Summary
Among their diverse roles as transcriptional regulators during development and cell fate specification, the RUNX transcription factors are best known for the parts they play in haematopoiesis. RUNX proteins are expressed throughout all haematopoietic lineages, being necessary for the emergence of the first haematopoietic stem cells to their terminal differentiation. Although much progress has been made since their discoveries almost two decades ago, current appreciation of RUNX in haematopoiesis is largely grounded in their lineage‐specifying roles. In contrast, the importance of RUNX to immunity has been mostly obscured for historic, technical and conceptual reasons. However, this paradigm is likely to shift over time, as a primary purpose of haematopoiesis is to resource the immune system. Furthermore, recent evidence suggests a role for RUNX in the innate immunity of non‐haematopoietic cells. This review takes a haematopoiesis‐centric approach to collate what is known of RUNX's contribution to the overall mammalian immune system and discuss their growing prominence in areas such as autoimmunity, inflammatory diseases and mucosal immunity.
Keywords: autoimmunity, haematopoiesis, immune system, mucosal immunity, RUNX transcription factors
Introduction: the RUNX family of transcription factors
The RUNX proteins are evolutionarily conserved transcription factors that share the Runt‐related domain for DNA binding and heterodimerization with a common partner, CBFβ.1 In diverse metazoans, RUNX proteins function as critical lineage determinants and in mammals are represented by RUNX1, 2 and 3 (also known as AML1/3/2, PEBP2αB/A/C, CBFα2/1/3). Mammalian RUNX genes share additional common features, which include the use of two promoters, termed P1 (distal) and P2 (proximal), through which different RUNX isoforms are derived.1 RUNX1 was originally identified as a frequent target of leukaemogenic chromosomal translocations in human acute myelogenous leukaemia.2 RUNX1 is required for the generation and maintenance of haematopoietic stem cells (HSC) and the differentiation of diverse haematopoietic lineages.3 RUNX2 is a master regulator of osteoblast differentiation and necessary for bone and cartilage development and maintenance.4 Haploinsufficiency in RUNX2, due largely to mutations in its DNA‐binding Runt‐domain, results in human cleidocranial dysplasia.1 The expression of RUNX3 is necessary for the differentiation of TrkC‐positive dorsal root ganglion neurons and observed in a range of epithelial tissues.1 Of relevance to this review, RUNX3 is expressed across haematopoietic lineages where its distribution overlaps significantly with RUNX1, while remaining distinct. In comparison, the expression of RUNX2 in haematopoietic lineages is less studied, except in specific contexts, while CBFβ isoforms are ubiquitously expressed across many tissues at approximately the same ratio.5, 6 As a result of their profound involvement in haematopoiesis and the maturation of cell lineages involved in virtually all facets of immunology, RUNX proteins hold important roles in host immunity. These functions will be highlighted and discussed in the following sections that describe RUNX's contribution to each major haematopoietic lineage.
RUNX and haematopoietic stem cells
The HSC are the multipotent stem cells from which all haematopoietic lineages are derived. Developmentally, the mammalian haematopoietic system can be demarcated into three discrete phases: (i) primitive haematopoiesis during embryogenesis, (ii) definitive haematopoiesis in late fetal development, and (iii) adult haematopoiesis. The importance of RUNX proteins to haematopoiesis was first revealed in the complete absence of definitive haematopoiesis in Runx1 knockout mice. The loss of Runx1 completely abolished the transition of the first definitive HSC from haemogenic endothelial cells at the aorta–gonad–mesonephros region.7, 8, 9, 10, 11, 12 Runx1 was also necessary for the maintenance of HSC in adult haematopoiesis, though not essential for their biogenesis. Several studies showed that conditional targeting of Runx1 in bone marrow (BM) HSC in adult mice by Mx1‐Cre resulted in defective T‐ and B‐lymphocyte development at various stages and a blockade of megakaryocyte maturation.13, 14, 15 Unexpectedly, some studies reported an initial expansion of the Runx1‐deficient HSC that was followed by their progressive exhaustion.13, 14, 15, 16, 17 These paradoxical phenotypes were attributed in part to the premature exit of HSC from its cellular niche because of the mis‐regulation of the chemokine receptor Cxcr4.17, 18 These HSC defects were strongly accentuated when Runx3 was concurrently deleted, suggesting that Runx proteins served overlapping functions in the homeostatic maintenance of HSC.19 Indeed, Runx1;Runx3 deletion in the BM led to profound differentiation and proliferative disorders across all haematopoietic lineages, eventually causing bone marrow failure or myeloproliferative disorder.19 Similarly, pan‐haematopoietic deletion of Cbfb severely impaired differentiation of all haematopoietic lineages and resulted in proliferative disorder in myeloid cells.20, 21 Interestingly, Mx1‐cre targeting of Cbfb did not cause lethal bone marrow failure observed in Runx1;Runx3 double knockout mice, concordant with a Cbfβ‐independent role for Runx1 and Runx3 in DNA repair.19
The role of RUNX in thymocyte differentiation
A major aspect of RUNX's contribution to immune function is in the differentiation and maturation of T cells, which has been investigated in detail through mouse genetics (reviewed in Collins et al.22). In all stages of T‐cell development, RUNX proteins are expressed in overlapping but distinct patterns and differential expression levels5 (Table 1). The expression of Runx proteins in the thymus goes beyond the haematopoietic system and extends to the thymic epithelial cells (Sela A & Abramson J, personal communication). Runx1 is most highly expressed in the thymic cortex where CD4− CD8− double‐negative (DN) thymocytes reside, reaching an apex at the DN3 stage before a sharp decline.5, 23, 24, 25 Thereafter, Runx1 expression is maintained in CD4+ CD8+ double‐positive (DP) and CD4+ and CD8+ single‐positive (SP) cells, with the expression in the CD4+ SP cells being higher.25, 26, 27 In line with this, the targeting of Runx1 in BM by Mx1‐Cre and thymocytes by lck‐Cre resulted in a maturation block of DN3 and DN4 thymocytes, respectively. Moreover, the ablation of Runx1 using Cd4‐cre disrupted DP to SP transition.13, 26 In human and mouse, these events coincide with the involvement of Runx1 in T‐cell receptor (TCR) ‐γδ and TCR‐αβ rearrangement, respectively (Fig. 1).28, 29, 30, 31 Runx1 orchestrates TCR rearrangement events by binding to the corresponding TCR chain enhancers and, in human Dδ2 to Dδ3 rearrangement, resulting in the recruitment of recombination activating gene 1 (RAG1) through physical interaction.32 Collectively, these studies firmly establish Runx1 as a key driver of early T‐cell development.
Table 1.
The roles of RUNX complex in the differentiation and effector functions of haematopoietic lineages
| Lineage | Differentiation step | RUNX/CBFβ | Description | Immune functions affected when RUNX/CBFβ is disrupted | References |
|---|---|---|---|---|---|
| T lymphocytes | Thymocytes | Runx1 | DN2 to DN3 transition by regulating IL7Rα, TCR‐γδ rearrangement | Defective TCR rearrangement and thymocyte maturation | 13, 14, 15, 26, 28, 29, 34 |
| CD4/CD8 | Runx3,1 | DP to CD8+ SP differentiation, TCR‐αβ rearrangement | Reduced CD8+ Tc/CTL numbers | 26, 30, 31, 33, 132 | |
| Runx1 | DP to CD4+ TCR‐αβ rearrangement | Reduced Il7r and survival | 132 | ||
| Th1/2 | Runx3 | Promotes Th1 phenotype in cooperation with T‐bet | IFN‐γ production, IL‐4 suppression | 37, 38 | |
| Treg |
Runx1 Runx1,3, Cbfβ |
Cooperates with Foxp3 in nTreg function Induction and function of iTreg |
Repression of IL‐2 Repression of IL‐2 Reduced immune tolerance especially in mucosal surfaces |
46
42, 43, 44, 45 42, 43 |
|
| Th17 | Runx1, 2 |
Promote Th17 differentiation by inducing RORγT and IL17 transcription Inhibits Th17 by cooperating with Foxp3 to suppress RORγT |
IL‐17 production | 55 | |
| Runx1, 3 | Promotes pathogenic Th17 and secretion of IFN‐γ with T‐bet | IL‐17and IFN‐γ | 57 | ||
| IEL | Runx3 | Necessary for CD8αα + expression. Cooperates with T‐bet to suppress Th‐POK | Accentuated Th17 differentiation | 64, 65, 66, 67 | |
| Tc/CTL | Runx3 | Necessary for CD8+ Tc differentiation | Reduced CTL activity | 26, 69,132 | |
| Regulates Eomes, granzyme, perforin and IFN‐γ expression | Reduced CTL activity | 39 | |||
| NKT | Runx1, Cbfβ | Needed for iNKT differentiation | Undetermined | 71, 72 | |
| Dermal dendritic T cells | Runx3 | Promotes maturation of γ3 thymocyte via CD103 and IL‐2Rβ | Loss of adult skin DETC | 75 | |
| B lymphocytes | Immature B cells | Runx1 Runx1/Cbfβ |
Early B‐cell maturation before Pre‐and Pro‐B stage Regulates pre‐B‐cell receptor Interacts with Ebf to activate mb‐1/cd79a Promote pre‐proB to pro‐B transition by inducing Ebf1 |
Defective B‐cell expansion and maturation Reduced IgM+ B cells and VH to DJH recombination |
13, 14, 15, 84, 85
15, 86 84 85 |
| B‐cell maturation | Runx3 | Cooperates with TGF‐β for activating germline Ig α promoter | Defective IgA class switching | 78, 79, 80, 81, 82 | |
| Runx1 | Promotes surface IgA expression in activated primary B cells | Defective IgA class switching | 82 | ||
| Runx3, Runx2 | Necessary for IgA expression in peripheral B cells | Reduced IgA production | 82 | ||
| Memory B cells | RUNX1 | Maintains undifferentiated state by silencing FCRL4 | Undetermined | 95 | |
| Primary B cells | RUNX1 | Suppresses proliferation of resting B cells | Undetermined | 88 | |
| RUNX3 | Immortalizes B cells via silencing of RUNX1 | Undetermined | 88, 89, 91, 92 | ||
| NK cells | NK differentiation | Cbfβ | Needed for NK1.1− CD122+ progenitor | Undetermined | 100, 101 |
| Runx3 |
Regulates CD122, Ly49 family, Mac‐1, CD43 and IFN‐γ
Needed for IL‐15‐induced NK cell proliferation and maturation |
Undetermined |
102, 103
104 |
||
| Uterine NK cells | Runx3 | Essential for IL‐15‐dependent uterine NK cells | Loss of uterine NK cells | 104 | |
| LTi cells | LTi cells | Runx1c, Cbfβ2 |
Necessary for early specification of LTi lineage Induction of Rorγt at anlagen |
Loss of Peyer's patches and peripheral LNs | 72 |
| Runx1 | Necessary in BM cells for LTi differentiation | Absent or defective Peyer's patches | 15 | ||
| Granulocytes | Dendritic cells | Cbfβ, Runx1,2 but not Runx3 | Required for Flt3+ DC progenitors | Loss of classical and plasmacytoid DC | 20 |
| Runx3 |
Cooperates with TGF‐β to suppress DC maturation Restricts DC migration by suppressing CCR7 expression |
Spontaneous DC maturation Allergic airway inflammation; severe gastritis |
48, 83
47, 48, 83 |
||
| Langerhans cells | Runx3 | Mediates TGF‐β‐induced Langerhans cell differentiation | Loss of Langerhans cells | 83, 105 | |
| Basophils | Runx1c | Required for generation of basophil progenitors | Attenuated basophil expansion and functions | 107 | |
| Monocytes/macrophage | RUNX1 | Cooperates with PU.1 to induce M‐CSFR | Reduced macrophage survival, differentiation and expansion | 108, 109 | |
| RUNX3,1 | Regulates LFA‐1/CD11a, CD11c, CD49d and ICAM‐3 | Undetermined | 110, 111, 112 | ||
| Microglia | Runx1 |
Runx1 promotes microglia maturation Runx1 is induced during nerve injury |
Runx1 restricts iNOS production in vitro | 115 | |
| Gastric epithelial cells | Runx3,Runx1 | Response to inflammation and infection | Secretion of IL23A | 129 |
Abbreviations: CTL, cytotoxic T lymphocytes; DC, dendritic cells; DN, double‐negative; DP, double‐positive; IFN, interferon; IL, interleukin; iNKT, invariant natural killer T; LN, lymph nodes; LTi, lymphoid tissue inducers; NK, natural killer; SP, single‐positive; TCR, T‐cell receptor; TGF, transforming growth factor; Th1, T helper type 1; Treg, regulatory T;
Figure 1.
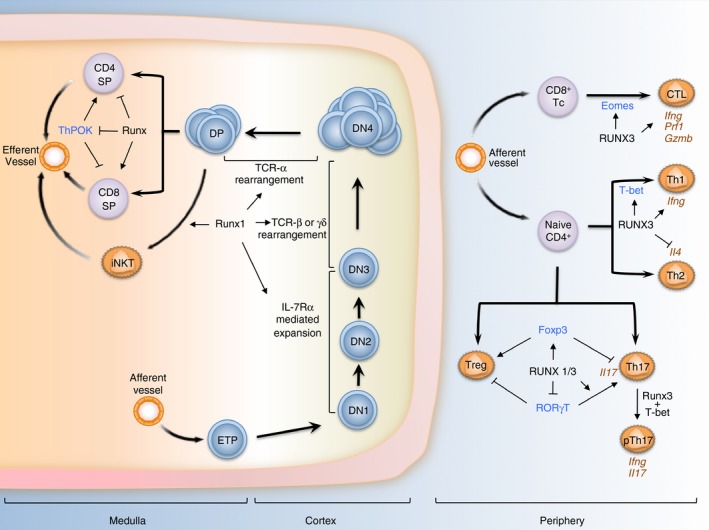
RUNX and T‐lymphocyte differentiation. In the thymic cortex, Runx1 is expressed in CD4− CD8− double‐negative (DN) thymocytes, reaching a maximum at DN3 before declining during DN3 to DN4 transition. Runx1 transcriptionally orchestrates interleukin 7 receptor α (IL‐7Rα) ‐mediated expansion, T cell receptor (TCR) γδ and TCR‐αβ rearrangement during these developmental stages. In addition, Runx1 is also a key factor for the differentiation of invariant natural killer T (iNKT) cells in the medulla cortex of the thymus. Following TCR‐mediated selection, Runx3 gains prominence and is a major driver of CD8+ T‐cell differentiation through the silencing of Cd4 and Thpok, master regulator of CD4+ differentiation. In the periphery, Runx3 promotes the maturation of CD8+ T cells into cytotoxic T lymphocytes (Tc/CTL) via its regulation of Eomes and key effector genes. In TCR‐activated CD4+ T cells, Runx3 cooperates with T‐bet to strengthen the T helper type 1 (Th1) phenotype by activating Ifng expression while suppressing Th2‐specific cytokine IL4. Runx proteins are also important in the differentiation and functions of regulatory T (Treg) cells through its regulation and interaction with Foxp3. Lastly, Runx proteins influence the development of Th17 in distinct ways. Runx could suppress or activate Rorγt depending on the presence of Foxp3, while interacting with RORγt to activate Il17 expression. Moreover, Runx1/3 are needed for the production of interferon‐γ (IFN‐γ) in a subset of Th17 cells important in the pathogenesis of autoinflammatory conditions. In this figure, key T lineage determinants that functionally interact with RUNX are labelled in blue and notable effector genes are labelled in brown. ETP denotes early thymocyte progenitors.
Compared with Runx1, Runx3 gains prominence later in T‐cell differentiation. It plays a dominant role in the specification of CD8+ cytotoxic T cells from immature CD4+ CD8+ DP cells. This is achieved through a number of mechanisms. First, Runx3, and also Runx1, bind to the silencer element in the Cd4 locus to suppress its expression.26, 33 Second, it binds to the silencer element of Thpok, a key CD4 lineage determinant.33 Runx3 also directs the activity of other participating transcription factors that regulate Cd4 and Thpok, such as TCF‐1 and LEF‐1.34 Lastly, the binding of Runx proteins to the Cd4 and Cd8 loci promotes their association and enables the long‐range epigenetic regulation that underlies their reciprocal expression patterns.35 In line with these important functions, the genetic ablation of the Runx complex resulted in the blockade of CD8+ cytotoxic T‐lymphocytes differentiation and a redirection of their development to a CD4+ CD8− phenotype.26, 33
RUNX in the differentiation of effector T‐cell subsets
Importantly, Runx1 and Runx3 are further involved in the maturation of naive CD4+ T cells into various effector T‐cell lineages following TCR activation and exposure to environmental cues. In detailed studies of these lineages, a recurring theme has been the functional co‐operation among Runx proteins and primary lineage‐specifying transcription factors.36 During T helper type 1 (Th1) differentiation, Runx3 expression increases with a corresponding reduction in Runx1 expression. Accordingly, Th1 differentiation and cytokine production were found to be impaired in Runx3‐deficient mice.37 Reprising its dualistic role in silencing Cd4 while activating Cd8, Runx3 cooperated with T‐bet, a Th1‐specific transcription factor, to fortify the Th1 phenotype. Together with Runx1, Runx3 concurrently activated the hallmark Th1 cytokine Ifng while suppressing the Th2‐specific cytokine Il4.37, 38, 39 Though not studied in detail, increase in Runx1 and decrease in Runx3 expression were observed during Th2 specification, suggesting a role for Runx1 in Th2 functions.37, 38
Regulatory T (Treg) cells co‐expressing CD4 and CD25 may arise spontaneously in the thymus (naturally occurring Treg or nTreg cells) or when induced by transforming growth factor‐β (TGF‐β), interleukin‐2 (IL‐2) and other signalling cues in the periphery (induced Treg or iTreg cells).40, 41 The Treg phenotype is driven by the transcription factor Foxp3. Runx proteins are essential for the maintenance of Foxp3 expression in nTreg cells and its induction during iTreg differentiation.42, 43, 44, 45 Furthermore, Runx proteins physically interact with Foxp3 to maintain the Treg genetic programme, which includes the repression of Il2.42, 45, 46 Consequently, the conditional ablation of Runx1, Runx3 or Cbfb in mice strongly disrupted nTreg and iTreg differentiation, maintenance and function.42, 43, 44 In particular, defects in Treg cells due to Runx/Cbfβ‐deficiency resulted in lymphoproliferative syndrome, hyper‐production of IgE, and autoimmunity in mucosal tissues, such as stomach and lung.42, 43 Of note, these phenotypes are reminiscent of the severe gastrointestinal and lung inflammation reported in a Runx3‐deficient mouse model, which had been attributed to spontaneous maturation of dendritic cells (DC).47, 48
In addition to the induction of iTreg cells, TGF‐β is also essential for the differentiation of Th17, a potent mediator of inflammation.41, 49, 50 This is dependent on concurrent stimulation by pro‐inflammatory cytokines, including IL‐6, tumour necrosis factor‐α (TNF‐α) and IL‐1β.51 Although Th17 cells are important in providing immunity against bacteria and fungus at mucosal surfaces, deregulation and hyperactivity of Th17 cells are linked strongly to the development of autoimmune diseases (reviewed in Ivanov et al.49 and Singh et al.52). As Runx proteins are important downstream mediators of TGF‐β, their involvement in the reciprocal induction of Treg and Th17 cells was extensively investigated.41, 50 Analogous to their regulation of Foxp3, Runx proteins act upstream of the Th17‐specifying transcription factor RORγt.53, 54, 55 Furthermore, Runx proteins physically interact with RORγt for the maximal induction of Il17, in a manner independent of their DNA‐binding activities.55 Remarkably, Foxp3 also interacts with RORγt, which leads to the suppression of Il17 transcription.55 A picture therefore emerges of a tripartite relationship between Runx, Foxp3 and RORγt in Th17 lineage specification and effector function, namely the secretion of IL‐17.55 It is now recognized that Th17‐associated autoimmune inflammation is mediated by a pathogenic subset of Th17 characterized by high IL‐23 receptor (IL23R) expression and the production of interferon‐γ (IFN‐γ), normally a Th1 cytokine.56 Recently, it was shown that IFN‐γ production by pathogenic IL23Rhigh Th17 cells required T‐bet and Runx1/3, hence linking Runx proteins in the pathogenesis of certain autoimmune conditions.57 Consistent with this, increased Runx1 and decreased Runx2 levels were associated with salt‐induced pathogenic Th17.58, 59, 60
In the intestinal mucosa where strong environmental cues such as TGF‐β and retinoic acid (RA) influence T‐cell differentiation, plasticity in the CD4+ T‐cell lineage helps to maintain homeostasis by preventing exaggerated responses to commensal microbiota.61, 62, 63 During their migration to the intestinal intraepithelial compartment, progenitors of intestinal epithelial lymphocytes (IEL) express Runx3 in response to IL‐15 signalling to endow a CD4+ CD8αα + phenoytpe.64, 65, 66 Moreover, Runx3 confers on CD4+ IEL cytotoxic T lymphocyte and innate‐like lymphocyte properties and attenuates Th17 differentiation in a TGF‐β‐ and RA‐dependent manner.65 As in the case of Th1 differentiation, Runx3 is positively regulated by T‐bet and partners it in executing the IEL differentiation programme, such as the down‐regulation of ThPOK.37, 66, 67
While the necessity of Runx3 for the specification and TCR‐mediated expansion of SP CD8+ cytotoxic T cell was a seminal discovery, the role of Runx proteins in the effector functions of mature CD8+ T cell awaits further investigation.26, 33, 68 Nevertheless, it is evident that Runx proteins are integral components of lineage specifying transcriptional circuit. In particular, Runx3 is requisite for the induction of T‐box protein Eomesodermin (Eomes) during the differentiation of cytotoxic T cells (Tc/CTL). Runx3‐deficient CD8+ Tc cells showed reduced cytolytic activity due to weakened TCR‐induced proliferation26, 69 and reduced expression of key Tc effector genes, including granzyme, perforin and IFN‐γ.39
Runx is important also in the development of natural killer T (NKT) cells, which are specialized T lymphocytes that are CD1d‐restricted and co‐express TCR‐αβ and NK maturation markers, like NK1.1 (CD161).70, 71, 72 In particular, Runx1 (together with RORγt) is indispensable for the differentiation of iNKT, a subclass of NKT cells with an invariant TCR‐α chain.71 These invariant NKT cells have been implicated in diverse immunological processes, including immune regulation, cytokine production, microbial immunity and autoimmunity.70, 73, 74 However, due to the lack of an NKT‐specific Cre mouse line, the precise contribution of Runx complex in NKT cell functions remains to be determined.
Lastly, Runx3 is critical for the development of dendritic epidermal T cells (DETC), a distinct skin‐associated intraepithelial γδ T cells marked by a dendritic morphology.75 Runx3 regulates IL‐2Rβ and CD103, which mediate IL‐2/IL‐15‐induced cell proliferation and the migration of thymic DETC to skin during late fetal development, respectively.75, 76 As the DETC are a major component of cutaneous immunity and homeostasis, the effects of their complete absence in Runx3‐deficient mice on immune modulation, surveillance and repair warrant further investigation.75, 77
RUNX in B‐cell determination and functions
A role for Runx proteins in B‐cell function was first revealed in a series of in vitro and ex vivo experiments, in which Runx3 was shown to cooperate with Smad proteins to mediate the induction of germline immunoglobulin α promoter by TGF‐β 78, 79, 80, 81, 82 (Fig. 2). This is a key event of IgA class switching when naive B cells are activated by antigen. In agreement with these observations, Runx3‐ and Runx2;Runx3‐deficient splenocytes displayed varying degrees of class switching defects ex vivo and in vivo.82, 83 In addition to Runx3, Runx1 is required for early B‐cell lineage specification in mouse. The targeting of Runx1 by Mx1‐Cre in adult BM resulted in a blockade of B220+ B‐cell differentiation from common lymphocyte progenitors.13, 14 This early involvement stemmed in part from Runx1's cooperation with the EBF transcription factor in its regulation of mb‐1 (also Cd79a), a component of the B cell receptor.84 Consequently, conditional ablation of Runx1 and Cbfb (but not Runx3) by mb1‐Cre blocked early B lymphopoiesis. Of note, pre‐proB to pro‐B transition was impaired, resulting in a loss of IgM+ B cells and reduced V H to DJ H recombination.85 Although not fully elucidated, Runx1 appears to be an integral component of the early B‐cell lineage specification circuit through its regulation of key lineage determinants, such as the pre‐B cell receptor.84, 85, 86
Figure 2.
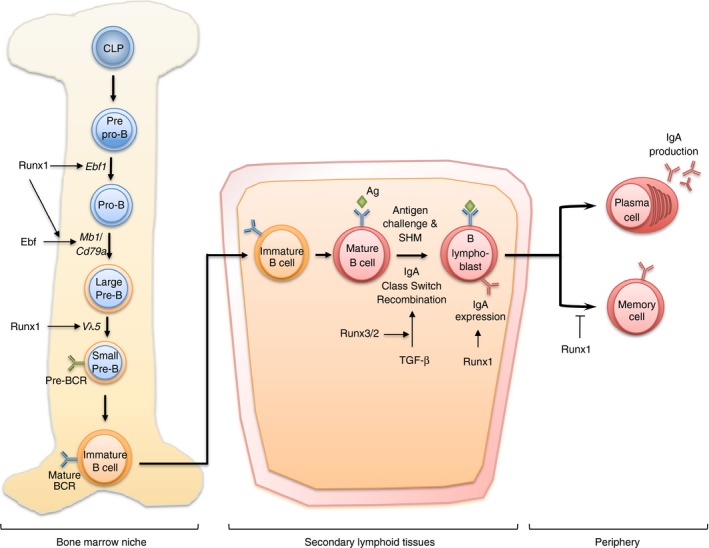
RUNX in B‐cell development and humoral immunity. In adult bone marrow, Runx1 is involved in the differentiation of B cells from common lymphoid progenitors (CLP). Specifically, Runx1 regulates and cooperates with Ebf for the transition of pre‐proB cells to pro‐B cells. Runx1 also takes part in the transition of large pre‐B cells to small pre‐B cells via V H to DJ H recombination as well as regulation of pre‐B cell receptor (BCR). Runx3 is necessary in the later stage of B‐cell development in secondary lymphoid tissues. Runx3 and Runx2 function downstream of transforming growth factor‐β (TGF‐β) ‐mediated IgA class switching, a key event in the development of B lymphoblasts. Runx1 is also needed to maintain high IgA expression on the surfaces of activated lymphoblasts. In human, RUNX1 further influences the differentiation of FCRL4+ memory B cells that normally reside in mucosal tissues.
In contrast, Runx3 gains prominence in later stages of B‐cell differentiation. An example of this was the strong induction of Runx3 observed in mouse B‐cell lines by TGF‐β in naive B cells during IgA class switching and the differentiation of effector B cells.78, 79, 80, 81, 87 Increased RUNX3 expression could also be observed following mitogenic or antigenic stimulation of human primary B cells, as well as during Epstein–Barr virus‐induced immortalization.88 As with T‐cell development, cross‐regulation between Runx proteins and their reciprocal expression during B‐cell differentiation has been reported.89 This is most readily observed in the suppression of RUNX1 by RUNX3 during Epstein–Barr virus immortalization of resting B cells to generate proliferating B lymphoblastoid cells.88, 90, 91 This is achieved by the silencing of RUNX1 distal P1 promoter through the VWRPY domain within RUNX3.92 Furthermore, RUNX3 is recruited by EBNA2 and EBNA3C to co‐occupy promoter and enhancer elements to modulate key regulatory proteins, such as CDKN2A/p14ARF and B‐cell maturation antigen (BCMA).91, 92, 93 BCMA plays an important role in the survival of B cells, particularly in mature memory B cells and plasma B cells.94
In humans, a distinct subpopulation of CD27‐independent FCRL4+ memory B cells resides in the palatine tonsils, crypt epithelium and intestine‐associated lymphoid tissues. The expression of RUNX1 was found to be correlated to the maintenance of these memory B cells in an undifferentiated FCRL4− state through its physical occupancy FCRL4 promoter.95 As in the case of naive B‐cell activation and Epstein–Barr virus immortalization, the maturation and expansion of FCRL4+ memory B cells coincided with a loss of RUNX1 expression.95 Taken together, RUNX1 appears to be necessary for the maintenance of immature and undifferentiated B cells whereas RUNX3 and RUNX2 expression are observed during terminal specialization of effector B cells.96
RUNX in lymphoid organogenesis
The Runx/Cbfβ complex has been implicated in the development of lymphoid tissue inducers (LTi) cells.72 These are specialized innate lymphoid cells expressing RORγt and IL‐7Rα in the absence of additional lineage markers. They are essential for the biogenesis of secondary lymphoid tissues, such as lymph nodes and Peyer's patches by initiating anlagen formation.97 Runx1c, a Runx1 isoform derived from the P1 promoter, and Cbfβ2 are involved in LTi differentiation at two distinct development stages.72 Specifically, Runx1c/Cbfβ2 are needed for the early specification of LTi differentiation and during the formation of lymph node anlagen.72 Consequently Runx1c/Cbfb2 knockout mice showed defective LTi differentiation and impaired lymphoid tissue organogenesis, particularly Peyer's patches and peripheral lymph nodes.72 Runx1c/Cbfβ2 are likely to mediate these functions through their regulation of Rorγt.72 This is reminiscent of the functional interaction between Runx/Cbfβ and RORγt during T helper and iNKT differentiation, providing a clear example of recurrent utilization of key transcriptional regulatory circuits during haematopoiesis.55, 71, 72
RUNX proteins in NK cell development
Natural killer cells are a distinct subset of cytotoxic lymphocyte lineage that play important roles in innate immunity, particularly against viral infection and tumour cells.98 They are unique in their ability to recognize target cells in the absence of antibodies or MHC molecules, relying instead on a separate set of receptors for immune recognition and self tolerance.99 The NK cells develop in multiple sites, including fetal and adult liver, BM, spleen and thymus. Their differentiation from the NK progenitor, marked by the expression of CD122 (IL‐2/IL‐15 receptor β chain) is reliant on IL‐15 signalling.98 The first evidence of Runx complex having a role in NK cells came in a hypomorphic Cbfβ mouse model, in which an early block in NK differentiation was observed. At low Cbfβ dosage, the fetal liver cells displayed a profound defect in their ability to generate NK1.1− CD122+ NK progenitor cells in ex vivo culture.100, 101 This was probably a result of the loss of Runx complex‐mediated CD122 transcription, thereby blocking the crucial IL‐15‐induced genetic programme.102 Runx proteins were shown to occupy and regulate the CD122 promoter in mouse NK progenitors, whereas a dominant negative Runx mutant suppressed CD122, Ly49 family, Mac‐1 and CD43 expression.102, 103 Due to its high expression, these studies implicated an important role for Runx3 during NK cell development.101, 102 Direct evidence was provided in a recent study of Runx3‐deficient mice. Although Runx3 was found to be largely dispensable for NK cell development and function in resting conditions, it was requisite for IL‐15‐induced NK cell proliferation and maturation in vivo and ex vivo.104 In particular, the loss of Runx3 resulted in the complete loss of IL‐15‐dependent uterine NK cells in pregnant mice.104 However, because of the limitation in current mouse models, it remains unresolved whether Runx proteins are further necessary for NK functions, such as IFN‐γ production and tumour immunity.101, 102, 104
RUNX in myeloid cell specification and functions
Relatively little is known of the contribution of RUNX to myeloid cell development and functions. Initial mouse studies showed that the deletion of Runx1 in adult HSC severely impaired megakaryocyte maturation but displayed no discernable defect in the differentiation of other myeloid lineages, including neutrophils, monocytes and erythrocytes.13, 14, 15 This comparatively mild phenotype is probably due to a greater degree of redundancy shared between Runx proteins in myeloid differentiation.19, 20, 21 Indeed, clearer phenotypes were observed upon the pan‐haematopoietic targeting of Cbfb by Vav1‐iCre, which disrupted the function of all Runx proteins. This led to severe reductions of classical and plasmacytoid DC in the spleen and peripheral tissues, including the lung and intestinal lamina propria.20 Cbfβ was necessary for the development of Flt3+ DC progenitors, as well as erythroid progenitors in the BM.20 Further analysis revealed that Runx1 was the primary driver of the Cbfβ knockout DC phenotype, with Runx2 playing a minor role later in DC maturation. However, no role was ascribed to Runx3, in contrast with earlier reports of aberrant DC development and spontaneous maturation in a Runx3 knockout mouse.20, 48, 83 Notwithstanding this discrepancy, Runx3 is needed for Langerhans cells, which are a distinct skin epidermal DC lineage derived from myeloid progenitors early in embryogenesis.83, 105 Here, Runx3 acts downstream of PU.1 to promote Langerhans cell differentiation induced by TGF‐β signalling.105
In addition to DC differentiation and maturation, RUNX3 regulates CD11c and chemokine expression in an isoform‐specific manner in vitro.106 As DC are central coordinators of adaptive immune responses and self‐tolerance, the importance of Runx proteins in their differentiation will have far‐reaching effects in the immune system. In addition to DC, a recent study showed that specific ablation of the Runx1c isoform in mice resulted in a drastic reduction in granulocytic basophils, owing to a block in the transition of granulocyte progenitors to basophil progenitors. Furthermore, Runx1c‐deficiency specifically attenuated basophil (but not mast cell) functions, including IgE‐mediated chronic allergic skin reaction, and their expansion in response to IL‐3 or nematodes.107
In monocytes and macrophages, RUNX1 cooperates with PU.1 to regulate macrophage colony‐stimulating factor (M‐CSF/CSF‐1) receptor, which is essential for the survival, differentiation and expansion of macrophages.108, 109 RUNX1 and RUNX3 are also important controllers of adhesive interactions through their regulation of integrin‐like lymphocyte function‐associated antigen 1 (LFA‐1)/CD11a, CD11c, CD49d and intercellular adhesion molecule 3 (ICAM‐3) in macrophage and monocytic cell lines.110, 111, 112 Of note, LFA‐1 and ICAM‐3 interaction contribute to the polarization of Th1 cells, highlighting the multifaceted contribution of RUNX to T‐cell differentiation and function.111 The microglia are unique phagocytic cells that reside within the parenchyma of the central nervous system (CNS), where they serve as inflammatory cells that safeguard the CNS against microbes and injuries. Akin to the Langerhans cells, microglia are derived from Runx1+ yolk sac primitive myeloid progenitors and are seeded in the CNS during embryonic and perinatal stages of development.113, 114, 115 Postnatally, Runx1 was reported to inhibit proliferation of immature microglia and promote their maturation.115 Interestingly, Runx1 expression was reactivated following injury to the nervous system, suggesting a role in the function of mature microglia in CNS surveillance and repair.115
RUNX in inflammation and autoimmunity
In the thymus, the induction of self‐tolerance by nTreg cells is essential for the maintenance of immune homeostasis, the loss of which would lead to autoimmunity.116 Given the heavy involvement of RUNX proteins in differentiation and function of nTreg and iTreg cells,42, 43, 44, 45, 46 as well as that of Th17 cells,53, 55 it is likely that the interference of RUNX function would contribute to autoimmune and inflammatory disease states. This possibility is supported by several extensive studies of genetic association in human autoimmune conditions. First, an intronic single nucleotide polymorphism within the PDCD1 locus that alters a RUNX binding site was associated with systemic lupus erythematosus in an ethnically diverse cohort. The alteration in RUNX site disrupted RUNX1 binding in vitro and led to aberrant expression of PDCD1, which is important to self‐tolerance and the suppression of hyperactivity in systemic lupus erythematosus.117 Of note, PDCD1 (also called PD‐1) is a negative regulator of tumour immunity and an important target of cancer immunotherapy.118 Similarly, a RUNX site variant between a PDZ‐domain phosphoprotein SLC9A3R1 and N‐acetyltransferase NAT9 was associated with psoriasis, a chronic inflammatory skin disorder.119 In addition, the RUNX1 locus and a RUNX site‐ablating single nucleotide polymorphism in the transporter gene SLC22A4 were found to be significantly associated with rheumatoid arthritis.120 It bears highlighting that the disease‐associated RUNX sites could function as the binding targets of all RUNX proteins as they recognize the same cognate sites.117, 119, 120 Consistent with this, a Runx3 knockout mouse model spontaneously developed an inflammatory bowel disease‐like condition characterized by infiltration of leucocytes with mixed Th1/Th2 response and hyperplastic mucosa.47 In human, the chromosomal region 1p36 where RUNX3 resides is a susceptibility region for inflammatory bowel disease.121, 122, 123 More recently, genome‐wide association studies identified variants in the RUNX3 locus to be associated with two subtypes of inflammatory bowel disease, namely coeliac disease and ulcerative colitis.124, 125 Lastly, single nucleotide polymorphisms within the RUNX1 locus and RUNX1 levels were also found associated with paediatric asthma.126 Together, the above findings suggest that alteration of RUNX functions could lead to dysregulated self‐tolerance, chronic lymphocyte hyperactivity and autoimmunity in organs.
Established and emerging roles of RUNX in mucosal immunity
Although direct evidence are only beginning to emerge, RUNX proteins are strongly implicated in innate and adaptive immunity in mucosal systems. RUNX family members play important roles in the development, maturation and effector functions of every major immune lineage within the mucosal system (illustrated in Fig. 3). Importantly, RUNX is functionally integrated into the overall immune response at multiple levels. An instance of this is in the biogenesis, organization and function of mucosal lymphoid tissues, like Peyer's patches.72 In addition to their formation, Runx proteins also participate in the activation of peripheral T and B cells within these tissues by regulating DC maturation for antigen presentation.20, 48, 83 Following activation, the terminal differentiation of naive peripheral T cells is coordinated by Runx proteins in partnership with various lineage‐defining transcription factors, into effector T helper and cytotoxic lineages.22, 127 Of particular relevance in the mucosal system is the role of Runx1/3 in the differentiation of IEL in response to environmental cues and pathogenic challenges.65, 66, 67 Runx1 is further involved in the development of iNKT lymphocytes, which orchestrate microbial immunity through cytokine production.71, 72 Concurrently, Runx3 and Runx2 are essential for IgA class switching in mature B cells, a crucial event in the configuration of mucosal immunity in response to microbial pathogens.128 Lastly, RUNX1 is implicated in the maturation of FCRL4+ memory B cells that reside in the gastrointestinal epithelial niche.95
Figure 3.
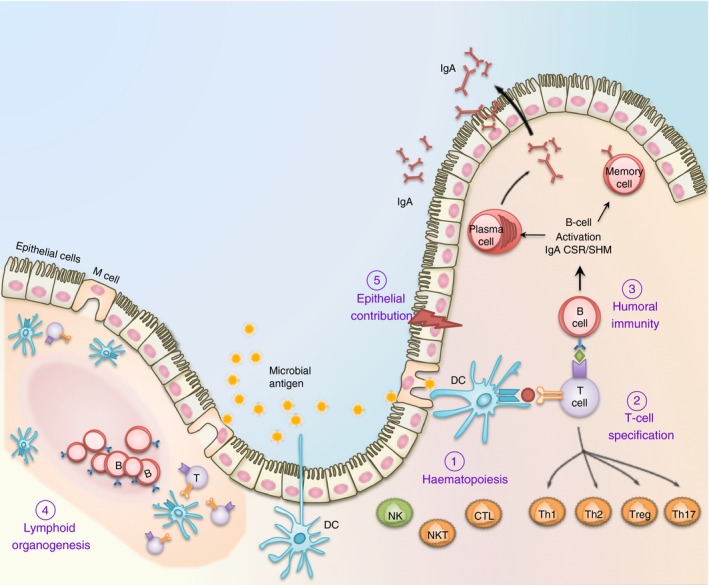
The multifaceted contribution of RUNX in mucosal immunity. (1) Runx proteins are essential for the differentiation and effector functions of diverse cell lineages that participate in the mucosal immune system. These include T cells, B cells and dendritic cells (DC) for adaptive immunity; as well as macrophages, natural killer (NK) cells and epithelial cells for innate immunity. (2) Runx1 and Runx3 are critically involved in the specification of CD8+ cytotoxic T (Tc) and naive CD4+ helper T (Th) lymphocytes before their entrance into the periphery. Following antigen engagement and T cell receptor (TCR) activation, Runx1/3 coordinate the terminal differentiation of CD4+ T cells into Th1, Th2, Th17 and Treg cells, and CD8+ T cells into cytotoxic T lymphocyte (CTL) effector T‐cell lineages. (3) In B lymphocytes, Runx proteins play a crucial role in transforming growth factor‐β (TGF‐β) ‐mediated IgA class switching following antigen engagement. Runx proteins are also necessary for the maximal surface expression of IgA on activated B lymphoblasts and the maintenance of specialized memory B (Bmem) cells distributed at the mucosa. (4) The Runx complex is required for the formation of anlagen that initiate Peyer's patches and peripheral lymph node biogenesis. These secondary lymphoid tissues are home to naive peripheral lymphocytes and dendritic cells and are necessary for maintaining surveillance and homeostasis. (5) The mucosal epithelium is in direct contact with the microbial load and functions as a physical as well as an immune barrier. RUNX proteins are functionally important in a diverse range of mucosal epithelial cells, including those in the lung and gastrointestinal tract. RUNX3 is necessary for the homeostasis of an intact mucosal epithelium by regulating the proliferation and apoptosis of epithelial cells.1 RUNX proteins may further contribute to innate immunity within the epithelium by regulating cytokine production (such as IL23A) during infection and inflammation.129
Though less understood, Runx proteins function through innate leukocytes and lymphocytes to mount innate immunity at the mucosa. Of note, Runx3 is necessary for the expansion and maturation of NK cells, which serve important cytotoxic functions independent of antigen presentation.104 Similarly, Runx1 is needed for the expansion of basophils and maximal anti‐parasitic and pro‐inflammatory activities.107 In macrophages and monocytes, RUNX transcriptionally regulates adhesion complexes to promote their migration to the sites of infection to perform their phagocytic functions.111, 112
In addition to haematopoietic cells, a major component of mucosal immunity is the epithelial cells that constitute the foremost barrier to the mucosal microbiota. The precise contribution of this cell type to mucosal immunity through antigen processing and cytokine production is currently under‐explored. Likewise, while RUNX proteins are important in a diverse range of epithelial tissues, the potential for their immune contribution has not been experimentally tested.1 Recently, RUNX3 was reported to transcriptionally regulate IL23A in gastric epithelial cells.129 IL23A is a subunit of IL‐23, a pro‐inflammatory cytokine best known for driving Th17 activities.130 Furthermore, RUNX3 strongly augmented the secretion of IL23A in the presence of the pro‐inflammatory cytokines TNF‐α and IL‐1, and the gastric pathogen Helicobacter pylori.129 These findings are conceptually significant as they provide evidence that RUNX proteins can further participate in innate immunity through epithelial cytokine production.
Concluding remarks
It is evident that RUNX proteins are profoundly involved in the development, organization and function of the mammalian immune system. Much of these are accomplished through their multifaceted contribution to definitive and adult haematopoiesis. In addition, RUNX is often recurrently involved in cell fate decision within a lineage in response to extracellular cues, through interplays with other primary lineage‐determining factors. This is most apparent during T‐cell differentiation, where an antagonistic interplay between Runx complex and Th‐POK determines the fate of CD8+ and CD4+ SP T cells. This is revisited in activated peripheral T cells, where Runx proteins concurrently promote one T helper phenotype while suppressing another through molecular interplays with T‐bet, FoxP3 or RORγt. Such observations implicate RUNX to be part of a finely tuned high‐order transcriptional circuit. Importantly, this circuit is functionally oriented and extends beyond haematopoietic lineage decision. For example, the RORγt–RUNX axis impacts diverse immune functions that encompass inflammation (via Th17 cells), commensal microbe tolerance (iNKT cells) and lymph node biogenesis (LTi cells). Such extensive involvement suggest that the RUNX complex is a primary building block of the immune system during evolution.131 Therefore, elucidating the spatial, temporal and functional contribution of RUNX to the dynamics and complexity of the immune system is an attractive goal. However, that RUNX functions are deeply woven into immune processes presents a significant technical challenge. Indeed, direct proof of their contribution to overall immunity awaits deeper investigations that combine lineage‐specific gene targeting with immunological challenges. It is hoped that this immune‐oriented survey of current knowledge will stimulate future studies of RUNX functions beyond haematopoiesis and into immunity.
Disclosures
The authors declare that they have no competing interests.
Acknowledgement
The authors are grateful to Motomi Osato, Chelsia Wang and Giselle Nah for helpful discussions and intellectual input. We thank Yeh‐Shiu Chu, Lee Koon Guan, Pah Pin Shang and Huajing Wang for feedback and assistance in the preparation of this manuscript.
References
- 1. Ito Y, Bae SC, Chuang LS. The RUNX family: developmental regulators in cancer. Nat Rev Cancer 2015; 15:81–95. [DOI] [PubMed] [Google Scholar]
- 2. Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 1996; 84:321–30. [DOI] [PubMed] [Google Scholar]
- 3. Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol 2008; 9:129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Komori T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res 2010; 339:189–95. [DOI] [PubMed] [Google Scholar]
- 5. de Bruijn MF, Speck NA. Core‐binding factors in hematopoiesis and immune function. Oncogene 2004; 23:4238–48. [DOI] [PubMed] [Google Scholar]
- 6. Kamachi Y, Ogawa E, Asano M, Ishida S, Murakami Y, Satake M et al Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J Virol 1990; 64:4808–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Q, Stacy T, Binder M, Marin‐Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA 1996; 93:3444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M et al Cbfa2 is required for the formation of intra‐aortic hematopoietic clusters. Development 1999; 126:2563–75. [DOI] [PubMed] [Google Scholar]
- 9. Yokomizo T, Ogawa M, Osato M, Kanno T, Yoshida H, Fujimoto T et al Requirement of Runx1/AML1/PEBP2αB for the generation of haematopoietic cells from endothelial cells. Genes Cells 2001; 6:13–23. [DOI] [PubMed] [Google Scholar]
- 10. Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 2009; 457:887–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 2010; 464:112–5. [DOI] [PubMed] [Google Scholar]
- 12. Chen MJ, Li Y, De Obaldia ME, Yang Q, Yzaguirre AD, Yamada‐Inagawa T et al Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell 2011; 9:541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T et al AML‐1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med 2004; 10:299–304. [DOI] [PubMed] [Google Scholar]
- 14. Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R et al Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 2005; 106:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Putz G, Rosner A, Nuesslein I, Schmitz N, Buchholz F. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene 2006; 25:929–39. [DOI] [PubMed] [Google Scholar]
- 16. Motoda L, Osato M, Yamashita N, Jacob B, Chen LQ, Yanagida M et al Runx1 protects hematopoietic stem/progenitor cells from oncogenic insult. Stem Cells 2007; 25:2976–86. [DOI] [PubMed] [Google Scholar]
- 17. Jacob B, Osato M, Yamashita N, Wang CQ, Taniuchi I, Littman DR et al Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood 2010; 115:1610–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang CQ, Jacob B, Nah GS, Osato M. Runx family genes, niche, and stem cell quiescence. Blood Cells Mol Dis 2010; 44:275–86. [DOI] [PubMed] [Google Scholar]
- 19. Wang CQ, Krishnan V, Tay LS, Chin DW, Koh CP, Chooi JY et al Disruption of Runx1 and Runx3 leads to bone marrow failure and leukemia predisposition due to transcriptional and DNA repair defects. Cell Rep 2014; 8:767–82. [DOI] [PubMed] [Google Scholar]
- 20. Satpathy AT, Briseno CG, Cai X, Michael DG, Chou C, Hsiung S et al Runx1 and Cbfβ regulate the development of Flt3+ dendritic cell progenitors and restrict myeloproliferative disorder. Blood 2014; 123:2968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang CQ, Chin DW, Chooi JY, Chng WJ, Taniuchi I, Tergaonkar V et al Cbfb deficiency results in differentiation blocks and stem/progenitor cell expansion in hematopoiesis. Leukemia 2015; 29:753–7. [DOI] [PubMed] [Google Scholar]
- 22. Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T‐cell lineage choice. Nat Rev Immunol 2009; 9:106–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Satake M, Nomura S, Yamaguchi‐Iwai Y, Takahama Y, Hashimoto Y, Niki M et al Expression of the Runt domain‐encoding PEBP2 α genes in T cells during thymic development. Mol Cell Biol 1995; 15:1662–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levanon D, Bernstein Y, Negreanu V, Ghozi MC, Bar‐Am I, Aloya R et al A large variety of alternatively spliced and differentially expressed mRNAs are encoded by the human acute myeloid leukemia gene AML1. DNA Cell Biol 1996; 15:175–85. [DOI] [PubMed] [Google Scholar]
- 25. North TE, Stacy T, Matheny CJ, Speck NA, de Bruijn MF. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells 2004; 22:158–68. [DOI] [PubMed] [Google Scholar]
- 26. Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T et al Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 2002; 111:621–33. [DOI] [PubMed] [Google Scholar]
- 27. Lorsbach RB, Moore J, Ang SO, Sun W, Lenny N, Downing JR. Role of RUNX1 in adult hematopoiesis: analysis of RUNX1‐IRES‐GFP knock‐in mice reveals differential lineage expression. Blood 2004; 103:2522–9. [DOI] [PubMed] [Google Scholar]
- 28. Hernandez‐Munain C, Krangel MS. Regulation of the T‐cell receptor δ enhancer by functional cooperation between c‐Myb and core‐binding factors. Mol Cell Biol 1994; 14:473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lauzurica P, Zhong XP, Krangel MS, Roberts JL. Regulation of T cell receptor δ gene rearrangement by CBF/PEBP2. J Exp Med 1997; 185:1193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wotton D, Ghysdael J, Wang S, Speck NA, Owen MJ. Cooperative binding of Ets‐1 and core binding factor to DNA. Mol Cell Biol 1994; 14:840–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCR α enhancer complex is dependent on LEF‐1‐induced DNA bending and multiple protein–protein interactions. Genes Dev 1995; 9:995–1008. [DOI] [PubMed] [Google Scholar]
- 32. Cieslak A, Le Noir S, Trinquand A, Lhermitte L, Franchini DM, Villarese P et al RUNX1‐dependent RAG1 deposition instigates human TCR‐δ locus rearrangement. J Exp Med 2014; 211:1821–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C et al Repression of the transcription factor Th‐POK by Runx complexes in cytotoxic T cell development. Science 2008; 319:822–5. [DOI] [PubMed] [Google Scholar]
- 34. Steinke FC, Yu S, Zhou X, He B, Yang W, Zhou B et al TCF‐1 and LEF‐1 act upstream of Th‐POK to promote the CD4+ T cell fate and interact with Runx3 to silence Cd4 in CD8+ T cells. Nat Immunol 2014; 15:646–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Collins A, Hewitt SL, Chaumeil J, Sellars M, Micsinai M, Allinne J et al RUNX transcription factor‐mediated association of Cd4 and Cd8 enables coordinate gene regulation. Immunity 2011; 34:303–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Djuretic IM, Cruz‐Guilloty F, Rao A. Regulation of gene expression in peripheral T cells by Runx transcription factors. Adv Immunol 2009; 104:1–23. [DOI] [PubMed] [Google Scholar]
- 37. Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T‐bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol 2007; 8:145–53. [DOI] [PubMed] [Google Scholar]
- 38. Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F et al Repression of interleukin‐4 in T helper type 1 cells by Runx/Cbf β binding to the Il4 silencer. J Exp Med 2007; 204:1749–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cruz‐Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG et al Runx3 and T‐box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med 2009; 206:51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell 2008; 133:775–87. [DOI] [PubMed] [Google Scholar]
- 41. Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M et al Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 2006; 441:235–8. [DOI] [PubMed] [Google Scholar]
- 42. Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H et al Indispensable role of the Runx1‐Cbfβ transcription complex for in vivo‐suppressive function of FoxP3+ regulatory T cells. Immunity 2009; 31:609–20. [DOI] [PubMed] [Google Scholar]
- 43. Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY. Runx‐CBFβ complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol 2009; 10:1170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klunker S, Chong MM, Mantel PY, Palomares O, Bassin C, Ziegler M et al Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J Exp Med 2009; 206:2701–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bruno L, Mazzarella L, Hoogenkamp M, Hertweck A, Cobb BS, Sauer S et al Runx proteins regulate Foxp3 expression. J Exp Med 2009; 206:2329–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T et al Foxp3 controls regulatory T‐cell function by interacting with AML1/Runx1. Nature 2007; 446:685–9. [DOI] [PubMed] [Google Scholar]
- 47. Brenner O, Levanon D, Negreanu V, Golubkov O, Fainaru O, Woolf E et al Loss of Runx3 function in leukocytes is associated with spontaneously developed colitis and gastric mucosal hyperplasia. Proc Natl Acad Sci USA 2004; 101:16016–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fainaru O, Shseyov D, Hantisteanu S, Groner Y. Accelerated chemokine receptor 7‐mediated dendritic cell migration in Runx3 knockout mice and the spontaneous development of asthma‐like disease. Proc Natl Acad Sci USA 2005; 102:10598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ivanov II, Zhou L, Littman DR. Transcriptional regulation of Th17 cell differentiation. Semin Immunol 2007; 19:409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO et al Transforming growth factor‐β induces development of the TH17 lineage. Nature 2006; 441:231–4. [DOI] [PubMed] [Google Scholar]
- 51. Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL‐17‐producing T cells. Immunity 2006; 24:179–89. [DOI] [PubMed] [Google Scholar]
- 52. Singh RP, Hasan S, Sharma S, Nagra S, Yamaguchi DT, Wong DT et al Th17 cells in inflammation and autoimmunity. Autoimmun Rev 2014; 13:1174–81. [DOI] [PubMed] [Google Scholar]
- 53. Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner‐Morton S et al Requirement for RORγ in thymocyte survival and lymphoid organ development. Science 2000; 288:2369–73. [DOI] [PubMed] [Google Scholar]
- 54. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ et al The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL‐17+ T helper cells. Cell 2006; 126:1121–33. [DOI] [PubMed] [Google Scholar]
- 55. Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORγt and Foxp3 regulate the differentiation of interleukin 17‐producing T cells. Nat Immunol 2008; 9:1297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ramesh R, Kozhaya L, McKevitt K, Djuretic IM, Carlson TJ, Quintero MA et al Pro‐inflammatory human Th17 cells selectively express P‐glycoprotein and are refractory to glucocorticoids. J Exp Med 2014; 211:89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang Y, Godec J, Ben‐Aissa K, Cui K, Zhao K, Pucsek AB et al The transcription factors T‐bet and Runx are required for the ontogeny of pathogenic interferon‐γ‐producing T helper 17 cells. Immunity 2014; 40:355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A et al Dynamic regulatory network controlling TH17 cell differentiation. Nature 2013; 496:461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y et al Induction of pathogenic TH17 cells by inducible salt‐sensing kinase SGK1. Nature 2013; 496:513–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA et al Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 2013; 496:518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Denning TL, Granger SW, Mucida D, Graddy R, Leclercq G, Zhang W et al Mouse TCRαβ +CD8αα intraepithelial lymphocytes express genes that down‐regulate their antigen reactivity and suppress immune responses. J Immunol 2007; 178:4230–9. [DOI] [PubMed] [Google Scholar]
- 62. Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M et al Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 2007; 317:256–60. [DOI] [PubMed] [Google Scholar]
- 63. Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol 2011; 11:445–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pobezinsky LA, Angelov GS, Tai X, Jeurling S, Van Laethem F, Feigenbaum L et al Clonal deletion and the fate of autoreactive thymocytes that survive negative selection. Nat Immunol 2012; 13:569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Reis BS, Rogoz A, Costa‐Pinto FA, Taniuchi I, Mucida D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4+ T cell immunity. Nat Immunol 2013; 14:271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Klose CS, Blatz K, d'Hargues Y, Hernandez PP, Kofoed‐Nielsen M, Ripka JF et al The transcription factor T‐bet is induced by IL‐15 and thymic agonist selection and controls CD8αα + intraepithelial lymphocyte development. Immunity 2014; 41:230–43. [DOI] [PubMed] [Google Scholar]
- 67. Reis BS, Hoytema van Konijnenburg DP, Grivennikov SI, Mucida D. Transcription factor T‐bet regulates intraepithelial lymphocyte functional maturation. Immunity 2014; 41:244–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx‐mediated commitment to the cytotoxic T cell lineage. Nat Immunol 2008; 9:1131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V et al Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci USA 2003; 100:7731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Monteiro M, Graca L. iNKT cells: innate lymphocytes with a diverse response. Crit Rev Immunol 2014; 34:81–90. [DOI] [PubMed] [Google Scholar]
- 71. Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L et al Genetic evidence supporting selection of the Vα14i NKT cell lineage from double‐positive thymocyte precursors. Immunity 2005; 22:705–16. [DOI] [PubMed] [Google Scholar]
- 72. Tachibana M, Tenno M, Tezuka C, Sugiyama M, Yoshida H, Taniuchi I. Runx1/Cbfβ2 complexes are required for lymphoid tissue inducer cell differentiation at two developmental stages. J Immunol 2011; 186:1450–7. [DOI] [PubMed] [Google Scholar]
- 73. Lehuen A, Lantz O, Beaudoin L, Laloux V, Carnaud C, Bendelac A et al Overexpression of natural killer T cells protects Vα14‐ Jα281 transgenic nonobese diabetic mice against diabetes. J Exp Med 1998; 188:1831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A et al Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012; 336:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Woolf E, Brenner O, Goldenberg D, Levanon D, Groner Y. Runx3 regulates dendritic epidermal T cell development. Dev Biol 2007; 303:703–14. [DOI] [PubMed] [Google Scholar]
- 76. Grueter B, Petter M, Egawa T, Laule‐Kilian K, Aldrian CJ, Wuerch A et al Runx3 regulates integrin α E/CD103 and CD4 expression during development of CD4–/CD8+ T cells. J Immunol 2005; 175:1694–705. [DOI] [PubMed] [Google Scholar]
- 77. Sharp LL, Jameson JM, Witherden DA, Komori HK, Havran WL. Dendritic epidermal T‐cell activation. Crit Rev Immunol 2005; 25:1–18. [DOI] [PubMed] [Google Scholar]
- 78. Shi MJ, Stavnezer J. CBF α3 (AML2) is induced by TGF‐β1 to bind and activate the mouse germline Ig α promoter. J Immunol 1998; 161:6751–60. [PubMed] [Google Scholar]
- 79. Hanai J, Chen LF, Kanno T, Ohtani‐Fujita N, Kim WY, Guo WH et al Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Cα promoter. J Biol Chem 1999; 274:31577–82. [DOI] [PubMed] [Google Scholar]
- 80. Pardali E, Xie XQ, Tsapogas P, Itoh S, Arvanitidis K, Heldin CH et al Smad and AML proteins synergistically confer transforming growth factor β1 responsiveness to human germ‐line IgA genes. J Biol Chem 2000; 275:3552–60. [DOI] [PubMed] [Google Scholar]
- 81. Zhang Y, Derynck R. Transcriptional regulation of the transforming growth factor‐β ‐inducible mouse germ line Ig α constant region gene by functional cooperation of Smad, CREB, and AML family members. J Biol Chem 2000; 275:16979–85. [DOI] [PubMed] [Google Scholar]
- 82. Watanabe K, Sugai M, Nambu Y, Osato M, Hayashi T, Kawaguchi M et al Requirement for Runx proteins in IgA class switching acting downstream of TGF‐β1 and retinoic acid signaling. J Immunol 2010; 184:2785–92. [DOI] [PubMed] [Google Scholar]
- 83. Fainaru O, Woolf E, Lotem J, Yarmus M, Brenner O, Goldenberg D et al Runx3 regulates mouse TGF‐β‐mediated dendritic cell function and its absence results in airway inflammation. EMBO J 2004; 23:969–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL et al Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb‐1 transcription. Nat Immunol 2004; 5:1069–77. [DOI] [PubMed] [Google Scholar]
- 85. Seo W, Ikawa T, Kawamoto H, Taniuchi I. Runx1‐Cbfβ facilitates early B lymphocyte development by regulating expression of Ebf1. J Exp Med 2012; 209:1255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Martensson A, Xie XQ, Persson C, Holm M, Grundstrom T, Martensson IL. PEBP2 and c‐myb sites crucial for λ5 core enhancer activity in pre‐B cells. Eur J Immunol 2001; 31:3165–74. [DOI] [PubMed] [Google Scholar]
- 87. Cerutti A. The regulation of IgA class switching. Nat Rev Immunol 2008; 8:421–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Spender LC, Cornish GH, Sullivan A, Farrell PJ. Expression of transcription factor AML‐2 (RUNX3, CBFα‐3) is induced by Epstein‐Barr virus EBNA‐2 and correlates with the B‐cell activation phenotype. J Virol 2002; 76:4919–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Spender LC, Whiteman HJ, Karstegl CE, Farrell PJ. Transcriptional cross‐regulation of RUNX1 by RUNX3 in human B cells. Oncogene 2005; 24:1873–81. [DOI] [PubMed] [Google Scholar]
- 90. Brady G, Elgueta Karstegl C, Farrell PJ. Novel function of the unique N‐terminal region of RUNX1c in B cell growth regulation. Nucleic Acids Res 2013; 41:1555–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhao B, Zou J, Wang H, Johannsen E, Peng CW, Quackenbush J et al Epstein–Barr virus exploits intrinsic B‐lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci USA 2011; 108:14902–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Brady G, Whiteman HJ, Spender LC, Farrell PJ. Downregulation of RUNX1 by RUNX3 requires the RUNX3 VWRPY sequence and is essential for Epstein–Barr virus‐driven B‐cell proliferation. J Virol 2009; 83:6909–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jiang S, Willox B, Zhou H, Holthaus AM, Wang A, Shi TT et al Epstein–Barr virus nuclear antigen 3C binds to BATF/IRF4 or SPI1/IRF4 composite sites and recruits Sin3A to repress CDKN2A. Proc Natl Acad Sci USA 2014; 111:421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. O'Connor BP, Raman VS, Erickson LD, Cook WJ, Weaver LK, Ahonen C et al BCMA is essential for the survival of long‐lived bone marrow plasma cells. J Exp Med 2004; 199:91–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ehrhardt GR, Hijikata A, Kitamura H, Ohara O, Wang JY, Cooper MD. Discriminating gene expression profiles of memory B cell subpopulations. J Exp Med 2008; 205:1807–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Whiteman HJ, Farrell PJ. RUNX expression and function in human B cells. Crit Rev Eukaryot Gene Expr 2006; 16:31–44. [DOI] [PubMed] [Google Scholar]
- 97. Withers DR. Lymphoid tissue inducer cells. Curr Biol 2011; 21:R381–2. [DOI] [PubMed] [Google Scholar]
- 98. Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol 2008; 9:503–10. [DOI] [PubMed] [Google Scholar]
- 99. Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol 2015; 15:243–54. [DOI] [PubMed] [Google Scholar]
- 100. Talebian L, Li Z, Guo Y, Gaudet J, Speck ME, Sugiyama D et al T‐lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFβ dosage. Blood 2007; 109:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Guo Y, Maillard I, Chakraborti S, Rothenberg EV, Speck NA. Core binding factors are necessary for natural killer cell development and cooperate with Notch signaling during T‐cell specification. Blood 2008; 112:480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ohno S, Sato T, Kohu K, Takeda K, Okumura K, Satake M et al Runx proteins are involved in regulation of CD122, Ly49 family and IFN‐γ expression during NK cell differentiation. Int Immunol 2008; 20:71–9. [DOI] [PubMed] [Google Scholar]
- 103. Hayashi K, Natsume W, Watanabe T, Abe N, Iwai N, Okada H et al Diminution of the AML1 transcription factor function causes differential effects on the fates of CD4 and CD8 single‐positive T cells. J Immunol 2000; 165:6816–24. [DOI] [PubMed] [Google Scholar]
- 104. Levanon D, Negreanu V, Lotem J, Bone KR, Brenner O, Leshkowitz D et al Transcription factor Runx3 regulates interleukin‐15‐dependent natural killer cell activation. Mol Cell Biol 2014; 34:1158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Chopin M, Seillet C, Chevrier S, Wu L, Wang H, Morse HC 3rd et al Langerhans cells are generated by two distinct PU.1‐dependent transcriptional networks. J Exp Med 2013; 210:2967–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Puig‐Kroger A, Aguilera‐Montilla N, Martinez‐Nunez R, Dominguez‐Soto A, Sanchez‐Cabo F, Martin‐Gayo E et al The novel RUNX3/p33 isoform is induced upon monocyte‐derived dendritic cell maturation and downregulates IL‐8 expression. Immunobiology 2010; 215:812–20. [DOI] [PubMed] [Google Scholar]
- 107. Mukai K, BenBarak MJ, Tachibana M, Nishida K, Karasuyama H, Taniuchi I et al Critical role of P1‐Runx1 in mouse basophil development. Blood 2012; 120:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhang DE, Hetherington CJ, Meyers S, Rhoades KL, Larson CJ, Chen HM et al CCAAT enhancer‐binding protein (C/EBP) and AML1 (CBFα2) synergistically activate the macrophage colony‐stimulating factor receptor promoter. Mol Cell Biol 1996; 16:1231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Himes SR, Cronau S, Mulford C, Hume DA. The Runx1 transcription factor controls CSF‐1‐dependent and ‐independent growth and survival of macrophages. Oncogene 2005; 24:5278–86. [DOI] [PubMed] [Google Scholar]
- 110. Dominguez‐Soto A, Relloso M, Vega MA, Corbi AL, Puig‐Kroger A. RUNX3 regulates the activity of the CD11a and CD49d integrin gene promoters. Immunobiology 2005; 210:133–9. [DOI] [PubMed] [Google Scholar]
- 111. Estecha A, Aguilera‐Montilla N, Sanchez‐Mateos P, Puig‐Kroger A. RUNX3 regulates intercellular adhesion molecule 3 (ICAM‐3) expression during macrophage differentiation and monocyte extravasation. PLoS ONE 2012; 7:e33313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Puig‐Kroger A, Sanchez‐Elsner T, Ruiz N, Andreu EJ, Prosper F, Jensen UB et al RUNX/AML and C/EBP factors regulate CD11a integrin expression in myeloid cells through overlapping regulatory elements. Blood 2003; 102:3252–61. [DOI] [PubMed] [Google Scholar]
- 113. Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res 1999; 117:145–52. [DOI] [PubMed] [Google Scholar]
- 114. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S et al Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010; 330:841–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zusso M, Methot L, Lo R, Greenhalgh AD, David S, Stifani S. Regulation of postnatal forebrain amoeboid microglial cell proliferation and development by the transcription factor Runx1. J Neurosci 2012; 32:11285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bowcock AM. The genetics of psoriasis and autoimmunity. Annu Rev Genomics Hum Genet 2005; 6:93–122. [DOI] [PubMed] [Google Scholar]
- 117. Prokunina L, Castillejo‐Lopez C, Oberg F, Gunnarsson I, Berg L, Magnusson V et al A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet 2002; 32:666–9. [DOI] [PubMed] [Google Scholar]
- 118. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF et al Safety, activity, and immune correlates of anti‐PD‐1 antibody in cancer. N Engl J Med 2012; 366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Helms C, Cao L, Krueger JG, Wijsman EM, Chamian F, Gordon D et al A putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is associated with susceptibility to psoriasis. Nat Genet 2003; 35:349–56. [DOI] [PubMed] [Google Scholar]
- 120. Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T et al An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet 2003; 35:341–8. [DOI] [PubMed] [Google Scholar]
- 121. Cho JH, Nicolae DL, Gold LH, Fields CT, LaBuda MC, Rohal PM et al Identification of novel susceptibility loci for inflammatory bowel disease on chromosomes 1p, 3q, and 4q: evidence for epistasis between 1p and IBD1. Proc Natl Acad Sci USA 1998; 95:7502–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Cho JH, Nicolae DL, Ramos R, Fields CT, Rabenau K, Corradino S et al Linkage and linkage disequilibrium in chromosome band 1p36 in American Chaldeans with inflammatory bowel disease. Hum Mol Genet 2000; 9:1425–32. [DOI] [PubMed] [Google Scholar]
- 123. Parkes M, Jewell D. Ulcerative colitis and Crohns disease: molecular genetics and clinical implications. Expert Rev Mol Med 2001; 2001:1–18. [DOI] [PubMed] [Google Scholar]
- 124. Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A et al Multiple common variants for celiac disease influencing immune gene expression. Nat Genet 2010; 42:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Weersma RK, Zhou L, Nolte IM, van der Steege G, van Dullemen HM, Oosterom E et al Runt‐related transcription factor 3 is associated with ulcerative colitis and shows epistasis with solute carrier family 22, members 4 and 5. Inflamm Bowel Dis 2008; 14:1615–22. [DOI] [PubMed] [Google Scholar]
- 126. Haley KJ, Lasky‐Su J, Manoli SE, Smith LA, Shahsafaei A, Weiss ST et al RUNX transcription factors: association with pediatric asthma and modulated by maternal smoking. Am J Physiol Lung Cell Mol Physiol 2011; 301:L693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Taniuchi I. Transcriptional regulation in helper versus cytotoxic‐lineage decision. Curr Opin Immunol 2009; 21:127–32. [DOI] [PubMed] [Google Scholar]
- 128. Cerutti A, Puga I, Cols M. Innate control of B cell responses. Trends Immunol 2011; 32:202–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hor YT, Voon DC, Koo JK, Wang H, Lau WM, Ashktorab H et al A role for RUNX3 in inflammation‐induced expression of IL23A in gastric epithelial cells. Cell Rep 2014; 8:50–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD et al IL‐23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med 2005; 201:233–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB et al Elephant shark genome provides unique insights into gnathostome evolution. Nature 2014; 505:174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med 2007; 204:1945–57. [DOI] [PMC free article] [PubMed] [Google Scholar]


