Abstract
A hallmark of RNA silencing is a class of approximately 22-nucleotide RNAs that are processed from double-stranded RNA precursors by Dicer. Accurate processing by Dicer is crucial for the functionality of microRNAs (miRNAs). The current model posits that Dicer selects cleavage sites by measuring a set distance from the 3′ overhang of the double-stranded RNA terminus. Here we report that human Dicer anchors not only the 3′ end but also the 5′ end, with the cleavage site determined mainly by the distance (~22 nucleotides) from the 5′ end (5′ counting rule). This cleavage requires a 5′-terminal phosphate group. Further, we identify a novel basic motif (5′ pocket) in human Dicer that recognizes the 5′-phosphorylated end. The 5′ counting rule and the 5′ anchoring residues are conserved in Drosophila Dicer-1, but not in Giardia Dicer. Mutations in the 5′ pocket reduce processing efficiency and alter cleavage sites in vitro. Consistently, miRNA biogenesis is perturbed in vivo when Dicer-null embryonic stem cells are replenished with the 5′-pocket mutant. Thus, 5′-end recognition by Dicer is important for precise and effective biogenesis of miRNAs. Insights from this study should also afford practical benefits to the design of small hairpin RNAs.
RNase III proteins have central roles in RNA silencing by processing double-stranded (ds)RNA precursors into small RNA duplexes1. Drosha cleaves a primary precursor of miRNA (pri-miRNA) to release a hairpin-shaped pre-miRNA2. Dicer cuts the pre-miRNA near the terminal loop and generates a short miRNA duplex3–7. Dicer also participates in small interfering RNA (siRNA) production from long RNA duplexes. One strand of the small RNA duplex is subsequently loaded onto the Argonaute protein to yield an active RNA-induced silencing complex8–10.
Precise selection of cleavage sites by RNase III enzymes is critical in miRNA biogenesis because alterations in the cleavage site can change the abundance and/or targeting specificity of the miRNA. To determine the cleavage site, Drosha and Dicer recognize certain RNA structures and cleave a fixed distance away from the structure. In the case of Drosha, its cofactor DGCR8 (also known as Pasha) binds to the base of the stem–loop structure and locates the catalytic site of Drosha ~11 base pairs (bp) away from the single-stranded (ss)RNA–dsRNA junction11. Thus, the ssRNA–dsRNA junction serves as the reference point for Drosha processing.
Dicer, on the other hand, is known to measure ~22 nucleotides away from the 3′ end of the open terminus of dsRNA helices12–14. The crystal structure of Dicer from Giardia intestinalis in its free state and biochemical analyses indicated that the PAZ domain of Dicer anchors the 3′ overhang of the dsRNA terminus, and that the dsRNA stem is placed along the positively charged protein extension to reach the catalytic centre of Dicer15,16. This spatial arrangement would enable Dicer to measure a fixed distance from the 3′ end of the terminus (the ‘3′ counting model’).
Recent studies have shown that certain pre-miRNAs are modified at their 3′ end in the cell. The most common type of pre-miRNA modification is addition of non-templated uridyl residues17–20. According to the current 3′ counting model, such 3′-end modifications are expected to shift the Dicer cleavage site towards the open terminus. This would change the seed sequences of miRNAs originating from the 3′ strand and might even alter strand selection21,22.
Human Dicer counts from the 5′-phosphorylated end
To understand the impact of pre-miRNA uridylation on Dicer processing, we prepared synthetic pre-let-7a-1 with extra uridine residues at the 3′ end (Fig. 1a, left). The RNA was labelled at the 5′ end with [γ-32P] ATP and incubated with immunopurified human Dicer. With the 3′-elongated substrates, we expected to observe a shift of the cleavage site, which would yield shorter products from the 5′ strand. Surprisingly, the size of the major cleavage products remained the same (22 nucleotides) (purple arrowheads), indicating that the pre-let-7a-1 variants were cleaved at the same site regardless of the 3′ extension. We observed similar cleavage patterns when pre-miR-16-1 variants were used (Supplementary Fig. 1a). Substrates labelled at the 3′ ends were also cleaved at the same sites, excluding the possibility that the 3′ extension was trimmed back by a contaminating nuclease (Supplementary Fig. 1b). This processing pattern was not influenced by the sequences of the nucleotides added to the 3′ overhang: addition of adenosine or cytidine instead of uridine gave comparable results (Supplementary Fig. 1a and data not shown).
Figure 1. Human Dicer counts 22 nucleotides from the 5′ end of RNA to locate the cleavage site.
a, Processing of pre-let-7a-1 (Let-7) with extra 3′ uridine tails using immunopurified human Dicer. Numbers on top of RNA substrates indicate the length of 3′ overhang. Purple and green arrowheads indicate the 5′ counting and 3′ counting cleavage products, respectively (this standard is used throughout the figures). Asterisks mark radio-labelled phosphates, throughout the figures. nt, nucleotide. b, Dicer processing of dsRNAs (ds-35) with extra 3′ uridine tails. Grey arrowhead indicates the cleavage product from the opposite end. c, The 5′ end of ds-35 was extended by 1 nucleotide (ds-35+5′C) and incubated with Dicer. d, Dicer processing of the ds-35 substrates with either a 5′-terminal phosphate or a hydroxyl group (cyan circle). Although the juxtaposed lanes are not contiguous, they are all from a single gel; this is true for all images with dashed lines throughout the figures.
We next examined duplex RNAs with varying 3′ overhangs (Fig. 1b, left). Like pre-miRNAs, the predominant products from the dsRNAs were 22 nucleotides in length in spite of the differences at the 3′ overhangs, indicating that human Dicer may not be dependent on the 3′ end for cleavage site selection. We noticed another group of minor products, which were shortened as the 3′ overhang was elongated (green arrowheads). This minor cleavage pattern is expected of the 3′ counting model. Small amounts of 3′ counting products were also generated from pre-miRNAs (Fig. 1a and Supplementary Fig. 1a).
To summarize, we observed two types of Dicer cleavage events occurring in parallel. In the type predominant for the substrates used here, the cleavage site does not change upon 3′ end elongation. In another type, the cleavage site is determined based on the distance from the 3′ end. These results indicate that in addition to the 3′ end, Dicer may recognize other structural feature(s) of the RNA substrate.
Substrates with a 2-nucleotide 3′ overhang were cleaved most uniformly and efficiently, indicating that Dicer binds to these canonical substrates most strongly by using both 3′-dependent and 3′-independent mechanisms. When the terminal structure deviates from the optimal 2-nucleotide 3′ overhang, either one of the two mechanisms seems to be used by Dicer, yielding the mixture of two distinct product populations.
We next questioned what determines the 3′-independent selection of the cleavage site. Because the cleavage site is always 22 nucleotides away from the 5′ end, we presumed that the 5′ end may have a role. To test this idea, dsRNA substrates were extended by adding one cytidine at the 5′ end (Fig. 1c). Dicer still yielded products of ~22 nucleotides, indicating that the cleavage site shifted by 1 nucleotide when the 5′ end was extended. Hence, Dicer measures a set distance from the 5′ end. We refer to this as the ‘5′ counting model’.
Because endogenous substrates of Dicer carry a 5′-terminal phosphate group, we tested whether the 5′ phosphate has a role in the recognition of the 5′ end. Dicer processing was performed with two sets of dsRNA substrates that carry either a 5′-terminal phosphate or a hydroxyl group (Fig. 1d). Both sets were labelled at the 3′ end of the opposite terminus to detect the cleavage products. The cleavage patterns of the two sets differed markedly. The phosphorylated dsRNAs followed the 5′ counting rule whereas the dsRNAs lacking the 5′ phosphate mainly obeyed the 3′ counting rule (Fig. 1d). We also noticed that the length of the products became more variable in the absence of the 5′ phosphate. Thus, Dicer interaction with the 5′-terminal phosphate helps precisely locate the enzyme on the substrate.
To investigate whether the 5′ counting rule can be generalized to other pre-miRNAs, we examined additional pre-miRNAs with 1–3 extra uridine residues at the 3′ end (Supplementary Fig. 2). Pre-miR-143, pre-miR-148b, pre-miR-27b and pre-miR-151 largely followed the 5′ counting rule, whereas pre-miR-200c showed a mixed pattern. Pre-miR-24-2 and pre-miR-142 complied mainly with the 3′ counting rule. Hence, although the 5′ counting applies to most pre-miRNAs tested, the relative contribution of the 5′ and 3′ ends seems to vary among pre-miRNAs. We noticed that pre-miRNAs following the 3′ counting rule are relatively stable at the stem termini, whereas pre-miRNAs following the 5′ counting rule have less stable structures at the terminal base pair (mismatch, G–U, or A–U pair) (Supplementary Fig. 2a). Thus, Dicer may require a flexible (thermodynamically unstable) 5′ terminus to efficiently recognize the 5′ end. To test this notion further, we changed Mg2+ concentrations in our processing assays because the Mg2+ ion is known to stabilize the dsRNA structure23. Mg2+ ions indeed had a significant influence on Dicer processing of pre-miR-24-2: the 3′ counting rule prevails at 4 mM whereas the 5′ counting rule predominates at 0.5 mM (Supplementary Fig. 3a). A similar observation was made when a duplex RNA with a 3-nucleotide overhang was used (Supplementary Fig. 3b, c). It is likely that at a low Mg2+ concentration, the terminal stem region tends to unwind, thereby facilitating the 5′-end recognition by Dicer. Given that the physiological concentration of free Mg2+ ions is estimated to be 0.5–1 mM24, the 5′ counting rule may apply to most 3′-modified pre-miRNAs in vivo, although we do not exclude the possibility that some pre-miRNAs with stable termini may follow the 3′ counting rule.
Conservation of the 5′ counting rule
As the 3′ counting model was proposed mainly from the work on Giardia Dicer15,16, we examined Giardia Dicer using our substrates (Fig. 2a, lanes 6–10). Unlike human Dicer, Giardia Dicer cleaved dsRNA substrates by strictly measuring from the 3′ end and yielded slightly larger products (24–26 nucleotides), as previously observed16. We also noticed that Giardia Dicer cleaves the blunt-ended substrate most efficiently, whereas human Dicer shows a strong preference for the 2-nucleotide 3′ overhang terminal structure. Thus, Giardia Dicer differs significantly from human Dicer in substrate recognition.
Figure 2. The 5′ counting rule is conserved in Drosophila Dicer-1 but not in Giardia Dicer.

a, Flag-tagged human Dicer and Giardia Dicer were immunopurified (IP) and incubated with ds-35 substrates. b, Immunopurified Drosophila Dicer-1–Loqs-PB complexes were incubated with Let-7 substrates. The ds-35 (+2) substrate was used as a negative control.
We next investigated the processing pattern of Drosophila Dicer-1. Dicer-1 acts in complex with the co-factor Loquacious-PB (Loqs-PB) for the processing of pre-miRNAs25–28, while another Dicer (Dicer-2) and its cofactor R2D2 are responsible for siRNA generation29. When pre-let-7a-1 variants were incubated with the Dicer-1–Loqs-PB complex, all variants were cleaved into 22-nucleotide products, without any detectable products following the 3′ counting (Fig. 2b). Taken together, the 5′-end recognition mechanism may be conserved in metazoans but not in organisms such as Giardia, which represents one of the earliest surviving branches of the eukaryotic phylogenetic tree.
Identification of the 5′-recognition pocket
To identify the motif that binds to the 5′ end, we selected putative RNA interacting residues (basic, polar) located around the PAZ domain and mutated them to alanines (Fig. 3a). The residues were selected on the basis of three criteria. First, we predicted the three-dimensional structure of the region encompassing the PAZ domain by applying I-TASSER simulation30 (Supplementary Fig. 4). Assuming that the 5′-end-binding residues are located ~20 Å away from the conserved 3′-end-binding pocket (3′ pocket), which is the expected distance between the 5′ and 3′ ends of a 2-nucleotide 3′ overhang structure14, we selected putative RNA interacting residues (R811, R986 and R993). Second, we solved the crystal structure of a human Dicer fragment spanning the ‘platform–PAZ–connector-helix’ domains (Supplementary Fig. 5 and manuscript in preparation). Interestingly, we found an inorganic phosphate that is coordinated to the side chains of R778, R780, R811 and H982, suggestive of a potential phosphate-binding pocket. The bound phosphate is located ~20 Å away from the 3′ pocket in the PAZ domain. R986 and R993 are in a disordered part of the structure (Supplementary Fig. 5a) but they are predicted to be in the vicinity of R778. Finally, in deciding on the residues for the mutagenesis study, we took into account phylogenetic conservation.
Figure 3. Residues required for the recognition of dsRNA terminus.
a, Domain organization of human Dicer. The mutated sites in the 5′ and 3′ pockets are shown as purple and green bars, respectively, and the mutations are listed. a.a., amino acids. b, Immunopurified wild-type and mutant Dicer proteins were incubated with ds-35 substrates. c, Amino acid sequences of the Dicer proteins from various species (At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Gi, Giardia intestinalis; Hs, Homo sapiens; Mm, Mus musculus; Sp, Schizosaccharomyces pombe; Xl, Xenopus laevis) are aligned using ClustalX program and the region spanning the 5′ pocket is presented. The 5′-interacting residues are indicated with boxes.
The mutants at R778/R780, R811 and R986/R993 produced a significantly smaller amount of 5′ counting products, indicating that these mutants are defective in 5′-end recognition (Supplementary Fig. 6a). When we combined these mutations to generate a ‘5′ mutant’ (R778A/R780A/R811A/H982A/R986A/R993A), the cleavage pattern clearly shifted to the 3′ counting one (Fig. 3b, lanes 6–10). The change in the cleavage pattern is highly specific to the identified residues; the point mutations at S984, H994 and W1014, which are located closely to the 5′-interacting residues, did not affect the cleavage pattern (Supplementary Fig. 6b). Thus, our results indicate that a basic motif composed of R778, R780, R811, R986 and R993 (5′ pocket) is required for 5′-end recognition. These amino acids are conserved in Drosophila Dicer-1 but not in Giardia Dicer (Fig. 3c).
As a control, we introduced mutations at Y926 and R927, which are conserved and located in the 3′ pocket of the PAZ domain (Fig. 3a). This mutant (3′ mutant) lost most of the 3′ counting products, indicating that the 3′ counting mechanism is disrupted in this mutant (Fig. 3b, lanes 11–15). This result is consistent with previous findings that 3′ counting is dependent on the interaction between the 3′ end and the PAZ domain14–16.
Overall processing efficiency was reduced in the 5′ mutant as well as in the 3′ mutant (Fig. 3b), consistent with the notion that human Dicer utilizes both ends for substrate binding. The substrate with a 2-nucleotide overhang was cleaved more heterogeneously by the 5′-mutant Dicer (21–23 nucleotides) compared to wild-type Dicer (22 nucleotides) (Fig. 3b, compare lanes 3 and 8), indicating that the 5′ mutant lacks precision in processing. Altogether, our in vitro data indicate that 5′-end recognition is important not only for 3′-modified pre-miRNAs but also for canonical substrates such as unmodified pre-miRNAs.
The 5′ pocket is required for miRNA biogenesis
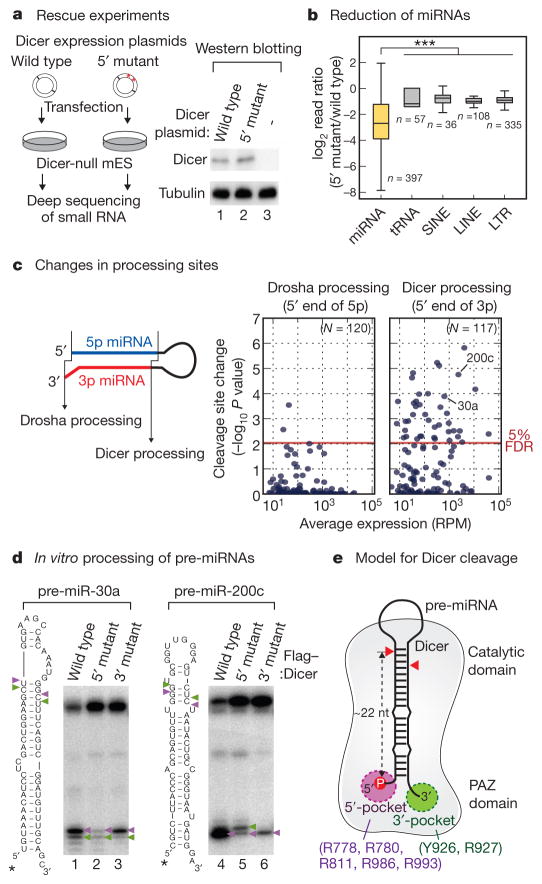
To evaluate the biological relevance of our findings, we introduced Dicer expression plasmids (wild type and 5′ mutant) into Dicer-null embryonic stem (ES) cells31. The small RNA populations from two biological replicates were sequenced (Fig. 4a and Supplementary Table 1). Wild-type Dicer successfully replenished the miRNA pool whereas the 5′ mutant showed significant defects. The overall miRNA abundance decreased in the 5′-mutant-expressing cells (Fig. 4b, Supplementary Fig. 7 and Supplementary Table 2), although some miRNA isoforms increased owing to cleavage site alterations (see later). Other RNA species such as transfer RNAs were unaffected, indicating that the differences are specific to miRNAs. The marked reduction of miRNA abundance was further confirmed by northern blotting (Supplementary Fig. 7d).
Figure 4. The 5′ pocket is critical for efficient and accurate cleavage in vivo.
a, Left, scheme for the Dicer rescue experiment using Dicer-null mouse ES cells. Right, western blotting using anti-Dicer antibody shows the comparable expression of wild-type and mutant Dicer. Tubulin was used as a loading control. b, Transcripts mapped to miRNA loci specifically decreased in the mutant library (P = 8.13×10−59, Mann–Whitney U-test, first replicate). Box represents the first and third quartiles and the internal bar indicates the median. Whiskers denote the lowest and highest values within 1.5 × interquartile range of the first and third quartiles, respectively. n represents the number of transcripts. c, Left, Drosha and Dicer cleavage sites were inferred from the 5′ end of 5p and 3p miRNAs, respectively. Right, the dissimilarity of cleavage pattern was quantified by Kullback–Leibler divergence (KLD), and statistical significance was measured using two-sample t-test. Red line corresponds to 5% false discovery rate (FDR). RPM, reads per million. d, Pre-miR-30a and pre-miR-200c were incubated with the same amount of wild-type or mutant Dicer protein. e, Double anchor model for Dicer processing.
We next examined the impact of the 5′-pocket mutation on processing site selection. As the 3′ ends of small RNAs are known to be frequently modified after Dicer processing17,20, we used the 5′ ends of miRNAs (or miRNAs*) to infer cleavage sites. Drosha creates the 5′ end of 5′-strand miRNAs (5p miRNAs) whereas Dicer makes the 5′ end of 3′-strand miRNAs (3p miRNAs) (Fig. 4c, left). When the wild-type and 5′-mutant libraries were compared, ~35% of miRNAs showed significant changes in Dicer cleavage sites (41 out of 117; below 5% false discovery rate) (Fig. 4c, right panel, Supplementary Figs 8 and 9 and Supplementary Table 2). In contrast, Drosha processing sites remained largely unchanged (Fig. 4c, left panel, and Supplementary Fig. 9), indicating that the differences in the small RNA population are due to the mutation in Dicer. The changes in Dicer cleavage sites often led to seed alterations and/or strand switches (Supplementary Table 3). The deep sequencing results are highly consistent with those from in vitro assays.
As further confirmation, we carried out in vitro processing of pre-miR-30a and pre-miR-200c, which showed significant changes in the 5′-mutant-expressing cells (Fig. 4c and Supplementary Fig. 10). The 5′-mutant Dicer was markedly impaired in both efficiency and accuracy of pre-miRNA processing in vitro (Fig. 4d). The 3′-pocket mutation reduced processing activity without significantly altering cleavage site selectivity (Fig. 4d). Taken together, the 5′ pocket is critical for efficient and precise generation of miRNA.
Discussion
This study provides new insight into the mechanism of Dicer processing (see Fig. 4e for a model). The basic 5′ pocket identified in our study is positioned in close proximity to the 3′ pocket on the same face of the Dicer protein. The 5′ and 3′ pockets of Dicer are positioned for the simultaneous accommodation of the 5′ and 3′ ends, respectively, of the substrate with a 2-nucleotide 3′ overhang. The 5′ pocket anchoring the 5′ phosphate is particularly important for securing Dicer in a fixed position, which enables Dicer to generate uniform products. Ongoing structural studies on RNA complexes of PAZ-containing fragments of metazoan Dicer will elucidate further the molecular basis of Dicer processing.
It is interesting to contemplate the evolutionary implications because the 5′-pocket motif is highly conserved among most miRNA-producing Dicer homologues. This motif is missing in Dicer from lower eukaryotes such as Giardia and fungi (Fig. 3c), which lack the miRNA pathway. Plant DCL1, the miRNA-producing enzyme, is only partially conserved in this region. Therefore, on the basis of the amino acid sequences, it is difficult to infer the existence of an orthologous motif. It is notable that, in Drosophila, the 5′-pocket motif seems to be conserved only in Dicer-1 (miRNA-producing enzyme) but not in Dicer-2 (siRNA-generating enzyme). The fact that the 5′ pocket is conserved only in Dicers with pre-miRNA processing activity implicates the 5′ counting mechanism as an important factor in miRNA maturation.
In the cell, 3′ ends of pre-miRNAs can be modified by exonucleases or nucleotidyl transferases17–20,32. It is yet unclear to what extent the 5′-pocket motif contributes to the processing of such modified pre-miRNAs in vivo, as we would need to determine exactly what fractions of pre-miRNAs undergo 3′-end modifications in vivo. Nonetheless, because the 5′ end of pre-miRNAs is generally more homogenous than the 3′ end33, it is tempting to speculate that the 5′ pocket has evolved to utilize the 5′ end, which is more reliable than the 3′ end. Using the 5′ end as a major reference point for positioning may ensure accurate processing of pre-miRNAs. In the case of the siRNA pathway, precise processing is not critical because, unlike miRNAs, any cleavage frame would result in functional siRNAs.
RNA interference (RNAi) in mammalian systems is commonly induced by expressing small hairpin RNAs (shRNAs) from an RNA polymerase II or III promoter, but the technology often suffers from inefficient and inaccurate Dicer processing34–36. On the basis of our findings, a hairpin with a 5′-terminal phosphate and a 2-nucleotide 3′ overhang should fit most optimally into the 5′ and 3′ pockets of Dicer. Also, it would be interesting to test whether a 5′ triphosphate could be efficiently accommodated into the 5′ pocket, as shRNAs driven by RNA polymerase III promoters bear a 5′ triphosphate. Understanding how human Dicer generates miRNAs will enable us to improve further the efficacy and safety of RNAi technology.
METHODS
Cell culture and transfection
HEK293T cells were grown in DMEM (Welgene) supplemented with 10% fetal bovine serum (Welgene). S2 cells were grown in HyClone SFX-Insect (Thermo Scientific) supplemented with 10% fetal bovine serum (Welgene). Dicer knockout mouse ES cells (a gift from G. J. Hannon) were grown on mouse CF-1 feeder cells or gelatin-coated dishes in knockout DMEM (Gibco, Invitrogen) supplemented with 15% fetal bovine serum (Gibco, Invitrogen), nonessential amino acids (Gibco, Invitrogen), 2 mM L-glutamine (Sigma), 0.1 mM 2-mercaptoethanol (Sigma) and 1,000 units ml−1 leukaemia inhibitory factor (Chemicon).
For HEK293T cells, transfection was carried out using the calcium-phosphate method. S2 cells were transfected using the DDAB method as previously described37.
Cloning and mutagenesis
Giardia Dicer cDNA was amplified from Giardia Dicer-pFastBac HTa plasmid (a gift from J. A. Doudna) by PCR using the following primers: 5′-GGATCCATGCATGCTTTGGGACACTG-3′; and 5′-GATATCGAGACTGCAGGCTCTAGATTCG-3′. PCR products were cloned into pGEM-T easy vector (Promega) and subsequently cloned into Flag-pcDNA3 vector (Invitrogen) at the BamHI and EcoRV site.
To introduce mutations into Dicer, QuickChange Site-Directed Mutagenesis Kit (Stratagene) was used. Mutated plasmids were confirmed by sequencing and subcloned into unmodified Flag-Dicer-pcDNA3.1 vector. The primer sequences used for the mutagenesis are provided in Supplementary Table 4.
Immunoprecipitation and in vitro Dicer processing
For immunoprecipitation of Flag-Dicer, HEK293T cells were grown on 10-cm or 15-cm dishes and harvested at 48 h after Flag-Dicer-pcDNA3.1 expression plasmid transfection. The cells were incubated with lysis buffer (500 mM NaCl, 1 mM EDTA, 20 mM Tris (pH 8.0), 1% Triton X-100) for 20 min on ice followed by sonication and centrifugation twice at 16,000g for 10 min at 4 °C. The supernatant was incubated with 10 μl of anti-Flag antibody conjugated to agarose beads (anti-Flag M2 affinity gel, Sigma) with constant rotation for 1 h at 4 °C. The beads were washed three times with lysis buffer and then four times with buffer D (200 mM KCl, 20 mM Tris (pH 8.0), 0.2 mM EDTA). The reactions were performed in a total volume of 30 μl in 2 mM MgCl2, 1 mM DTT, 1 unit μl−1 ribonuclease inhibitor (Takara), 5′-end-labelled pre-miRNA of 1×104 to 1×105 c.p.m. and 15 μl of the immunopurified proteins in buffer D. The reaction mixture was incubated at 37 °C for 60–90 min. RNA was purified from the reaction mixture by phenol extraction and separated on 15% urea polyacrylamide gel. Along with Decade marker (Ambion), synthetic hsa-let-7a RNA (22 nucleotides) was 5′-end labelled and used as a size marker, because the 20-nucleotide RNA in Decade marker is often degraded to 18–19 nucleotides as we previously reported11.
For preparation of Drosophila Dicer-1, S2 cells confluent in a 10-cm dish were transfected with Myc-Loquacious-PB-pRmHa3 expression plasmid (a gift from M. C. Siomi). We used the Loqacious-PB immunoprecipitates instead of the Dicer-1 immunoprecipitates in this experiment because the ectopic expression level of Dicer-1 was too low. After 1 day, 1 mM CuSO4 was added to the medium and cells were collected 2 days after CuSO4 treatment. The cells were incubated with lysis buffer for 30 min on ice, followed by sonication and centrifugation at 16,000g for 10 min at 4 °C. The supernatants were pre-cleared by incubation with 10 μl Protein A-Sepharose bead (GE Healthcare) for 2 h. Then, pre-cleared extract was incubated with 20 μl Protein A-Sepharose bead bound to anti-Myc antibody, 9E10, for 2 h at 4 °C. The beads were washed three times with lysis buffer and then four times with buffer D and used for in vitro Dicer processing.
Preparation of substrates
Pre-let-7a-1, pre-miR-16-1, pre-miR-24-2 (mouse), pre-miR-142, pre-miR-143, pre-miR-200c and pre-miR-30a were synthesized by ST Pharm. The sequences are presented in the figures. The pre-miRNA substrates with different 3′-overhang lengths and pre-miR148b, pre-miR-27b and pre-miR-151 were generated by ligating two synthetic ssRNAs as described previously32. The sequences of RNA used for ligation are listed in Supplementary Table 5. The RNAs were labelled at the 5′ end with T4 polynucleotide kinase (T4 PNK, Takara) and [γ-32P] ATP. Sequences of all endogenous pre-miRNAs used in our analysis are listed in Supplementary Table 6.
For preparation of dsRNA substrates, a synthetic ssRNA was labelled at the 5′ end with [γ-32P] ATP and T4 PNK. After phenol extraction, the labelled RNA was annealed to the complementary RNA by heating at 90 °C for 2 min and incubating at 30 °C for 2 h. In Fig. 1d, one strand of RNA was ligated to [α-32P] pCp and treated with calf intestinal alkaline phosphatase (Takara) to generate the labelled 3′ end with a hydroxyl group. To attach a phosphate group at the 5′ end, 3′-end-labelled RNAs were incubated with cold ATP and T4 PNK (Takara). Phenol extraction of RNA was performed after each reaction. Then, the labelled RNA was annealed to the RNA as described earlier.
In vitro addition of uridine residues to pre-miRNAs
A terminal nucleotidyl transferase, TUT4, is able to add 1–3 nucleotides of uridine residues at the 3′ end of pre-miRNA in the absence of Lin28 protein in vitro (I. Heo et al., unpublished data). For this reaction, Flag-TUT4 expression plasmid was transfected in HEK293T cells using the calcium-phosphate method. After 48 h, total cell extract was prepared in buffer D by sonication and centrifugation at 16,000g for 10 min at 4 °C. Thirty microlitres of reaction mixture contains 15 μl total cell extract (10 μg), 3.2 mM MgCl2, 1 mM DTT, 0.25 mM UTP, 1 unit μl−1 ribonuclease inhibitor (Ambion), and 5′-end-labelled pre-miRNA of 1×104 to 1×105 c.p.m. The reaction mixture was incubated at 37 °C for 15 min. After phenol extraction, the uridylated pre-miRNAs were gel purified and used for in vitro Dicer processing.
Three-dimensional structure prediction of Dicer fragment
The three-dimensional structure of Dicer fragment containing the PAZ domain was predicted by I-TASSER simulation30 (http://zhanglab.ccmb.med.umich.edu/I-TASSER) with amino acid sequences 751–1070. Crystal structure of PAZ domains from Dicer (PDB accession code 2FFL) and Argonautes (PDB codes 1U04, 3DLB, 1R4K), together with RumA, a 23S ribosomal RNA methyltransferase (PDB code 1UWV), were used as templates for the comparative modelling. Among the five models predicted from the server, the one with a high C score (−2.36) and an organized structure was chosen.
Structure-based identification of the 5′ pocket in human Dicer
Diffraction quality crystals were grown for the complex of Dicer ‘platform–PAZ–connector-helix’ cassette (residues 755–1055) and a self-complementary AGCGAAUUCGCUU duplex (underlined segment forms duplex) in phosphate-containing solution. The crystals of the complex belonged to space group I222, diffracted to 2.6 Å, and the structure of the complex was refined to Rwork = 19.7 and Rfree = 23.7. In this structure, inorganic phosphate, which is anchored by basic residues (Arg 778, Arg 780, Arg 811 and His 982), reveals the potential 5′-phosphate-binding pocket (Supplementary Fig. 5).
Dicer rescue experiments
For transfection, Dicer knockout mouse ES cells were separated from feeder cells and 1,500,000 cells were seeded on gelatin-coated 6-well plates one day before transfection. Ten micrograms of plasmids (wild-type Dicer-pCK or 5′-mutant Dicer-pCK) were added to each well along with 10 μl of Lipofectamine 2000, according to the manufacturer’s protocol (Invitrogen). Protein and RNA was extracted at 48 h after transfection. To determine the protein levels, western blotting was performed using anti-Dicer and anti-tubulin (Abcam) antibodies. Expression of RNA was confirmed by northern blotting using the following probes: mmu-miR-293 (5′-ACACTACAAACTCTGCGGCACT-3′); mmu-miR-101a (5′-TTCAGTTATCACAGTACTGTA-3′); mmu-miR-16 (5′-CGCCAATATTTACGTGCTGCTA-3′); and tRNA-Lys-AAG (5′-GAGATTAAGAGTCTCATGCTC-3′).
To prepare small RNA cDNA libraries, RNA was extracted using TRIzol reagent (Invitrogen) or mirVana miRNA isolation kit (Ambion) and separated on 15% urea-PAGE. RNA of 17–26 nucleotides in length was gel purified and ligated to the 3′ adaptor using truncated T4 RNA Ligase2 (NEB) in ATP-free conditions. Subsequently, the ligation product was gel purified and ligated to the 5′ adaptor using T4 RNA Ligase1 (NEB). The final ligation product was gel purified and used for reverse transcription using SuperScript II (Invitrogen). The cDNA was PCR-amplified with Phusion DNA polymerase (NEB). The resulting libraries were sequenced using Illumina Genome Analyser II.
Sequence analysis
The essential workflow for early sequence analysis was performed as previously described38 with few modifications. After removing sequence reads including very low quality bases (<10 in phred quality), the 3′ adaptor sequence was trimmed from the reads using a 5′-free variant of the Smith–Waterman algorithm (scoring parameters: 2 for match, −3 for mismatch, −3 for linear gap). Then, we dropped short (≤17 nucleotides) or repetitive sequences (0.7 and 1.5 for mono- or dinucleotide entropy of each sequence). The filtered sequences were aligned to Illumina adaptor and primer sequences using the BWA short-read aligner39 with 4 of allowed maximum edit distance, then matched reads were removed from further analysis. In the same way, the remaining sequences were aligned to the mouse genome mm9 assembly, which is downloaded from the University of California at Santa Cruz (UCSC; http://genome.cse.ucsc.edu/). Annotations for aligned regions were retrieved using in-house software from RefSeq, RepeatMasker and miRBase (downloaded from UCSC or miRBase on April 8, 2011). Software used in data processing and analysis can be downloaded from http://www.narrykim.org/s/park-dicer-2011/.
Analysis of cleavage site change
We first selected miRBase stem–loops that are relatively unaffected by reads aligned to multiple miRNA loci to avoid artefacts from over- or underestimated read counts. Stem–loops with more than 90% reads aligned to a single stem–loop in every single sequencing lane were chosen for the later steps. Kullback–Leibler divergence (KLD) was used to quantify cleavage site change (difference of 5′-end position frequency) between two sequencing samples. To measure statistical significance of cleavage site change, Student’s t-test was performed for KLDs between wild-type and 5′-mutant Dicer rescued samples, and KLDs between wild-type Dicer rescued samples and J140, mouse embryo at 7.5 day20 or R1. Multiple testing correction was applied using the Benjamini–Hochberg method41.
Supplementary Material
Acknowledgments
We are grateful to G. Hannon for Dicer-null mouse ES cells; J. Doudna for Giardia Dicer cDNA; and M. Siomi for Drosophila Dicer-1, Dicer-2, Loqs-PB and R2D2 constructs. We also thank the members of the V.N.K. laboratory, particularly C. Joo, M.-J. Yoon, K.-H. Yeom and A. Cho for discussions and technical help. The V.N.K. laboratory was supported by the Creative Research Initiatives Program (2010000021) and National Honor Scientist Program (20100020415) through the National Research Foundation and the BK21 Fellowships (J.-E.P. and H.C.) from the Ministry of Education, Science and Technology. Research in the D.J.P. laboratory was supported by the National Institutes of Health.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions J.-E.P., I.H. and D.J. performed biochemical and cell biological experiments. H.C. carried out bioinformatic analyses. Y.T., D.K.S. and D.J.P. performed structural studies. J.-E.P., I.H. and V.N.K. designed the study and wrote the paper.
Small RNA sequencing data were deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE27903.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Ketting RF, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 5.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 6.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicerin the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 7.Knight SW, Bass BL. A role for the RNase III enzymeDCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 9.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 10.Mourelatos Z, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 12.Vermeulen A, et al. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 15.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nature Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 16.MacRae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 17.Burroughs AM, et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Ye C, Ramirez D, Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS ONE. 2009;4:e7566. doi: 10.1371/journal.pone.0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 23.Serra MJ, et al. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. RNA. 2002;8:307–323. doi: 10.1017/s1355838202024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunther T. Concentration, compartmentation and metabolic function of intracellular free Mg2+ Magnes Res. 2006;19:225–236. [PubMed] [Google Scholar]
- 25.Jiang F, et al. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1–Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyoshi K, Miyoshi T, Hartig JV, Siomi H, Siomi MC. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA. 2010;16:506–515. doi: 10.1261/rna.1952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Förstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 30.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both microRNAs and their miRNA* strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nature Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 35.Silva J, Chang K, Hannon GJ, Rivas FV. RNA-interference-based functional genomics in mammalian cells: reverse genetics coming of age. Oncogene. 2004;23:8401–8409. doi: 10.1038/sj.onc.1208176. [DOI] [PubMed] [Google Scholar]
- 36.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nature Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 37.Han K. An efficient DDAB-mediated transfection of Drosophila S2 cells. Nucleic Acids Res. 1996;24:4362–4363. doi: 10.1093/nar/24.21.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.