Abstract
Size is a primary feature of biological systems that varies at many levels, from the organism to its constituent cells and subcellular structures. Amphibians populate some of the extremes in biological size and have provided insight into scaling mechanisms, upper and lower size limits, and their physiological significance. Body size variation is a widespread evolutionary tactic among amphibians, with miniaturization frequently correlating with direct development that occurs without a tadpole stage. The large genomes of salamanders lead to large cell sizes that necessitate developmental modification and morphological simplification. Amphibian extremes at the cellular level have provided insight into mechanisms that accommodate cell-size differences. Finally, how organelles scale to cell size between species and during development has been investigated at the molecular level, because subcellular scaling can be recapitulated using Xenopus in vitro systems.
From the smallest known vertebrate (the 7-mm frog Paedophryne amanuensis) to large genomes (e.g., 121 pg in the Gulf Coast waterdog), size extremes in amphibians provide insight into the biology of size at all levels.
Size is a fundamental biological feature that impacts physiology at all levels, from organism to organ to cell to subcellular structures/organelles. One basic aspect of size is its absolute value, which has upper and lower limits because of functional requirements. For example, a vertebrate organ, such as an eye or an inner ear, may require a minimum number of cells, or a minimum physical size, to operate. Importantly, surface area and volume scale differently with size, and this also has physiological consequences at both the organism and cellular levels, affecting basic processes, such as desiccation and diffusion. A second important feature of size is scaling relationships, as the overall size of an organism or tissue is determined both by cell size and cell number. At the subcellular level, size scaling may or may not occur depending on the organelle, as absolute values are constrained by the nature and flexibility of constituent molecular building blocks. For example, whereas the size of the nucleus varies significantly and scales with cell size, organelle transport vesicles are of more uniform size owing to the conserved structure of their coat proteins. Extremes in amphibian size and scaling relationships derive primarily from dramatic variations in genome size, and provide instructive examples of size relationships, underlying molecular mechanisms, and above all the remarkable flexibility and power of evolution to adapt biological function across a wide range of size scales.
AMPHIBIAN BODY SIZE LIMITS
Body size is one of the most significant organismal traits because it influences so many biological attributes. These include development, physiology, such as locomotion and reproductive biology, behavior, such as feeding, and ecology, including habitat and relationships with other species. Living amphibians consist of three clades: Anura (frogs and toads), Caudata or Urodela (salamanders, of which newts are one type) and Gymnophiona (caecillians—legless, snake-like organisms). Amphibians range in length over 250-fold. At one extreme is the smallest known vertebrate at 7 mm long, the frog Paedophryne amanuensis (Rittmeyer et al. 2012), whereas the Goliath frog (Conraua goliath) can grow up to 33 cm, and the Chinese salamander (Andrias davidianis) to 1.8 m (Frost 2014).
Different body sizes come with distinct advantages and disadvantages. Large amphibians have fewer predators, a lower metabolic rate, and they can more easily maintain their temperature and hydration than small amphibians. What establishes the upper limit to amphibian size is unknown, and, in some cases, amphibians increase in size throughout adulthood, a phenomenon called indeterminate growth, which is discussed in Hariharan et al. (2015). Leaving environmental issues, such as food and space, aside, the maximum size for land vertebrates is limited by allometric scaling laws. For example, whereas the cross-sectional strength of the skeleton increases as the square of the length, weight increases as the cube. So, weight increases faster than strength and, although bone mass can increase, at some maximum size, a large land animal can no longer support itself (Hokkanen 1986). Although lower metabolic rates enable large species to survive longer without food, more food and space are required, making habitat and environmental range crucial. The largest amphibian that ever existed is thought to be Prionosuchus, which reached an estimated length of 9 m in the middle Permian period (270 million years ago), occupying the ecological niche of crocodiles and alligators in what is now northeastern Brazil (Fox and Hutchinson 1991).
At 7 mm, the lower size limit for amphibians is indeed miniscule. Miniaturization has been documented in all three clades, and occurred independently many times during evolution. Although their size makes the smallest amphibians vulnerable to more predators, including insects (Rittmeyer et al. 2012), it also enables them to hide more easily, exploit alternate food sources, use physically smaller niches, and attain reproductive ability at an earlier age (Zimkus et al. 2012). In contrast, the endothermic metabolism of birds and mammals dictates a larger minimum adult body size. A major disadvantage to small body size is susceptibility to desiccation, given their high surface-area-to-volume ratio. Miniaturized frogs, therefore, inhabit tropical wet-forest leaf litter or dense, moist moss.
What are the mechanisms that allow amphibians to evolve different body sizes? To become bigger, growth rates and/or the period of growth must increase. Underlying molecular mechanisms have not been studied in amphibians, but among dogs, which display the greatest diversity in size among land vertebrates, insulin-like growth-factor signaling has been strongly implicated (Sutter et al. 2007; Hoopes et al. 2012). More interesting perhaps is animal miniaturization. Miniature species often bear a strong resemblance to juveniles, resulting from either precocious cessation of growth or reduction in growth rate. Again, the molecular mechanisms are unknown, but extreme size reduction is accompanied by morphological novelty, as the distinct functions of different body parts and organs necessitate different scaling relationships (Hanken and Thorogood 1993). For example, the inner ear scales smaller at a lower rate than the body as a whole. To compensate, miniaturized amphibians have rearranged the adjacent skull and jaw (Hanken 1983). Thus, miniaturization has important physiological consequences including reduced function or even loss of organs, loss of digits, and simplification of other external structures. A lower size limit may, therefore, be dictated by a number of different organ systems that require a minimum size to function properly. In addition, small species tend to have fewer offspring, because organs fill most of the body cavity with little space leftover for eggs (Hedges 2008), making miniature species less adaptable to environmental challenges. However, evolution to different body sizes may also allow species to populate new habitats and promote successful radiation and coexistence with other species. Thus, size variation is a widespread evolutionary tactic among amphibians, with the scaling features of essential body systems setting both upper and lower size limits.
CELL–ORGANISM SCALING RELATIONSHIPS, PHYSIOLOGY AND DEVELOPMENT
How does organism size relate to the size of its constituent cells? Although cell size varies widely depending on the tissue and developmental state of an organism, among mammals, it is primarily the number of cells that differs among different-sized species. This correlates with the relatively low variation in DNA content among mammalian cells, in the range of 1–4 pg/nucleus (Gregory 2001a). In contrast, especially large variation has been noted in salamanders, with genome sizes ranging from 14 to 120 pg/nucleus. Corresponding changes in cell size occur, but body size is not always correlative. For example, one salamander species of the genus Thorius possesses a large genome (25 pg) and large cells, but is small (Sessions and Larson 1987; Hanken and Thorogood 1993). However, in certain frog species, such as Xenopus laevis and Xenopus tropicalis, body size scales with genome size and cell size. X. laevis is allotetraploid (a hybrid species with both parental genomes present in gametes: 36 chromosomes) and larger (∼10 cm adults), whereas X. tropicalis is diploid (20 chromosomes) and smaller (∼4 cm adults). Scaling at the organismal and genome levels is accompanied by differences in the size of the egg as well as that of subcellular structures formed in egg extracts, including nuclei and mitotic spindles (discussed below) (Levy and Heald 2012; Edens and Levy 2014b). Despite their size differences, the close phylogenetic relationship between these two species allows the production of hybrid embryos by cross-fertilization (Burki 1985; Narbonne et al. 2011). Interestingly, fertilization of large X. laevis eggs with X. tropicalis sperm gives rise to swimming tadpoles and even frogs that are of intermediate size between the two species, providing a unique opportunity to explore the contribution of genome and maternal components to cell and organism size. In contrast, embryos of the reverse hybrid, small X. tropicalis eggs fertilized with X. laevis sperm, die as late blastulae. It is not yet clear whether the difference in viability stems from size relationships or is caused by lack of maternally derived species-specific factors (Narbonne et al. 2012). Perhaps a large egg can accommodate a genome smaller than normal, whereas a small egg cannot tolerate a larger set of chromosomes. Exploring the origin of incompatibility and cause of death in these hybrids may shed light on the importance of scaling cell size to genome size.
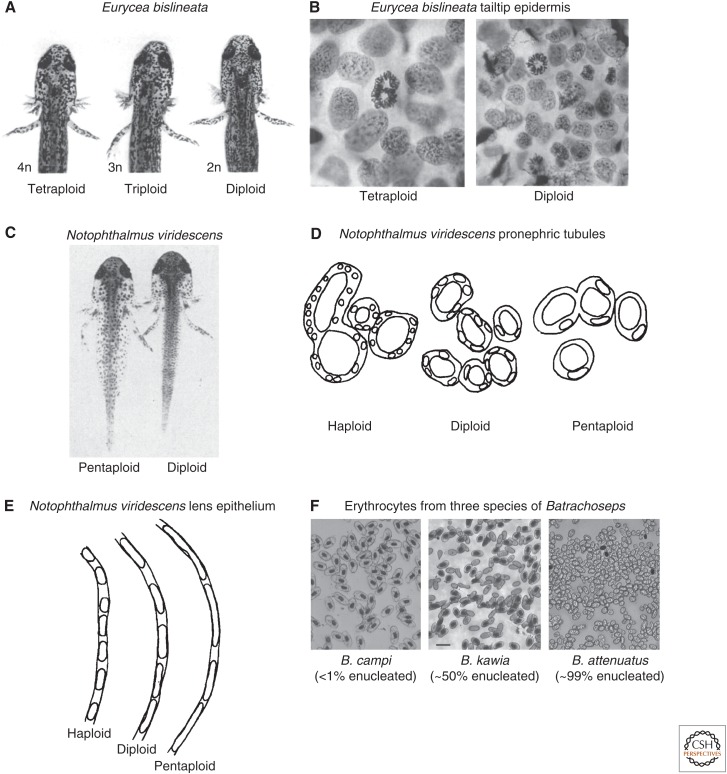
What are the developmental consequences of different cell–organism scaling relationships? Species that are the same physical size but possess different genome and cell sizes likely also differ in ways that significantly affect morphogenesis, growth, and adult morphology. If animal size is held constant, then the larger the cells, the fewer their number, which provides fewer building blocks and requires morphological modification and/or simplification (Fig. 1A–E). For example, polyploid newt larvae (Notophthalmus viridescens) possess large cells that pose a challenge in the kidney, where they must undergo more dramatic shape changes than in diploid animals to form kidney tubules and ducts of normal dimensions. Similarly, the thickness of the lens epithelium of the eye is unchanged in polyploids where fewer cells undergo more extensive flattening (Fankhauser 1945a). Salamanders of the genus Hydromantes have the largest genome of any terrestrial amphibian, and also possess one of the most simplified brains of any vertebrate. The relationship between cell size and brain morphology is discussed at length in Roth and Walkowiak (2015).
Figure 1.
Changes in cell size are accompanied by modification of cell and tissue architecture. (A) Eurycea bislineata (northern two-lined salamander) larvae of differing ploidy are shown. Polyploids occur spontaneously in nature at a frequency of 5%–10% for triploids and <1% for tetraploids. With increasing ploidy, cell sizes increase and cell numbers decrease, so that, ultimately, animal size remains roughly constant. In this image, one notes fewer and larger pigment cells in the head of the tetraploid as compared with the diploid. (From Fankhauser 1939; reprinted, with permission, from Oxford University Press © 1939.) (B) Nuclei in the tetraploid epidermal cells are much larger than in the diploid, and, by inference, cell size is also greater. It is evident from the metaphase cells that chromosome number is much greater in the tetraploid. (From Fankhauser 1939; reprinted, with permission, from Oxford University Press © 1939.) (C) The pentaploid Notophthalmus viridescens (eastern newt) larva at 5.5 wk of age appears similar to the diploid, except for the altered size and number of pigment cells. Spontaneous pentaploids are found in nature at a frequency of <1%. Polyploidy can also be induced by heat treatment (Fankhauser and Watson 1942). (From Fankhauser 1945b; reprinted, with permission, from The University of Chicago Press © 1945.) (D) Polyploid Notophthalmus viridescens larvae that arose spontaneously in the laboratory were fixed, sectioned, and diagramed to show tissue morphology. Although boundaries between adjacent cells are not apparent, the spacing of nuclei approximates cell sizes and positions. Cross-sections of pronephric tubules from 35- to 40-d-old larvae are shown. Tubule sizes and wall diameters are roughly the same in all three animals despite large differences in cell size and number. Increasing ploidy necessitates more dramatic cell shape changes to maintain normal tissue morphology. At some ploidy extreme, cell size must be too large to accommodate the requisite shape changes; indeed, morphological defects become apparent in highly polyploid animals (Fankhauser 1945b). (From Fankhauser 1945a; reprinted, with permission, from Wiley-Liss, A Wiley Company © 1945.) (E) The morphology of the epithelium covering the outer half of the lens was examined for the same animals described in D. The thickness of the epithelium is the same independent of ploidy, requiring cells to take on a much more elongated and flattened morphology with increasing ploidy. In contrast to the pronephric tubules, in the lens epithelium, the shape of the nuclei must also change to maintain normal tissue thickness. (From Fankhauser 1945a; reprinted, with permission, from Wiley-Liss, A Wiley Company © 1945.) (F) Photomicrographs of erythrocytes from three species of Batrachoseps salamanders are shown. Batrachoseps campi is a nonattenuate species and most of its erythrocytes are nucleated, whereas the other two smaller species show varying degrees of enucleation. One explanation for this adaptation in the miniaturized species is to facilitate circulation by reducing erythrocyte size. Scale bar, 40 µm (A-F). (From Mueller et al. 2008; reprinted, with permission, from Elsevier © 2008.)
Scaling of cell and tissue size has also been investigated in the circulatory system. Erythrocyte sizes have been most thoroughly documented because of the facility with which blood can be collected and analyzed. At the upper end of the spectrum among vertebrate somatic cells, erythrocytes from Amphiuma tridactylum, an aquatic salamander, measure 66 × 37 × 15 µm at their greatest thickness (Hartman and Lessler 1964; Gregory 2011). Some of the smallest recorded amphibian cells are erythrocytes from X. tropicalis and Pyxicephalus adspersus with diameters ∼6 µm, similar to typical mammalian erythrocytes (Horner and Macgregor 1983; Gregory 2011). Across a number of urodele and anuran species, red blood cell size scales with capillary diameter. This trend extends to mammals that have much thinner capillaries and more deformable red blood cells that lack nuclei (Safranyos et al. 1983; Snyder and Sheafor 1999). Interestingly, five clades of salamanders in the family Plethodontidae that have large genomes and small body sizes have evolved high levels of enucleated blood cells, which is highly unusual in nonmammalian vertebrate species, and is likely a response to rheological problems associated with the circulation of large blood cells in these small animals (Fig. 1F) (Mueller et al. 2008).
In cases where cell size scales with animal size, the lower limit to cell size (discussed below) dictates that miniature amphibians face a similar problem of low cell numbers and must adapt. Although development to the tadpole stage appears quite similar comparing X. laevis and X. tropicalis, a tiny related frog Hymenochirus (2.5 cm) may have reached a cell number low enough to affect its development, but this has not been directly shown. Interestingly, Hymenochirus tadpoles with a body length of 2.6 mm are the smallest known vertebrate predators (Deban and Olson 2002). However, it appears that development in many amphibians is overall quite tolerant to changes in cell number. Decreasing cell numbers in Xenopus by blocking cell division or removing many totipotent blastula cells yields embryos that contain fewer cells but possess normal morphology (Cooke 1973, 1981). Thus, situations that generate small cell numbers—miniaturization or large genomes—can be accommodated in development but frequently require developmental modification and morphological simplification.
Other support for a potential link between organism size and mechanisms of development comes from the observation that of the 29 smallest frogs, 24 (83%) lack a larval tadpole stage and develop directly (Rittmeyer et al. 2012). Like frog miniaturization, direct development has arisen independently on multiple occasions (Duellman and Trueb 1986), and is associated with adoption of a terrestrial life style. The eggs of frogs that develop directly require a very high yolk content and most are >3 mm in diameter, as compared with 1.3- to 1.8-mm-diameter eggs common among frogs with tadpoles (Fig. 2). To compensate for reduced fitness associated with large eggs and to increase reproductive capacity, some species of the African genus Nectophrynoides are oviductally viviparous, which means that developing embryos are retained in the oviduct. Epithelial cells secrete a highly nutritious material that is orally ingested by the fetuses and supports their growth, allowing clutch sizes of more than 100 froglets (Wake 1980, 1993).
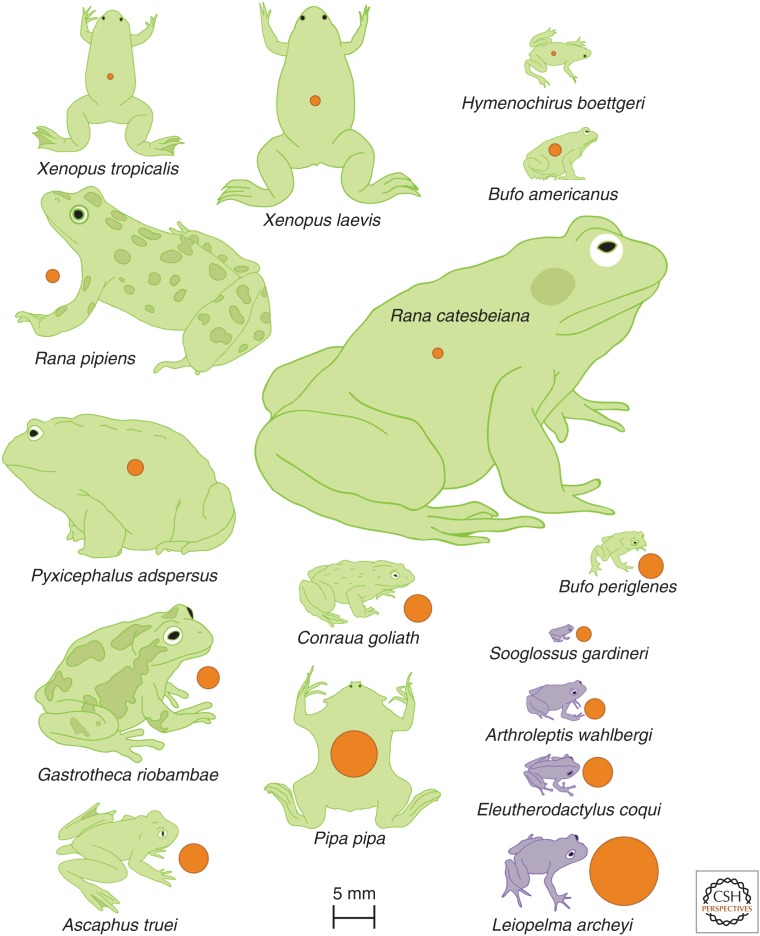
Figure 2.
Relationship between egg diameter and snout-vent length of newly transformed froglets of various species. Direct developers (purple) generally have larger eggs and smaller froglets, with egg and froglet size scaling among species. In comparison, egg and froglet size do not correlate for biphasic developers with tadpoles (green). Pipa pipa, a biphasic frog with a large egg, develops from a nonfeeding tadpole under a layer of skin on the mother’s back. Despite their size differences at metamorphosis, Rana pipiens and Rana catesbeiana reach similar sizes as adults. Conraua goliath and Pyxicephalus adspersus become the largest adult frogs and can exceed 25 cm in length. Embryos of the Andean marsupial tree frog Gastrotheca riobambae develop in the female’s dorsal pouch. Its tailed appendage, an extension of the cloaca of males, makes Ascaphus truei distinct, and improves breeding success by minimizing loss of sperm in the turbulent streams inhabited by this species. (Adapted from data in Callery 2006.)
What is the contribution of a larval (tadpole) stage to frog size? There is wide variation in tadpole size and it correlates with egg size, but not always with postmetamorphic frog size (Fig. 2). For example, the frog Pseudis paradoxa was so named because of the discrepancy in size between its large tadpole and its relatively small adult, and an early study confusing size with age had the frog transforming into the tadpole during ontogeny (Gans 1956; Emerson 1988). The main function of the tadpole is to feed and increase body mass. Thus, growth is biphasic, and body size can increase during the tadpole stage, postmetamorphosis, or both. Environmental factors like predators and population density affect how soon metamorphosis occurs, but size at transformation is characteristic of a species, indicating a genetic component to metamorphic size determination, which is implemented through thyroid hormone signaling (Laudet 2011). Transgenic X. laevis tadpoles overexpressing growth hormone were larger than normal, but underwent metamorphosis at the same time as their wild-type siblings, generating large frogs with symptoms reminiscent of acromegaly (Huang and Brown 2000). Interestingly, biphasic growth is lost in frogs that develop directly, as the embryonic and metamorphic periods overlap and growth occurs more continuously.
In summary, at the level of the organism, amphibians illustrate extreme flexibility in the size of constituent cells, which has led to interesting adaptations in tissues and during embryonic development. Differences in genome size correlate with cell-size changes, but the underlying mechanisms are unknown (discussed in Mueller 2015). Diverse organism–cell-size relationships in amphibians provide a unique opportunity to study the physiological consequences and constraints of differences in cell size and number, and the mechanisms by which overall animal size is determined.
UPPER AND LOWER CELL-SIZE LIMITS AND MECHANISMS
There appear to be intrinsic limitations to cell size, and amphibian extremes have provided insight into cell-size regulation and mechanisms that accommodate cell-size differences. Determinants of minimal cell size include a cell volume large enough to contain the organelles and biomolecules essential for viability, as well as surface area sufficient to accommodate receptors necessary for sensing, signaling, and attachment. As cell size increases, volume increases as the cube of the length while surface area lags behind, scaling as the square, assuming roughly spheroid or ellipsoid cell morphology. As a consequence, large cells might lack sufficient surface area to take up nutrients required to sustain their metabolism. Eggs and cells of early amphibian embryos that are hundreds of micrometers in size exceed the theoretical upper limit, and some amphibian eggs are over 6 mm in size (Collazo and Keller 2010), orders of magnitude greater than the largest somatic cells. The solution to this potential size problem is that large eggs are packed with sufficient proteins and membranes to generate a few thousand cells during rapid cleavage divisions, as well as intracellular yolk platelets that sustain the embryo until the swimming tadpole stage, or froglet in the case of direct development (Jorgensen et al. 2009). An interesting consequence of the embryonic cleavage divisions is that components preloaded in the egg become limiting as cellularization proceeds, providing mechanisms to regulate developmental timing as discussed in Amodeo and Skotheim (2015). Researchers have long taken advantage of Xenopus eggs that can be obtained in large amounts and are stockpiled with cellular contents by preparing cytoplasmic egg extracts that recapitulate cell-cycle events and organelle assembly in vitro (Chan and Forbes 2006; Maresca and Heald 2006).
Intracellular diffusion poses a problem for large cells. For example, the whole-cell reorganization that occurs when a cell divides requires synchronous activation of the mitotic kinase Cdk1 throughout the cell to induce downstream events, including chromosome condensation, spindle formation, and cytokinesis. For a typical somatic cell with a radius of 10 µm, it takes only seconds for activated Cdk1 to spread throughout the cytoplasm, whereas diffusion across a 600-µm-diameter egg would require 2 h. The solution to this problem is the generation of “trigger waves” of Cdk1 activation through a positive feedback loop of kinase activation by posttranslational modification. Mitotic kinase activity was observed to spread at ∼40 times the rate of diffusion through a Teflon tube containing Xenopus egg extract (Chang and Ferrell 2013). Another mechanism is directed transport of molecular cargos by motor proteins along cytoskeletal tracks of filamentous actin and microtubules, for example, to achieve polarized localization of RNA developmental cues in amphibian oocytes (Marracci et al. 2011; Gagnon et al. 2013). Surprisingly, gravity is also relevant to intracellular organization in large cells and even organelles. Xenopus oocyte nuclei possess an actin array that maintains organization of large ribonucleoprotein complexes in the face of gravity, and disruption of this network results in sedimentation and fusion of nucleoli and histone locus bodies (Brangwynne 2013; Feric and Brangwynne 2013).
Biomechanical properties of the cell division machinery face challenges at the extremes of cell size. For example, spindles in the large cells of early frog embryos show an upper size limit, which may be set by the intrinsic length scale of their constituent microtubules (Wühr et al. 2008; Marshall et al. 2012). Nevertheless, the microtubule cytoskeleton mediates large-scale chromosome movements at mitotic exit, as discussed in Mitchison et al. (2015). Assembly properties of the actomyosin contractile ring could also be important, limiting the size of cells able to undergo successful cytokinesis. This problem has been overcome in large amphibian eggs >2–3 mm in diameter by a process of irregular, partial cell divisions, termed meroblastic cleavages (Collazo and Keller 2010; Marshall et al. 2012), and maximal amphibian egg size is likely constrained by the processes of cleavage and gastrulation. At the other extreme, small cells would face problems with cell division if chromosome length exceeded cell length, preventing their proper segregation, or if components necessary to form a functional cell division apparatus became limiting at small volumes. A lower spindle size limit was observed in encapsulated droplets of Xenopus extract, as compartments smaller than 20 µm in diameter failed to support meiotic spindle assembly (Good et al. 2013). In addition to limiting components, another possible explanation is that microtubule dynamics become compromised at small length scales, as illustrated by centrosome-positioning defects observed in small microfabricated chambers in which microtubules buckle when they contact the chamber periphery (Faivre-Moskalenko and Dogterom 2002). Indeed, abnormal mitotic cell size and shape can alter spindle assembly and positioning and interfere with proper chromosome capture (Cadart et al. 2014).
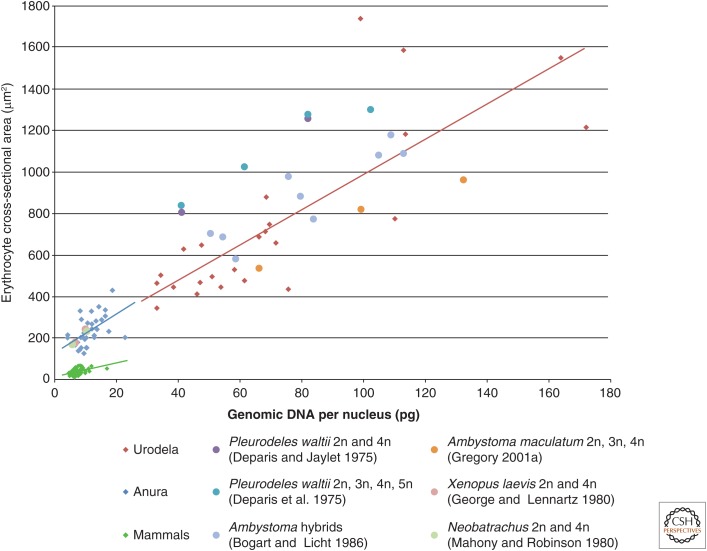
A major factor determining cell size is genome size (Sessions and Larson 1987; Cavalier-Smith 2005). It has long been observed that genome and cell size scale, and this is particularly striking in the case of amphibians (Fig. 3) (Horner and Macgregor 1983; Gregory 2001a,b). In general, amphibian genomes are larger than those of mammals, with the largest amphibian genome measuring 121 pg (Gulf Coast waterdog Necturus lewisi) (Gregory et al. 2007). Interestingly, species with the largest genomes, such as amoeba, are unicellular, suggesting that multicellularity imposes an upper limit to genome size (Gregory 2014). Hybridization and whole genome duplication are common in amphibians and frequently lead to increased cell size as documented in a variety of polyploid species (Fankhauser 1939, 1945a,b; Pollack and Koves 1977; Winklbauer and Hausen 1985; Mable et al. 2011). The wide range of genome and cell sizes among amphibians enable studies of the consequences for organismal physiology. For example, metabolic rate scales inversely with genome size (Gregory 2003; Vinogradov and Anatskaya 2006), and cell size affects development and tissue architecture as described above (Fig. 1).
Figure 3.
Relationship between red blood cell size and genome size among amphibians and mammals. Diamonds show genome and erythrocyte sizes for different species of urodela (red), anura (blue), and mammals (green), using data obtained from online sources. (From Gregory et al. 2007; with permission from Oxford University Press © 2007; and Gregory 2014.) Best-fit lines for these three groups are shown. Included here are only those species for which both genome size and dry red blood cell area have been reported. Unless otherwise noted, data along the x-axis reflects diploid genome size (i.e., twice the C-value). Amphibian erythrocytes and genomes scale to significantly larger sizes than mammals, which appear to have a less steep curve. However, this may not be the best comparison because mammalian red blood cells lack nuclei. Hybridization and whole genome duplication are common in amphibians and may explain why amphibian genomes and cell sizes span a much larger size range than other animals. The circles on the plot show data from the same amphibian species with differing ploidy. In these cases, erythrocyte sizes were either reported or estimated from images in the literature (Deparis and Jaylet 1975; Deparis et al. 1975; George and Lennartz 1980; Mahony and Robinson 1980; Bogart and Licht 1986; Gregory 2001a). Interestingly, the polyploid data scale similarly to the diploid species. Among amphibians, erythrocyte size correlates with cell sizes in other tissues (Kozlowski et al. 2010), and the data plotted here are consistent with ploidy effects on cell size in other tissue types, including the Rana pipiens spinal cord (Pollack and Koves 1977), X. laevis lateral line (Winklbauer and Hausen 1985), and multiple different organs in newts and salamanders (Fankhauser 1939, 1945a,b).
What are the underlying mechanisms that regulate cell size and do cells actively sense their size? In principle, cell size is determined by the balanced regulation of cell division and cell growth. Potential mechanisms linking genome size and cell size are discussed in Roth and Walkowiak (2015), Amodeo and Skotheim (2015), and Mueller (2015). In turn, cell growth, division, and differentiation status determine the size of tissues and organs. The urodele amphibians are particularly fascinating in this respect, being the only adult vertebrates capable of limb regeneration. Upon amputation or tissue removal, postmitotic cells beneath the wound epidermis dedifferentiate and reenter the cell cycle to allow for tissue growth to regenerate a limb of the correct size (Brockes 1997; Roensch et al. 2013).
SUBCELLULAR SIZE REGULATION IN AMPHIBIANS
How do the sizes of intracellular organelles and structures vary to accommodate differences in cell size? General mechanisms of subcellular size control are discussed in detail in Marshall (2015) and Reber and Goehring (2015). Amphibians, and Xenopus frogs in particular, have provided unique approaches to elucidate scaling mechanisms, both among species with different-sized cells as well as during development when cleavage divisions rapidly give rise to smaller and smaller cells (Fig. 4). Here, we focus on intracellular structures that have been shown to exhibit size scaling, namely, the interphase nucleus, mitotic spindle, and mitotic chromosomes. Much less is known about scaling of other organelles in amphibians, like the endoplasmic reticulum (ER) and Golgi. Some structures, such as vesicles, are more uniform in size and less likely to be influenced by cell size and instead may scale to the size of their cargoes (Jin et al. 2012).
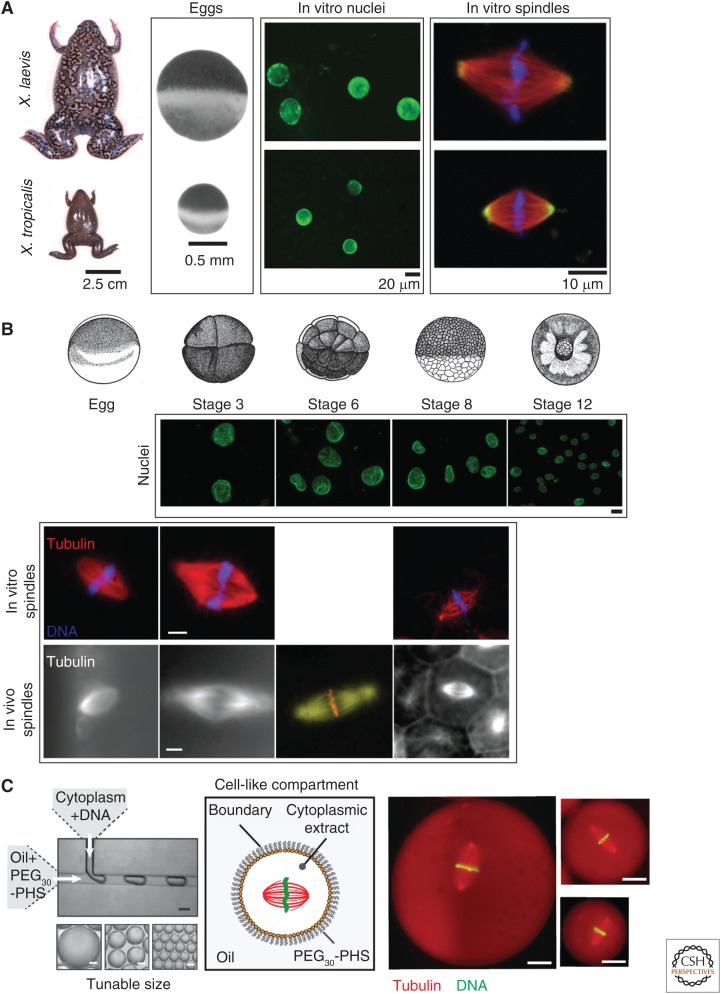
Figure 4.
Intracellular scaling in Xenopus. (A) Examples of Xenopus interspecies organelle scaling are shown. X. laevis and X. tropicalis frogs and eggs differ in size. Nuclei and meiotic spindles assembled de novo in egg extracts recapitulate differences in the sizes of these two subcellular structures. Nuclei assembled in interphase egg extract were stained for the nuclear pore complex (green). (From Levy and Heald 2010; reprinted, with permission, from the authors.) Spindles assembled in meiotic egg extract were visualized for tubulin (red), DNA (blue), and katanin (green). (From Loughlin et al. 2011; adapted, with permission, from Elsevier © 2011.) (B) Examples of Xenopus developmental organelle scaling are shown. Drawings of different stage X. laevis embryos are from data in Nieuwkoop and Faber (1967). Nuclei were isolated from Xenopus embryos arrested in late interphase with cycloheximide, to ensure complete karyomere fusion in early stage embryos. Nuclei were visualized as in A. Scale bar, 20 µm. (From Levy and Heald 2010; adapted, with permission, from Elsevier © 2010.) In vitro spindles were reconstituted in X. laevis egg or embryo extracts and visualized for tubulin (red) and DNA (blue). In vivo X. laevis spindles were imaged by tubulin immunofluorescence (gray) and shows tubulin (yellow) and DNA (red) staining. (All spindle images from Wilbur and Heald 2013; adapted and made available using a Creative Commons Attribution License, except for the stage 6 in vivo spindle image from Wühr et al. 2008; adapted, with permission, from Elsevier © 2008.) Scale bar, 10 µm. (C) Scaling of spindle length by limiting cytoplasmic volume was shown using a microfluidic encapsulation technique. Cytoplasm refers to X. laevis egg extract and DNA refers to demembranated X. laevis sperm. The boundaries of the in vitro assembled cell-like compartments consist of PEG30-PHS (polyhydroxystearate), and droplet size was tuned by varying dimensions and flow rates within the microfluidic devices. Spindles assembled in small droplets are smaller than in large droplets, mirroring spindle length scaling that occurs in X. laevis embryos during early development. Scale bar, 20 µm. (From Good et al. 2013; adapted, with permission, from the authors.) Similar results were reported in Hazel et al. (2013).
The size of the nucleus is intimately linked to that of the cell and genome across many different species, and this is certainly true for amphibians (Wilson 1925; Horner and Macgregor 1983; Gregory et al. 2007; Gregory 2011). Genome size and the degree of chromatin compaction likely define a lower limit for nuclear size. However, it is also clear that genome size is not the only determinant of nuclear size. Dramatic changes in nuclear size during embryonic development and cell differentiation are generally independent of altered ploidy (Butler et al. 2009; Edens et al. 2013). Different cell types within the same multicellular organism also show differences in nuclear size, although some of this variability may be attributable to cell-type-specific differences in the quantity of nuclear DNA, as discussed in Gillooly et al. (2015). Cancer cells also frequently show nuclear enlargement independent of large-scale changes in karyotype (Tapon et al. 2001; Cremer et al. 2003; Zink et al. 2004; Jevtic and Levy 2014; Jevtic et al. 2014). With respect to chromatin compaction, genome size expansion has been accompanied by evolutionary changes in histone 2A sequences that mediate increased compaction of larger genomes, potentially impacting nuclear size (Macadangdang et al. 2014).
If genome size sets a minimum to nuclear size, are there upper limits? There may be mechanical limits to nuclear size, for instance, large nuclei expanding in Xenopus egg extract will rupture (Levy and Heald 2010). Nuclei may be enlarged in cells requiring increased nuclear function. Cancer cells with large nuclei show increased metabolism, gene expression, and rates of cell proliferation. Whether these changes in nuclear function are a cause or consequence of altered nuclear size is an open question. Regulation of nuclear size might alter chromatin organization and gene expression, contributing to cell-type specification during development and cell differentiation, or impact the concentrations and diffusion rates of nuclear macromolecules, thereby affecting reaction and assembly kinetics. For example, altering nuclear size and the nucleocytoplasmic ratio in early Xenopus embryos affects the timing of the midblastula transition (Jevtic and Levy 2015).
What are the mechanisms that regulate nuclear size? Since the early 20th century, it was known that the nucleocytoplasmic ratio, the ratio of nuclear to cytoplasmic volumes, is relatively constant (Wilson 1925). Classic Xenopus studies supported the idea that cytoplasmic components regulate nuclear size. For instance, injection of isolated HeLa nuclei into X. laevis oocytes caused the nuclei to expand (Gurdon 1976). More recently, nuclear scaling between different-sized frog species was recapitulated using egg extracts from X. laevis and X. tropicalis (Levy and Heald 2010). Developmental nuclear scaling has also been studied in Xenopus, as nuclear size decreases during early embryogenesis (Gerhart 1980; Montag et al. 1988; Levy and Heald 2010). A simplified mechanism for nuclear scaling arising from these Xenopus studies is that limiting import through nuclear pores, perhaps of structural components, such as nuclear lamins, restricts nuclear growth. It seems reasonable that structural components of the nucleus determine nuclear size, and limiting component models for the regulation of nuclear size have been advanced (Goehring and Hyman 2012). However, more dynamic mechanisms may be at play, for instance, through balanced rates of assembly and disassembly (Marshall 2002, 2008; Edens and Levy 2014a). Furthermore, extranuclear structures and cell-cycle events also contribute to the regulation of nuclear size and morphology (Edens et al. 2013; Jevtic et al. 2014).
The mitotic spindle is another example of a dynamic macromolecular structure that scales to cell size. Mechanisms of spindle size regulation have been elucidated in a variety of different Xenopus systems, as discussed in detail in Mitchison et al. (2015) (Brown et al. 2007; Wühr et al. 2008; Loughlin et al. 2011; Good et al. 2013; Hazel et al. 2013; Wilbur and Heald 2013). One common mechanism is that microtubule depolymerization kinetics contribute to spindle length scaling. It has also become apparent that spindle architecture and assembly mechanisms differ between large and small spindles (Wilbur and Heald 2013). One factor contributing to these differences was recently identified as the spindle assembly factor TPX2, which is present at threefold higher levels in X. tropicalis egg extracts compared with X. laevis (Helmke and Heald 2014). Increasing TPX2 levels in X. laevis to match those in X. tropicalis not only reduced spindle length but also caused rearrangement of spindle microtubules, leading to a more X. tropicalis–like spindle morphology. Spindle size, morphology, and dynamics also vary in different cell types within the same species to facilitate chromosome segregation. For example, X. laevis neural epithelial cells show reduced microtubule density at the spindle midzone and more dramatic chromosome movements during anaphase compared with other cell types, and this is regulated by levels of the microtubule-binding protein PRC1 (Kieserman et al. 2008).
Studies of chromosome scaling during Xenopus development show that mitotic chromosome sizes do not scale with cell size in the early embryo, but become progressively smaller through the blastula and neurula stages (Micheli et al. 1993; Kieserman and Heald 2011). Compared with scaling of the nucleus and spindle by cytoplasmic factors, developmental mitotic chromosome scaling seems to be intrinsically determined by chromosome architecture and regulated expression of chromatin proteins, although the relevant scaling factors remain to be identified. Interestingly, whereas increasing nuclear size in egg extracts before mitosis did not affect mitotic chromosome size unless the DNA replicated (Kieserman and Heald 2011), inhibiting nuclear growth resulted in shorter, thicker mitotic chromosomes, suggesting that mitotic chromosome size may be modulated by increasing but not decreasing nuclear DNA density (Hara et al. 2013). Overall, Xenopus developmental scaling of mitotic chromosome size mirrors spindle scaling as cells become small enough that the distance that chromosomes can separate is constrained (Wühr et al. 2008; Field and Lenart 2011; Kieserman and Heald 2011; Levy and Heald 2012).
CONCLUDING REMARKS
Because of their wide-ranging sizes at the organismal, genome, cellular, and subcellular levels, amphibians have provided powerful approaches to study size relationships and the effects of large genome and cell sizes as well as animal miniaturization on physiology and development. Xenopus extracts remain the only system to recapitulate complex physiological processes in vitro and have facilitated the molecular identification of subcellular scaling factors. More generally, research in Xenopus has led to the discovery of fundamental principles and mechanisms of cell biology and development including key cell-cycle regulators and the developmental reprogramming that underlies animal cloning, as well as a variety of genes and mechanisms operating during embryogenesis and patterning (Gurdon et al. 1958; Harland and Grainger 2011). Many of the key findings relate to size and growth mechanisms, and frog systems are likely to continue to drive the field of biological size control. Other species of amphibians provide novel opportunities to investigate direct development and regeneration (Brockes 1997; Callery 2006; Roensch et al. 2013; Simon and Tanaka 2013). Stay tuned as unique amphibian systems continue to teach us valuable lessons about conserved mechanisms of biological size and scaling.
ACKNOWLEDGMENTS
D.L.L. is supported by R15GM106318. R.H. is supported by R01GM098766. We thank Ray Keller, David Wake, Karen White, Jeremy Wilbur, and Iswar Hariharan for comments on the manuscript, and Favian Hernandez for preparing the artwork in Figure 2.
Footnotes
Editors: Rebecca Heald, Iswar K. Hariharan, and David B. Wake
Additional Perspectives on Size Control in Biology: From Organelles to Organisms available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- *.Amodeo AA, Skotheim JM. 2015. Cell-size control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogart JP, Licht LE. 1986. Reproduction and the origin of polyploids in hybrid salamanders of the genus Ambystoma. Can J Genet Cytol 28: 605–617. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP. 2013. Phase transitions and size scaling of membrane-less organelles. J Cell Biol 203: 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes JP. 1997. Amphibian limb regeneration: Rebuilding a complex structure. Science 276: 81–87. [DOI] [PubMed] [Google Scholar]
- Brown KS, Blower MD, Maresca TJ, Grammer TC, Harland RM, Heald R. 2007. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J Cell Biol 176: 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki E. 1985. The expression of creatine kinase isozymes in Xenopus tropicalis, Xenopus laevis laevis, and their viable hybrid. Biochem Genet 23: 73–88. [DOI] [PubMed] [Google Scholar]
- Butler JT, Hall LL, Smith KP, Lawrence JB. 2009. Changing nuclear landscape and unique PML structures during early epigenetic transitions of human embryonic stem cells. J Cell Biochem 107: 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadart C, Zlotek-Zlotkiewicz E, Le Berre M, Piel M, Matthews HK. 2014. Exploring the function of cell shape and size during mitosis. Dev Cell 29: 159–169. [DOI] [PubMed] [Google Scholar]
- Callery EM. 2006. There’s more than one frog in the pond: A survey of the Amphibia and their contributions to developmental biology. Semin Cell Dev Biol 17: 80–92. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2005. Economy, speed and size matter: Evolutionary forces driving nuclear genome miniaturization and expansion. Ann Bot 95: 147–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RC, Forbes DI. 2006. In vitro study of nuclear assembly and nuclear import using Xenopus egg extracts. Methods Mol Biol 322: 289–300. [DOI] [PubMed] [Google Scholar]
- Chang JB, Ferrell JE Jr. 2013. Mitotic trigger waves and the spatial coordination of the Xenopus cell cycle. Nature 500: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo A, Keller R. 2010. Early development of Ensatina eschscholtzii: An amphibian with a large, yolky egg. Evodevo 1: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J. 1973. Morphogenesis and regulation in spite of continued mitotic inhibition in Xenopus embryos. Nature 242: 55–57. [DOI] [PubMed] [Google Scholar]
- Cooke J. 1981. Scale of body pattern adjusts to available cell number in amphibian embryos. Nature 290: 775–778. [DOI] [PubMed] [Google Scholar]
- Cremer M, Kupper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. 2003. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol 162: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deban SM, Olson WM. 2002. Suction feeding by a tiny predatory tadpole. Nature 420: 41–42. [DOI] [PubMed] [Google Scholar]
- Deparis P, Jaylet A. 1975. Studies on the origin of various blood cell lines in the salamander Pleurodeles waltlii (author’s translation). J Embryol Exp Morphol 33: 665–683. [PubMed] [Google Scholar]
- Deparis P, Beetschen JC, Jaylet A. 1975. Red blood cells and hemoglobin concentration in normal diploid and several types of polyploid salamanders. Comp Biochem Physiol A Comp Physiol 50: 263–266. [DOI] [PubMed] [Google Scholar]
- Duellman WE, Trueb L. 1986. Biology of amphibians. McGraw-Hill, New York. [Google Scholar]
- Edens LJ, Levy DL. 2014a. cPKC regulates interphase nuclear size during Xenopus development. J Cell Biol 206: 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens LJ, Levy DL. 2014b. Size scaling of subcellular organelles and structures in Xenopus laevis and Xenopus tropicalis. In Xenopus development (ed. Kloc M, Kubiak JZ), pp. 325–345. Wiley, Hoboken, NJ. [Google Scholar]
- Edens LJ, White KH, Jevtic P, Li X, Levy DL. 2013. Nuclear size regulation: From single cells to development and disease. Trends Cell Biol 23: 151–159. [DOI] [PubMed] [Google Scholar]
- Emerson SB. 1988. The giant tadpole of Pseudis paradoxa. Biol J Linn Soc Lond 34: 93–104. [Google Scholar]
- Faivre-Moskalenko C, Dogterom M. 2002. Dynamics of microtubule asters in microfabricated chambers: The role of catastrophes. Proc Natl Acad Sci 99: 16788–16793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser G. 1939. Polyploidy in the salamander, Eurycea bislineata. J Hered 30: 379–388. [Google Scholar]
- Fankhauser G. 1945a. Maintenance of normal structure in heteroploid salamander larvae, through compensation of changes in cell size by adjustment of cell number and cell shape. J Exp Zool 100: 445–455. [DOI] [PubMed] [Google Scholar]
- Fankhauser G. 1945b. The effects of changes in chromosome number on amphibian development. Q Rev Biol 20: 20–78. [Google Scholar]
- Fankhauser G, Watson RC. 1942. Heat-induced triploidy in the newt, Triturus viridescens. Proc Natl Acad Sci 28: 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Brangwynne CP. 2013. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat Cell Biol 15: 1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Lenart P. 2011. Bulk cytoplasmic actin and its functions in meiosis and mitosis. Curr Biol 21: R825–R830. [DOI] [PubMed] [Google Scholar]
- Fox CB, Hutchinson P. 1991. Fishes and amphibians from the late Permian Pedra de Fogo formation of northern Brazil. Palaeontology 34: 561–573. [Google Scholar]
- Frost D. 2014. Amphibian species of the world: An online reference. Version 6.0. Electronic database accessible at research.amnh.org/herpetology/amphibia/index.html American Museum of Natural History, New York. [Google Scholar]
- Gagnon JA, Kreiling JA, Powrie EA, Wood TR, Mowry KL. 2013. Directional transport is mediated by a dynein-dependent step in an RNA localization pathway. PLoS Biol 11: e1001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans C. 1956. Frogs and paradoxes. Animaland 23: 1–4. [Google Scholar]
- George SA, Lennartz MR. 1980. Methods for determining ploidy in amphibians: Nucleolar number and erythrocyte size. Experientia 36: 687–688. [Google Scholar]
- Gerhart JC. 1980. Mechanisms regulating pattern formation in the amphibian egg and early embryo. In Biological regulation and development (ed. Goldberger RF), pp. 133–316. Plenum, New York. [Google Scholar]
- *.Gillooly JF, Hein A, Damiani R. 2015. Nuclear DNA content varies with cell size across human cell types. Cold Spring Harb Perspect Biol 7: a019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Hyman AA. 2012. Organelle growth control through limiting pools of cytoplasmic components. Curr Biol 22: R330–R339. [DOI] [PubMed] [Google Scholar]
- Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. 2013. Cytoplasmic volume modulates spindle size during embryogenesis. Science 342: 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. 2001a. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol Rev Camb Philos Soc 76: 65–101. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2001b. The bigger the C-value, the larger the cell: Genome size and red blood cell size in vertebrates. Blood Cells Mol Dis 27: 830–843. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2003. Variation across amphibian species in the size of the nuclear genome supports a pluralistic, hierarchical approach to the C-value enigma. Biol J Linn Soc Lond 79: 329–339. [Google Scholar]
- Gregory TR. 2011. Cell size database. www.genomesize.com/cellsize. [Google Scholar]
- Gregory TR. 2014. Animal genome size database. www.genomesize.com. [Google Scholar]
- Gregory TR, Nicol JA, Tamm H, Kullman B, Kullman K, Leitch IJ, Murray BG, Kapraun DF, Greilhuber J, Bennett MD. 2007. Eukaryotic genome size databases. Nucleic Acids Res 35: D332–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB. 1976. Injected nuclei in frog oocytes: Fate, enlargement, and chromatin dispersal. J Embryol Exp Morphol 36: 523–540. [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischberg M. 1958. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 182: 64–65. [DOI] [PubMed] [Google Scholar]
- Hanken J. 1983. Miniaturization and its effects on cranial morphology in plethodontid salamanders, genus Thorius (Amphibia, Plethodontidae). II: The fate of the brain and sense organs and their role in skull morphogenesis and evolution. J Morphol 177: 255–268. [DOI] [PubMed] [Google Scholar]
- Hanken J, Thorogood P. 1993. Evolution and development of the vertebrate skull: The role of pattern formation. Trends Ecol Evol 8: 9–15. [DOI] [PubMed] [Google Scholar]
- Hara Y, Iwabuchi M, Ohsumi K, Kimura A. 2013. Intranuclear DNA density affects chromosome condensation in metazoans. Mol Biol Cell 24: 2442–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hariharan IK, Wake DB, Wake MH. 2015. Indeterminate growth: Could it represent the ancestral condition? Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM, Grainger RM. 2011. Xenopus research: Metamorphosed by genetics and genomics. Trends Genet 27: 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman FA, Lessler MA. 1964. Erythrocyte measurements in fishes, amphibia, and reptiles. Biol Bull 126: 83–88. [Google Scholar]
- Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. 2013. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science 342: 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB. 2008. At the lower size limit in snakes: Two new species of threadsnakes (Squamata: Leptotyphlopidae: Leptotyphlops) from the Lesser Antilles. Zootaxa 1841: 1–30. [Google Scholar]
- Helmke KJ, Heald R. 2014. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J Cell Biol 206: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokkanen JE. 1986. The size of the largest land animal. J Theor Biol 118: 491–499. [DOI] [PubMed] [Google Scholar]
- Hoopes BC, Rimbault M, Liebers D, Ostrander EA, Sutter NB. 2012. The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm Genome 23: 780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner HA, Macgregor HC. 1983. C value and cell volume: Their significance in the evolution and development of amphibians. J Cell Sci 63: 135–146. [DOI] [PubMed] [Google Scholar]
- Huang H, Brown DD. 2000. Overexpression of Xenopus laevis growth hormone stimulates growth of tadpoles and frogs. Proc Natl Acad Sci 97: 190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtic P, Levy DL. 2014. Mechanisms of nuclear size regulation in model systems and cancer. Adv Exp Med Biol 773: 537–569. [DOI] [PubMed] [Google Scholar]
- Jevtic P, Levy DL. 2015. Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Curr Biol 25: 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtic P, Edens LJ, Vukovic LD, Levy DL. 2014. Sizing and shaping the nucleus: Mechanisms and significance. Curr Opin Cell Biol 28C: 16–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgartel C, Schekman R, Rape M. 2012. Ubiquitin-dependent regulation of COPII coat size and function. Nature 482: 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Steen JA, Steen H, Kirschner MW. 2009. The mechanism and pattern of yolk consumption provide insight into embryonic nutrition in Xenopus. Development 136: 1539–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieserman EK, Heald R. 2011. Mitotic chromosome size scaling in Xenopus. Cell Cycle 10: 3863–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieserman EK, Glotzer M, Wallingford JB. 2008. Developmental regulation of central spindle assembly and cytokinesis during vertebrate embryogenesis. Curr Biol 18: 116–123. [DOI] [PubMed] [Google Scholar]
- Kozlowski J, Czarnoleski M, Francois-Krassowska A, Maciak S, Pis T. 2010. Cell size is positively correlated between different tissues in passerine birds and amphibians, but not necessarily in mammals. Biol Lett 6: 792–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudet V. 2011. The origins and evolution of vertebrate metamorphosis. Curr Biol 21: R726–R737. [DOI] [PubMed] [Google Scholar]
- Levy DL, Heald R. 2010. Nuclear size is regulated by importin α and Ntf2 in Xenopus. Cell 143: 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R. 2012. Mechanisms of intracellular scaling. Annu Rev Cell Dev Biol 28: 113–135. [DOI] [PubMed] [Google Scholar]
- Loughlin R, Wilbur JD, McNally FJ, Nedelec FJ, Heald R. 2011. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 147: 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable BK, Alexandrou MA, Taylor M.I. 2011. Genome duplication in amphibians and fish: An extended synthesis. J Zool 284: 151–182. [Google Scholar]
- Macadangdang BR, Oberai A, Spektor T, Campos OA, Sheng F, Carey MF, Vogelauer M, Kurdistani SK. 2014. Evolution of histone 2A for chromatin compaction in eukaryotes. eLife 3: e02792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony MJ, Robinson ES. 1980. Polyploidy in the Australian leptodactylid frog genus Neobatrachus. Chromosoma 81: 199–212. [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Heald R. 2006. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol Biol 322: 459–474. [DOI] [PubMed] [Google Scholar]
- Marracci S, Michelotti V, Casola C, Giacoma C, Ragghianti M. 2011. Daz- and Pumilio-like genes are asymmetrically localized in Pelophylax (Rana) oocytes and are expressed during early spermatogenesis. J Exp Zool B Mol Dev Evol 316: 330–338. [DOI] [PubMed] [Google Scholar]
- Marshall W. 2002. Size control in dynamic organelles. Trends Cell Biol 12: 414–419. [DOI] [PubMed] [Google Scholar]
- Marshall WF. 2008. Engineering design principles for organelle size control systems. Semin Cell Dev Biol 19: 520–524. [DOI] [PubMed] [Google Scholar]
- *.Marshall WF. 2015. Subcellular size. Cold Spring Harb Perspect Biol 7: a019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Young KD, Swaffer M, Wood E, Nurse P, Kimura A, Frankel J, Wallingford J, Walbot V, Qu X, et al. 2012. What determines cell size? BMC Biol 10: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli G, Luzzatto AR, Carri MT, de Capoa A, Pelliccia F. 1993. Chromosome length and DNA loop size during early embryonic development of Xenopus laevis. Chromosoma 102: 478–483. [DOI] [PubMed] [Google Scholar]
- *.Mitchison TJ, Ishihara K, Ngyuen P, Wühr M. 2015. Size scaling of microtubule assemblies in early Xenopus embryos. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag M, Spring H, Trendelenburg MF. 1988. Structural analysis of the mitotic cycle in pre-gastrula Xenopus embryos. Chromosoma 96: 187–196. [DOI] [PubMed] [Google Scholar]
- *.Mueller RL. 2015. Genome biology and the evolution of cell-size diversity. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RL, Gregory TR, Gregory SM, Hsieh A, Boore JL. 2008. Genome size, cell size, and the evolution of enucleated erythrocytes in attenuate salamanders. Zoology (Jena) 111: 218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne P, Simpson DE, Gurdon JB. 2011. Deficient induction response in a Xenopus nucleocytoplasmic hybrid. PLoS Biol 9: e1001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne P, Halley-Stott RP, Gurdon JB. 2012. On the cellular and developmental lethality of a Xenopus nucleocytoplasmic hybrid. Commun Integr Biol 5: 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. 1967. Normal table of Xenopus laevis (Daudin). North-Holland, Amsterdam. [Google Scholar]
- Pollack ED, Koves J. 1977. Compensatory responses in the development of the brachial lateral motor column in triploid Rana pipiens. Anat Rec 188: 173–179. [DOI] [PubMed] [Google Scholar]
- *.Reber S, Goehring NW. 2015. Intracellular scaling mechanisms. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittmeyer EN, Allison A, Grundler MC, Thompson DK, Austin CC. 2012. Ecological guild evolution and the discovery of the world’s smallest vertebrate. PLoS ONE 7: e29797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roensch K, Tazaki A, Chara O, Tanaka EM. 2013. Progressive specification rather than intercalation of segments during limb regeneration. Science 342: 1375–1379. [DOI] [PubMed] [Google Scholar]
- *.Roth G, Walkowiak W. 2015. The influence of genome and cell size on brain morphology in amphibians. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safranyos RG, Ellis CG, Tyml K, Groom AC. 1983. Heterogeneity of capillary diameters in skeletal muscle of the frog. Microvasc Res 26: 151–156. [DOI] [PubMed] [Google Scholar]
- Sessions SK, Larson A. 1987. Developmental correlates of genome size in plethodontid salamanders and their implication for genome evolution. Evolution 41: 1239–1251. [DOI] [PubMed] [Google Scholar]
- Simon A, Tanaka EM. 2013. Limb regeneration. Wiley Interdiscip Rev Dev Biol 2: 291–300. [DOI] [PubMed] [Google Scholar]
- Snyder GK, Sheafor BA. 1999. Red blood cells: Centerpiece in the evolution of the vertebrate circulatory system. Amer Zool 39: 189–198. [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, et al. 2007. A single IGF1 allele is a major determinant of small size in dogs. Science 316: 112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105: 345–355. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE, Anatskaya OV. 2006. Genome size and metabolic intensity in tetrapods: A tale of two lines. Proc Biol Sci 273: 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake MH. 1980. The reproductive biology of Nectophrynoides malcolmi (Amphibia: Bufonidae), with comments on the evolution of reproductive modes in the genus Nectophrynoides. Copeia 1980: 193–209. [Google Scholar]
- Wake MH. 1993. Evolution of oviductal gestation in amphibians. J Exp Zool 266: 394–413. [Google Scholar]
- Wilbur JD, Heald R. 2013. Mitotic spindle scaling during Xenopus development by kif2a and importin α. eLife 2: e00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EB. 1925. The karyoplasmic ratio. In The cell in development and heredity, pp. 727–733. Macmillan, New York. [Google Scholar]
- Winklbauer R, Hausen P. 1985. Development of the lateral line system in Xenopus laevis. III: Development of the supraorbital system in triploid embryos and larvae. J Embryol Exp Morphol 88: 183–192. [PubMed] [Google Scholar]
- Wühr M, Chen Y, Dumont S, Groen AC, Needleman DJ, Salic A, Mitchison TJ. 2008. Evidence for an upper limit to mitotic spindle length. Curr Biol 18: 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimkus BM, Lawson L, Loader SP, Hanken J. 2012. Terrestrialization, miniaturization and rates of diversification in African puddle frogs (Anura: Phrynobatrachidae). PLoS ONE 7: e35118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. 2004. Nuclear structure in cancer cells. Nat Rev Cancer 4: 677–687. [DOI] [PubMed] [Google Scholar]