Summary
Protein kinase C (PKC) form a key family of enzymes involved in signalling pathways that specifically phosphorylates substrates at serine/threonine residues. Phosphorylation by PKC is important in regulating a variety of cellular events such as cell proliferation and the regulation of gene expression. In the immune system, PKCs are involved in regulating signal transduction pathways important for both innate and adaptive immunity, ultimately resulting in the expression of key immune genes. PKCs act as mediators during immune cell signalling through the immunological synapse. PKCs are traditionally known to be cytoplasmic signal transducers and are well embedded in the signalling pathways of cells to mediate the cells' response to a stimulus from the plasma membrane to the nucleus. PKCs are also found to transduce signals within the nucleus, a process that is distinct from the cytoplasmic signalling pathway. There is now growing evidence suggesting that PKC can directly regulate gene expression programmes through a non‐traditional role as nuclear kinases. In this review, we will focus on the role of PKCs as key cytoplasmic signal transducers in immune cell signalling, as well as its role in nuclear signal transduction. We will also highlight recent evidence for its newly discovered regulatory role in the nucleus as a chromatin‐associated kinase.
Keywords: chromatin, epigenetics, immune system, protein kinase C, signal transduction
Abbreviations
- BAF60c
Brg1/Brm‐associated factor 60c
- Bcl10
B‐cell leukemia/lymphoma 10
- BCR
B‐cell receptor
- Btk
Bruton's tyrosine kinase
- CARMA1
caspase recruitment domain family (CARD)‐containing membrane‐associated guanylate kinase (MAGUK) protein 1
- CIITA
class II transactivator
- CREB
cAMP response element‐binding protein
- DAG
diacylglycerol
- ERα
oestrogen receptor α
- GLK
germinal centre kinase (GCK)‐like kinase (MAP4K3)
- H1
histone H1
- H2B
histone H2B
- H3
histone H3
- HEXIM1
hexamethylene‐bis‐acetamide‐induced mRNA‐encoded proteins 1
- IFN
interferon
- IKK
inhibitor of κB (IκB) kinase
- IL
interleukin
- IκB
inhibitor of κB
- K
lysine
- Ki‐1/57
57‐000 MW human protein antigen recognized by the CD30 antibody Ki‐1
- MALT1
mucosa‐associated lymphoid tissue 1
- MyD88
myeloid differentiation primary‐response protein 88
- NF‐κB
nuclear factor κB
- NLS
nuclear localization signal
- P
proline
- PCAF
p300/CREB‐binding protein‐associated factor
- PKA
protein kinase A
- PKC
protein kinase C
- PKG
protein kinase G
- S
serine
- S/T‐P‐S/T
SPT
- SATB1
special AT‐rich binding protein 1
- STAT
signal transducer and activator of transcription
- T
threonine
- TAK1
transforming‐growth‐factor–activated kinase 1
- TCR
T‐cell receptor
- Th
T helper
- TIR
Toll–IL‐1 receptor
- TIRAP
Toll–IL‐1 receptor domain‐containing adaptor protein
- TLR
Toll‐like receptor
- TRAF6
tumour necrosis factor receptor‐associated factor 6
- TRAM
TRIF‐related adaptor molecule
- TRIF
TIR domain‐containing adaptor inducing interferon‐β
- Y
Tyrosine
Introduction
Protein kinase C (PKC) is a key family of enzymes involved in signalling pathways that specifically phosphorylates substrates at serine/threonine residues, influencing a variety of cellular events such as cell proliferation and the regulation of gene expression.1, 2 PKC is a subfamily of AGC (PKA, PKG and PKC) kinases, incorporating 10 kinase members that share a highly conserved catalytic kinase domain, and a less conserved regulatory domain responsible for binding to activators and to anchoring proteins.2 Isoforms of PKC can be divided into three sub‐classes of serine/threonine kinases (classical, novel and atypical) according to structural motifs and activation requirements (Fig. 1).1 Classical (also called conventional) PKC (cPKC) isoforms, including α, β (I and II) and γ, contain motifs for diacylglycerol (DAG) and calcium‐dependent phospholipid binding, and so require both DAG and calcium for activation to occur.
Figure 1.
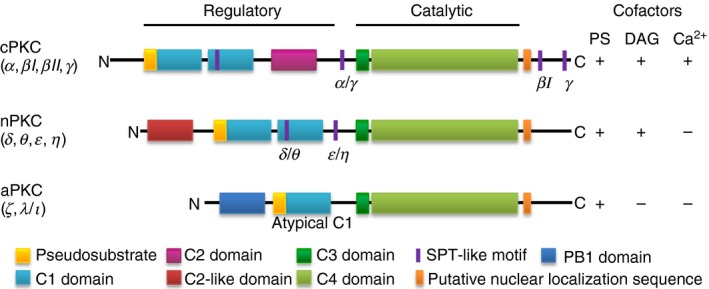
Schematic diagram of the primary structure of protein kinase C (PKC) family members. All PKC isoforms have the pseudosubstrate (yellow) and the C1 domain (blue) for diacylglycerol (DAG) (except for atypical PKCs) and phosphatidylserine (PS) binding in the regulatory region located at the N‐terminal. They also have a catalytic domain, C3 (dark green) and C4 (light green), in the C‐terminal region. Conventional PKCs (cPKC) have a C2 domain (pink) for the calcium (Ca2+) dependent binding of anionic lipids such as phosphatidylinositol 4,5‐biphosphate (PIP 2). Novel PKCs (nPKC) have a C2‐like domain (red), which cannot bind Ca2+ or PIP 2. Atypical PKCs (aPKC) have an atypical C1 domain that cannot bind DAG and Phox/Bem domain 1 (PB1), which allows protein interactions. All PKC isoforms also have the putative nuclear localization sequence and all except for aPKCs show the presence of SPT‐like motif. Different PKC isoforms have SPT‐like motif present at different locations within their structure.
Unlike cPKCs, in novel PKCs (nPKCs – δ, ε, η and θ), the C2 domain (also known as the V1 domain) is located at the N‐terminus of the C1 domain and lacks the aspartic acid residues necessary for coordinating calcium ions.3 Furthermore, the same lipids activate cPKCs and nPKCs but they can activate cPKCs only in the presence of calcium, this is in part due to the higher affinity of the C1 domain of nPKCs to DAG.4 In contrast, atypical PKC isoforms (aPKCs – ζ and λ/ι) contain a single C1 domain and are therefore incapable of binding DAG and do not bind calcium. Atypical PKC contain a single zinc‐finger motif within the C1 domain that can be bound by zinc‐finger proteins.5
While PKCs act to phosphorylate substrates, the enzyme itself requires three ordered phosphorylations in order to be catalytically competent.2 The first, rate‐limiting phosphorylation occurs on the activation loop at T500 by phosphoinositide‐dependent kinase, 3‐phosphoinositide dependent protein kinase‐1.2 This then prompts the rapid phosphorylation of a turn motif at T641, which results in autophosphorylation at S660 on the hydrophobic motif. The fully phosphorylated PKCs are maintained in a catalytically inactive form mainly by intramolecular interactions such as that of the pseudosubstrate domain until the binding of cofactors such as phosphatidylserine, DAG and calcium, to the regulatory modules. All PKC enzymes are allosterically activated by phosphatidylserine, which binds to the C1 domain, but its affinity for membrane phospholipids is increased by the binding of DAG to the C1 domain and calcium‐binding to the C2 domain, depending on the class of PKCs. As a result of the cofactor binding, the pseudosubstrate domain is released from the kinase core, allowing PKC to phosphorylate target substrates.6
The vast amount of published data presented on PKCs describes their function as cytoplasmic signal transducers, incorporated into the pathways of every mammalian cell to serve as an intermediatory between membrane binding and nuclear events.1, 7, 8 PKCs are expressed in numerous tissue and cell types and although most PKC isoforms are ubiquitously expressed, some isoforms are expressed in a tissue‐specific and cell‐specific manner (Table 1). In the immune system, PKCs are important mediators of immune cell signalling through the immunological synapse. All PKC isoforms are expressed by immune cells, with the exception of PKCγ which is preferably expressed by the central nervous system.9 Although ubiquitously expressed, the expression pattern and levels of each PKC isoform are cell‐type specific, highlighting their specific function and non‐redundant roles in the immune system. Recent data suggest that PKCs have additional, non‐traditional roles as nuclear kinases, whereby these enzymes have the capacity to directly regulate gene expression programmes. In this review, we will focus on the role of specific PKCs as key cytoplasmic signal transducers in well‐characterized immune cell signalling pathways in the innate and adaptive immune system. We will also discuss the role of PKC in the context of nuclear signal transduction and highlight recent evidence for its newly discovered regulatory role in the nucleus as a chromatin‐associated kinase, a function that appears to be evolutionarily conserved.
Table 1.
Protein kinase C (PKC) isoform cell‐specific expression and defects in knockout mice
| PKC | Tissue expressiona | Knockout mouse phenotype |
|---|---|---|
| PKCα | Ubiquitous, T cells, plasmacytoid dendritic cells (pDC) | T‐cell activation and T‐cell immunity defects148 |
| PKCβ | Ubiquitous, B cells and mast cells | B‐cell signalling and survival defects; mast cells defects56, 149 |
| PKCδ | Ubiquitous, B cells, mast cells, macrophages | B‐cell homeostasis defects150 |
| PKCε | Ubiquitous | Macrophage activation defect16 |
| PKCη | Ubiquitous, T cells and macrophages | T‐cell homeostasis and regulatory T cell function defects151, 152 |
| PKCθ | T cells, mast cells, platelets, skeletal muscle | T‐cell activation defects37, 39 |
| PKCζ | Ubiquitous | B‐cell receptor signalling defects; T helper type 2 response defects153 |
| PKCλ/ι | Ubiquitous | Embryonic lethal154 |
PKCγ is preferably expressed in the brain.
PKC as cytoplasmic signal transducers
The PKCs are involved in regulating signal transduction pathways important for both innate and adaptive immunity, ultimately resulting in the expression of key immune genes. Different PKC isoforms are involved in distinct signalling pathways, with selective functions in a cell‐specific manner. This review will focus on the Toll‐like receptor (TLR) signalling in the innate system, T‐cell receptor (TCR) signalling and B‐cell receptor (BCR) signalling in the adaptive immune system as these pathways are very well characterized signalling pathways known in the literature. Furthermore, PKCs are known to be key signalling molecules in these pathways. Although different PKC isozymes are involved in these signalling pathways, we will highlight the key PKC isozymes implicated for each pathway in this review.
PKCε in TLR4 signalling
Toll‐like receptors play a key role in the innate immune system by defending the host against microbial infection.10 TLRs are a family of pattern recognition receptors that are present in many cell types, with most of the expression by macrophages, neutrophils and dendritic cells.11 The best‐studied TLR ligand is lipopolysaccharide, a component of the Gram‐negative bacteria. It activates the innate immune system through binding with TLR4 to initiate two intracellular pathways: MyD88 (myeloid differentiation primary‐response protein 88)‐dependent and MyD88‐independent pathways (Fig. 2a).12 The MyD88‐dependent pathway requires TIRAP [Toll–interleukin‐1 (IL‐1) receptor (TIR) domain‐containing adaptor protein] to link MyD88 to TLR4 receptor, leading to the expression of inflammatory cytokines such as IL‐1, IL‐6 and tumour necrosis factor α. In contrast, the MyD88‐independent pathway is mediated by TRIF [TIR domain‐containing adaptor inducing interferon‐β (IFN‐β)] and TRAM (TRIF‐related adaptor molecule) to induce expression of type I IFNs.13 It is beyond the scope of this review to cover these pathways in detail hence we refer readers to the multiple reviews that explain the TLR4 signalling pathway in depth.10, 11, 14, 15
Figure 2.
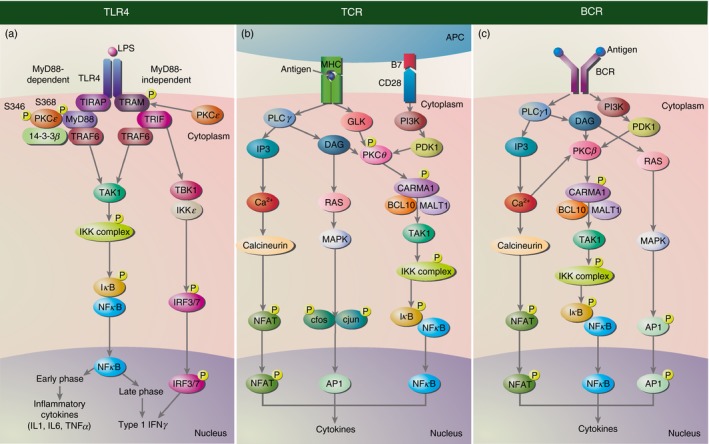
Protein kinase C (PKC) involvement in cytoplasmic signal transduction pathways in the immune system. (a) Protein kinase Cε (PKC ε) is an important player in the Toll‐like receptor 4 (TLR4) signalling pathway during macrophage activation. Binding of lipopolysaccharide (LPS) to the TLR4 initiates the activation of two intracellular pathways: myeloid differentiation primary‐response protein 88 (MyD88)‐dependent and MyD88‐independent pathways. In the MyD88‐dependent pathway, Toll–interleukin (IL)‐1 receptor (TIR) domain‐containing adaptor (TIRAP) links MyD88 to the TLR4 receptor. PKC ε is then recruited to TLR4 via MyD88 and phosphorylated on serine 346/368. This phosphorylation leads to binding with 14‐3‐3β and the formation of a complex with TLR4, TIRAP, MyD88 and TNF receptor‐associated factor 6 (TRAF6) as well. In the MyD88‐independent pathway, TRIF‐related adaptor molecule (TRAM) is phosphorylated by PKC ε allowing TLR4 to link with TRIF. TRIF then recruits kinases such as transforming‐growth‐factor‐activated kinase 1 (TAK1) (through TRAF6), TANK‐binding kinase 1 (TBK1) and inhibitor of κB (IkB) kinase epsilon (IKK ε) for activation of nuclear factor κB (NF‐κB) and interferon regulatory factor (IRF)‐3/7 signalling pathways to produce inflammatory cytokines and Type I interferons. (b) In T cell receptor (TCR) signalling, activation occurs when an antigen is presented by the antigen‐presenting cell (APC) on the MHC is complexed with the TCR together with the binding of co‐stimulatory molecules CD28 and B7. This leads to the activation of calcium signalling pathways and mitogen‐activated protein kinase (MAPK) pathways through phospholipase Cγ (PLC γ), GCK‐like kinase (GLK) activation and 3‐phosphoinositide dependent protein kinase‐1 (PDK1) activation through phosphoinositide 3‐kinase (PI3K). Diacylglycerol (DAG) generated through PLC γ1 activation binds to protein kinase Cθ (PKC θ), which is also phosphorylated by PDK1 and GLK. Activation of PKC θ leads to the activation of NF‐κB signalling pathways. (c) B cells are activated upon antigen binding to the B cell receptor (BCR). Similar pathways to TCR are activated but the key PKC isoform involved is PKC β. In addition, calcium (Ca2+) generated from PLC γ1 activation also acts to activate protein kinase Cβ (PKC β). For both TCR and BCR signalling, the signalling pathways activated leads to the recruitment and activation of nuclear transcription factors [NF‐κB, activator protein 1 (AP1) and nuclear factor of activated T cells (NFAT)] to then produce cytokines.
Although PKC isoforms are involved at many levels of the TLR signalling cascade, we will focus on the involvement of PKCε in TLR4 signalling as PKCε is implicated as an important player in the TLR4 signalling pathway during macrophage activation.7 The role of PKCε in host defence against bacterial infection was revealed through studies in PKCε knockout mice, where mice lacking PKCε have a diminished response to lipopolysaccharide stimulation, characterized by low levels of several cytokines, namely tumour necrosis factor‐α and IL‐1β.16 Other studies show PKCε playing a role in both the MyD88‐dependent and MyD88‐independent pathways of TLR4 signalling (Fig. 2a).17, 18 In the MyD88‐dependent pathway, lipopolysaccharide stimulation leads to PKCε recruitment to TLR4 and phosphorylation on S346 and S368 via MyD88.17 These phosphorylations lead to binding with 14‐3‐3β, which is also MyD88 dependent. The phosphorylation event is important for downstream signalling as cells expressing mutant PKCε S346A/S368A were unable to activate nuclear factor‐κB (NF‐κB) upon TLR induction. This suggests that PKCε not only needs to be phosphorylated for its ability to bind to 14‐3‐3β, but it also exists as a complex with TLR, MyD88 and 14‐3‐3β to regulate gene expression. In the MyD88‐independent pathway, PKCε is required for TLR4 activation via the TRAM substrate as phosphorylation of TRAM is disrupted in PKCε‐deficient cells.18 TRAM is localized to the plasma membrane in the unstimulated state but upon lipopolysaccharide stimulation, it is phosphorylated by PKCε on a serine residue near the N‐terminal end.14 This phosphorylation dissociates TRAM from the membrane, allowing it to then link TLR4 with TRIF.7 The TLR4–TRAM–TRIF complex is necessary for activation of further downstream NF‐κB and IFN regulatory factor‐3/7 signalling pathways.19
The innate immunity provided by the TLR pathway is also required for the adaptive response by T‐cell activation against antigens.20, 21, 22 Cross‐talk in signalling pathways is not uncommon but the specificity of the response depends on the stimulus provided and the cell type engaged to generate the appropriate immune response.
PKCθ in TCR signalling
The cells of the adaptive immune system form the immunological synapse with antigen‐presenting cells to activate signal transduction pathways that induce gene expression programmes for lymphocyte function. In T cells, activation of T cells occurs when the TCR recognizes an antigen presented by the antigen‐presenting cells. This leads to T‐cell differentiation and is a process involving the activation of multiple pathways including PKC signalling (Fig. 2b). Following TCR–antigen‐presenting cell complex formation, PKC localizes to the immunological synapse and subsequently stimulates the recruitment and activation of nuclear transcription factors (such as NF‐κB, activator protein 1 and nuclear factor of activated T cells) required for induction of immune effector genes.8 These effector genes are then expressed in a rapid and transient manner to produce cytokines, chemokines, cell surface molecules and growth factors, important for T‐cell proliferation and differentiation. The details of the T‐cell activation pathways have been reviewed extensively elsewhere.23, 24, 25, 26, 27, 28 Although other members of the PKC family can also be found in the immunological synapse of different T‐cell subsets,29 PKCθ is the most prominently studied PKC in TCR signalling since the discovery of its selective recruitment to the immunological synapse in effector T cells.30 Furthermore, it is selectively expressed in T cells within the haematopoietic cell population.31 Upon TCR activation, PKCθ localizes to the central supramolecular activation cluster of the immunological synapse at the plasma membrane.32 The membrane translocation of PKCθ requires association with co‐stimulatory molecule CD28 with the V3 domain of PKCθ.33, 34, 35 The ability of PKCθ to segregate correctly to the central supramolecular activation cluster is dependent on the presence of CD28 as activated T cells from CD28‐deficient mice were unable to form the mature immunological synapse with PKCθ, forming a diffuse pattern throughout the synapse instead.33 The specific localization of PKCθ to the immunological synapse is critical for an effective T‐cell activation and this translocated PKCθ is also enzymatically active.30 Interestingly, the activity of PKCθ is also regulated by the intracellular redox state, in which the oxidized inactive form of PKCθ is recruited to the plasma membrane in naive T cells.36
The role of PKCθ in regulating T‐cell function was initially characterized using PKCθ‐knockout mice (Table 1).37, 38, 39 Further studies on PKCθ‐deficient T cells reveal that PKCθ performs different functions depending on the T‐cell subpopulations. For example, PKCθ is required for a T helper type 2 (Th2) cell but not Th1 cell in vivo immune response against helminth infection and allergic airway inflammation.40 However, contrasting studies in mouse experimental autoimmune encephalomyelitis show impaired Th1 responses in PKCθ‐deficient mice suggesting that PKCθ is important for regulating both Th1 and Th2 responses but in an antigen‐dependent and organ‐specific manner.41, 42 These studies have also shown that PKCθ is required for the Th17‐dependent development of experimental autoimmune encephalomyelitis, implicating PKCθ in controlling Th17 differentiation.41, 42 According to Kwon et al.,43 PKCθ up‐regulates signal transducer and activator of transcription 3 (STAT3) under Th17 priming conditions upon TCR stimulation with PMA. PKCθ promotes the activation of STAT3 by regulating the association of activator protein 1 and NF‐κB transcription factors to STAT3 promoter.43 More recently, PKCθ was found to be essential in suppressing Th1‐typical genes such as Stat4, Tbet and Ifng during Th17 immune activation, as a way to stabilize the Th17 cell phenotype.44 PKCθ is also involved in regulating immune memory. In a study on antiviral responses by CD8+ T‐cell responses, PKCθ is required for antigen recall responses upon in vitro infection by lymphocytic choriomeningitis virus and influenza virus.45, 46 Furthermore, the efficient and timely recruitment of PKCθ to the immunological synapse is critical for memory T‐cell development.47
Interestingly, PKCθ also localizes to the immunological synapse in effector T cells to positively regulate cell function but activation of regulatory T cells sequesters PKCθ away from the immunological synapse, leading to negative regulation of induced regulatory T cells.48, 49 This negative regulation by PKCθ involves inhibiting differentiation of induced regulatory T cells through the AKT/Foxo1/3a pathway.50 Hence, PKCθ plays a role in regulating the T‐cell immune response through maintaining the equilibrium of T‐cell subpopulations. However, the precise mechanism by which it deciphers the signals received within each cell subset to perform the cell‐type‐specific function is yet to be elucidated.
PKCβ in BCR signalling
Like T cells, B‐cell activation leads to production of regulatory cytokines and chemokines to eliminate pathogens. However, B cells also present antigen to T cells and specialize in producing high‐affinity antibodies and long‐lived memory cells, generating rapid and long‐lasting protection against secondary exposure to the same pathogen.51 Activation of B cells occurs when an antigen binds to the BCR, initiating phosphorylation events by Src‐family kinases as well as Syk and Bruton's tyrosine kinase (Btk)/Tec family kinases. This signals the organized assembly kinases and adaptor proteins forming the signalosome and activating multiple signalling cascades (Fig. 2c).52 Secondary messengers such as DAG and inositol‐1,4,5‐triphosphate are generated as part of the BCR activation signalling cascade to initiate Ca2+ and PKC downstream signalling pathways, respectively, leading to activation of transcription factors (Myc, nuclear factor of activated T cells, NF‐κB, activator protein 1) critical for B‐cell function.53 While B cells express multiple isoforms of PKC (α, β, δ, ε, η, ζ, and λ), PKCβ is the key PKC isoform that is important in BCR signalling.54, 55, 56, 57, 58
The role for PKCβ in regulating B‐cell functions was first discovered through PKCβ gene knockout mice, which were shown to have impaired B‐cell activation, inability to proliferate upon BCR stimulation and defects in T‐cell‐independent immune responses (Table 1).58 The immunodeficiency traits exhibited by these PKCβ knockout mice are similar to those seen in Btk‐deficient or X‐linked immunodeficient mice, suggesting that Btk and PKCβ may be linked in BCR signalling.59 Indeed, Btk has been shown to be important for NF‐κB activation upon BCR engagement and is regulated by PKCβ in a negative feedback mechanism.60, 61, 62 Specifically, PKCβ directly phosphorylates Btk to down‐regulate Btk kinase activity and alter its membrane localization in BCR signalling.62 This result was also confirmed using a PKCβ‐selective inhibitor, where inhibiting PKCβ kinase activity leads to an increase in Btk kinase activity to enhance calcium mobilization, so implicating PKCβ in the regulation of calcium release upon BCR activation.63
Another study used N‐ethyl‐N‐nitrosurea‐induced mutagenesis to generate PKCβ mutant mice (Tilcara) that have heterozygous mis‐sense mutations.64 The Tilcara mutant mice have single amino acid substitutions in conserved residues within the kinase domain of PKCβ. This results in an S552P mutation that is close to a docking site for the pseudosubstrate domain. These mice show impaired T‐cell independent antibody responses, as seen in the PKCβ gene knockout mice.58 Another effect of the Tilcara mutation is loss of active PKCβI but not PKCβII in B‐cell protein lysates in homozygous mutants, implying that the region of mutation is important for PKCβI function. While the Tilcara mutant mice do not show as significant an effect phenotypically compared to PKCβ knockout mice, the S552P substitution occurs at an evolutionarily conserved residue, hence there could be changes occurring at the transcriptional level. It would be of great interest to examine how mutation at this conserved residue can affect BCR signalling on a genotypic level.
PKCβ is also involved in forming the BCR signalosome upon BCR engagement (Fig. 2c). PKCβ phosphorylates CARMA1 [caspase recruitment domain family (CARD)‐containing membrane‐associated guanylate kinase (MAGUK) protein 1] on S668 to lead to the formation and recruitment of the CARMA1, B‐cell leukaemia/lymphoma 10 (Bcl10), and mucosa‐associated lymphoid tissue 1 (MALT1) complex to lipid rafts to form part of the BCR signalosome.65 PKCβ is also required in recruiting inhibitor of κB (IκB) kinase (IKK) to the CARMA1–Bcl10–MALT1 complex.56 For IKK activation to occur, the adaptor protein CARMA1 is directly phosphorylated by PKCβ, which then brings IKK and another protein kinase transforming‐growth factor‐activated kinase 1 (TAK1) close together.66, 67 This allows TAK1 to phosphorylate IKK leading to its activation.66 As a result, IκB is phosphorylated by IKK to lead to activation of NF‐κB, hence demonstrating the critical role that PKCβ plays in BCR‐dependent NF‐κB signalling.
Hence, from all the examples shown in the different immune cells, PKC signalling pathways converge with other signalling pathways in the nucleus to regulate inducible gene transcription. Furthermore, only a specific isozyme has been discussed in this review but there are other isozymes that participate in each of the signalling pathways, in which crosstalk between the different isozymes could occur. Undoubtedly, the PKC signalling pathway is important in transducing signals in the cytoplasm upon immune cell activation but PKCs are also found in the nucleus, suggesting a dual role by PKC as nuclear signal transducers as well as cytoplasmic signal transducers.
PKC as nuclear signal transducers
While the mechanism of PKC signalling in the cytoplasm through the plasma membrane is very well characterized, relatively less is known about PKC signalling within the nucleus. Since the discovery of nuclear PKC in the nuclei of rat liver,68 there is a growing body of evidence to support the presence of PKC in the nucleus, with specific expression of the isoforms depending on the cell type and differential distribution within subnuclear compartments.69, 70, 71 The PKCs present in the cell nucleus are either translocated from the cytoplasm upon activation or exist constitutively within the nucleus.72 In the immune system, translocation of PKC to the nucleus appears to be the main mechanism in which nuclear PKC regulates immune cell differentiation. This translocation occurs upon stimulus with differentiation agonists such as macrophage colony‐stimulating factor, hexamethylene‐bis‐acetamide, vitamin D3, anti‐trans retinoic acid, PMA and nerve growth factor (reviewed in ref. 71). Granulocyte–macrophage colony‐forming cells treated with macrophage colony‐stimulating factor show increased PKCα levels and stimulated its translocation to the nucleus, leading to macrophage differentiation.73 Similarly, hexamethylene‐bis‐acetamide‐induced differentiation of Friend erythroleukaemia cells requires the localization of PKCα to the nucleus as PKCα‐antisense transfection prevented cell differentiation.74 Also, the translocation of specific PKC isoforms is dependent on the stimulus used within the same cells.75 This can be seen in human promyelocytic leukaemia HL‐60 cell line, where vitamin D3 exposure leads to increased levels of PKCζ isoform while anti‐trans retinoic acid treatment leads to an increase in PKCα and PKCζ isoform in the nucleus.76, 77 In contrast, HL‐60 cells stimulated with PMA leads to the accumulation of PKCδ within the nucleus.78
As mentioned earlier, PKC is activated by second messengers generated from immune cell activation such as calcium and/or DAG depending on the PKC isoform. Interestingly, there are distinct second messenger signalling pathways in the cytoplasm and nucleus. Calcium released in the cytoplasm causes cytoplasmic PKC to translocate to the plasma membrane while calcium that is released within the nucleus translocates nuclear PKC to the nuclear envelope.79 Also, there are separate pools of DAG produced by phospholipids localized within the cytoplasm or the nucleus.80 Furthermore, stimulus‐dependent production of DAG leads to selective translocation of specific PKC isoforms to the nucleus. For example, in HL‐60 cells, differentiation signals lead to DAG production in the nucleus by phospholipase D causing PKCα nuclear translocation. When proliferation stimulus is used, nuclear DAG is produced by phosphatidylinositol (4,5) biphosphate leading to PKCβII nuclear migration.81 Hence, PKC in the cytoplasm and the nucleus are differentially regulated depending on its localization. Although PKC isoforms have been detected in the nucleus of immune cells in the resting state, it is unclear how it is being retained in the nucleus and whether it is structurally and functionally different from the translocated PKCs. It is postulated that PKC‐binding proteins may play a role not only in the retention of PKC in the nucleus but also in the translocation of PKC into the nucleus.75, 82
Nuclear translocation signals for nucleocytoplasmic shuttling
Proteins can be transported between the cytoplasm and the nucleus through nuclear pores located within the nuclear envelope. The ability of proteins to translocate into or out of the cell nucleus involves nuclear translocation signals such as nuclear localization signals (NLS) or nuclear export signals, respectively.83 It can also occur through binding with proteins that have NLS as a way to enter the nucleus. The best characterized NLSs are classical NLS motifs, which can have a monopartite or bipartite motif.84 PKC isoforms do not have the canonical NLS but contain a nuclear targeting motif that is similar to the classical bipartite NLS, forming a putative NLS that is conserved across the different PKC isoforms (Fig. 1).85, 86 In line with this, a putative NLS motif was proven to be functional for nuclear import of PKCδ to initiate apoptosis.86 Upon induction with apoptotic agents in ParC5 cells, PKCδ is phosphorylated at Y64 and Y155, causing a conformational change to expose the NLS and allow the nuclear import factor, importin‐α, to bind and facilitate nuclear import of PKCδ.87 Atypical PKC was also shown to contain both a functional NLS and nuclear export signals within the zinc‐finger domain, allowing for rapid shuttling between the nucleus and cytoplasm.88 Similarly, aPKC is phosphorylated at Y256 upon nerve growth factor stimulation in PC12 cells, and bound by importin‐α for nuclear translocation.89 Hence, there seem to be similarities in the mechanism of translocation between PKC isoforms but this is quite likely to occur in a stimulus‐specific and cell context‐dependent manner.
Translocation of PKC into the nucleus occurs through mechanisms that are different from the proteins with canonical NLS.90, 91 For example, PKCα requires an intact cytoskeleton to translocate into the nucleus but a protein with canonical NLS is unaffected when the cytoskeleton is disrupted.90 In addition, PKCα does not require nuclear import factors p97/importin/karyopherin β or GTP to transport into the nucleus, unlike proteins with canonical NLS.91 Interestingly, aPKCs have isoform‐specific regulation of nucleocytoplasmic shuttling. Even though both PKCζ and PKCι contain the same NLS, only PKCζ require the NLS for nuclear translocation while the ability of PKCι to translocate is independent of the NLS.92 In contrast, using chimeric proteins, Seidl et al.92 showed that PKCι requires the hinge region for nuclear localization but the hinge region of PKCζ excludes it from the nucleus. Undoubtedly, the requirement of the PKC NLS and the associated protein domains as well as nuclear import factors plays a role in nucleocytoplasmic shuttling of PKC. What is interesting is how different isoforms each have specific requirements for each of those features, which leads to isoform‐specific localization of PKC, and hence isoform‐specific regulation of nuclear function.
In addition to the NLS, another motif, the S/T‐P‐S/T (SPT‐like) motif, was suggested to perform like a general NLS for signalling proteins that do not have the canonical NLS, implying that PKC could potentially be regulated by the SPT‐like motif.93 Using protein sequence alignment, Sutcliffe et al.94 showed that novel and conventional PKC isoforms were predicted to have SPT‐like motif present. The SPT‐like motif appears to reside close to where the regulatory and kinase/catalytic domains are and deletions in the regulatory or kinase domains of PKC are shown to affect nuclear translocation.95, 96 Mutation studies substituting the SPT motif in PKCθ with an alternative motif that cannot be phosphorylated impaired the localization of PKCθ to the nucleus, suggesting that phosphorylation of SPT is required for nuclear translocation.94 Interestingly, mutating the putative NLS motif has no effect on the distribution of PKC in the cell, suggesting that the SPT‐like motif may be an alternative NLS motif used by signalling kinases that do not have the canonical NLS for nuclear transport.94 Hence, it would be of interest to examine how PKC activation can affect the configuration of the protein folding/structure and as a consequence the exposure of the nuclear localization motifs for regulation.
The fact that there are numerous substrates described for nuclear PKC suggests that PKC could potentially regulate nuclear functions like gene transcription (reviewed in refs. 97, 98). Indeed, some of the nuclear PKC substrates include chromatin factors such as histone (H1, H2B, H3), RNA polymerase II, transcription factors Fos and cAMP response element‐binding protein (CREB), as well as architectural proteins such as high mobility group proteins, implicating PKC in directly regulating gene transcription in the nucleus.
PKC as nuclear chromatin regulators
Gene transcription is dynamically regulated through the chromatin structure by various epigenetic factors. The fundamental unit of chromatin is the nucleosome: 147 base pairs of DNA wrapped around a histone octamer (two sets of H2A, H2B, H3 and H4) often with a linker histone (H1) present.99, 100 The accessibility of gene regulatory regions is dependent on whether the chromatin is in the open (permissive) or closed (repressive) state, so leading to either gene activation or repression, respectively. Control of chromatin accessibility hence gene expression involves transcriptional regulatory complexes such as transcription factors, histone variants, chromatin remodelling complexes and histone modifiers (reviewed in refs 101, 102, 103).
The earliest evidence of a signalling kinase involvement in directly regulating chromatin is in yeast where the key signalling kinase Hog1 was found to physically associated with promoter regions upon osmotic stress.104 Since then, more evidence has revealed the role of signalling kinases as epigenetic regulators that can modify transcription factors, histones as well as histone modifiers in the nucleus (reviewed in ref. 105). This includes the PKC signalling family, which has been shown to have a role in directly regulating gene transcription through either phosphorylating specific histone residues, histone modifiers, transcription factors as well as other transcriptional regulatory factors or forming part of a transcriptional regulatory complex (Fig. 3, Table 2).
Figure 3.
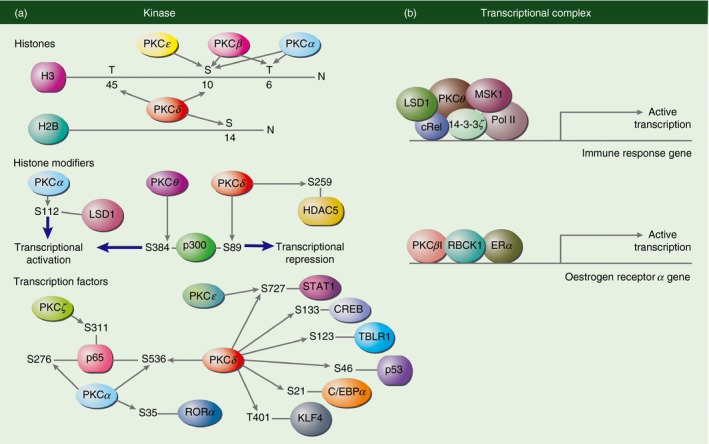
Protein kinase C (PKC) has a role as chromatin regulator in the nucleus. (a) PKC can act as a kinase to phosphorylate histones, histone modifiers or transcription factors at specific serine and/or threonine residues leading to transcriptional activation or repression. Both PKC α and PKC β can phosphorylate histone H3 at threonine (T)6 and serine (S)10 while PKC ε phosphorylates S10 only. PKC δ phosphorylates H3S10, H3T45 and H2BS14. PKC α phosphorylates S112 on lysine specific demethylase 1 (LSD1) leading to transcriptional activation. While PKC θ phosphorylates S384 on p300 causes gene activation, PKC δ phosphorylation on S389 of p300 leads to transcriptional repression. PKC δ can also phosphorylate S259 on histone deacetylase 5 (HDAC5). As for transcription factors, PKC α phosphorylates S276 and S536 on p65 and S35 on retinoic acid‐related orphan nuclear receptor α (ROR α). PKC ζ phosphorylates S311 on p65. PKC δ phosphorylates T401 on Krüppel‐like factor 4 (KLF4), S21 on CCAAT/enhancer‐binding protein α (C/EBP α), S46 on p53, S123 on transducin β‐like 1X‐linked receptor 1 (TBLR4), S133 on cAMP response element‐binding protein (CREB) and S727 on signal transducer and activator of transcription 1 (STAT1). PKC ε can also phosphorylate STAT1 at S727. (b) PKC can also form a transcriptional complex on a gene promoter. For example, PKC θ forms a transcriptional complex with LSD1, mitogen and stress‐activated protein kinase 1 (MSK1), RNA polymerase II (Pol II), 14‐3‐3ζ and cRel on immune response gene promoters to lead to transcriptional activation. PKC βI is recruited to the oestrogen receptor a promoter as part of a regulatory complex with RanBP‐type and C3HC4‐type zinc finger containing 1 (RBCK1) and oestrogen receptor α (ER α) to regulate ER α promoter gene expression.
Table 2.
Chromatin‐associated protein kinase C (PKC) substrates
| Kinase | Histone | Histone modifier | Transcription factor | Other chromatin regulatory factors | Phosphorylated residue | Regulatory role |
|---|---|---|---|---|---|---|
| PKCα | H3 | S10 | ND | |||
| H3 | T6 | Txn activation | ||||
| HDAC6 | Txn activation | |||||
| LSD1 | S112 | Txn activation | ||||
| p65 | S276/S536 | Txn activation | ||||
| RORα | S35 | Txn repression | ||||
| Sp1 | S‐P | Txn activation | ||||
| PKCβ | H3 | S10 | Txn activation | |||
| CBP | Txn activation | |||||
| PKCβI | H3 | T6 | Txn activation | |||
| PKCβII | PCAF | Txn activation | ||||
| HMGB1 | Cytokine secretion | |||||
| PKCδ | H2B | S14 | Pro‐apoptotic | |||
| H3 | S10 | Pro‐apoptotic | ||||
| H3 | T45 | Pro‐apoptotic | ||||
| HDAC5 | S259 | ND | ||||
| p300 | S89 | Txn repression | ||||
| C/EBPα | S21 | Txn activation | ||||
| CREB | S133 (I) | Txn activation | ||||
| KLF4 | T401 | Txn repression | ||||
| p53 | S46 | Txn activation | ||||
| p65 | S536 (I) | Txn activation | ||||
| STAT1 | S727 | Txn activation | ||||
| TBLR1 | S123 | Txn activation | ||||
| DNMT1 | Proteasomal degradation | |||||
| hnRNPK | S302 | Pro‐apoptotic | ||||
| TIF1β | S473 | Txn repression | ||||
| PKCε | H1 | Anchoring protein | ||||
| H3 | S10 | Txn activation | ||||
| STAT1 | S727 | Txn activation | ||||
| Hsp90β | Txn activation | |||||
| PKCθ | MSK‐1, LSD1 (C) | Pol II, 14‐3‐3ζ (C) | Txn activation | |||
| p300 | S384 | Txn activation | ||||
| CREB | Txn activation | |||||
| Ki‐1/57 | T‐P | Subcellular localization | ||||
| PKCζ | p65 | S311 | Txn activation | |||
| CDP/Cut | Txn repression | |||||
| Ki‐1/57 | T/S‐P | Subcellular localization | ||||
| SHP‐T55 | T55 | Txn repression | ||||
| PKCζ/λ | BAF60c‐S247 | S247 | Txn activation | |||
| PKC | H2A | ND | ||||
| SATB1 | Txn activation | |||||
| HEXIM1 | S158 | PTEFb activation | ||||
| HMGB1 | DNA bending | |||||
| Ki‐1/57 | Subcellular localization | |||||
| Pol II | Pol II activation |
BAF60c, Brg1/Brm‐associated factor 60c; C, transcriptional complex; C/EBPα, CCAAT/enhancer‐binding protein α; CBP, CREB‐binding protein; CDP/Cut, Cut‐like homeodomain protein; CREB, cAMP response element‐binding protein; H1, histone H1; H2B, histone H2B; H3, histone H3; HDAC, histone deacetylase; HEXIM1, hexamethylene bisacetamide‐(HMBA)‐induced mRNA‐encoded proteins 1; HMGB1, high mobility group box 1; hnRNPK, heterogeneous nuclear ribonucleoprotein K; Hsp90β, heat‐shock protein 90 β; I, interaction; Ki‐1/57, 57‐kDa human protein antigen recognized by the CD30 antibody Ki‐1; KLF4, Krüppel‐like factor 4; LSD1, lysine‐specific demethylase 1; MSK1, mitogen and stress‐activated protein kinase 1; ND, not determined; P, phosphorylation; PCAF, p300/CBP‐associated factor; Pol II, RNA polymerase II; RORα, retinoic acid‐related orphan nuclear receptor α; S, serine; SATB1, special AT‐rich binding protein 1; SHP, small heterodimer partner; Sp1, specificity protein 1; STAT1, signal transducer and activator of transcription 1; T, threonine; TBLR1, transducin β‐like 1X‐linked receptor 1; TIFb, transcriptional intermediary factor 1β; Txn, transcriptional.
PKCδ as a histone kinase in immune‐mediated apoptosis
Protein kinase Cδ regulates the immune system through the events of mitochondrial‐dependent apoptosis by phosphorylating histones at apoptotic histone residues such as H2BS14, H3S10 and H3T45 (Fig. 3).106, 107, 108, 109 In B cells, the nuclear localization of a catalytically active PKCδ leads to phosphorylation of H2BS14 to result in apoptosis of resting B cells.107 Similarly, in Sjögren's syndrome, interaction of B cells with human salivary gland cells causes translocation and activation of PKCδ into the nucleus to phosphorylate H2BS14 leading to a B‐cell‐mediated apoptosis of epithelial cells.106 Using PKCδ small interfering RNAs in Jurkat T cells, Park et al.109 showed that phosphorylation of H3S10 by PKCδ is required for DNA damage‐induced apoptosis. In vitro studies in neutrophils showed that phosphorylation of H3T45 increases significantly in apoptotic cells and that this phosphorylation is mediated by PKCδ.108 These systems focused on the pro‐apoptotic function of PKCδ as a result of cellular stress but PKCδ can also be anti‐apoptotic depending on the cell type. Specifically, PKCδ promotes the survival of cancer cells through established anti‐apoptotic pathways including NF‐κB.110 However, whether the anti‐apoptotic activity by PKCδ is regulated through histone phosphorylation is yet to be studied.
PKCθ association with histone modifiers in T‐cell activation
In addition to phosphorylating histones, PKC can also interact with and phosphorylate histone modifiers (Table 2). Specifically in the immune system, PKCθ not only has a cytoplasmic signalling role during TCR activation, it is also present in the nucleus of T cells to regulate transcription of inducible immune genes.94, 111, 112 In stimulated human Jurkat T cells, PKCθ associates with the N‐terminal transcriptional activation domain of p300 and phosphorylates S384 on p300 leading to its transcriptional activation.111 In a study by Sutcliffe et al., 112 PKCθ does not appear to phosphorylate histones directly but instead forms a complex with RNA polymerase II, histone kinase mitogen and stress‐activated protein kinase 1, adaptor protein 14‐3‐3ζ and histone demethylase lysine‐specific demethylase 1 at the proximal promoter of inducible genes in human Jurkat T cells (Fig. 3). The assembly of this complex is dependent on NF‐κB as inhibiting NF‐κB impaired PKCθ, RNA polymerase II and lysine‐specific demethylase 1 recruitment to immune gene promoters upon TCR activation.94 This formation of an active transcriptional complex on key inducible genes can also be seen in breast cancer stem cells, where PKCθ acts as a molecular switch that regulates epithelial to mesenchymal transition.112, 113 Interestingly, inhibiting PKCθ in T cells leads to increased expression of microRNAs, small non‐coding RNAs responsible for post‐transcriptional repression of gene expression.112 This negative regulation of microRNA genes by PKCθ involves NF‐κB, both of which may be part of a repressive complex on microRNA genes to repress its transcription.94 This nuclear activity further strengthens the notion that PKCθ is a key enzyme in the regulation of T‐cell activation. So far only PKCθ is shown to associate and phosphorylate histone modifiers in the immune system. Given the key role that histone modifiers play in regulating inducible gene expression, it can be postulated that other PKC isoforms could potentially regulate histone modifiers through their kinase activity in the immune context.
Direct regulation of transcription factors by PKCs
Protein kinase C signalling in the cytoplasm activates downstream signalling cascades leading to transcription factor activation, and hence gene transcription (Fig. 2). In the nucleus, PKC plays a more direct role in regulating transcription factors through phosphorylating the factors directly or regulating interactions of transcriptional regulatory factors to lead to transcriptional activation of genes (Table 2). For example, monocyte induction by macrophage colony‐stimulating factor leads to phosphorylation of NF‐κB p65 at S276 by PKCα, which is important for transcriptional activation of NF‐κB.114 In dendritic cells, PKCζ phosphorylation of p65 at S311 prevents binding of histone methyltransferase glucagon‐like peptide to p65 at monomethylated K310, allowing expression of p65 target genes.115 Similarly, IL‐32β mediates PKCδ phosphorylation of CCAAT/enhancer‐binding protein α at S21 to prevent binding of the CCAAT/enhancer‐binding protein α to IL‐10 promoter, hence activating the gene in human myeloid cells.116
Upon heat‐shock induction, PKCε phosphorylates STAT1 at S727 in Jurkat T cells to activate the heat‐shock protein 90β (hsp90β) gene.117 While in macrophages, induction by IFN‐γ leads to STAT1 phosphorylation by PKCδ at S727 to promote class II transactivator (CIITA) gene expression.118 A similar study shows that CIITA gene expression in B cells is controlled by CREB phosphorylation by PKCδ.119 The phosphorylation of CREB by PKCδ occurs at S133 upon B‐cell activation.120 Whereas in T cells, PKCθ is involved in the phosphorylation and binding of CREB to the IL‐2 promoter.121 In human Jurkat T cells, phosphorylation by PKC can regulate the interaction of special AT‐rich binding protein 1 (SATB1) with either histone deacetylase 1 or p300/CREB‐binding protein‐associated factor (PCAF).122 In the basal state, PKC‐phosphorylated SATB1 at S185 associates with histone deacetylase 1, so acting as a repressor, but upon activation, dephosphorylated SATB1 associates with PCAF, leading to activation of the IL‐2 gene. These studies show that nuclear PKCs directly regulate transcription factors through phosphorylation for transcriptional activation in the immune system. However, phosphorylation of transcription factors by PKCs can also lead to gene repression123, 124 but this has not been shown in the immune context.
Other PKC isoforms as chromatin regulators in non‐immune systems
At present, there are no reported epigenetic activities by PKCγ and PKCη. Although not published to have epigenetic roles in immune cells, PKCα and PKCβ have been shown to act as histone kinases in other studies (Table 2).125, 126 A screen with 97 kinases using HeLa cells showed that PKCα, PKCβI and PKCβII can phosphorylate H3T6 but not H3T3, H3S10 or H3T11.126 This is in line with a study using high‐resolution nuclear magnetic resonance spectroscopy, where PKCα and both PKCβ isoforms could phosphorylate H3T6 and H3S10 but in a mutually exclusive manner (Fig. 3).125 Using prostate cancer cells, Metzger et al.126 showed that PKCβI phosphorylates H3T6, which blocks H3K4 demethylation by lysine‐specific demethylase 1 during androgen receptor‐dependent gene activation. In breast cancer cells, PKCβI is recruited to the oestrogen receptor α (ERα) promoter B as part of a regulatory complex with RanBP‐type and C3HC4‐type zinc finger containing 1 and ERα to regulate ERα gene expression (Fig. 3).127 This recruitment of PKCβI is associated with the presence of PKCβI‐dependent H3K4me2 and H3T6 phosphorylation though a direct causal effect has not been demonstrated in this model.127 Given that PKCβ is specifically expressed in B cells, it can be postulated that PKCβ could potentially regulate immune gene expression as an epigenetic enzyme.
We have discussed examples of PKC isoforms functioning as epigenetic enzymes in neutrophils (PKCδ), macrophages (PKCα, PKCδ), myeloid cells (PKCδ), dendritic cells (PKCζ), B cells (PKCδ) and T cells (PKCδ, PKCε, PKCθ). Given the defects seen in immune cell homeostasis, activation and function in various PKC knockout studies (Table 1), it is highly possible that PKC isoforms that are not known to have an epigenetic function could potentially regulate the expression of key immune genes as epigenetic enzymes.
Therapeutic potential of PKCs in disease
As PKCs play a ubiquitous role in cell signalling, it is not surprising that dysregulation of PKC leads to disease such as diabetes, cancer, cardiovascular disease, dermatological disease, psychiatric diseases, neurological conditions and immune‐mediated diseases.128 The two main PKC isoforms implicated in human autoimmunity and inflammation diseases are PKCδ and PKCθ.128 PKCδ deficiency is linked with autoimmune diseases involving B‐cell development immunodeficiency.129, 130 While there are currently no drugs in development against PKCδ in autoimmune diseases, a peptide inhibitor of PKCδ, delcasertib (δV1‐1), shows promising results in myocardial infarction with reduction of infarct size when administered in patients.131 Furthermore, two published patents reported the potential use of antisense oligonucleotides against PKCδ to regulate its expression in infectious and autoimmune disease.132 Hence, there are tools currently available that can be applied for therapeutic studies of PKCδ in the immune context.
While there are a number of PKC regulators in clinical trials for various diseases, the main research of autoimmune PKC activity and drug targeting is in T cells due to the pivotal role that T cells play in the immune response.128 There is a growing body of evidence that PKCθ is indispensable for critical T‐cell functions such as protective responses to pathogens. Hence PKCθ is less likely to result in off‐target immunosuppression.133 As a result, PKCθ, is highly studied as a potential therapeutic target.9 The most advanced PKCθ inhibitor to be developed is sotrastaurin (AEB071), which is currently undergoing clinical trials for the treatment of psoriasis and organ transplantation.134, 135, 136, 137, 138 Another promising pharmaceutical PKCθ inhibitor is compound 27, for which oral administration in mice led to a decreased IL‐2 response.139 Furthermore, similar compounds of PKCθ inhibitors developed by the same group were developed and tested in in vivo mouse models of colitis and multiple sclerosis and the mice treated with the PKCθ inhibitors showed improved general well‐being and reduced disease severity.140 Undoubtedly, these findings highlight the importance and great potential for development of small molecule inhibitors of PKCs in treating immune‐mediated diseases.
Conclusion
The review highlights the contrasting roles of PKC in the cytoplasm compared with the nucleus. In the cytoplasm, active PKC bind membrane components to act as signalling molecules and phosphorylate different substrates to mediate activation of transcription factors required for immune gene expression via the signalling cascade. The phosphorylated substrates that may be anchored to membrane fractions by adaptor proteins and/or directly bind active PKC once phosphorylated, may be released to the cytoplasm and act in the cytoplasm or they may translocate to the nucleus upon phosphorylation. In the nucleus, PKCs directly phosphorylate histones/transcriptional regulatory factors or form complexes that associate with chromatin. Whereas the mechanism of PKC as a cytoplasmic signalling transducer is extensively studied, its role in the nucleus either as a signal transducer or chromatin regulator is not as well‐studied.
Different activation mechanisms and subcellular localization of PKC can determine different functions but what remains to be determined is how translocation to subcellular locations is determined upon different stimuli.71, 75 Although local increases of DAG and calcium are shown to affect PKC translocation, it is unclear if this translocation is a diffusion mediated process due to local increases of specific lipids or calcium.79, 80, 81 Alternatively, is translocation mediated by active transport aided by anchoring proteins such as receptors for activated C‐kinases? The receptors for activated C‐kinases anchor proteins bind directly with the C2 domain of activated PKC but this has only been shown for PKCβII and PKCε.141, 142 There are other proteins described as forming protein–protein interactions with PKC be it adapter proteins, anchor proteins or PKC substrates and these have great potential for therapeutic target development (reviewed in ref. 128).
It will also be interesting to uncover how the nuclear PKC and cytoplasmic PKC interpret external stimuli. Is there a different mechanism of activation that relays the message allowing them to perform their cell‐specific function depending on their location within the cell? Also, current examples of PKC as epigenetic enzymes are found only in the adaptive immune system. There is no evidence as yet of PKC as an epigenetic enzyme in innate immune response but this is highly probable as there is evidence of PKC translocating to the nucleus in human monocytes.143 Following from there, are the PKCs in the nucleus the same as the one in cytoplasm; i.e. do they perform the same function and are recycled or are there specific nuclear PKCs and cytoplasmic PKCs? Examples from another protein kinase family, mitogen‐activated protein kinases, show that cytosolic and nuclear mitogen‐activated protein kinases are regulated differently, and so a potentially similar mechanism could be present for PKCs.144 Cleaved forms of PKC may be a further avenue of nuclear and cytoplasmic function differentiation as catalytic fragments of cleaved PKCδ and PKCβII isoforms have been suggested to accumulate in the nucleus.145, 146 Hence, nuclear translocation or retention could be mediated through cleavage of PKC isoforms, in which the catalytic domain could be retained for further phosphorylation or interaction with other proteins. In addition, phosphorylation of PKC is a key mechanism regulating PKC function. It remains to be determined which substrate for phosphorylation is critical for different outcomes depending on the context the isoform is in.
It would be essential to uncover the difference between nuclear and cytoplasmic PKCs as these differences could be used to design specific drug targets. Most existing PKC kinase inhibitors target the common catalytic ATP pocket and hence are not isozyme‐selective. There are also isozyme‐specific inhibitors and these tend to target PKC binding to receptors for activated C‐kinases, but not all PKCs have been shown to interact with receptors for activated C‐kinases and the key interaction sites are not known.128 Hence, it is important to understand how and when PKCs translocate to the nucleus to enable the design of specific modulators of protein–protein interactions. This was shown in a proof of concept study where a peptide was developed that specifically inhibited the nuclear translocation of extracellular signal‐regulated kinase 1/2 kinase to combat its function to induce cell proliferation in melanoma cells.147 Structural studies of the nuclear PKC domains and the contribution of the nuclear import receptors will enhance our understanding of the mode of action of these kinases. This provides a strong basis for the design of high‐affinity inhibitors capable of specifically blocking the nuclear translocation and function of PKCs in an isoform‐specific manner. Specific inhibition of substrate interactions with specific PKC isoforms could pave the way for more targeted therapeutics. As discussed, dysregulation of PKC leads to the development of disease and particularly with PKC being involved in multiple signalling pathways, it is even more critical to uncover how PKC functions in its different roles to build on the knowledge and understanding for future therapeutic target development.
Disclosure
The authors declare no conflict of interest.
Acknowledgements
This work was supported by NHMRC Project Grant (APP1025718) awarded to SR and also by a UC PDF Fellowship awarded to PSL in SR's laboratory.
References
- 1. Keenan C, Long A, Kelleher D. Protein kinase C and T cell function. Biochim Biophys Acta 1997; 1358:113–26. [DOI] [PubMed] [Google Scholar]
- 2. Newton AC. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev 2001; 101:2353–64. [DOI] [PubMed] [Google Scholar]
- 3. Nalefski EA, Falke JJ. The C2 domain calcium‐binding motif: structural and functional diversity. Protein Sci 1996; 5:2375–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giorgione JR, Lin JH, McCammon JA, Newton AC. Increased membrane affinity of the C1 domain of protein kinase Cδ compensates for the lack of involvement of its C2 domain in membrane recruitment. J Biol Chem 2006; 281:1660–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Puls A, Schmidt S, Grawe F, Stabel S. Interaction of protein kinase C zeta with ZIP, a novel protein kinase C‐binding protein. Proc Natl Acad Sci U S A 1997; 94:6191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Newton AC. Protein kinase C: structure, function, and regulation. J Biol Chem 1995; 270:28495–8. [DOI] [PubMed] [Google Scholar]
- 7. Loegering DJ, Lennartz MR. Protein kinase C and toll‐like receptor signaling. Enzyme Res 2011; 2011:537821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Isakov N, Altman A. Protein kinase Cθ in T cell activation. Annu Rev Immunol 2002; 20:761–94. [DOI] [PubMed] [Google Scholar]
- 9. Altman A, Kong KF. Protein kinase C inhibitors for immune disorders. Drug Discov Today 2014; 19:1217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee MS, Kim YJ. Signaling pathways downstream of pattern‐recognition receptors and their cross talk. Annu Rev Biochem 2007; 76:447–80. [DOI] [PubMed] [Google Scholar]
- 11. West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll‐like receptors. Annu Rev Cell Dev Biol 2006; 22:409–37. [DOI] [PubMed] [Google Scholar]
- 12. Miyake K. Innate recognition of lipopolysaccharide by Toll‐like receptor 4‐MD‐2. Trends Microbiol 2004; 12:186–92. [DOI] [PubMed] [Google Scholar]
- 13. Yamamoto M, Sato S, Hemmi H et al TRAM is specifically involved in the Toll‐like receptor 4‐mediated MyD88‐independent signaling pathway. Nat Immunol 2003; 4:1144–50. [DOI] [PubMed] [Google Scholar]
- 14. Gay NJ, Gangloff M. Structure and function of Toll receptors and their ligands. Annu Rev Biochem 2007; 76:141–65. [DOI] [PubMed] [Google Scholar]
- 15. Kang JY, Lee JO. Structural biology of the Toll‐like receptor family. Annu Rev Biochem 2011; 80:917–41. [DOI] [PubMed] [Google Scholar]
- 16. Castrillo A, Pennington DJ, Otto F, Parker PJ, Owen MJ, Boscá L. Protein kinase Cε is required for macrophage activation and defense against bacterial infection. J Exp Med 2001; 194:1231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Faisal A, Saurin A, Gregory B, Foxwell B, Parker PJ. The scaffold MyD88 acts to couple protein kinase Cε to Toll‐like receptors. J Biol Chem 2008; 283:18591–600. [DOI] [PubMed] [Google Scholar]
- 18. McGettrick AF, Brint EK, Palsson‐McDermott EM, Rowe DC, Golenbock DT, Gay NJ, Fitzgerald KA, O'Neill LA. Trif‐related adapter molecule is phosphorylated by PKCε during Toll‐like receptor 4 signaling. Proc Natl Acad Sci U S A 2006; 103:9196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fitzgerald KA, Rowe DC, Barnes BJ et al LPS‐TLR4 signaling to IRF‐3/7 and NF‐κB involves the toll adapters TRAM and TRIF. J Exp Med 2003; 198:1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Medzhitov R, Preston‐Hurlburt P, Janeway CA Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 1997; 388:394–7. [DOI] [PubMed] [Google Scholar]
- 21. Kawai T, Akira S. Toll‐like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011; 34:637–50. [DOI] [PubMed] [Google Scholar]
- 22. Schnare M, Barton GM, Holt AC, Takeda K, Akira S, Medzhitov R. Toll‐like receptors control activation of adaptive immune responses. Nat Immunol 2001; 2:947–50. [DOI] [PubMed] [Google Scholar]
- 23. Acuto O, Cantrell D. T cell activation and the cytoskeleton. Annu Rev Immunol 2000; 18:165–84. [DOI] [PubMed] [Google Scholar]
- 24. Diehn M, Alizadeh AA, Rando OJ, Liu CL, Stankunas K, Botstein D, Crabtree GR, Brown PO. Genomic expression programs and the integration of the CD28 costimulatory signal in T cell activation. Proc Natl Acad Sci U S A 2002; 99:11796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Favero J, Lafont V. Effector pathways regulating T cell activation. Biochem Pharmacol 1998; 56:1539–47. [DOI] [PubMed] [Google Scholar]
- 26. Smith‐Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol 2009; 27:591–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fooksman DR, Vardhana S, Vasiliver‐Shamis G et al Functional anatomy of T cell activation and synapse formation. Annu Rev Immunol 2010; 28:79–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yokosuka T, Saito T. The immunological synapse, TCR microclusters, and T cell activation. Curr Top Microbiol Immunol 2010; 340:81–107. [DOI] [PubMed] [Google Scholar]
- 29. Kong K‐F, Altman A. In and out of the bull's eye: protein kinase Cs in the immunological synapse. Trends Immunol 2013; 34:234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Monks CR, Kupfer H, Tamir I, Barlow A, Kupfer A. Selective modulation of protein kinase C‐theta during T‐cell activation. Nature 1997; 385:83–6. [DOI] [PubMed] [Google Scholar]
- 31. Baier G, Telford D, Giampa L, Coggeshall KM, Baier‐Bitterlich G, Isakov N, Altman A. Molecular cloning and characterization of PKC θ, a novel member of the protein kinase C (PKC) gene family expressed predominantly in hematopoietic cells. J Biol Chem 1993; 268:4997–5004. [PubMed] [Google Scholar]
- 32. Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three‐dimensional segregation of supramolecular activation clusters in T cells. Nature 1998; 395:82–6. [DOI] [PubMed] [Google Scholar]
- 33. Huang J, Lo PF, Zal T, Gascoigne NR, Smith BA, Levin SD, Grey HM. CD28 plays a critical role in the segregation of PKC θ within the immunologic synapse. Proc Natl Acad Sci U S A 2002; 99:9369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kong KF, Yokosuka T, Canonigo‐Balancio AJ, Isakov N, Saito T, Altman A. A motif in the V3 domain of the kinase PKC‐θ determines its localization in the immunological synapse and functions in T cells via association with CD28. Nat Immunol 2011; 12:1105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yokosuka T, Kobayashi W, Sakata‐Sogawa K, Takamatsu M, Hashimoto‐Tane A, Dustin ML, Tokunaga M, Saito T. Spatiotemporal regulation of T cell costimulation by TCR‐CD28 microclusters and protein kinase C θ translocation. Immunity 2008; 29:589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. von Essen MR, Kongsbak M, Levring TB et al PKC‐θ exists in an oxidized inactive form in naive human T cells. Eur J Immunol 2013; 43:1659–66. [DOI] [PubMed] [Google Scholar]
- 37. Sun Z, Arendt CW, Ellmeier W et al PKC‐θ is required for TCR‐induced NF‐κB activation in mature but not immature T lymphocytes. Nature 2000; 404:402–7. [DOI] [PubMed] [Google Scholar]
- 38. Anderson K, Fitzgerald M, Dupont M et al Mice deficient in PKC θ demonstrate impaired in vivo T cell activation and protection from T cell‐mediated inflammatory diseases. Autoimmunity 2006; 39:469–78. [DOI] [PubMed] [Google Scholar]
- 39. Pfeifhofer C, Kofler K, Gruber T, Tabrizi NG, Lutz C, Maly K, Leitges M, Baier G. Protein kinase C θ affects Ca2+ mobilization and NFAT cell activation in primary mouse T cells. J Exp Med 2003; 197:1525–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marsland BJ, Soos TJ, Spath G, Littman DR, Kopf M. Protein kinase C θ is critical for the development of in vivo T helper (Th)2 cell but not Th1 cell responses. J Exp Med 2004; 200:181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salek‐Ardakani S, So T, Halteman BS, Altman A, Croft M. Protein kinase Cθ controls Th1 cells in experimental autoimmune encephalomyelitis. J Immunol 2005; 175:7635–41. [DOI] [PubMed] [Google Scholar]
- 42. Tan SL, Zhao J, Bi C et al Resistance to experimental autoimmune encephalomyelitis and impaired IL‐17 production in protein kinase C θ‐deficient mice. J Immunol 2006; 176:2872–9. [DOI] [PubMed] [Google Scholar]
- 43. Kwon MJ, Ma J, Ding Y, Wang R, Sun Z. Protein kinase C‐θ promotes Th17 differentiation via upregulation of Stat3. J Immunol 2012; 188:5887–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wachowicz K, Hermann‐Kleiter N, Meisel M, Siegmund K, Thuille N, Baier G. Protein kinase C θ regulates the phenotype of murine CD4+ Th17 cells. PLoS ONE 2014; 9:e96401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marsland BJ, Nembrini C, Schmitz N, Abel B, Krautwald S, Bachmann MF, Kopf M. Innate signals compensate for the absence of PKC‐θ during in vivo CD8+ T cell effector and memory responses. Proc Natl Acad Sci U S A 2005; 102:14374–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marsland BJ, Kopf M. T‐cell fate and function: PKC‐θ and beyond. Trends Immunol 2008; 29:179–85. [DOI] [PubMed] [Google Scholar]
- 47. Teixeiro E, Daniels MA, Hamilton SE, Schrum AG, Bragado R, Jameson SC, Palmer E. Different T cell receptor signals determine CD8+ memory versus effector development. Science 2009; 323:502–5. [DOI] [PubMed] [Google Scholar]
- 48. Gupta S, Manicassamy S, Vasu C, Kumar A, Shang W, Sun Z. Differential requirement of PKC‐θ in the development and function of natural regulatory T cells. Mol Immunol 2008; 46:213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zanin‐Zhorov A, Ding Y, Kumari S et al Protein kinase C‐θ mediates negative feedback on regulatory T cell function. Science 2010; 328:372–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ma J, Ding Y, Fang X, Wang R, Sun Z. Protein kinase C‐θ inhibits inducible regulatory T cell differentiation via an AKT‐Foxo1/3a‐dependent pathway. J Immunol 2012; 188:5337–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harwood NE, Batista FD. New insights into the early molecular events underlying B cell activation. Immunity 2008; 28:609–19. [DOI] [PubMed] [Google Scholar]
- 52. Harwood NE, Batista FD. Early events in B cell activation. Annu Rev Immunol 2010; 28:185–210. [DOI] [PubMed] [Google Scholar]
- 53. Packard TA, Cambier JC. B lymphocyte antigen receptor signaling: initiation, amplification, and regulation. F1000Prime Rep 2013; 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mischak H, Kolch W, Goodnight J, Davidson WF, Rapp U, Rose‐John S, Mushinski JF. Expression of protein kinase C genes in hemopoietic cells is cell‐type‐ and B cell‐differentiation stage specific. J Immunol 1991; 147:3981–7. [PubMed] [Google Scholar]
- 55. Brick‐Ghannam C, Ericson ML, Schelle I, Charron D. Differential regulation of mRNAs encoding protein kinase C isoenzymes in activated human B cells. Hum Immunol 1994; 41:216–24. [DOI] [PubMed] [Google Scholar]
- 56. Su TT, Guo B, Kawakami Y et al PKC‐β controls I κB kinase lipid raft recruitment and activation in response to BCR signaling. Nat Immunol 2002; 3:780–6. [DOI] [PubMed] [Google Scholar]
- 57. Saijo K. Protein kinase C β controls nuclear factor κB activation in B cells through selective regulation of the IκB kinase α . J Exp Med 2002; 195:1647–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Leitges M, Schmedt C, Guinamard R, Davoust J, Schaal S, Stabel S, Tarakhovsky A. Immunodeficiency in protein kinase cβ‐deficient mice. Science 1996; 273:788–91. [DOI] [PubMed] [Google Scholar]
- 59. Fruman DA, Satterthwaite AB, Witte ON. Xid‐like phenotypes: a B cell signalosome takes shape. Immunity 2000; 13:1–3. [DOI] [PubMed] [Google Scholar]
- 60. Petro JB, Rahman SM, Ballard DW, Khan WN. Bruton's tyrosine kinase is required for activation of IκB kinase and nuclear factor κB in response to B cell receptor engagement. J Exp Med 2000; 191:1745–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bajpai UD, Zhang K, Teutsch M, Sen R, Wortis HH. Bruton's tyrosine kinase links the B cell receptor to nuclear factor κB activation. J Exp Med 2000; 191:1735–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kang SW, Wahl MI, Chu J et al PKCβ modulates antigen receptor signaling via regulation of Btk membrane localization. EMBO J 2001; 20:5692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Venkataraman C, Chen XC, Na S, Lee L, Neote K, Tan SL. Selective role of PKCβ enzymatic function in regulating cell survival mediated by B cell antigen receptor cross‐linking. Immunol Lett 2006; 105:83–9. [DOI] [PubMed] [Google Scholar]
- 64. Teh CE, Horikawa K, Arnold CN et al Heterozygous mis‐sense mutations in Prkcb as a critical determinant of anti‐polysaccharide antibody formation. Genes Immun 2013; 14:223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shinohara H, Maeda S, Watarai H, Kurosaki T. IκB kinase β‐induced phosphorylation of CARMA1 contributes to CARMA1 Bcl10 MALT1 complex formation in B cells. J Exp Med 2007; 204:3285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shinohara H, Yasuda T, Aiba Y, Sanjo H, Hamadate M, Watarai H, Sakurai H, Kurosaki T. PKC β regulates BCR‐mediated IKK activation by facilitating the interaction between TAK1 and CARMA1. J Exp Med 2005; 202:1423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sommer K, Guo B, Pomerantz JL, Bandaranayake AD, Moreno‐Garcia ME, Ovechkina YL, Rawlings DJ. Phosphorylation of the CARMA1 linker controls NF‐κB activation. Immunity 2005; 23:561–74. [DOI] [PubMed] [Google Scholar]
- 68. Capitani S, Girard PR, Mazzei GJ, Kuo JF, Berezney R, Manzoli FA. Immunochemical characterization of protein kinase C in rat liver nuclei and subnuclear fractions. Biochem Biophys Res Commun 1987; 142:367–75. [DOI] [PubMed] [Google Scholar]
- 69. Beckmann R, Lindschau C, Haller H, Hucho F, Buchner K. Differential nuclear localization of protein kinase C isoforms in neuroblastoma × glioma hybrid cells. Eur J Biochem 1994; 222:335–43. [DOI] [PubMed] [Google Scholar]
- 70. Disatnik MH, Buraggi G, Mochly‐Rosen D. Localization of protein kinase C isozymes in cardiac myocytes. Exp Cell Res 1994; 210:287–97. [DOI] [PubMed] [Google Scholar]
- 71. Martelli AM, Sang N, Borgatti P, Capitani S, Neri LM. Multiple biological responses activated by nuclear protein kinase C. J Cell Biochem 1999; 74:499–521. [PubMed] [Google Scholar]
- 72. Raben DM, Jarpe MB, Leach KL. Nuclear lipid metabolism in NEST: Nuclear Envelope Signal Transduction. J Membr Biol 1994; 142:1–7. [DOI] [PubMed] [Google Scholar]
- 73. Whetton AD, Heyworth CM, Nicholls SE, Evans CA, Lord JM, Dexter TM, Owen‐Lynch PJ. Cytokine‐mediated protein kinase C activation is a signal for lineage determination in bipotential granulocyte macrophage colony‐forming cells. J Cell Biol 1994; 125:651–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mallia CM, Aguirre V, McGary E, Tang Y, Scandurro AB, Liu C, Noguchi CT, Beckman BS. Protein kinase cα is an effector of hexamethylene bisacetamide‐induced differentiation of Friend erythroleukemia cells. Exp Cell Res 1999; 246:348–54. [DOI] [PubMed] [Google Scholar]
- 75. Buchner K. The role of protein kinase C in the regulation of cell growth and in signalling to the cell nucleus. J Cancer Res Clin Oncol 2000; 126:1–11. [DOI] [PubMed] [Google Scholar]
- 76. Zauli G, Visani G, Bassini A et al Nuclear translocation of protein kinase C‐α and ‐ζ isoforms in HL‐60 cells induced to differentiate along the granulocytic lineage by all‐trans retinoic acid. Br J Haematol 1996; 93:542–50. [DOI] [PubMed] [Google Scholar]
- 77. Bertolaso L, Gibellini D, Secchiero P et al Accumulation of catalytically active PKC‐ζ into the nucleus of HL‐60 cell line plays a key role in the induction of granulocytic differentiation mediated by all‐trans retinoic acid. Br J Haematol 1998; 100:541–9. [DOI] [PubMed] [Google Scholar]
- 78. Owen PJ, Johnson GD, Lord JM. Protein kinase C‐δ associates with vimentin intermediate filaments in differentiated HL60 cells. Exp Cell Res 1996; 225:366–73. [DOI] [PubMed] [Google Scholar]
- 79. Echevarria W, Leite MF, Guerra MT, Zipfel WR, Nathanson MH. Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat Cell Biol 2003; 5:440–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Irvine RF. Nuclear lipid signaling. Sci STKE 2002; 2002:re13. [DOI] [PubMed] [Google Scholar]
- 81. Neri LM, Bortul R, Borgatti P, Tabellini G, Baldini G, Capitani S, Martelli AM. Proliferating or differentiating stimuli act on different lipid‐dependent signaling pathways in nuclei of human leukemia cells. Mol Biol Cell 2002; 13:947–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rosenberger U, Lehmann I, Weise C, Franke P, Hucho F, Buchner K. Identification of PSF as a protein kinase Cα‐binding protein in the cell nucleus. J Cell Biochem 2002; 86:394–402. [DOI] [PubMed] [Google Scholar]
- 83. Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev 2001; 65:570–94, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. McLane LM, Corbett AH. Nuclear localization signals and human disease. IUBMB Life 2009; 61:697–706. [DOI] [PubMed] [Google Scholar]
- 85. Malviya AN, Block C. A bipartite nuclear targeting motif in protein kinase C? Trends Biochem Sci 1992; 17:176. [DOI] [PubMed] [Google Scholar]
- 86. DeVries TA, Neville MC, Reyland ME. Nuclear import of PKCδ is required for apoptosis: identification of a novel nuclear import sequence. EMBO J 2002; 21:6050–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Adwan TS, Ohm AM, Jones DNM, Humphries MJ, Reyland ME. Regulated binding of importin‐α to protein kinase Cδ in response to apoptotic signals facilitates nuclear import. J Biol Chem 2011; 286:35716–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Perander M, Bjørkøy G, Johansen T. Nuclear import and export signals enable rapid nucleocytoplasmic shuttling of the atypical protein kinase C λ . J Biol Chem 2001; 276:13015–24. [DOI] [PubMed] [Google Scholar]
- 89. White WO, Seibenhener ML, Wooten MW. Phosphorylation of tyrosine 256 facilitates nuclear import of atypical protein kinase C. J Cell Biochem 2002; 85:42–53. [PubMed] [Google Scholar]
- 90. Schmalz D, Kalkbrenner F, Hucho F, Buchner K. Transport of protein kinase C α into the nucleus requires intact cytoskeleton while the transport of a protein containing a canonical nuclear localization signal does not. J Cell Sci 1996; 109(Pt 9):2401–6. [DOI] [PubMed] [Google Scholar]
- 91. Schmalz D, Hucho F, Buchner K. Nuclear import of protein kinase C occurs by a mechanism distinct from the mechanism used by proteins with a classical nuclear localization signal. J Cell Sci 1998; 111(Pt 13):1823–30. [DOI] [PubMed] [Google Scholar]
- 92. Seidl S, Braun UB, Leitges M. Functional comparison of protein domains within aPKCs involved in nucleocytoplasmic shuttling. Biol Open 2012; 1:436–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chuderland D, Konson A, Seger R. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol Cell 2008; 31:850–61. [DOI] [PubMed] [Google Scholar]
- 94. Sutcliffe EL, Li J, Zafar A et al Chromatinized protein kinase C‐θ: can it escape the clutches of NF‐κB? Front Immunol 2012; 3:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wagner S, Harteneck C, Hucho F, Buchner K. Analysis of the subcellular distribution of protein kinase Cα using PKC‐GFP fusion proteins. Exp Cell Res 2000; 258:204–14. [DOI] [PubMed] [Google Scholar]
- 96. Eldar H, Livneh E. Phosphorylation of p90 and p52 in response to phorbol‐esters in Swiss/3T3 cells overexpressing protein kinase C‐α . Mol Biol Cell 1992; 3:1049–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Buchner K. Protein kinase C in the transduction of signals toward and within the cell nucleus. Eur J Biochem 1995; 228:211–21. [PubMed] [Google Scholar]
- 98. Martelli AM, Evangelisti C, Nyakern M, Manzoli FA. Nuclear protein kinase C. Biochim Biophys Acta 2006; 1761:542–51. [DOI] [PubMed] [Google Scholar]
- 99. Kornberg RD, Lorch Y. Twenty‐five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999; 98:285–94. [DOI] [PubMed] [Google Scholar]
- 100. Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature 2000; 408:877–81. [DOI] [PubMed] [Google Scholar]
- 101. Lim PS, Li J, Holloway AF, Rao S. Epigenetic regulation of inducible gene expression in the immune system. Immunology 2013; 139:285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lim PS, Shannon MF, Hardy K. Epigenetic control of inducible gene expression in the immune system. Epigenomics 2010; 2:775–95. [DOI] [PubMed] [Google Scholar]
- 103. Lee TI, Young RA. Transcription of eukaryotic protein‐coding genes. Annu Rev Genet 2000; 34:77–137. [DOI] [PubMed] [Google Scholar]
- 104. Alepuz PM, Jovanovic A, Reiser V, Ammerer G. Stress‐induced map kinase Hog1 is part of transcription activation complexes. Mol Cell 2001; 7:767–77. [DOI] [PubMed] [Google Scholar]
- 105. Baek SH. When signaling kinases meet histones and histone modifiers in the nucleus. Mol Cell 2011; 42:274–84. [DOI] [PubMed] [Google Scholar]
- 106. Varin MM, Guerrier T, Devauchelle‐Pensec V, Jamin C, Youinou P, Pers JO. In Sjogren's syndrome, B lymphocytes induce epithelial cells of salivary glands into apoptosis through protein kinase C δ activation. Autoimmun Rev 2012; 11:252–8. [DOI] [PubMed] [Google Scholar]
- 107. Mecklenbrauker I, Kalled SL, Leitges M, Mackay F, Tarakhovsky A. Regulation of B‐cell survival by BAFF‐dependent PKCδ‐mediated nuclear signalling. Nature 2004; 431:456–61. [DOI] [PubMed] [Google Scholar]
- 108. Hurd PJ, Bannister AJ, Halls K et al Phosphorylation of histone H3 Thr‐45 is linked to apoptosis. J Biol Chem 2009; 284:16575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Park CH, Kim KT. Apoptotic phosphorylation of histone H3 on Ser‐10 by protein kinase Cδ . PLoS ONE 2012; 7:e44307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Basu A, Pal D. Two faces of protein kinase Cδ: the contrasting roles of PKCδ in cell survival and cell death. ScientificWorldJournal 2010; 10:2272–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Granja AG, Perkins ND, Revilla Y. A238L inhibits NF‐ATc2, NF‐κB, and c‐Jun activation through a novel mechanism involving protein kinase C‐θ‐mediated up‐regulation of the amino‐terminal transactivation domain of p300. J Immunol 2008; 180:2429–42. [DOI] [PubMed] [Google Scholar]
- 112. Sutcliffe EL, Bunting KL, He YQ et al Chromatin‐associated protein kinase C‐θ regulates an inducible gene expression program and microRNAs in human T lymphocytes. Mol Cell 2011; 41:704–19. [DOI] [PubMed] [Google Scholar]
- 113. Zafar A, Wu F, Hardy K et al Chromatinized protein kinase C‐θ directly regulates inducible genes in epithelial to mesenchymal transition and breast cancer stem cells. Mol Cell Biol 2014; 34:2961–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wang Y, Mo X, Piper MG, Wang H, Parinandi NL, Guttridge D, Marsh CB. M‐CSF induces monocyte survival by activating NF‐κB p65 phosphorylation at Ser276 via protein kinase C. PLoS ONE 2011; 6:e28081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Levy D, Kuo AJ, Chang Y et al Lysine methylation of the NF‐κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF‐κB signaling. Nat Immunol 2011; 12:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kang JW, Park YS, Kim MS et al Interleukin (IL)‐32β‐mediated CCAAT/enhancer‐binding protein α (C/EBPα) phosphorylation by protein kinase Cδ (PKCδ) abrogates the inhibitory effect of C/EBPα on IL‐10 production. J Biol Chem 2013; 288:23650–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Cheng MB, Zhang Y, Zhong X et al Stat1 mediates an auto‐regulation of hsp90β gene in heat shock response. Cell Signal 2010; 22:1206–13. [DOI] [PubMed] [Google Scholar]
- 118. Kwon MJ, Yao Y, Walter MJ, Holtzman MJ, Chang CH. Role of PKCδ in IFN‐γ‐inducible CIITA gene expression. Mol Immunol 2007; 44:2841–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kwon MJ, Soh JW, Chang CH. Protein kinase C δ is essential to maintain CIITA gene expression in B cells. J Immunol 2006; 177:950–6. [DOI] [PubMed] [Google Scholar]
- 120. Blois JT, Mataraza JM, Mecklenbrauker I, Tarakhovsky A, Chiles TC. B cell receptor‐induced cAMP‐response element‐binding protein activation in B lymphocytes requires novel protein kinase Cδ . J Biol Chem 2004; 279:30123–32. [DOI] [PubMed] [Google Scholar]
- 121. Solomou EE, Juang YT, Tsokos GC. Protein kinase C‐θ participates in the activation of cyclic AMP‐responsive element‐binding protein and its subsequent binding to the ‐180 site of the IL‐2 promoter in normal human T lymphocytes. J Immunol 2001; 166:5665–74. [DOI] [PubMed] [Google Scholar]
- 122. Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo . Mol Cell 2006; 22:231–43. [DOI] [PubMed] [Google Scholar]
- 123. Lee JM, Kim IS, Kim H et al RORα attenuates Wnt/β‐catenin signaling by PKCα‐dependent phosphorylation in colon cancer. Mol Cell 2010; 37:183–95. [DOI] [PubMed] [Google Scholar]
- 124. Zhang XH, Zheng B, Gu C, Fu JR, Wen JK. TGF‐β1 downregulates AT1 receptor expression via PKC‐δ‐mediated Sp1 dissociation from KLF4 and Smad‐mediated PPAR‐iγ association with KLF4. Arterioscler Thromb Vasc Biol 2012; 32:1015–23. [DOI] [PubMed] [Google Scholar]
- 125. Liokatis S, Stutzer A, Elsasser SJ et al Phosphorylation of histone H3 Ser10 establishes a hierarchy for subsequent intramolecular modification events. Nat Struct Mol Biol 2012; 19:819–23. [DOI] [PubMed] [Google Scholar]
- 126. Metzger E, Imhof A, Patel D et al Phosphorylation of histone H3T6 by PKCβI controls demethylation at histone H3K4. Nature 2010; 464:792–6. [DOI] [PubMed] [Google Scholar]
- 127. Gustafsson Sheppard N, Heldring N, Dahlman‐Wright K. Estrogen receptor‐α, RBCK1, and protein kinase C β1 cooperate to regulate estrogen receptor‐α gene expression. J Mol Endocrinol 2012; 49:277–87. [DOI] [PubMed] [Google Scholar]
- 128. Mochly‐Rosen D, Das K, Grimes KV. Protein kinase C, an elusive therapeutic target? Nat Rev Drug Discov 2012; 11:937–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Salzer E, Santos‐Valente E, Klaver S et al B‐cell deficiency and severe autoimmunity caused by deficiency of protein kinase C δ . Blood 2013; 121:3112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kuehn HS, Niemela JE, Rangel‐Santos A et al Loss‐of‐function of the protein kinase C δ (PKCδ) causes a B‐cell lymphoproliferative syndrome in humans. Blood 2013; 121:3117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Chen L, Hahn H, Wu G et al Opposing cardioprotective actions and parallel hypertrophic effects of δ PKC and ε PKC. Proc Natl Acad Sci U S A 2001; 98:11114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sanchez‐Bautista S, Nicolas FE. Recent patents concerning modulators of protein kinase C. Recent Pat DNA Gene Seq 2013; 7:74–81. [DOI] [PubMed] [Google Scholar]
- 133. Zhang EY, Kong KF, Altman A. The yin and yang of protein kinase C‐ζ (PKCζ): a novel drug target for selective immunosuppression. Adv Pharmacol 2013; 66:267–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. He X, Koenen HJ, Smeets RL et al Targeting PKC in human T cells using sotrastaurin (AEB071) preserves regulatory T cells and prevents IL‐17 production. J Invest Dermatol 2014; 134:975–83. [DOI] [PubMed] [Google Scholar]
- 135. Sommerer C, Zeier M. AEB071–a promising immunosuppressive agent. Clin Transplant 2009; 23(Suppl. 21):15–8. [DOI] [PubMed] [Google Scholar]
- 136. Skvara H, Dawid M, Kleyn E et al The PKC inhibitor AEB071 may be a therapeutic option for psoriasis. J Clin Invest 2008; 118:3151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Budde K, Sommerer C, Becker T et al Sotrastaurin, a novel small molecule inhibiting protein kinase C: first clinical results in renal‐transplant recipients. Am J Transplant 2010; 10:571–81. [DOI] [PubMed] [Google Scholar]
- 138. Friman S, Arns W, Nashan B et al Sotrastaurin, a novel small molecule inhibiting protein‐kinase C: randomized phase II study in renal transplant recipients. Am J Transplant 2011; 11:1444–55. [DOI] [PubMed] [Google Scholar]
- 139. Jimenez JM, Boyall D, Brenchley G et al Design and optimization of selective protein kinase C ζ (PKCζ) inhibitors for the treatment of autoimmune diseases. J Med Chem 2013; 56:1799–810. [DOI] [PubMed] [Google Scholar]
- 140. Curnock A, Bolton C, Chiu P et al Selective protein kinase Cθ (PKCθ) inhibitors for the treatment of autoimmune diseases. Biochem Soc Trans 2014; 42:1524–8. [DOI] [PubMed] [Google Scholar]
- 141. Ron D, Luo J, Mochly‐Rosen D. C2 region‐derived peptides inhibit translocation and function of beta protein kinase C in vivo . J Biol Chem 1995; 270:24180–7. [DOI] [PubMed] [Google Scholar]
- 142. Csukai M, Chen CH, De Matteis MA, Mochly‐Rosen D. The coatomer protein β'‐COP, a selective binding protein (RACK) for protein kinase Cε . J Biol Chem 1997; 272:29200–6. [DOI] [PubMed] [Google Scholar]
- 143. Arruda S, Ho JL. IL‐4 receptor signal transduction in human monocytes is associated with protein kinase C translocation. J Immunol 1992; 149:1258–64. [PubMed] [Google Scholar]
- 144. Wang Y, Schramek H, Dunn MJ. Cytosolic and nuclear mitogen‐activated protein kinases are regulated by distinct mechanisms. Exp Cell Res 1996; 225:382–8. [DOI] [PubMed] [Google Scholar]
- 145. Chiarini A, Whitfield JF, Armato U, Dal Pra I. Protein kinase C‐β II Is an apoptotic lamin kinase in polyomavirus‐transformed, etoposide‐treated pyF111 rat fibroblasts. J Biol Chem 2002; 277:18827–39. [DOI] [PubMed] [Google Scholar]
- 146. DeVries‐Seimon TA, Ohm AM, Humphries MJ, Reyland ME. Induction of apoptosis is driven by nuclear retention of protein kinase C δ . J Biol Chem 2007; 282:22307–14. [DOI] [PubMed] [Google Scholar]
- 147. Plotnikov A, Flores K, Maik‐Rachline G et al The nuclear translocation of ERK1/2 as an anticancer target. Nat Commun 2015; 6:6685. [DOI] [PubMed] [Google Scholar]
- 148. Pfeifhofer C, Gruber T, Letschka T et al Defective IgG2a/2b class switching in PKC α −/− mice. J Immunol 2006; 176:6004–11. [DOI] [PubMed] [Google Scholar]
- 149. Nechushtan H, Leitges M, Cohen C, Kay G, Razin E. Inhibition of degranulation and interleukin‐6 production in mast cells derived from mice deficient in protein kinase Cβ . Blood 2000; 95:1752–7. [PubMed] [Google Scholar]
- 150. Miyamoto A, Nakayama K, Imaki H et al Increased proliferation of B cells and auto‐immunity in mice lacking protein kinase Cδ . Nature 2002; 416:865–9. [DOI] [PubMed] [Google Scholar]
- 151. Fu G, Hu J, Niederberger‐Magnenat N et al Protein kinase C η is required for T cell activation and homeostatic proliferation. Sci Signal 2011; 4:ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Kong KF, Fu G, Zhang Y et al Protein kinase C‐η controls CTLA‐4‐mediated regulatory T cell function. Nat Immunol 2014; 15:465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Martin P, Duran A, Minguet S, Gaspar ML, Diaz‐Meco MT, Rennert P, Leitges M, Moscat J. Role of ζ PKC in B‐cell signaling and function. EMBO J 2002; 21:4049–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Soloff RS, Katayama C, Lin MY, Feramisco JR, Hedrick SM. Targeted deletion of protein kinase C λ reveals a distribution of functions between the two atypical protein kinase C isoforms. J Immunol 2004; 173:3250–60. [DOI] [PubMed] [Google Scholar]


