Abstract
The ratio of nonsynonymous to synonymous substitution rates (ω) is often used to measure the strength of natural selection. However, ω may be influenced by linkage among different targets of selection, that is, Hill–Robertson interference (HRI), which reduces the efficacy of selection. Recombination modulates the extent of HRI but may also affect ω by means of GC-biased gene conversion (gBGC), a process leading to a preferential fixation of G:C (“strong,” S) over A:T (“weak,” W) alleles. As HRI and gBGC can have opposing effects on ω, it is essential to understand their relative impact to make proper inferences of ω. We used a model that separately estimated S-to-S, S-to-W, W-to-S, and W-to-W substitution rates in 8,423 avian genes in the Ficedula flycatcher lineage. We found that the W-to-S substitution rate was positively, and the S-to-W rate negatively, correlated with recombination rate, in accordance with gBGC but not predicted by HRI. The W-to-S rate further showed the strongest impact on both dN and dS. However, since the effects were stronger at 4-fold than at 0-fold degenerated sites, likely because the GC content of these sites is farther away from its equilibrium, ω slightly decreases with increasing recombination rate, which could falsely be interpreted as a consequence of HRI. We corroborated this hypothesis analytically and demonstrate that under particular conditions, ω can decrease with increasing recombination rate. Analyses of the site-frequency spectrum showed that W-to-S mutations were skewed toward high, and S-to-W mutations toward low, frequencies, consistent with a prevalent gBGC-driven fixation bias.
Keywords: gBGC, Hill–Robertson interference, dN/dS, divergence, diversity, rate of molecular evolution
Introduction
Estimation of nucleotide substitution rates in protein coding sequences allows investigating the processes that drive gene sequence evolution. In particular, the ratio of nonsynonymous (dN) to synonymous (dS) substitution rates (commonly referred to as ω) is a widely used measure that provides information on the strength of natural selection acting on the evolution of protein-coding sequences (Yang and Swanson 2002). However, other processes can also affect substitution rates, including recombination (Webster and Hurst 2012). Everything else being equal, two genes subject to similar selection pressure can yield contrasting ω estimates if they are located in different recombination environments. This is partly because the rate of recombination will affect the extent and character of linked selection, of which the local rate of recombination is a strong determinant. More specifically, linkage between targets of selection reduces the efficacy of selection, known as Hill–Robertson interference (HRI) (Hill and Robertson 1966). It slows down the fixation rate of beneficial variants and thereby the rate of adaptive evolution, resulting in reduced ω. At the same time, linkage among sites hinders the action of purifying selection and thus increases the fixation rate of slightly deleterious mutations, resulting in increased ω. Given that a predominant part of nonsynonymous mutations are deleterious, a negative relationship between recombination rate and ω may be expected (Haddrill et al. 2007; Betancourt et al. 2009; Hurst 2009; Campos et al. 2014).
Another widespread phenomenon by which recombination rate may affect substitution rates is GC-biased gene conversion (gBGC). gBGC is a process that induces a fixation bias for “strong” nucleotides (S; strong in the sense of the number of hydrogen bonds between base pairs, i.e., three between G and C) over “weak” nucleotides (W; two hydrogen bonds between A and T). More precisely, it acts on sites in the neighborhood of recombination-initiating double-strand breaks (DSBs) that are heterozygous for a strong and a weak nucleotide. These heterozygous sites induce mismatches in heteroduplex DNA, which is formed as part of the repair mechanism of DSBs, that will be repaired more frequently in favor of the G:C allele (Marais 2003; Mancera et al. 2008; Lesecque et al. 2013). Importantly, the nonrandom increase in frequency of G:C alleles is not caused by a difference in fitness of strong relative to weak alleles. However, gBGC resembles the action of natural selection as it influences the probability of fixation of mutations and therefore has direct implications for inferences of natural selection. gBGC can cause an increased probability of fixation of mildly deleterious mutations in coding sequences creating potentially significant negative fitness effects (Duret and Galtier 2009; Glemin 2010; Lartillot 2013a; Lachance and Tishkoff 2014) as well as false inference of positive selection (Berglund et al. 2009; Galtier et al. 2009; Ratnakumar et al. 2010). Even though signatures of gBGC are pervasive in many taxa (Romiguier et al. 2010; Escobar et al. 2011; Muyle et al. 2011; Pessia et al. 2012; Lartillot 2013b; Weber et al. 2014; Lassalle et al. 2015; Wallberg et al. 2015), the effect of gBGC on ω has predominantly been investigated in mammals where a greater impact on nonsynonymous substitutions than on synonymous substitutions is suggested to increase ω (Galtier et al. 2009; Ratnakumar et al. 2010; Kostka et al. 2012). However, since the relative effect of gBGC on the two substitution classes depends on a multitude of factors, it might generally affect ω in more complex ways (Capra and Pollard 2011; Lartillot 2013a).
The combined effect of HRI and gBGC mediated through recombination rate variation and their relative importance on estimates of ω is not immediately obvious. One way of dissecting how recombination affects the inference of selection and disentangling the role of HRI and gBGC is to separately analyze substitutions varying in the degree to which they may be influenced by gBGC (Berglund et al. 2009). Specifically, the rate of weak-to-strong (W-to-S) substitutions can be expected to show a positive, and the rate of strong-to-weak (S-to-W) substitutions a negative, correlation with recombination rate (Duret and Arndt 2008). In turn, this means that correlations between ω and recombination rate may differ between different mutation categories given the interplay between selection and gBGC.
Here, we analyze the relationship between recombination rate and the rate of molecular evolution in an avian system, and how this relationship is affected by HRI and gBGC. Of particular relevance is that the rate of recombination shows significant variation across the avian genome, more so than in many other vertebrate lineages and that the landscape of recombination rate variation has remained relatively stable over evolutionary time scales, such that signatures of different processes have had time to build up (Mugal et al. 2013; Kawakami et al. 2014). Moreover, GC content at putatively neutrally evolving sites has not yet reached its equilibrium state in avian genomes and typically evolving toward a higher GC content (Nabholz et al. 2011; Weber et al. 2014). It has therefore been suggested that gBGC has had a profound effect on avian genome evolution (Webster et al. 2006; Backstrom et al. 2013; Mugal et al. 2013; Weber et al. 2014). These characteristics represent striking differences to other vertebrate lineages, in particular to primates where GC content at putatively neutrally evolving sites is on average declining (Duret et al. 2006; Romiguier et al. 2010). In this study, we benefit from a well-annotated genome sequence of high assembly quality of the collared flycatcher (Ficedula albicollis). This, along with the access to a detailed recombination rate map and polymorphism data from whole-genome resequencing of population samples of this species, gives us unusual power to study how recombination modulates sequence evolution in an avian system. Our results suggest that ω can be a misleading measure for making inference of selection if gBGC is not properly accounted for and that HRI has a minor influence on gene sequence evolution in this avian system.
Results
The Impact of Recombination Rate, GC Content, and Exon Density on Rates of Molecular Evolution
To investigate patterns of substitution and their interplay with different genomic properties, we aligned 8,423 one-to-one orthologous gene sequences from the genomes of collared flycatcher (F. albicollis), zebra finch (Taeniopygia guttata), and chicken (Gallus gallus). We binned genes according to the rate of recombination into 21 bins and concatenated exons within each of the bins to reduce noise in the estimates of substitution rates. We estimated dN, dS, and ω in the flycatcher lineage using the phylogeny ((chicken)(zebra finch, flycatcher)). To investigate the impact of HRI and gBGC on these estimates, we performed a multiple linear regression analysis with three different candidate explanatory variables: 1) pedigree-based recombination rate, the primary parameter of interest, 2) exon density, as a proxy of the density of targets of selection, which impacts the strength of HRI, and 3) GC content at 4-fold degenerated sites (GC4) as a proxy of long-term recombination rate, since recombination may impact GC content through the process of gBGC (Meunier and Duret 2004; Duret and Galtier 2009). Indeed, there was a strong correlation between recombination rate and GC4 (Pearson correlation coefficient r = 0.854, P = 8.69·10−7).
The multiple linear regression analysis revealed no significant relationships between dN and any of the candidate explanatory variables. On the other hand, dS showed a positive relationship with GC4 and a slightly negative relationship with exon density (table 1). We would not expect any of these variables to affect the putatively neutral substitution rate (i.e., dS) via the action of HRI. Strong gBGC could explain the observed relationship between dS and GC4 since gBGC increases the fixation probability of GC alleles, which would lead to a simultaneous increase of substitution rate and GC content. Alternatively, selection on codon usage could lead to a relationship between GC content and dS. However, there is no clear evidence for selection on codon usage in birds (Rao et al. 2011; Wang et al. 2014). We also found a significant negative relationship between ω and GC4 (table 1), which could indicate the action of HRI. However, given that dS is affected by GC4, caution might be required to interpret the relationship between GC4 and ω as the action of HRI. By a more detailed analysis, we demonstrate below that this would actually be a false conclusion.
Table 1.
Linear Regression t Values and P Values for dN, dS, and ω as a Function of Recombination Rate, Exon Density, and GC4.
|
dN |
dS |
ω |
||||
|---|---|---|---|---|---|---|
| Explanatory variable | t | P | t | P | t | P |
| Recombination rate | 0.61 | 5.5·10−1 | 0.14 | 9.0·10−1 | 0.75 | 4.6·10−1 |
| GC4 | 1.96 | 7.0·10−2 | 6.78 | 3.2·10−6 | −2.14 | 4.7·10−2 |
| Gene density | −0.49 | 6.3·10−1 | −2.55 | 2.1·10−2 | 0.81 | 4.3·10−1 |
| Multiple R2 | 0.63 | 6.0·10−4 | 0.92 | 1.4·10−9 | 0.38 | 4.0·10−2 |
The Impact of gBGC on Rates of Molecular Evolution
To explore the possible role of gBGC in determining rates of molecular evolution, we applied a strand-symmetric model (Lobry 1995) implemented in the BppML package of the Bio++ suite program (Dutheil and Boussau 2008). This model allowed us to estimate specific substitution rates for the four different mutation categories X-to-Y, corresponding to the substation rates X-to-Y for any of the four possible (W, S) combinations, namely strong-to-strong (S-to-S), S-to-W, W-to-S, and weak-to-weak (W-to-W), where weak bases are A and T and strong bases are G and C. To distinguish between nonsynonymous and synonymous substitutions, we separately estimated substitution rates for 0-fold and 4-fold degenerated sites. Provided that gBGC affects substitution rates and that the rate of recombination reflects the extent of gBGC, we make the following predictions: 1) There should be a strong and positive correlation between the W-to-S substitution rate and recombination rate, as gBGC increases the fixation probability of S alleles. 2) There should be a negative correlation between the S-to-W substitution rate and recombination rate, as gBGC decreases the fixation probability of W alleles. Since gBGC might in principle affect both nonsynonymous and synonymous substitutions, these correlations are expected both for 0-fold and 4-fold degenerated sites. 3) We should a priori not expect recombination rate to correlate with the S-to-S or the W-to-W substitution rate. Since gBGC affects both nonsynonymous and synonymous substitutions in the same way, we expect that these predictions hold for both 0-fold and 4-fold degenerated sites.
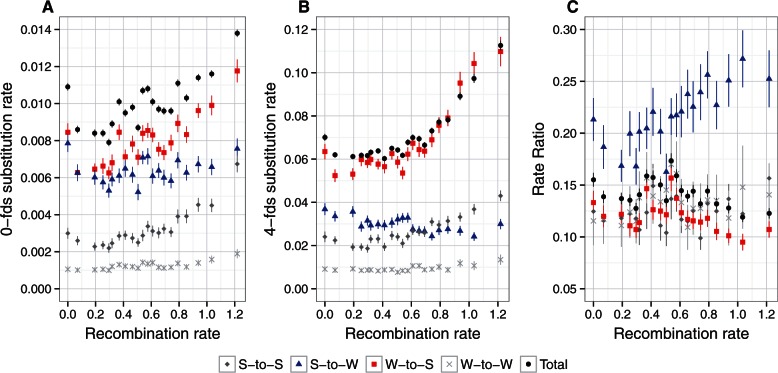
We tested these predictions by estimating substitution rates for each of the four mutation categories in the 21 recombination rate bins. We observed a strong positive correlation between the W-to-S substitution rate and recombination rate for both 0-fold and, in particular, 4-fold degenerated sites, which robustly points toward gBGC as an important process in determining substitution rates (fig. 1, table 2). The first prediction was thus met. In accordance with the second prediction, the S-to-W substitution rate was negatively correlated with recombination rate at 4-fold degenerated sites. At 0-fold degenerated sites, the correlation between the S-to-W substitution rate and recombination rate was not statistically significant (table 2), potentially due to a reduced effect of gBGC on nonsynonymous changes because of interaction with selection. Finally, we found significant positive correlations between both the S-to-S and the W-to-W substitution rate and recombination rate for 0-fold as well as 4-fold degenerated sites. This is unexpected under a model of gBGC favoring the fixation of W over S alleles, which should neither affect the S-to-S nor the W-to-W substitution rate. On the other hand, this could indicate that recombination is mutagenic or that these correlations may arise indirectly as a result of a correlation between recombination rate and another parameter that we did not consider. Interestingly, similar correlations for S-to-S and W-to-W substitutions have been previously described in humans (Duret and Arndt 2008).
Fig. 1.
Relationship between recombination rate and substitution rates for different mutation categories: S-to-S (dark gray); S-to-W (blue); W-to-S (red); W-to-W (light gray); total (substitution rates estimated by codeml; black). Mean recombination rate per bin was log10 transformed, after adding a constant of 1. (A) Substitution rate for 0-fold degenerated sites (0-fds). (B) Substitution rate for 4-fold degenerated sites (4-fds). (C) Substitution rate ratio calculated as the ratio of the 0-fold and 4-fold substitution rates.
Table 2.
Pairwise Pearson Correlation Coefficients and P Values between Recombination Rate and Substitution Rates, and the Ratio of 0-Fold to 4-Fold Substitution Rates, for Different Mutation Categories.
| Substitution Rate | 0-Fold | P | 4-Fold | P | Ratio | P |
|---|---|---|---|---|---|---|
| S-to-S | 0.83 | 2.8·10−6 | 0.87 | 2.7·10−7 | 0.36 | 1.1·10−1 |
| S-to-W | 0.24 | 3.0·10−1 | −0.70 | 4.9·10−4 | 0.77 | 4.0·10−5 |
| W-to-S | 0.78 | 3.2·10−5 | 0.85 | 1.4·10−6 | −0.45 | 4.3·10−2 |
| W-to-W | 0.78 | 3.0·10−5 | 0.68 | 6.7·10−4 | 0.28 | 3.0·10−1 |
| Totala | 0.72 | 2.2·10−4 | 0.82 | 4.8·10−6 | −0.43 | 4.9·10−2 |
aTotal substitution rate estimated by codeml.
Figure 1 demonstrates that the rate of W-to-S substitution was far higher than any of the rates for the other three mutation categories. This applies particularly for 4-fold degenerated sites, where the relative contribution of the W-to-S substitution rate to the total rate was on average 46%, whereas S-to-W substitutions contribute only 25%. As a consequence, the W-to-S substitution rate largely governed dS. Indeed, the relationship between dS and recombination rate was very similar to that between the W-to-S substitution rate and recombination rate (fig. 1B, table 2). The same was true for 0-fold degenerated sites where the relative contributions of W-to-S and S-to-W were 46% and 30%, respectively (fig. 1A). In conclusion, our results show that W-to-S substitutions largely determine both the nonsynonymous and the synonymous substitution rate. The W-to-S substitution rate increases with the rate of recombination, consistent with the hypothesis that strong gBGC raises the total substitution rate.
Estimates of ω over the four mutation classes combined showed a marginally significant negative correlation with recombination rate and the same was observed for the W-to-S substitution rate ratio (fig. 1C). On the contrary, the S-to-W rate ratio was strongly positively correlated with recombination rate, whereas S-to-S and W-to-W rate ratios showed no correlation with recombination rate (fig. 1C, table 2). Interestingly, we found a difference in dN and ω estimates between the nonrecombining bin and the bins with low recombination in all four mutation categories (fig. 1A and C). This difference may reflect the role of HRI in nonrecombining regions. In contrast, if HRI was determining patterns of ω throughout the whole range of recombination rate variation, we would expect a negative correlation between dN and recombination rate, which was not observed in our data. On the contrary, we observed a positive correlation between dN and recombination rate, which argues for a stronger impact of gBGC in determining dN.
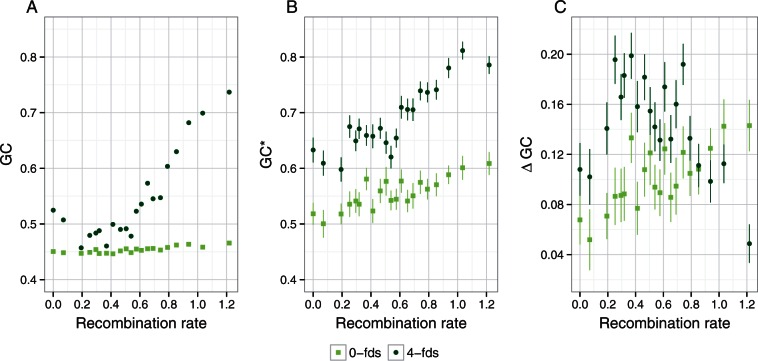
If recombination governs the rates of molecular evolution by means of gBGC, this should be reflected in the evolution of the GC content (Galtier et al. 2001; Meunier and Duret 2004; Duret and Arndt 2008). Specifically, the equilibrium GC content (GC*), which represents the GC content that would be reached at equilibrium if estimated substitutions rates where invariable over time, can be used as a proxy for the strength of gBGC. The current GC content on the other hand reflects the accumulated effect of gBGC over the past. Provided that the recombination landscape is evolutionary stable, such that signatures of gBGC can accumulate over time, both current GC content and GC* are expected to be positively correlated with recombination rate. Indeed, we observe that recombination rate is strongly positively correlated with current GC content (fig. 2A) (r = 0.84, P = 1.95·10−06 and r = 0.854, P = 8.69·10−07 for 0-fold and 4-fold degenerated sites, respectively) as well as the GC* (fig. 2B) (r = 0.841, P = 1.8·10−06 and r = 0.892 P = 5.62·10−08 for 0-fold and 4-fold degenerated sites, respectively). Moreover, we found that the difference between GC* and current GC content (ΔGC) is positive (GC* > GC) in all recombination bins (fig. 2C). As ΔGC measures the distance of the current GC content to its equilibrium value, this suggests that GC content is still increasing and that gBGC leads to an overall increase of substitution rates. It is true for both 0-fold and 4-fold degenerated sites, where 4-fold degenerated sites are further away from their equilibrium than 0-fold degenerated sites (ΔGC4 > ΔGC0). The greater distance of the current GC content to its equilibrium at 4-fold degenerated sites when compared with 0-fold degenerated sites could explain why synonymous substitutions are more strongly affected by gBGC than nonsynonymous substitutions.
Fig. 2.
Relationship between recombination rate and GC content for 0-fold degenerated sites (light green) and 4-fold degenerated sites (dark green). Mean recombination rate per bin was log10 transformed, after adding a constant of 1. (A) Current GC content. (B) Equilibrium GC content (GC*). (C) ΔGC, calculated as GC* − GC current.
The Impact of gBGC on Patterns of Diversity
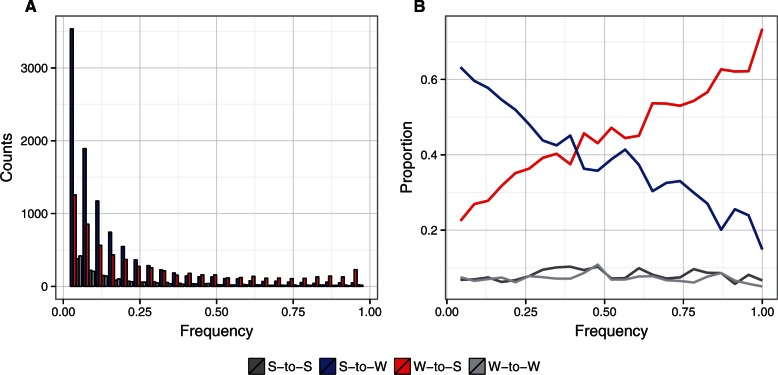
Besides their influence on the rates of molecular evolution, HRI and gBGC can affect patterns of nucleotide diversity. HRI reduces the efficacy of selection in low recombination regions, which is expected to lead to a positive correlation between recombination rate and neutral diversity irrespective of the mutation category (Campos et al. 2014). Moreover, HRI is not expected to affect the site frequency spectrum (SFS) of the four mutation categories in different directions. On the other hand, the impact of gBGC on patterns of diversity should differ between mutation categories. Importantly, gBGC skews the SFS of W-to-S mutations toward high frequencies and of S-to-W mutations toward low frequencies (Capra et al. 2013; Lachance and Tishkoff 2014; Wallberg et al. 2015). To better distinguish between HRI and gBGC, we therefore also analyzed diversity levels and the SFS of the four different mutation categories. We investigated nucleotide diversity in protein-coding genes based on data from whole-genome resequencing of 20 collared flycatchers from an allopatric population. Using the same set of 8,423 genes that were used for substitution rate estimates, we analyzed a total number of 8,460,789 sites (6,934,131 0-fold and 1,526,658 4-fold degenerated sites), of which we identified 33,581 as polymorphic (14,354 0-fold and 19,227 4-fold degenerated sites); this represents an unusually large data set for coding sequence polymorphisms from a natural population. We calculated Watterson’s θ (θW) and the unfolded SFS for each of the S-to-S, S-to-W, W-to-S, and W-to-W mutation categories and distinguished 0-fold from 4-fold degenerated sites. The SFS showed a right skew and higher proportion of high-frequency derived variants for the W-to-S class, and a higher proportion of low-frequency derived alleles for the S-to-W class, which points to a strong impact of gBGC on patterns of diversity. This holds true both at 4-fold (fig. 3A) and 0-fold degenerated sites (supplementary fig. S1A, Supplementary Material online). Illustrations of the relative SFS for each class clearly visualize a strong shift in the relative proportion of S-to-W and W-to-S derived mutations (fig. 3B and supplementary fig. S1B, Supplementary Material online).
Fig. 3.
The site frequency spectra (SFS) for different mutation categories at 4-fold degenerated sites. S-to-S (dark gray); S-to-W (blue); W-to-S (red); and W-to-W (light gray). (A) SFS based on derived allele counts from 19,227 polymorphic sites. (B) Proportion of each mutation category for a given derived allele frequency. For 0-fold degenerated sites, see Supplementary fig. S1, Supplementary Material online).
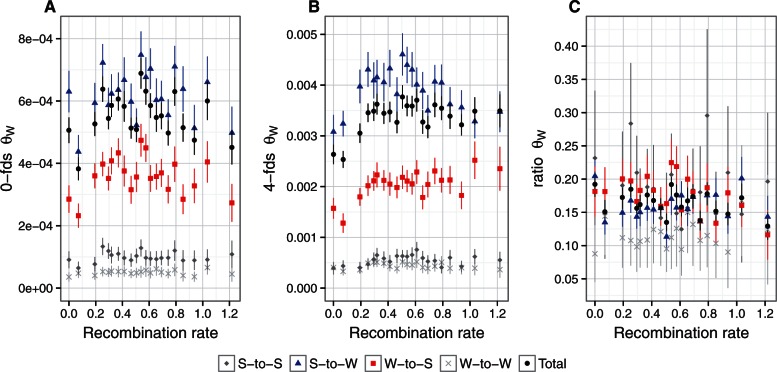
Estimates of θW showed no significant relationship with recombination rate at 0-fold degenerated sites for any mutational class or for total θW (fig. 4A, table 3). For 4-fold degenerated sites, there was a positive and statistically significant correlation only between W-to-S-specific θW and recombination rate (fig. 4B, table 3), compatible with an influence of gBGC on patterns of diversity. The W-to-S-specific ratio of θW at 0-fold and 4-fold degenerate sites showed a negative correlation with recombination, which should at least in part be driven by the positive correlation between W-to-S-specific θW and recombination rate. We found no correlation between W-to-W-specific and S-to-S-specific θW estimates with recombination rate (fig. 4, table 3).
Fig. 4.
Relationship between recombination rate and diversity estimates (θW) for different mutation categories: S-to-S (dark gray); S-to-W (blue); W-to-S (red); W-to-W (light gray); total θW (black). Mean recombination rate per bin was log10 transformed, after adding a constant of 1. (A) θW for 0-fold degenerated sites. (B) θW for 4-fold degenerated sites. (C) the ratio of θW at 0-fold to 4-fold degenerated sites.
Table 3.
Pairwise Pearson Correlation Coefficients and P Values between Recombination Rate and Diversity (θW) for Different Mutation Categories.
| θW | 0-fold | P | 4-fold | P | Ratio | P |
|---|---|---|---|---|---|---|
| S-to-S | 0.04 | 8.6·10−1 | 0.30 | 1.8·10−1 | −0.21 | 3.5·10−1 |
| S-to-W | 0.07 | 7.8·10−1 | −0.11 | 6.3·10−1 | 0.19 | 4.1·10−1 |
| W-to-S | 0.07 | 7.6·10−1 | 0.70 | 4.4·10−4 | −0.54 | 1.1·10−2 |
| W-to-W | 0.08 | 7.3·10−1 | −0.09 | 7.0·10−1 | 0.20 | 3.9·10−1 |
| Total | 0.08 | 7.4·10−1 | 0.59 | 5.0·10−3 | −0.47 | 3.4·10−2 |
Finally, estimates of θW indicate that S-to-W mutations are the most prevalent mutations across all recombination bins (fig. 4A and B). We estimated the mutational bias (Rμ) for S-to-W and W-to-S as the ratio of the rate of singletons of each mutation category. The average Rμ at 4-fold degenerated sites across all recombination bins was 3.41. This suggests that the observed substitution bias for W-to-S is not driven by a mutational bias but a fixation bias.
Theoretical Insights on the Interaction between gBGC and Natural Selection
To better understand the consequences of recombination via gBGC on rates of molecular evolution, we analytically described the impact of gBGC on substitution rates at neutrally evolving sites (reflecting synonymous substitution rates) as well as sites evolving under natural selection (reflecting nonsynonymous substitution rates). The strength of gBGC was modeled by the coefficient of gBGC (b), which correlates linearly with the recombination rate and affects the probability of fixation similar to the coefficient of selection (s) (for details, see Materials and Methods). Thus, similar to the action of selection, the degree by which b affects the rate of molecular evolution depends directly on Ne. The larger Ne the stronger is the effect of gBGC, which can be expressed as the population scaled coefficient of gBGC, B (B = 4 Ne b).
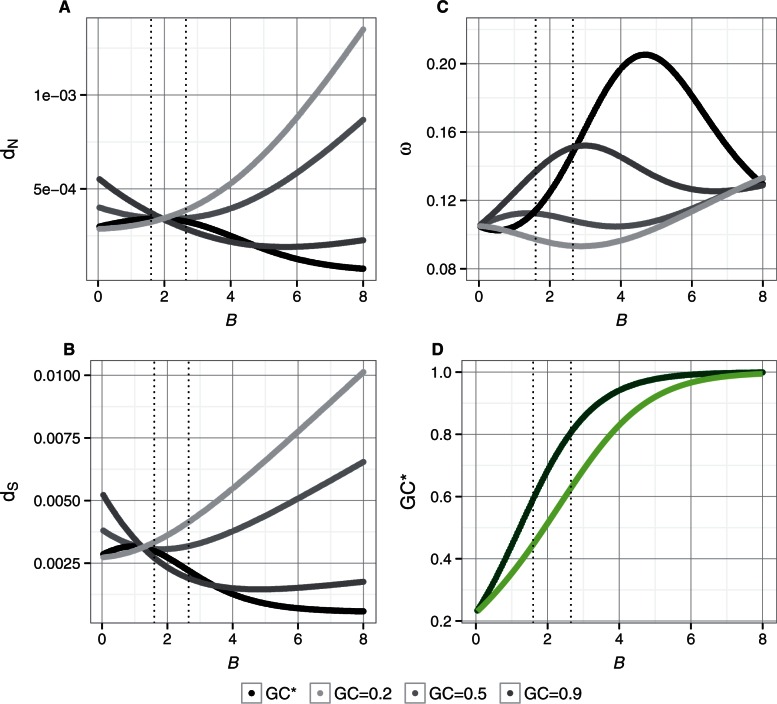
We allowed B to vary from 0 to 8 to cover strengths of gBGC corresponding to a wide range of GC*, encompassing the range found in the flycatcher lineage (fig. 5D). The mutation rate was approximated by the rate of singletons at 4-fold degenerated sites and therefore differed between mutation categories (fig. 4B). To model molecular evolution at sites evolving under natural selection, an estimate of the distribution of fitness effects (DFE) is required, knowledge of which is limited in birds. We therefore made the simplifying assumption that the DFE is represented by three categories: 1) lethal mutations, that is, mutations that are immediately removed by selection and do not appear as polymorphic sites; 2) slightly deleterious mutations, and 3) slightly advantageous mutations. We approximated the proportion of lethal mutations as 1 – [the ratio of the total singleton rate at 0-fold degenerated sites to the total singleton rate at 4-fold degenerated sites], which was 0.78. For the remaining two categories, 90% of the mutations were assigned to be slightly deleterious, while only 10% were assigned to be slightly advantageous. The strength of the population scaled selection coefficient was assumed to be the same for deleterious and advantageous mutations (|Ne s| ≤ 1). These parameters led to lower GC* at selected than at neutrally evolving sites, consistent with the pattern found in the flycatcher lineage. To explore the impact of current GC content (or, more specifically, of ΔGC) on patterns of molecular evolution we applied four different scenarios: 1) the current GC is at equilibrium (GC = GC*, i.e., ΔGC = 0) and synonymous and nonsynonymous sites have their own equilibria (as shown in fig. 5D), 2) current GC = 0.2 and ΔGC > 0 regardless of the value of B, 3) current GC = 0.5 and ΔGC is either positive or negative, and 4) current GC = 0.9 and ΔGC < 0 except for extremely high values of B where GC* is close to 1. Specifically, this allowed us to assess how dN, dS, and ω can vary with the strength of gBGC depending on how far the GC content is from the equilibrium value.
Fig. 5.
Analytical description of the effect of gBGC on dN, dS, ω and GC* at different GC contents (GC = GC* [black], GC = 0.2 [light gray], GC = 0.5 [gray], GC = 0.9 [dark gray]) at both neutrally evolving sites and sites evolving under natural selection. (A) dN. (B) dS. (C) ω. (D) GC*; 0-fold degenerated sites (light green) and 4-fold degenerated sites (dark green). Dashed vertical lines indicate the range of observed values of GC* across recombination bins (fig. 2B).
The model shows that both dN and dS will increase with increasing B whenever the current GC content is lower than GC* (ΔGC > 0), as found in the flycatcher lineage. On the contrary, if the current GC content is higher than the GC*, dN and dS may decrease (fig. 5A and B). Figure 5 shows that depending on the current GC content and how far it is from GC*, the difference in the impact of gBGC on dN and dS can create both positive and negative relationships between B and ω (fig. 5D), and thus between recombination rate and ω. On the basis of this model, we argue that gBGC may lead to reduced ω in high recombining regions under the conditions that ΔGC is positive and higher at neutrally evolving sites than in sites evolving under natural selection, as found in the flycatcher lineage (fig. 2C). Therefore, the impact of gBGC on the total nonsynonymous and synonymous substitution rates is not only governed by the GC content but even more so by the difference between the current GC content and GC*. The effect of gBGC on substitution rates of each mutational class is visualized in supplementary figure S2, Supplementary Material online.
Discussion
We explored the impact of recombination rate variation via HRI and gBGC on inferences of natural selection in an avian model system. Specifically, we addressed the question if HRI and/or gBGC govern the relationship between recombination rate and ω. Since in mammals gBGC has previously been found to increase ω (Berglund et al. 2009; Galtier et al. 2009; Ratnakumar et al. 2010), the weak negative relationship found in the flycatcher lineage seems to point toward a prominent role of HRI in this lineage. However, estimation of lineage-specific substitution rates for 0-fold and 4-fold degenerated sites for four different mutation categories that are differently affected by gBGC provided evidence for a strong impact of gBGC on rates of molecular evolution. In contrast, the role of HRI in determining genome wide patterns of molecular evolution seemed comparatively weak. Analyses of patterns of diversity, including the SFS, of different mutation categories revealed similar conclusions.
In an effort to explain the discrepancies between our findings and earlier studies, we identified ΔGC as an important determinant of the impact of gBGC on both dN and dS. It follows that the relative strength of gBGC on nonsynonymous and synonymous substitutions, as given by differences in ΔGC between 0-fold and 4-fold degenerated sites, determines the impact of gBGC on ω. In the flycatcher genome, the GC content is below the equilibrium value, and both ΔGC4 and ΔGC0 are positive. ΔGC4 is on average larger than ΔGC0, suggesting a stronger impact of gBGC on synonymous than on nonsynonymous substitutions. In line with this notion, our analytical description of the gBGC process shows that if neutrally evolving sites are further away from their equilibrium than sites evolving under natural selection, ω can decrease with increasing recombination rate. This seems at first glance surprising since previous studies have reported that gBGC will in most scenarios have a greater impact on nonsynonymous substitutions than on synonymous substitutions, leading to an increase of ω with increasing recombination rate (Galtier and Duret 2007; Duret and Galtier 2009; Galtier et al. 2009; Ratnakumar et al. 2010). For example, Galtier et al. (2009) also analytically described gBGC and showed that in primate lineages gBGC increases ω under most conditions. However, this study did not explore the possibility of ΔGC4 being larger than ΔGC0 and did not allow for both to be positive, as here observed in the avian data.
Our analysis of the rate of molecular evolution in the flycatcher lineage demonstrates that gBGC may influence inferences of selection in unexpected ways. Specifically, a higher impact of gBGC on synonymous than on nonsynonymous substitutions may not lead to the expected increase in ω as found in primates where GC4 is on average declining but on the contrary lead to a decrease in ω. This problem should be common to organisms in which base composition evolves toward higher GC content. We therefore strongly advise against interpreting observed relationships between gene sequence evolution and recombination without properly accounting for gBGC. For example, a recent study on the rates of molecular evolution in two passerine bird lineages (great tit, Parus major, and zebra finch) proposed that the observed negative relationship between ω and recombination rate was owing to a large effect of HRI (Gossmann et al. 2014). However, this study only vaguely investigated the impact of gBGC on patterns of ω. Although we observed a weak negative correlation between recombination rate and ω, our results widely support the hypothesis that the reduction in ω in high recombination regions is mainly the result of a strong impact of gBGC, which affects nonsynonymous and synonymous substitutions to a different extent.
In conclusion, our work stresses the importance of investigating different groups of organisms to gain a better understanding of the general mechanism by which gBGC influences rates of molecular evolution. As shown here, differences in the recombination landscape, Ne and ΔGC among species may lead to different signatures of gBGC on dN, dS and ω. In birds, the generally high but at the same time heterogeneous rate of recombination, along with a stable recombination landscape and large Ne, may have allowed gBGC to show a particularly strong impact on substitution rates.
Materials and Methods
Sequence Data
We retrieved putative 1:1 orthologous genes of collared flycatcher, zebra finch, and chicken through the Biomart retrieval tool in Ensembl release 73 (Kasprzyk 2011; Flicek et al. 2014). Codon-based alignments were generated using PRANK (v.130410) (Loytynoja and Goldman 2005). The Heads-or-Tails (HoT) algorithm implemented in the program Guidance was used to calculate alignment confidence scores that reflect alignment uncertainties (Landan and Graur 2007; Penn et al. 2010). Using default settings, misaligned columns were discarded. We excluded genes if the length of the coding sequence was shorter than 200 bp, if the genomic location was unknown, sex linked, or in microchromosomes with less than 5 Mb of assembled sequence (chromosomes LGE22, 25 and Fal35; due to limited amount of data) according to the FicAlb1.5 assembly version of the collared flycatcher genome (Kawakami et al. 2014). We also excluded genes that had overlapping transcripts as a result of antisense transcription and genes with premature STOP codons.
Estimation of Genomic Features
We obtained recombination rate estimates in cM/Mb for nonoverlapping 200 kb windows of the collared flycatcher genome from Kawakami et al. (2014). Recombination rate values for each ortholog were assigned by mapping genes to these windows. When a gene covered two or more windows, we calculated a weighted average of the recombination rates in the corresponding windows. The same approach was used for assigning exon density values (the proportion of coding base pairs in the assembled sequence in each window). Genes were grouped and concatenated into 21 bins based on their recombination rate. Every bin contained approximately the same number of genes (403 or 404) except for bin 0 that contained all autosomal genes with recombination rate estimates of 0 cM/Mb (348 genes). Mean values of recombination rate in the 20 nonzero categories ranged from 0.176 cM/Mb to 15.535 cM/Mb.
Estimation of Rates of Molecular Evolution
We used the codeml program in the phylogenetic analysis by maximum likelihood (PAML4.7) package (Yang 1997) to estimate rates of nonsynonymous (dN) and synonymous (dS) substitutions for each recombination bin. We used a free-ratio-model (model = 1) to estimate the flycatcher lineage-specific dN, dS, and their ratio (ω), assuming a constant ω over all sites and different equilibrium nucleotide frequencies for each codon position (F3x4) (Yang and Swanson 2002). To assess the potential role of gBGC in the evolution of base composition and substitution rates, we used a strand-symmetric model (Lobry 1995) implemented in the package BppML in the Bio++ suite program (Dutheil and Boussau 2008). This model is nonreversible and thereby allows for the estimation of substitution rates of S-to-S, S-to-W, W-to-S, and W-to-W mutation categories. Flycatcher branch-specific estimates for concatenated genes per recombination bin were made separately for 0-fold and 4-fold degenerated sites. The model also allowed us to estimate the branch-specific GC*, including GC0* and GC4* (GC* of 0-fold and 4-fold degenerated sites, respectively). Analyses using 2nd and 3rd codon positions instead of 0-fold and 4-fold degenerated sites provided similar results (not shown).
Estimation of Diversity and SFS Analysis
We retrieved single-nucleotide polymorphism data from 20 individuals of an Italian population of collared flycatchers (Burri et al. 2015) to investigate potential signatures of gBGC on the SFS. Briefly, whole-genome resequencing of unrelated birds was done using Illumina technology. Reads were mapped to the collared flycatcher genome assembly version FicAlb1.5 (Kawakami et al. 2014) using Burrows-Wheeler Aligner 0.7.4 (Li and Durbin 2009), and variant calling was performed using the Genome Analysis Toolkit (GATK) 2.8-1 (McKenna et al. 2010). We filtered polymorphic sites according to a minimum mapping quality of 20 and minimum variant quality of 15. Additionally, we required at least 12 genotypes with minimum coverage of 5x per individual. We then randomly choose 24 alleles from the genotypes passing the filtering criteria to constantly obtain the same sample size for each site.
We assigned polymorphic sites into one of the different mutation categories described above (S-to-S, S-to-W, W-to-S, and W-to-W). We polarized variant sites using the genome sequence of two outgroup species, Ficedula parva and Ficedula hyperythra (Burri et al. 2015). We defined the ancestral state when the same allele was fixed in at least two of the three species. To ensure low error rates in polarization, we removed sites where more than one species was polymorphic or had missing data. Nonbiallelic positions and codons with more than one polymorphic site were discarded. We estimated θW per site separately for 0-fold and 4-fold degenerated sites for each of the four mutation categories as the ratio of the number of polymorphic sites of each class to the product of the harmonic mean of the sample size and the number of total potential sites for that class (i.e., S-to-W-specific θW was defined as the number of S-to-W polymorphic sites divided by the product of the harmonic mean and the total number of “strong” or G/C sites passing the filtering criteria). We calculated the unfolded SFS separately for 0-fold and 4-fold degenerated sites to compare the distributions of derived allele counts of the different polymorphism classes.
To test for potential differences in mutation rate across different recombination environments, we calculated the rate of singletons for different mutation categories as the ratio of the number of singletons of each class to the total number of potential sites for that class. We also examined the mutational bias (Rμ) between different mutational categories by considering the ratio of S-to-W to W-to-S singleton rates at 4-fold degenerated sites.
Statistical Analyses
We used R version 3.0.1 (R Core Team 2013) to perform all statistical testing. Multiple linear regression analyses were performed with recombination rate, exon density, and GC4 as explanatory variables, and dN, dS, and ω as response variables. Explanatory variables were transformed to minimize the skew in their distribution. Specifically, recombination rate values were log10 transformed after adding 1 to all values, while GC4 and exon density were transformed by using square root transformation. These values were then Z-transformed, which means scaling the data to standardize the mean value to 0 and standard deviation to 1. We computed Pearson correlation coefficients to assess correlation between variables.
The 95% confidence intervals (CIs) were obtained by generating 100 bootstrap replicates of aligned sequences for each recombination bin. For each of these replicates, we estimated the parameters of interest. The standard error (SE) for each parameter was estimated as the standard deviation of the resampling distribution. We then estimated the CIs as the 2.5th and 97.5th percentiles of the Student t distribution. For substitution rates estimated with codeml, we relied on the SE provided by the software and estimated CIs as specified above.
A Mathematical Model of gBGC
Let uX → Y represent the substitution rate from X-to-Y, where the pair (X, Y) represents any of the four combinations of W and S. Then substitution rate uX → Y can be expressed as a function of the effective population size Ne, the particular mutation rate μX → Y and the probability of fixation pX → Y
| (1) |
Consider a mutation at a selectively neutral site, where the probability of fixation of the segregating polymorphism is not influenced by natural selection. Then gBGC influences the dynamics of the segregating polymorphism just like directional selection (Nagylaki 1983). However, gBGC only impacts the probability of fixation of W-to-S and S-to-W mutations but not the other types of mutations. So the probability of fixation of W-to-W and S-to-S mutations is 1/2Ne, while the probability of fixation of W-to-S mutations is
| (2) |
and that of S-to-W mutations is
| (3) |
where b represents the coefficient of gBGC. On the other hand, for a mutation under selection, the dynamics of the segregating polymorphism might be influenced by an interplay of selection and gBGC. The probability of fixation of W-to-W and S-to-S mutations remains unaffected by gBGC,
| (4) |
where s represents the selection coefficient and ϕ(s) represents the DFE. The probability of fixation of W-to-S mutations is affected by gBGC,
| (5) |
as well as that of S-to-W mutations,
| (6) |
Now, let u represent the overall substitution rate per site. Then u can be expressed as the sum of the different categories of nucleotide substitution rates weighted by their respective opportunities of mutation, that is, the GC content (xGC),
| (7) |
Combining equations (1)–(3) and (7) for the neutral scenario leads to
| (8) |
For a scenario that invokes natural selection we combine equations (1) and (4)–(7), which leads to
| (9) |
Further, GC* can be expressed as a function of the substitution rates from W-to-S and S-to-W
| (10) |
Combining equations (1)–(3) and (10) for the neutral scenario leads to
| (11) |
Combining equations (1), (4)–(6), and (10) for a scenario that invokes natural selection leads to
| (12) |
Supplementary Material
Supplementary figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Sylvain Glémin and the members of the Ellegren lab for helpful discussions. They are also thankful to two anonymous reviewers for helpful comments. This work was supported by the Swedish Research Council (grant numbers 2010-5650 and 2013-8271); the European Research Council (AdG 249976); and the Knut and Alice Wallenberg Foundation. Computations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX).
References
- Backstrom N, Zhang Q, Edwards SV. 2013. Evidence from a house finch (Haemorhous mexicanus) spleen transcriptome for adaptive evolution and biased gene conversion in passerine birds. Mol Biol Evol. 30:1046–1050. [DOI] [PubMed] [Google Scholar]
- Berglund J, Pollard KS, Webster MT. 2009. Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 7:45–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt AJ, Welch JJ, Charlesworth B. 2009. Reduced effectiveness of selection caused by a lack of recombination. Curr Biol. 19:655–660. [DOI] [PubMed] [Google Scholar]
- Burri R, Nater A, Kawakami T, Mugal CF, Olason PI, Smeds L, Suh A, Dutoit L, Bures S, Garamszegi LZ, et al. Forthcoming 2015. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. Advance Access published September 9, 2015, doi: 10.1101/gr.196485.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos JL, Halligan DL, Haddrill PR, Charlesworth B. 2014. The relation between recombination rate and patterns of molecular evolution and variation in Drosophila melanogaster. Mol Biol Evol. 31:1010–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Pollard KS. 2011. Substitution patterns are GC-biased in divergent sequences across the metazoans. Genome Biol Evol. 3:516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Hubisz MJ, Kostka D, Pollard KS, Siepel A. 2013. A model-based analysis of GC-biased gene conversion in the human and chimpanzee genomes. PLoS Genet. 9:e1003684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Arndt PF. 2008. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 4:e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Eyre-Walker A, Galtier N. 2006. A new perspective on isochore evolution. Gene 385:71–74. [DOI] [PubMed] [Google Scholar]
- Duret L, Galtier N. 2009. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 10:285–311. [DOI] [PubMed] [Google Scholar]
- Dutheil J, Boussau B. 2008. Non-homogeneous models of sequence evolution in the bio++ suite of libraries and programs. BMC Evol Biol. 8:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar JS, Glemin S, Galtier N. 2011. GC-biased gene conversion impacts ribosomal DNA evolution in vertebrates, angiosperms, and other eukaryotes. Mol Biol Evol. 28:2561–2575. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2014. Ensembl 2014. Nucleic Acids Res. 42:D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Duret L. 2007. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 23:274–277. [DOI] [PubMed] [Google Scholar]
- Galtier N, Duret L, Glemin S, Ranwez V. 2009. GC-biased gene conversion promotes the fixation of deleterious amino acid changes in primates. Trends Genet. 25:1–5. [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L. 2001. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics 159:907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S. 2010. Surprising fitness consequences of GC-biased gene conversion: I. Mutation load and inbreeding depression. Genetics 185:939–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann TI, Santure AW, Sheldon BC, Slate J, Zeng K. 2014. Highly variable recombinational landscape modulates efficacy of natural selection in birds. Genome Biol Evol. 6:1061–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Halligan DL, Tomaras D, Charlesworth B. 2007. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 8:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. 1966. The effect of linkage on limits to artificial selection. Genet Res. 8:269–294. [PubMed] [Google Scholar]
- Hurst LD. 2009. Genetics and the understanding of selection. Nat Rev Genet. 10:83–93. [DOI] [PubMed] [Google Scholar]
- Kasprzyk A. 2011. Biomart: driving a paradigm change in biological data management. Database (Oxford) 2011:bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Smeds L, Backstrom N, Husby A, Qvarnstrom A, Mugal CF, Olason P, Ellegren H. 2014. A high-density linkage map enables a second-generation collared flycatcher genome assembly and reveals the patterns of avian recombination rate variation and chromosomal evolution. Mol Ecol. 23:4035–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka D, Hubisz MJ, Siepel A, Pollard KS. 2012. The role of GC-biased gene conversion in shaping the fastest evolving regions of the human genome. Mol Biol Evol. 29:1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance J, Tishkoff SA. 2014. Biased gene conversion skews allele frequencies in human populations, increasing the disease burden of recessive alleles. Am J Hum Genet. 95:408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landan G, Graur D. 2007. Heads or tails: a simple reliability check for multiple sequence alignments. Mol Biol Evol. 24:1380–1383. [DOI] [PubMed] [Google Scholar]
- Lartillot N. 2013a. Interaction between selection and biased gene conversion in mammalian protein-coding sequence evolution revealed by a phylogenetic covariance analysis. Mol Biol Evol. 30:356–368. [DOI] [PubMed] [Google Scholar]
- Lartillot N. 2013b. Phylogenetic patterns of GC-biased gene conversion in placental mammals and the evolutionary dynamics of recombination landscapes. Mol Biol Evol. 30:489–502. [DOI] [PubMed] [Google Scholar]
- Lesecque Y, Mouchiroud D, Duret L. 2013. GC-biased gene conversion in yeast is specifically associated with crossovers: molecular mechanisms and evolutionary significance. Mol Biol Evol. 30:1409–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassalle F, Perian S, Bataillon T, Nesme X, Duret L, Daubin V. 2015. GC-content evolution in bacterial genomes: the biased gene conversion hypothesis expands. PLoS Genet. 11:e1004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR. 1995. Properties of a general model of DNA evolution under no-strand-bias conditions. J Mol Evol. 41:326–330. [DOI] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 102:10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G. 2003. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 19:330–338. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J, Duret L. 2004. Recombination drives the evolution of GC-content in the human genome. Mol Biol Evol. 21:984–990. [DOI] [PubMed] [Google Scholar]
- Mugal CF, Arndt PF, Ellegren H. 2013. Twisted signatures of GC-biased gene conversion embedded in an evolutionary satable karyotype. Mol Biol Evol. 30:1700–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A, Serres-Giardi L, Ressayre A, Escobar J, Glemin S. 2011. GC-biased gene conversion and selection affect GC content in the Oryza genus (rice). Mol Biol Evol. 28:2695–2706. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Künstner A, Wang R, Jarvis ED, Ellegren H. 2011. Dynamic evolution of base composition: causes and consequences in avian phylogenomics. Mol Biol Evol. 28:2197–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki T. 1983. Evolution of a finite population under gene conversion. Proc Natl Acad Sci USA. 80:6278–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. 2010. Guidance: a web server for assessing alignment confidence scores. Nucleic Acids Res. 38:W23–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessia E, Popa A, Mousset S, Rezvoy C, Duret L, Marais GAB. 2012. Evidence for widespread GC-biased gene conversion in eukaryotes. Genome Biol Evol. 4:787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing . Vienna (Austria): R foundation for statistical computing. Available from: http://www.R-project.Org/. [Google Scholar]
- Ratnakumar A, Mousset S, Glemin S, Berglund J, Galtier N, Duret L, Webster MT. 2010. Detecting positive selection within genomes: the problem of biased gene conversion. Philos Trans R Soc B. 365:2571–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao YS, Wu GZ, Wang ZF, Chai XW, Nie QH, Zhang XQ. 2011. Mutation bias is the driving force of codon usage in the Gallus gallus genome. DNA Res. 18:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJP, Galtier N. 2010. Contrasting GC-content dynamics across 33 mammalian genomes: relationship with life-history traits and chromosome sizes. Genome Res. 20:1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A, Glemin S, Webster MT. 2015. Extreme recombination frequencies shape genome variation and evolution in the honeybee, Apis mellifera. PLoS Genet. 11:e1005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZJ, Zhang JL, Yang W, An N, Zhang P, Zhang GJ, Zhou Q. 2014. Temporal genomic evolution of bird sex chromosomes. BMC Evol Biol. 14:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CC, Boussau B, Romiguier J, Jarvis ED, Ellegren H. 2014. Evidence for GC-biased gene conversion as a driver of between-linege differences in avian base composition. Genome Biol. 15:549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, Axelsson E, Ellegren H. 2006. Strong regional biases in nucleotide substitution in the chicken genome. Mol Biol Evol. 23:1203–1216. [DOI] [PubMed] [Google Scholar]
- Webster MT, Hurst LD. 2012. Direct and indirect consequences of meiotic recombination: implications for genome evolution. Trends Genet. 28:101–109. [DOI] [PubMed] [Google Scholar]
- Yang ZH. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 13:555–556. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Swanson WJ. 2002. Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol Biol Evol. 19:49–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.