Abstract
Populations of an organism living in marked geographical or evolutionary isolation from other populations of the same species are often termed subspecies and expected to show some degree of genetic distinctiveness. The common chimpanzee (Pan troglodytes) is currently described as four geographically delimited subspecies: the western (P. t. verus), the nigerian-cameroonian (P. t. ellioti), the central (P. t. troglodytes) and the eastern (P. t. schweinfurthii) chimpanzees. Although these taxa would be expected to be reciprocally monophyletic, studies have not always consistently resolved the central and eastern chimpanzee taxa. Most studies, however, used data from individuals of unknown or approximate geographic provenance. Thus, genetic data from samples of known origin may shed light on the evolutionary relationship of these subspecies. We generated microsatellite genotypes from noninvasively collected fecal samples of 185 central chimpanzees that were sampled across large parts of their range and analyzed them together with 283 published eastern chimpanzee genotypes from known localities. We observed a clear signal of isolation by distance across both subspecies. Further, we found that a large proportion of comparisons between groups taken from the same subspecies showed higher genetic differentiation than the least differentiated between-subspecies comparison. This proportion decreased substantially when we simulated a more clumped sampling scheme by including fewer groups. Our results support the general concept that the distribution of the sampled individuals can dramatically affect the inference of genetic population structure. With regard to chimpanzees, our results emphasize the close relationship of equatorial chimpanzees from central and eastern equatorial Africa and the difficult nature of subspecies definitions.
Keywords: genetic differentiation, isolation by distance, structure, microsatellites, genotyping
INTRODUCTION
Understanding the distribution of genetic variation within an endangered species such as the common chimpanzee, Pan troglodytes, is crucial for elucidating its evolutionary history (Morin et al., 1994; Stone et al., 2002; Gonder et al., 2006; Becquet et al., 2007), especially because there is an almost complete absence of a fossil record (McBrearty and Jablonski, 2005). In addition, knowledge of population relationships might also facilitate the use of the limited resources available for conservation efforts (Schonewald-Cox et al., 1983; Avise, 1996) and help in guiding breeding programs of chimpanzees kept in captivity (reviewed in Witzenberger and Hochkirch, 2011; Hvilsom et al., 2013).
Arguing on the basis of morphological or genetic evidence and consideration of geographic distribution, researchers have separated Pan troglodytes into three different subspecies: the western chimpanzee (P. t. verus), the central chimpanzee (P. t. troglodytes), and the eastern chimpanzee (P. t. schweinfurthii) (e.g. Hill, 1969; Morin et al., 1994; Groves, 2001; Becquet et al., 2007). Later, the existence of a fourth subspecies, the Nigerian-Cameroonian chimpanzee, P. t. ellioti (Oates et al., 2009), originally termed P. t. vellerosus, was suggested (Gonder et al., 1997). The validity of that taxon was supported by studies utilizing mitochondrial DNA (mtDNA) (Gonder et al., 1997; Gagneux et al., 1999; Gonder et al., 2006; Bjork et al., 2010), microsatellite loci (Gonder et al., 2011), autosomal sequence data (Bowden et al., 2012) as well as complete genomes (Prado-Martinez et al., 2013). Thus, the currently most widely accepted taxonomy of the common chimpanzee splits them into four geographically defined subspecies (Fig. 1), separated from each other from west to east by the Dahomey gap, the Sanaga River and the Ubangi River.
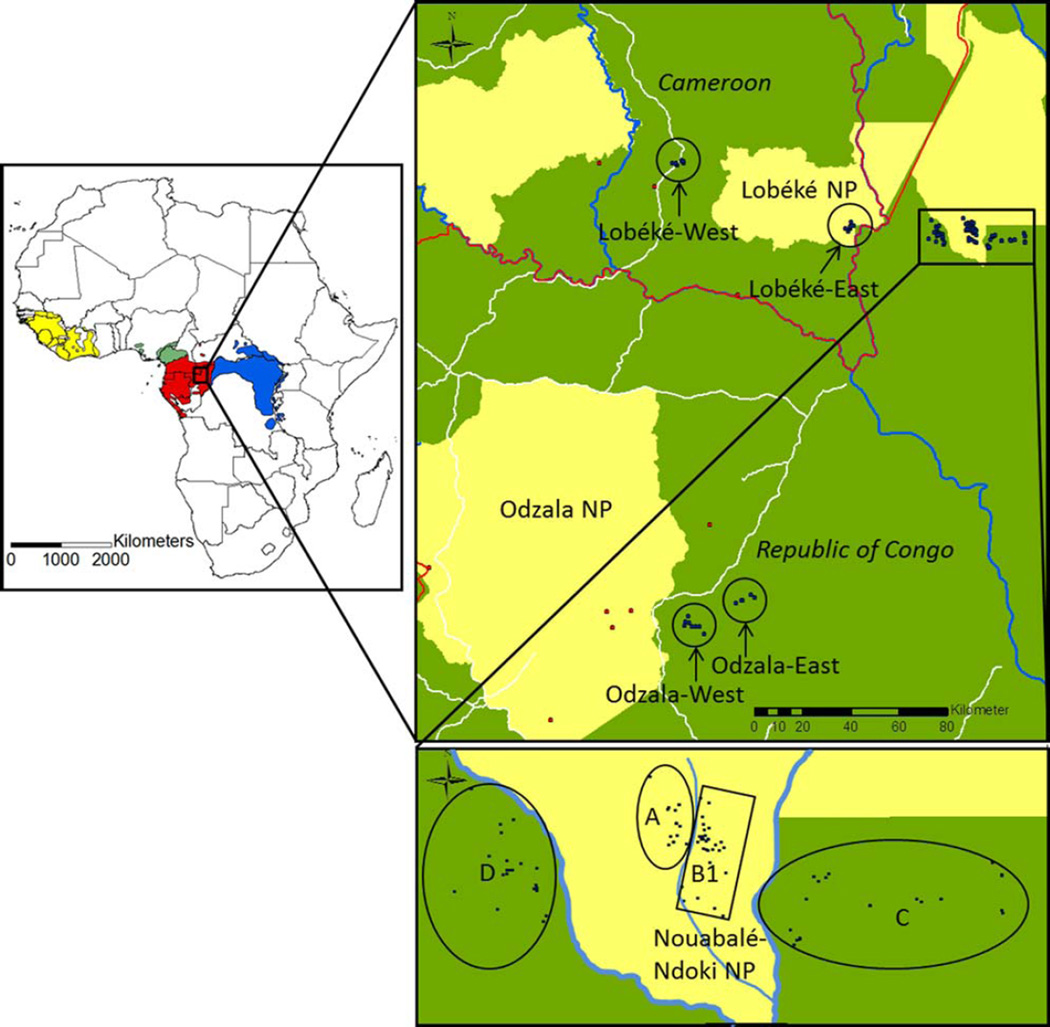
Fig. 1.
The map of Africa on the left side indicates the ranges of chimpanzee subspecies (yellow: P. t. verus, green: P. t. ellioti, red: P. t. troglodytes, blue: P. t. schweinfurthii). The zoomed in maps on the right illustrate the region sampled for the present study with blue and red dots depicting sampling locations. The red dots refer to geographically outlying samples that were not assigned to a certain group for calculations of IBD and genetic differentiation (see Methods). National borders are in red, national parks in yellow, rivers in blue and roads in white. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
However, this classification is not without controversy. Fischer et al. (2006) generally questioned the subspecies concept in Pan troglodytes. Using nuclear nonrepetitive DNA sequences they found an extent of genetic differentiation among subspecies comparable to that seen among human populations. They speculated that a more geographically-informed sampling would reveal a pattern of isolation by distance (IBD) as has been described for within-continental variation in humans (Serre and Pääbo, 2004; Rosenberg et al., 2005; Lao et al., 2008; Tishkoff et al., 2009). IBD describes the situation in which individuals found closer together are genetically more similar than those further apart, and it arises when the distance an individual may disperse is smaller than the continuous distribution of the organism (Wright, 1943). To detect such a cline in genetic variation one needs to use samples of known geographic origin sampled according to a pattern that is as little clumped and as widespread as possible. However, most studies that detected clearly separated subspecies of Pan troglodytes used, at least partly, samples with unknown exact origins or rather clustered sampling schemes (Becquet et al., 2007; Bowden et al., 2012; Prado-Martinez et al., 2013).
Although not challenging the concept of chimpanzee subspecies in general, some authors have suggested that central and eastern chimpanzees do not represent geographically separated subspecies, but might rather form one taxon of equatorial chimpanzees. Specifically, morphological studies found a close dental (Pilbrow, 2006) and cranial (Guy et al., 2003) similarity between P. t. troglodytes and P. t. schweinfurthii. Studies describing the distribution of genetic variation within chimpanzees could potentially evaluate the evolutionary independence of these taxa by investigating if monophyletic clades of central and eastern chimpanzees are found. Gagneux et al. (1999), analyzing mtDNA sequences of 154 eastern and 24 central chimpanzees, found the eastern clade to be nested within the central clade, thereby raising the question if these two subspecies should not be regarded as one clade of equatorial chimpanzees. Later studies including more mtDNA haplotypes again did not find consistent support for monophyly of P. t. troglodytes and P. t. schweinfurthii (Gagneux et al., 2001; Gonder et al., 2006). In addition, in the study by Gonder et al. (2006) no fixed nucleotide differences distinguishing the haplotypes of central and eastern chimpanzees were detected.
These findings were, however, based on the use of a single genetic locus. A more comprehensive determination of the phylogeographic history of a species might be obtained by use of multiple autosomal loci. Gonder et al. (2011) characterized 94 chimpanzees from all four subspecies at 27 autosomal microsatellite loci. In accordance with their mtDNA results (Gonder et al., 2006) central and eastern chimpanzees were found as one population of equatorial chimpanzees. Even though some structuring into three different subpopulations was visible within equatorial chimpanzees, the genetic differentiation between these was low and insignificant. Thus, they speculated that, until recently, the equatorial chimpanzees displayed a geographical gradient of allele frequencies with ongoing gene flow between groups (Gonder et al., 2011). Fischer et al. (2011) investigated the genetic variation in chimpanzees and bonobos using DNA sequences from 15 autosomal regions totaling some 1,50,000 base pairs. Central and eastern chimpanzees did not form monophyletic groups for any of the 15 loci.
Multiple studies (Becquet et al., 2007; Fischer et al., 2011; Gonder et al., 2011; Bowden et al., 2012) that aimed at determining the genetic structure of chimpanzees have made use of Bayesian based clustering algorithms implemented in software such as STRUCTURE (Pritchard et al., 2000). This approach has the advantage of not requiring a priori assignment of individuals into delineated populations. However, datasets should be tested for an underlying pattern of IBD before relying on its results, because there is considerable evidence that IBD alone or in combination with a clumped sampling pattern can lead clustering algorithms to overestimate the number of populations. In simulations conducted by Schwartz and McKelvey (2009), STRUCTURE correctly inferred the existence of only one population in datasets with gradual genetic variation when the samples were randomly distributed. However, when they modeled a clumped sampling scheme they accordingly detected multiple clusters using STRUCTURE. By using subsamples of an extensive empirical dataset from Rosenberg et al. (2002), Serre and Pääbo (2004) revealed that in humans, whose genetic variation exhibits typically clinal variation in allele frequencies (Cavalli-Sforza et al., 1994), clusters are less apparent when the sampling is more evenly distributed geographically (but see also Rosenberg et al., 2005). Moreover, Frantz et al. (2009), showed that even with a random sampling pattern all Bayesian programs (including STRUCTURE) detected more than one cluster in datasets that did not contain any genetic discontinuities, but did contain high levels of IBD. Thus, results obtained by STRUCTURE reported in studies that also found IBD or did not test for IBD should be treated with caution.
For the present study noninvasively collected fecal samples of 185 central and published genotypes from 283 eastern chimpanzees of known geographic origin were used. The origins of the individuals were as continuously distributed as was feasible and included central and eastern chimpanzees from their western- and eastern-most range, respectively. We generated genotypes at 12 microsatellite loci and analyzed them by applying multiple methods. We used a Bayesian based clustering algorithm implemented in STRUCTURE as well as assessments of genetic differentiation, including an analysis of IBD, to ask whether our data support a distinction between central and eastern chimpanzees as in the traditional chimpanzee taxonomy or rather if these may be better described as one taxon of equatorial chimpanzees.
METHODS
Study area and sample collection
Fecal samples from wild central chimpanzees were obtained within and near Nouabalé-Ndoki National Park (NP), Odzala NP (both Republic of Congo), and Lobéké NP (Cameroon) (Fig. 1). Samples were collected noninvasively between 1999 and 2013 and stored in RNAlater (samples from Lobéké) or processed by short term storage in ethanol followed by preservation with silica gel (samples from Odzala and the Nouabalé-Ndoki NP) (Nsubuga et al., 2004).
DNA extraction, quantification, and amplification
We extracted DNA from fecal samples using the QIAamp Stool kit (QIAGEN) following manufacturer’s instructions with slight modifications (Nsubuga et al., 2004). To estimate the amount of amplifiable DNA in the extracts we used a real-time quantitative PCR with Maxima SYBR® Green (Thermo Scientific) as Master Mix and c-myc_E3_F1U1 plus c-myc_E3_R1U1 (Morin et al., 2001) as primers to amplify a portion of the c-myc gene. Standard curves were constructed from serially diluted human genomic DNA (BIO-35025, Bioline). Initially, three independent amplifications from each extract were performed at 12 microsatellite loci (D3s2459, D3s3038, D4s1627, D5s1470, D6s1056, D7s817, D7s2204, D10s676, D11s2002, D12s66, D14s306, D18s536), using a two-step multiplex PCR (Arandjelovic et al., 2009). For sex determination we also amplified a segment of the X–Y homologous amelogenin gene in a one-step PCR (Bradley et al., 2001).
We also used 30 DNA extracts from chimpanzee fecal samples collected in a similar manner in Loango National Park, Gabon (Boesch et al., 2007). These extracts were previously genotyped at D3s3038, D5s1470, D6s1056, D10s676, D11s2002, D14s306 and typed for sex (Arandjelovic et al., 2011). We performed the second step of the two-step multiplex PCR (Arandjelovic et al., 2009) to further genotype them at D3s2459, D4s1627, D7s817, D7s2204, D12s66, D18s536.
The ABI PRISM 3100 Genetic Analyzer was used for electrophoresis of the PCR products. The sizes of the alleles relative to an internal size standard were determined using GeneMapper Software version 3.7 (Applied Biosystems). We calculated allelic dropout rates to assess how many observations from independent reactions are necessary to confirm homozygosity (Morin et al., 2001). Heterozygous genotypes were always confirmed with 99% certainty by observing each allele two or more times (Taberlet et al., 1996). For each extract up to six independent reactions were analyzed per locus.
Discrimination of individuals and distinguishing gorilla and chimpanzee samples
We used CERVUS 3.0 (Kalinowski et al., 2007) to calculate PIDsib (Waits et al., 2001), the probability that samples with matching genotypes come from siblings rather than from the same individual. Matching genotypes were combined into a consensus genotype when the PIDsib was <0.01. In the rare cases where two genotypes matched but the PIDsib was >0.01 the less complete genotype was removed from further analyses.
In the field, fecal samples from gorillas are occasionally mistakenly identified as chimpanzee feces, and vice versa. We therefore checked for incorrectly identified samples by genotyping 10 samples of known gorilla origin at the 12 microsatellite loci typically analyzed in chimpanzees (see above) and performing STRUCTURE analysis of them in combination with the purported chimpanzee samples as in Arandjelovic et al. (2010). After attributing samples to the correct species, we added those samples to our dataset that were genetically identified as chimpanzees and removed samples that were genetically identified as gorillas.
Data analysis
In addition to the central chimpanzees that were either completely (individuals from Republic of Congo and Cameroon) or partly (individuals from Loango, Gabon) genotyped for the present study, we made use of previously published genotypes of 283 eastern chimpanzees from six communities (Langergraber et al., 2009; Langergraber et al., 2011) (Table 1 and Fig. 2). Using CERVUS 3.0 (Kalinowski et al., 2007) we analyzed the dataset consisting of 185 central and 283 eastern chimpanzees to check for the existence of null alleles within four geographically distinct subsets: (i) Loango central chimpanzees, (ii) north-eastern central chimpanzees, (iii) Kibale eastern chimpanzees, and (iv) Budongo eastern chimpanzees. GENEPOP 4.2 (Raymond and Rousset, 1995) was used to perform the exact tests of Guo and Thompson (1992) for deviations from Hardy-Weinberg equilibrium (HWE) as well as the test of composite linkage disequilibrium (LD) (Weir, 1996) within the data subsets. We found null allele frequency estimates >0.1 for D7s2204 within the Budongo and the north-eastern central chimpanzee subsets. We thus tested for HWE within each subset both before and after removal of D7s2204. The Loango, Budongo, and Kibale subsets did not deviate significantly from HWE either before or after removal of D7s2204 (P > 0.05 after Bonferroni correction for multiple tests). The north-eastern central chimpanzee subset, however, was only in HWE after exclusion of D7s2204. Two pairs of loci (D3s3038 + D10s676 and D6s1056 + D4s1627) were in LD (P < 0.05 after Bonferroni correction for multiple tests) across all data subsets. True LD, however, can only occur between loci that are at the same chromosome. We thus conducted all downstream analyses excluding D7s2204 and using the remaining eleven loci.
TABLE 1.
Overview about groups between which genetic differentiation measured as FST was determined
| Subspecies | Country | Area | Group/Community | N |
|---|---|---|---|---|
| Central chimpanzees | Gabon | Loango NP | Loango | 30 |
| Cameroon | Lobéké NP | Lobéké-West | 12 | |
| Lobéké-East | 8 | |||
| Congo | Odzala NP | Odzala-West | 8 | |
| Odzala-East | 5 | |||
| Nouabalé Ndoki NP | A | 25 | ||
| B1 | 35 | |||
| C | 31 | |||
| D | 21 | |||
| Eastern chimpanzees | Uganda | Budongo Forest Reserve | Busingiro | 41 |
| Kasokwa | 5 | |||
| Sonso | 29 | |||
| Kibale NP | Kanyantale | 49 | ||
| Kanyawara | 40 | |||
| Ngogo | 119 | |||
| Totals | 4 countries | 6 areas | 15 groups | 458 individuals |
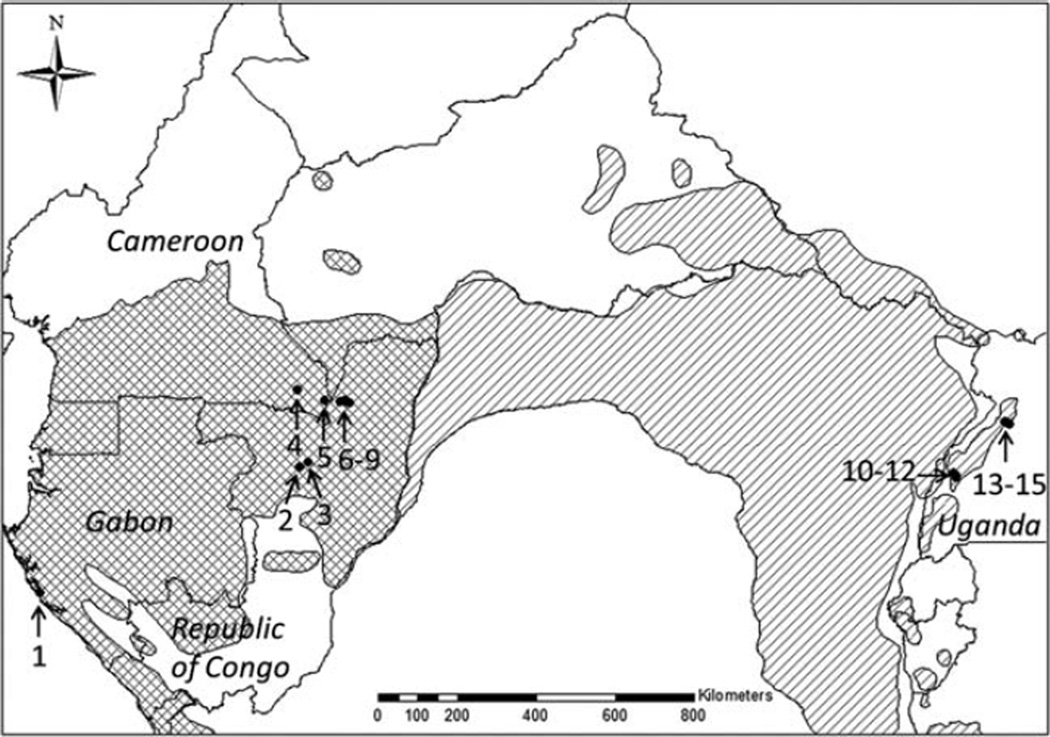
Fig. 2.
Location of chimpanzee groups (black dots) between which genetic differentiation was measured. Countries in which these groups can be found are indicated. Cross hatch depicts central and simple hatch depicts eastern chimpanzee range. 1: Loango; 2: Odzala-West; 3: Odzala-East; 4: Lobéké-West; 5: Lobéké-East; 6–9: Nouabalé-Ndoki NP groups (A, B1, C, D); 10–12: Kibale communities (Kanyantale, Kanyawara, Ngogo); 13–15: Budongo communities (Busingiro, Kasokwa, Sonso).
We used STRUCTURE 2.3.3 (Pritchard et al., 2000) to infer subdivision of populations. In STRUCTURE we used the admixture model, correlated allele frequencies, a burn-in period of 100,000 steps and then 500,000 steps of data collection. We varied the number of clusters K from one to eight and conducted 10 independent iterations at each K. We present the results of those runs that had the highest estimated logarithm of the probability of the data at each K. We applied different methods to determine the optimum value for K. First, we used Harvester Web v0.6.93 (Earl and vonHoldt, 2011) to calculate ΔK (Evanno et al., 2005), a measure of second-order rate of change in the likelihood of K. Second, we looked for the K at which the likelihood distribution began to plateau (Pritchard et al., 2000) and third we investigated for which K value K + 1 no longer refined the clusters (i.e. at K + 1 the clusters distinguished at K were no longer split according to geography) (Tishkoff et al., 2009).
To measure the genetic differentiation between different groupings of individuals, a priori delineation of groups is required. For the majority of central chimpanzees we did not know the community to which they belonged. Because chimpanzee communities occupy limited territories, we defined nine groupings of spatially clumped individuals within the central chimpanzees: Loango, Odzala-West, Odzala-East, Lobéké-West, Lobéké-East, and four from the Nouabalé Ndoki National Park (A, B1, C, D), excluding seven geographically outlying samples from Odzala and three from Lobéké. Each grouping represents a geographic grouping which is separated from the other groupings either by distance (Loango, Lobéké-West, Lobéké-East) or by watercourses running between them (A, B1, C, D) (Figs. 1 and 2). For the eastern chimpanzees community membership was known and we accordingly considered each of the six eastern chimpanzee communities to represent a distinct group. For each of the central chimpanzee groupings the sizes of their minimum convex polygons (MCPs) were determined using ESRI® ArcMap™ 9.2. They ranged from 5.1 km2 (Loango) to 64.7 km2 (C), which are broadly similar to the estimated territories of known eastern chimpanzee communities with sizes from ~7 km2 at the Sonso community (Newton-Fisher, 2003) up to ~35 km2 at the Ngogo community (Mitani et al., 2010). A total of 15 chimpanzee groups (nine central and six eastern) were defined (Table 1 and Fig. 2). Using Arlequin 3.5. (Excoffier and Lischer, 2010) we calculated the FST values between all 105 possible pairs of groups and correlated them with the corresponding geographic distances (measured as natural logarithm of distance in meters). For the calculations of the geographic distances, we assessed the positions of the MCP centroids of the central chimpanzee groupings using ESRI® ArcMap™ 9.2. Regarding the eastern chimpanzee groups we obtained the positions of the MCP centroids for Ngogo and Kanyantale (S. Amsler, personal communication), whereas central points within each territory were used for Busingiro, Sonso, Kasokwa, and Kanyawara (J. Moore, personal communication). We then plotted a regression line and conducted a Mantel test in R (Development Core Team, 2012) and considered its slope significantly positive if there was a less than 5% probability of obtaining a slope greater than the observed one in 10,000 permutations. This allowed us to see if the variation in genetic distance was consistent with a pattern of IBD encompassing both subspecies.
If the classification of central and eastern chimpanzees as distinct subspecies reflects long-term, independent evolutionary histories of these two taxa, one predicts that pairwise comparisons between-subspecies should show a higher genetic differentiation than those within-subspecies. Therefore we investigated if there are within-subspecies pairs of groups that show higher genetic differentiation than between-subspecies pairs of groups. To further elucidate how varying sampling schemes could lead to varying conclusions about the genetic structure of a population and to test whether previous reports of the subdivision of equatorial chimpanzees into central and eastern chimpanzees (e.g., Hill, 1969; Morin et al., 1994; Bjork et al., 2010; Prado-Martinez et al., 2013) might have been biased by a clumped sampling scheme, we simulated an increasingly clumped sampling and investigated if that leads to a decreasing number of pairs that exhibit a higher genetic differentiation within than between-subspecies. We began by determining genetic differentiation between (i) all nine central chimpanzee and six eastern chimpanzee groups (the least clumped scheme possible with our available data) and then successively decreased the number of included groups to obtain an increasingly clumped pattern: (ii) eight central chimpanzee groups (excluding Loango) and all six eastern chimpanzee groups; (iii) six central chimpanzee groups (excluding Loango, Odzala-West and Odzala-East) and all six eastern chimpanzee groups; (iv) six central chimpanzee groups (excluding Loango, Odzala-West and Odzala-East) and three eastern chimpanzee groups (excluding the three Budongo communities).
RESULTS
Genotype analysis
We genotyped 278 putative central chimpanzee fecal samples at 12 autosomal microsatellite loci. These samples yielded 242 usable genotypes representing 158 unique individuals. An analysis of these genotypes using STRUCTURE in comparison with known gorilla DNAs typed at the same loci showed that 15 of the purported chimpanzee DNAs were actually from gorillas, leaving a total of 143 different chimpanzees. Field researchers thus correctly identified chimpanzee dung in 227/242 = 94% of cases. In a reciprocal analysis of genotypes purportedly from gorillas and typed as part of a complementary study (Fünfstück et al., 2014), we found 16 samples were derived from chimpanzees. These 16 samples were secondarily genotyped at microsatellite loci used here. Four samples were identical to already existing genotypes and 12 represented additional individuals. The final dataset therefore contained 155 different central chimpanzee genotypes, which were on average 88% complete (Supporting Information Table 1). These 155 individuals consisted of 74 females (F) and 78 males (M) while the sex could not be determined for three individuals. Twenty individuals were from Odzala (13F, 7M), 23 from Lobéké (10F, 10M, 3 unknown sex), and 112 (51F, 61M) from Nouabalé-Ndoki.
The average DNA concentration of the 225 quantified extracts was 237.4 pg/μl (median: 108.0 pg/μl; SD: 573.4 pg/μl; range: 3.3–6063 pg/μl). Considering results from individual PCRs, allelic dropout rates over all loci were 0.31 for extracts with the lowest DNA amounts (≤5 pg/μl), 0.24 for extracts with intermediate DNA amounts (5–10 pg/μl) and 0.04 for extracts with the highest DNA amounts (>10 pg/μl). Thus, four independent observations of homozygosity were needed to confirm homozygous genotypes with 99% certainty in samples with DNA concentrations ≤5 pg/μl (0.31^4 = 0.009), whereas three independent observations were sufficient for all other extracts (0.24^3 = 0.01 for 5–10 pg/μl and 0.04^3 = 0.00006 for >10 pg/μl).
We also genotyped 30 extracts that were known to originate from different central chimpanzees (Arandjelovic et al., 2011) at six additional autosomal microsatellite loci. These six-loci genotypes were 72.2% complete. However, the genotypes of the other six-loci done by Arandjelovic et al. (2011) were 99.4% complete leading to 85.8% complete genotypes over all 12 loci (Supporting Information Table 1).
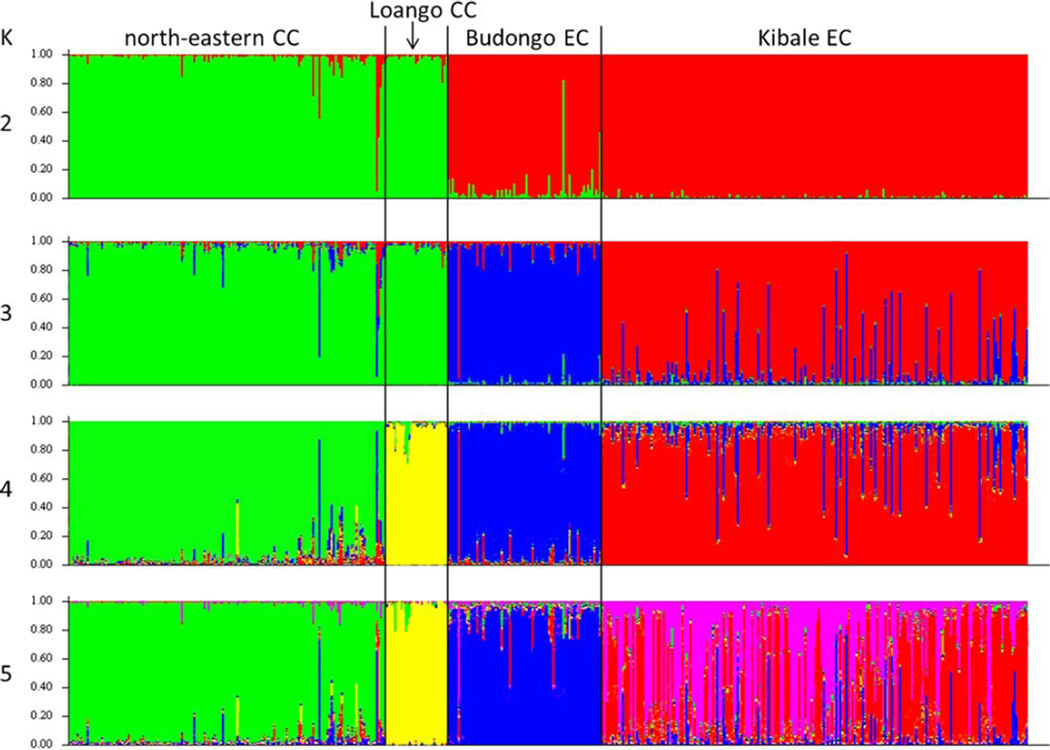
We conducted analyses using STRUCTURE to examine the distribution of individuals into clusters. Different methods of inferring the optimal value of K produced different inferences. The first method (using ΔK) supported K = 2, the second one (using the K at which the likelihood distribution begun to plateau) produced ambiguous results but suggested K = 3 and the third one (using the K at which K + 1 no longer refined the clusters) found K = 4. The STRUCTURE results for K = 2 to K = 5 are shown (Fig. 3). At K = 2 the analysis of 185 central and 283 eastern chimpanzees produced distinct clusters for central and eastern chimpanzees. At K = 3 the central chimpanzees formed one cluster and the eastern chimpanzees were further subdivided into individuals from Budongo and Kibale, whereas at K = 4 the central chimpanzees were divided into a Loango and a north-eastern cluster and the eastern chimpanzees were divided into Budongo and Kibale clusters. Finally, at K = 5, the clusters distinguished at K = 4 were no longer split. Instead the individuals from the Kibale cluster were admixed and found to have ancestry from two different sources.
Fig. 3.
STRUCTURE analysis including central chimpanzees from the north-eastern region (present study) (bars 1–155) and from Loango, Gabon (Arandjelovic et al., 2011 and present study) (bars 156–185) as well as eastern chimpanzees from Budongo, Uganda (Langergraber et al., 2011) (bars 186–260) and from Kibale NP, Uganda (Langergraber et al., 2009) (bars 261–468). Colors represent the inferred ancestry from K ancestral populations. Ten runs were conducted at each K. Shown are the results for K = 2 to K = 5, presenting those runs that had the highest estimated logarithm of the probability of the data. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Genetic differentiation
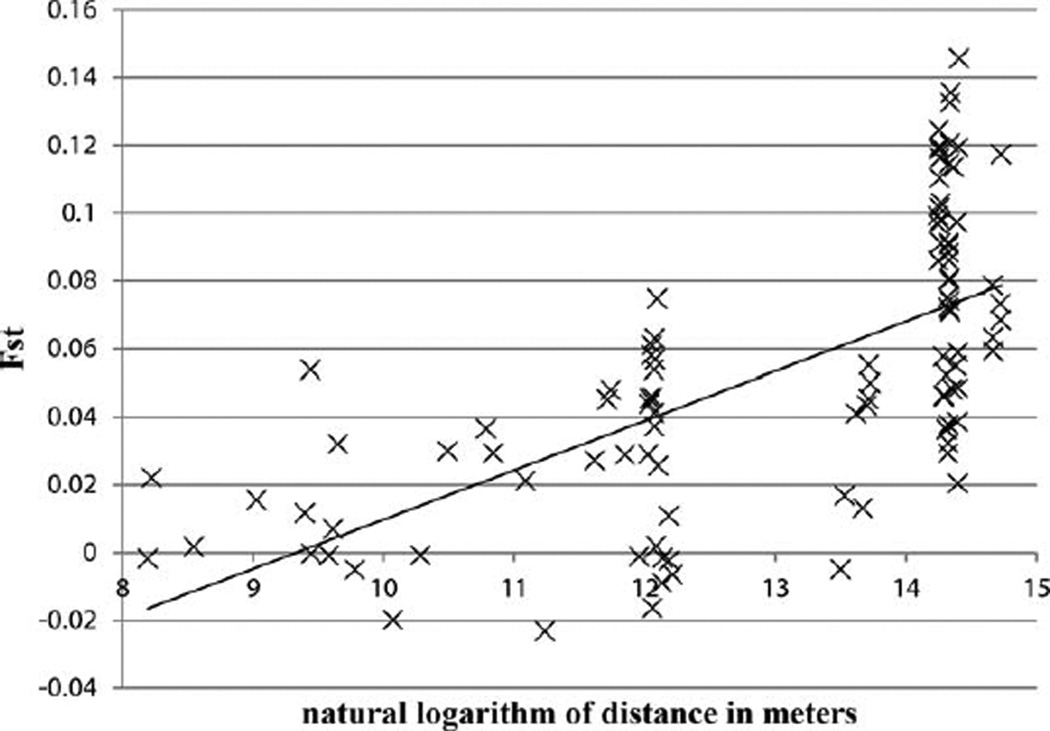
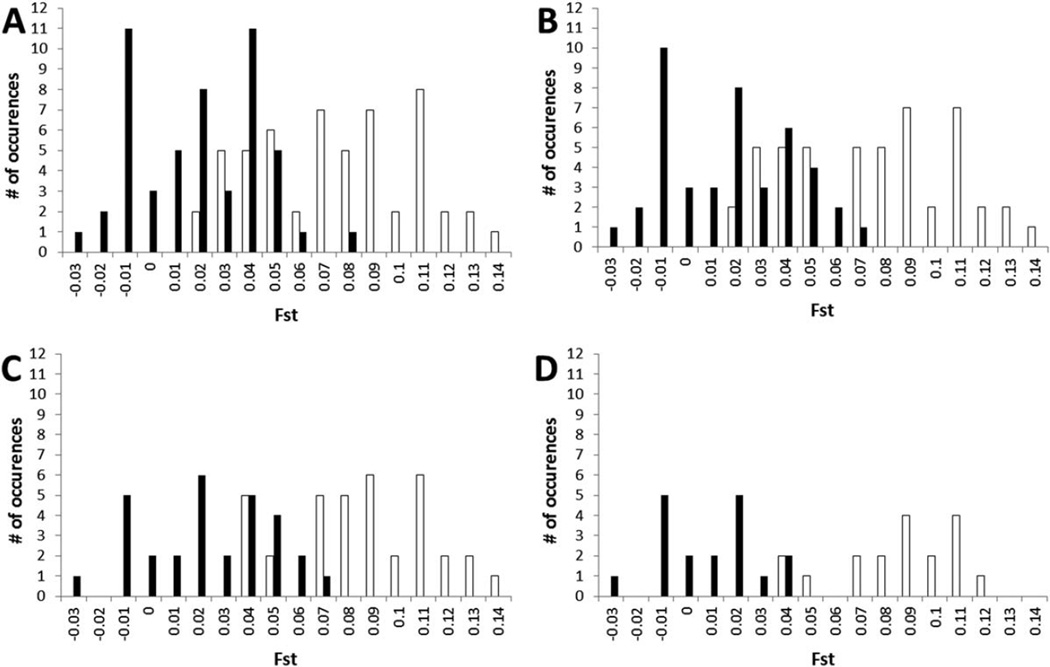
While the results of our STRUCTURE analysis seem to indicate that equatorial chimpanzees can be divided into at least two demes that seemingly correspond to central and eastern “subspecies,” pairwise comparisons between the 15 groups of both putative subspecies showed that genetic differentiation increased significantly with geographic distance, a pattern suggestive of strong IBD effects (Fig. 4). The FST values of pairwise comparisons ranged from slightly negative values, suggesting no differentiation, to 0.1456 between Lobéké-West and Kasokwa (Table 2). The average FST values between two groups of central chimpanzees, two groups of eastern chimpanzees and a central versus an eastern chimpanzee group were 0.0164, 0.0426 and 0.0802, respectively. When we used all nine central and six eastern chimpanzee groups for calculations of genetic differentiation, we found that 54.90% (28/51) of all within-subspecies comparisons exhibited higher values of FST than the least differentiated between-subspecies comparison (Table 2, Fig. 5). When we excluded the geographically outlying Loango central chimpanzee sample from the analysis, this value changed only very little to 55.81% (24/43). To further investigate the effects of increasingly limited sampling, we next excluded Odzala-West and Odzala-East from the central chimpanzee data, yielding into a proportion of 26.67% (8/30). Finally, by the omission of the three Budongo eastern chimpanzee communities, that value dropped again to just 5.56% (1/18). That is, there is a clear trend that successively omitting more geographically distant populations from our sample results in fewer within-subspecies comparisons exhibiting higher FST values than the least differentiated between-subspecies pair, therefore reiterating the importance of IBD in explaining the genetic differentiation among central and eastern equatorial populations of chimpanzees.
Fig. 4.
Genetic distances measured as Fst plotted against the natural logarithm of straight line distance in meters. Each point represents one pairwise comparison between the fifteen groups. The linear equation for the regression line was y = 0.0146x − 0.1358 with r = 0.651 and P = 0.0001.
TABLE 2.
Genetic differentiation (measured as FST) of pairwise comparisons (below diagonal) and associated levels of significance (above diagonal) between nine central and six eastern chimpanzee groups
| A | B1 | C | D | L-E | L-W | O-W | O-E | Lo | Bus | Kas | Son | Ngo | Kt | Kw | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | n.s. | n.s. | n.s. | *** | *** | n.s. | ** | *** | *** | *** | *** | *** | *** | *** | |
| B1 | −0.0018 | n.s. | * | ** | *** | n.s. | ** | *** | *** | *** | *** | *** | *** | *** | |
| C | −0.0051 | −0.0009 | n.s. | * | *** | n.s. | * | *** | *** | *** | *** | *** | *** | *** | |
| D | −0.0003 | 0.0070 | 0.0009 | ** | *** | n.s. | * | *** | *** | *** | *** | *** | *** | *** | |
| L-E | 0.0366 | 0.0294 | 0.0212 | 0.0300 | n.s. | n.s. | n.s. | n.s. | *** | *** | *** | *** | *** | *** | |
| L-W | 0.0451 | 0.0479 | 0.0288 | 0.0272 | −0.0232 | n.s. | n.s. | *** | *** | *** | *** | *** | *** | *** | |
| O-W | −0.0024 | 0.0109 | −0.0065 | −0.0016 | −0.0164 | −0.0090 | n.s. | n.s. | *** | *** | * | *** | *** | ** | |
| O-E | 0.0372 | 0.0413 | 0.0256 | 0.0290 | −0.0012 | 0.0019 | −0.0199 | n.s. | *** | ** | *** | ** | ** | ** | |
| Lo | 0.0453 | 0.0552 | 0.0499 | 0.0435 | 0.0132 | 0.0411 | −0.0050 | 0.0168 | *** | *** | *** | *** | *** | *** | |
| Bus | 0.0883 | 0.0906 | 0.0751 | 0.0808 | 0.0481 | 0.0588 | 0.0386 | 0.0550 | 0.0732 | * | *** | *** | *** | *** | |
| Kas | 0.1326 | 0.1206 | 0.1145 | 0.1356 | 0.11354 | 0.1456 | 0.1194 | 0.0973 | 0.1174 | 0.0321 | ** | *** | *** | ** | |
| Son | 0.0861 | 0.0913 | 0.0721 | 0.0708 | 0.0481 | 0.0485 | 0.0204 | 0.0550 | 0.0684 | 0.0221 | 0.0538 | *** | *** | *** | |
| Ngo | 0.1016 | 0.1102 | 0.0859 | 0.0911 | 0.0460 | 0.0716 | 0.0323 | 0.0364 | 0.0633 | 0.0439 | 0.0539 | 0.0580 | n.s. | *** | |
| Kt | 0.1163 | 0.1196 | 0.0967 | 0.1030 | 0.0464 | 0.0741 | 0.0378 | 0.0363 | 0.0593 | 0.0457 | 0.0747 | 0.0630 | 0.0018 | *** | |
| Kw | 0.1187 | 0.1245 | 0.0992 | 0.0979 | 0.0577 | 0.0802 | 0.0293 | 0.052434 | 0.0787 | 0.0455 | 0.0568 | 0.0609 | 0.0155 | 0.0117 |
(with n.s. = not significant,
P < 0.05,
P < 0.01,
P < 0.001) between nine central and six eastern chimpanzee groups
Pairwise comparisons within subspecies and between subspecies are in italic and normal font, respectively.
Bus = Busingiro; Kt = Kanyantale; Kw = Kanyawara; Kas = Kasokwa; Lo = Loango; L-E = Lobéké-East; L-W = Lobéké-West; Ngo = Ngogo; O-E = Odzala-East; O-W = Odzala-West; Son = Sonso.
Fig. 5.
Number of pairwise comparisons (y-axes) within different ranges of genetic differentiation measured as FST (x-axes). Black bars and white bars represent within and between subspecies comparisons, respectively. Groups included into the bar plots were as follows. Top left (A): nine central and six eastern chimpanzee groups. Top right (B): eight central (excluding Loango) and six eastern chimpanzee groups. Bottom left (C): Six central (excluding Loango, Odzala-West and Odzala-East) and six eastern chimpanzee groups. Bottom right (D): Six central (excluding Loango, Odzala-West and Odzala-East) and three eastern chimpanzee (excluding the three Budongo communities) groups.
DISCUSSION
In this study we investigated the population structure of central and eastern chimpanzees by subjecting the microsatellite genotypes of 185 central and 283 eastern chimpanzees of known geographic origin to multiple analyses. First, we applied a Bayesian based clustering algorithm implemented in STRUCTURE. Second, we tested for IBD using correlations between Euclidean and genetic distances. Finally, we compared the genetic differentiation between groups drawn from the same subspecies and between groups drawn from different subspecies. Such use of various methods is highly recommended for attempting to obtain an unbiased, accurate and comprehensive view about the genetic structure of the populations under study (Frantz et al., 2009; Schwartz and McKelvey, 2009; Fünfstück et al., 2014).
In STRUCTURE, each method of inferring the optimal value for K suggested different amounts of clusters. Determining ΔK suggested two populations, consistent with a division of the sample into eastern and central chimpanzees. Contrary to that, the likelihood distribution began to plateau at K = 3, suggesting one cluster of central chimpanzees and two clusters of eastern chimpanzees. Finally, the clusters distinguished at K = 4, were no longer split at K = 5, arguing for four clusters of geographically clumped individuals: (i) central chimpanzees from Loango, (ii) north-eastern central chimpanzees, (iii) eastern chimpanzees from Budongo, and (iv) eastern chimpanzees from Kibale (Fig. 3). Thus, the analyses conducted in STRUCTURE seem to support the existence of two to four genetic clusters within equatorial chimpanzees. These STRUCTURE results, however, might lead to a wrong or at least biased perception of the population structure within equatorial chimpanzees for several reasons. First, the fact that different methods for inferring the exact amount of populations found different solutions, makes it difficult to find definitive answers about the genetic stratification within central and eastern chimpanzees. Second, due to its problems in dealing with clinal genetic variation, the algorithm used by STRUCTURE is prone to overestimate the number of populations when there is an underlying pattern of IBD, especially when the sampling scheme is clumped (Frantz et al., 2009; Schwartz and McKelvey, 2009). By comparing geographic and genetic distances, we found highly significant evidence for IBD and our STRUCTURE analysis also identified several individuals that had less than 75% ancestry within one cluster meaning that they appeared admixed and not attributable to a single cluster. Such high proportions of admixed individuals are commonly detected by STRUCTURE if the distribution of genetic variation is characterized by a pattern of IBD (Pritchard et al., 2003). Third, and probably most important, although our sampling was as less clumped as feasible, there were still large geographical gaps between some sampling locations (Fig. 2). In the clustering solutions that were supported by our STRUCTURE results, the suggested populations consisted of individuals that were geographically clumped together and the geographic distances between samples from different clusters were much larger than those between samples from the same cluster. We thus believe that, even if STRUCTURE did not overestimate the amount of populations due to IBD, it simply depicts the lack of sampling at geographic gaps between the clusters. That is, the results of STRUCTURE reflect the chosen sampling scheme. It might well be that a geographically more continuous sampling would not reveal clearly distinct genetic clusters within equatorial chimpanzees. In sum, these results suggest that the geographic distance shapes the pattern of genetic variation observed in our sample and those geographic clusters are inconsistently inferred.
To evaluate if another approach suggests a primary division of equatorial chimpanzee genetic variation into two units, we calculated pairwise genetic differentiations between groups from the same subspecies and between groups from different subspecies. If a taxonomical distinction as different subspecies reflects a long period of independent evolution, one would expect that within-subspecies comparisons exhibit lower estimates of genetic variation than between-subspecies comparisons. Moreover, if the previously reported distinction between central and eastern chimpanzees is real, and not an artifact of clumped sampling, the proportion of within-subspecies comparisons showing higher genetic differentiation than the least differentiated between-subspecies comparison should be independent of the sampling scheme. Contrary to these predictions we found that a substantial proportion (54.90%) of all within-subspecies comparisons exhibited higher values of FST than the least differentiated between-subspecies comparison when all groups were included (which represents the least clumped scheme). After stepwise exclusion of groups in order to simulate an increasingly clumped sampling scheme, there was a clear trend of obtaining reduced proportions of within-subspecies comparisons exceeding between-subspecies comparisons with a final value of only 5.56% in the most clumped scheme. Two main conclusions can be inferred from these results. The high proportions found in the least clumped schemes argue for an incomplete division with recent admixture between central and eastern chimpanzees, whereas the second point demonstrates how much a certain sampling pattern affects the resulting estimates of genetic variation and hence also the conclusions drawn from it.
Previous studies that found a clear distinction between central and eastern chimpanzees, although sometimes employing extensive datasets (Becquet et al., 2007; Prado-Martinez et al., 2013), were often relying on samples of unknown exact origin, a small number of analyzed individuals or a combination of both (Gonder et al., 1997; Bjork et al., 2010; Becquet et al., 2007; Prado-Martinez et al., 2013). In contrast, our study investigates the population structure of central and eastern chimpanzees across large parts of their range by analyzing microsatellite genotypes of spatially explicit and widely distributed samples. Compared with the analysis of mtDNA sequence data, the use of microsatellites has the advantage that the latter are representing selectively neutral multi-locus markers, which are biparentally inherited. We are therefore confident that our results represent a reliable description of the population structure of equatorial chimpanzees. We argue that instead of the traditional taxonomy with two clearly distinct subspecies of central and eastern chimpanzees, the population structure of equatorial chimpanzees might be better described by clinal genetic variation across their range with ongoing gene flow and recent admixture rather than a strong distant split. That leads to an encouraging agreement with other studies on chimpanzee population structure that also did not find evidence for central and eastern chimpanzees constituting clearly separated subspecies (Gagneux et al., 1999, 2001; Gonder et al., 2006, 2011; Fischer et al., 2011). As our sampling design also allowed us to determine to which extent a clumped sampling can bias the results, we recommend that further studies should aim to obtain spatially explicit samples in a preferably even less clumped sampling pattern. We speculate that analysis of such datasets will demonstrate yet more clearly that equatorial chimpanzees are characterized by IBD and that previously found subdivisions might have been artifacts caused by an incomplete sampling scheme. Likewise to the situation in humans, where traditionally the existence of separate races was proposed, classical primate taxonomists were often obsessed with finding distinct subspecies (Hill, 1969). In humans, a more continuous sampling revealed a pattern of IBD rather than typological races (Serre and Pääbo, 2004; Lao et al., 2008; Tishkoff et al., 2009). Our results indicate that also in other primate taxa an obvious line between distinct subspecies may be difficult to draw.
From a conservation point of view, chimpanzees play an extraordinarily important role, because they are not only the closest living relatives of humans, but also act as a flagship species, umbrella species and environmental indicator species (Wrangham et al., 2008). Moreover, through their role as seed-dispersers they are important in maintaining an intact forest ecosystem (Balcomb and Chapman, 2003). Both central and eastern chimpanzees are categorized as endangered by the IUCN (Walsh et al., 2003; Wilson et al., 2009) and their abundance has severely declined over the last decades (Walsh et al., 2003; Hicks et al., 2010; Plumptre et al., 2010). Our results, indicating a mostly contiguous population of equatorial chimpanzees, might argue for large-scale oriented conservation efforts considering central and eastern chimpanzees as one group. However, the range of both subspecies together spans over more than 1500 km from north to south and more than 2000 km from east to west (Fig. 2). Many chimpanzees live in small populations within fragmented forest patches and the low connectivity between these populations is considered as one of the leading threats to the species existence (Plumptre et al., 2010). Thus, efforts to maintain and repair the interconnectedness of these fragments and mitigate detrimental impacts such as inbreeding are warranted. Corridor conservation is a relatively new field in equatorial Africa, but given the expansion of natural resource extraction industries in forestry and mining sectors and the development of associated hard infrastructures (Laporte et al., 2007) it will likely be necessary on behalf of conserving chimpanzees in the Congo Basin. The prospects for such strategies remain high given there still persist extensive chimpanzee populations in central and eastern Africa that have as yet to be disturbed by present day anthropogenic impacts (Morgan and Sanz, 2003; Hicks et al., 2014). Further, Junker et al. (2012), by relating ape presence information to environmental and human impact factors, predicted that geographical connectivity of patches within the Congo Basin of suitable habitat are promising, especially in the north-eastern part of the Republic of Congo and the eastern parts of the Democratic Republic of Congo. Future efforts integrating the relevance of these findings to conservation of local chimpanzee populations in terms of genetic viability and dispersal would improve prioritization scenarios.
Supplementary Material
ACKNOWLEDGMENTS
For permission to carry out research in Republic of Congo, the authors thank the Ministère de l’Économie Forestière et Développement Durable; the Ministère de la Recherche Scientifique et de l’Innovation Technologique; and the Ministère de la Santé. For permission to collect samples in Cameroon, the authors thank the Ministère de la Recherche Scientifique et de l’Innovation (MINRESI) as well as the Ministère des Forêts et de la Faune (MINFOF). The authors thank the Agence Nationale des Parcs Nationaux (ANPN) and the Centre National de la Recherche Scientifique et Technique (CENAREST) of Gabon for permission to conduct their research in Loango National Park as well as the staff and the SIV team from PRESICA for logistical support in Cameroon. They also thank African Parks Network for facilitation of the work. They thank A. Abraham for laboratory assistance, C. Stephens, and R. Mundry for statistical assistance. They thank two anonymous referees for helpful comments. They thank L. Rabanal, L. Mackaga, E. R. Guizard, N. Tagg, B. Graw, E. Fairet, M. Gregoire, L. Rankin S. Kaba, M. J. Akongo, E. B. Ngouembe, and G. Bounga for sample collection. All work presented herein was conducted in compliance with appropriate animal care regulations and national laws and also in compliance with the American Association of Physical Anthropologists Code of Ethics.
Grant sponsor: Max Planck Society, the Arcus Foundation, the Paul G. Allen Family Foundation, the Dunemere Foundation, the Bradley L. Goldberg Foundation, the Société pour la Conservation et le Développement (SCD), the Wildlife Conservation Society (WCS).; Grant sponsor: National Institutes of Health; Grant number: R01 AI50529.; Grant sponsor: Agence Nationale de Recherches sur le SIDA, France; Grant number: ANRS 12255.
Abbreviations
- HWE
Hardy-Weinberg equilibrium
- IBD
isolation by distance
- LD
linkage disequilibrium
- MCP
minimum convex polygon
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Arandjelovic M, Guschanski K, Schubert G, Harris TR, Thalmann O, Siedel H, Vigilant L. Two-step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Mol Ecol Resour. 2009;9:28–36. doi: 10.1111/j.1755-0998.2008.02387.x. [DOI] [PubMed] [Google Scholar]
- Arandjelovic M, Head J, Kühl H, Boesch C, Robbins MM, Maisels F, Vigilant L. Effective non-invasive genetic monitoring of multiple wild western gorilla groups. Biol Conserv. 2010;143:1780–1791. [Google Scholar]
- Arandjelovic M, Head J, Rabanal LI, Schubert G, Mettke E, Boesch C, Robbins MM, Vigilant L. Non-invasive genetic monitoring of wild central chimpanzees. PLoS ONE. 2011;6:e14761. doi: 10.1371/journal.pone.0014761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC. Introduction: the scope of conservation genetics. In: Avise JC, Hamrick J, editors. Conservation genetics: case histories from nature. New York: Chapman & Hall; 1996. pp. 1–9. [Google Scholar]
- Balcomb SR, Chapman CA. Bridging the gap: influence of seed deposition on seedling recruitment in a primate-tree interaction. Ecol Monogr. 2003;73:625–642. [Google Scholar]
- Becquet C, Patterson N, Stone AC, Przeworski M, Reich D. Genetic structure of chimpanzee populations. PLoS Genet. 2007;3:e66. doi: 10.1371/journal.pgen.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork A, Liu W, Wertheim JO, Hahn BH, Worobey M. Evolutionary history of chimpanzees inferred from complete mitochondrial genomes. Mol Biol Evol. 2010;28:615–623. doi: 10.1093/molbev/msq227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesch C, Head J, Tagg N, Arandjelovic M, Vigilant L, Robbins MM. Fatal chimpanzee attack in Loango National Park, Gabon. Int J Primatol. 2007;28:1025–1034. [Google Scholar]
- Bowden R, MacFie TS, Myers S, Hellenthal G, Nerrienet E, Bontrop RE, Freeman C, Donnelly P, Mundy NI. Genomic tools for evolution and conservation in the chimpanzee: Pan troglodytes ellioti is a genetically distinct population. PLoS Genet. 2012;8:e1002504. doi: 10.1371/journal.pgen.1002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley BJ, Chambers KE, Vigilant L. Accurate DNA-based sex identification of apes using non-invasive samples. Conserv Genet. 2001;2:179–181. [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton, NJ: Princeton University Press; 1994. [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2011;4:359–361. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fischer A, Pollack J, Thalmann O, Nickel B, Pääbo S. Demographic history and genetic differentiation in apes. Curr Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Fischer A, Prüfer K, Good JM, Halbwax M, Wiebe V, André C, Atencia R, Mugisha L, Ptak SE, Pääbo S. Bonobos fall within the genomic variation of chimpanzees. PloS One. 2011;6:e21605. doi: 10.1371/journal.pone.0021605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz AC, Cellina S, Krier A, Schley L, Burke T. Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? J Appl Ecol. 2009;46:493–505. [Google Scholar]
- Fünfstück T, Arandjelovic M, Morgan DB, Sanz C, Breuer T, Stokes EJ, Reed P, Olson SH, Cameron K, Ondzie A, Peeters M, Kühl HS, Cipolletta C, Todd A, Masi S, Doran-Sheehy DM, Bradley BJ, Vigilant L. The genetic population structure of wild western lowland gorillas (Gorilla gorilla gorilla) living in continuous rain forest. Am J Primatol. 2014;76:868–878. doi: 10.1002/ajp.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux P, Gonder MK, Goldberg TL, Morin PA. Gene flow in wild chimpanzee populations: what genetic data tell us about chimpanzee movement over space and time. Philos Trans R Soc B Biol Sci. 2001;356:889–897. doi: 10.1098/rstb.2001.0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux P, Wills C, Gerloff U, Tautz D, Morin PA, Boesch C, Fruth B, Hohmann G, Ryder OA, Woodruff DS. Mitochondrial sequences show diverse evolutionary histories of African hominoids. Proc Natl Acad Sci. 1999;96:5077–5082. doi: 10.1073/pnas.96.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder MK, Disotell TR, Oates JF. New genetic evidence on the evolution of chimpanzee populations and implications for taxonomy. Int J Primatol. 2006;27:1103–1127. [Google Scholar]
- Gonder MK, Locatelli S, Ghobrial L, Mitchell MW, Kujawski JT, Lankester FJ, Stewart C-B, Tishkoff SA. From the Cover: evidence from Cameroon reveals differences in the genetic structure and histories of chimpanzee populations. Proc Natl Acad Sci. 2011;108:4766–4771. doi: 10.1073/pnas.1015422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder MK, Oates JF, Disotell TR, Forstner MR, Morales JC, Melnick DJ. A new West African chimpanzee subspecies? Nature. 1997;388:337. doi: 10.1038/41005. [DOI] [PubMed] [Google Scholar]
- Groves C. Primate taxonomy. Washington, DC: Smithsonian Institution Press; 2001. [Google Scholar]
- Guo SW, Thompson EA. Performing exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Guy F, Brunet M, Schmittbuhl M, Viriot L. New approaches in hominoid taxonomy: morphometrics. Am J Phys Anthropol. 2003;121:198–218. doi: 10.1002/ajpa.10261. [DOI] [PubMed] [Google Scholar]
- Hicks TC, Darby L, Hart J, Swinkels J, January N, Menken S. Trade in orphans and bushmeat threatens one of the Democratic Republic of the Congo’s most important populations of eastern chimpanzees (Pan troglodytes schweinfurthii) Afr Primates. 2010;7:1–18. [Google Scholar]
- Hicks TC, Tranquilli S, Kuehl H, Campbell G, Swinkels J, Darby L, Boesch C, Hart J, Menken SBJ. Absence of evidence is not evidence of absence: discovery of a large, continuous population of Pan troglodytes schweinfurthii in the Central Uele region of northern DRC. Biol Conserv. 2014;171:107–113. [Google Scholar]
- Hill WCO. The nomenclature, taxonomy, and distribution of chimpanzees. In: Bourne GH, editor. The chimpanzee. Vol. 1. Basel: Karger; 1969. pp. 22–49. [Google Scholar]
- Hvilsom C, Frandsen P, Børsting C, Carlsen F, Sallé B, Simonsen BT, Siegismund HR. Understanding geographic origins and history of admixture among chimpanzees in European zoos, with implications for future breeding programmes. Heredity. 2013;110:586–593. doi: 10.1038/hdy.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker J, Blake S, Boesch C, Campbell G, Toit L du, Duvall C, Ekobo A, Etoga G, Galat-Luong A, Gamys J, Ganas-Swaray J, Gatti S, Ghiurghi A, Granier N, Hart J, Head J, Herbinger I, Hicks TC, Huijbregts B, Imong IS, Kuempel N, Lahm S, Lindsell J, Maisels F, McLennan M, Martinez L, Morgan B, Morgan D, Mulindahabi F, Mundry R, N’Goran KP, Normand E, Ntongho A, Okon DT, Petre C-A, Plumptre A, Rainey H, Regnaut S, Sanz C, Stokes E, Tondossama A, Tranquilli S, Sunderland-Groves J, Walsh P, Warren Y, Williamson EA, Kuehl HS. Recent decline in suitable environmental conditions for African great apes. Divers Distrib. 2012;18:1077–1091. [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Langergraber K, Mitani J, Vigilant L. Kinship and social bonds in female chimpanzees (Pan troglodytes) Am J Primatol. 2009;71:840–851. doi: 10.1002/ajp.20711. [DOI] [PubMed] [Google Scholar]
- Langergraber K, Schubert G, Rowney C, Wrangham R, Zommers Z, Vigilant L. Genetic differentiation and the evolution of cooperation in chimpanzees and humans. Proc R Soc B Biol Sci. 2011;278:2546–2552. doi: 10.1098/rspb.2010.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao O, Lu TT, Nothnagel M, Junge O, Freitag-Wolf S, Caliebe A, Balascakova M, Bertranpetit J, Bindoff LA, Comas D, Holmlund G, Kouvatsi A, Macek M, Mollet I, Parson W, Palo J, Ploski R, Sajantila A, Tagliabracci A, Gether U, Werge T, Rivadeneira F, Hofman A, Uitterlinden AG, Gieger C, Wichmann H-E, Rüther A, Schreiber S, Becker C, Nürnberg P, Nelson MR, Krawczak M, Kayser M. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- Laporte NT, Stabach JA, Grosch R, Lin TS, Goetz SJ. Expansion of industrial logging in Central Africa. Science. 2007;316:1451–1451. doi: 10.1126/science.1141057. [DOI] [PubMed] [Google Scholar]
- McBrearty S, Jablonski NG. First fossil chimpanzee. Nature. 2005;437:105–108. doi: 10.1038/nature04008. [DOI] [PubMed] [Google Scholar]
- Mitani JC, Watts DP, Amsler SJ. Lethal intergroup aggression leads to territorial expansion in wild chimpanzees. Curr Biol. 2010;20:R507–R508. doi: 10.1016/j.cub.2010.04.021. [DOI] [PubMed] [Google Scholar]
- Morgan D, Sanz C. Naïve encounters with chimpanzees in the Goualougo Triangle, Republic of Congo. Int J Primatol. 2003;24:369–381. [Google Scholar]
- Morin PA, Chambers KE, Boesch C, Vigilant L. Quantitative polymerase chain reaction analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzees (Pan troglodytes verus) Mol Ecol. 2001;10:1835–1844. doi: 10.1046/j.0962-1083.2001.01308.x. [DOI] [PubMed] [Google Scholar]
- Morin PA, Moore JJ, Chakraborty R, Jin L, Goodall J, Woodruff DS. Kin selection, social structure, gene flow, and the evolution of chimpanzees. Science. 1994;265:1193–1201. doi: 10.1126/science.7915048. [DOI] [PubMed] [Google Scholar]
- Newton-Fisher NE. The home range of the Sonso community of chimpanzees from the Budongo Forest, Uganda. Afr J Ecol. 2003;41:150–156. [Google Scholar]
- Nsubuga AM, Robbins MM, Roeder AD, Morin PA, Boesch C, Vigilant L. Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Mol Ecol. 2004;13:2089–2094. doi: 10.1111/j.1365-294X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- Oates JF, Groves CP, Jenkins PD. The type locality of Pan troglodytes vellerosus (Gray, 1862), and implications for the nomenclature of West African chimpanzees. Primates. 2009;50:78–80. doi: 10.1007/s10329-008-0116-z. [DOI] [PubMed] [Google Scholar]
- Pilbrow V. Population systematics of chimpanzees using molar morphometrics. J Hum Evol. 2006;51:646–662. doi: 10.1016/j.jhevol.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Plumptre A, Rose R, Nangendo G, Williamson EA, Didier K, Hart F, Mulindahabi F, Hicks C, Griffin B, Ogawa H, Nixon S, Pintea L, Vosper A, McLennan M, Amsini F, McNeilage A, Makana JR, Kanamori M, Hernandez A, Piel A, Stewart F, Moore J, Zamma K, Nakamura M, Kamenya S, Idani G, Sakamaki T, Yoshikawa M, Greer D, Tranquilli S, Beyers R, Hashimoto C, Furuichi T, Bennett E. Gland, Switzerland: IUCN; 2010. Status survey and conservation action plan for the Eastern chimpanzee (Pan troglodytes schweinfurthii), 2010–2020; p. 48. [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O’Connor TD, Santpere G, Cagan A, Theunert C, Casals F, Laayouni H, Munch K, Hobolth A, Halager AE, Malig M, Hernandez-Rodriguez J, Hernando-Herraez I, Prüfer K, Pybus M, Johnstone L, Lachmann M, Alkan C, Twigg D, Petit N, Baker C, Hormozdiari F, Fernandez-Callejo M, Dabad M, Wilson ML, Stevison L, Camprubí C, Carvalho T, Ruiz-Herrera A, Vives L, Mele M, Abello T, Kondova I, Bontrop RE, Pusey A, Lankester F, Kiyang JA, Bergl RA, Lonsdorf E, Myers S, Ventura M, Gagneux P, Comas D, Siegismund H, Blanc J, Agueda-Calpena L, Gut M, Fulton L, Tishkoff SA, Mullikin JC, Wilson RK, Gut IG, Gonder MK, Ryder OA, Hahn BH, Navarro A, Akey JM, Bertranpetit J, Reich D, Mailund T, Schierup MH, Hvilsom C, Andrés AM, Wall JD, Bustamante CD, Hammer MF, Eichler EE, Marques-Bonet T. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Wen W, Falush D. [Accessed on July 13, 2004];Documentation for STRUCTURE software: version 2. 2003 Available at: http://seq.ege.fcen.uba.ar/materias/ecomolecular/Material/An%C3%A1lsis%20Bayesiano%20Metapoblaciones.%20STRUCTURE/Structure2.1/Help%20Files/readme.pdf.
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HW, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Schonewald-Cox CM, Chambers SM, MacBryde B, Thomas WL. Genetics and conservation: a reference for managing wild animal and plant populations. Menlo Park, California: Benjamin/Cummings Publishing Company; 1983. [Google Scholar]
- Schwartz MK, McKelvey KS. Why sampling scheme matters: the effect of sampling scheme on landscape genetic results. Conserv Genet. 2009;10:441–452. [Google Scholar]
- Serre D, Pääbo S. Evidence for gradients of human genetic diversity within and among continents. Genome Res. 2004;14:1679–1685. doi: 10.1101/gr.2529604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone AC, Griffiths RC, Zegura SL, Hammer MF. High levels of Y-chromosome nucleotide diversity in the genus Pan. Proc Natl Acad Sci. 2002;99:43–48. doi: 10.1073/pnas.012364999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo J-M, Doumbo O, Ibrahim M, Juma AT, Kotze MJ, Lema G, Moore JH, Mortensen H, Nyambo TB, Omar SA, Powell K, Pretorius GS, Smith MW, Thera MA, Wambebe C, Weber JL, Williams SM. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waits LP, Luikart G, Taberlet P. Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol Ecol. 2001;10:249–256. doi: 10.1046/j.1365-294x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- Walsh P, Abernathy K, Bermejo M, Beyers R, De Wachter P, Akou ME, Huijbregts B, Mambounga DI, Toham AK, Kilbourn AM, Lahm SA, Latour S, Maisels F, Mbina C, Mihindou Y, Obiang SN, Effa EN, Starkey MP, Telfer P, Thibault M, Tutin CEG, White LJT, Wilkie DS. Catastrophic ape decline in western equatorial Africa. Nature. 2003;422:611–614. doi: 10.1038/nature01566. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis II. Sunderland, Massachusetts: Sinauer; 1996. [Google Scholar]
- Wilson ML, Balmforth Z, Cox D, et al. Pan troglodytes ssp. schweinfurthii. 2009 IUCN red list of threatened species. Version 2009.1. [Accessed 17 March 2010];2009 Available at: www.iucnredlist.org.
- Witzenberger KA, Hochkirch A. Ex situ conservation genetics: a review of molecular studies on the genetic consequences of captive breeding programmes for endangered animal species. Biodivers Conserv. 2011;20:1843–1861. [Google Scholar]
- Wrangham RW, Hagel G, Leighton M, Marshall AJ, Waldau P, Nishida T. The great ape world heritage species project. In: Stoinski TS, Steklis HD, Mehlman PT, editors. Conservation in the 21st century: gorillas as a case study. New York: Springer Science and Business Media, LLC; 2008. pp. 282–296. [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.