Abstract
Objectives
We aimed to identify the genetic relatedness of multiple-drug resistance (MDR) in Acinetobacter baumannii clinical isolates recovered from a hospital in Los Angeles.
Methods
Twenty one MDR A. baumannii isolates were collected and their antibiotic susceptibility were determined according to the CLSI guidelines. Genes coding for antibiotic resistance were identified by PCR and their identities were confirmed by DNA sequencing. Clonal relationships were studied by pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST).
Results
MDR consistently correlated with the presence of oxacillinases, mostly in the form of plasmid-mediated OXA-23 enzyme which were detected in 12 (57.1%) isolates. GES-type carbapenemases were found in 20 (95.2%) strains, AAC in all 21 (100%) strains, PER in 7 (33.3%) strains and ISAba1 has been detected in 16 (76.2%) isolates. The association between ISAba1 and resistant genes confirms insertion elements as a source of β-lactamase production. Of the 21 clinical isolates, 5 were found to be related to sequence type-1 (ST1) and 16 to ST2 as analyzed by MLST. PFGE demonstrated that the majority of clinical isolates are highly related (>85%).
Conclusions
This study supports a more complete understanding of genotyping of antibiotic resistance for better assessment of MDR strains transmission.
Keywords: A. baumannii, β-lactamase, MLST, PFGE
1. Background
Acinetobacter baumannii has emerged as a predominant cause of healthcare-associated infections (including those seen in wounded soldiers) both in the United States and world-wide.1–10 A. baumannii infections include pneumonia (especially ventilator associated pneumonia), wound infections, urinary tract infections, septicemia and surgical site infections.11–13 Risk factors for A. baumannii infections, especially in elderly individuals, include; patients having underlying diseases (e.g. diabetes), immune suppression, burns, trauma, invasive medical procedures, mechanical ventilation, catheters, previous antibiotic treatments and extended hospital stay.14 Of great concern is the recent rise in the frequency of multiple drug resistant (MDR) and extremely drug resistant (XDR)- A. baumannii infections.2,11,12 The percentage of A. baumannii infections caused by MDR strains (defined as resistance to ≥1 agent in at least three antimicrobial categories) and XDR strains (defined as resistance to all available antibiotics except colistin and tigecycline) has increased from <4% in 2000 to 60–70% in 2010.14 Infections caused by MDR A. baumannii are associated with longer hospitalization, greater healthcare costs, greater morbidity, and >60% mortality for bloodstream infections as compared to drug-susceptible strains.15–18 XDR infections are treatable only with second-line agents, such as tigecycline and colistin which are associated with clinical failure, development of resistance and nephrotoxicity.16,19–28 Further, pandrug-resistant infections (PDR) are resistant to every FDA approved antibiotic, and are hence untreatable. Because of the difficulty in treating MDR, XDR and PDR A. baumannii infections, surveillance of A. baumannii isolates represent the cornerstone of prevention and control of these infections. In the current study, we aimed at characterizing the resistance mechanisms and determining the genetic relatedness of clinical isolates recovered from Harbor-UCLA Medical Center at Los Angeles County.
2. Methods
2.1. Bacterial Identifications
Twenty four clinical strains of A. baumannii obtained from in-patients Harbor-UCLA Medical Center (HUMC) of which 21 isolates were investigated and identified to the species level by using Vitek2 (BioMérieux Vitek Systems Inc., USA) and MicroScan (WalkAway System, Siemens, USA) systems utilizing biochemical methods.
2.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was determined for all HUMC strains of A. baumannii by automated broth microdilution method (Vitek2) (Vitek AMS; BioMérieux Vitek Systems Inc., USA) and the results were analyzed and interpreted using clinical breakpoints according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.29 The antibiotics tested were: Amikacin, amoxicillin/clavulanic acid, ampicillin/sulbactam, ampicillin, cefazolin, cefepime, cefotaxime, ceftazidime, ceftriaxone, cefuroxime, cefoxitin, cefpodoxime, cephalothin, ceftriazone, ciprofloxacin, gentamicin, imipenem, meropenem, levofloxacin, nitrofurantoin, norfloxacin, tetracycline, tobramycin, trimethoprim/sulfamethoxazole, piperacillin/tazobactam, pipercillin, tigecycline, colistin and tigecycline. Extended spectrum β-lactamase (ESBL) production was confirmed by Vitek2 analyzer, Microscan and disk diffusion tests. Minimum inhibitory concentration (MICs) of quinolones, fluoroquinolones and β-lactams including carbapenems were determined using the E-test method (CLSI 2012).29 Isolates that showed resistance to at least three classes of antibiotics were considered as MDR strains, whereas isolates showing resistance to all antibiotics except for colistin and tigecycline were considered as XDR strains.
2.3. Identification of Housekeeping Genes
Bacterial DNA was extracted using QIAquick PCR Purification Kit (Qiagen, USA). Primers used for polymerase chain reaction (PCR) amplifications of seven housekeeping genes in A. baumannii are listed in Table 1.
Table 1.
Gene primers used for housekeeping genes detection by PCR in genes in clinical A. baumannii isolates in MLST studies.
| Gene | Primer Sequence | Amplicon Size |
|---|---|---|
|
cpn60-F cpn60-R |
ACTGTACTTGCTCAAGC TTCAGCGATGATAAGAAGTGG |
405 bp |
|
fusA-F fusA-R |
ATCGGTATTTCTGCKCACATYGAT CCAACATACKYTGWACACCTTTGTT |
633 bp |
|
gltA-F gltA-R |
AATTTACAGTGGCACATTAGGTCCC GCAGAGATACCAGCAGAGATACACG |
483 bp |
|
pyrG-F pyrG-R |
GGTGTTGTTTCATCACTAGGWAAAGG ATAAATGGTAAAGAYTCGATRTCACCMA |
297 bp |
|
recA-F recA-R |
CCTGAATCTTCYGGTAAAAC GTTTCTGGGCTGCCAAACATTAC |
372 bp |
|
rplB-F rplB-R |
GTAGAGCGTATTGAATACGATCCTAACC CACCACCACCRTGYGGGTGATC |
330 bp |
|
rpoB-F rpoB-R |
GGTCCTGGTGGTTTAACACG CGAATAACGATACGAGAAGCA |
456 bp |
2.4. Antibiotic Resistance Genes by PCR and DNA Sequencing
The presence of resistant genes (Table 2) was investigated by PCR using GoTaq Green Master Mix (Promega, USA). PCR was conducted in a GeneAmp 9700 system (Perkin-Elmer, Illinois, USA) using the conditions specified for each primer as corresponding to the reference source. PCR was performed for metallo-β-lactamase and ESBL-encoding genes including bla SIM (Seoul imipenemase), bla VIM (Verona integron-encoded metallo- β-lactamases), bla VEB (Vietnamese extended-spectrum- β-lactamase) and bla IMP (Imipenemase), bla TEM-1 (Temoneira) and bla SHV (Sulfhydryl variable), bla CTX-M-like (Cefotaximase-Munchen),30 bla NDM-1 (New-Delhi metallo- β-lactamase),31 qnrA, qnrB and qnrS (Quinolone resistance genes),32 aac (N-acetyltransferase),33 gyrA (DNA gyrase subunit A) and parC (Topoisomerase IV subunit C),34 AmpC (class C β-lactamases),35 bla PER (Pseudomonas extended resistance),36 bla GES (Guiana extended spectrum),36 OXA-(oxacillinases)-encoding genes including bla OXA-51-like, bla OXA-58-like, bla OXA-48-like, bla OXA-23-like, bla OXA-24-like genes,37,38 IS (insertion sequence),39 and ISAba1.40 Amplified PCR products were purified with Qiagen purification kit (Qiagen, USA) according to the manufacturer's instructions and both strands were sequenced by automated AB13100 DNA sequencer (Applied BioSystems) system. The BLAST program of the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov) was used to search and compare databases for similar nucleotide acid sequences.
Table 2.
Gene primers used for PCR amplification of antibiotic resistance genes in clinical A. baumannii isolates.
| Gene | Amplicon Size | Tm °C | Primer Sequence |
|---|---|---|---|
| OXA-58 | 599bp | 52 | AAGTATTGGGGCTTGTGCTG (Forward) CCCCTCTGCGCTCTACATAC (Reverse) |
| OXA-51 | 353bp | 52 | TAATGCTTTGATCGGCCTTG (Forward) TGGATTGCACTTCATCTTGG (Reverse) |
| OXA-24 | 246bp | 52 | GGTTAGTTGGCCCCCTTAAA (Forward) AGTTGAGCGAAAAGGGGATT(Reverse) |
| OXA-23 | 501bp | 52 | GATCGGATTGGAGAACCAGA (Forward) ATTCTGACCGCATTTCCAT(Reverse) |
| OXA-48 | 438bp | 62 | GCGTGGTTAAGGATGAACAC (Forward) CATCAAGTTCAACCCAACCG (Reverse) |
| CTXM | 550bp | 60 | CGCTTTGCGATGTGCAG (Forward) ACCGCGATATCGTTGGT (Reverse) |
| CTXM2 | 896bp | 55 | CGGAATTCATGATGACTCAGAGCATTCG (Forward) GCTCTAGATTATTGCATCAGAAACCGTG (Reverse) |
| PER | 900bp | 43 | ATGAATGTCATTATAAAAGC (Forward) AATTTGGGCTTAGGGCAGAA (Reverse) |
| VEB | 600bp | 55 | CGACTTCCATTTCCCGATGC (Forward) GGACTCTGCAACAAATACGC (Reverse) |
| QnrA | 580bp | 54 | AGAGGATTTCTCACGCCAGG (Forward) TGCCAGGCACAGATCTTGAC (Reverse) |
| QnrS | 428bp | 54 | GCAAGTTCATTGAACAGGGT (Forward) TCTAAACCGTCGAGTTCGGCG (Reverse) |
| QnrB | 264bp | 54 | GGMATHGAAATTCGCCACTG (Forward) TTTGCYGYYCGCCAGTCGAA (Reverse) |
| TEM | 850bp | 42 | GAGTATTCAACATTTCCGTGTC (Forward) TAATCAGTGAGGCACCTATCTC (Reverse) |
| GES | 846bp | 55 | ATGCGCTTCATTCACGCAC (Forward) CTATTTGTCCGTGCTCAGGA (Reverse) |
| SHV | 861bp | 55 | ATGCGTTATWTTCGCCTGTGT (Forward) TTAGCGTTGCCAGTGCTCG (Reverse) |
| CTX | 554bp | 60 | TCTTCCAGAATAAGGAATCCC (Forward) CCGTTTCCGCTATTACAAAC (Reverse) |
| GyrA6 | 620bp | 56 | CGACCTTGCGAGAGAAAT (Forward) GTTCCATCAGCCCTTCAA (Reverse) |
| ParCF43 | 964bp | 53 | AGCGCCTTGCGTACATGAAT (Forward) GTGGTAGCGAAGAGGTGGTT (Reverse) |
| AAC | 482bp | 55 | TTGCGATGCTCTATGAGTGGCTA (Forward) CTCGAATGCCTGGCGTGTTT (Reverse) |
| SIM | 570bp | 65 | TACAAGGGATTCGGCATCG (Forward) TAATGGCCTGTTCCCATGTG (Reverse) |
| IMP | 232bp | 60 | GGAATAGAGTGGCTTAAYTC (Forward) TCGGTTTAAYAAAACAACCACC (Reverse) |
| VIM | 390bp | 62 | GATGGTGTTTGGTCGCATA (Forward) CGAATGCGCAGCACCAG (Reverse) |
| NDM | 621bp | 65 | GGTTTGGCGATCTGGTTTTC (Forward) CGGAATGGCTCATCACGATC (Reverse) |
| KPC | 798bp | 58 | CGTCTAGTTCTGCTGTCTTG (Forward) CTTGTCATCCTTGTTAGGCG (Reverse) |
| IS | 615bp | 56 | GTGCCCAAGGGGAGTGTATG (Forward) ACYTTACTGGTRCTGCACAT (Reverse) |
| ISAbaI | 389bp | 57 | ATGCAGCGCTTCTTTGCAGG (Forward) AATGATTGGTGACAATGAAG (Reverse) |
2.5. Pulsed-Field Gel Electrophoresis
Pulsed-Field Gel Electrophoresis (PFGE) analysis was based on techniques described elsewhere.41 PFGE of ApaI-digested genomic DNA (Promega, UK) from each HUMC strain was performed to detect the relationships among the clinical isolates of A. baumannii. After PFGE, the gels were stained with ethidium bromide and scanned. The analysis of gels was performed using BioNumerics software version 7.1 (Applied Maths, Ghent, Belgium). This software facilitates the development of the algorithms necessary for the comparison of profiles of isolates based on the Dice coefficient and the hierarchic unweighted pair arithmetic average algorithm. Cluster analysis and phylogenetic trees were subsequently analyzed with an optimization of 1.0% and a tolerance of 0.7%. Isolates were considered to belong to the same PFGE clone if their Dice similarity index was >85%.
2.6. Multi Locus Sequence Typing
Multi-locus sequence typing (MLST) was based on a sequence analysis of the internal fragments of seven housekeeping genes: cpn60 (60-KDa chaperonin), fusA (elongation factor EF-G), gltA (citrate synthase), pyrG (CTP synthase), recA (homologous recombination factor), rplB (50S ribosomal protein L2), rpoB (RNA polymerase subunit B). The MLST scheme including amplification and sequencing primers, allele sequences and sequence types (STs) were available at Institute Pasteur’s MLST web site (http://www.pasteur.fr/recherche/genopole/PF8/mlst/references_Abaumannii.html). The housekeeping genes for the MLST scheme were selected on the basis of their sequence availability in GenBank and prior studies of the phylogenetic relationships for the genus Acinetobacter and their presence in other MLST schemes available for other bacterial species. PCR primers were chosen from previous studies or were designed for amplification of the seven selected genes (Table 1).
All PCR amplifications were carried out using GoTaq Green Matser Mix (Promega, USA) under the following conditions: 35 cycles (denaturation at 94°C for 30 sec, annealing at 50°C for 30 sec, and extension at 72°C for 30 sec) proceeded by a 2 min denaturation at 94°C and followed by a 5 min extension at 72°C. PCR products were directly purified from the reaction mixture with the QIAquick PCR purification kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's recommendations. Sequencing of internal DNA fragments of 297 bp to 633 bp of the selected housekeeping genes was performed using ABI Prism 377 sequencer using the ABI Prism BigDye terminator cycle sequencing ready reaction kit V3.1 (PE Applied Biosystems, Foster City, CA) according to the manufacturer's recommendations. PCR primers were used for sequencing on both strands. Sequence data were aligned by CLUSTALW (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
2.7. Plasmid Curing Experiments
Plasmid curing procedure was performed for five selected HUMC isolates (HUMC-1, HUMC-3, HUMC-4, HUMC-10, HUMC-19) of A. baumannii using 47°C as a growing temperature. The five isolates were selected to represent the presence or absence of resistance genes and ISAba1. Plasmid curing using temperature as a curing agent were examined after 3 days incubation at 47C followed by antibiotic sensitivity testing.
3. Results
3.1. Antimicrobial Susceptibilities to A. baumannii Isolates
A total of 21 clinical samples were analyzed in 2011. The sources of the isolates included 9 samples of respiratory secretions (42.9%), 4 samples of sputum (19%), 2 samples from abdominal secretions (9.5%), 2 samples from wounds (9.5%) and one sample each from other sites (4.8%) including urine, bronchoalveolar lavage, foot and groin samples. Antibiotic sensitivity testing revealed that all the isolates were resistant to ceftriaxone, ceftazidime, cefotaxime, ciprofloxacin, cefepime, gentamicin, levofloxacin, tetracycline, ticarcillin/K clavulanate, whereas two isolates were found to be sensitive to meropenem, eight to imipenem, three to amikacin, four to ampicillin/sulbactam, one to each of trimethoprim/sulfamethoxazole tobramycin. All clinical isolates of A. baumannii were sensitive to colistin, and tigecycline with the exception of one and two strains, respectively. Susceptibility testing results of the studied clinical isolates are summarized in Table 3.
Table 3.
Susceptibility profiles of HUMC strains. Isolates were designated susceptible (S), intermediate (I), or resistant (R) according to CLSI antibiotic breakpoint guidelines. Minimum inhibitory concentrations (MIC) were shown in brackets.
| Antibiotic/Isolate | A/S | AK | CAZ | CP | CPE | GM | IMP | LVX | MER | T/S | TE | TIM | TO | CST | TGC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HUMC-1 | I (16/8) | S (≤16) | R (>32) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | R (>2/38) | I (8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-3 | R (>16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | R (>2/38) | I (8) | R (>64) | R (>8) | S (≤2) | R (4) |
| HUMC-4 | I (16/8) | S (≤16) | R (>16) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | R (>2/38) | I (8) | R (>64) | S (≤4) | S (≤2) | I (2) |
| HUMC-5 | R (>16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | S (≤4) | R (>4) | I (8) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-6 | R (>16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | S (≤4) | R (>4) | I (8) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-9 | S (≤8/4) | S (≤16) | R (>16) | R (>2) | R (>16) | R (>8) | I (8) | R (>4) | R (>8) | R (>2/38) | I (8) | R (>64) | R (>8) | S (≤2) | I (2) |
| HUMC-10 | S (≤8/4) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | S (≤4) | R (>4) | S (≤4) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-11 | R (>16/8) | R (>32) | R (>16) | R (>2) | I (16) | R (>8) | S (≤4) | R (>4) | I (8) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-12 | R (>16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | S (≤4) | R (>4) | S (≤4) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-13 | R (>16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | S (≤4) | R (>4) | I (8) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-14 | R (>16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | S (≤4) | R (>4) | I (8) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-15 | I (16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | R (>2/38) | I (8) | R (>64) | R (>8) | S (≤2) | R (4) |
| HUMC-16 | I (16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | S (≤2/38) | I (8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-17 | I (16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-18 | I (16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | R (>2/38) | I (8) | R (>64) | R (>8) | S (≤2) | I (2) |
| HUMC-19 | S (≤8/4) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-20 | I (16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-21 | I (16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | R (>2/38) | R (>8) | R (>64) | R (>8) | R (>2) | S (≤1) |
| HUMC-22 | I (16/8) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | R (>2/38) | I (8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-23 | S (≤8/4) | R (>32) | R (>16) | R (>2) | R (>16) | R (>8) | R (>8) | R (>4) | R (>8) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
| HUMC-24 | R (>16/8) | R (>32) | R (>16) | R (>2) | I (16) | R (>8) | S (≤4) | R (>4) | I (8) | R (>2/38) | R (>8) | R (>64) | R (>8) | S (≤2) | S (≤1) |
Abbreviations: A/S, Ampicillin/Sulbactam; AK, Amikacin; CAZ, Ceftazidime; CP, Ciprofloxacin; CPE, Cefepime; GM, Gentamicin; IMP, Imipenem; LVX, Levofloxacin; MER, Meropenem; T/S, Trimethoprim/Sulfamethoxazole; TE, Tetracycline; TIM, Ticarcillin/K Clavulanate; TO, Tobramycin; CST, Colistin; TGC, Tigecycline.
3.2. Characterization of Carbapenemases and other β-Lactamase Genes
We identified the presence of oxacillinases, mostly in the form of plasmid-mediated OXA-23 enzyme which were detected in 12 (57.1%) isolates as well as β-lactamase resistant genes in 7 isolates harboring PER (33.3%), 21 isolates harboring AAC (100%) and 20 isolates harboring GES (95.2%) -type enzymes (Table 4). MDR consistently correlated with the presence of oxacillinases, mostly in the form of plasmid-mediated OXA-51 and OXA-23 enzymes which were detected in 21 (100%) and 13 (61.9%) of the clinical isolates collected, respectively. None of the isolates harbored OXA-58, OXA-24 or OXA-48. ISAba1 was detected in 16 (76.2%) isolates. None of the clinical isolates harbored KPC, IMP, VIM, SIM, NDM, or QNR-type genes (Table 4).
Table 4.
Antibiotic resistance genes results detected by PCR in HUMC clinical strains. Abbreviations: (−) denotes negative, (+) denotes positive PCR reaction.
| Gene/Isolate | OXA-51 | OXA-23 | PER | GES | AAC | ISAba1 |
|---|---|---|---|---|---|---|
| HUMC-1 | + | + | − | + | + | + |
| HUMC-3 | + | + | − | + | + | − |
| HUMC-4 | + | − | + | + | + | + |
| HUMC-5 | + | − | + | + | + | + |
| HUMC-6 | + | − | + | + | + | + |
| HUMC-9 | + | + | − | + | + | + |
| HUMC-10 | + | − | − | + | + | + |
| HUMC-11 | + | − | + | + | + | + |
| HUMC-12 | + | − | − | + | + | + |
| HUMC-13 | + | − | + | + | + | + |
| HUMC-14 | + | − | + | + | + | + |
| HUMC-15 | + | + | − | + | + | − |
| HUMC-16 | + | + | − | + | + | + |
| HUMC-17 | + | + | − | + | + | + |
| HUMC-18 | + | + | − | + | + | − |
| HUMC-19 | + | + | − | − | + | + |
| HUMC-20 | + | + | − | + | + | + |
| HUMC-21 | + | + | − | + | + | − |
| HUMC-22 | + | + | − | + | + | − |
| HUMC-23 | + | + | − | + | + | + |
| HUMC-24 | + | − | + | + | + | + |
3.3. Frequency of Insertion Sequences for Different Enzymes
The frequency of insertion sequences presence in all of the clinical isolates of A. baumannii harboring antibiotic resistance genes were found to be 7 out of 12 (58.3%) in OXA-23, 7 out of 7 (100%) in PER, 15 out of 20 (75%) in GES and 17 out of 21 (81%) in AAC (Table 4).
3.4. Molecular Genotyping of A. baumannii Clinical Isolates
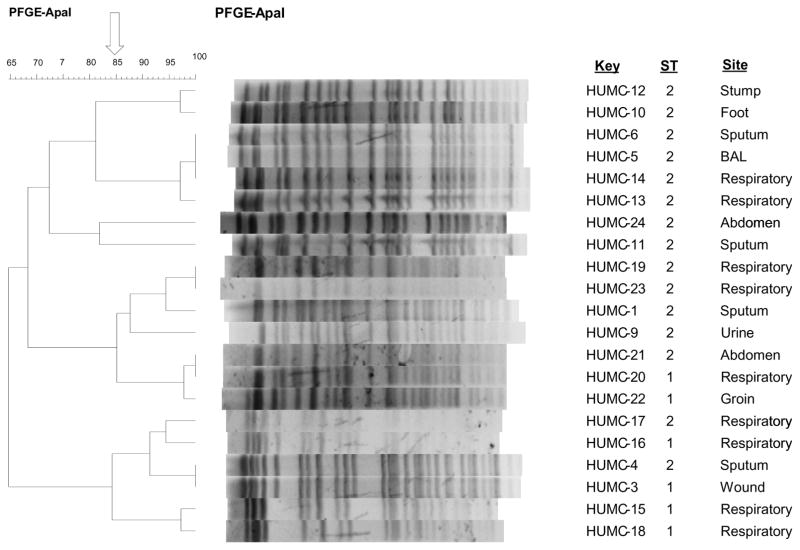
Genotyping analysis of the 21 clinical isolates by PFGE revealed the circulation of different PFGE types. We found high clonal relationship among all the typed strains. All the isolates were MDR to at least three antimicrobial groups. PFGE analysis demonstrated that the majority of clinical isolates are highly related (≥85%). MLST studies have shown that 5 (23.8%) clinical isolates of A. baumannii were found to be related to sequence type-1 (ST1) and 16 (76.2%) belong to sequence type-2 (ST2) (Table 5). The results of PFGE and MLST are summarized in Figure 1, along with the information of the specimen original sources.
Table 5.
Allele and sequence number results of HUMC clinical isolates as analyzed by MLST.
| HUMC Strains | Allele Number | ST | ||||||
|---|---|---|---|---|---|---|---|---|
| Cpn60 | fusA | gltA | pyrG | recA | rplB | rpoB | ||
| HUMC-1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-3 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | 1 |
| HUMC-4 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-5 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-9 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-10 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-11 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-12 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-13 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-14 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-15 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | 1 |
| HUMC-16 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | 1 |
| HUMC-17 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-18 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | 1 |
| HUMC-19 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-20 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-21 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-22 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | 1 |
| HUMC-23 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| HUMC-24 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
Figure 1.
Dendrogram representing PFGE profiles, MLST results and site of infection of HUMC clinical isolates.
3.5. Plasmid Curing of A. baumannii Clinical Isolates
Our results have shown that the five clinical isolates cured for plasmids, did not lose their ability for resistance to antimicrobial agents. Plasmids curing using temperature as a curing agent failed even after 3 days incubation at 47°C (Table S1).
4. Discussion
Antimicrobial resistance in Enterobacteriaceae has emerged as a major clinical problem in recent years.42,43 Drug resistance among this group of bacteria is mainly caused by the emergence and proliferation of extended-spectrum β-lactamases,44 fluoroquinolone resistance,45 and the dissemination of multiple drug-resistant (MDR) and carbapenem resistant strains.46,47 Data from the National Healthcare Safety Network at the CDC showed high rates of carbapenem resistance among A. baumannii throughout USA with increased incidence in hospital-associated infections especially those of ventilated acquired pneumonia, central line associated bloodstream infections, catheter associated urinary tract infections and surgical site infections.48 Understanding the fundamental mechanisms that underline Acinetobacter infections including the original sources of the infecting strains, resistance patterns, their clonality and geographical spread are critical for the development of appropriate infection control measures and more efficient treatment strategies.
We used two typing methods of PFGE and MLST to detect the molecular epidemiology of A. baumannii isolates from Harbor-UCLA Medical Center in Los Angeles County.49,50 By using MLST we have shown that clinical isolates of A. baumannii belonged to two main clones; ST1 and ST2. The high clonal relationship in PFGE analysis between HUMC strains as reflected by ≥85% is in agreement with the MLST studies which showed more than 76.2% of the tested isolates belonged to ST2. It is prudent to mention that MLST is a high resolution molecular tool for discriminating between closely related bacterial species.50 The first MLST scheme for A. baumannii was published by Bartual et al.51 and by Diancourt et al.52 at the Pasteur Institute (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html), which we have used in this study. Further, MLST approaches to genotype A. baumannii isolates, which are based on sequencing regions of housekeeping genes,53 are reproducible and portable facilitating comparison among laboratories worldwide. While MLST is an expensive typing method due to the need for DNA sequencing, selective use of this technique can substantially enhance our understanding of molecular epidemiology across different hospitals and geographic locations. In contrast, although PFGE analysis is highly discriminatory, it is not suitable for inter-laboratory comparisons unless the procedures are meticulously standardized,49 and the interpretation of the pulsed field results may be a challenge in non-outbreak situations.50 However, genotyping by methods like PFGE allows investigation of clonal spread and can be used to identify the source of the original infection. Therefore, whenever possible the use of both methods for genotyping is advisable. Equally important, our previous work using PFGE analysis on 5 isolates of MDR A. baumannii demonstrated that all these isolates were genetically different from the drug susceptible A. baumannii ATCC 17978.54
Previous genome sequencing studies have shown that MDR A. baumannii strains causing infection are related to one another with extensive variation in gene content even among strains that were very closely related phylogenetically and epidemiologically.55 Several mechanisms contribute to this diversity, including transfer of mobile genetic elements and mobilization of insertion sequences.55 In addition, widespread genetic variation among clinical isolates from the same hospital and/or patient reinforces the need for molecular diagnostic testing and genomic analysis to determine resistance profiles, rather than to rely primarily on strain typing and antimicrobial resistance phenotypes for molecular epidemiological studies.55
The role of insertion sequences is important to understand the expression of carbapenemases in A. baumannii (e.g., ISAba1). It has been reported that insertion sequences play a role in the expression of the carbapenem-hydrolyzing β-lactamases.40 In this study, we have shown that ISAba1 was detected in almost 76.2% of the clinical isolates. We also showed that not all isolates harboring ISAba1 were resistant to carbapenems (Table 4). These findings are concordant with the fact that resistance to carbapenems is mainly caused by the OXA-type enzymes, including plasmid-encoded β-lactamases (OXA-23, OXA-40, and OXA-58).38 Studies have shown that chromosomally encoded OXA β-lactamase (OXA-51-like) can confer resistance to carbapenems in A. baumannii when the genetic environment around the gene promoted its expression.38,56 Our findings are in agreement with other studies in which we show that almost all A. baumannii strains possess chromosomally encoded OXA β-lactamase (OXA-51 like). The high frequency of insertion sequences for different types of enzymes, oxacillinases and carbapenemases series in A. baumannii could be due to differences in antibiotic treatment given to those patients. Our plasmid curing studies have shown that clinical strains did not lose their ability for resistance to antimicrobial agents, indicating a high stability of acquired plasmids.
The two clonal lineages of MDR A. baumannii identified in our study have been found in other States as well.46 In the United States, an outbreak of MDR A. baumannii in Houston was caused by clonal complex 92 (an ST-2 type).57 In addition, a survey of bacterial isolates collected from 52 U.S. hospitals over 6 years also showed the preponderance of the same clone.58 Other studies has revealed good correlation between antibiotic susceptibility profiles and genetic fingerprints from clinical A. baumannii isolates from nosocomial outbreaks and the mechanisms of antibiotic resistance.59 Molecular epidemiology of clinical isolates of A. baumannii identified in New York, Pennsylvania, Florida, Missouri, Nevada, and California revealed the predominance of CC92 among carbapenem non-susceptible isolates in US hospitals, suggesting that they constitute part of the global epidemic driven by this clonal complex belonging to EUII.46 Worldwide, it is not surprising to detect common STs. For examples, ST-1 and ST-2 have been previously identified in the Middle East including Saudi Arabia, Lebanon and Yemen.60–62 Moreover, ST-1 and ST-2 are known as endemic strains in European countries including Spain, Italy, France and Greece,63–66 Asia including Japan and Taiwan,67,68 and even the Scandinavian countries like in Denmark.69
There is no doubt that early pathogen identification, followed by the right antibiotic treatment may reduce the prevalence of antibiotic resistance in A. baumannii. Therefore, future studies will focus on characterizing the composite transposable elements in detail. Though, the clinical isolates collected in this study may not represent the overall epidemiology of MDR A. baumannii since it all originated from one hospital in Los Angeles County, we plan to further research the epidemiological analyses of this organism beside other MDR and XDR organisms in other hospitals for continuous surveillance of these strains in the United States.
5. Conclusion
Two distinct clones of MDR or XDR A. baumannii were identified at Harbor UCLA Medical Center in Los Angeles County. The epidemiological data obtained suggested that the increase in the number of A. baumannii infections in that hospital was caused by these two clones. MLST studies maybe more accurate in distinguishing between A. baumannii isolates than PFGE typing. This study supports a more complete understanding of genotyping of antibiotic resistance for better assessment of MDR and/or XDR strains transmission. Continuous surveillance is needed for monitoring the spread of these strains equipped with multiple drug resistance mechanisms.
Supplementary Material
Highlights.
Oxacillinases correlated with 90% of MDR A. baumannii from a single hospital
GES-type carbapenemases and ISAba1 were also detected.
Insertion elements are likely behind resistance via β-lactamase production
Majority of the strains belonged to sequence type-2
Genotyping of resistance aids in understanding of MDR A. baumannii transmission
Acknowledgments
This work was supported by Public Health Service grants 1R21AI119339-01 to ASI. The authors would like to extend sincere appreciation to Dr. Brad Spellberg for providing the clinical information of the isolates at Harbor-UCLA Medical Center and Mr. Shady Farran at Research Core Facility of the Health Sciences Centre, Kuwait University (Project No. SRUL02/13), Mrs. Qudsiya Electricwala and Mrs. Leina Ibrahim at Faculty of Allied Health Sciences, Kuwait University for their help and assistance.
Research described in this manuscript was conducted in part at the research facilities of the Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center.
Footnotes
Competing Interests:
The authors declare that there are no competing interests.
Authors’ Contributions:
SS, AD, and ASI conceived and designed the experiments. SS, LV and MB performed the experiments. SS, LV and ASI analyzed the data. SS and ASI wrote the paper. AD, LV and MB revised the paper. All authors have approved the final article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51:3471–84. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maragakis LL, Tucker MG, Miller RG, Carroll KC, Perl TM. Incidence and prevalence of multidrug-resistant Acinetobacter using targeted active surveillance cultures. JAMA. 2008;299:2513–4. doi: 10.1001/jama.299.21.2513. [DOI] [PubMed] [Google Scholar]
- 3.Maragakis LL, Winkler A, Tucker MG, et al. Outbreak of multidrug-resistant Serratia marcescens infection in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2008;29:418–23. doi: 10.1086/587969. [DOI] [PubMed] [Google Scholar]
- 4.Paterson DL. The epidemiological profile of infections with multidrug-resistant Pseudomonas aeruginosa and Acinetobacter species. Clin Infect Dis. 2006;43(Suppl 2):S43–8. doi: 10.1086/504476. [DOI] [PubMed] [Google Scholar]
- 5.Aronson NE, Sanders JW, Moran KA. In harm's way: infections in deployed American military forces. Clin Infect Dis. 2006;43:1045–51. doi: 10.1086/507539. [DOI] [PubMed] [Google Scholar]
- 6.Harman DR, Hooper TI, Gackstetter GD. Aeromedical evacuations from Operation Iraqi Freedom: a descriptive study. Mil Med. 2005;170:521–7. doi: 10.7205/milmed.170.6.521. [DOI] [PubMed] [Google Scholar]
- 7.Hinsley DE, Phillips SL, Clasper JS. Ballistic fractures during the 2003 Gulf conflict early prognosis and high complication rate. J R Army Med Corps. 2006;152:96–101. doi: 10.1136/jramc-152-02-06. [DOI] [PubMed] [Google Scholar]
- 8.Murray CK, Roop SA, Hospenthal DR, et al. Bacteriology of war wounds at the time of injury. Mil Med. 2006;171:826–9. doi: 10.7205/milmed.171.9.826. [DOI] [PubMed] [Google Scholar]
- 9.Murray CK, Yun HC, Griffith ME, et al. Recovery of multidrug-resistant bacteria from combat personnel evacuated from Iraq and Afghanistan at a single military treatment facility. Mil Med. 2009;174:598–604. doi: 10.7205/milmed-d-03-8008. [DOI] [PubMed] [Google Scholar]
- 10.Perez F, Hujer AM, Hulten EA, et al. Antibiotic resistance determinants in Acinetobacter spp and clinical outcomes in patients from a major military treatment facility. Am J Infect Control. 2010;38:63–5. doi: 10.1016/j.ajic.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talbot GH, Bradley J, Edwards JE, Jr, Gilbert D, Scheld M, Bartlett JG. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin Infect Dis. 2006;42:657–68. doi: 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 12.Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 13.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–82. doi: 10.1128/CMR.00058-07. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dizbay M, Tunccan OG, Sezer BE, Hizel K. Nosocomial imipenem-resistant Acinetobacter baumannii infections: epidemiology and risk factors. Scand J Infect Dis. 2010;42:741–6. doi: 10.3109/00365548.2010.489568. [DOI] [PubMed] [Google Scholar]
- 15.Sunenshine RH, Wright MO, Maragakis LL, et al. Multidrug-resistant Acinetobacter infection mortality rate and length of hospitalization. Emerg Infect Dis. 2007;13:97–103. doi: 10.3201/eid1301.060716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doi Y, Husain S, Potoski BA, McCurry KR, Paterson DL. Extensively drug-resistant Acinetobacter baumannii. Emerg Infect Dis. 2009;15:980–2. doi: 10.3201/eid1506.081006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lautenbach E, Synnestvedt M, Weiner MG, et al. Epidemiology and impact of imipenem resistance in Acinetobacter baumannii. Infect Control Hosp Epidemiol. 2009;30:1186–92. doi: 10.1086/648450. [DOI] [PubMed] [Google Scholar]
- 18.Metan G, Sariguzel F, Sumerkan B. Factors influencing survival in patients with multi-drug-resistant Acinetobacter bacteraemia. Eur J Intern Med. 2009;20:540–4. doi: 10.1016/j.ejim.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Gordon NC, Wareham DW. A review of clinical and microbiological outcomes following treatment of infections involving multidrug-resistant Acinetobacter baumannii with tigecycline. J Antimicrob Chemother. 2009;63:775–80. doi: 10.1093/jac/dkn555. [DOI] [PubMed] [Google Scholar]
- 20.Karageorgopoulos DE, Falagas ME. Current control and treatment of multidrug-resistant Acinetobacter baumannii infections. Lancet Infect Dis. 2008;8:751–62. doi: 10.1016/S1473-3099(08)70279-2. [DOI] [PubMed] [Google Scholar]
- 21.Gardiner D, Dukart G, Cooper A, Babinchak T. Safety and efficacy of intravenous tigecycline in subjects with secondary bacteremia: pooled results from 8 phase III clinical trials. Clin Infect Dis. 2010;50:229–38. doi: 10.1086/648720. [DOI] [PubMed] [Google Scholar]
- 22.Park YK, Jung SI, Park KH, et al. Independent emergence of colistin-resistant Acinetobacter spp. isolates from Korea. Diagn Microbiol Infect Dis. 2009;64:43–51. doi: 10.1016/j.diagmicrobio.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Adams MD, Nickel GC, Bajaksouzian S, et al. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gardiner D, Dukart G, Cooper A, Babinchak T. Safety and efficacy of intravenous tigecycline in subjects with secondary bacteremia: pooled results from 8 phase III clinical trials. Clin Infect Dis. 2009;53:3628–34. doi: 10.1086/648720. [DOI] [PubMed] [Google Scholar]
- 24.Falagas ME, Rafailidis PI, Matthaiou DK, Virtzili S, Nikita D, Michalopoulos A. Pandrug-resistant Klebsiella pneumoniae, Pseudomonas aeruginosa and Acinetobacter baumannii infections: characteristics and outcome in a series of 28 patients. Int J Antimicrob Agents. 2008;32:450–4. doi: 10.1016/j.ijantimicag.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Levin AS, Barone AA, Penco J, et al. Intravenous colistin as therapy for nosocomial infections caused by multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Clin Infect Dis. 1999;28:1008–11. doi: 10.1086/514732. [DOI] [PubMed] [Google Scholar]
- 26.Oliveira MS, Prado GV, Costa SF, Grinbaum RS, Levin AS. Ampicillin/sulbactam compared with polymyxins for the treatment of infections caused by carbapenem-resistant Acinetobacter spp. J Antimicrob Chemother. 2008;61:1369–75. doi: 10.1093/jac/dkn128. [DOI] [PubMed] [Google Scholar]
- 27.Valencia R, Arroyo LA, Conde M, et al. Nosocomial outbreak of infection with pan-drug-resistant Acinetobacter baumannii in a tertiary care university hospital. Infect Control Hosp Epidemiol. 2009;30:257–63. doi: 10.1086/595977. [DOI] [PubMed] [Google Scholar]
- 28.Pogue JM, Mann T, Barber KE, Kaye KS. Carbapenem-resistant Acinetobacter baumannii: epidemiology, surveillance and management. Expert Rev Anti Infect Ther. 2013 Apr;11(4):383–93. doi: 10.1586/eri.13.14. Review. [DOI] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. CLSI; Wayne, PA: 2012. p. 501. Document M100-S21. [Google Scholar]
- 30.Blanco M, Alonso MP, Nicolas-Chanoine MH, Dahbi G, Mora A, Blanco JE, López C, Cortés P, Llagostera M, Leflon-Guibout V, Puentes B, Mamani R, Herrera A, Coira MA, García-Garrote F, Pita JM, Blanco J. Molecular epidemiology of Escherichia coli producing extended-spectrum beta-lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J Antimicrob Chemother. 2009;63(6):1135–41. doi: 10.1093/jac/dkp122. Epub 2009 Apr 7. [DOI] [PubMed] [Google Scholar]
- 31.Poirel L, Lagrutta E, Taylor P, Pham J, Nordmann P. Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob Agents Chemother. 2010;54(11):4914–6. doi: 10.1128/AAC.00878-10. Epub 2010 Sep 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. 2007;60(2):394–7. doi: 10.1093/jac/dkm204. Epub 2007 Jun 8. [DOI] [PubMed] [Google Scholar]
- 33.Coelho A, González-López JJ, Miró E, Alonso-Tarrés C, Mirelis B, Larrosa MN, Bartolomé RM, Andreu A, Navarro F, Johnson JR, Prats G. Characterization of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int J Antimicrob Agents. 2010;36(1):73–8. doi: 10.1016/j.ijantimicag.2010.03.005. Epub 2010 Apr 13. [DOI] [PubMed] [Google Scholar]
- 34.Giraud E, Brisabois A, Martel JL, Chaslus-Dancla E. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counter selection of highly fluoroquinolone-resistant strains in the field. Antimicrob Agents Chemother. 1999;43(9):2131–7. doi: 10.1128/aac.43.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–62. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poirel L, Bonnin RA, Nordmann P. Genetic support and diversity of acquired extended-spectrum β-lactamases in Gram-negative rods. Infect Genet Evol. 2012;12(5):883–93. doi: 10.1016/j.meegid.2012.02.008. Epub 2012 Mar 3. [DOI] [PubMed] [Google Scholar]
- 37.Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob Agents Chemother. 2010;54(1):24–38. doi: 10.1128/AAC.01512-08. Epub 2009 Aug 31. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans BA, Amyes SG. OXA β-lactamases. Clin Microbiol Rev. 2014;27(2):241–63. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiratisin P, Apisarnthanarak A, Saifon P, Laesripa C, Kitphati R, Mundy LM. The emergence of a novel ceftazidime-resistant CTX-M extended-spectrum beta-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn Microbiol Infect Dis. 2007;58(3):349–55. doi: 10.1016/j.diagmicrobio.2007.02.005. Epub 2007 Apr 20. [DOI] [PubMed] [Google Scholar]
- 40.Mugnier PD, Poirel L, Nordmann P. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J Bacteriol. 2009;191(7):2414–8. doi: 10.1128/JB.01258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3(1):59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 42.Bao L, Peng R, Ren X, Ma R, Li J, Wang Y. Analysis of some common pathogens and their drug resistance to antibiotics. Pak J Med Sci. 2013;29(1):135–9. doi: 10.12669/pjms.291.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dashti AA, Vali L, El-Shazly S, Jadaon MM. The characterization and antibiotic resistance profiles of clinical Escherichia coli O25b-B2-ST131 isolates in Kuwait. BMC Microbiol. 2014;14:214. doi: 10.1186/s12866-014-0214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma M, Pathak S, Srivastava P. Prevalence and antibiogram of Extended Spectrum β-Lactamase (ESBL) producing Gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. J Clin Diagn Res. 2013;7(10):2173–7. doi: 10.7860/JCDR/2013/6460.3462. Epub 2013 Oct 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sjölund-Karlsson M, Howie R, Rickert R, Krueger A, Tran TT, Zhao S, Ball T, Haro J, Pecic G, Joyce K, Fedorka-Cray PJ, Whichard JM, McDermott PF. Plasmid-mediated quinolone resistance among non-Typhi Salmonella enterica isolates, USA. Emerg Infect Dis. 2010;16(11):1789–91. doi: 10.3201/eid1611.100464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams-Haduch JM, Onuoha EO, Bogdanovich T, Tian GB, Marschall J, Urban CM, Spellberg BJ, Rhee D, Halstead DC, Pasculle AW, Doi Y. Molecular epidemiology of carbapenem-nonsusceptible Acinetobacter baumannii in the United States. J Clin Microbiol. 2011;49(11):3849–54. doi: 10.1128/JCM.00619-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee GC, Lawson KA, Burgess DS. Clinical epidemiology of carbapenem-resistant enterobacteriaceae in community hospitals: a case-case-control study. Ann Pharmacother. 2013;47(9):1115–21. doi: 10.1177/1060028013503120. [DOI] [PubMed] [Google Scholar]
- 48.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK National Healthcare Safety Network Team; Participating National Healthcare Safety Network Facilities. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008 Nov;29(11):996–1011. doi: 10.1086/591861. Erratum in: Infect Control Hosp Epidemiol 2009; 30(1)107. [DOI] [PubMed] [Google Scholar]
- 49.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol. 2005;43(9):4328–35. doi: 10.1128/JCM.43.9.4328-4335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh A, Goering RV, Simjee S, Foley SL, Zervos MJ. Application of molecular techniques to the study of hospital infection. Clin Microbiol Rev. 2006;19(3):512–30. doi: 10.1128/CMR.00025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43(9):4382–90. doi: 10.1128/JCM.43.9.4382-4390.2005. Erratum in: J Clin Microbiol 2007 Jun; 45(6):2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One. 2010;5(4):e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urwin R, Maiden MC. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 2003;11(10):479–87. doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 54.Luo G, Spellberg B, Gebremariam T, Lee H, Fu Y, French SW, Ibrahim AS. Diabetic murine models for Acinetobacter baumannii infection. J Antimicrob Chemother. 2012 doi: 10.1093/jac/dks050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright MS, Haft DH, Harkins DM, Perez F, Hujer KM, Bajaksouzian S, Benard MF, Jacobs MR, Bonomo RA, Adams MD. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. M Bio. 2014;5(1):e00963–13. doi: 10.1128/mBio.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Opazo A, Vali L, Al Obaid K, Dashti AA, Amyes SG. Novel genetic structure harbouring blaPER-1 in ceftazidime-resistant Acinetobacter baumannii isolated from Kuwait. Int J Antimicrob Agents. 2014;43(4):383–4. doi: 10.1016/j.ijantimicag.2014.01.001. Epub 2014 Jan 21. [DOI] [PubMed] [Google Scholar]
- 57.Shelburne SA, 3rd, Singh KV, White AC, Jr, Byrne L, Carmer A, Austin C, Graviss E, Stager C, Murray BE, Atmar RL. Sequential outbreaks of infections by distinct Acinetobacter baumannii strains in a public teaching hospital in Houston, Texas. J Clin Microbiol. 2008 Jan;46(1):198–205. doi: 10.1128/JCM.01459-07. Epub 2007 Nov 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wisplinghoff H, et al. MLST-based molecular epidemiology of clinical Acinetobacter baumannii bloodstream isolates from 52 hospitals in the US, 2010. Abstr. C2-590. Abstr. 50th Intersci Conf Antimicrob. Agents Chemother. [Google Scholar]
- 59.Valentine SC, Contreras D, Tan S, Real LJ, Chu S, Xu HH. Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in Los Angeles County, California. J Clin Microbiol. 2008 Aug;46(8):2499–507. doi: 10.1128/JCM.00367-08. Epub 2008 Jun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aly M, Tayeb HT, AlJohani SM, Alyamani EJ, Aldughaishem F, Alabdulkarim I, et al. Genetic diversity of OXA-51-like genes among multidrug-resistant Acinetobacter baumannii in Riyadh, Saudi Arabia. Eur J Clin Microbiol Infect Dis. 2014;33:1223–1228. doi: 10.1007/s10096-014-2068-0. [DOI] [PubMed] [Google Scholar]
- 61.Rafei R, Dabboussi F, Hamze M, Eveillard M, Lemarié C, Gaultier MP, Mallat H, Moghnieh R, Husni-Samaha R, Joly-Guillou ML, Kempf M. Molecular analysis of Acinetobacter baumannii strains isolated in Lebanon using four different typing methods. PLoS One. 2014 Dec 26;9(12):e115969. doi: 10.1371/journal.pone.0115969. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bakour S, Alsharapy SA, Touati A, Rolain JM. Characterization of Acinetobacter baumannii clinical isolates carrying bla (OXA-23) carbapenemase and 16S rRNA methylase armA genes in Yemen. Microb Drug Resist. 2014;20:604–609. doi: 10.1089/mdr.2014.0018. [DOI] [PubMed] [Google Scholar]
- 63.Villalón P, Valdezate S, Medina-Pascual MJ, Rubio V, Vindel A, Saez Nieto JA. Clonal diversity of nosocomial epidemic Acinetobacter baumannii strains isolated in Spain. J Clin Microbiol. 2011;49:875–882. doi: 10.1128/JCM.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mezzatesta ML, Caio C, Gona F, Cormaci R, Salerno I, Zingali T, et al. Carbapenem and multidrug resistance in Gram-negative bacteria in a single centre in Italy: considerations on in vitro assay of active drugs. Int J Antimicrob Agents. 2014;44:112–116. doi: 10.1016/j.ijantimicag.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 65.Jeannot K, Diancourt L, Vaux S, Thouverez M, Ribeiro A, Coignard B, Courvalin P, Brisse S. Molecular epidemiology of carbapenem non-susceptible Acinetobacter baumannii in France. PLoS One. 2014 Dec 17;9(12):e115452. doi: 10.1371/journal.pone.0115452. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gogou V, Pournaras S, Giannouli M, Voulgari E, Piperaki ET, Zarrilli R, Tsakris A. Evolution of multidrug-resistant Acinetobacter baumannii clonal lineages: a 10 year study in Greece (2000–09) J Antimicrob Chemother. 2011 Dec;66(12):2767–72. doi: 10.1093/jac/dkr390. Epub 2011 Sep 19. [DOI] [PubMed] [Google Scholar]
- 67.Matsui M, Suzuki S, Yamane K, Suzuki M, Konda T, Arakawa Y, Shibayama K. Distribution of carbapenem resistance determinants among epidemic and non-epidemic types of Acinetobacter species in Japan. J Med Microbiol. 2014 Jun;63(Pt 6):870–7. doi: 10.1099/jmm.0.069138-0. Epub 2014 Mar 5. [DOI] [PubMed] [Google Scholar]
- 68.Chen CM, Ke SC, Li CR, Chang CC. The comparison of genotyping, antibiogram, and antimicrobial resistance genes between carbapenem-susceptible and -resistant Acinetobacter baumannii. Comp Immunol Microbiol Infect Dis. 2014 Dec;37(5–6):339–46. doi: 10.1016/j.cimid.2014.10.002. Epub 2014 Oct 17. [DOI] [PubMed] [Google Scholar]
- 69.Hammerum AM, Hansen F, Skov MN, Stegger M, Andersen PS, Holm A, Jakobsen L, Justesen US. Investigation of a possible outbreak of carbapenem-resistant Acinetobacter baumannii in Odense, Denmark using PFGE, MLST and whole-genome-based SNPs. J Antimicrob Chemother. 2015 Jul;70(7):1965–8. doi: 10.1093/jac/dkv072. Epub 2015 Mar 19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.