Abstract
Protein O-GlcNAcylation, which is controlled by O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), has emerged as an important posttranslational modification that may factor in multiple diseases. Until recently, it was assumed that OGT/OGA protein expression was relatively constant. Several groups, including ours, have shown that OGT and/or OGA expression changes in several pathologic contexts, yet the cis and trans elements that regulate the expression of these enzymes remain essentially unexplored. Here, we used a reporter-based assay to analyze minimal promoters and leveraged in silico modeling to nominate several candidate transcription factor binding sites in both Ogt (i.e. the gene for OGT protein) and Mgea5 (i.e. the gene for OGA protein). We noted multiple E2F binding site consensus sequences in both promoters. We performed chromatin immunoprecipitation in both human and mouse cells and found that E2F1 bound to candidate E2F binding sites in both promoters. In HEK293 cells, we overexpressed E2F1, which significantly reduced OGT and MGEA5 expression. Conversely, E2F1-deficient mouse fibroblasts had increased Ogt and Mgea5 expression. Of the known binding partners for E2F1, we queried whether retinoblastoma 1 (Rb1) might be involved. Rb1-deficient mouse embryonic fibroblasts showed increased levels of Ogt and Mgea5 expression, yet overexpression of E2F1 in the Rb1-deficient cells did not alter Ogt and Mgea5 expression, suggesting that Rb1 is required for E2F1-mediated suppression. In conclusion, this work identifies and validates some of the promoter elements for mouse Ogt and Mgea5 genes. Specifically, E2F1 negatively regulates both Ogt and Mgea5 expression in an Rb1 protein-dependent manner.
Keywords: E2F transcription factor; gene transcription; O-GlcNAcylation; O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT); retinoblastoma protein (pRb, RB); O-GlcNAcase
Introduction
The posttranslational O-linked N-acetylglucosamine (O-GlcNAc)2 modification occurs in a variety of cellular proteins and is known to be an important metabolic sensor in all metazoans. Alterations in O-GlcNAc levels occur in several diseases, including heart failure, diabetes, cancer, and neurodegenerative disorders (1–5). Evidence suggests that changes in O-GlcNAc levels play a critical role in protein activity, stability, and localization (6–9). Using the substrate uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), the enzyme O-GlcNAc transferase (OGT) adds the GlcNAc moiety to serine and threonine residues, whereas O-GlcNAcase (OGA) removes it. Embryonic lethality and developmental delay observed by genetic deletion of Ogt or Mgea5 demonstrates the critical role of these enzymes in O-GlcNAc cycling (10, 11). We (1), along with others (12–14), have observed alterations in the expression level of OGT and OGA protein along with concomitant protein O-GlcNAc modification in various pathological conditions. We recently reported that the up-regulation of miR-539 could be a mechanism for decreased OGA level in the infarcted mouse heart (15). Another study showed that increases in O-GlcNAc levels by pharmacological inhibition of OGA increased OGA expression through the O-GlcNAc modification of RNA polymerase II, whereas OGT levels decreased (13). Thus, changes in OGT/OGA protein expression occur; however, molecular mechanisms that regulate the expression of these enzymes remain to be elucidated.
To characterize the promoter region of Ogt and Mgea5, we used a luciferase reporter system and identified the minimal promoter region based on the reporter activity. The transcription factors that bind to the delineated promoter regions were identified using the MatInspector program (16). Among the list of transcription factors, the E2F transcription factor binding sites were observed in both Ogt and Mgea5 promoter regions. The E2F family members (E2F1 to E2F8) form heterodimers with dimerization partner members and regulate gene transcription. Among the E2Fs, E2F1 to -3 are often referred to as activators, and E2F4 and -5 are often referred to as repressors (17). The tumor suppressor retinoblastoma (Rb) pocket proteins (pRb/Rb1, p107, and p130) are also known to bind to E2F transcription factors in a phosphorylation-dependent manner and control its function (18–20). E2F1, in particular, is known to play an important role in activating the expression of a wide spectrum of genes that are involved in the cell cycle, nucleotide biosynthesis, and apoptosis (21–24). E2F1 directly represses several genes, including human TERT (telomerase reverse transcriptase) and Mcl-1 (anti-apoptotic protein myeloid cell leukemia sequence 1) (25, 26). Interestingly, E2F1 is involved in the repression of key genes that regulate energy homeostasis and mitochondrial function (27). These results suggest that E2F1 can activate or repress various target genes. Here, we identified the minimal promoter sequences of mouse Ogt and Mgea5 that confer transcriptional activity. Taken together, our results demonstrate the basal promoter element of Ogt and Mgea5 and identified E2F1 transcription factor in their regulation.
Experimental Procedures
Cell Culture
HEK293, H9c2, and 3T3 L1 cells were grown in DMEM containing 10% fetal bovine serum and penicillin/streptomycin. The HEK293 and H9c2 cells were plated overnight at 70% confluence and then used for transient transfection and reporter assays. Dr. Douglas C. Dean (University of Louisville) provided wild type and E2F1-deficient (E2F1−/−) mouse adult fibroblasts (MAFs), and Dr. Brian F. Clem (University of Louisville) supplied wild type and Rb1-deficient (Rb1−/−) mouse embryonic fibroblasts (MEFs). Both MAF and MEF cells were maintained in DMEM containing 10% fetal bovine serum.
Promoters and Plasmids
To characterize the core promoter region of mouse Ogt and Mgea5, we used a luciferase reporter system to analyze an about 1000-bp sequence upstream of the putative transcriptional start site (TSS). A series of 5′ truncations were made to further narrow down the promoter activity. These included the 5′-flanking regions of full-length Ogt having SacI and XhoI restriction enzyme sites (−1419 to +233 bp, −698 to +233 bp, −294 to +233 bp, −178 to +233 bp, −105 to +233 bp, and −66 to +233 bp) and the 5′-flanking regions of Mgea5 having KpnI and HindIII restriction enzyme sites (−1034 to +181 bp; −582 to +181 bp; −407 to +181 bp, and −48 to +181 bp). These 5′-flanking regions were PCR-amplified with specific primers (Table 1) using mouse genomic DNA and then cloned into a pGL3-basic luciferase reporter vector (Promega). E2F binding sites in the pGL3-Ogt (−178 to +233 bp) and pGL3-Mgea5 (−407 to +181 bp) promoter constructs were mutated using specific primers with base substitution at the conserved core sites (Table 2). The PureYieldTM plasmid miniprep system (Promega) was used for small scale plasmid extraction, and the sequence was verified. Large scale plasmid purification for downstream transfection experiments was carried out using the QIAfilterTM plasmid midi kit (Qiagen). The pCMVHA E2F1 was obtained from Addgene (plasmid 24225), and the pCMVHA plasmid vector was used as the transfection control (22).
TABLE 1.
Primers and their sequences used to clone the truncated 5′ regions of Ogt and Mgea5 promoters, ChIP assay, and quantitative PCR analysis
| Primers | Sequences |
|---|---|
| Cloning primers | |
| OGT-TPF1 (−1419 to +233 bp) | 5′-ACGCACTAATCAAGAAGGCC-3′ |
| OGT-TPF2 (−698 to +233 bp) | 5′-GAGGAAAAAGGCATCCTCTT-3′ |
| OGT-TPF3 (−294 to +233 bp) | 5′-CGTACTTTAGCGCTTGCCAA-3′ |
| OGT-TPF4 (−178 to +233 bp) | 5′-GAGCCCTTAACCCGGAGTT-3′ |
| OGT-TPF5 (−105 to +233 bp) | 5′-CTTGGGCGGAAGGTTACGTA-3′ |
| OGT-TPF6 (−66 to +233 bp) | 5′-ACAGTCACTTGGTCGGTAGT-3′ |
| OGT-TPR | 5′-GGAGCTTCTTGAGATAGAAC-3′ |
| MGEA5-TPF1 (−1034 to +181 bp) | 5′-CCCTTACGCACACCGCACACTC-3′ |
| MGEA5-TPF2 (−582 to +181 bp) | 5′-AGAAGTATCGTCCCGGCGCCT-3′ |
| MGEA5-TPF3 (−407 to +181 bp) | 5′-AATGGTACGTCCCACCGCCG-3′ |
| MGEA5-TPF4 (−48 to +181 bp) | 5′-AGGCAGCGGCTGACAAA-3′ |
| MGEA5-TPR | 5′-TGGCTCTCCTTCTGCACCAT-3′ |
| ChIP primers | |
| mmuOGT-CP-F | 5′-CGGTAGTCTTGTTACGCGCT-3′ |
| mmuOGT-CP-R | 5′-TCTACTGCCGCCACTACTGA-3′ |
| mmuOGA-CP-F | 5′-AAAATGGTACGTCCCACCG-3′ |
| mmuOGA-CP-R | 5′-GCTTCCTGTTTATCCGCACT-3′ |
| huOGT-CP-F | 5′-AAAACGTATCCGTCCCGTGT-3′ |
| huOGT-CP-R | 5′-TGTTACAATCAAGTGCCCGC-3′ |
| huMGEA5-CP-F | 5′-GTAATGGTCCGTCCGTCCC-3′ |
| huMGEA5-CP-R | 5′-CGCTTCCTGTTTATCCGCACT-3′ |
| qPCR primers | |
| OGT-F | 5′-CCTGGGTCGCTTGGAAGA-3′ |
| OGT-R | 5′-TGGTTGCGTCTCAATTGCTTT-3′ |
| MGEA5-F | 5′-CAAGTTGCACACAGTGGAGCTAA-3′ |
| MGEA5-R | 5′-AAAGAGGGTGCAGCAACTAAGG-3′ |
| 18S-F | 5′-CGAACGTCTGCCCTATCAACTT-3′ |
| 18S-R | 5′-ACCCGTGGTCACCATGGT-3′ |
TABLE 2.
Primers and their sequences used to mutate the E2F binding sites in the Ogt (−178 bp/+233 bp) and Oga (−407 bp/+181 bp) reporter plasmids
| Primers | Sequences |
|---|---|
| OGT-E2F mut-F1 | 5′-CGGTCGAGGAGCCCTCGATCCCATTTCAAGACAGTGA-3′ |
| OGT-E2F mut-R1 | 5′-TCACTGTCTTGAAATGGGATCGAGGGCTCCTCGACCG-3′ |
| OGT-E2F mut-F2 | 5′-AGTCACTTGGTCGGTAGTCTTGTTAATCGCTACCTCCGCGG-3′ |
| OGT-E2F mut-R2 | 5′-CCGCGGAGGTAGCCATTAACAAGACTACCGACCAAGTGACT-3′ |
| OGT-E2F mut-F3 | 5′-AGTCATGTTTGTCGCAATGGACCTTATCCGGAAGGTTACGTAAGAG-3′ |
| OGT-E2F mut-R3 | 5′-CTCTTACGTAACCTTCCGGATAAGGTCCATTGCGACAAACATGACT-3′ |
| OGT-E2F mut-F4 | 5′-CATGCGCTCTCTACAGTCATGATCGATCCAATGGACCTTATCCGGAAGG-3′ |
| OGT-E2F mut-R4 | 5′-CCTTCCGGATAAGGTCCATTGGATCGATCATGACTGTAGAGAGCGCATG-3′ |
| MGEA5-E2F mut-F1 | 5′-GCAGGGCTCCGGGCCTTCATCCCAACGGACCGAGG-3′ |
| MGEA5-E2F mut-R1 | 5′-CCTCGGTCCGTTGGGATGAAGGCCCGGAGCCCTGC-3′ |
| MGEA5-E2F mut-F2 | 5′-GAGCCCCGGAGGCATCCGTTCGGAGCCGC-3′ |
| MGEA5-E2F mut-R2 | 5′-GCGGCTCCGAACGGATGCCTCCGGGGCTC-3′ |
| MGEA5-E2F mut-F3 | 5′-GGAGGACGAGCGAAGCTACCTCCTCGGTCCTCC-3′ |
| MGEA5-E2F mut-R3 | 5′-GGAGGACCGAGGAGGTAGCTTCGCTCGTCCTCC-3′ |
| MGEA5-E2F mut-F4 | 5′-GCAATGGGCTTATCCCAAGGAAAAAAGGGGGAGCCGGG-3′ |
| MGEA5-E2F mut-R4 | 5′-CCCGGCTCCCCCTTTTTTCCTTGGGATAAGCCCATTGC-3′ |
| MGEA5-E2F mut-F5 | 5′-GTGTTCACGGCGCCCTTTGTCCTTAAACTCCCCTAGCAAATAAAG-3′ |
| MGEA5-E2F mut-R5 | 5′-CTTTATTTGCTAGGGGAGTTTAAGGACAAAGGGCGCCGTGAACAC-3′ |
| MGEA5-E2F mut-F6 | 5′-ACCTCCGCGGTGTTCACGATCCCCTTTGTCCTTTTTCTC-3′ |
| MGEA5-E2F mut-R6 | 5′-GAGAAAAAGGACAAAGGGGATCGTGAACACCGCGGAGGT-3′ |
Transcription Factor Binding Sites Predictions
Transcription factor binding sites for the delineated Ogt and Mgea5 promoter regions were identified using the MatInspector program. Only predicted transcription factor binding sites with a core matrix similarity of ≥0.95 and an overall matrix similarity of ≥0.90 were considered significant.
Transient Transfection and Luciferase Reporter Assay
HEK293 cells plated in 12-well plates at 70% confluence were transiently transfected with the full-length and 5′ truncations of the Ogt and Mgea5 promoters (500 ng/well) using Lipofectamine 2000 transfection reagent according to the manufacturer's guidelines (Thermo Fisher Scientific). To determine the effect of E2F1, wild type and mutated E2F site 5′-flanking constructs of Ogt (−178 to +233 bp) and Mgea5 (−407 to +181 bp) were cotransfected with E2F1 expression plasmid (pCMVHA E2F1; 500 ng/well) or control plasmid (pCMVHA). The pRL-CMV Renilla luciferase control vector (Promega) was used as the transfection control (20 ng/well). Firefly and Renilla luciferase reporter activities were assessed using the Dual-Luciferase reporter assay system (Promega) 24 h after transfection. Luminescence was detected using the Glomax luminometer (Promega), and the resulting measurements from the firefly luciferase were normalized to the Renilla luciferase. Relative light units were calculated using readouts from the pGL3-basic (promoterless) vector.
Quantitative Real-time PCR Analysis
Total RNA was isolated using TRIzol® reagent according to the manufacturer's instructions (Thermo Fisher Scientific). cDNA synthesis from the extracted RNA was performed using the High Capacity cDNA synthesis kit (Thermo Fisher Scientific). Relative levels of Mgea5 and OGT mRNA were examined with specific primers (Table 1) using FastSYBR Green (Applied Biosystems) and normalized to levels of 18S mRNA. All quantitative RT-PCRs were performed in quadruplicate, and expression level was calculated using the comparative CT (ΔΔCT) method.
Western Blotting
The whole cell lysate from the HEK293, MEF, and MAF cells was separated in 4–12% NuPAGE BisTris gels by electrophoresis, transferred to PVDF membrane, and probed for E2F1, Rb, OGA (200 ng/ml; sc-193, sc-50, and sc-135093, Santa Cruz Biotechnology, Inc.), OGT (500 ng/ml; O6264, Sigma), O-GlcNAc (3 μg/ml; CTD 110.6, Covance), and α-tubulin (500 ng/ml; Sigma) followed by secondary antibody (Santa Cruz Biotechnology) using a standard protocol as described earlier (15). The membranes were exposed with WesternPlus ECL substrate, and the densitometry analysis was performed on a Fuji LAS-3000 bioimaging analyzer.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed using a ChIP assay kit (Millipore) according to the manufacturer's instructions. In brief, 1 × 106 3T3 L1 and HEK293 cells were cross-linked with 1% formaldehyde for 10 min at 37 °C and washed twice with ice-cold PBS containing protease inhibitor mixture (Sigma). The nuclei were isolated using SDS lysis buffer and sonicated to shear the DNA into fragments of 500 bp to 1 kb and diluted in ChIP dilution buffer. The chromatin complexes were immunoprecipitated overnight in a rotator at 4 °C with E2F1 (sc-193 X), polymerase II (sc-899 X), and control IgG (sc-2029) antibodies (Santa Cruz Biotechnology). The immune complexes were incubated with Protein A-agarose/salmon sperm DNA for 1 h at 4 °C with constant rotation and washed sequentially with low salt buffer, high salt buffer, and LiCl buffer. Finally, they were eluted with elution buffer and reverse cross-linked by heating to 65 °C for 4 h followed by incubation with proteinase K at 45 °C for 1 h. The reverse cross-linked input chromatin was used as the control. The DNA was purified using the GenEluteTM PCR cleanup kit (Sigma) and was then subjected to PCR using specific primers (Table 1).
Statistical Analysis
Statistical analyses were conducted using GraphPad Prism version 5.0d software. Student's paired t test was used for comparisons between two groups, or analysis of variance was performed when comparisons were made among three or more groups along with Bonferroni's post hoc analysis. The results were expressed as mean ± S.D., and p < 0.05 was accepted as statistically significant.
Results
Identification of Promoter Regions of Ogt and Mgea5
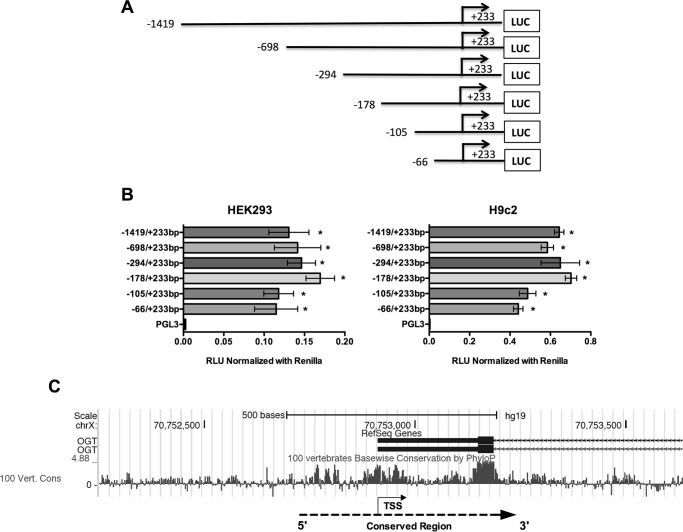
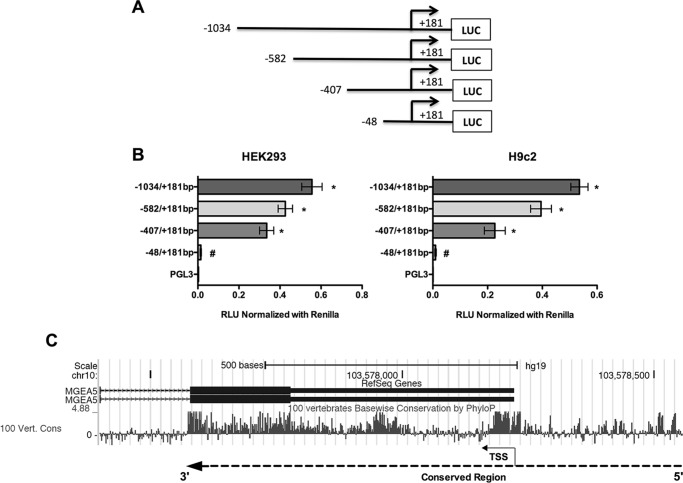
To identify the DNA sequences important for promoter activity of the Ogt and Mgea5 genes, a series of reporter constructs with 5′-flanking regions of mouse Ogt (Chr X; NC_000086.7) and Mgea5 (Chr19; NC_000085.6) were cloned into pGL3-basic luciferase reporter plasmid (Figs. 1A and 2A). Each plasmid having the truncated promoter was transfected into HEK293 and H9c2 cells and analyzed for luciferase activity. From the Ogt 5′-flanking region, the fragment from −66 to +233 bp showed significant reporter activity, and the highest activity was observed with the −178 to +233 bp fragment (Fig. 1B). In the Mgea5 5′-flanking region, the fragment from −48 to +181 bp showed significant reporter activity, which was further enhanced in the −407 to +181 bp fragment (Fig. 2B). These findings suggest that the −66 to +233 bp region in Ogt and the −48 to +181 bp region in Mgea5 are core promoter regions required for expression of these genes. Further 5′-truncations revealed distal promoter elements. Phylogenetic conservation analysis revealed that the regions of Ogt around −200 to +300 bp and around −300 to +600 bp of Mgea5 are highly conserved among vertebrates (Figs. 1C and 2C).
FIGURE 1.
Schematic diagram for the Ogt promoter fragments spanning from −1419 to +233 bp. A, the 5′-truncated DNA fragments were cloned upstream of the firefly luciferase (LUC) reporter gene in the pGL3-basic vector. B, the truncated fragments were transfected into HEK293 and H9c2 cells, and the luciferase activity was measured 24 h post-transfection. The results are presented as relative luciferase units (RLU) (firefly luciferase/Renilla luciferase). Significant promoter activity was observed from −66 to +233 bp. All data are presented as the mean ± S.D. (error bars) from triplicate measurements (p < 0.001). C, phylogenetic conservation analysis of the human OGT promoter (around 1000 bp) using the UCSC Genome Browser (version hg19) showed that the region around −200 to +300 bp is highly conserved among vertebrate species. Positive peaks indicate conserved bases, and the height of the peak denotes level of conservation. Dotted line and arrow, conserved region and gene orientation, respectively.
FIGURE 2.
Schematic diagram for the Mgea5 promoter fragments spanning from −1034 to +181 bp. Mgea5/MGEA5 is the official name of the gene encoding the OGA protein. A, the 5′-truncated DNA fragments were cloned upstream of the firefly luciferase (LUC) reporter gene in the pGL3-basic vector. B, the truncated fragments were transfected in HEK293 and H9c2 cells, and the luciferase activity was measured 24 h post-transfection. The results are presented as relative luciferase units (RLU) (firefly luciferase/Renilla luciferase). Significant promoter activity was observed from −48 to +181 bp. All data are presented as the mean ± S.D. (error bars) from triplicate measurements (p < 0.005). C, phylogenetic conservation analysis of the human MGEA5 promoter (around 1000 bp) using the UCSC Genome Browser (version hg19) showed that the region around −300 to +600 bp of MGEA5 is highly conserved among vertebrate species. Positive peaks indicate conserved bases, and the height of the peak denotes the level of conservation. Dotted line and arrow, conserved region and gene orientation, respectively.
E2F1 Binds to Ogt and Mgea5 Promoters
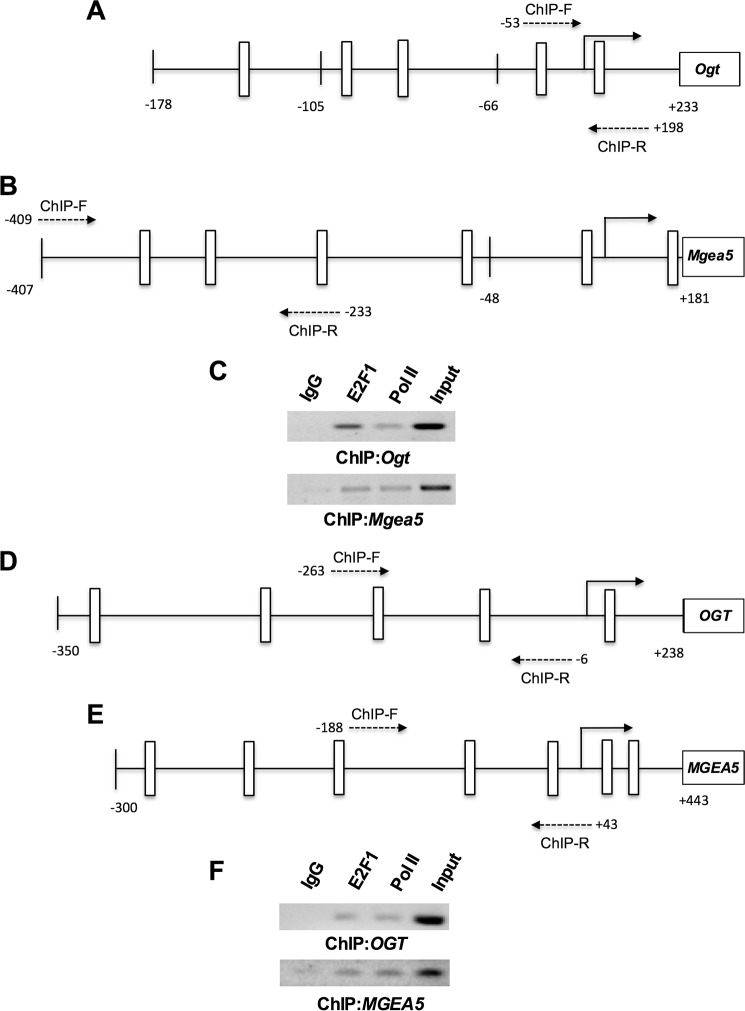
Comprehensive analysis of transcription factors binding to the promoter region of mouse Ogt (−178 to +233 bp) and Mgea5 (−407 to +181 bp) was performed with the MatInspector algorithm. Several ubiquitous transcription factors, such as USF (upstream transcription factor), Sp (specificity protein), AP (activating protein), and E2F transcription factors, were predicted to bind both Ogt and Mgea5 promoter elements. From those, multiple E2F family transcription factor binding sites were observed, leading us to further evaluate the role of E2F transcription factors in Ogt and Mgea5 expression (Fig. 3, A and B). The analysis of transcription factor binding sites in human OGT (−350 to +238 bp) and MGEA5 (−300 to +443) promoter regions showed multiple E2F transcription factor binding sites (Fig. 3, D and E). Among the E2F transcription factors, E2F1 plays an important role in various cellular processes, including metabolism. To determine the transcriptional regulation of the Ogt and Mgea5 promoters by E2F1, we cotransfected the luciferase promoter reporter containing wild type or mutated E2F binding sites (Ogt, 178 to +233 bp; Mgea5, −407 to +181 bp) along with pCMVHA E2F1 or pCMVHA control plasmids. Overexpression of E2F1 significantly decreased luciferase activity in both the wild type Ogt and Mgea5 reporter plasmids but not when E2F binding sites were mutated. We also observed significantly elevated luciferase activity in the mutants compared with their wild type reporter plasmids (Fig. 4, A and B). To further verify the binding of E2F1 to the mouse and human Ogt/OGT and Mgea5/MGEA5 promoter motifs in vivo, ChIP analysis was performed. DNA recovered from the ChIP assay was subjected to PCR amplification containing E2F1 binding sites. As shown in Fig. 3 (C and F), E2F1 was identified to bind directly to both the OGT and MGEA5 promoters in vivo.
FIGURE 3.
E2F binds to Ogt and Mgea5 promoter. A, B, D, and E, E2F transcription factors binding sites for the delineated mouse Ogt (−178/+233 bp) and Mgea5 promoter region (−407/+181 bp) and the human OGT (−350/+238 bp) and MGEA5 promoter region (−300/+443 bp) were identified using the MatInspector program, and the schematic diagram shows possible E2F transcription factor binding sites (white bar). C and F, ChIP analysis was done by incubating a 3T3 L1 and HEK293 chromatin complex with E2F1 antibody, and the PCR analysis for OGT and Mgea5 promoter region showed direct binding of E2F1.
FIGURE 4.
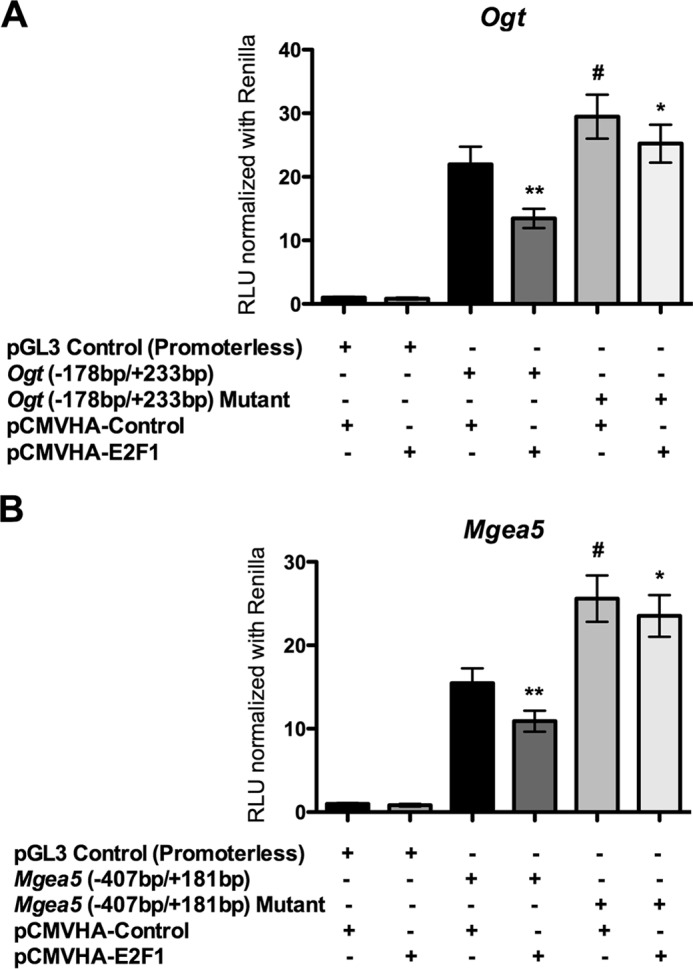
E2F1 reduced reporter activity in truncated Ogt (A) and Mgea5 (B) plasmids. The wild type and mutated E2F binding site pGL3 reporter plasmids of Ogt (−178/+233 bp) and Mgea5 (−407/+181 bp) were cotransfected with E2F1 expression plasmid in HEK293 cells and analyzed for reporter activity. The luciferase assay showed a significant decrease in the reporter activity of wild type Ogt (n = 4; p < 0.01) and Mgea5 (n = 4; p < 0.01) with E2F1 overexpression. The mutation in the E2F binding sites showed significantly increased reporter activity in both Ogt (n = 4; p < 0.05) and Mgea5 (n = 4; p < 0.01), whereas the E2F1 overexpression did not significantly alter luciferase activity of mutant reporter plasmids. RLU, relative luciferase units. Error bars, S.D.
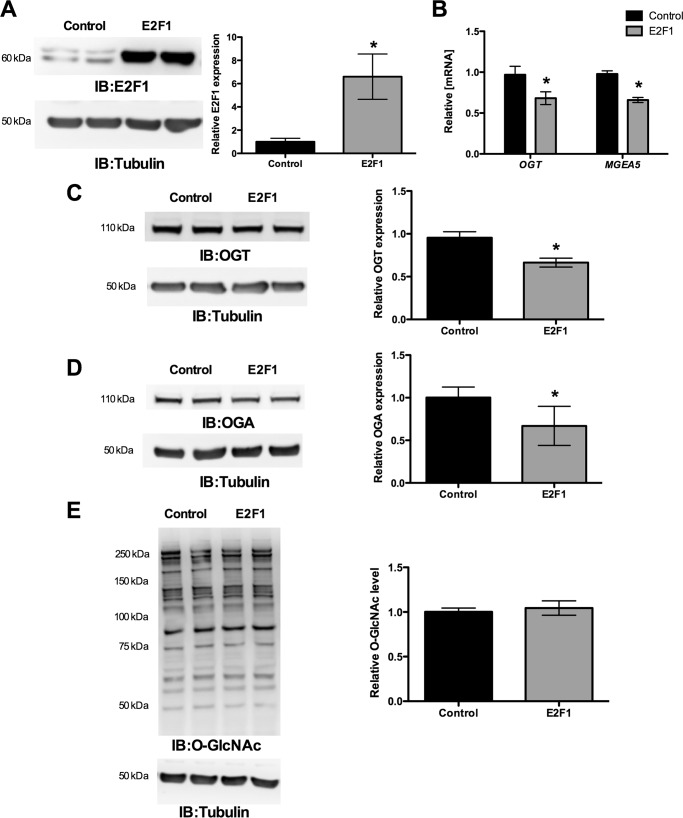
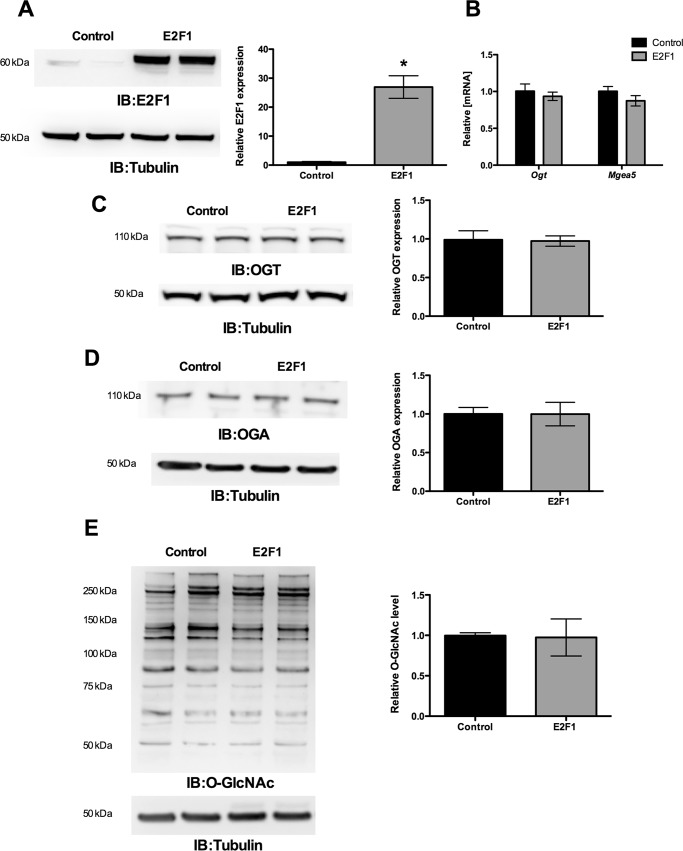
Ectopic Expression of E2F1 Decreased OGT and MGEA5 Expression
To evaluate the effect of the E2F1 transcription factor, the E2F1 expression plasmid construct was transiently transfected in HEK293 cells and analyzed for OGT and OGA expression. Fig. 5A shows significantly increased E2F1 expression upon transfection. The qPCR results for OGT and MGEA5 showed significantly decreased mRNA levels at 12 h compared with the control vector (Fig. 5B). Western blot analysis was performed after 48 h of transfection and showed decreased OGT and OGA protein expression compared with the control vector (Fig. 5, C and D). These results indicate that E2F1 represses OGT and MGEA5 expression at both mRNA and protein levels. We observed that the O-GlcNAc level was not altered upon changes in the OGT and OGA level by E2F1 expression (Fig. 5E).
FIGURE 5.
E2F1 suppresses OGT and OGA (MGEA5) expression. E2F1 was overexpressed in HEK293 cells, and the expression of OGT and MGEA5 mRNA and protein levels were measured. A, increased E2F1 expression was observed upon transfecting the pCMV-E2F1 expression plasmid. B, qPCR analysis showed that ectopic expression of E2F1 significantly decreased OGT and MGEA5 mRNA level after 12 h of transfection (n = 4; p < 0.005). C and D, Western blot analysis (IB) showed a significant decrease in OGT (n = 4; p < 0.01) and OGA (n = 4; p < 0.05) protein level after a 48-h transfection. E, proteins from E2F1-overexpressing and control HEK293 cells were probed for O-GlcNAc Western blot that showed the same level of O-GlcNAcylation. Error bars, S.D.
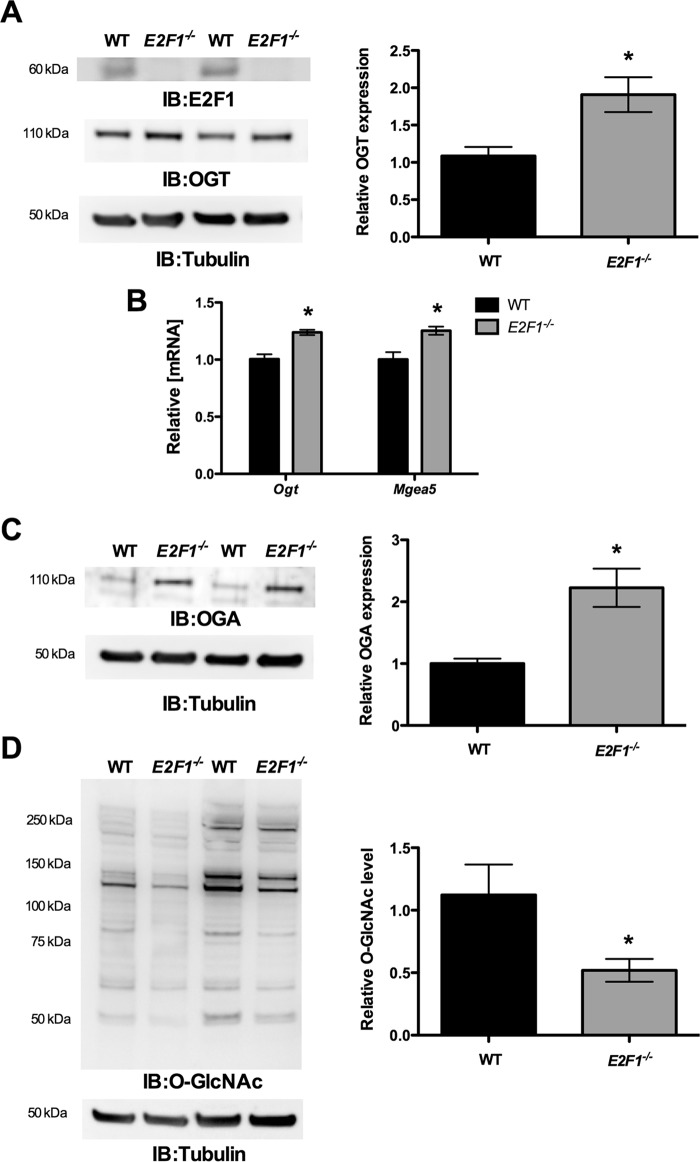
Deletion of E2F1 Increased Ogt and Mgea5 Expression
To further examine the effect of E2F1 on OGT and OGA expression, we used MAFs that lack E2F1. Western blot analysis confirmed the absence of E2F1 in the E2F1−/− MAFs (Fig. 6A). WT and E2F1−/− MAFs were grown in normal culture media to around 70% confluence, and the RNA and protein were analyzed for Ogt and Mgea5 expression. The qPCR analysis showed significantly elevated Ogt and Mgea5 mRNA expression in E2F1−/− cells compared with WT MAFs (Fig. 6B). Similarly, immunoblotting revealed significantly increased levels of both OGT and OGA protein in E2F1−/− cells compared with WT MAFs (Fig. 6, A and C). Interestingly, we observed significantly reduced O-GlcNAcylation in the E2F1−/− cells compared with WT cells (Fig. 6D).
FIGURE 6.
Knockdown of E2F1 increased OGT and OGA expression. Wild type and E2F1−/− MAFs were cultured and analyzed for Ogt and Mgea5 expression. A, Western blot analysis confirmed the lack of E2F1 expression in E2F1−/− MAFs. A–C, qPCR and Western blot analysis (IB) showed significantly increased expression of Ogt and Mgea5 mRNA (n = 4; p < 0.005) and protein levels (n = 3; p < 0.05) in E2F1−/− compared with WT MAFs. Error bars, S.D. *, p < 0.05.
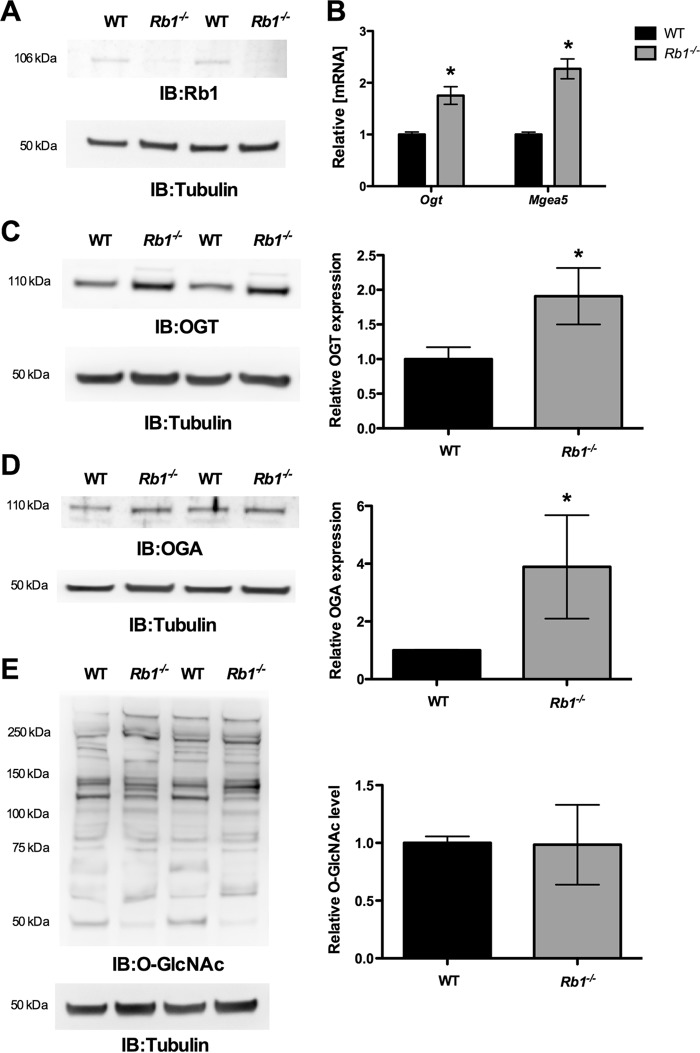
Rb1 Required for E2F1-mediated Ogt and Mgea5 Repression
To determine the role of Rb1 in Ogt and Mgea5 expression, we assessed Ogt and Mgea5 levels in wild type and Rb1-deficient MEFs. Western blot analysis confirmed the loss of Rb1 expression in Rb1−/− MEFs (Fig. 7A). The qPCR analysis showed significantly elevated Ogt and Mgea5 mRNA expression in Rb1−/− cells compared with WT cells (Fig. 7B). Western blot analysis showed a significant increase in both OGT and OGA protein levels in Rb1−/− cells compared with WT cells (Fig. 7, C and D); however, global protein O-GlcNAcylation was not altered in the Rb1−/− cells compared with WT cells (Fig. 7E). To further analyze whether Rb1 is required for E2F1-mediated repression, we overexpressed E2F1 in the Rb1−/− cells and evaluated the expression of Ogt and Mgea5. Fig. 8A shows a significant increase expression of E2F1 in Rb1−/− cells that were transiently transfected with the E2F1 plasmid. The mRNA and protein levels of both Ogt and Mgea5 were not altered with the expression of E2F1 in Rb1−/− cells compared with control plasmid-transfected cells (Fig. 8, B, C, and D). Western blot analysis showed unaltered protein O-GlcNAcylation in the E2F1-transfected Rb1−/− cells compared with control (Fig. 8E).
FIGURE 7.
Loss of Rb1 increased Ogt and Mgea5 expression. MEFs that lack Rb1 were compared with the WT cells for the expression of Ogt and Mgea5. A, Western blot analysis (IB) showed significant loss of Rb1 in Rb1−/− cells compared with WT MEFs. B, significant increase in the Ogt (n = 4; p < 0.001) and Mgea5 (n = 4; p < 0.0001) mRNA level was observed in Rb1−/− MEFs. C and D, Western blot analysis showed a significant increase in the OGT (n = 3; p < 0.005) and OGA (n = 3; p < 0.05) protein level, whereas the O-GlcNAc level was not changed (E). Error bars, S.D. *, p < 0.05.
FIGURE 8.
E2F1 did not suppress OGT and OGA expression in the absence of Rb1. Rb1−/− MEFs were transfected with E2F1 and analyzed for the expression of Ogt and Mgea5. A, Western blot analysis (IB) showed significant expression of E2F1 expression in Rb1−/− MEFs (n = 4; p < 0.0001). B–D, Ogt and Mgea5 mRNA (n = 4) and protein levels (n = 3) were not changed upon E2F1 expression in Rb1−/− cells compared with control. E, proteins from E2F1-overexpressing and control Rb1−/− cells were probed for O-GlcNAc Western blot that showed the same level of O-GlcNAcylation. Error bars, S.D. *, p < 0.05.
Discussion
The dynamic reversible O-GlcNAc modification is a key regulator of various cellular processes from cellular signaling to development. O-GlcNAcylation is a cytoprotective response in various cell types, including cancer cells under metabolic and oxidative stress (4, 28, 29). In addition to changes in the glucose flux, alterations in OGT/OGA protein expression occur in chronic diseases, such as heart failure, diabetes, cancer, and neurodegenerative disorders (12, 15, 30, 31). Thus, delineation of promoter elements for Ogt and Mgea5 and evaluation of the transcription factors by which these promoter elements are regulated remain an unmet need.
In the present study, conserved putative transcription binding motifs were analyzed between humans and mice, which showed conserved E2F transcription factor binding sites in both Ogt and Mgea5. An in vivo ChIP assay demonstrated the direct binding of E2F1 transcription factor to the Ogt and Mgea5 promoters. Overexpression of E2F1 could repress exogenous luciferase reporter activity and endogenous protein levels. Constitutive knock-out of E2F1 or Rb1 led to increased expression of Ogt and Mgea5. Taken together, our results demonstrated the basal promoter elements of Ogt and Mgea5 and highlighted the potential role of E2F1 transcription factor in their regulation.
In addition, we report Ogt and Mgea5 promoter elements and the DNA sequences required for their basal expression. The Ogt promoter has a TATA box (TATAAC) immediately upstream of the TSS (−13 bp) in the proximal region, whereas the Mgea5 promoter does not. That truncation with the TATA box significantly increased reporter activity suggests that the TATA box facilitates transcription of Ogt. On the other hand, the significant reporter activity of the TATA-less region (−48/+181 bp) in the Mgea5 promoter element showed that the TATA box might not be required for the basal transcription of Mgea5. We noticed that the Mgea5 core promoter is rich in G/C content. Genome-wide analysis of mammalian promoters revealed that only a minority (<10%) of genes have a classical TATA box (32). In the present study, we defined the regions from −66 to +233 bp of Ogt and from −48 to +181 bp of Mgea5 as the core promoters. In addition, we analyzed possible transcription factors bound to the promoter regions and identified E2F1 as a repressor for both Ogt and Mgea5.
A myriad of cellular proteins involved in critical functions like mitosis, DNA repair, development, and differentiation are controlled by ubiquitously expressed E2F family members (33–35). As a metabolic/stress sensor/signal, protein O-GlcNAcylation is a key part of numerous cellular processes, making it critical to understand how Ogt and Mgea5 are regulated by E2F transcription factors that have shared DNA binding elements. Previous ChIP on chip analysis in different cell types showed common consensus sequences (TTTSSCGC or BKTSSCGS, where S represents C or G, B is C or G or T, and K is G or T) for E2F transcription factors in many targets (36, 37). Ogt and Mgea5 genes are predicted to have E2F transcription factor binding sites in their core and distal promoter elements, and in vivo ChIP analysis confirmed the binding of E2F1 to Ogt and Mgea5 promoter regions.
In general, E2F1 is classified as an activator protein that increases the transcription of genes. In our study, overexpression of E2F1 showed decreased levels of OGT and OGA protein expression, which suggests that E2F1 may act as a repressor in Ogt and Mgea5 regulation. It is well documented that association of retinoblastoma proteins (pRB/Rb1, p107, and p130) with E2F inhibits the expression of E2F-regulated genes by blocking its binding to promoters or by recruitment of repressors like histone deacetylases and methyltransferase (38, 39). In our present study, we observed a significant reduction in the reporter activity by overexpression of E2F1 with Ogt and Mgea5 promoter reporters. In addition, mutation of E2F binding sites in the reporter constructs increased reporter activity, which suggest that E2F1 has a suppressive effect on Ogt and Mgea5 promoters. The significantly reduced OGT and Mgea5 transcript and protein levels upon ectopic E2F1 expression confirm that E2F1 plays an important role in the regulation of Ogt and Mgea5 expression. Recently, Wells et al. (40) reported that during the G1 phase of the cell cycle, E2F1-associated pRB is O-GlcNAc modified during hypophosphorylated conditions. Others reported that O-GlcNAcylation was increased in a serum-stimulated cell cycle and deceased upon release from G2/M arrest (11). Taken together, these findings suggest that the activity of E2F1 could be regulated by O-GlcNAcylated or phosphorylated pRB, thereby tightly regulating Ogt and Mgea5 expression. Increased expression of Ogt and Mgea5 (mRNA and protein) in the E2F1−/− MAFs and Rb1−/− MEFs confirmed that both E2F1 and Rb1 play an important role in regulating Ogt and Mgea5 expression. It is interesting to note the significant reduction in O-GlcNAcylation in E2F1−/− MAFs, which could be ascribed to higher expression of OGA protein (2.2-fold) over that of OGT (1.8-fold) compared with WT cells. Because E2F1 deficiency could have other effects that indirectly affect O-GlcNAcylation, the aforementioned explanation remains speculative. Although global protein O-GlcNAcylation is not changed in Rb1−/− MEFs, the Rb1−/− MEFs exhibit apparently different patterns of O-GlcNAcylation compared with WT MEFs (Fig. 7E), which might be due to alteration in the specific protein expression or O-GlcNAcylation per se. Further studies are required to clarify whether and how Rb1 association interferes with E2F1 activity on Ogt and Mgea5 expression under various stress and cellular states and to evaluate whether the alteration of O-GlcNAcylation affects E2F1 and its interaction with other associated proteins.
In conclusion, we characterized the promoter elements of mouse Ogt and Mgea5 and identified E2F1 as an important transcription factor that regulates Ogt and Mgea5 expression by binding directly to their promoter regions. It is important to determine the effects of other E2F family members and to evaluate the transcription factors that regulate Ogt and Mgea5 expression. Our findings offer new insights into transcriptional regulation of Ogt and Mgea5, which may be explored to further our understanding of disease context-specific pathophysiology.
Author Contributions
S. P. J. conceived the concept of the paper. S. P. J., T. H., K. U. H., and S. M. designed the study. S. P. J. and S. M. designed experiments and wrote the paper. S. M. carried out the majority of the experiments and analyzed the data. S. D. provided technical assistance in Western blot and qPCR experiments. All authors had access to the results and approved the final version of the manuscript.
Acknowledgments
We are grateful to Dr. Douglas C. Dean (University of Louisville) for providing wild type and E2F1−/− MAFs and Dr. Brian F. Clem (University of Louisville) for supplying wild type and Rb1−/− MEFs. We are also grateful for the expert technical assistance of Bethany W. Long and Linda T. Harrison.
This work was supported by National Institutes of Health Grants R01 HL083320, R01 HL094419, P20 GM103492, and P01 HL078825 (to S. P. J.) and American Heart Association (Great Rivers Affiliate) Predoctoral Fellowship 14PRE19710015 (to S. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- O-GlcNAc
- O-linked N-acetylglucosamine
- OGA
- O-GlcNAcase protein
- MGEA5
- meningioma expressed antigen 5 (the gene for O-GlcNAcase)
- OGT/Ogt
- O-GlcNAc transferase protein/gene
- E2F
- E2 transcription factor
- HEK
- human embryonic kidney
- MAF
- mouse adult fibroblast
- MEF
- mouse embryonic fibroblast
- Rb1
- retinoblastoma 1
- TSS
- transcriptional start site
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- qPCR
- quantitative PCR.
References
- 1. Watson L. J., Facundo H. T., Ngoh G. A., Ameen M., Brainard R. E., Lemma K. M., Long B. W., Prabhu S. D., Xuan Y. T., and Jones S. P. (2010) O-Linked β-N-acetylglucosamine transferase is indispensable in the failing heart. Proc. Natl. Acad. Sci. U.S.A. 107, 17797–17802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Slawson C., Copeland R. J., and Hart G. W. (2010) O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci. 35, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yuzwa S. A., and Vocadlo D. J. (2014) O-GlcNAc and neurodegeneration: biochemical mechanisms and potential roles in Alzheimer's disease and beyond. Chem. Soc. Rev. 43, 6839–6858 [DOI] [PubMed] [Google Scholar]
- 4. Ma Z., and Vosseller K. (2014) Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J. Biol. Chem. 289, 34457–34465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hart G. W. (2014) Minireview series on the thirtieth anniversary of research on O-GlcNAcylation of nuclear and cytoplasmic proteins: nutrient regulation of cellular metabolism and physiology by O-GlcNAcylation. J. Biol. Chem. 289, 34422–34423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu P., Shimoji S., and Hart G. W. (2010) Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 584, 2526–2538 [DOI] [PubMed] [Google Scholar]
- 7. Facundo H. T., Brainard R. E., Watson L. J., Ngoh G. A., Hamid T., Prabhu S. D., and Jones S. P. (2012) O-GlcNAc signaling is essential for NFAT-mediated transcriptional reprogramming during cardiomyocyte hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 302, H2122–H2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zachara N. E., and Hart G. W. (2006) Cell signaling, the essential role of O-GlcNAc!. Biochim. Biophys. Acta 1761, 599–617 [DOI] [PubMed] [Google Scholar]
- 9. Srikanth B., Vaidya M. M., and Kalraiya R. D. (2010) O-GlcNAcylation determines the solubility, filament organization, and stability of keratins 8 and 18. J. Biol. Chem. 285, 34062–34071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shafi R., Iyer S. P., Ellies L. G., O'Donnell N., Marek K. W., Chui D., Hart G. W., and Marth J. D. (2000) The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang Y. R., Song M., Lee H., Jeon Y., Choi E. J., Jang H. J., Moon H. Y., Byun H. Y., Kim E. K., Kim D. H., Lee M. N., Koh A., Ghim J., Choi J. H., Lee-Kwon W., Kim K. T., Ryu S. H., and Suh P. G. (2012) O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell 11, 439–448 [DOI] [PubMed] [Google Scholar]
- 12. Perez-Cervera Y., Dehennaut V., Aquino Gil M., Guedri K., Solórzano Mata C. J., Olivier-Van Stichelen S., Michalski J. C., Foulquier F., and Lefebvre T. (2013) Insulin signaling controls the expression of O-GlcNAc transferase and its interaction with lipid microdomains. FASEB J. 27, 3478–3486 [DOI] [PubMed] [Google Scholar]
- 13. Zhang Z., Tan E. P., VandenHull N. J., Peterson K. R., and Slawson C. (2014) O-GlcNAcase expression is sensitive to changes in O-GlcNAc homeostasis. Front. Endocrinol. 5, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Park K., Saudek C. D., and Hart G. W. (2010) Increased expression of β-N-acetylglucosaminidase in erythrocytes from individuals with pre-diabetes and diabetes. Diabetes 59, 1845–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muthusamy S., DeMartino A. M., Watson L. J., Brittian K. R., Zafir A., Dassanayaka S., Hong K. U., and Jones S. P. (2014) MicroRNA-539 is up-regulated in failing heart and suppresses O-GlcNAcase expression. J. Biol. Chem. 289, 29665–29676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., and Werner T. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 17. Attwooll C., Lazzerini Denchi E., and Helin K. (2004) The E2F family: specific functions and overlapping interests. EMBO J. 23, 4709–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du W., and Pogoriler J. (2006) Retinoblastoma family genes. Oncogene 25, 5190–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cobrinik D. (2005) Pocket proteins and cell cycle control. Oncogene 24, 2796–2809 [DOI] [PubMed] [Google Scholar]
- 20. Kong L. J., Chang J. T., Bild A. H., and Nevins J. R. (2007) Compensation and specificity of function within the E2F family. Oncogene 26, 321–327 [DOI] [PubMed] [Google Scholar]
- 21. Fajas L., Landsberg R. L., Huss-Garcia Y., Sardet C., Lees J. A., and Auwerx J. (2002) E2Fs regulate adipocyte differentiation. Dev. Cell 3, 39–49 [DOI] [PubMed] [Google Scholar]
- 22. Lukas J., Petersen B. O., Holm K., Bartek J., and Helin K. (1996) Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol. Cell. Biol. 16, 1047–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stevens C., and La Thangue N. B. (2003) E2F and cell cycle control: a double-edged sword. Arch. Biochem. Biophys. 412, 157–169 [DOI] [PubMed] [Google Scholar]
- 24. Wu Z., Zheng S., and Yu Q. (2009) The E2F family and the role of E2F1 in apoptosis. Int. J. Biochem. Cell Biol. 41, 2389–2397 [DOI] [PubMed] [Google Scholar]
- 25. Crowe D. L., Nguyen D. C., Tsang K. J., and Kyo S. (2001) E2F-1 represses transcription of the human telomerase reverse transcriptase gene. Nucleic Acids Res. 29, 2789–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Croxton R., Ma Y., Song L., Haura E. B., and Cress W. D. (2002) Direct repression of the Mcl-1 promoter by E2F1. Oncogene 21, 1359–1369 [DOI] [PubMed] [Google Scholar]
- 27. Blanchet E., Annicotte J. S., Lagarrigue S., Aguilar V., Clapé C., Chavey C., Fritz V., Casas F., Apparailly F., Auwerx J., and Fajas L. (2011) E2F transcription factor-1 regulates oxidative metabolism. Nat. Cell Biol. 13, 1146–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zafir A., Readnower R., Long B. W., McCracken J., Aird A., Alvarez A., Cummins T. D., Li Q., Hill B. G., Bhatnagar A., Prabhu S. D., Bolli R., and Jones S. P. (2013) Protein O-GlcNAcylation is a novel cytoprotective signal in cardiac stem cells. Stem cells 31, 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hardivillé S., and Hart G. W. (2014) Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 20, 208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taylor R. P., Parker G. J., Hazel M. W., Soesanto Y., Fuller W., Yazzie M. J., and McClain D. A. (2008) Glucose deprivation stimulates O-GlcNAc modification of proteins through up-regulation of O-linked N-acetylglucosaminyltransferase. J. Biol. Chem. 283, 6050–6057 [DOI] [PubMed] [Google Scholar]
- 31. Förster S., Welleford A. S., Triplett J. C., Sultana R., Schmitz B., and Butterfield D. A. (2014) Increased O-GlcNAc levels correlate with decreased O-GlcNAcase levels in Alzheimer disease brain. Biochim. Biophys. Acta 1842, 1333–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C. A., Taylor M. S., Engström P. G., Frith M. C., Forrest A. R., Alkema W. B., Tan S. L., Plessy C., Kodzius R., Ravasi T., Kasukawa T., Fukuda S., Kanamori-Katayama M., Kitazume Y., Kawaji H., Kai C., Nakamura M., Konno H., Nakano K., Mottagui-Tabar S., Arner P., Chesi A., Gustincich S., Persichetti F., Suzuki H., Grimmond S. M., Wells C. A., Orlando V., Wahlestedt C., Liu E. T., Harbers M., Kawai J., Bajic V. B., Hume D. A., and Hayashizaki Y. (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38, 626–635 [DOI] [PubMed] [Google Scholar]
- 33. Stevaux O., and Dyson N. J. (2002) A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14, 684–691 [DOI] [PubMed] [Google Scholar]
- 34. Ouseph M. M., Li J., Chen H. Z., Pécot T., Wenzel P., Thompson J. C., Comstock G., Chokshi V., Byrne M., Forde B., Chong J. L., Huang K., Machiraju R., de Bruin A., and Leone G. (2012) Atypical E2F repressors and activators coordinate placental development. Dev. Cell 22, 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo R., Chen J., Zhu F., Biswas A. K., Berton T. R., Mitchell D. L., and Johnson D. G. (2010) E2F1 localizes to sites of UV-induced DNA damage to enhance nucleotide excision repair. J. Biol. Chem. 285, 19308–19315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rabinovich A., Jin V. X., Rabinovich R., Xu X., and Farnham P. J. (2008) E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res. 18, 1763–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Prüss M., Reuter I., and Schacherer F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 28, 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu Z., Luo R. Z., Peng H., Huang M., Nishmoto A., Hunt K. K., Helin K., Liao W. S., and Yu Y. (2006) E2F-HDAC complexes negatively regulate the tumor suppressor gene ARHI in breast cancer. Oncogene 25, 230–239 [DOI] [PubMed] [Google Scholar]
- 39. Valdez C. D., Davis J. N., Odeh H. M., Layfield T. L., Cousineau C. S., Berton T. R., Johnson D. G., Wojno K. J., and Day M. L. (2011) Repression of androgen receptor transcription through the E2F1/DNMT1 axis. PLoS One 6, e25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wells L., Slawson C., and Hart G. W. (2011) The E2F-1 associated retinoblastoma-susceptibility gene product is modified by O-GlcNAc. Amino Acids 40, 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]