SUMMARY
Epigenomics has grown exponentially, providing a better understanding of the mechanistic aspects of new and old phenomena originally described through genetics, as well as providing unexpected insights into the way chromatin modulates the genomic information. In this overview, some of the advances are selected for discussion and comment under six topics: (1) histone modifications, (2) weak interactions, (3) interplay with external inputs, (4) the role of RNA molecules, (5) chromatin folding and architecture, and, finally, (6) a view of the essential role of chromatin transactions in regulating the access to genomic DNA.
Without distinct histone modifications, all nucleosomes would be more or less structurally identical. These modifications are essential for determining whether, when, and how the underlying DNA sequence is read.
Overview
The size and scope of this new Cold Spring Harbor Laboratory Press Epigenetics collection gives a tangible sense of the vast expansion of the field in the 7 years since the first edition. Not all of it goes under the tag of epigenetic in the strict sense of involving long-lasting, heritable responses in the gene-expression machinery. Epigenomics has revealed that chromatin is not just a way to package genomic DNA—it is a structure that provides the genome with a large range of variables that can be exploited to tune and articulate the function of the underlying DNA. Thus, in chromatin a set of complex interactions intervenes between the genetic material and the transcriptional readout.
The expansion has been both qualitative and quantitative, driven by the emergence of technologies for genome-wide analysis: first with the broad availability of genomic tiling microarrays and then of the next-generation parallel-sequencing technologies. We can now view the distribution of chromatin proteins, nucleosomes, histone modifications, and transcriptional activity, not just at an individual site but over the entire genome. It is true that the results are generally averages over a large number of cells, but, in some cases, the analysis of chromatin activities in single cells has become possible. Massively parallel sequencing has also made possible modes of analysis that were previously unthinkable. We can evaluate how often a given sequence is represented in the transcriptional output of the genome, and we can determine how much time a given genomic sequence spends in the vicinity of any other sequence using one of the currently available chromatin conformation capture (3C) approaches.
The broad availability of high-throughput technologies applied to the analysis of genomic structures and their transactions has sent colossal volumes of data pouring into computer storage. The DNA sequence data stored at the European Bioinformatics Institute has grown from a few terabases in 2008 to >200 terabases in 2012. The genomic data now exceeds 2 petabytes and is set to double in a year’s time (Marx 2013). What has this deluge of information produced? A word of caution first: The vast mass of chromatin-related data is highly heterogeneous in format, structure, degree of documentation, ease of access, and, above all, I suspect, quality. Given the vertiginous pace of research and publications, there are likely many incorrect results scattered in the literature owing to unexamined assumptions, inappropriate applications, insufficient controls, and hasty conclusions. Antibodies, an indispensable resource, are both extraordinary enablers and treacherous tools. Most researchers are aware of the need for rigorous testing for specificity. But criteria that are sufficient for one application are not necessarily adequate for another. This is particularly true when the antibodies are used for chromatin immunoprecipitation. The sheer volume of data means that the resulting inconsistencies may often go unnoticed for a long time because investigators analyze their results primarily with respect to the specific concerns that prompted their experiments. Although sophisticated bioinformatics analysis has been applied, in some cases, to extract features and patterns out of the data, the majority of the information is far from fully used and will likely remain so. New technological advances will probably mean that new data sets will supersede those of today in the databases of the future.
In attempting to assess the advances brought on by this vast accumulation of data, I have highlighted a few developments that give food for thought, and I end with the synthesis of a global chromatin accessibility hypothesis. Topics discussed include the following.
Histone modifications, the chromatin complexes that write, read, and erase them, and the interplay between intra- and internucleosomal interactions can produce long-lasting chromatin states.
Weak interactions along the epigenome provide an opportunistic or searching mode by which chromatin associations can occur.
A context is provided for chromatin regulation—the interplay with external inputs.
The pervasive role of RNA molecules is discussed: The dark energy of the nucleus is beginning to be not so dark.
The importance of chromatin folding and nuclear localization is discussed.
Finally, I will spend several pages discussing what seems to me to be the antithesis that underlies so many aspects of chromatin transactions: the conflict between the need to prevent and the imperative of allowing access to the underlying DNA.
These topics do not, by any means, exhaust what can be said about progress in chromatin research and epigenomics. For that, a whole collection would be needed. And, indeed, a whole collection is what you have in your hands.
1. Contextualizing some advances
1.1. Chromatin
1.1.1. Histone Posttranslational Modifications: Instructors or Bystanders?
The effort to catalog, classify, and interpret the significance of the histone modifications, chromatin complexes, and individual factors with which chromatin is thickly encrusted has fascinated a large number of researchers and led to a new era in molecular genetics. The task now is to understand their role in modulating the multiple activities that operate on the genomic material.
Many questions have been raised about the relationship between the molecular machinery that manipulates chromatin and histone modifications that are associated with the various kinds of chromatin transactions. Do these modifications carry information that extends the genetically encoded information? Do they instruct activities or simply accompany or facilitate these activities? Are they self-maintaining or do they fade more or less rapidly after the events with which they are associated? In essence, are they causes or consequences of mechanisms that act on the underlying DNA (Henikoff and Shilatifard 2011)? There is probably no single answer; each modification may play somewhat different roles depending on the context.
If histone modifications carry information, it is clearly not the same kind as genetic information. Unlike sequence-based information, this “epigenetic” information is not meant to be stable over the long term. Chromatin marks such as histone or DNA methylation are the products of activities operating on the chromatin. They carry information about the activities that have resulted in the placement of the methylation marks, but also affect subsequent activities. Many characteristic epigenetic states such as those found in heterochromatic regions, Polycomb-repressed regions, or genes silenced by DNA methylation are reset during the course of differentiation. It came, nevertheless, as something of a shock to many researchers to discover that there existed demethylases that could remove even the most treasured marks of epigenetic repression, histone H3K9 methylation, associated with heterochromatin as well as histone H3K27 methylation associated with Polycomb silencing. This was just unfolding at the time of the first edition of this collection. Now, even cytosine methylation on the DNA has a means of being actively erased via the Tet enzymes (Kohli and Zhang 2013). The consequent implication is that epigenetic silencing is not forever; it could be and is constantly tinkered with both at the level of the histone marks and complexes that write and read them. This does not mean that epigenetic states cannot be maintained for long periods, through many rounds of cell division and, in some cases, transgenerationally. It means, however, that an epigenetic state cannot simply be equated to the presence of a particular mark. The mechanisms that put chromatin marks in place, maintain them, and respond to them are more complex and dynamic than had previously been thought.
The fact that histone modifications can endure through multiple cell cycles means that they can serve as a mark of the antecedent events that produced them and, therefore, influence the activities of a chromatin region as a function of its history. We now know that various complexes that interact with chromatin (chromatin complexes) include structural domains that recognize specific histone marks (commonly referred to as readers) and that these interactions govern the function of many molecular machines that operate on chromatin. In some cases, the presence of a histone mark is recognized by the same chromatin complex that deposits that mark and this can stimulate further deposition of the same mark on neighboring nucleosomes. This feed-forward effect means that histone marks such as H3K9 and H3K27 methylation can promote and help to maintain their own presence, thereby explaining their “epigenetic” persistence from one cell cycle to the next.
In the case of H3K9 methylation, the protein that binds to the H3K9me3 mark, HP1, is associated with the H3K9 methyltransferase, together helping to maintain the histone modification and spread heterochromatin. The complex containing both partners not only recognizes H3K9 methylation in surrounding chromatin but is also stimulated by its presence (Al-Sady et al. 2013). Interestingly, heterochromatin-associated H3K9 trimethylation appears in some circumstances to be able to maintain itself once it is generated de novo. For instance, once a domain of H3K9 trimethylation was generated at a GAL4 binding site, through the expression of a transgenic HP1 fused with a GAL4 DNA-binding domain (GAL4-HP1), the methylated and repressed state were then maintained even after the GAL4-HP1 was no longer produced (Hathaway et al. 2012). This implies that H3K9 methylation set by GAL4-HP1 is sufficient to recruit endogenous HP1 and ensures further methylation, a key feature of Epigenetics. Thus, HP1 binding is tightly linked to methylation activity in heterochromatin formation, more so than the corresponding activities of Polycomb complexes PRC1 and PRC2; a Polycomb-repressed domain cannot maintain itself, at least in Drosophila, when the Polycomb-response element (PRE) is deleted (Busturia et al. 1997). The extensive H3K27 trimethylation and H2A ubiquitination associated with it are important contributors, but not sufficient to continue recruiting the Polycomb complexes. The long-term persistence of the Polycomb repressed state requires a recruiting element, in addition to the histone modifications, and the absence of strong antagonizing activities (see, however, Secs. 1.2 and 2.5).
A surprise of the last few years has been the discovery that many nonhistone chromatin proteins are targets of posttranslational modifications—in some cases, placed by the same chromatin factors that modify histones. For instance, p53 is acetylated by CBP; it is also methylated by SMYD at K370 to prevent its binding to DNA or methyltransferase SET7/9 at K372, which prevents methylation at K370 in response to DNA damage (Ivanov et al. 2007). Do such modifications convey information? Of course they do; they switch the function of the target protein. Another surprising case is retinoic acid receptor α, which can be methylated by EZH2, targeting it for degradation by the proteasome. EZH2 acts, in this case, without the participation of the other components of the PRC2 complex that are normally required for methyltransferase activity (Lee et al. 2012). These effects remind us that some of the well-known chromatin factors can have additional effects that are entirely independent of chromatin, including roles outside of the nucleus (Su et al. 2005).
1.1.2. Nucleosome Marks and Chromatin Differentiation
Nucleosomes can carry a bewildering number of histone modifications simultaneously. Unfortunately, there is no evidence that histone marks have simple instructive combinatorial properties. In general, chromatin complexes distinguish nucleosomes by the presence or absence of one particular histone mark, and each mark has its proper interacting protein or “reader.” There are some very interesting exceptions. For example, phosphorylation of H3S10 or H3S28 prevents the binding of chromodomains to adjacent methylated H3K9 or H3K27, respectively, and may constitute a toggle or binary switch to rapidly neutralize the role of these methylations. Another example of the combinatorial action of histone marks is the way certain “reader” proteins require a specific combination of marks with the right spacing and position. Double acetylation marks recognized by proteins containing two bromodomains provide such an example. Another interesting example is the reading of multiple histone marks by the DNA repair protein 53BP, which binds to nucleosomes containing both H4K20me1/2 and H2AK15ub marks through a combination of its Tudor domain and a carboxy-terminal ubiquitin-dependent recruitment domain (Fradet-Turcotte et al. 2013). The question these examples raise is do they represent ways to extend the repertoire of “meaningful” marks on a nucleosome? Or are they ways to integrate multiple inputs that control a chromatin process?
We tend to focus on the functional consequences that a nucleosome and interacting protein reader can have. Ultimately, however, what a given stretch of chromatin can do also depends on how it folds locally to allow interactions between the nucleosomes, and proteins bound to them and between trans-acting factors and the DNA. This appears to be the role of the HMG (high-mobility group) proteins, small proteins with a highly negatively charged carboxy-terminal tail. HMG proteins are abundant components of chromatin that are generally thought to have accessory “architectural” roles, binding to DNA and helping it to fold in particular structures important for the function of chromatin regulators. But, HMG proteins are also targets of posttranslational modifications including acetylation, methylation, phosphorylation, and probably others. Is there a whole new world of chromatin marks hidden there? In the case of HMGA1, for example, acetylation at K64 by CBP is said to destabilize the interferon-β enhanceosome, leading to transcriptional turnoff, whereas acetylation of K70 by PCAF/GCN5 (CBP-associated factor) has the opposite effect (see Zhang and Wang 2010). Furthermore, HMG proteins, with or without posttranslational modifications, could have an important role in modulating modifications of the histones. If so, the complexities of chromatin modifications through posttranslational modifications are enormously multiplied. So far, the differential distribution of HMGs and their modifications in the genome, and the relationship between the histone modifications involved in HMG association and resultant gene activity, have not been studied systematically.
1.2. The Power of Weak Interactions and Their Role in Chromatin Scanning
Classically, strong interactions between DNA-binding proteins and their cognate DNA are viewed as the efficient way to target and regulate specific genomic sites predictably, given sufficient nuclear concentrations. However, efficient use of specificity for genome regulation requires a strategy that relies on weak interactions. Weak interactions do not imply lack of interactions; on the contrary, potential binding sites are frequently visited (or perhaps “sampled” is a better word). But, in the absence of other events that might stabilize the interaction, the binding is weak and transient. This kind of interaction is typical of a specific DNA-binding protein with nonspecific DNA sequences and, indeed, constitutes the searching phase during which the protein spends much time weakly bound to nonspecific sequences. When the protein finds a consensus or near-consensus sequence, the interaction then becomes stronger and takes longer to dissociate. These dynamics were recognized a long time ago by Peter von Hippel (von Hippel et al. 1974), who showed that weak interactions reduce the three-dimensional volume that a protein needs to explore and find its specific binding sites.
The binding of chromatin complexes to chromatin is, in general, of the weak and transient variety because interactions with histones or histone modifications are, at best, in the micromolar range of affinities. However, in the nucleus, all nucleosomes, except for those in highly condensed regions, are, in principle, accessible for interaction. In most cases, such transient interactions do not necessarily leave a trace detectable by chromatin immunoprecipitation (ChIP) or similar approaches unless the complex has an enzymatic activity such as a methyltransferase or deacetylase activity, which can mark the genome with a change in histone modifications. But when a factor or complex reaches an environment in which additional interactions are possible, such as binding to other histone marks, interactions with a specific DNA-binding protein, or cooperative interactions with other factors, the interactions stabilize a longer residence time and we call that binding to a specific site.
Weak binding is essential for all regulated processes and is enshrined in the concept of mass-action equilibria. Any molecule in the nucleus can potentially interact with any other. We tend to lose track of this fact when we think, for example, that RNA polymerase binds only at promoters. It also binds, with greater or lesser affinity, to any other accessible site. If it binds long enough or frequently enough, it will produce some kind of transcript (see the argument in Sec. 2). Thus, when a chromatin region becomes accessible, this can produce unwanted transcripts. Various mechanisms are available to control this, from exosome surveillance of the transcripts, to H3K36 methylation and recruitment of repressive complexes in regions that become too accessible because of active transcription (Li et al. 2007).
In addition to strong protein binding, cooperative binding, or binding via “readers” of histone marks, an important emerging issue is that relatively weak or transient interactions produce an opportunistic surveillance strategy for targeting chromatin complexes. A variant PRC1 complex illustrates the point: We know it binds specifically via the CXXC zinc finger motif of its KDM2B component to mediate repression at a subset of unmethylated CpG (cytosine and guanine separated by a phosphate) island target genes, rather than via chromobox homolog protein (CBX), the more conventional PRC1 component that reads H3K27me3 marks. But, interestingly, Farcas et al. (2012) also detected a much weaker presence of the complex at most CpG islands. Klose et al. (2013) argue that this is part of an opportunistic strategy of recruitment through weak interactions at all such sites, rather than targeting to a specific subset of sites. At any given site, if any cooperating factors are present, they can stabilize encounters for which weak interactions have created the opportunity. I will argue in the second half of this essay that such genome-wide scanning activities may be an underlying prerequisite for many basic chromatin functions.
1.3. The Interplay of Epigenetic Mechanisms and the Environment
We have seen that histone modifications alone cannot be responsible for long-lived epigenetic states. These modifications can come and go and are, in many cases, the target of activities that remove them. It is rather the whole mechanism with which they are associated that can generate long-lived states. We are beginning to get glimpses of the fact that epigenetic mechanisms are far more complex and likely to involve the interplay of the genome, physiological state of the organism, nervous system, and other environmental input.
One of the most remarkable stories to emerge in the past decade is the role that chromatin modifications play in extending the effects of early newborn experiences in rats to later functions in the adult via epigenetic programming in the hypothalamus. Specifically, Weaver, Szyf, and collaborators (Weaver et al. 2004, 2005; McGowan et al. 2009) found that maternal licking behavior is important to reduce alarm in rat pups. Inadequate maternal licking resulted in stressed and insecure rats that grew up to be stressed, insecure, and aggressive. This was traced to chromatin alterations in the genes associated with glucocorticoid response, a major stress response pathway in rats and man. Thus, early experience caused long-lasting phenotypes (epigenetic changes) affecting the expression of a pathway important for stress behavior. How the chromatin changes resulting from early stress are maintained over a period of years in the lifetime of an individual is not entirely clear. This pathway also has a remarkable transgenerational effect: The stressed and aggressive adult female behavior, resulting from the epigenomic programming occurring after birth as a consequence of negligent maternal nurturing, propagates to the next generation through their inadequate maternal licking behavior to their offspring. This stress syndrome is thus transmitted over multiple generations by a combination of epigenetic memory, behavior, and what might be called cultural inheritance. It is not hard to see how this interplay might be relevant to a variety of behavioral patterns such as autism spectrum disorders (see, e.g., LaSalle 2013). This is just a taste of what research on the interplay between behavior and chromatin modifications in the central nervous system is likely to reveal in many other aspects of behavior. And, in principle, a similar interplay can be expected between chromatin modifications and other inputs such as metabolic, nutritional, and physiological states, environmental conditions, and chemical or immunological challenges.
1.4. The Pervasive Roles of Nuclear RNA Molecules
Another major theme in the past decade of research is the new-found ascendancy of RNA molecules. RNAs are far from the subservient intermediary between genomic DNA and proteins envisioned by Francis Crick’s central dogma. Deep sequencing of the transcriptome has revealed virtually ubiquitous transcription. Far from being mere genomic deserts, many intergenic regions are actively transcribed to produce RNA molecules that lack appreciable protein-coding potential (noncoding RNAs or ncRNAs) and whose functions are still, for the most part, unknown. Indeed, the expansion in the field has led to the emergence of a variety of new names to refer to the different types of ncRNAs with different functions (see Fig. 26 of Allis et al. 2014).
I will not attempt to summarize the well-known roles of small RNAs in directing cleavage of RNA molecules, modulating translation, heterochromatin formation, and transcriptional silencing. I will focus, instead, on the role of long noncoding RNAs (lncRNAs) that has generated much excitement in the epigenetic field. Some lncRNAs have been found to be critical components of chromatin complexes that regulate chromatin states and, therefore, transcription itself. These discoveries have now led to the surmise that many, if not all, lncRNAs have similar roles.
Enough is known at this time to show that the role of lncRNAs such as Xist, Tsix, HOTAIR, Air, and enhancer RNAs (eRNAs) described in this collection are varied. Some act in cis, perhaps by being captured as they are being transcribed, and are important in X chromosome inactivation, allele-specific regulation, and imprinting. Others act in trans. They may recognize DNA sequence motifs in some way, possibly by forming DNA–RNA hybrids, and serve a targeting role for RNA-binding proteins. Or they may act as scaffolds to assemble multiple chromatin-modifying complexes to activate or silence target genes. Overall, they seem to be highly versatile and powerful components of the nuclear arsenal of tools that regulate transcriptional activity, even though they themselves are the direct products of transcription.
Are all genomic sequences, thus, “functional” in some way? Although it is true that some ncRNAs are potentially involved in important chromatin processes, this has not been shown for the vast majority. Ubiquitous ncRNA functionality is not supported by the fact that many sequences are not evolutionarily conserved. However, sequence conservation is not necessarily required—often it is the transcriptional activity itself that is important. In other cases, the functional properties of an ncRNA depend on very short motifs and secondary structure rather than on extensive sequence conservation. Recent work has shown that sequence specificity is not involved in the binding of the PRC2 complex to lncRNAs such as HOTAIR (Davidovich et al. 2013; Kaneko et al. 2013). Rather, PRC2 has high nonspecific, but size-dependent, affinities for all RNA including nascent transcripts. This, in turn, implies that we need a different way of thinking about the role of such lncRNAs and suggests that the local concentration of RNA molecules is important in controlling the availability of complexes such as PRC2.
A particularly surprising role of RNA molecules and an application of the concept of small RNAs as guides for sequence-specific targeting of chromatin complexes is the clustered regularly interspaced short palindromic repeats (CRISPR)-mediated mechanism recently discovered in bacteria (see the review by Brouns 2012) and now increasingly applied to eukaryotic genomic engineering. The CRISPR system stores snippets of DNA sequence from previous invading DNA in a context that can produce corresponding short CRISPR RNAs (scRNAs). These act as guides for CRISPR-associated (Cas) proteins to target and cleave homologous DNA. Although widespread in bacteria, where they are used primarily for host defense, CRISPR-like mechanisms have not yet been discovered in mammals. However, the ease and versatility of the CRISPR targeting mechanism has attracted many researchers seeking ways to direct sequence-specific activities of all sorts to a desired genomic target. Modifications or chimeric fusions of a Cas protein can be used to direct, not only nucleolytic cleavage, but transcriptional activation, repression, chromatin modifications, fluorescent tags, or any other desired activity, including just plain binding, to a specific genomic sequence using an appropriate short scRNA. Although the major application of the CRISPR/Cas technique so far has been to edit (insert or delete) sequences into the genome, its versatility is just beginning to be exploited. It would not be surprising if this powerful technique will find varieties of additional uses in combination with the rich armamentarium of micro RNA- and RNA interference (RNAi)-related mechanisms. Potentially, even more powerful, given the ability to store a true memory of earlier DNA-based events, is the mechanism by which new CRISPR spacer sequences are acquired from foreign DNA. Little is known about the molecular details of this process, but could it be harnessed to “teach” a genome?
1.5. Getting Around in the Nucleus
Another important development in the past 10 years has been the growing awareness that larger-scale chromatin domains and nuclear localization play crucial roles in the regulation of the genome. The nucleus is not just an amorphous bag of chromatin: There are chromosomes, centromeres, and telomeres; ribosomal RNA genes assemble a nucleolus; and certain chromatin regions are more likely to be in contact with the nuclear envelope.
Through numerous genome-wide and locus-specific studies, mostly using 3C approaches (explained in Gibcus and Dekker 2013; Dekker and Misteli 2014), we have seen now that different parts of the genome tend to associate with one another in long-range interactions on different scales of distance along the chromatin fiber. There are local interactions that form loops allowing enhancers to contact one another and their cognate promoters on a scale of tens of kilobases. The function of these interactions seems evident: to bring different regulatory regions controlling one or a few genes into contact with the corresponding promoters. Larger chromatin domains, of the order of a few megabases, contain many genes among which there is much internal looping. The function of this kind of interaction is much less clear in most cases other than in special regions such as the mammalian X-chromosome inactivation center or immunoglobulin genes. On a larger scale, chromatin from a given chromosome tends to occupy a distinct chromosomal territory, but individual regions clearly loop out of the territory into a common space and often associate with other regions with similar properties on the same or different chromosomes. These are structures such as transcription factories, with which active genes are thought to associate for a few rounds of transcription at a time, although their existence is still debated (Zhang et al. 2013; Zhao et al. 2014), or Polycomb bodies, in which chromatin domains repressed by Polycomb mechanisms often associate. Chromatin domains such as promyelocytic leukemia bodies, regions associated with the nuclear lamina, and certain heterochromatin regions may be other examples.
Surprisingly, two types of factors have turned out to be involved in many, perhaps all, such interactions: insulator proteins and cohesin complexes. Although the designation insulator protein is now very likely irrevocably affixed, these proteins have, in fact, turned out to be organizers of chromatin architecture, which, in some cases, prevents an enhancer or silencer from acting on neighboring regions, but more frequently functions to juxtapose enhancers and promoters. The role of cohesin complexes may be related to that of insulator proteins in some cases, but is much less well understood although surprising connections continue to be reported. How transcription factories or Polycomb bodies assemble is still very unclear, but it is likely that insulator proteins and cohesin play a role there as well. It is surprising, in this respect, that mammalian genomes contain only one reported insulator protein, CTCF, whereas Drosophila contains several. And, although a close link is apparent between mammalian CTCF and cohesin, such a link, if it exists, is much less direct in Drosophila. Mechanistically, we will need much more work to understand how insulator proteins function and what controls their activities. In particular, it remains unclear how a given insulator-binding site distinguishes which other sites to interact with and which not, particularly if, as appears to be the case in mammals, there really is just one DNA-binding insulator protein: CTCF. Do insulator protein modifications such as small ubiquitin-related modifier (SUMO)ylation (MacPherson et al. 2009), or perhaps interactions with ncRNA as suggested by the reported involvement of Argonaute proteins (Moshkovich et al. 2011), “customize” CTCF according to specific functions?
2. Building a global chromatin accessibility hypothesis
2.1. The Accessibility Problem: The Difficulty of Reading Genomic DNA
Ultimately, the distinction of one bit of chromatin from another depends on the local DNA sequence. Wrapping the DNA into nucleosomes makes the DNA sequence difficult to access, and if it cannot be accessed, one nucleosome cannot be distinguished from another. Histone modifications help to restore a functional differentiation. The presence of nucleosomes and nature of the histone modifications they carry produce chromatin states that determine whether, when, or how the sequence is read by DNA-binding proteins. Unless DNA-binding proteins have access to the DNA sequence, the apparatus that binds to chromatin and acts on it would not know where to go and, most importantly, the genes and other sequence information could not be read.
In the second part of this essay, I want to consider the question of the accessibility of genomic DNA, apart from cooperative binding or the remodeling that follows the binding of pioneer factors (elaborated in Sec. 2.3); this is a problem that underlies many of the questions concerning histone modifications—their multiplicities and dynamics. I propose an argument that begins to integrate the mass of information about histone modifications from the vantage point of DNA accessibility.
Textbook accounts explain the wrapping of the DNA into nucleosomes as a means of packaging eukaryotic genomes into the nucleus. The wrapping of 147 bp of DNA into a nucleosome clearly reduces the volume occupied, although more by reducing the degrees of freedom than by actually decreasing the space occupied by the double helix. DNA can be compacted much more than it generally is in a eukaryotic nucleus, as shown by the packing of a bacteriophage genome into a capsid or of the human genome in a sperm nucleus. The difference is that the bacteriophage DNA is not available except by releasing the entire content of the capsid, and neither is much, or all, of the sperm genome. The eukaryotic genome in a somatic cell, instead, is available or specifically addressable in part (i.e., euchromatin), and the heterochromatic complement could be said to be accessible at least part of the time. What makes the difference, of course, is that the eukaryotic genome is partitioned into myriads of small packets (i.e., nucleosomes). These can be further packaged hierarchically into higher-order structures that, potentially, can be folded or unfolded locally. In other words, an arbitrary fraction of the genome down to single nucleosomes can, in principle, be opened up individually, allowing access to its DNA content. To do this, however, a nucleosome has to be differentiated from its neighbors or its DNA content has to be searchable at least intermittently.
2.2. Chromatin as a Response to DNA Concentration in the Nucleus
Although space is limiting in the nucleus and compaction of at least part of the genomic DNA, an imperative, far more important tension exists: a tension between the need for controlled genomic accessibility to transcription and the need to reduce the accessibility to most of the genomic DNA. Packaging of a large genome in the nucleus is a necessity, but perhaps far more important is the imperative to hide most of the DNA so that it is not easily accessible to the protein machinery that needs to act on it. All mechanisms that distinguish one genomic site from another, except those based on bulk physical or topological properties, must be able to recognize specific nucleotide sequence motifs, short enough to be readable and bindable by a single protein with sufficient discrimination to minimize the noise resulting from inappropriate binding. The ratio between signal and noise (i.e., specific versus nonspecific binding) is critical for carrying out any kind of gene regulation. This discrimination is lost if the concentration of nonspecific DNA is so great that any DNA-binding protein would spend its time bound inappropriately to the wrong sequences. The key to specificity in the nucleus is therefore intrinsically tied to the need to mask most of the genomic DNA so as to make it unavailable for binding to the regulatory and transcriptional machinery. However, the differentiation of chromatin and specific activities of the genetic information depend ultimately on the local DNA sequence. Packaging into nucleosomes not only prevents access to the DNA sequence, but, by itself, it would reduce the genome to an undifferentiated collection of more or less structurally identical nucleosomes. Viewed in this light, it is clear then that a large part of eukaryotic gene regulation must consist of ways to (1) remove or remodel nucleosomes so as to make the underlying DNA available for DNA-binding proteins; (2) do this in a sequence-specific way or at least produce sequence-specific results; and (3) develop a way to mark nucleosomes or nucleosomal domains so as to restore some specificity for the action of regulatory proteins (e.g., acetylating histone tails to make certain nucleosomes easier to displace or remodel).
2.3. Nucleosome Density
It is well known that the density of nucleosomes is important for retaining regulatory specificity. If insufficient histones are produced, nucleosomal density is reduced. In Drosophila, this causes loss of heterochromatic silencing and suppression of position-effect variegation (Moore et al. 1983). In yeast and mammalian cells, histone insufficiency causes derepression of many conditionally expressed genes (Han and Grunstein 1988; Lenfant et al. 1996; Wyrick et al. 1999; Celona et al. 2011; Gossett and Lieb 2012). Interestingly, a reduced nucleosome density changes the occupancy (the frequency with which a position is occupied) rather than the distribution of nucleosomes. This is because certain DNA sequences favor the formation of nucleosomes by more easily wrapping around the histone core than others. In addition, the more DNA becomes accessible, the more DNA-binding proteins can bind to their preferred sequences and compete with nucleosome formation.
As shown long ago, initially by Drosophila histone gene deletion experiments, nucleosome density is a necessary precondition for heterochromatic silencing. If the nucleosome density is too low, the DNA becomes too accessible to DNA-binding proteins, in particular, RNA polymerase, and increasing evidence shows that indiscriminate access results in indiscriminate transcription. Transcriptional activity is associated with many other nucleosome-modifying activities, particularly histone acetylation, which prevents the establishment of the heterochromatic state and promotes further accessibility. A similar effect is obtained in the presence of a normal histone gene complement if the concentration of a gene activator is increased (Ahmad and Henikoff 2001). These effects show that without nucleosomes the ability to repress transcription is lost and the ability of nucleosomes to prevent access is in competition with the concentration-dependent binding of the transcriptional machinery. What is lost is not only the ability to repress, but also the control of transcriptional activation in that the requirement for activators to produce transcription is at least partly absolved if RNA polymerase no longer needs the help of various remodeling activities to access the DNA sequence. Furthermore, although the control of access is a major component of the repressive activities of heterochromatin, in normal cells there are windows of opportunity to access even the heterochromatic DNA sequence, and a sufficiently high concentration of a DNA-binding activator can exploit these to bind to its sequence and produce local derepression. In many cases, then, a major limiting factor in the control of transcription is access of RNA polymerase to DNA. Many genes, particularly in higher eukaryotes, have developed ways to ensure that RNA polymerase is preloaded, often transcriptionally initiated, but arrested (paused polymerase) and ready to respond to transcriptional signals that allow it to elongate. In most cases, this requires the access of DNA-binding proteins that configure the nucleosomes around the promoter site. Here too, however, the pausing is dependent on the need of additional factors to overcome nucleosomal obstacles to elongation.
When the DNA is fully occupied by nucleosomes, or at least when histone levels do not limit the nucleosome density, most of the DNA sequence is not directly accessible to DNA-binding proteins. It has been shown that some transcription factors are better able than others to bind to nucleosomal DNA sequences, at least when the binding sites are close to one edge of the nucleosome (Zaret and Carroll 2011). Such “pioneer” factors can gain a toehold by binding to the DNA entering the nucleosome even in a compacted chromatin structure, evict linker histone H1, and invoke nucleosome remodeling machines to unravel the DNA and expose it for the binding of other enhancer binding factors in a multistage process (Li et al. 2010).
2.4. Roaming Activities
Special features may allow certain sequence-specific binding proteins to find their binding sites, possibly by profiting opportunistically from transient opening of the chromatin structure. In general, however, access to DNA requires the help of remodeling machines. Thus, to allow access to nucleosomal DNA without prior sequence information, we would need to hypothesize roaming activities that survey the genomic chromatin and periodically turn it over, so to speak, temporarily opening access to the underlying sequence. At the same time, to prevent such access and ensure regulated opening, we might expect an opposing activity. Are there any known chromatin marks or chromatin activities that might support this hypothesis?
Two features have been identified that are characteristic of sites in which DNA has to be maintained in an accessible state: One is the binding of the CBP histone acetylase or its close relative p300, whether or not accompanied by steady-state enrichment in histone H3K27 acetylation. The other is the chromatin mark H3K4me1, whose role in accessibility is not well understood. These features are characteristically found at enhancer sites, in which CBP is thought to be recruited by most enhancer-binding factors (Heintzman et al. 2007; Xi et al. 2007; Visel et al. 2009). They are also found at promoters and wherever DNA-binding proteins find access to the genomic DNA. These sites have also been found to be hot spots of active nucleosome turnover, detectable by the deposition of nucleosomes containing the histone variant H3.3, often together with the histone variant H2A.Z. This combination of variants is less stable than normal, and easier to remodel or turn over (Jin et al. 2009). The fact that acetylation is not always detected at enhancer sites, despite the presence of CBP, suggests that the acetylated nucleosomes are those that have been displaced to create the nucleosome-free region, which is occupied by DNA-binding proteins.
CBP is often associated with a histone-remodeling activity (e.g., Drosophila Brahma, ortholog of human SNF2L2) and UTX, one of two known histone H3K27 demethylases, but the only one found in Drosophila (Tie et al. 2012). UTX is an essential component of the Trithorax-related (TRR) H3K4 methyltransferase (or MLL3 and MLL4 in mammals), which is the source of H3K4me1 and sometimes H3K4me2, found at enhancers, promoters, and other protein-bound DNA-binding sites (Herz et al. 2012). We might wonder what a H3K27 demethylase might be doing at these sites, but there is a strong connection; CBP is responsible for H3K27 acetylation and this activity is blocked by the simple presence of preexisting H3K27 methylation, without need to recruit repressive complexes of any kind.
2.5. Ubiquitous H3K27 Methylation
H3K27 methylation is, in fact, ubiquitous in the genome. It is produced by the Polycomb repressive complex 2 (PRC2), whose methyltransferase subunit in Drosophila is E(z) (ortholog of mammalian Ezh1 and Ezh2). PRC2 is responsible for mono-, di-, trimethylated H3K27. The trimethylated state is the one that has received the most attention because it is the one associated with Polycomb-repressed genes. However, its most abundant product is not H3K27me3, which in somatic cells constitutes some 5%–10% of total histone H3, but H3K27me2, which is found in a staggering 50%–60% of all H3 (Peters et al. 2003; Ebert et al. 2004; Jung et al. 2010; Voigt et al. 2012). Dimethylation is therefore the major activity of PRC2 in flies as in man. Kinetic studies (McCabe et al. 2012) show, in fact, that although PRC2 monomethylates and dimethylates rapidly, trimethylation is enzymatically more difficult and probably occurs in vivo primarily where PRC2 is stably bound. H3K27me2 is also the most abundant and broadly distributed type of histone modification, found everywhere except in regions that are enriched in H3K27me3 or undergoing transcriptional activity. The reason for the former is obvious. The reason for the latter (i.e., its depletion in transcribed regions) is most likely twofold: (1) nucleosomes are less densely distributed in such regions because of increased instability and turnover, and (2) transcriptionally active regions are targeted by the UTX H3K27 demethylase, producing H3K27me1 and H3K27me0. Very similar results have been recently reported for mouse embryonic stem cells (Ferrari et al. 2014). In these cells, H3K27me3, me2, and me1 constitute, respectively, 7%, 70%, and 4% of total H3, whereas H3K27ac is 2% and unmodified H3K27 is 16%. H3K27me2 is confined to transcriptionally inactive regions, except those that have stably bound PRC2, whereas H3K27me1 is found only in transcriptionally active regions. Most likely then, H3K27me2 is actively removed by demethylation, with H3K27me1 as an intermediate to complete demethylation. Not surprisingly, transcriptionally active regions are also enriched in UTX, the only known H3K27 demethylase in Drosophila. Removal of H3K27 methylation in active regions is required for H3K27 acetylation, which is generally found in the 5′ region of active transcription units and enhancers. But this leaves the twin questions: Why is there H3K27 methylation in the first place? And why is H3K27 acetylation specifically needed?
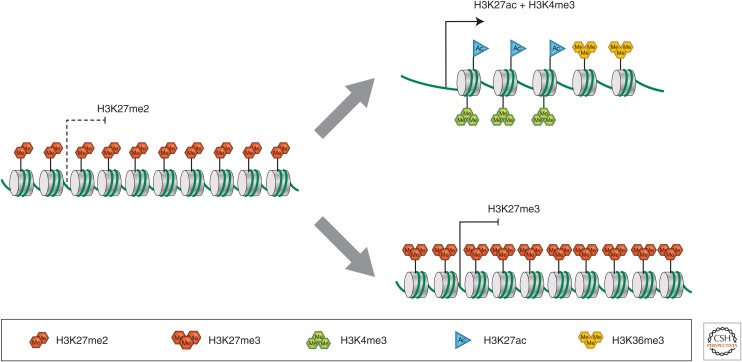
Unlike H3K27me3, which is found mainly at genomic sites that can stably recruit the PRC2 complex, the activity that produces H3K27me2 must target the whole genome. Although it might be associated with the replication fork, it is most likely explained by the transient interaction of free PRC2 with nucleosomes by a hit-and-run mechanism. It is not a completely random mechanism, however. The methylation activity of PRC2 is modulated by several inputs from the surrounding chromatin. One of these is dependent on a hydrophobic pocket in the PRC2 subunit extra sex combs (ESC)/Eed, which binds methylated H3K27 (Margueron et al. 2009). When this binding occurs, it effects a conformational shift in the catalytic subunit E(z) that greatly stimulates its methylation activity. Although H3K27me3 binds more strongly, H3K27me2 also binds to the aromatic pocket. Therefore, the presence of H3K27me2 or H3K27me3 in surrounding nucleosomes promotes the methylation of newly deposited nucleosomes. Mutations in the ESC hydrophobic pocket drastically reduce the global level of both H3K27me3 and H3K27me2. Other mechanisms that modulate PRC2 activity probably contribute, although they have not been tested in vivo. Thus, the nucleosome density surrounding a target nucleosome appears also to stimulate methylation activity (Yuan et al. 2012), whereas the presence of H3K4me3 or H3K36me2/me3 on the target nucleosome reduces the methylation activity of PRC2 (Schmitges et al. 2011; Yuan et al. 2011). As a consequence, regions that already contain H3K27 methylation are better targets for PRC2, whereas regions that have a lower nucleosome density or nucleosomes that bear H3K4me3 or H3K36me2/me3, all marks of transcriptional activity, are poor targets. The discovery of these several devices in PRC2 and other methyltransferases has made it clear that feedback and feed-forward mechanisms can be incorporated in chromatin-modifying machines to both self-renew a chromatin mark and avoid regions marked with certain other histone modifications. It is worth pointing out here that these mechanisms not only help to maintain Polycomb repression from one cell cycle to the next through the maintenance of H3K27me3, but, by modulating the deposition of H3K27me2, they also provide a memory of transcriptional activity. Regions that have been recently transcribed have lower nucleosome density and are enriched in the H3K4me3 and H3K36me2/me3 marks that favor renewed transcriptional activity and, at the same time, inhibit H3K27 methylation. In other words, in the case of a globally distributed histone mark such as H3K27me2, the absence of the mark is itself a mark that carries information of previous transcriptional activity (Fig. 1).
Figure 1.
Transcriptional memory of the chromatin state. The schematic drawing illustrates some key changes in the chromatin marks associated with a chromatin region that has recently been transcribed or becomes stably repressed by Polycomb mechanisms. A region that has not been recently transcribed is marked by heavy H3K27me2. A region recently transcribed has lost H3K27me2, but instead gained H3K27ac and H3K4me3 marks in the promoter-proximal part and H3K36me3 (which, in turn, recruits deacetylating complexes) to control the excessive access allowed by the loss of H3K27me2. Regions that can recruit stable binding of Polycomb complexes PRC1 and PRC2 acquire H3K27me3. For simplicity, other histone marks are not shown.
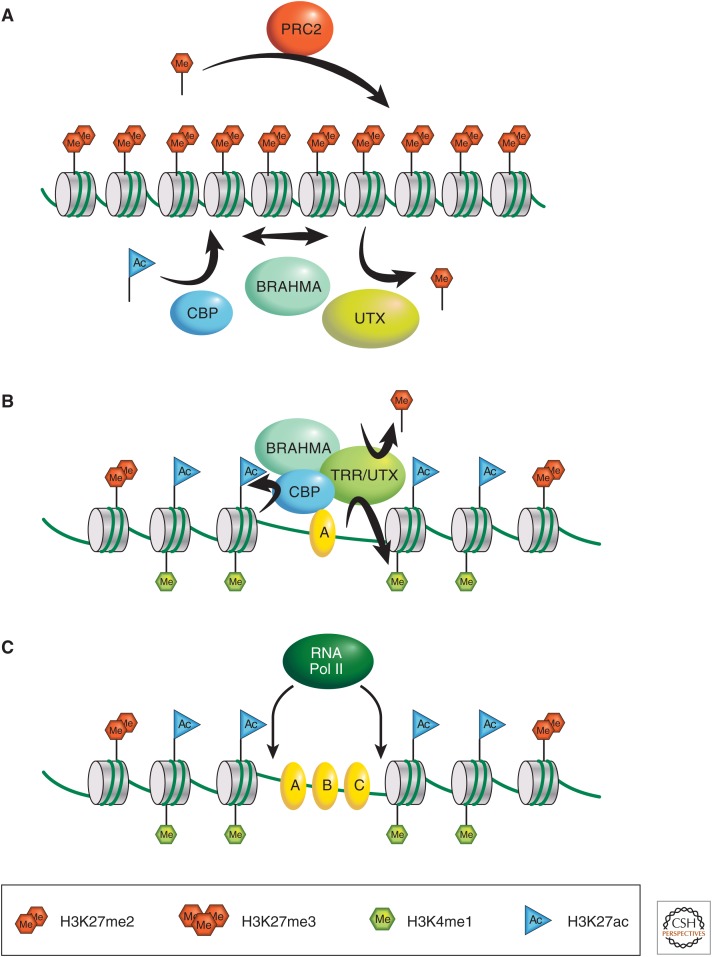
Figure 2.
A model for the control of DNA accessibility in chromatin. (A) The model proposes that antagonistic roaming activities transiently interact with genomic chromatin: one, caused by PRC2, deposits the H3K27me2 mark. Another removes this methylation mark and remodels nucleosomes, allowing transient access to the DNA sequence. These activities are attributed to UTX, CBP, and BRAHMA. (B) A DNA-binding factor A binds to its cognate binding motif in the DNA, transiently made accessible, and recruits stable binding of CBP together with a remodeling activity (BRAHMA) and the TRR/MLL3,4 complex containing UTX. These activities remove H3K27 methylation, depositing instead the H3K27ac and H3K4me1 marks. (C) The remodeling activity provides stable access to the DNA, leading to the binding of additional factors B and C to an enhancer region (or other regulatory element on the DNA). The region of DNA made accessible can also be opportunistically targeted by RNA polymerase, which may produce short transcripts from both DNA strands.
PRC2, therefore, provides a global mechanism to mark chromatin according to its recent usage. But what does the H3K27me2 mark do and how is it interpreted? We have become accustomed to thinking of histone modifications as marks that are “read” by chromatin proteins that possess appropriate binding domains. This is possible, but unlikely for H3K27me2. The “reader” approach is suitable for marks that distinguish a region from the rest of the chromatin. A global mark such as H3K27me2 would bind the “reader” virtually everywhere. A more economical interpretation is that, rather than being “read,” the presence of the mark provides both the reading and response at the same time; H3K27 methylation preempts the lysine so that it cannot be acetylated. H3K27ac is a mark associated with the 5' region of active genes. As we have seen, it is also potentially present at all sites containing CBP—that is, sites such as enhancers that involve the access of DNA-binding proteins to DNA. In principle, monomethylation of H3K27 would serve the same purpose. H3K27me1 has been often considered to be associated, not with repression, but with transcriptional activity. H3K27me1 is found in transcriptionally active genes most likely because these are sites in which H3K27me2 is demethylated by UTX. The monomethylated state is most likely a stage in demethylation or remethylation.
2.6. The Accessibility Hypothesis
A hypothesis that would integrate these various findings is that access to the DNA content of nucleosomes is a major limiting factor in controlling all sequence-specific activity in the genome. Control of the access to DNA is therefore a key regulatory principle. According to the hypothesis (Fig. 2), access is provided intermittently by a roaming nucleosome remodeling activity that “turns over” nucleosomes, temporarily making their DNA accessible. For some reason that is not clear at this stage, this activity involves H3K27 acetylation, perhaps transiently, and is blocked by preventing H3K27 acetylation. In general, this acetylation is constantly removed by roaming histone deacetylases, as has been shown in yeast (Vogelauer et al. 2000). The remodeling that provides accessibility is counteracted by a roaming PRC2 activity that dimethylates H3K27 genome-wide; this PRC2-mediated H3K27 dimethylation thereby preempts this position and blocks its acetylation. The H3K27 dimethylation can be removed by the UTX demethylase, whose main activity is therefore to remove the block to acetylation and allow more stable access to the DNA. This is needed at sites such as enhancers, promoters, PREs, and others in which multiple DNA-binding proteins need to see the nucleotide sequence. The binding of these factors recruits stable CBP and is associated with remodeling activity whose longer-term presence displaces nucleosomes, producing a nucleosome-depleted region. Such regions are typically hypersensitive to DNaseI treatment and found associated with enhancers, promoters, PREs, sites whose edges are also enriched for histone H3.3 relative to their surroundings. This, as analyzed by Mito et al. (2007), is caused by nucleosome replacement, which means that nucleosomes found there are not formed as part of the replicative process, but are due to continuous turnover.
2.7. Transcription of Nucleosome-Free Regions and the RNAi Response
The accessibility hypothesis requires that sites of nucleosomal depletion or remodeling, or any region that is not densely populated by nucleosomes, will have a high probability of binding RNA polymerase on an opportunistic basis and producing some transcriptional products. The amount and length of such transcripts is likely to be very variable and dependent on the sequence, the vicinity to some enhancer-like activity, and probably many other factors. There should be little strand specificity for these transcriptional starts, which, therefore, are likely to result in production of RNA from both strands and thus would be targeted by RNAi mechanisms.
Promoters, which have a short nucleosome-depleted region, are well known to produce short transcripts from both strands within a region of a few hundred nucleotides surrounding the transcription start called TSSa-RNAs (Seila et al. 2008; Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009; Taft et al. 2009). Promoter regions have evolved ways to minimize the production of RNA from both strands by selecting a high frequency of polyadenylation signals such that productive elongation occurs predominantly in the direction downstream from the gene TSS (Almada et al. 2013; Ntini et al. 2013). Enhancers are also the source of transcripts, the so-called enhancer RNAs or eRNAs, from both strands (De Santa et al. 2010; Kim et al. 2010; Ørom and Shiekhattar 2013). DNA damage sites, where nucleosomes are removed for a considerable length surrounding a double-strand break, also produce transcripts from both strands. These RNAs are now known to be processed by Dicer and Drosha and required for the binding of ATM, a kinase that phosphorylates the histone variant H2AX to initiate the formation of DNA damage repair foci (Francia et al. 2012). RNAs produced from all such nucleosome-depleted regions are by-products of the nucleosome remodeling processes occurring at these sites. They need not have particular function, but it should not be surprising to find that they have acquired a function at certain sites.
Regions that are partially depleted of nucleosomes or become too easily accessible to RNA polymerase are prone to initiate transcription, which is not strand-specific. In the general case, therefore, most such accessible regions would produce RNA transcripts from both strands. One possible consequence of bidirectional transcription is the recruitment of the RNAi machinery. The bidirectional transcripts produced from DNA damage sites clearly recruit components of the RNAi machinery (Francia et al. 2012). RNAi proteins such as Dicer2 and AGO2 are associated with active promoters that produce small bidirectional RNAs (Cernilogar et al. 2011). It has been claimed that the RNAi protein, AGO2, associates with a variety of sites that are expected to be depleted of nucleosomes, including CTCF-binding sites, promoters, and PREs (Moshkovich et al. 2011). It is not clear what the function of AGO2 might be in these cases, but its loss leads to a decrease in insulator activity or Polycomb repression (Grimaud et al. 2006; Lei and Corces 2006).
RNAi mechanisms are often thought to be protectors of genome integrity against attacks by viruses or proliferating transposons. In the nucleus, they result in the recruitment of histone H3K9 methylation, the binding of heterochromatin proteins such as HP1 and histone deacetylases, and the stabilization of nucleosomes, in essence, the opposite of the process that opened up the chromatin and produced the bidirectional transcripts at enhancers, promoters, etc. The connection between DNA accessibility and the RNAi response is, I suggest, not accidental. Regions that are partially depleted of nucleosomes or become too easily accessible to RNA polymerase are prone to initiate transcription, which is not strand-specific. In the general case, therefore, most such accessible regions would produce RNA transcripts from both strands. If these RNAs recruit the RNAi response, this response is endemic and inseparable from the fundamental necessity of gaining access to the genomic DNA. It could be argued, therefore, that the RNAi response might be, in its basic form, a way to recruit proteins that stabilize nucleosomes (HP1, linker histone, histone deacetylases), restore nucleosomal occupation, or restore nucleosomal stability to sites that, for whatever reason, might have transiently become open. The fact that the RNAi response has become a valuable protection against invading genetic elements would not be incompatible with the even more basic function of keeping the genomic DNA covered and ensuring that transiently opened regions do not get out of hand.
2.8. PRC2 and Heterochromatin
RNAi mechanisms are thought to be important for the establishment of heterochromatin. This has been worked out in detail for the fission yeast Schizosaccharomyces pombe, but many aspects of this relationship apply to Drosophila and mammalian heterochromatin formation. The arguments presented above help to understand why E(z) has been found to play a role in the efficient establishment of heterochromatin and, in fact, is known as a suppressor of position-effect variegation in Drosophila (Laible et al. 1997). This role has been a puzzle for many years because there is no specific presence of E(z) or H3K27me3 in heterochromatin. This role is better understood in terms of accessibility. In Drosophila, the early embryonic stages are a time of extremely rapid and synchronous nuclear divisions. These slow down by the 14th cycle (3 h postfertilization), but the chromatin produced must now be the target of a massive H3K27 methylation effort. This is accomplished thanks to correspondingly massive amounts of PRC2 components that are deposited in the egg during oogenesis. By the time nuclear proliferation slows down and heterochromatin first becomes detectable, global dimethylation of H3K27 must be in place. At this stage, it is important to suppress H3K27 acetylation, remodeling, and adventitious transcriptional activity to allow the RNAi and other mechanisms to initiate and maintain heterochromatin. Access to DNA is never completely prevented even in heterochromatin, as shown by the fact that strong activators can prevent heterochromatic silencing of a reporter gene (Ahmad and Henikoff 2001), but the absence of H3K27me2 would certainly result in a level of access to activators and RNA polymerase that would interfere with the establishment of heterochromatic silencing.
2.9. Effects of Loss of PRC2
If H3K27 methylation plays such a global genomic role, loss of PRC2 function would surely have major consequences—increasing pervasive transcription, among others. Unfortunately, it has not been possible yet to separate the global H3K27 dimethylation function from the Polycomb-related and more specific H3K27 trimethylation. Loss of PRC2 is an early embryonic lethal both in mammals and Drosophila, and it produces embryos with classical homeotic derepression phenotypes (Struhl and Brower 1982). Loss of Polycomb repression of Hox genes and many other developmentally important genes would certainly be sufficient to account for lethality. In addition, it would make it difficult to determine whether any other effects should be attributed to indirect consequences of derepression or tloss of H3K27 dimethylation. Nevertheless, loss of PRC2 activity is not cell lethal. Mammalian embryonic stem cells with knockouts of Ezh2 or Eed are viable, although unable to differentiate. In mouse embryonic stem cells lacking PRC2 function, H3K27 acetylation appears at new sites together with H3K4me1, forming a signature typical of poised enhancer regions (Ferrari et al. 2014). This suggests that many normally silent regions become accessible and transcriptionally active. Activation of new transcription sites not normally associated with H3K27me3 was also observed. An increase in the accessibility of chromatin to RNA polymerase was also observed in Drosophila embryos lacking maternal and zygotic ESC, an essential component of PRC2 (Chopra et al. 2011). Promoters of thousands of genes became occupied by RNA polymerase II, whether or not they were transcriptionally activated.
Mutations affecting EZH2 and PRC2 activity are associated with a variety of aggressive cancers but, strangely, both hyperactivity and loss of activity appear to be oncogenic. The interpretation has generally been that these effects are mediated by the hyperrepression or derepression of Polycomb target genes, and this is undoubtedly true, at least in part. For example, genes that block cell cycle progression, such as INK4A/B, are regulated by Polycomb mechanisms and hyperrepression would remove brakes to cell proliferation. Because much evidence supports a cancer-promoting role of PRC2 activity, the discovery that loss of PRC2 function can also promote cancers, such as myeloid leukemia, has been puzzling (Hock 2012; Simon et al. 2012; Tamagawa et al. 2013).
A particularly interesting case is that of the recently characterized mutations converting K27 to methionine in histone H3 or H3.3 genes. This mutation was found associated with a particularly malignant glioblastoma, in which it has a dominant effect totally out of proportion with the relatively small fraction of total histone H3 that is produced by the mutated histone H3 gene copy (Chan et al. 2013; Lewis et al. 2013). The methionine at position 27 mimics, in part, the K27 methylation, but lacks the moderating positive charge that the amine nitrogen retains even when trimethylated. As a consequence, the EZH2 catalytic domain binds, but does not readily release, the H3K27M peptide. Although this has not been directly shown, one consequence might be that the PRC2 complex becomes effectively sequestered and unavailable, causing H3K27 to become undermethylated genome-wide. Attention has been focused on the partial loss of H3K27me3 and consequent derepression of Polycomb target genes. I suggest a somewhat different interpretation. Genome-wide H3K27 dimethylation would be even more powerfully affected because it is strongly dependent on a hit-and-run mechanism and, therefore, on the pool of free PRC2. The loss of H3K27 methylation would be expected to derepress a large number of genes whose silencing depends on the inability of activators and RNA polymerase II to access the promoter. More important, perhaps, is that transcription may start anywhere, including within gene bodies, producing partial proteins that would have unexpected neomorphic effects.
These observations support the global accessibility hypothesis, at least in part, but are a long way from providing substantial proof. They suggest, nevertheless, that the abundance and ubiquitousness of H3K27 dimethylation are not without a significance that could help to understand the way in which different chromatin modifications jostle with one another, the interplay that has provided the raw material from which evolution has shaped the chromatin landscape and its functions.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, Danny Reinberg, and Monika Lachner
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this subject collection.
- Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project (Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, Gingeras TR). 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S. 2001. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104: 839–847. [DOI] [PubMed] [Google Scholar]
- *.Allis CD, Jenuwein T, Reinberg D. 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739. [DOI] [Google Scholar]
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. 2013. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B, Madhani HD, Narlikar GJ. 2013. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol Cell 51: 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ. 2012. A Swiss army knife of immunity. Science 337: 808–809. [DOI] [PubMed] [Google Scholar]
- Busturia A, Wightman CD, Sakonju S. 1997. A silencer is required for maintenance of transcriptional repression throughout Drosophila development. Development 124: 4343–4350. [DOI] [PubMed] [Google Scholar]
- Celona B, Weiner A, Di Felice F, Mancuso FM, Cesarini E, Rossi RL, Gregory L, Baban D, Rossetti G, Grianti P, et al. 2011. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol 9: e1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Sardo FL, Saxena A, Miyoshi K, Siomi H, et al. 2011. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K-M, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, et al. 2013. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Hendrix DA, Core LJ, Tsui C, Lis JT, Levine M. 2011. The polycomb group mutant esc leads to augmented levels of paused Pol II in the Drosophila embryo. Mol Cell 42: 837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. 2013. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol 20: 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dekker J, Misteli T. 2014. Long-range chromatin interactions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei C-L, Natoli G. 2010. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G. 2004. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev 18: 2973–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. 2012. KDM2B links the Polycomb repressive complex 1 (PRC1) to recognition of CpG islands. eLife 1: e00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stützer A, Fischle W, Bonaldi T, Pasini D. 2014. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell 53: 49–62. [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CCY, Huang H, Landry M-C, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, et al. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499: 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d’Adda di Fagagna F. 2012. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488: 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. 2013. The hierarchy of the 3D genome. Mol Cell 49: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossett AJ, Lieb JD. 2012. In vivo effects of histone H3 depletion on nucleosome occupancy and position in Saccharomyces cerevisiae. PLoS Genet 8: e1002771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. 2006. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell 124: 957–971. [DOI] [PubMed] [Google Scholar]
- Han M, Grunstein M. 1988. Nucleosome loss activates yeast downstream promoters in vivo. Cell 55: 1137–1145. [DOI] [PubMed] [Google Scholar]
- Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. 2012. Dynamics and memory of heterochromatin in living cells. Cell 149: 1447–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu YT, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinctive and predictive chromain signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. 2011. Histone modification: Cause or cog? Trends Genet 27: 389–396. [DOI] [PubMed] [Google Scholar]
- Herz H-M, Mohan M, Garruss AS, Liang K, Takahashi Y-H, Mickey K, Voets O, Verrijzer CP, Shilatifard A. 2012. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev 26: 2604–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H. 2012. A complex Polycomb issue: The two faces of EZH2 in cancer. Genes Dev 26: 751–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov GS, Ivanova T, Kurash J, Ivanov A, Chuikov S, Gizatullin F, Herrera-Medina EM, Rauscher F III, Reinberg D, Barlev NA. 2007. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol 27: 6756–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HR, Pasini D, Helin K, Jensen ON. 2010. Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol Cell Proteomics 9: 838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. 2013. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20: 1258–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N. 2013. Chromatin sampling—An emerging perspective on targeting polycomb repressor proteins. PLoS Genet 9: e1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. 2013. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502: 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. 1997. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J 16: 3219–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM. 2013. Epigenomic strategies at the interface of genetic and environmental risk factors for autism. J Hum Genet 58: 396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Lee JS, Kim H, Kim K, Park H, Kim J-Y, Lee SH, Kim IS, Kim J, Lee M, et al. 2012. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase vomplex. Mol Cell 48: 572–586. [DOI] [PubMed] [Google Scholar]
- Lei EP, Corces VG. 2006. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet 38: 936–941. [DOI] [PubMed] [Google Scholar]
- Lenfant F, Mann RK, Thomsen B, Ling X, Grunstein M. 1996. All four core histone N-termini contain sequences required for the repression of basal transcription in yeast. EMBO J 15: 3974–3985. [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. 2007. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21: 1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Hu G, Stokes D, Wang Y-H, Reinberg D. 2010. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol Cell 38: 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. 2009. The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol 29: 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ III, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the Polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. 2013. Biology: The big challenges of big data. Nature 498: 255–260. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A III, Diaz E, et al. 2012. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492: 108–112. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. 2009. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 12: 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. 2007. Histone replacement marks the boundaries of cis-regulatory domains. Science 315: 1408–1411. [DOI] [PubMed] [Google Scholar]
- Moore GD, Sinclair DA, Grigliatti TA. 1983. Histone gene multiplicity and position effect variegation in Drosophila melanogaster. Genetics 105: 327–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. 2011. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev 25: 1686–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntini E, Järvelin AI, Bornholdt J, Chen Y, Boyd M, Jørgensen M, Andersson R, Hoof I, Schein A, Andersen PR, et al. 2013. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol 20: 923–928. [DOI] [PubMed] [Google Scholar]
- Ørom UA, Shiekhattar R. 2013. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154: 1190–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589. [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. 2008. Divergent transcription from active promoters. Science 322: 1849–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux SB, Boucher GV, Chagnon P, Drouin S, Lambert RL, et al. 2012. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev 26: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Brower D. 1982. Early role of the esc+ gene product in the determination of segments in Drosophila. Cell 31: 285–292. [DOI] [PubMed] [Google Scholar]
- Su I-H, Dobenecker M-W, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wülfing C, Tarakhovsky A. 2005. Polycomb group protein Ezh2 controls actin polymerization and cell signaling. Cell 121: 425–436. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest ARR, Grimmond SM, Schroder K, et al. 2009. Tiny RNAs associated with transcription start sites in animals. Nat Genet 41: 572–578. [DOI] [PubMed] [Google Scholar]
- Tamagawa H, Oshima T, Numata M, Yamamoto N, Shiozawa M, Morinaga S, Nakamura Y, Yoshihara M, Sakuma Y, Kameda Y, et al. 2013. Global histone modification of H3K27 correlates with the outcomes in patients with metachronous liver metastasis of colorectal cancer. Eur J Surg Oncol 39: 655–661. [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Conrad PA, Scacheri PC, Harte PJ. 2012. Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol Cell Biol 32: 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Wu J, Suka N, Grunstein M. 2000. Global histone acetylation and deacetylation in yeast. Nature 408: 495–498. [DOI] [PubMed] [Google Scholar]
- Voigt P, LeRoy G, Drury WJ III, Zee BM, Son J, Beck DB, Young NL, Garcia BA, Reinberg D. 2012. Asymmetrically modified nucleosomes. Cell 151: 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P, Revzin A, Gross C, Wang A. 1974. Nonspecific DNA binding of genome regulating proteins as a biological control mechanism: 1. The lac operon: Equilibrium aspects. Proc Natl Acad Sci 71: 4808–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. 2004. Epigenetic programming by maternal behavior. Nat Neurosci 7: 847–854. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. 2005. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: Altering epigenetic marking later in life. J Neurosci 25: 11045–11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421. [DOI] [PubMed] [Google Scholar]
- Xi S, Zhu H, Xu H, Schmidtmann A, Geiman TM, Muegge K. 2007. Lsh controls Hox gene silencing during development. Proc Natl Acad Sci 104: 14366–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. 2011. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286: 7983–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, et al. 2012. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337: 971–975. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev 25: 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang Y. 2010. HMG modifications and nuclear function. Biochim Biophys Acta 1799: 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wong C-H, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, et al. 2013. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 504: 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZW, Roy R, Gebhardt JC, Suter DM, Chapman AR, Xie XS. 2014. Spatial organization of RNA polymerase II inside a mammalian cell nucleus revealed by reflected light-sheet superresolution microscopy. Proc Natl Acad Sci 111: 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]