrast to the pancytopenic phenotype observed in Ctnnb1CAosb mice, narrowing of the bone marrow cavity in osteopetrosis mouse models does not affect peripheral blood parameters 53, 54. Thus, compromised HSCs function, leukemogenesis and mortality observed in Ctnnb1CAosb mice occur independent of osteopetrosis.
The mechanisms through which osteoblasts affect hematopoiesis are now being elucidated. As they emerge, they suggest a variety of signals that can affect different aspects of hematopoiesis. A functional interaction between osteoblasts and HSCs that involve engagement of Notch1/Jag1 signaling promotes HSC proliferation 11, 12. One involving non-canonical Wnt signaling maintains quiescent long-term HSCs(55), whereas, inactivation of Wnt signaling in osteoblasts disrupts stem cell quiescence, leading to a loss of self-renewal potential through a Shh-mediated pathway 56. Recently, disruption of HIF signaling in osteoprogenitors was shown to directly modulate erythropoiesis 57. These studies underscore the importance of understanding the entire spectrum of functions of osteoblasts in regulating HSC activity and indicate that identifying the mechanisms through which such functions occur can have implications in hematological diseases.
Targeting the hematopoietic niche is currently a new strategy for eradicating persistent and drug-resistant leukemia stem cells 58–60. Our observations show that a new niche- specific interaction between activated β-catenin and FoxO1 which takes place in osteoblasts is sufficient to initiate a complex phenotype of disordered hematopoiesis that is reminiscent of AML in humans. In view of these data, we propose that targeting of FoxO signaling may inhibit osteoblast-induced AML or render the osteoblastic niche hostile to CML or AML cells in other models.
Supplementary Material
Acknowledgments
The authors are grateful to Dr. Makoto Taketo for providing the Catnb+/lox(ex3) mice and to Dr Ronald A. DePinho for providing the FoxO1fl mice. We are also thankful to Steven Shikhel for critical reading of the manuscript, to Alexandra Tarasenko for technical assistance, and to the histology and metabolic unit facility of the Diabetes and Endocrinology Research Center (DERC, supported by NIDDK DK063608-07) and the Molecular Pathology facility of the Herbert Irving Cancer Center of Columbia University Medical Center for help with histological analysis. This work was supported by the National Institutes of Health (R01 AR054447, R01 AR055931 and P01 AG032959 to SK).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary information is available at Leukemia’s website.
Reference List
- 1.Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Lo CC, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 6.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6:251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler IG, Barbier V, Nowlan B, Jacobsen RN, Forristal CE, Patton JT, et al. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat Med. 2012;18:1651–1657. doi: 10.1038/nm.2969. [DOI] [PubMed] [Google Scholar]
- 9.Levesque JP, Helwani FM, Winkler IG. The endosteal ‘osteoblastic’ niche and its role in hematopoietic stem cell homing and mobilization. Leukemia. 2010;24:1979–1992. doi: 10.1038/leu.2010.214. [DOI] [PubMed] [Google Scholar]
- 10.Oh IH, Kwon KR. Concise review: multiple niches for hematopoietic stem cell regulations. Stem Cells. 2010;28:1243–1249. doi: 10.1002/stem.453. [DOI] [PubMed] [Google Scholar]
- 11.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 13.El-Badri NS, Wang BY, Cherry, Good RA. Osteoblasts promote engraftment of allogeneic hematopoietic stem cells. Exp Hematol. 1998;26:110–116. [PubMed] [Google Scholar]
- 14.Nilsson SK, Johnston HM, Whitty GA, Williams B, Webb RJ, Denhardt DT, et al. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood. 2005;106:1232–1239. doi: 10.1182/blood-2004-11-4422. [DOI] [PubMed] [Google Scholar]
- 15.Yoshihara H, Arai F, Hosokawa K, Hagiwara T, Takubo K, Nakamura Y, et al. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 2007;1:685–697. doi: 10.1016/j.stem.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Janzen V, Fleming HE, Riedt T, Karlsson G, Riese MJ, Lo CC, et al. Hematopoietic stem cell responsiveness to exogenous signals is limited by caspase-3. Cell Stem Cell. 2008;2:584–594. doi: 10.1016/j.stem.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayack SR, Wagers AJ. Osteolineage niche cells initiate hematopoietic stem cell mobilization. Blood. 2008;112:519–531. doi: 10.1182/blood-2008-01-133710. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Wang LD, Wagers AJ. Dynamic niches in the origination and differentiation of haematopoietic stem cells. Nat Rev Mol Cell Biol. 2011;12:643–655. doi: 10.1038/nrm3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan CK, Chen CC, Luppen CA, Kim JB, DeBoer AT, Wei K, et al. Endochondral ossification is required for haematopoietic stem-cell niche formation. Nature. 2009;457:490–494. doi: 10.1038/nature07547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu JY, Purton LE, Rodda SJ, Chen M, Weinstein LS, McMahon AP, et al. Osteoblastic regulation of B lymphopoiesis is mediated by Gs{alpha}-dependent signaling pathways. Proc Natl Acad Sci U S A. 2008;105:16976–16981. doi: 10.1073/pnas.0802898105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fulzele K, Krause DS, Panaroni C, Saini V, Barry KJ, Liu X, et al. Myelopoiesis is regulated by osteocytes through Gsalpha-dependent signaling. Blood. 2013;121:930–939. doi: 10.1182/blood-2012-06-437160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katayama Y, Battista M, Kao WM, Hidalgo A, Peired AJ, Thomas SA, et al. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124:407–421. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 23.Walkley CR, Olsen GH, Dworkin S, Fabb SA, Swann J, McArthur GA, et al. A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor gamma deficiency. Cell. 2007;129:1097–1110. doi: 10.1016/j.cell.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim YW, Koo BK, Jeong HW, Yoon MJ, Song R, Shin J, et al. Defective Notch activation in microenvironment leads to myeloproliferative disease. Blood. 2008;112:4628–4638. doi: 10.1182/blood-2008-03-148999. [DOI] [PubMed] [Google Scholar]
- 25.Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krause DS, Fulzele K, Catic A, Sun CC, Dombkowski D, Hurley MP, et al. Differential regulation of myeloid leukemias by the bone marrow microenvironment. Nat Med. 2013;19:1513–1517. doi: 10.1038/nm.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kode A, Manavalan JS, Mosialou I, Bhagat G, Rathinam CV, Luo N, et al. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature. 2014;506:240–244. doi: 10.1038/nature12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- 29.Almeida M, Han L, Martin-Millan M, O’Brien CA, Manolagas SC. Oxidative stress antagonizes Wnt signaling in osteoblast precursors by diverting beta-catenin from T cell factor- to forkhead box O-mediated transcription. J Biol Chem. 2007;282:27298–27305. doi: 10.1074/jbc.M702811200. [DOI] [PubMed] [Google Scholar]
- 30.Iyer S, Han L, Bartell SM, Kim HN, Gubrij I, de CR, et al. Sirtuin1 (Sirt1) Promotes Cortical Bone Formation by Preventing beta (beta)-Catenin Sequestration by FoxO Transcription Factors in Osteoblast Progenitors. J Biol Chem. 2014;289:24069–24078. doi: 10.1074/jbc.M114.561803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iyer S, Ambrogini E, Bartell SM, Han L, Roberson PK, de CR, et al. FOXOs attenuate bone formation by suppressing Wnt signaling. J Clin Invest. 2013;123:3409–3419. doi: 10.1172/JCI68049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sykes SM, Lane SW, Bullinger L, Kalaitzidis D, Yusuf R, Saez B, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146:697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002;224:245–251. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- 35.Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rached MT, Kode A, Silva BC, Jung DY, Gray S, Ong H, et al. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. J Clin Invest. 2010;120:357–368. doi: 10.1172/JCI39901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan Y, Lin MH, Tian X, Cheng HT, Gridley T, Shen J, et al. gamma-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev Cell. 2004;7:731–743. doi: 10.1016/j.devcel.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 38.de BJ, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 39.Rossert J, Eberspaecher H, de CB. Separate cis-acting DNA elements of the mouse pro-alpha 1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J Cell Biol. 1995;129:1421–1432. doi: 10.1083/jcb.129.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, Cardiff RD, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 41.Cuenco GM, Nucifora G, Ren R. Human AML1/MDS1/EVI1 fusion protein induces an acute myelogenous leukemia (AML) in mice: a model for human AML. Proc Natl Acad Sci U S A. 2000;97:1760–1765. doi: 10.1073/pnas.030421197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rached MT, Kode A, Xu L, Yoshikawa Y, Paik JH, DePinho RA, et al. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab. 2010;11:147–160. doi: 10.1016/j.cmet.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambrogini E, Almeida M, Martin-Millan M, Paik JH, DePinho RA, Han L, et al. FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab. 2010;11:136–146. doi: 10.1016/j.cmet.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Santamaria CM, Chillon MC, Garcia-Sanz R, Perez C, Caballero MD, Ramos F, et al. High FOXO3a expression is associated with a poorer prognosis in AML with normal cytogenetics. Leuk Res. 2009;33:1706–1709. doi: 10.1016/j.leukres.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 46.Naka K, Hoshii T, Muraguchi T, Tadokoro Y, Ooshio T, Kondo Y, et al. TGF-beta-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010;463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- 47.Zhu J, Garrett R, Jung Y, Zhang Y, Kim N, Wang J, et al. Osteoblasts support B-lymphocyte commitment and differentiation from hematopoietic stem cells. Blood. 2007;109:3706–3712. doi: 10.1182/blood-2006-08-041384. [DOI] [PubMed] [Google Scholar]
- 48.Taichman RS, Reilly MJ, Emerson SG. Human osteoblasts support human hematopoietic progenitor cells in in vitro bone marrow cultures. Blood. 1996;87:518–524. [PubMed] [Google Scholar]
- 49.Colmone A, Amorim M, Pontier AL, Wang S, Jablonski E, Sipkins DA. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 2008;322:1861–1865. doi: 10.1126/science.1164390. [DOI] [PubMed] [Google Scholar]
- 50.Stier S, Ko Y, Forkert R, Lutz C, Neuhaus T, Grunewald E, et al. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med. 2005;201:1781–1791. doi: 10.1084/jem.20041992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sneddon JB, Werb Z. Location, location, location: the cancer stem cell niche. Cell Stem Cell. 2007;1:607–611. doi: 10.1016/j.stem.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyamoto K, Yoshida S, Kawasumi M, Hashimoto K, Kimura T, Sato Y, et al. Osteoclasts are dispensable for hematopoietic stem cell maintenance and mobilization. J Exp Med. 2011;208:2175–2181. doi: 10.1084/jem.20101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lowell CA, Niwa M, Soriano P, Varmus HE. Deficiency of the Hck and Src tyrosine kinases results in extreme levels of extramedullary hematopoiesis. Blood. 1996;87:1780–1792. [PubMed] [Google Scholar]
- 54.Gowen M, Lazner F, Dodds R, Kapadia R, Feild J, Tavaria M, et al. Cathepsin K knockout mice develop osteopetrosis due to a deficit in matrix degradation but not demineralization. J Bone Miner Res. 1999;14:1654–663. doi: 10.1359/jbmr.1999.14.10.1654. [DOI] [PubMed] [Google Scholar]
- 55.Sugimura R, He XC, Venkatraman A, Arai F, Box A, Semerad C, et al. Noncanonical Wnt signaling maintains hematopoietic stem cells in the niche. Cell. 2012;150:351–365. doi: 10.1016/j.cell.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaniel C, Sirabella D, Qiu J, Niu X, Lemischka IR, Moore KA. Wnt-inhibitory factor 1 dysregulation of the bone marrow niche exhausts hematopoietic stem cells. Blood. 2011;118:2420–2429. doi: 10.1182/blood-2010-09-305664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rankin EB, Wu C, Khatri R, Wilson TL, Andersen R, Araldi E, et al. The HIF Signaling Pathway in Osteoblasts Directly Modulates Erythropoiesis through the Production of EPO. Cell. 2012;149:63–74. doi: 10.1016/j.cell.2012.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 59.Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat Med. 2006;12:1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- 60.Reynaud D, Pietras E, Barry-Holson K, Mir A, Binnewies M, Jeanne M, et al. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell. 2011;20:661–673. doi: 10.1016/j.ccr.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.