Abstract
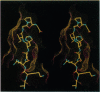
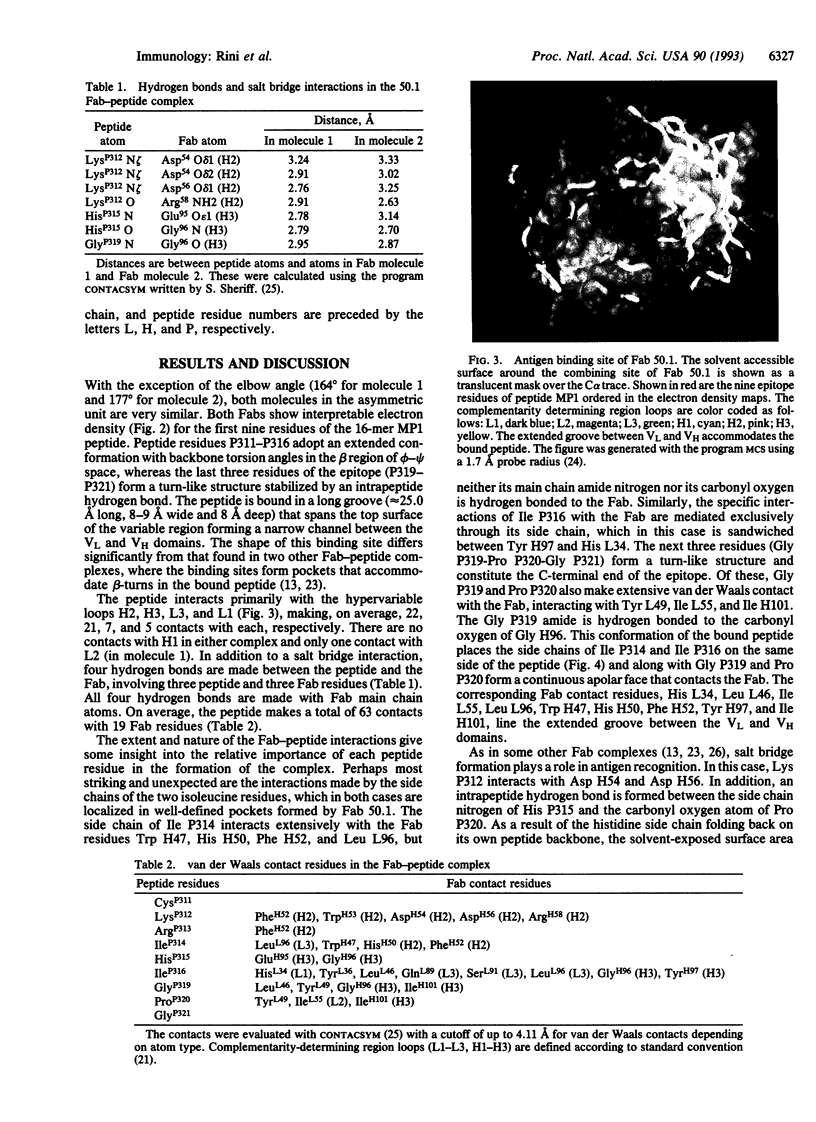
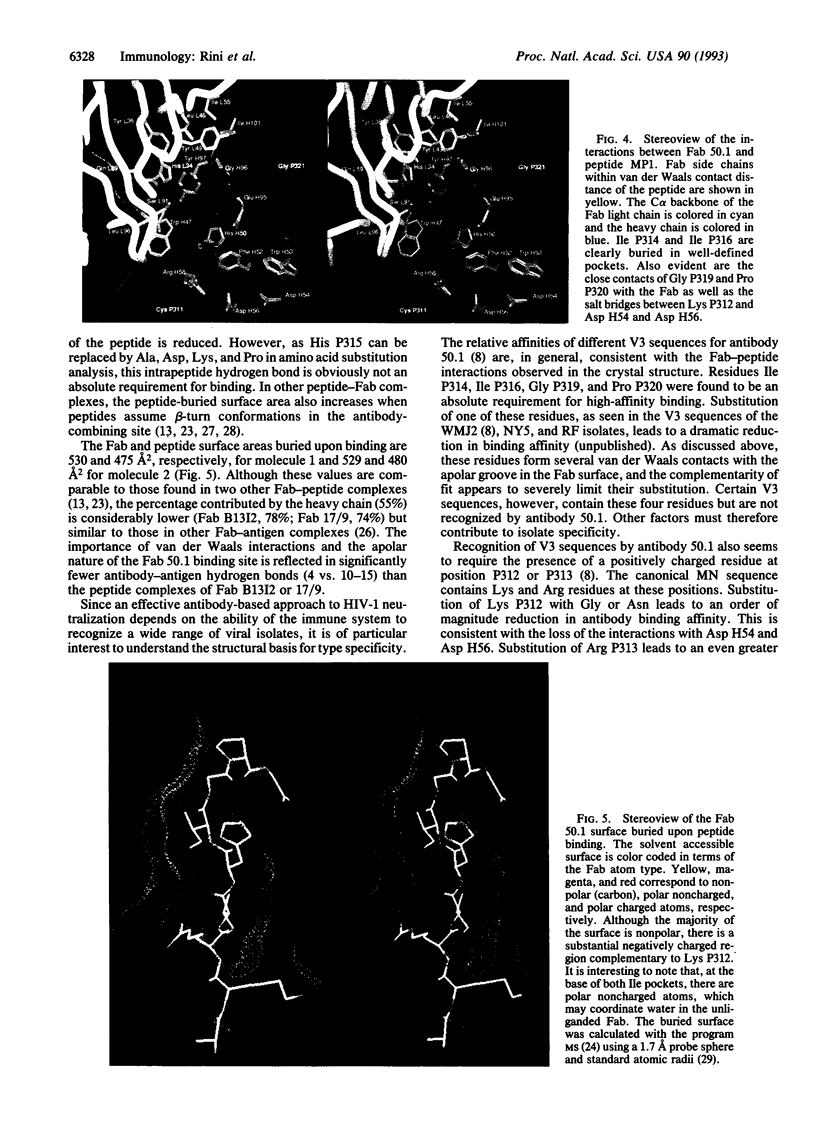
The crystal structure of the Fab fragment of a human immunodeficiency virus type 1 (HIV-1) neutralizing monoclonal antibody Fab has been determined at 2.8 A resolution in complex with a linear 16-residue peptide from the third hypervariable region (V3) of gp120. The first 9 residues of the peptide are ordered in the electron density maps, and their conformation is in partial agreement with the beta-strand-type II beta-turn structure predicted for this portion of the V3 loop. Notably, several of the peptide residues that are well conserved among different HIV-1 isolates contact a nonpolar 25-A-long groove in the antibody-combining site. The largely extended structure of the peptide differs from the beta-turns seen as the primary determinants in other published anti-peptide Fab structures. Analysis of the specific Fab-peptide interactions only partially explains the MN isolate specificity shown by this antibody.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brünger A. T., Krukowski A., Erickson J. W. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr A. 1990 Jul 1;46(Pt 7):585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar K., Profy A. T., Dyson H. J. Solution conformational preferences of immunogenic peptides derived from the principal neutralizing determinant of the HIV-1 envelope glycoprotein gp120. Biochemistry. 1991 Sep 24;30(38):9187–9194. doi: 10.1021/bi00102a009. [DOI] [PubMed] [Google Scholar]
- Davies D. R., Padlan E. A., Sheriff S. Antibody-antigen complexes. Annu Rev Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- Emini E. A., Schleif W. A., Nunberg J. H., Conley A. J., Eda Y., Tokiyoshi S., Putney S. D., Matsushita S., Cobb K. E., Jett C. M. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature. 1992 Feb 20;355(6362):728–730. doi: 10.1038/355728a0. [DOI] [PubMed] [Google Scholar]
- Garcia K. C., Ronco P. M., Verroust P. J., Brünger A. T., Amzel L. M. Three-dimensional structure of an angiotensin II-Fab complex at 3 A: hormone recognition by an anti-idiotypic antibody. Science. 1992 Jul 24;257(5069):502–507. doi: 10.1126/science.1636085. [DOI] [PubMed] [Google Scholar]
- Gelin B. R., Karplus M. Side-chain torsional potentials: effect of dipeptide, protein, and solvent environment. Biochemistry. 1979 Apr 3;18(7):1256–1268. doi: 10.1021/bi00574a022. [DOI] [PubMed] [Google Scholar]
- Goudsmit J., Back N. K., Nara P. L. Genomic diversity and antigenic variation of HIV-1: links between pathogenesis, epidemiology and vaccine development. FASEB J. 1991 Jul;5(10):2427–2436. doi: 10.1096/fasebj.5.10.2065891. [DOI] [PubMed] [Google Scholar]
- Javaherian K., Langlois A. J., LaRosa G. J., Profy A. T., Bolognesi D. P., Herlihy W. C., Putney S. D., Matthews T. J. Broadly neutralizing antibodies elicited by the hypervariable neutralizing determinant of HIV-1. Science. 1990 Dec 14;250(4987):1590–1593. doi: 10.1126/science.1703322. [DOI] [PubMed] [Google Scholar]
- Javaherian K., Langlois A. J., McDanal C., Ross K. L., Eckler L. I., Jellis C. L., Profy A. T., Rusche J. R., Bolognesi D. P., Putney S. D. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6768–6772. doi: 10.1073/pnas.86.17.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRosa G. J., Davide J. P., Weinhold K., Waterbury J. A., Profy A. T., Lewis J. A., Langlois A. J., Dreesman G. R., Boswell R. N., Shadduck P. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science. 1990 Aug 24;249(4971):932–935. doi: 10.1126/science.2392685. [DOI] [PubMed] [Google Scholar]
- Nara P. L., Smit L., Dunlop N., Hatch W., Merges M., Waters D., Kelliher J., Gallo R. C., Fischinger P. J., Goudsmit J. Emergence of viruses resistant to neutralization by V3-specific antibodies in experimental human immunodeficiency virus type 1 IIIB infection of chimpanzees. J Virol. 1990 Aug;64(8):3779–3791. doi: 10.1128/jvi.64.8.3779-3791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palker T. J., Clark M. E., Langlois A. J., Matthews T. J., Weinhold K. J., Randall R. R., Bolognesi D. P., Haynes B. F. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1932–1936. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985 Jan 24;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Rini J. M., Schulze-Gahmen U., Wilson I. A. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science. 1992 Feb 21;255(5047):959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- Rusche J. R., Javaherian K., McDanal C., Petro J., Lynn D. L., Grimaila R., Langlois A., Gallo R. C., Arthur L. O., Fischinger P. J. Antibodies that inhibit fusion of human immunodeficiency virus-infected cells bind a 24-amino acid sequence of the viral envelope, gp120. Proc Natl Acad Sci U S A. 1988 May;85(9):3198–3202. doi: 10.1073/pnas.85.9.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield R. L., Fieser T. M., Lerner R. A., Wilson I. A. Crystal structures of an antibody to a peptide and its complex with peptide antigen at 2.8 A. Science. 1990 May 11;248(4956):712–719. doi: 10.1126/science.2333521. [DOI] [PubMed] [Google Scholar]
- Stura E. A., Stanfield R. L., Fieser G. G., Silver S., Roguska M., Hincapie L. M., Simmerman H. K., Profy A. T., Wilson I. A. Crystallization, sequence, and preliminary crystallographic data for an antipeptide Fab 50.1 and peptide complexes with the principal neutralizing determinant of HIV-1 gp120. Proteins. 1992 Dec;14(4):499–508. doi: 10.1002/prot.340140410. [DOI] [PubMed] [Google Scholar]
- White-Scharf M. E., Potts B. J., Smith L. M., Sokolowski K. A., Rusche J. R., Silver S. Broadly neutralizing monoclonal antibodies to the V3 region of HIV-1 can be elicited by peptide immunization. Virology. 1993 Jan;192(1):197–206. doi: 10.1006/viro.1993.1022. [DOI] [PubMed] [Google Scholar]
- Wilmot C. M., Thornton J. M. Analysis and prediction of the different types of beta-turn in proteins. J Mol Biol. 1988 Sep 5;203(1):221–232. doi: 10.1016/0022-2836(88)90103-9. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Rini J. M., Fremont D. H., Fieser G. G., Stura E. A. X-ray crystallographic analysis of free and antigen-complexed Fab fragments to investigate structural basis of immune recognition. Methods Enzymol. 1991;203:153–176. doi: 10.1016/0076-6879(91)03009-6. [DOI] [PubMed] [Google Scholar]
- Yeates T. O., Rini J. M. Intensity-based domain refinement of oriented but unpositioned molecular replacement models. Acta Crystallogr A. 1990 May 1;46(Pt 5):352–359. doi: 10.1107/s0108767389013073. [DOI] [PubMed] [Google Scholar]