Abstract
Recent developments in diffusion magnetic resonance imaging (MRI) make it a promising tool for non-invasive mapping of the spatial organization of axonal and dendritic networks in gray matter regions of the brain. Given the complex cellular environments, in which these networks reside, evidence on the capability of diffusion MRI-based tractography to study these networks is still lacking. In this study, we used a localized diffusion MRI approach to acquire high spatial and angular resolution images of the live mouse hippocampus. The diffusion MRI and tractography results were compared with histology and the Allen mouse brain connectivity atlas using a multi-step image registration pipeline. The results demonstrated that in vivo diffusion MRI data at 0.1 mm isotropic resolution revealed the organization of axonal and dendritic networks in the hippocampus and the tractography results shared remarkable similarity with the viral tracer data in term of their spatial projection patterns. Quantitative analysis showed significant correlations between tractography- and tracer-based projection density measurements in the mouse hippocampus. These findings suggest that high-resolution diffusion MRI and tractography can reveal macroscopic neuronal projections in the mouse hippocampus and are important for future development of advanced tractography methods.
Keywords: Diffusion MRI, in vivo, tractography, mouse hippocampus, Allen mouse brain connectivity atlas
1. Introduction
Mapping structural connectivity of the brain is critical for understanding its structural and functional organization. An array of imaging techniques have been used to dissect structural connectivity from the synaptic level using electron micrograph (Helmstaedter et al., 2013; Takemura et al., 2013) up to the system level using magnetic resonance imaging (MRI) (Lazar, 2010; Van Essen et al., 2013). The last fifteen years have witnessed rapid development in diffusion MRI-based tract reconstruction, or tractography (Mori and van Zijl, 2002; Tournier et al., 2007; Tuch et al., 2002; Wedeen et al., 2008). To date, it is the primary tool for non-invasive mapping of large white matter tracts in the brain and an important component of the Human Connectome Project (Toga et al., 2012; Van Essen et al., 2013).
With rapid advances in MRI techniques, high-resolution diffusion MRI data from the human brain are increasingly available (Gaggl et al., 2014; Setsompop et al., 2013; Ugurbil et al., 2013), and increasing efforts have been made towards tracing small neuronal connections deep in gray matter regions, e.g., small white matter tracts and axonal projections in the hippocampus and cortex (Dell’Acqua et al., 2013; Gomez et al., 2015; Kurniawan et al., 2014; Yassa et al., 2010; Zeineh et al., 2012), in order to map the organization of various anatomical units in these regions. Along these fine connections, tractography faces a vastly different landscape from those in large white matter tracts, as the proximity of small axonal bundles to neurons, astrocytes, and adjacent axonal bundles makes it challenging to determine their trajectories and microstructural properties. Even though the strengths and limitations of diffusion MRI tractography have been investigated in large white matter tracts with tracer-based histological data (Choe et al., 2012; Dauguet et al., 2007; Dyrby et al., 2007; Leergaard et al., 2010; Seehaus et al., 2013; Thomas et al., 2014), evidence on its capability to resolve small neuronal connections in gray matter regions remains scarce.
In this study, we examined the mouse hippocampus and the spatial organization of its axonal and dendritic networks using in vivo high-resolution diffusion MRI. We chose the hippocampus because it plays an important role in memory, spatial navigation, and emotion (Eichenbaum et al., 1996; Strange et al., 2014), and contains an intrinsic network between its subfields with distinct functions (van Strien et al., 2009). Modem viral tracing techniques, such as those used by the Allen Mouse Brain Connectivity Atlas (AMBCA) (Oh et al., 2014), have generated extensive tracer-based neuronal connectivity data in the mouse brain, including the hippocampus. With our recently developed localized high-resolution diffusion MRI technique (Wu et al., 2014) that can “zoom-in” a selected region in the live mouse brain to reach a high spatial resolution, and advanced image processing techniques to co-register histological and MRI data, we examined the capability of high-resolution diffusion MRI by direct comparison of diffusion MRI tractography data and co-registered anterograde viral tracing data from AMBCA.
2. Materials and Methods
2.1 In vivo high-resolution diffusion magnetic resonance imaging
All experimental procedures were approved by the Animal Use and Care Committee at the Johns Hopkins University School of Medicine. Twelve adult mice (C57BL/6, three-month old, female) from the Jackson Laboratory (Bar Harbor, ME) were used in this study. In vivo MRI was performed on a horizontal 11.7 Tesla MR scanner (Bruker Biospin, Billerica, MA, USA) with a triple-axis gradient system. Images were acquired using a quadrature volume excitation coil (72 mm diameter, for excitation) and a receive-only planar surface coil (15 mm diameter). The planar surface coil was placed on top of the head, above the region containing the right hippocampus. During imaging, the mice were anesthetized with isoflurane (1–1.5 %) together with air and oxygen mixed at a 3:1 ratio via a vaporizer. The mice were restrained in an animal holder with ear pins and a bite bar. Respiration was monitored via a pressure sensor (SAII, Stony Brook, NY, USA) and maintained at 40–60 breaths per minute. After imaging, animals recovered within five minutes.
To locate the hippocampus in each mouse brain, multi-slice T2-weighted images were first acquired as an anatomical reference. In this study, the region for localized imaging, or field of excitation (FOE), was set to cover the right hippocampus and surrounding regions, which had a spatial dimension of 8 mm (dorsoventral, the x axis) x 5 mm (mediolateral, the y axis) x 5 mm (rostrocaudal from bregma 0 to -5 mm, the z axis). To achieve localized imaging, spatially selective excitation radio frequency (RF) pulses were designed based on a linear class of large tip-angle (LCLTA) pulses (Pauly et al., 1989), and calculated in Matlab (Mathworks, Natick, MA, USA), with a pulse duration of 2.5 ms and a maximum pulse amplitude of 15 μT, to excite a rectangular FOE in the x–y plane. The performance of the spatially selective excitation RF pulses had been evaluated in our previous report (Wu et al., 2014).
Localized high-resolution diffusion MRI data were acquired using a modified 3D diffusion-weighted gradient- and spin-echo (DW-GRASE) sequence (Aggarwal et al., 2010; Wu et al., 2014) with a spatially selective excitation pulse and a slice-selective refocusing pulse (Mao et al., 1988) that restricted the imaging slab in the z-direction. The sequence acquired 20 echoes after each excitation using a double-sampled EPI readout (Yang et al., 1996). A twin-navigator scheme (Mori and van Zijl, 1998) was implemented to correct motion-induced phase errors, and no respiration trigger was used. The diffusion MRI parameters were: echo time (TE)/repetition time (TR) = 21/500ms; two signal averages; field of view (FOV) = 9.6 mm × 5.6 mm × 5 mm, resolution = 0.1 mm × 0.1 mm × 0.1 mm; four non-diffusion weighted image (b0); 30 (n = 5) or 60 (n=7) diffusion directions (Jones et al., 1999); gradient pulse duration = 4 ms; diffusion separation = 12 ms; and b = 2500 s/mm2 (Alexander and Barker, 2005). It took less than 2 minutes to acquire a single diffusion-weighted image. The total scan time was 63 and 118 minutes for the 30- and 60-direction high angular resolution diffusion imaging (HARDI) (Frank, 2001; Tuch et al., 2003) data, respectively. Raw data from the scanner were Fourier transformed after zero-padding (to 50 μm isotropic resolution) with navigator-based phase corrections in Matlab. Images at lower spatial resolution (0.15 mm, 0.2 mm, and 0.3 mm isotropic) than the actual resolution were obtained by cropping the raw data before zero-padding (also padded to 50 μm isotropic resolution). Using the log-linear fitting method implemented in DTIStudio (http://www.mristudio.org), diffusion tensor was calculated at each pixel along with the apparent diffusion coefficient (ADC) and fractional anisotropy (FA) (Basser et al., 1994).
2.2 Registration of MRI data to the Allen Reference Atlas (ARA)
Projection mapping images from 18 tracer experiments (with injection sites in the hippocampus, listed in supplementary Table S1) were downloaded from the AMBCA (http://connectivity.brain-map.org/). These images were labeled using injected recombinant adeno-associated virus (AAV) tracers expressing enhanced green fluorescent protein (EGFP) and acquired using serial two-photon microscopy (Ragan et al., 2012). A series of image registration procedures (supplementary Fig. S1(A)) were taken to co-register the MRI and tracer data, which were both registered to the Allen Reference Atlas (ARA) (Goldowitz, 2010). Since the localized high-resolution HARDI data only covered partial brain volume, an in vivo whole mouse brain MRI atlas from our previous study (Wu et al., 2013) was used to bridge the localized HARDI datasets and the ARA. First, the whole brain MRI atlas was aligned to the ARA (both re-sampled to 50 μm isotropic resolution) using landmark-based rigid transformation (Diffeomap, www.mristudio.org), followed by intensity-based 12-degree affine transformation and large deformation diffeomorphic metric mapping (LDDMM) (Ceritoglu et al., 2009), utilizing the similar but inverted tissue contrasts between the ARA and fractional anisotropy (FA) image from the MRI atlas. Second, the HARDI datasets were aligned to the transformed whole brain MRI atlas (obtained from the previous step and cropped to match the localized volume in the HARDI datasets) through landmark-based rigid transformation, affine transformation, and LDDMM. The registration accuracy between the ARA and the HARDI datasets were evaluated by measuring the distances between 15 landmarks manually placed throughout the hippocampus in the two datasets (Fig. S1(B)). The average registration error was 0.28 ± 0.14 mm as evaluated by two independent raters. After the registration steps, structural segmentations in the ARA (including 17 hippocampal subfields) were transferred into the HARDI datasets (Fig. S1(C)). Because the hippocampus is a C-shaped structure, we used the septotemporal axis (van Strien et al., 2009) in addition to the rostrocaudal, dorsoventral, and mediolateral axes to locate sub-regions of the hippocampus (Fig. 1A).
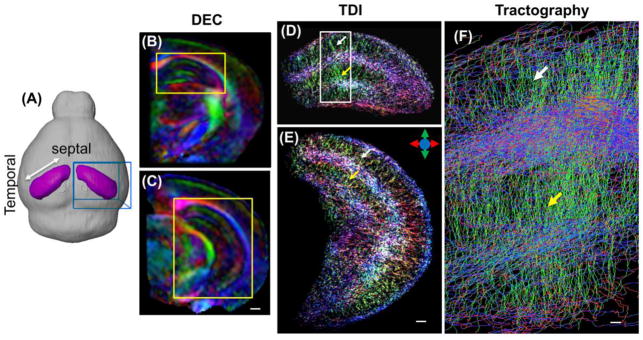
Figure 1.
In vivo high-resolution diffusion MRI of the mouse hippocampus. (A) Images were acquired from a localized imaging volume (the blue box) that contained the right hippocampus (purple) in the mouse brain. (B–C) Coronal direction encoded colormap (DEC) images based on diffusion tensor reconstruction show unique tissue contrasts in the mouse hippocampus. Scale bar = 500 μm. (D–E) Corresponding whole hippocampus tract-density images (TDI) show detailed microstructural organization patterns, such as the radiating patterns in the stratum radiatum and molecular layer of the dentate gyrus. Scale bar = 250 μm. The same patterns can also be visualized from the streamline tractography result (F) from the region in the white box in D. Scale bar = 50 μm. The white and yellow arrows indicate the locations of the stratum radiatum and molecular layer of the dentate gyrus, respectively. The color scheme used here are: red: the left-right axis; green: superior-inferior axis, blue: the anterior-posterior axis, as illustrated by the color arrows in E.
2.3 Diffusion MRI-based tractography
Using MRtrix (http://www.brain.org.au/software/mrtrix/) (Tournier et al., 2012), fiber orientation distribution (FOD) at each voxel was estimated using the constrained spherical deconvolution (CSD) method (Tournier et al., 2007) with a maximum harmonic order of 6. Both deterministic and probabilistic fiber-tracking (Jones and Pierpaoli, 2005; Lazar and Alexander, 2005) was performed based on the FOD data with a step size of 0.005 mm, a minimal length of 0.5 mm, and a maximum angle of 45° between steps. Tracking terminated when the FOD amplitude became less than 0.005, or when fibers exited the manually segmented hippocampal region. The method has been demonstrated in postmortem mouse brain specimens to provide the most sensitive detection of white matter pathways (Moldrich et al., 2010). Whole hippocampus tract density images (TDIs) at a grid size of 10 μm isotropic were generated based on short streamlines (0.4–1 mm in length) from random seed points over the hippocampal region to visualize the microstructural organization (Calamante et al., 2012). Specific fiber streamlines were generated from specific regions of interest (ROIs) as seed regions, which were defined according to the viral tracer injection sites in the corresponding tracer experiments. The resulting seed-based probabilistic streamlines were used to generate TDIs at a grid size of 50 μm isotropic. The tract density values equal the fractions of fibers in a voxel compared to the total number of fibers in the entire volume. TDIs generated from individual mice were averaged to obtain group-averaged TDI, and then registered to the ARA using the same transformation that was used to align the HARDI data.
In each of the 17 hippocampal subfield structural delineations in the ARA, the tracer-based projection density was defined as the fraction of tracer-projected pixels to the total number of pixels in a subfield (quantitative data directly obtained from AMBCA). Similarly, tractography-based projection density was calculated as the fraction of detected voxels with TDI values greater than 0.001 (0.1% of the total fibers within the right hippocampus) to the total number of voxels in each structural region. The threshold was determined by comparing the tractography and tracer data (e.g. in Fig. 4). The projection intensity was also examined with the tracer-based projection intensity defined as the averaged viral tracer intensity in the detected pixels within a subfield and the tractography-based FOD amplitude and tract density values (Willats et al., 2014) in the detected voxels within a subfield.
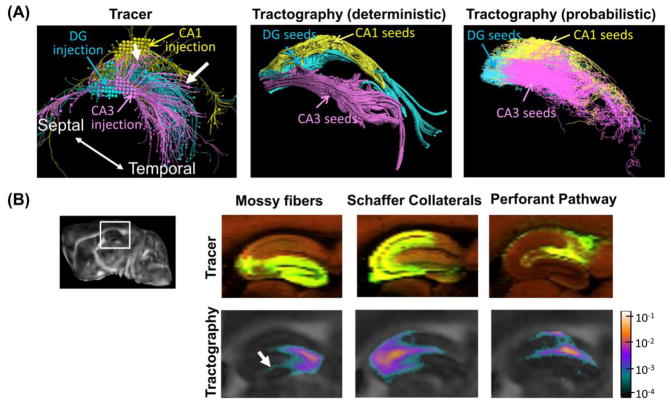
Figure 4.
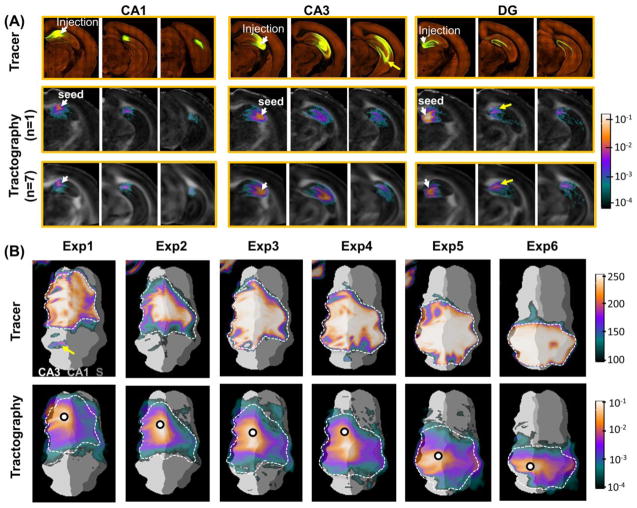
Visual comparisons of tracer and tractography based reconstructions of mouse intra-hippocampal neuronal projections. (A) Three-dimensional views of the projections reconstructed from the rAAV tracer projections from injection sites in CA1, CA2, and dentate gyrus (DG) (images downloaded directly from AMBCA), in comparison with the results of deterministic and probabilistic tractography from similar seed regions. The white arrows indicate the projections from the CA3 injection site to CA1. (B) Sagittal views of projections within the mossy fiber, Shaffer collateral, and performant pathways revealed by rAAV tracer (top row) and corresponding tract density images generated from seed points in the dente gyrus (DG), CA3, and CA1 (bottom row) in a selected animal. The injection sites for the tracer results are located in the DG, CA3, and entorhinal cortex. The tract density images are overlaid on the fractional anisotropy (FA) images, and the pixel values represent the local fiber counts normalized by the number of total fibers reconstructed within the right hippocampus. The relative location of the enlarged region is shown in the upper left sagittal whole brain FA image. The white arrow indicates the location where tract densities decrease rapidly.
2.4 Generation of flattened map of the mouse hippocampus
To better visualize the tractography results in the hippocampus, we used landmark-based LDDMM to flatten the CA1, CA2, CA3 regions and subiculum using the technique described in (Joshi and Miller, 2000). A set of 168 landmarks was manually placed throughout the right hippocampus except the dentate gyrus in the ARA. A corresponding set of landmarks was defined in a 3D rectangular grid, using the boundary between CA1 and CA2 regions along the septotemporal axis as the vertical axis and preserving the length along the transverse axis in the hippocampus. The diffeomorphic mappings generated by landmarked-based LDDMM were then applied to the tracer and tractography data separately to generate flattened maps similar to (Ishizuka et al., 1990).
2.5 Statistical analysis
For each seed region (sources), tractography-based projection densities were compared with tracer-based projection densities in the segmented hippocampal subfields (targets). Linear regression between the density values was analyzed in GraphPad (http://graphpad.com), and the goodness of fit is evaluated by R2 and p-value from an F-test of the regression. The same analysis was used to compare the tractography-based and trace-based projection intensity values. We also performed multiple linear regressions on tractography and tracer-based project densities using with the average length of tracts between source and target regions as an additional variable in the following linear model:
| Equation 1 |
where Y is the tractography-based projection density, X1 is the tracer-based projection density, X2 is the average length of tracts between two regions, and β is a constant. 2D connectivity maps were constructed by arranging the source-target projection density pairs in a matrix, and the cross correlation between the 2D connectivity maps was also calculated.
2.6 Immunohistochemistry
Histological sections on the spatial organization of axons and dendrites in the hippocampus were obtained from a separate group of adult C57BL/6 mice (three month old, female) (Wu et al., 2014). Briefly, mouse brains were cryo-protected in a 30% sucrose solution in 0.1M PBS after perfusion fixation with 4% paraformaldehyde. The specimens were cryostat-sectioned at 40 mm along the axial axis. Representative sections of the hippocampus were stained with Anti-Pan-Axonal Neurofilament Marker (SMI312R, Covance, Princeton, NJ, USA, 1:2000) for axons and Anti-Microtubule-associated Protein 2 (Anti-MAP2, M1406, Sigma-Aldrich, St. Louis, MO, USA, 1:1000) for dendrites. High-resolution images were captured using a Zeiss Observer.Z1 microscope equipped with an AxioCam MRc camera at 20X.
3. Results
3.1 High-resolution diffusion MRI revealed microstructural organization in the mouse hippocampus
In vivo high-resolution HARDI data of the mouse hippocampus acquired from a localized imaging volume (as defined in Fig. 1A) provided rich tissue contrasts that reflected the laminar organization of axons and dendrites (Fig. 1B and C). Based on tractography results, the radiating dendritic processes in the stratum radiatum and molecular layer of the dentate gyrus (DG) can be appreciated in the track density images (Fig. 1D and E) and the enlarged streamline view (Fig. 1F).
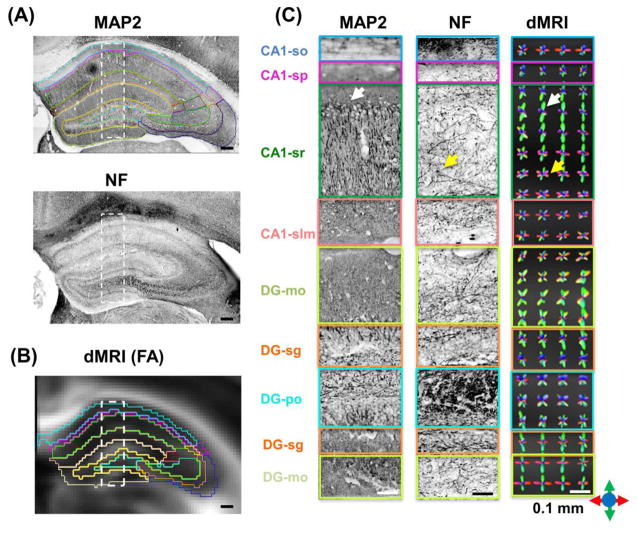
Comparisons between fiber orientation distribution (FOD) maps computed from the HARDI data and microtubule-associated protein 2 (MAP2) and neurofilament (NF) stained histological sections further demonstrated that high-resolution diffusion MRI was able to extract the basic organization of the axonal and dendritic networks in the hippocampus (Fig. 2). For example, in the upper three quarters of the CA1 stratum radiatum (CA1-sr), the radially orientated (green) FOD lobes dominated and ran parallel to the densely populated dendritic processes as shown in the MAP2 stained section. The lower one quarter of the region and the underlying stratum lacunosum-moleculare (CA1-slm) were mostly populated by transversely orientated (red) FOD lobes running in the same direction as the axons in the Schaffer collaterals, as shown by the NF-stained section and previous tracer based studies (Ishizuka et al., 1990). Tractography results based on the high-resolution diffusion MRI data provided a direct visualization of the organization observed in the FOD maps (Fig. 3A). This organization pattern, however, became less well defined in the FOD maps and tract density images at lower spatial resolution (Fig. S2 in the supplementary materials).
Figure 2.
Diffusion MRI-based reconstruction of the organization of axonal and dendritic networks in the mouse hippocampus. (A) Microtubule-associate protein 2 (MAP2) and neurofilament (NF) stained coronal sections (20X) of the mouse hippocampus. Scale bars = 200 μm. (B) Diffusion MRI-derived fractional anisotropy (FA) image from a comparable coronal location overlaid with structural boundaries from the Allen Mouse Brain Atlas after registration. Scale bar = 200 μm. (C) Enlarged MAP2 and NF stained images from the rectangular boxes in (A), with the hippocampal subfields delineated based on the Allen Mouse Brain Atlas. Scale bars = 50 μm. (D) Fiber orientation distribution (FOD) maps derived from diffusion MRI data at 0.1 isotropic resolution (Scale bars = 50 μm. At each pixel, the size and orientation of the lobes reflect the probability of water molecule diffusion along this orientation. The color scheme used here are: red: the left-right axis; green: superior-inferior axis, blue: the anterior-posterior axis, as illustrated by the color arrows in the lower right corner. Abbreviations are: so: stratum oriens; sp: stratum pyramidale; sr: stratum radiatum; slm: stratum lacunosum-moleculare; sg: granule cell layer; mo: molecular layer; po: polymorphic layer;
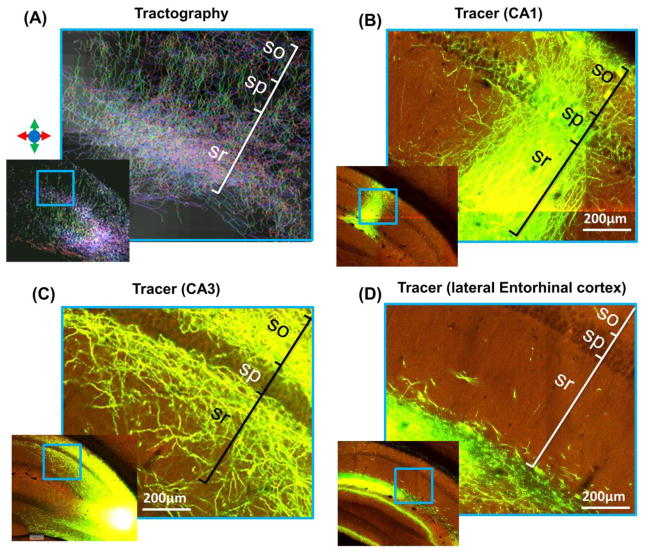
Figure 3.
High magnification tract density and tracer-based projection images of the CA1 region. (A) Tract density image shows transverse (blue and purple) and radial (green) streamlines in the CA1 region. The color scheme in the tract density images is indicated with colored arrows. The approximate locations of the stratum oriens (so), stratum pyramidale (sp), and stratum radiatum (sr) are marked according to the ARA. Projection images from the AMBCA reveal the neuronal projections of CA1 neurons (B), CA3 neurons (C), and neurons in the lateral entorhinal cortex (D), which have distinct trajectories within the CA1 region.
The axonal and dendritic networks in the mouse hippocampus consist of numerous neuronal projections, and the rich AMBCA datasets allowed us to examine these projections based on their origins. We found that neuronal projections from different hippocampal subfields often possessed distinct trajectories. For example, most local projections from CA1 neurons were radially oriented (Fig. 3B), and the transverse projections from CA3 (Fig. 3C) and entorhinal cortex (Fig. 3D) passed through different layers in the CA1 region. These distinct features could provide additional clues for tractography to tease out these neuronal projections from the collection shown in Fig. 3A.
3.2 Macroscopic organization of hippocampal neuronal projections revealed by tractography and tracer
Three-dimensional reconstructions of neuronal projections based on tracer data from injection sites in the CA1, CA3, and DG regions and tractography results from similar seed regions showed matching spatial patterns (Fig. 4A). For example, both tracer and tractography results from a site in the CA3 region (magenta streamlines in Fig. 4A) showed temporally directed projections. The probabilistic tractography results further showed projections towards the CA1 region (indicated by the white arrow), which were also present in the tracer results. Three prominent neuronal pathways in the rodent hippocampus, the mossy fibers, Schaffer collaterals, and Perforant pathways (Amaral and Witter, 1995), were identified in the tracer results and, to some degree, in the tractography-based tract density images as well (Fig. 4B). Compared to the tracer results, the tractography results showed rapid decreases in tract density with increasing distances from the seed regions. For example, the tract densities from the seed region in the DG decreased quickly as they entered the CA3 region (indicated by the white arrow in Fig. 4B).
Remarkable similarities in spatial distribution were found between tracer-labeled projection mapping images from AMBCA and co-registered tract density maps from in vivo tractography in the mouse hippocampus. Both single subject and population averaged tract density maps generated from a CA1 seed region (corresponds to the position of the tracer injection site) showed projections along the rostrocaudal axis within the CA1 field, which agreed with the tracer projection images (Fig. 5A). Tract density maps from a CA3 seed region showed projections between the CA3 region and CA1 region, presumably through the Shaffer collaterals. At lower spatial or angular resolutions, the Schaffer collaterals could not be well reconstructed (supplementary Fig. S2). However, the tractography results did not reach the ventral portion of the caudal hippocampus (indicated by the yellow arrow). The tractography results from the rostral DG region extended rostrocaudally along the V-shaped DG field to the subiculum, but some fibers crossed the hippocampal fissure and entered the CA1 region (indicated by the yellow arrow). Population averaged tractography results showed that these patterns were consistent among different subjects (n = 7).
Figure 5.
Diffusion MRI-based tractography of the mouse intra-hippocampal neuronal projections. (A) Tracer projection mapping images from the Allen mouse brain connectivity atlas (top row) with injection sites in rostral CA1, CA3, and dentate gyrus (DG), respectively, and diffusion MRI tractography results (tract density images) generated from corresponding tracking seeds. The tract density images (TDI) were overlaid on the fractional anisotropy (FA) images, and the pixel values represent the local fiber count normalized by the number of total fibers reconstructed within the right hippocampus. Both individual (n = 1, middle row) and group averaged (n = 7, bottom row) TDIs are shown here. The white arrows indicate the injection sites of the tracer experiments and the corresponding seed regions used for tractography. (B) Flattened tracer projection maps (top row) with six different injection sites in the CA3 field (specified in supplementation Table S1), and corresponding group averaged (n = 7) TDIs (bottom row) with seed points labeled by black circles. The dashed curves are used to outline areas with high tracer intensities and overlaid on the TDIs for comparison.
To obtain a global view of the projection patterns, Fig. 5B showed the flattened projection maps based on viral tracing and probabilistic tractography, with injection sites/seed regions (circled) in the CA3 field. Both maps indicated that the neuronal projections spread from seeds along the septotemporal axis (the vertical axis in Fig. 5B), and also crossed into nearby CA1 and CA2 fields. The dashed curves depicted the boundaries of tracer projections, which were then transferred to the tractography-based projection maps. While the intensities of the tracer-based projection maps were rather homogenous within the boundaries, the intensities in the tractography-based projection maps decreased rapidly from seed regions towards the boundaries, suggesting that the distribution intensities may not match well. The regions with a track density above 0.001 (0.1% of the total tracts found in the right hippocampus) approximated the boundaries defined in the tracer data.
3.3 Correlations between projection densities based on tractography and tracer results
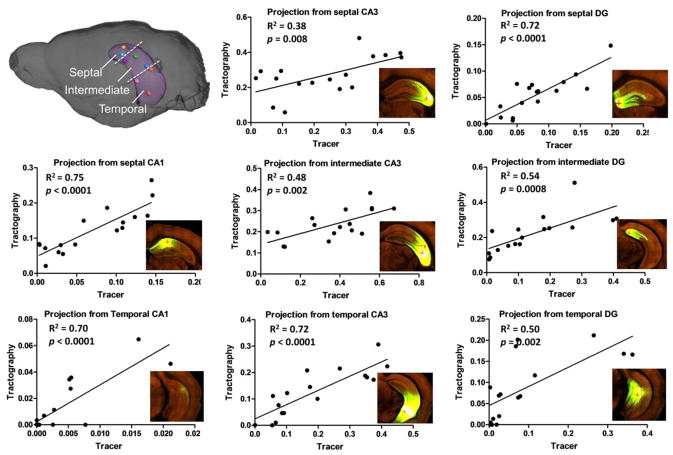
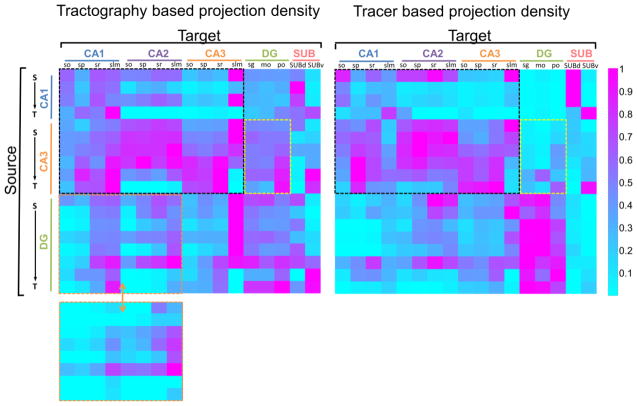
After aligning the HARDI data to the ARA, segmentation of hippocampal subfields defined in the ARA was transferred to the HARDI data (supplementary Fig. S1) and used to examine the projections from injection sites to individual subfields. Mean FA, ADC, and tract density values in the 17 hippocampal subfields were reported in Table 1. The similarity between the tractography and tracer based projections was quantitatively investigated using projection density, which measures the fraction of a target region connected to the seed region. Based on the results of eight representative tracer experiments, which had distinct injection sites across the hippocampus (Fig. 5 inset), the tractography-based (n = 7) and tracer-based projection density measurements showed significant correlations for all injection sites (p < 0.01) (Fig. 6), with the strongest correlation for injection sites in the CA1 regions (R2 ≥ 0.70) and moderate correlations for injection sites in the CA3 region (R2 ≥ 0.38). The correlations between the two projection density measurements were slightly improved when the average tract distances between the injection site and target regions were included as a variable for analysis (Table 2). We also examined the relationships between tracer-based projection intensity (averaged tracer intensity of detected pixels in each subfield), which indicated the strength of connection, and FOD amplitude and tract density values, but found no significant correlation between these measurements (supplementary Fig. S3). When pooling all the projection density data into a source / target connectivity map (Fig. 7), both tractography density and tracer density connectivity maps showed higher projection densities along the diagonals than the off-diagonals, indicating stronger connectivity between neighboring regions than distant regions. The two connectivity maps had a cross-correlation of 0.66, and the connectivity matrices within the dashed box that represented CA1-to-CA3 connection shared a similarity of 0.76. Mismatches also existed, for example, the spurious connection in the lower-left corner of the tractography density map (in the green dashed box in Fig. 7) indicated false positive fiber tracts between DG and CA1 across the hippocampal fissure. This false positive connection was corrected by manually excluding the hippocampal fissure during tractography (lower left inset in Fig. 7).
Table 1.
Fractional anisotropy (FA), apparent diffusion coefficient (ADC), and track density of the 17 hippocampal subfields obtained from the in vivo diffusion MRI data (mean ± standard deviation, n = 7). The track density was calculated as the ratio of fiber tracts in each subfield versus all fiber tracks in the hippocampus by placing tracking seek points randomly all over the hippocampus.
| Structure | FA | ADC (× 10−4 mm2/s) | Track Density (%) |
|---|---|---|---|
| CA1-so | 0.309 ± 0.003 | 5.28 ± 0.03 | 0.113 ± 0.005 |
| CA1-sp | 0.240 ± 0.002 | 5.24 ± 0.04 | 0.110 ± 0.002 |
| CA1-sr | 0.225 ± 0.002 | 5.25 ± 0.04 | 0.181 ± 0.005 |
| CA1-slm | 0.199 ± 0.002 | 5.22 ± 0.05 | 0.256 ± 0.009 |
| CA2-so | 0.324 ± 0.006 | 5.42 ± 0.02 | 0.131 ± 0.005 |
| CA2-sp | 0.308 ± 0.005 | 5.56 ± 0.03 | 0.134 ± 0.006 |
| CA2-sr | 0.281 ± 0.005 | 5.53 ± 0.01 | 0.162 ± 0.007 |
| CA2-slm | 0.231 ± 0.005 | 5.33± 0.06 | 0.190 ± 0.013 |
| CA3-so | 0.356 ± 0.003 | 5.21 ± 0.04 | 0.090 ± 0.006 |
| CA3-sp | 0.292 ± 0.003 | 5.03 ± 0.04 | 0.112 ± 0.006 |
| CA3-sr | 0.218 ± 0.003 | 5.07 ± 0.05 | 0.153 ± 0.008 |
| CA3-slm | 0.217 ± 0.003 | 5.23 ± 0.03 | 0.185 ± 0.012 |
| DG-sg | 0.266 ± 0.004 | 5.04 ± 0.08 | 0.168 ± 0.008 |
| DG-mo | 0.278 ± 0.006 | 5.48 ± 0.01 | 0.148 ± 0.008 |
| DG-po | 0.236 ± 0.001 | 4.83 ± 0.06 | 0.227 ± 0.009 |
| SUBd | 0.420 ± 0.003 | 5.21 ± 0.04 | 0.185 ± 0.004 |
| SUBv | 0.309 ± 0.006 | 5.12 ± 0.09 | 0.156 ± 0.008 |
Abbreviations are: so: stratum oriens; sp: stratum pyramidale; sr: stratum radiatum; slm: stratum lacunosum-moleculare; sg: granule cell layer; mo: molecular layer; po: polymorphic layer; DG: dentate gyrus; SUBd: dorsal subiculum; SUBv: ventral subiculum.
Figure 6.
Quantitative comparisons of projection densities based on eight viral tracer experiments with injection sites throughout the hippocampus and corresponding group-averaged tractography results (n = 7). In each sub-graph, the data represent projection densities in the 17 hippocampal subfields from one injection site / tracking seed (n = 1 for the tracer data, n = 7 for the tractography data), located in the septal, intermediate and temporal sections of the CA1, CA3, and DG, as illustrated in the upper-left inset. The horizontal axis represents the projection density from tracer data, and the vertical axis represents the projection density from tractography results.
Table 2.
R2 values of single and multiple linear regressions between tracer and tractography-based projection density measurements for several hippocampal regions.
| R2 | septal CA1 | tempora l CA1 | septal CA3 | intermedi ate CA3 | temporal CA3 | septal DG | intermed iate DG | temporal DG |
|---|---|---|---|---|---|---|---|---|
| Single regression (α1) | 0.75** | 0.70** | 0.38* | 0.48* | 0.72* | 0.72** | 0.54* | 0.50** |
| Multiple regression (α1/α2) | 0.76** | 0.70** | 0.38* | 0.62* | 0.83* | 0.72** | 0.57* | 0.70** |
The significance of the correlation is indicated by * for p<0.01 and ** for p< 0.001.
Figure 7.
Connectivity matrices consist of projection densities from the 18 group-averaged tractography results (left, n = 7) and corresponding single subject tracer data (right). The columns represent the 17 hippocampal subfields (targets), and the rows represent the 18 source regions arranged according to their septotemporal positions (S→T) in CA1, CA3 and DG. The black dashed boxes in both matrices highlight the CA1-CA3 connectivity. The yellow boxes highlight the projections from CA3 to DG, and the orange boxes highlight the projects from DG to CA1 and CA2. The bottom-left inset shows the tractography-based projection densities from DG to CA1/CA2 after removing fibers crossing the hippocampal fissure. Abbreviation: so: stratum oriens; sp: stratum pyramidale; sr: stratum radiatum; slm: stratum lacunosum-moleculare; sg: granule cell layer; mo: molecular layer; po: polymorphic layer; SUBd: dorsal subiculum; SUBv: ventral subiculum.
4. Discussion
It has been well recognized that diffusion MRI tractography is fundamentally different from cell tracing using chemical or viral tracers. The resolution of two-photon microscopy data in the AMBCA (0.35 μm in-plane with 100 μm interval) allows the tracing of single axons and provides direct evidence of neuronal projections at the cellular level. In comparison, the resolution of diffusion MRI in this study (100 μm isotropic) confines us to the ensemble average of all structures in each voxel, which contains a large number of axons, dendrites, and other structures, many of which are not labeled in a particular AMBCA dataset, e.g., retrograde connections and connections from other sources (e.g., Fig. 3). Instead, diffusion MRI data and tractography results provides the basic organization of the entire axonal and dendritic networks, without revealing microscopic or functional details. Furthermore, the labeling fluorescent proteins used in AMBCA are distributed relatively uniformly throughout the axons and dendrites of infected neurons via active transport, whereas tractography infers potential macroscopic connections indirectly by following locally estimated water molecule diffusion profiles based on diffusion MRI signals (Jones et al., 2013; Thomas et al., 2014). Because of these differences in resolution, specificity, and mechanisms to propagate, comparison between the AMBCA and tractography results is not always straightforward and should be limited to the macroscopic scale.
Both visual (Figs. 4–5) and quantitative comparisons (Figs. 6–7) demonstrated considerable similarities between the tractography and tracer-based results. One potential structural basis of these similarities may be the existence of multiple axonal collaterals in the hippocampus and their distinct trajectories. This has been demonstrated in Fig. 3 and may exist in finer scales. For example, axons originated from neurons at different locations in the CA3 region project to the CA1 region via several distinct axonal collaterals (Ishizuka et al., 1990). The interstitial diffusion of rAAV tracer may also contribute to the similarities in the macroscopic projection patterns, although that the diameters of infected areas (from 400 to 1000 μm with an average volume of 0.24 mm3) (Oh et al., 2014) were small compared to the spatial patterns observed in this study. One interesting finding is that there existed significant correlations between tractography and tracer-based projection densities but no apparent link between the projection intensities (Fig. S3 in Supplementary Materials). These results suggest that the current tractography technique may be used to study the spatial distribution of neuronal projections, indirectly supporting the notion of using connectivity to parcellate gray matter structures (Beckmann et al., 2009; Klein et al., 2007), but not connectivity strength.
There were also marked differences between tractography and tracer-based results. The most noticeable difference is the rapid decrease of tract density values in the tractography results compared to tracer-based projection images (Fig. 4B and Fig. 5). In a major white matter tract with no crossing or branching fibers, the estimated local diffusion profiles often have one dominant direction, and tract densities along the tract remains relatively constant as the reconstructed tracts mostly propagate without terminating and remain packed together. In comparison, the estimated diffusion profiles in the mouse hippocampus usually have more than one dominant directions. For instance, the dendritic processes in the CA1 stratum radiatum mix with axons in the Shaffer collaterals, and the estimated FODs have two dominant directions reflecting this organization (Fig. 2). Without additional information to pick the correct path, the tractography method used here propagated along all potential paths, and sometimes, terminated, resulted in the decreased tract densities as observed in Fig. 4B and Fig. 5. This, combined with accumulation of errors in the estimated FODs due to imaging noises, make it challenging for tractography to reconstruct long pathways in the hippocampus, e.g., the projections from rostral CA3 to caudoventral hippocampus (Fig. 5A, yellow arrow). Multiple linear regression analysis further showed that physical distances of a particular neuronal projection affect the correlation between tractography and tracer-based projection density measurements.
In some cases, tractography-based projection densities were higher than tracer-based projection densities between two regions. For example, due to the unidirectional Mossy fiber projection from the DG to CA3 and less projections vice versa, the tracer data in Fig. 7 showed overall lower projection density from CA3 to DG (yellow box) than from DG to CA3, whereas the corresponding tractography data showed relatively high CA3 to DG projection. This was mainly caused by the fact that the rAAV is an anterograde tracer and only processes of the infected neurons were detected in the tracer-based results, whereas tractography does not distinguish anterograde or retrograde connections and may include projections originated from neurons outside the injection site. Another type of difference was mainly caused by the limited imaging resolution of diffusion MRI. For example, false positives were found between the CA1 and nearby DG (Fig. 7B), crossing the hippocampal fissure. One possible solution is to exclude tractography results that cross the hippocampal fissure based on anatomical knowledge and structural segmentations from the ARA. Applying prior knowledge to aid tractography is a common practice in diffusion tractography to reduce false positives (Huang et al., 2004) and can be conveniently implemented with the co-registered ARA structural segmentations.
In this study, we used the spherical deconvolution-based probabilistic tractography technique, which was shown to have high sensitivity but limited specificity (Thomas et al., 2014), to probe neuronal projections between sub-regions of the mouse hippocampus. The performance of tractography hinge on how accurately the estimated diffusion profiles capture the underlying neural networks. Previous high-resolution diffusion MRI studies of post-mortem brain specimens have demonstrated unique sensitivity in identifying small tracts with spatial resolution up to 0.1 mm in the human brain (Aggarwal et al., 2013; Dell’Acqua et al., 2013; Kleinnijenhuis et al., 2013; Leuze et al., 2014; McNab et al., 2009; Takahashi et al., 2011). Ex vivo specimens, however, may not accurately represent tissue microstructures under normal physiological conditions due to the significant microstructural changes associated with death and chemical fixation in post-mortem samples (Shepherd et al., 2009), e.g., changes in membrane permeability and ratio between intra-cellular and extra-cellular spaces, and their effects on observed diffusion MRI signals. It is not clear whether in vivo diffusion MRI can be equally effective in extracting microstructural information considering the limited imaging resolution and scan time. Our results from the mouse hippocampus demonstrated that the FODs estimated based on in vivo high-resolution diffusion MRI followed the basic spatial organization of dendritic and axonal networks in the hippocampus, as shown in Fig. 2. With more sophisticated diffusion acquisition schemes, e.g., multi-shell HARDI (Descoteaux et al., 2011) and diffusion spectrum imaging (Wedeen et al., 2008), and diffusion signal modeling (Wang et al., 2011; Zhang et al., 2012) that can potentially separate diffusion MRI signals from the axonal and dendritic compartments from signals from other compartments, our capability to infer axonal and dendritic organization in gray matter regions from diffusion MRI signals may be further enhanced.
The tractography results from data acquired with different spatial and angular resolutions also suggest that spatial resolution is important for dissecting small neuronal projections as the complexity of the cellular composition inside each voxel potentially reduces as the size of the voxel decreases, making it easier for existing methods to estimate diffusion profiles that correspond to underlying connections. While it can take formidably long time to scan an entire brain at high resolution, the localized imaging approach used in this study achieved both high spatial and angular resolution in vivo (0.1 mm isotropic resolution and 60 directions in 2 hours). Similar approach has been recently shown to reach 0.4 mm resolution in the human brain (Gaggl et al., 2014). While this approach is not applicable for studying long-range neural connections, it can be useful to study regional connections or particular segments of large connections.
In summary, we demonstrated in vivo mapping of macroscopic neuronal projections in the mouse hippocampus using high-resolution diffusion MRI and tractography. Using an advanced image-processing pipeline, the tractography results were examined with co-registered tracer data from the AMBCA on the intra-hippocampal neuronal projections. The results suggested that diffusion tractography can be potentially used to probe small pathways in the gray matter regions. Combing the existing knowledge from tracer study with state-of-the-art diffusion MRI techniques could potentially expand our ability to investigate the integrity of gray matter connections in vivo, and their longitudinal changes in normal aging (Raz et al., 2005; Reuter-Lorenz, 2002) or pathological changes in neurological diseases (De Lacoste and White, 1993; Toga and Thompson, 2014).
Supplementary Material
Highlights.
High-resolution diffusion MRI of the mouse hippocampus.
The organization of the neuronal networks can be visualized in vivo.
Tractography and viral tracer results showed comparable spatial patterns.
Tractography-based projection densities correlate with tracer-based results.
Acknowledgments
We would like to thank Dr. Dominik Reisinger for acquiring the immunohistological images shown in Fig. 2. We would also like to thank Dr. Solange P. Brown in the department of neuroscience at the Johns Hopkins University School of Medicine for her suggestions and comments on the manuscript. This work was supported by Howard Hughes Medical Institute (HHMI) International Student Research Fellowship (DW), National Institute of Health (NIH) NIH R01 NS070909 (JZ), and NIH R01 HD074593 (JZ), and Maryland Stem Cell Research Fund TEDCO 2014-MSCRFE-0714 (JZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal M, Mori S, Shimogori T, Blackshaw S, Zhang JY. Three-Dimensional Diffusion Tensor Microimaging for Anatomical Characterization of the Mouse Brain. Magnetic Resonance in Medicine. 2010;64:249–261. doi: 10.1002/mrm.22426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal M, Zhang J, Pletnikova O, Crain B, Troncoso J, Mori S. Feasibility of creating a high-resolution 3D diffusion tensor imaging based atlas of the human brainstem: a case study at 11.7 T. Neuroimage. 2013;74:117–127. doi: 10.1016/j.neuroimage.2013.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DC, Barker GJ. Optimal imaging parameters for fiber-orientation estimation in diffusion MRI. Neuroimage. 2005;27:357–367. doi: 10.1016/j.neuroimage.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Amaral D, Witter M. Hippocampal formation. In: Paxinos G, editor. The rat nervous system. Academic Press; San Diego: 1995. [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann M, Johansen-Berg H, Rushworth MFS. Connectivity-Based Parcellation of Human Cingulate Cortex and Its Relation to Functional Specialization. Journal of Neuroscience. 2009;29:1175–1190. doi: 10.1523/JNEUROSCI.3328-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamante F, Tournier JD, Kurniawan ND, Yang Z, Gyengesi E, Galloway GJ, Reutens DC, Connelly A. Super-resolution track-density imaging studies of mouse brain: comparison to histology. Neuroimage. 2012;59:286–296. doi: 10.1016/j.neuroimage.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Ceritoglu C, Oishi K, Li X, Chou MC, Younes L, Albert M, Lyketsos C, van Zijl PC, Miller MI, Mori S. Multi-contrast large deformation diffeomorphic metric mapping for diffusion tensor imaging. Neuroimage. 2009;47:618–627. doi: 10.1016/j.neuroimage.2009.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe AS, Stepniewska I, Colvin DC, Ding Z, Anderson AW. Validation of diffusion tensor MRI in the central nervous system using light microscopy: quantitative comparison of fiber properties. Nmr in Biomedicine. 2012;25:900–908. doi: 10.1002/nbm.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauguet J, Peled S, Berezovskii V, Delzescaux T, Warfield SK, Born R, Westin CF. Comparison of fiber tracts derived from in-vivo DTI tractography with 3D histological neural tract tracer reconstruction on a macaque brain. Neuroimage. 2007;37:530–538. doi: 10.1016/j.neuroimage.2007.04.067. [DOI] [PubMed] [Google Scholar]
- De Lacoste MC, White CL., 3rd The role of cortical connectivity in Alzheimer’s disease pathogenesis: a review and model system. Neurobiol Aging. 1993;14:1–16. doi: 10.1016/0197-4580(93)90015-4. [DOI] [PubMed] [Google Scholar]
- Dell’Acqua F, Bodi I, Slater D, Catani M, Modo M. MR diffusion histology and micro-tractography reveal mesoscale features of the human cerebellum. Cerebellum. 2013;12:923–931. doi: 10.1007/s12311-013-0503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descoteaux M, Deriche R, Le Bihan D, Mangin JF, Poupon C. Multiple q-shell diffusion propagator imaging. Med Image Anal. 2011;15:603–621. doi: 10.1016/j.media.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Dyrby TB, Sogaard LV, Parker GJ, Alexander DC, Lind NM, Baare WF, Hay-Schmidt A, Eriksen N, Pakkenberg B, Paulson OB, Jelsing J. Validation of in vitro probabilistic tractography. Neuroimage. 2007;37:1267–1277. doi: 10.1016/j.neuroimage.2007.06.022. [DOI] [PubMed] [Google Scholar]
- Eichenbaum H, Schoenbaum G, Young B, Bunsey M. Functional organization of the hippocampal memory system. Proc Natl Acad Sci U S A. 1996;93:13500–13507. doi: 10.1073/pnas.93.24.13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank LR. Anisotropy in high angular resolution diffusion-weighted MRI. Magn Reson Med. 2001;45:935–939. doi: 10.1002/mrm.1125. [DOI] [PubMed] [Google Scholar]
- Gaggl W, Jesmanowicz A, Prost RW. High-resolution reduced field of view diffusion tensor imaging using spatially selective RF pulses. Magn Reson Med. 2014;72:1668–1679. doi: 10.1002/mrm.25092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldowitz D. Allen Reference Atlas. A Digital Color Brain Atlas of the C57BL/6J Male Mouse - by H. W. Dong. Genes, Brain and Behavior. 2010;9:128–128. [Google Scholar]
- Gomez J, Pestilli F, Witthoft N, Golarai G, Liberman A, Poltoratski S, Yoon J, Grill-Spector K. Functionally defined white matter reveals segregated pathways in human ventral temporal cortex associated with category-specific processing. Neuron. 2015;85:216–227. doi: 10.1016/j.neuron.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature. 2013;500:168–174. doi: 10.1038/nature12346. [DOI] [PubMed] [Google Scholar]
- Huang H, Zhang JY, van Zijl PCM, Mori S. Analysis of noise effects on DTI-based tractography using the brute-force and multi-ROI approach. Magnetic Resonance in Medicine. 2004;52:559–565. doi: 10.1002/mrm.20147. [DOI] [PubMed] [Google Scholar]
- Ishizuka N, Weber J, Amaral DG. Organization of intrahippocampal projections originating from CA3 pyramidal cells in the rat. J Comp Neurol. 1990;295:580–623. doi: 10.1002/cne.902950407. [DOI] [PubMed] [Google Scholar]
- Jones DK, Horsfield MA, Simmons A. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magn Reson Med. 1999;42:515–525. [PubMed] [Google Scholar]
- Jones DK, Knosche TR, Turner R. White matter integrity, fiber count, and other fallacies: the do’s and don’ts of diffusion MRI. Neuroimage. 2013;73:239–254. doi: 10.1016/j.neuroimage.2012.06.081. [DOI] [PubMed] [Google Scholar]
- Jones DK, Pierpaoli C. Confidence mapping in diffusion tensor magnetic resonance imaging tractography using a bootstrap approach. Magnetic Resonance in Medicine. 2005;53:1143–1149. doi: 10.1002/mrm.20466. [DOI] [PubMed] [Google Scholar]
- Joshi SC, Miller MI. Landmark matching via large deformation diffeomorphisms. IEEE Trans Image Process. 2000;9:1357–1370. doi: 10.1109/83.855431. [DOI] [PubMed] [Google Scholar]
- Klein JC, Behrens TEJ, Robson MD, Mackay CE, Higham DJ, Johansen-Berg H. Connectivity-based parcellation of human cortex using diffusion MRI: Establishing reproducibility, validity and observer independence in BA 44/45 and SMA/pre-SMA. Neuroimage. 2007;34:204–211. doi: 10.1016/j.neuroimage.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Kleinnijenhuis M, Zerbi V, Kusters B, Slump CH, Barth M, van Cappellen van Walsum AM. Layer-specific diffusion weighted imaging in human primary visual cortex in vitro. Cortex. 2013;49:2569–2582. doi: 10.1016/j.cortex.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Kurniawan ND, Richards KL, Yang Z, She D, Ullmann JF, Moldrich RX, Liu S, Yaksic JU, Leanage G, Kharatishvili I, Wimmer V, Calamante F, Galloway GJ, Petrou S, Reutens DC. Visualization of mouse barrel cortex using ex-vivo track density imaging. Neuroimage. 2014;87:465–475. doi: 10.1016/j.neuroimage.2013.09.030. [DOI] [PubMed] [Google Scholar]
- Lazar M. Mapping brain anatomical connectivity using white matter tractography. Nmr in Biomedicine. 2010;23:821–835. doi: 10.1002/nbm.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar M, Alexander AL. Bootstrap white matter tractography (BOOT-TRAC) Neuroimage. 2005;24:524–532. doi: 10.1016/j.neuroimage.2004.08.050. [DOI] [PubMed] [Google Scholar]
- Leergaard TB, White NS, de Crespigny A, Bolstad I, D’Arceuil H, Bjaalie JG, Dale AM. Quantitative histological validation of diffusion MRI fiber orientation distributions in the rat brain. PLoS One. 2010;5:e8595. doi: 10.1371/journal.pone.0008595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuze CW, Anwander A, Bazin PL, Dhital B, Stuber C, Reimann K, Geyer S, Turner R. Layer-specific intracortical connectivity revealed with diffusion MRI. Cereb Cortex. 2014;24:328–339. doi: 10.1093/cercor/bhs311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Mareci TH, Andrew ER. Experimental-Study of Optimal Selective 180-Degrees Radiofrequency Pulses. Journal of Magnetic Resonance. 1988;79:1–10. [Google Scholar]
- McNab JA, Jbabdi S, Deoni SCL, Douaud G, Behrens TEJ, Miller KL. High resolution diffusion-weighted imaging in fixed human brain using diffusion-weighted steady state free precession. Neuroimage. 2009;46:775–785. doi: 10.1016/j.neuroimage.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Moldrich RX, Pannek K, Hoch R, Rubenstein JL, Kurniawan ND, Richards LJ. Comparative mouse brain tractography of diffusion magnetic resonance imaging. Neuroimage. 2010;51:1027–1036. doi: 10.1016/j.neuroimage.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, van Zijl PC. A motion correction scheme by twin-echo navigation for diffusion-weighted magnetic resonance imaging with multiple RF echo acquisition. Magn Reson Med. 1998;40:511–516. doi: 10.1002/mrm.1910400403. [DOI] [PubMed] [Google Scholar]
- Mori S, van Zijl PC. Fiber tracking: principles and strategies - a technical review. Nmr in Biomedicine. 2002;15:468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang QX, Lau C, Kuan L, Henry AM, Mortrud MT, Ouellette B, Nguyen TN, Sorensen SA, Slaughterbeck CR, Wakeman W, Li Y, Feng D, Ho A, Nicholas E, Hirokawa KE, Bohn P, Joines KM, Peng HC, Hawrylycz MJ, Phillips JW, Hohmann JG, Wohnoutka P, Koch C, Bernard A, Dang C, Jones AR, Zeng HK, Gerfen CR. A mesoscale connectome of the mouse brain. Nature. 2014;508:207-+. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly J, Nishimura D, Macovski A. A linear class of large-tip-angle selective excitation pulses. Journal of Magnetic Resonance (1969) 1989;82:571–587. [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nature Methods. 2012;9:255–U248. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz N, Lindenberger U, Rodrigue KM, Kennedy KM, Head D, Williamson A, Dahle C, Gerstorf D, Acker JD. Regional brain changes in aging healthy adults: General trends, individual differences and modifiers. Cereb Cortex. 2005;15:1676–1689. doi: 10.1093/cercor/bhi044. [DOI] [PubMed] [Google Scholar]
- Reuter-Lorenz PA. New visions of the aging mind and brain. Trends in Cognitive Sciences. 2002;6:394–400. doi: 10.1016/s1364-6613(02)01957-5. [DOI] [PubMed] [Google Scholar]
- Seehaus AK, Roebroeck A, Chiry O, Kim DS, Ronen I, Bratzke H, Goebel R, Galuske RA. Histological validation of DW-MRI tractography in human postmortem tissue. Cereb Cortex. 2013;23:442–450. doi: 10.1093/cercor/bhs036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Kimmlingen R, Eberlein E, Witzel T, Cohen-Adad J, McNab JA, Keil B, Tisdall MD, Hoecht P, Dietz P, Cauley SF, Tountcheva V, Matschl V, Lenz VH, Heberlein K, Potthast A, Thein H, Van Horn J, Toga A, Schmitt F, Lehne D, Rosen BR, Wedeen V, Wald LL. Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. Neuroimage. 2013;80:220–233. doi: 10.1016/j.neuroimage.2013.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd TM, Ozarslan E, King MA, Mareci TH, Blackband SJ. Structural insights from high-resolution diffusion tensor imaging and tractography of the isolated rat hippocampus. Neuroimage. 2006;32:1499–1509. doi: 10.1016/j.neuroimage.2006.04.210. [DOI] [PubMed] [Google Scholar]
- Shepherd TM, Thelwall PE, Stanisz GJ, Blackband SJ. Aldehyde fixative solutions alter the water relaxation and diffusion properties of nervous tissue. Magn Reson Med. 2009;62:26–34. doi: 10.1002/mrm.21977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange BA, Witter MP, Lein ES, Moser EI. Functional organization of the hippocampal longitudinal axis. Nature Reviews Neuroscience. 2014;15:655–669. doi: 10.1038/nrn3785. [DOI] [PubMed] [Google Scholar]
- Takahashi E, Dai G, Rosen GD, Wang R, Ohki K, Folkerth RD, Galaburda AM, Wedeen VJ, Ellen Grant P. Developing neocortex organization and connectivity in cats revealed by direct correlation of diffusion tractography and histology. Cereb Cortex. 2011;21:200–211. doi: 10.1093/cercor/bhq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, Zhao T, Horne JA, Fetter RD, Takemura S, Blazek K, Chang LA, Ogundeyi O, Saunders MA, Shapiro V, Sigmund C, Rubin GM, Scheffer LK, Meinertzhagen IA, Chklovskii DB. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci U S A. 2014;111:16574–16579. doi: 10.1073/pnas.1405672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Clark KA, Thompson PM, Shattuck DW, Van Horn JD. Mapping the human connectome. Neurosurgery. 2012;71:1–5. doi: 10.1227/NEU.0b013e318258e9ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. Connectopathy in ageing and dementia. Brain. 2014;137:3104–3106. doi: 10.1093/brain/awu276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: Non-negativity constrained super-resolved spherical deconvolution. Neuroimage. 2007;35:1459–1472. doi: 10.1016/j.neuroimage.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Connelly A. MRtrix: Diffusion tractography in crossing fiber regions. International Journal of Imaging Systems and Technology. 2012;22:53–66. [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Wedeen VJ. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med. 2002;48:577–582. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40:885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- Ugurbil K, Xu J, Auerbach EJ, Moeller S, Vu AT, Duarte-Carvajalino JM, Lenglet C, Wu X, Schmitter S, Van de Moortele PF, Strupp J, Sapiro G, De Martino F, Wang D, Harel N, Garwood M, Chen L, Feinberg DA, Smith SM, Miller KL, Sotiropoulos SN, Jbabdi S, Andersson JL, Behrens TE, Glasser MF, Van Essen DC, Yacoub E Consortium WUMH. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage. 2013;80:80–104. doi: 10.1016/j.neuroimage.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K Consortium WUMH. The WU-Minn Human Connectome Project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Strien NM, Cappaert NL, Witter MP. The anatomy of memory: an interactive overview of the parahippocampal-hippocampal network. Nature Reviews Neuroscience. 2009;10:272–282. doi: 10.1038/nrn2614. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Q, Haldar JP, Yeh FC, Xie M, Sun P, Tu TW, Trinkaus K, Klein RS, Cross AH, Song SK. Quantification of increased cellularity during inflammatory demyelination. Brain. 2011;134:3590–3601. doi: 10.1093/brain/awr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WY, Dai G, Pandya DN, Hagmann P, D’Arceuil H, de Crespigny AJ. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage. 2008;41:1267–1277. doi: 10.1016/j.neuroimage.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Willats L, Raffelt D, Smith RE, Tournier JD, Connelly A, Calamante F. Quantification of track-weighted imaging (TWI): characterisation of within-subject reproducibility and between-subject variability. Neuroimage. 2014;87:18–31. doi: 10.1016/j.neuroimage.2013.11.016. [DOI] [PubMed] [Google Scholar]
- Wu D, Reisinger D, Xu JD, Fatemi SA, van Zijl PCM, Mori S, Zhang JY. Localized diffusion magnetic resonance micro-imaging of the live mouse brain. Neuroimage. 2014;91:12–20. doi: 10.1016/j.neuroimage.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Xu J, McMahon MT, van Zijl PC, Mori S, Northington FJ, Zhang J. In vivo high-resolution diffusion tensor imaging of the mouse brain. Neuroimage. 2013;83:18–26. doi: 10.1016/j.neuroimage.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QX, Posse S, LeBihan D, Smith MB. Double-sampled echo-planar imaging at 3 tesla. Journal of Magnetic Resonance Series B. 1996;113:145–150. doi: 10.1006/jmrb.1996.0167. [DOI] [PubMed] [Google Scholar]
- Yassa MA, Muftuler LT, Stark CE. Ultrahigh-resolution microstructural diffusion tensor imaging reveals perforant path degradation in aged humans in vivo. Proc Natl Acad Sci U S A. 2010;107:12687–12691. doi: 10.1073/pnas.1002113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineh MM, Holdsworth S, Skare S, Atlas SW, Bammer R. Ultra-high resolution diffusion tensor imaging of the microscopic pathways of the medial temporal lobe. Neuroimage. 2012;62:2065–2082. doi: 10.1016/j.neuroimage.2012.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012;61:1000–1016. doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.