Abstract
DNMT1, the major maintenance DNA methyltransferase in animals, helps to regulate gene expression, genome imprinting, and X-chromosome inactivation. We report on the crystal structure of a productive covalent mouse DNMT1(731-1602)–DNA complex containing a central hemimethylated CpG site. The methyl group of methylcytosine is positioned within a shallow hydrophobic concave surface, whereas the cytosine on the target strand is looped out and covalently anchored within the catalytic pocket. The DNA is distorted at the hemimethylated CpG step, with side chains from catalytic and recognition loops inserting through both grooves to fill an intercalation-type cavity associated with a dual base flip-out on partner strands. Structural and biochemical data establish how a combination of active and autoinhibitory mechanisms ensures the high fidelity of DNMT1-mediated maintenance DNA methylation.
In mammals, DNA methylation predominantly occurs on the carbon-5 position of cytosines within CpG dinucleotides (1–4) and is critically maintained by DNMT1 during DNA replication (5). DNMT1 is composed of a C-terminal methyltransferase domain and an N-terminal regulatory domain, linked by a conserved (Gly-Lys)n repeat (fig. S1A). The N-terminal domain contains sequences that mediate interactions of DNMT1 with other proteins (6, 7), a nuclear localization sequence, a target recognition sequence that localizes DNMT1 to the DNA replication fork (8), a zinc finger CXXC (Cys-X-X-Cys) domain that specifically recognizes unmethylated CpG DNA (9, 10), and a pair of bromo-adjacent homology (BAH) domains (11). The methyltransferase domain of DNMT1 is further folded into two subdomains, designated as the catalytic core and the target recognition domain (TRD).
We have determined the crystal structure of a productive complex of truncated mouse DNMT1 (mDNMT1) with DNA (Fig. 1A), which contrasts with our previously determined autoinhibitory structure of the mDNMT1-DNA complex (12). The complex contains mDNMT1(731-1602), which encompasses both BAH domains and the methyltransferase domain, bound to DNA within a (mCpG)-(fCpG) dinucleotide context. The complementary 12-mer DNA duplex is composed of a central 5-methylcytosine (mC)–containing hemi-mCpG step on the parental strand, positioned opposite to a 5-fluorocytosine (5fC)–containing fCpG step (13) on the target strand. The complex was produced by enzymatically cross-linking the 5fC on the DNA target strand to the reactive cysteine positioned in the catalytic pocket of mDNMT1 (fig. S1B).
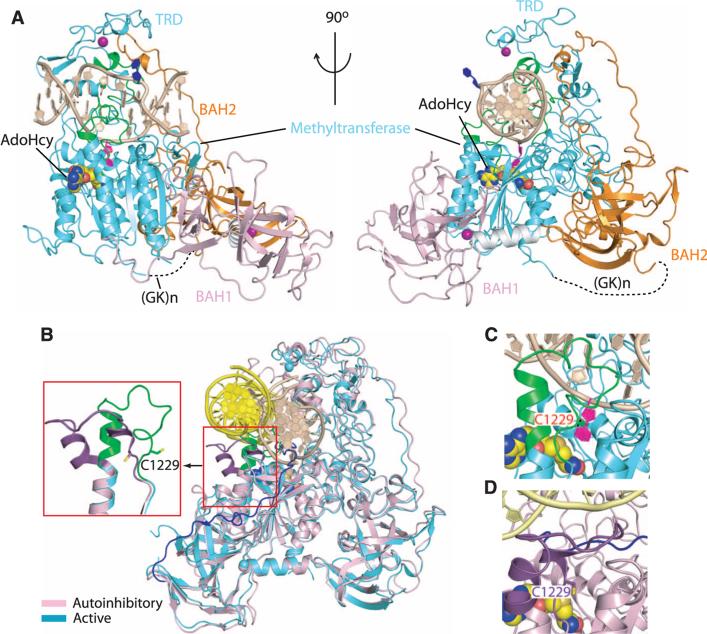
Fig. 1.
Structural overview of mDNMT1(731-1602) bound covalently to a hemi-mCpG site. (A) Ribbon representation of the covalent complex in two orthogonal views. The BAH1, BAH2, and methyltransferase domains are colored light pink, orange, and cyan, respectively; DNA and zinc ions are barley and purple, respectively. The disordered (Gly-Lys)n linker [(GK)n] is shown as black dashed lines, the 5-methyl group from mC6 is green, the flipped-out target fC7′ is purple, the flipped-out C from parental strand is blue, the catalytic loop and TRD loops 1 and 2 are green, and the bound AdoHcy is in a space-filling representation. (B) Structural superposition of the covalent mDNMT1(731-1602)–DNA complex with the autoinhibited mDNMT1(650-1602)–DNA complex. The bound DNA in the covalent complex is in beige with the protein in cyan; they are yellow and cyan in the autoinhibited complex. The catalytic loop (expanded view in inset) is colored purple in the autoinhibited complex and green in the covalent complex. The CXXC-BAH1 domain linker is in dark blue in the autoinhibited complex. The inset highlights the transition from a straight helix (green) in the productive covalent complex to a kinked helix (purple) in the autoinhibited complex. (C) Close-up view of the catalytic loop (in green) in the covalent mDNMT1 (731-1602)–DNA complex (this study). (D) Close-up view of the catalytic loop (in dark brown) in the autoinhibited mDNMT1(650-1602)–DNA complex (PDB: 3PT6). The side chain of residue Cys1229 is shown in stick representation.
Superposition of the structures of the productive (this study) and autoinhibited (12) mDNMT1-DNA complexes demonstrates that the largest conformational change is centered within the catalytic loop of mDNMT1 (Fig. 1B and fig. S2). In the productive covalent complex, the catalytic loop (in green) is followed by a straight α helix (green helical segment in inset of Fig. 1B) and penetrates into the DNA minor groove (Fig. 1C), forming extensive protein-DNA contacts (see below), whereas in the autoinhibitory structure of the complex, the α helix following the catalytic loop has a kink at the N-terminal end (purple helical segment in inset of Fig. 1B), and the catalytic loop (in purple) is excluded from the DNA minor groove by the autoinhibitory linker (in blue) (Fig. 1D).
mDNMT1(731-1602)–mediated cytosine methylation kinetics on DNA 14-mer substrates containing single central hemi-mCpG versus unmodified CpG sites (fig. S3A) yielded kinetic (kcat and kcat/KM) parameters similar to those previously measured for mDNMT1(717-1602) (fig. S3B and table S1) (12); the relative velocity of methylation of hemi-mCpG DNA substrate was greater than that of its CpG DNA counterpart by a factor of ~10 (fig. S3C).
The DNA duplex (Fig. 2A) is embedded in the catalytic cleft of mDNMT1, with the fC7′ of the target strand looped out of the helix and inserted into the catalytic pocket of the methyltransferase domain, where it is methylated and anchored through covalent bond formation with the reactive cysteine (Cys1229) (Fig. 2B and fig. S4A). The fC7′ is surrounded by strictly conserved residues from the catalytic core (fig. S4), as initially observed for bacterial M.HhaI-DNA (14) and M.HaeIII-DNA (15) complexes, and is in proximity to the S-adenosyl homocysteine (AdoHcy) (fig. S5).
Fig. 2.
Insertion of amino acid side chains into the intercalation-like distortion of the bound DNA and base-specific recognition at the hemi-mCpG site in the covalent mDNMT1-DNA complex. (A) Sequencing and numbering system of the hemi-mCpG–containing 12-mer duplex. (B) Stabilization of the flipped-out target cytosine fC7′ (light blue ball) in the active site of the enzyme. (C) The 5-methyl group (green ball) of mC6 is anchored within a hydrophobic concave surface of mDNMT1. (D) Insertion of side chains of Met1235 and Lys1537 into the intercalation-type space, and buttressing by indole ring of Trp1512. The covalent bond between the sulfur atom of Cys1229 and the C6 atom of the target cytosine fC7′ is shown as a dark line. (E) End-on view of Met1235 and Lys1537 side chain insertion in the covalent complex. (F) Direct and water-mediated (labeled with purple W) hydrogen bond interactions target the Watson-Crick mC6-G6′ base pair from both grooves. (G) Stabilization of the repositioned guanine G7 through hydrogen-bonding interactions. (H) Schematic tabulation of direct and water-mediated hydrogen bond and electrostatic interactions between mDNMT1 and DNA in the covalent complex. Abbreviations for amino acids: C, Cys; E, Glu; F, Phe; G, Gly; H, His; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; W, Trp; Y, Tyr.
Although the CXXC domain and the CXXC-BAH1 linker together play an autoinhibitory role in preventing methylation of unmethylated CpG sites by DNMT1 (12), DNMT1 retains considerable substrate preference toward hemi-mCpG steps in the absence of these two elements (fig. S3), which suggests that the methyltransferase domain by itself encodes capabilities for evaluating the methylation status of the substrate.
In the current structure of the productive mDNMT1-DNA complex, the methyl group attached to the C-5 position of mC6 on the parental strand is directed toward a hydrophobic segment within the TRD domain (Fig. 2C). In particular, the indole ring of a tryptophan (Trp1512) slides into the DNA major groove upon complex formation and is partially stacked with the base of mC6, and, together with other hydrophobic residues (Cys1501, Leu1502, Leu1515, and Met1535), forms a shallow concave surface harboring the 5-methyl group of mC6 (Fig. 2C). This observation establishes the molecular basis for intrinsic preference of DNMT1 toward hemi-mCpG DNA substrates.
The formation of the productive mDNMT1-DNA complex involves infiltration of the DNA from both major (two TRD loops) and minor (catalytic loop) grooves (Fig. 1A and fig. S6), with the TRD domain undergoing concerted movement toward the DNA major groove by about 2 to 3 Å upon complex formation (Fig. 1B). We observe a large helical distortion around the central hemi-mCpG site resulting in the opening of an intercalation-type cavity 7.5 Å in length along the DNA helical axis (fig. S7, A and B). The side chain of Met1235 from the catalytic loop inserts into the DNA from the minor groove and occupies the space vacated by the extruded fC7′ on the target strand (Fig. 2, D and E). The side chain of Lys1537 from TRD loop 2 inserts into the DNA from the major groove and occupies the adjoining empty space on the parental strand (Fig. 2, D and E), which results from translation of the stacked orphan guanine (G7) by one step along the DNA helix toward the 3′ end, thereby flipping out the immediate downstream residue C8 (Fig. 2D). The antiparallel alignment of the side chains of Met1235 and Lys1537 is stabilized by a single hydrogen bond and further buttressed by the indole ring of Trp1512 (Fig. 2, D and E), with these distortions (fig. S8A) distinct from helical distortions reported for bacterial DNA methyltransferases (fig. S8, B and C). The mC6-G6′ Watson-Crick pair (Fig. 2F) and G7 of the noncanonical G7•G8′ pair (Fig. 2G) that bracket the intercalation site are recognized by elements from both the TRD and the catalytic core (Fig. 2H; stereo views in fig. S9, A and B).
The catalytic loop (residues 1227 to 1243) inserts into and spans both strands of the minor groove (Fig. 3A), making a network of direct and water-mediated contacts with mC6 and G7 on the parental strand, fC7′ on the target strand, and the phosphate backbone on either side of fC7′ (stereo view in fig. S10). In addition, both TRD loop 1 (residues 1501 to 1516) and loop 2 (residues 1530 to 1537) insert into and span both strands of the major groove centered about the hemi-mCpG step (fig. S6). TRD loop 1 contacts the phosphate backbone of the parental strand (Fig. 3B; stereo view in fig. S11), whereas TRD loop 2 is involved in base-specific recognition of the hemi-mCpG step and the DNA cavity (Fig. 3C; stereo view in fig. S12). Both TRD loops participate in formation of the hydrophobic concave surface that recognizes the 5-methyl group of mC6. Other DNA-interacting residues in the methyltransferase domain include Arg1272, Arg1314, Ser1421, Arg1427, Arg1492, and Thr1527 (Fig. 2H and fig. S13).
Fig. 3.
Insertion of the catalytic loop and a pair of TRD loops into the grooves centered about the hemi-mCpG site in the covalent mDNMT1-DNA complex. (A) Catalytic loop–DNA interactions. (B) TRD loop 1–DNA interactions. The imidazole ring of His1504 bridges the zinc finger with the backbone phosphate of mC6. (C) TRD loop 2–DNA interactions. (D) Intermolecular hydrogen-bond interactions between side chains of a segment of the BAH2 loop and DNA in the complex. (E) Methylation activities of mDNMT1(731-1602) and its mutants involving amino acids that line the mC-recognizing hydrophobic surface, or insert into the intercalation-type cavity, and catalytic and BAH2 loop residues involved in substrate recognition. Methylation activities were monitored after reaction on hemi-mCpG DNA substrate (black bars) or unmodified CpG DNA substrate (white bars) for 10 min. Error bars represent SD calculated from two measurements.
Formation of the covalent mDNMT1-DNA complex is further accompanied by the conformational change within the BAH2 domain, whose helical tip moves toward the DNA along with the TRD (fig. S14). As a result, the side chains of three residues (Ser981, Tyr983, and Lys985) immediately C-terminal to the tip, which are disordered in both free mDNMT1 and the autoinhibited mDNMT1-DNA complex, become ordered and engage in interactions with the phosphate backbone of the DNA target strand (Fig. 3D).
Guided by the structural analysis, we mutated a number of key and evolutionarily conserved residues (fig. S15) to monitor their impact on DNA methylation. First, we mutated residues lining the shallow concave surface that harbors the 5-methyl group of mC6. We observe that the Trp1512 → Ala mutant is essentially catalytically dead (Fig. 3E and fig. S16, A and B), consistent with the observation by a previous study (16) and the role of Trp1512 in base-stacking with the 5-methylcytosine. All other single mutants had their methylation rates reduced by factors of 2 to 4 for hemi-mCpG DNA substrate (Fig. 3E and fig. S16A) and by factors of 3 to 4 on unmethylated CpG substrate (Fig. 3E and fig. S16B); these findings suggest that the overall hydrophobic environment generated by a cluster of residues, rather than individual residues, is likely to play a dominant role in dictating the substrate specificity of mDNMT1.
Among residues that insert into the DNA intercalation site, the Met1235 → Ala mutant lost major catalytic activity on both hemi-mCpG and CpG DNA substrates (Fig. 3E and fig. S16, C and D). By contrast, the Lys1537 → Ala mutant exhibited a factor of 2 reduction in methylation rate for hemi-mCpG DNA substrate but an unanticipated factor of 2 increase in methylation rate on unmethylated CpG substrate (Fig. 3E and fig. S16, C and D). We have no definitive explanation for the latter result, but it is conceivable that such altered enzymatic behavior could arise from the impact of the Lys1537 → Ala mutation on the side chain orientations of Trp1512 and mC6 (Fig. 2C), which in turn could influence the substrate recognition.
Additional methylation kinetics were also performed for the Arg1237 → Ala mutant and for the Ser981 → Ala, Tyr983 → Ala, Lys985 → Ala triple mutant. In accordance with the structural analysis, both mutations show reduced catalytic activities upon DNA methylation (Fig. 3E and fig. S16, E and F). Taken together, our enzymatic methylation assays on selective mDNMT1 mutants appear to support structure-based molecular insights into substrate recognition by DNMT1.
Maintenance of DNA methylation patterns during cell division is crucial to animal development. DNMT1 is tethered to the DNA replication fork and is the major player in maintaining DNA methylation patterns during replication. The current structure of the productive mDNMT1-DNA complex, when combined with our earlier structure of the corresponding autoinhibited complex (12), establishes that two distinct but mutually supportive mechanisms are used by DNMT1 to ensure faithful maintenance of DNA methylation patterns after replication. As unmethylated CpG dinucleotides emerge from the replication complex, such sites are protected from de novo methylation by the autoinhibitory mechanism. On the other hand, as hemimethylated CpG sites emerge from the replication complex, specific recognition by the TRD domain of DNMT1 makes such sites optimal targets for maintenance DNA methylation. These two mechanisms, working in tandem and mutually supportive of each other, together with anticipated regulation by other proteins in the replication complex (such as UHRF1), ensure the fidelity of DNMT1-mediated maintenance DNA methylation.
Supplementary Material
Acknowledgments
Supported by funds from the Abby Rockefeller Mauze Trust and STARR and Maloris Foundations (D.J.P.). We thank T. Bestor and M. Goll for helpful discussions, and the staff of the NE-CAT beamlines at the Advanced Photon Source, Argonne National laboratory, for access and assistance with data collection. The structure of the complex has been deposited under accession no. 4DA4 in the Protein Data Bank.
References and Notes
- 1.Cheng X, Blumenthal RM. Structure. 2008;16:341. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goll MG, Bestor TH. Annu. Rev. Biochem. 2005;74:481. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 3.Law JA, Jacobsen SE. Nat. Rev. Genet. 2010;11:204. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki MM, Bird A. Nat. Rev. Genet. 2008;9:465. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 5.Li E, Bestor TH, Jaenisch R. Cell. 1992;69:915. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 6.Chuang LS, et al. Science. 1997;277:1996. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 7.Rountree MR, Bachman KE, Baylin SB. Nat. Genet. 2000;25:269. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 8.Leonhardt H, Page AW, Weier HU, Bestor TH. Cell. 1992;71:865. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 9.Lee JH, Voo KS, Skalnik DG. J. Biol. Chem. 2001;276:44669. doi: 10.1074/jbc.M107179200. [DOI] [PubMed] [Google Scholar]
- 10.Pradhan M, et al. Biochemistry. 2008;47:10000. doi: 10.1021/bi8011725. [DOI] [PubMed] [Google Scholar]
- 11.Callebaut I, Courvalin JC, Mornon JP. FEBS Lett. 1999;446:189. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 12.Song J, Rechkoblit O, Bestor TH, Patel DJ. Science. 2011;331:1036. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osterman DG, DePillis GD, Wu JC, Matsuda A, Santi DV. Biochemistry. 1988;27:5204. doi: 10.1021/bi00414a039. [DOI] [PubMed] [Google Scholar]
- 14.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. Cell. 1994;76:357. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 15.Reinisch KM, Chen L, Verdine GL, Lipscomb WN. Cell. 1995;82:143. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 16.Takeshita K, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9055. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.