Abstract
The oral Gram-negative anaerobic bacterium Porphyromonas gingivalis is an important pathogen involved in chronic periodontitis. Among its virulence factors, the major extracellular proteinases, Arg-gingipain and Lys-gingipain, are of interest given their abilities to degrade host proteins and process other virulence factors. Gingipains possess C-terminal domains (CTDs) and are translocated to the cell surface or into the extracellular milieu by the type IX secretion system (T9SS). Gingipains contribute to the colonial pigmentation of the bacterium on blood agar. In this study, Omp17, the PGN_0300 gene product, was found in the outer membrane fraction. A mutant lacking Omp17 did not show pigmentation on blood agar and showed reduced proteolytic activity of the gingipains. CTD-containing proteins were released from bacterial cells without cleavage of the CTDs in the omp17 mutant. Although synthesis of the anionic polysaccharide (A-LPS) was not affected in the omp17 mutant, the processing of and A-LPS modification of CTD-containing proteins was defective. PorU, a C-terminal signal peptidase that cleaves the CTDs of other CTD-containing proteins, was not detected in any membrane fraction of the omp17 mutant, suggesting that the defective maturation of CTD-containing proteins by impairment of Omp17 is partly due to loss of function of PorU. In the mouse subcutaneous infection experiment, the omp17 mutant was less virulent than the wild type. These results suggested that Omp17 is involved in P. gingivalis virulence.
INTRODUCTION
Periodontal disease, a chronic inflammatory disease that causes the destruction of periodontal tissues and alveolar bone (1), is one of the most frequently occurring infectious diseases in humans (2). The anaerobic Gram-negative bacterium Porphyromonas gingivalis, an etiologically important agent of periodontal disease (3), possesses a number of virulence factors, including fimbriae, hemagglutinins, lipopolysaccharides, and proteinases. P. gingivalis has two major types of cysteine proteinases, Arg-specific gingipain and Lys-specific gingipain, which are the products of 3 separate genes: rgpA, rgpB, and kgp (4, 5). Gingipains are highly active extracellular and surface proteinases and are of particular importance because they can destroy various components of periodontal tissue, including extracellular matrix proteins, cytokines, complement proteins, antibodies, and proteinase inhibitors (6–9). It was demonstrated that individual gingipain-deficient strains had significantly reduced virulence in comparison to that of the parental strain in murine models (10–12). The kgp and rgpA genes encode polyproteins that comprise the signal peptide, propeptide, proteinase, and adhesion domains and the C-terminal domain (CTD). The rgpB gene encodes a protein that comprises the signal peptide, propeptide, and proteinase domains and the CTD. Kgp and Rgp are synthesized in the cytoplasm as preproenzymes and are translocated across the inner membrane via a signal peptide targeting the Sec apparatus. Then, they are translocated across the outer membranes via the Por secretion system (PorSS)/type IX secretion system (T9SS) (13–15). Subsequently, Kgp and Rgp are either secreted into the extracellular milieu as mature proteinases or are located on the cell surface as complexes noncovalently associated with adhesion domain proteins (14).
The T9SS is widely distributed in the Bacteroidetes phylum, but it is not found in Bacteroides fragilis and Bacteroides thetaiotaomicron (14, 16, 17). The P. gingivalis T9SS proteins PorK, PorL, PorM, PorN, PorW, PorT, and Sov share similarity in amino acid sequence with the Flavobacterium johnsoniae gliding motility proteins GldK, GldL, GldM, GldN, SprE, SprT, and SprA, respectively (14, 18–20). In F. johnsoniae, disruption of the sprT gene disrupts translocation of the gliding motility protein SprB and secretion of chitinase, suggesting that the T9SS is linked to the gliding motility of bacteria in the Bacteroidetes phylum (14).
The P. gingivalis genome encodes approximately 34 CTD-containing proteins (21). CTD-containing proteins are also found in predicted proteins of other bacteria in the Bacteroidetes phylum, including Prevotella intermedia and Tannerella forsythia (22–24). In P. gingivalis, the CTD-containing proteins, including RgpA, RgpB, Kgp, TapA, and HBP35 and the proteins encoded by PGN_0335 (CPG70), and PGN_0898 (peptidyl-arginine deiminase, PAD), are translocated across the outer membrane by the T9SS (13, 16, 25).
The CTD region functions as a recognition signal for the T9SS; it also plays a role in the glycosylation of CTD-containing proteins after T9SS-dependent translocation and removal of the CTD region. The cysteine protease PorU (PGN_0022, PG0026) proteolytically removes the CTD from RgpB to produce the mature 48-kDa, soluble form of RgpB (26). HBP35 and TapA are modified by anionic lipopolysaccharide (A-LPS) and are anchored to the bacterial cell surface (13, 25). In addition, a study using a green fluorescent protein-CTD fusion indicated that the CTDs of CPG70, PAD, HBP35, and RgpB play roles in T9SS-dependent translocation and glycosylation (13). A-LPS modification has been proposed to be an anchoring mechanism for linking RgpB to the outer membrane. The deacylase LptO (PGN_0023, PG0027) also is involved in the A-LPS modification of CTD-containing proteins (27). However, the structure of the processing and modification system for CTD-containing proteins has yet to be elucidated.
In general, outer membrane (OM) proteins (OMPs) are synthesized in the cytoplasm with cleavable signal peptides at their N termini and are translocated across the inner membrane through Sec translocation (28). After the signal peptides are processed, OMPs are translocated across the periplasmic space and are inserted into the OM by the β-barrel assembly machinery (BAM) complex (29). Since the OMPs contain amphipathic, antiparallel β-strands that form a barrel structure, the OMPs are prone to aggregation in the periplasm, an aqueous cellular compartment, and need to be conveyed by chaperone proteins, including SurA (peptidyl-prolyl isomerase), Skp (OmpH), and DegP (HtrA) (30). P. gingivalis possesses homologs of SurA, Skp (OmpH), and DegP (HtrA). In the P. gingivalis ATCC 33277 genome, PGN_1550 and PGN_1552 were annotated as encoding SurA and PGN_0391 and PGN_0637 as encoding DegP. PGN_0300 and PGN_0301 were predicted to encode OmpH-like proteins. However, to our knowledge, a relationship between these chaperones and CTD-containing proteins has not yet been reported. We hypothesized that OmpH-like proteins might contribute to the maturation or modification of CTD-containing proteins in P. gingivalis. In this study, we constructed a ΔPGN_0300 mutant to demonstrate our hypothesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used are listed in Table S1 in the supplemental material. Escherichia coli strains were grown aerobically in Luria-Bertani (LB) medium (Nacalai Tesque) at 37°C. P. gingivalis cells were grown anaerobically (10% CO2, 10% H2, and 80% N2), using an anaerobic cabinet (Whitley Workstation DG250; Microbiology International), at 37°C in enriched brain heart infusion (BHI) broth (4), on enriched tryptic soy (TS) agar (4), and on blood agar prepared by adding hemolyzed defibrinated sheep blood (Nippon Bio-Test Laboratories, Inc.) to enriched TS agar at 5%. Antibiotics were used at the following concentrations: ampicillin (Ap; 100 μg/ml for E. coli), erythromycin (Em; 10 μg/ml for P. gingivalis), and tetracycline (Tc; 0.7 μg/ml for P. gingivalis).
Strain construction.
The construction and complementation of P. gingivalis deletion mutants were performed as previously described (4) and are detailed in the supplemental material. The construction of an E. coli strain expressing His6-tagged PorU (Asn800 to Asn1140) is also described in the supplemental material. Primers used in this study are listed in Table S2 in the supplemental material.
Reverse transcription-PCR (RT-PCR).
Total RNA was isolated from P. gingivalis cells using the TRIzol plus RNA purification kit (Life Technologies) according to the manufacturer's instructions. The RNA samples were then incubated with DNase I (TaKaRa Bio) at 37°C for 1 h. DNA contamination was checked by PCR. Two micrograms of total RNA was reverse transcribed into cDNA with a random hexamer primer using the SuperScript III first-strand synthesis system (Life Technologies), and the resulting cDNAs were then used for PCR amplification. The primer sets used were as follows: PGN_0299PF and PGN_0299PR for PGN_0299; PGN_0299PF and PGN_0300PR for the intergenic region between PGN_0299 and PGN_0300; PGN_0300PF and PGN_0300PR for PGN_0300; PGN_0300PF and PGN_0301PR for the intergenic region between PGN_0300 and PGN_0301; and PGN_0301PF and PGN_0301PR for PGN_0301.
Hemagglutination.
The hemagglutination assay was performed as previously described (31). Overnight cultures of P. gingivalis cells grown in enriched BHI broth were centrifuged, washed 3 times with phosphate-buffered saline (PBS), and suspended in PBS at an optical density at 620 nm of 1.3. The bacterial suspensions were then diluted in a 2-fold series with PBS, applied to each well of a round-bottom 96-well microplate, mixed with an equal volume of 1% sheep erythrocytes suspended in PBS, and incubated at room temperature for 3 h.
Protease activity assay.
Kgp and Rgp activity assays were performed as previously described (32) and are detailed in the supplemental material.
Subcellular fractionation.
Subcellular fractionation of P. gingivalis cells was performed according to the method of Sato et al. (14) and is detailed in the supplemental material.
Antibodies.
A peptide derived from the amino acid sequence (Ile93 to Lys106) of PGN_0300 with an N-terminal cysteine residue, CIVKKEQQASELKRK, and conjugated to keyhole limpet hemocyanin was purchased from Operon Biotechnologies. To raise antiserum against the PGN_0300 protein, guinea pigs were immunized with the PGN_0300 peptide by EveBioscience Co., Ltd. PorU-His6 protein was mixed with TiterMax gold (TiterMax), and the mixtures were injected into mice (BALB/c) subcutaneously to obtain anti-PorU antiserum. An anti-Kgp rabbit polyclonal antibody (33), an anti-Rgp mouse polyclonal antibody (33), and an anti-HBP35 rabbit polyclonal antibody (34) were used to detect Kgp, RgpA and RgpB, and HBT35, respectively. A monoclonal antibody (MAb), 1B5, that reacts with A-LPS was kindly provided by M. A. Curtis (35, 36).
Western blotting.
SDS-PAGE was performed according to the method of Laemmli (37). Protease inhibitors (leupeptin and Nα-p-tosyl-l-lysine chloromethyl ketone TLCK) were added to Laemmli solubilizing buffer to avoid proteolysis by endogenous proteases. Separated proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Immobilon P; Millipore), blocked with 5% skim milk in TBST buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) for 1 h at room temperature, and then incubated with primary antibodies overnight at 4°C. After 2 washes with TBST, the blots were incubated with horseradish peroxidase-conjugated secondary antibody IgG for 1 h at room temperature and visualized using ChemiDoc MP (Bio-Rad) after 4 washes with TBST.
Tiling microarray analysis.
Tiling microarray analysis was performed as previously described (14) and is detailed in the supplemental material.
MS analysis and database search for protein identification.
Proteins were identified by peptide mass fingerprinting (PMF) after in-gel tryptic digestion as previously described (14). A gel plug containing proteins was subjected to the following procedures: washing with 50% (vol/vol) acetonitrile, washing with 100% acetonitrile, reduction with 10 mM dithiothreitol (DTT), alkylation with 55 mM iodoacetamide, washing/dehydration with 50% (vol/vol) acetonitrile, and digestion for 10 h with 10 μg/ml trypsin. The resulting peptides were extracted from the gel plug with 0.1% (vol/vol) trifluoroacetic acid–50% (vol/vol) acetonitrile. Digests were spotted onto a matrix-assisted laser desorption ionization (MALDI) target using α-cyano-4-hydroxycinnamic acid as a matrix. Spectra were acquired on a 4800 MALDI tandem time of flight (TOF/TOF) analyzer (Applied Biosystems). Data analysis and MS database searching were performed using GPS Explorer and MASCOT software.
Mouse virulence assay.
The levels of virulence of the P. gingivalis strain W83 and its Δomp17 mutant were determined by mouse subcutaneous infection experiments (25, 38, 39). Bacterial cells were grown at 37°C until an optical density at 590 nm of 1.0 was reached. The cells were harvested and then resuspended and adjusted to a concentration of approximately 1 × 1012 CFU/ml in the enriched BHI broth. Female BALB/c mice (10 weeks of age) were challenged with subcutaneous injections of 0.1 ml of bacterial suspension at two sites on the depilated dorsal surface (0.2 ml per mouse). Injected mice were examined daily for survival. Three sets of experiments were carried out. A total of 15 mice for each group were used.
For data analysis, Kaplan-Meier plots were constructed and the log-rank test was used to evaluate the differences in mean survival rates in three experiments between mice infected with the W83 parent strain and those infected with the mutant strain.
All animal care and experimental procedures were performed in accordance with the Guidelines for Animal Experimentation of Nagasaki University with approval from the Institutional Animal Care and Use Committee.
Statistical analysis.
Statistical analysis of the data was performed using Student's t test, except for the mouse virulence assay.
RESULTS
Localization of PGN_0300 protein.
The P. gingivalis genome possesses two ompH (encoding outer membrane protein H)-like coding sequences (CDSs), PGN_0300 and PGN_0301. The amino acid sequence of the PGN_0300 protein exhibited 22.5% identity with that of PGN_0301 protein (see Fig. S1 in the supplemental material). We analyzed the amino acid sequences of both proteins using the TMHMM Server software, version 2.0 (http://www.cbs.dtu.dk/services/TMHMM//). The results showed that the PGN_0300 protein contained one transmembrane helix at the N terminus (Arg7 to Leu29) and predicted a membrane protein, while the PGN_0301 protein did not contain a transmembrane helix. The SignalP 3.0 Server software (http://www.cbs.dtu.dk/services/SignalP-3.0/) predicted that both the PGN_0300 and the PGN_0301 protein have an N-terminal signal peptide region.
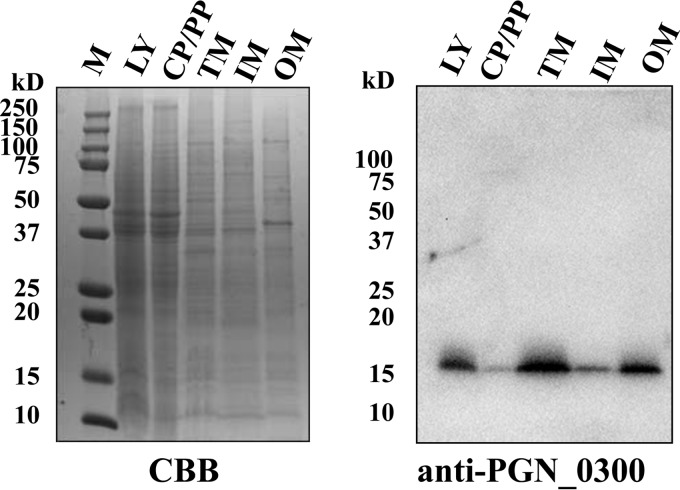
To determine whether the PGN_0300 protein is a membrane protein, cells of the wild-type strain were fractionated into cytoplasm-periplasm, inner membrane, and outer membrane fractions and then were subjected to SDS-PAGE and Western blotting with anti-PGN_0300 protein antiserum (Fig. 1). A 17-kDa immunoreactive protein was detected mainly in the outer membrane fraction and not in the cytoplasm-periplasm fraction. These results suggested that the PGN_0300 protein predominantly localizes to the outer membrane. The mature PGN_0300 protein (148 amino acids) is predicted to be a 17.2-kDa protein, which is in very close agreement with the observed mass (17 kDa). We designated the PGN_0300 protein as outer membrane protein 17 (Omp17) and the PGN_0300 protein-encoding gene as omp17.
FIG 1.
Localization of PGN_0300. P. gingivalis ATCC 33277 cells were subjected to fractionation, followed by SDS-PAGE and immunoblot analysis with anti-PGN_0300 antiserum. M, marker; LY, whole-cell lysate; CP/PP, cytoplasm plus periplasm; TM, total membrane; IM, inner membrane; OM, outer membrane; CBB, Coomassie brilliant blue staining.
Construction of an Omp17-deficient mutant using gene-directed mutagenesis.
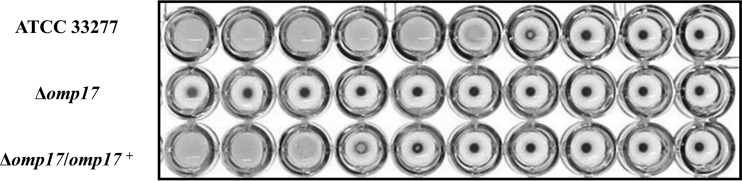
To investigate the role of Omp17, we constructed an Omp17 mutant in which omp17 was replaced with the ermF cassette. The Δomp17 strain showed no pigmentation on blood agar plates (see Fig. S2 in the supplemental material) and no hemagglutination (Fig. 2). To confirm the relationship between omp17 and colonial pigmentation and hemagglutination, we constructed a complemented strain in which the wild-type omp17 gene was introduced into the PGN_1045 locus of the Δomp17 strain. The resulting Δomp17/omp17+ strain showed the same colonial pigmentation and hemagglutination as the wild-type strain (Fig. 2; see also Fig. S2).
FIG 2.
Hemagglutination. P. gingivalis cells were grown in enriched BHI broth, washed with PBS, and resuspended in PBS at an optical density at 620 nm of 1.3. The suspension and a series of 2-fold dilutions were applied, from left to right, to the wells of a microtiter plate and mixed with sheep erythrocyte suspension.
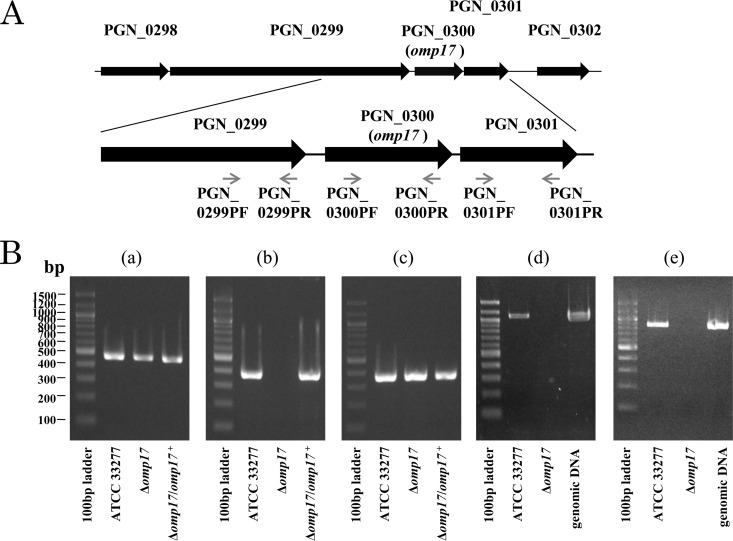
In the P. gingivalis ATCC 33277 genome, the upstream and downstream ends of omp17 are separated from the PGN_0299 CDS by a 57-bp space and the PGN_0301 CDS by a 35-bp space, respectively. This fact suggests that the CDSs PGN_0299, PGN_0300 (omp17), and PGN_0301 form an operon. To test this possibility, we performed RT-PCR using gene-specific primers for the PGN_0299 CDS, omp17, and the PGN_0301 CDS (Fig. 3). We were able to amplify fragments using both of the primer sets PGN_0299PF-PGN_0300PR and PGN_0300PF-PGN_0301PR, suggesting that the PGN_0299 CDS, omp17, and the PGN_0301 CDS form an operon. To confirm whether these CDSs comprise an operon, microarray analysis using a custom tiling DNA array chip with the genome sequence of P. gingivalis ATCC 33277 was performed. The tiling DNA array analysis revealed that these three CDSs formed an operon with the CDSs PGN_0296 to PGN_0298 (see Fig. S3 in the supplemental material). Using the primer set PGN_0300PF-PGN_0300PR, we were able to amplify a PCR fragment using the ATCC 33277 and Δomp17/omp17+ strain genomes but not using the Δomp17 strain genome. When both of the primer sets PGN_0301PF-PGN_0301PR and PGN_0299PF-PGN_0299PR were used, we successfully amplified PCR fragments using the Δomp17 strain genome, as well as the ATCC 33277 genome. These results ruled out the possibility of a polar effect of ermF. Taken together, we concluded that omp17 was correctly knocked out and that the expression of the PGN_0299 and PGN_0301 genes was unaffected at the transcriptional level in the Δomp17 strain.
FIG 3.
Gene organization of the PGN_0300 (omp17) regions of the P. gingivalis ATCC 33277 chromosome. Black arrows indicate the direction of gene transcription. PGN_0299 is separated from omp17 by 57 bp, and omp17 is separated from PGN_0301 by 35 bp. PCR primers were designed to detect the presence of mRNA spanning PGN_0299, omp17, and PGN_0301 (small gray arrows). (B) RT-PCR products indicate that omp17 is cotranscribed with PGN_0299 (d) and PGN_0301 (e). Individual gene and operon products were amplified with unique primer sets. The primer sets used were PGN_0299PF and PGN_0299PR (a), PGN_0300PF and PGN_0300PR (b), PGN_0301PF and PGN_0301PR (c), PGN_0299PF and PGN_0300PR (d), and PGN_0300PF and PGN_0301PR (e). The genomic DNA from P. gingivalis ATCC 33277 was also employed in the experiments whose results are shown in panels d and e.
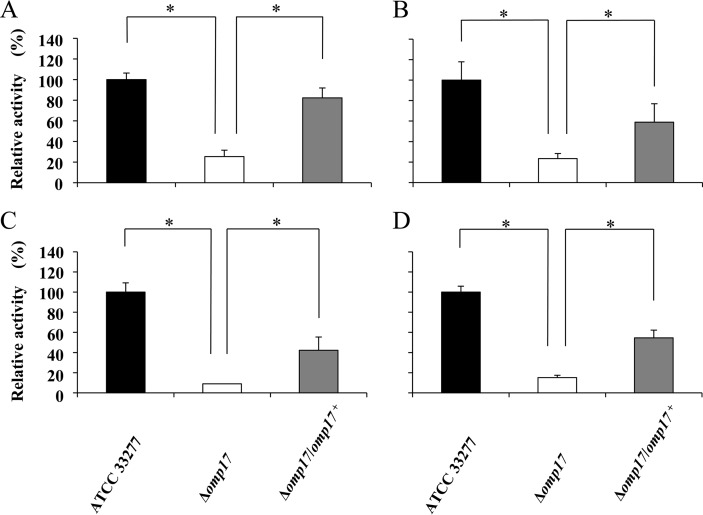
Reduction of gingipain activities in the omp17 mutant.
Several studies have indicated that the cell surface activities of the proteinases Kgp and Rgp are associated with colonial pigmentation on blood agar plates (31, 40, 41). We therefore determined the Kgp and Rgp activities in intact cells and culture supernatants of the Δomp17 strain. The activities of Kgp and Rgp in the Δomp17 strain decreased significantly both in intact cells and in the vesicle-containing culture supernatants. The activities of Kgp and Rgp were moderately restored in the Δomp17/omp17+ strain (Fig. 4). We determined the mRNA levels of kgp, rgpA, and rgpB by quantitative real-time PCR. The mRNA levels of kgp, rgpA, and rgpB in the Δomp17 strain were comparable to those in the wild-type strain, suggesting that a decrease of Rgp and Kgp activities of the Δomp17 strain cannot be explained by altered expression of rgpA, rgpB, and kgp at the transcriptional level (data not shown).
FIG 4.
Rgp and Kgp activities in the omp17 mutant. P. gingivalis cells were grown anaerobically in enriched BHI medium at 37°C for 36 h. Kgp (A and C) and Rgp (B and D) activities of the cell lysates (A and B) and the culture supernatants, including vesicles (C and D), were measured. The statistical significance of the changes was evaluated using a two-sided t test. Error bars indicate standard deviations calculated from the enzyme activity. *, P < 0.05.
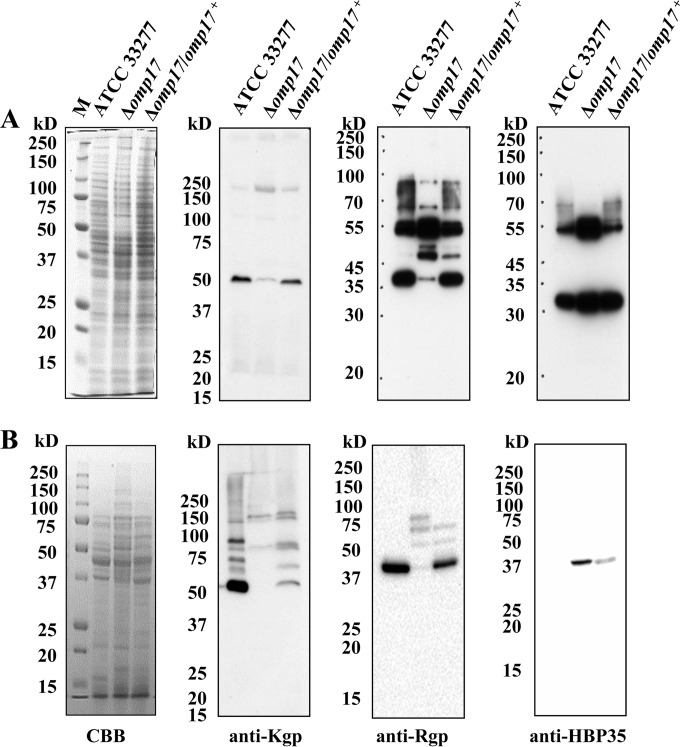
Western blotting using anti-Kgp, anti-Rgp, and anti-HBP35 antibodies.
kgp and rgpA, comprising 5,193-bp and 5,118-bp CDSs, respectively, encode polyproteins that consist of four segments: signal peptide, propeptide, proteinase, and adhesin domains (42, 43). The C-terminal adhesin domains comprise four subdomains (Hgp44/A1, Hgp15 [HbR]/A2, Hgp17/A3, and Hgp27/A4) that are involved in hemagglutination and hemoglobin binding (44–46). The rgpB gene comprises a 2,208-bp CDS and lacks most of the adhesin domain (47). Cultures of the Δomp17 and the wild-type strain were separated into bacterial cells and vesicle-free culture supernatants, and were subjected to Western blotting with anti-Kgp, anti-Rgp, and anti-HBP35 antibodies (Fig. 5). Kgp, RgpAB, and HBP35 belong to CTD-containing proteins that are secreted by T9SS (14). In the wild-type strain, the 190-kDa proprotein and the 50-kDa processed protein that are immunoreactive to anti-Kgp antibody were detected in the whole-cell lysate of the wild-type strain, as well as in the lysates of the Δomp17 and Δomp17/omp17+ strains. However, the 50-kDa processed protein was abundant in the wild-type strain and in the Δomp17/omp17+ strain, whereas the 190-kDa proprotein was detected in the Δomp17 strain. The quantities and molecular masses of anti-Rgp antibody-immunoreactive proteins in the whole-cell lysate were almost the same between the wild-type and the Δomp17/omp17+ strain. The 43-kDa processed protein was the most prominent in the wild-type strain and in the Δomp17/omp17+ strain, whereas the 43-kDa protein was scarcely detected and the 48-kDa protein band was abundant in the Δomp17 strain. In addition, diffuse bands of 70 to 90 kDa that were immunoreactive to anti-Rgp antibody were absent in the whole-cell lysate of the Δomp17 strain.
FIG 5.
Immunoblot analysis of CTD-containing proteins in P. gingivalis cell lysate and culture supernatant. The whole-cell lysate (A) and culture supernatant (B) of P. gingivalis strains were analyzed by SDS-PAGE, followed by immunoblot analyses using anti-Kgp, anti-Rgp, and anti-HBP35 antibodies. M, marker; CBB, Coomassie brilliant blue staining.
In the vesicle-free culture supernatant of the Δomp17 strain, the 140-kDa and 80-kDa intermediate proteins were mainly immunoreactive to anti-Kgp antibody. However, the 50-kDa processed protein, which was most prominent in the wild-type strain, did not react with anti-Kgp antibody. Using anti-Rgp antibody, we detected 3 weak protein bands with molecular masses of 90, 70, and 50 kDa in the Δomp17 strain, whereas the 43-kDa Rgp protein was the most prominent one in the wild-type strain and in the Δomp17/omp17+ strain. No protein was detected in the wild-type strain with anti-HBP35 antibody, whereas a 40-kDa protein was detected in the Δomp17 strain.
These results suggested that CTD-containing proteins were not correctly processed and maturated in the Δomp17 strain.
Identification of proteins in the culture supernatant of the omp17 mutant.
The results described above suggested that the immature forms of CTD-containing proteins were released into the medium in the Δomp17 strain. To test this possibility, proteins in the vesicle-free supernatants of the wild-type and Δomp17 strains were subjected to MALDI-TOF mass analysis. Ten and 17 proteins in the wild-type strain and the Δomp17 strain, respectively, were successfully analyzed (Fig. 6), and the results are shown in Table 1. In the Δomp17 strain, several protein bands that were absent in the wild-type strain were detected. Of these protein bands, no. 11 was identified as PGN_1728 (Kgp), no. 13 and 22 were identified as PGN_0335, and no. 12 and 17 were identified as PGN_1094 and PGN_1476, respectively. All of these proteins were assigned as CTD-containing proteins by sequence analysis. The size of band no. 11 was 120 kDa, which is different from that of mature Kgp, suggesting that this protein was an unprocessed form of Kgp. Indeed, the peptide map showed that this protein contained the propeptide. The peptide map fingerprinting (PMF) data of the protein bands showed that the proteins in bands no. 13 (PGN_0335), no. 17 (PGN_1476), and no. 22 (PGN_0335) contained the CTD (48) (see Fig. S4 in the supplemental material). The cleavage site for the CTD in PGN_1094 is unknown. However, the peptide map showed that the protein in band no. 12 might also contain a CTD, because the sizes of the CTDs were around 70 amino acids. These results indicate that predicted CTD-containing proteins in the culture supernatant of the Δomp17 strain contained CTD and were released from the bacterial cells in the Δomp17 strain.
FIG 6.
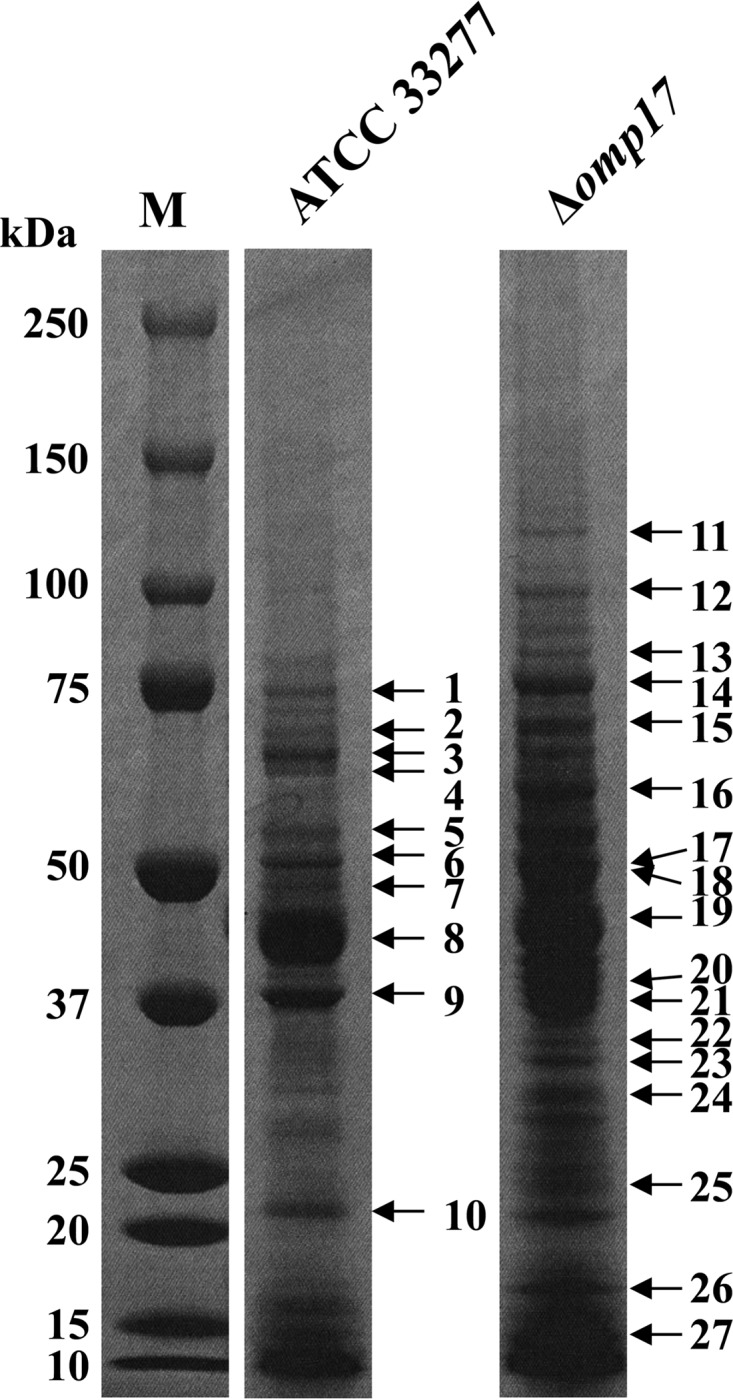
Peptide map fingerprinting (PMF) analyses of proteins in the P. gingivalis culture supernatant. The vesicle-free supernatants of P. gingivalis strain ATCC 33277 and the Δomp17 strain were separated by SDS-PAGE, and the resulting gel was stained with Coomassie brilliant blue. Bands indicated by numbered arrows to the right are identified in Table 1.
TABLE 1.
Identification of protein bands shown in Fig. 6
| Strain | Band | Locus tag | Scoreb | Protein |
|---|---|---|---|---|
| Wild type | 1 | PGN_1349 | 159 | Dipeptidyl aminopeptidase |
| 2 | PGN_1800 | 67 | Urocanate hydratase | |
| 3 | PGN_1349 | 117 | Dipeptidyl aminopeptidase | |
| 4 | PGN_0914 | 98 | Peptidase M24 family | |
| 5 | PGN_1434 | 112 | M20_peptidase d-aminoacyl-histidine dipeptidase | |
| 6 | PGN_0727 | 127 | 4-hydroxybutyryl-CoA dehydratase | |
| 7 | PGN_0294 | 76 | Receptor antigen B (RagB) | |
| 8 | PGN_1367 | 144 | Glutamate dehydrogenase | |
| 9 | PGN_0180 | 70 | FimA | |
| 10 | PGN_0558 | 111 | Outer-membrane hemin utilization (HmuY) | |
| Δomp17 strain | 11 | PGN_1728a | 114 | Lysine-specific cysteine proteinase (Kgp) |
| 12 | PGN_1094a | 129 | Glycine dehydrogenase | |
| 13 | PGN_0335a | 95 | Conserved hypothetical protein with zinc carboxypeptidase domain (CPG70) | |
| 14 | PGN_1349 | 129 | Probable dipeptidyl aminopeptidase | |
| 15 | PGN_1800 | 79 | Urocanate hydratase | |
| 16 | PGN_1452 | 141 | GroEL | |
| 17 | PGN_1476a | 116 | Hypothetical protein | |
| 18 | PGN_1315 | 116 | Hypothetical protein | |
| 19 | PGN_1367 | 95 | Glutamate dehydrogenase | |
| 20 | PGN_0876 | 141 | TPRc domain protein | |
| 21 | PGN_0876 | 62 | TPR domain protein | |
| 22 | PGN_0335a | 92 | Conserved hypothetical protein with zinc carboxypeptidase domain (CPG70) | |
| 23 | PGN_1695 | 184 | Fructose-1,6-bisphosphate aldolase | |
| 24 | PGN_1164 | 76 | Prokaryotic protein of unknown function (DUF849) | |
| 25 | PGN_1587 | 89 | Elongation factor Ts | |
| 26 | PGN_2037 | 84 | DNA-binding protein from starved cells (Dps) | |
| 27 | PGN_1167 | 75 | Hypothetical protein |
CTD-containing protein.
MASCOT score.
TPR, tetratricopeptide repeat.
There is a possibility that the smaller amounts of protein in the vesicle-free supernatant of the wild type were due to proteolytic activities of gingipains. To estimate the contribution of gingipains, the vesicle-free supernatant of the gingipain-deficient (rgpA, rgpB, and kgp) mutant strain KDP136 (41) was subjected to MALDI-TOF mass analysis (see Fig. S5 in the supplemental material). Only five proteins in strain KDP136 were successfully analyzed, suggesting that the smaller amounts of protein in the vesicle-free supernatant of the wild-type strain were not due to activities of gingipains or the quantities of proteins released from cells in the Δomp17 strain. It is notable that two of the five proteins in this analysis contained CTD, but the reason why these two proteins contained CTD is unknown at this time.
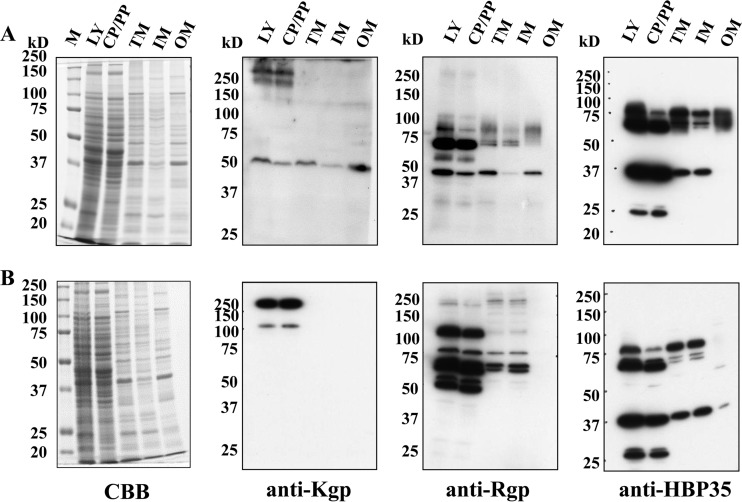
Subcellular localization of Kgp, Rgp, and HBP-35 in the omp17 mutant.
The 50-kDa Kgp protein and the 43-kDa Rgp protein were detected with anti-Kgp antibody and anti-Rgp antibody, respectively, in the whole-cell lysates of the wild-type and Δomp17 strains, and they were detected in the vesicle-free culture supernatant of wild-type strain but not in that of the Δomp17 strain (Fig. 5). These results suggested that the subcellular localization of Kgp and Rgp in the Δomp17 strain was different from their localization in the wild-type strain. Thus, we attempted to determine the subcellular localization of Kgp, Rgp, and HBP35 in the Δomp17 strain (Fig. 7). The 50-kDa Kgp protein was detected in the outer membrane fraction of the wild-type strain. In addition, the protein with a molecular mass of more than 250 kDa and the 190-kDa proprotein were detected with anti-Kgp antibody in both the cytoplasm and the periplasm fraction in the wild-type strain. In the Δomp17 strain, the 50-kDa protein was not detected with anti-Kgp antibody in any fraction, while the 190-kDa proprotein and 120-kDa proprotein were detected with the anti-Kgp antibody in the cytoplasm and periplasm fractions.
FIG 7.
Subcellular localization of CTD-containing proteins. P. gingivalis ATCC 33277 (A) and the Δomp17 strain (B) were subjected to fractionation, followed by SDS-PAGE and immunoblot analysis with anti-Kgp, anti-Rgp, and anti-HBP35 antibodies. M, marker; CP/PP, cytoplasm plus periplasm; TM, total membrane; IM, inner membrane; OM, outer membrane; CBB, Coomassie brilliant blue staining.
The 43-kDa Rgp protein, along with diffuse bands of 60 to 80 kDa, was detected in the outer membrane fraction in the wild-type strain. In the Δomp17 strain, no protein reacted with anti-Rgp antibody in the outer membrane fraction. In the Δomp17 strain, the 120-kDa proprotein, in addition to the 70-kDa and 50-kDa proproteins, strongly reacted with anti-Rgp antibody in the cytoplasm-plus-periplasm fraction.
Diffuse bands of 60 to 80 kDa were detected with anti-HBP35 antibody in the outer membrane fraction of the wild-type strain. The Δomp17 strain showed several faint protein bands that were immunoreactive to anti-HBP35 antibody in the outer membrane fraction.
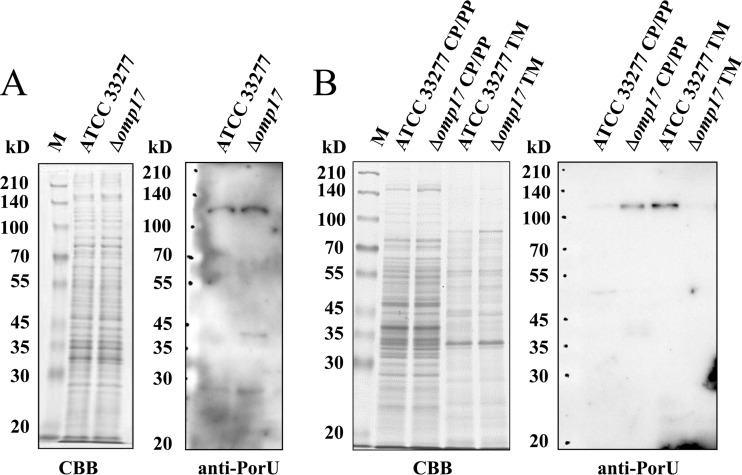
Loss of PorU in the membrane fractions of the omp17 mutant.
Since MALDI-TOF mass analysis indicated that in the Δomp17 strain, immature forms of CTD-containing proteins were released into the medium without cleavage of the CTD, it is possible that PorU is inactive in the Δomp17 strain. To address this possibility, whole-cell lysates, proteins in the cytoplasm-and-periplasm fraction, and proteins in the membrane fraction of wild-type cells and the Δomp17 strain were subjected to Western blotting with anti-PorU antibody. As shown by the results in Fig. 8, anti-PorU antibody reacted with the 120-kDa protein from whole-cell lysates of both the wild-type strain and the Δomp17 strain with equal intensity. Interestingly, anti-PorU antibody reacted with the 120-kDa protein in the membrane fraction of the wild-type strain but not with this fraction of the Δomp17 strain. It clearly reacted with the 120-kDa protein in the cytoplasm and the periplasm fraction of the Δomp17 strain. These data suggest that PorU was not inserted into or translocated across the outer membrane in the omp17 deletion mutant.
FIG 8.
PorU protein is not localized to the membrane fractions of the Δomp17 strain. Whole-cell lysates (A) and the cytoplasm-plus-periplasm and membrane fractions (B) of P. gingivalis ATCC 33277 and the Δomp17 strain were subjected to SDS-PAGE and immunoblot analysis with anti-PorU. M, marker; CP/PP, cytoplasm plus periplasm; TM, total membrane; CBB, Coomassie brilliant blue staining.
Detection of A-LPS in the omp17 mutant.
It is thought that A-LPS glycosylation is required by several CTD-containing proteins in order to anchor to the cell surface after secretion by T9SS. Using MAb 1B5, which recognizes a glycan epitope of anionic polysaccharide bound to lipid A (A-LPS), diffuse bands were detected by Western blot analysis in the Δomp17 strain, as well as in the wild-type and the complemented strain (Fig. 9). However, the bands from the Δomp17 strain showed a lower molecular mass range than did the bands from the wild-type strain, suggesting that A-LPS was synthesized but did not bind to CTD-containing proteins in the Δomp17 strain.
FIG 9.
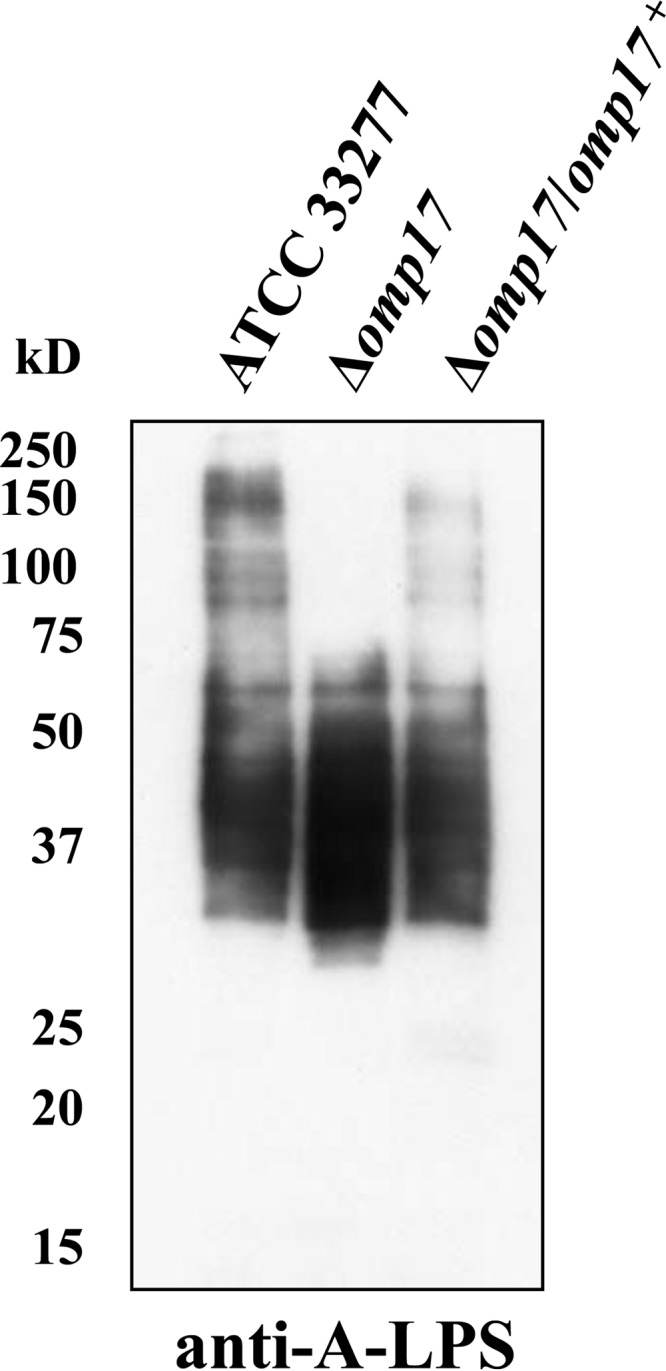
Immunoblot analyses of P. gingivalis cell lysate using MAb 1B5 (anti-A-LPS antibody).
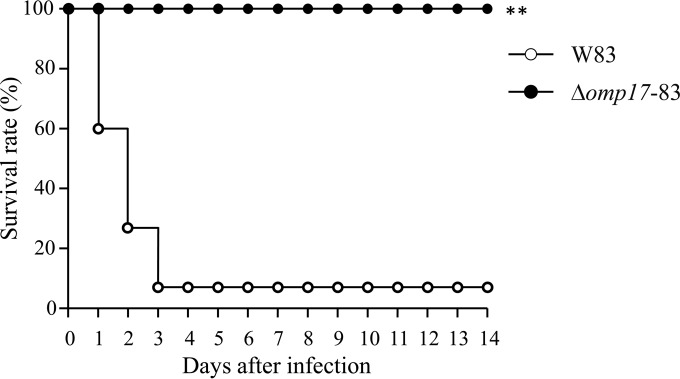
Contribution of omp17 to virulence of P. gingivalis.
BALB/c mice were challenged with subcutaneous injections of bacterial suspension (2 × 1011 CFU per animal), and their survival was monitored for 14 days. About 93.3% of the mice challenged with W83 died within 3 days (Fig. 10). In contrast, mice inoculated with the Δomp17 strain did not die during a 14-day period. The gingipains are major virulence factors of P. gingivalis (10–12), and the activities of gingipains were significantly reduced in the Δomp17 strain. Taken together, the results suggested that omp17 was involved in P. gingivalis virulence and that this involvement might be an indirect effect through the activities of gingipains.
FIG 10.
Survival rates of mice challenged with P. gingivalis strain W83 and the Δomp17 strain (Δomp17-83). BALB/c mice were subcutaneously inoculated with P. gingivalis cells injected into the dorsal surface (2 × 1011 CFU), and then their survival was monitored daily for up to 14 days. The animal experiment, in which five mice were used for each bacterial strain, was performed three times. **, P < 0.05 versus the corresponding values for the wild-type littermates, as determined with the log-rank test.
DISCUSSION
In this study, we demonstrated that an omp17 (PGN_0300) deletion mutant (Δomp17 strain) showed no pigmentation on blood agar plates and no hemagglutination, had reduced Kgp and Rgp activities, and was clearly less virulent than the wild type in mouse subcutaneous infection experiments.
Wild-type strains of P. gingivalis display black pigmentation on blood agar plates. The black pigmentation is caused by the accumulation of μ-oxo heme dimer on the cell surface (49). Numerous nonpigmented mutants have previously been isolated and characterized (31, 50–57). Colonial pigmentation on blood agar plates has been shown to be linked with hemagglutination and the activities of major proteinases. It has been shown that Kgp can degrade hemoglobin protein, which holds heme molecules, and that this degradation correlates with the inability to pigment, as a mutant with the kgp gene was found to display a nonpigmentation phenotype on blood agar plates (12, 40).
Previous studies have indicated that porT (32) and porR (53) are involved in the pigmentation phenotype in P. gingivalis. The porT mutation eliminates gingipain activities on the cell surface or in culture supernatants. In the porT mutant, CTD-containing proteins, including gingipains, are not secreted and accumulate in the periplasmic space. The PorT mutants are defective in the secretion of proteins targeted to the T9SS (14). Conversely, in the porR mutant, gingipain activities on the cell surface are significantly reduced but are retained in the culture supernatant. CTD-containing proteins are secreted from the periplasm to the cell surface, suggesting that the molecules involved in biosynthesis of A-LPS and/or in anchoring the gingipains on the cell surface are defective in the PorR-type mutants. Defects in sugar biosynthesis pathways imposed by inactivation of the vim locus or porR result in the release of non-A-LPS-modified RgpB into the culture medium (53–55, 58, 59).
In addition to the porT- and porR-type mutations, inactivation of lptO (PGN_0023) results in the accumulation of A-LPS and unmodified CTD-containing proteins in the periplasm (27). During the process of crossing the outer membrane and attaching to the cell surface, the CTDs of the CTD-containing proteins are cleaved by the C-terminal signal peptidase PorU (PGN_0022 in ATCC 33277 and PG0026 in strain W83) (26). The cleaved CTD-containing proteins then are attached to A-LPS, resulting in localization of the proteins on the cell surface. P. gingivalis porU deletion mutants and mutants lacking the predicted catalytic Cys residue secrete and accumulate CTD-containing proteins on the cell surface with their CTDs intact and without the A-LPS modification (26). In these cases, the CTD was not released into the surrounding medium in PG0026 mutant strains; moreover, the size of A-LPS was shifted to a lower molecular mass (26).
Components of the T9SS, PorK, PorL, PorM, PorN, PorV, and PorW were localized in the membrane fraction both in the wild-type strain and in the Δomp17 strain, suggesting that the function of the T9SS was not affected by the disruption of omp17 (data not shown). However, PorU was not detected in the membrane fraction but accumulated in the cytoplasm-and-periplasm fraction in the Δomp17 strain. In this mutant, the CTD-containing proteins Kgp, Rgp, and HBP35 were not detected in their processed forms in the outer membrane fraction (Fig. 7B), and their unprocessed forms, containing the CTD region, were released into the medium (Fig. 5 and Table 1). Moreover, the antibodies against Rgp and HBP35 detected diffuse bands, which were A-LPS-modified forms of these proteins, in the outer membrane fraction from the wild-type strain but did not detect bands in any subcellular fraction of the omp17 mutant (Fig. 5 and 7). In addition, A-LPS was present in lower-molecular-mass forms in the omp17 mutant than in the wild-type strain (Fig. 9), which is consistent with the phenotype of the porU mutant. The failure to process and modify CTD-containing proteins in the omp17 mutant was due, at least partly, to the loss of function of PorU. Indeed, PorU was synthesized but was not transported to the outer membrane in the Δomp17 strain.
Gram-negative bacteria possess several chaperone molecules, including SurA, Skp (OmpH), DegP (HtrA), FkpA, and LolA, in the periplasmic space. It is thought that the roles of these chaperone molecules are to stabilize the nonnative conformations of OMPs, to prevent their aggregation, and to facilitate their folding (reviewed in reference 60). As a chaperone, Skp is involved in the assembly of OMPs and assists the early steps of β-barrel folding of OMPs (61, 62). In an E. coli skp null mutant, envelope protein levels are dramatically reduced (63). Mutations in both skp and surA have a bacteriostatic effect and lead to filamentation (63).
In the P. gingivalis ATCC 33277 genome, the PGN_0300 (omp17) and PGN_0301 CDSs were predicted to encode OmpH-like domain proteins. The results of the analysis using TMHMM Server version 2.0 indicated that the PGN_0301 protein did not possess any transmembrane region, like the Skp protein of E. coli, which localizes in the periplasmic space. It is notable that we have not yet been able to construct a strain with a deletion of the PGN_0301 CDS, suggesting that PGN_0301 generates a protein that is essential for P. gingivalis cells.
On the other hand, Omp17 was suggested to be an OMP in our analysis. OmpH-like proteins are exposed on the surface in several bacteria, such as Flavobacterium psychrophilum (64) and Yersinia pseudotuberculosis (65); however, the functions of OmpH-like proteins have not been elucidated. Based on its localization, Omp17 may function differently from the Skp protein of E. coli, but the PGN_0301 protein may possess a function similar to that of E. coli's Skp. It is feasible that Omp17 is involved in the translocation of PorU to the outer membrane, as a molecular chaperone.
If Omp17 worked as a molecular chaperone, another possibility would be raised. The CTD functions as a recognition signal for the T9SS, and glycosylation of CTD-containing proteins occurs after the removal of the CTD region. Zhou et al. demonstrated that cleavage of the CTD is not dependent on a specific residue and that recognition of the cleavage site is independent of the local amino acid sequence (66). They proposed that the volume and conformation of the residues surrounding the cleavage site are required during the outer membrane translocation step to allow for efficient cleavage by PorU. Furthermore, the length of the junction between the CTD and the adjacent Ig-like subdomain has a critical influence on posttranslational glycan modification of the protein, suggesting that a conformational motif is required for digesting the CTDs and for further modification of CTD-containing proteins.
Although we did not determine the conformation of CTD-containing proteins, there was a possibility that these proteins had an alternative conformation that impaired efficient processing of the CTD. We could not rule out this possibility. Further work will be required to test this hypothesis and to reveal the roles of Omp17 and PGN_0301, including whether they possess chaperone activity.
In conclusion, our findings indicate that Omp17 contributes to the processing and modification of CTD-containing proteins, possibly by reducing the levels of PorU in the outer membrane. To our knowledge, this is the first case of a nonpigmented mutant of P. gingivalis that was not caused by blocked secretion, loss of function in the gene encoding the CTD cleavage enzyme, or the absence of A-LPS on the cell surface.
Supplementary Material
Funding Statement
This work was supported in part by a Grant-in-Aid for Science Research from the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan and by the Association for Research on Lactic Acid Bacteria.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01308-15.
REFERENCES
- 1.Page RC, Offenbacher S, Schroeder HE, Seymour GJ, Kornman KS. 1997. Advances in the pathogenesis of periodontitis: summary of developments, clinical implications and future directions. Periodontol 2000 14:216–248. doi: 10.1111/j.1600-0757.1997.tb00199.x. [DOI] [PubMed] [Google Scholar]
- 2.Armitage GC. 1996. Periodontal diseases: diagnosis. Ann Periodontol 1:37–215. doi: 10.1902/annals.1996.1.1.37. [DOI] [PubMed] [Google Scholar]
- 3.Holt SC, Ebersole JL. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000 38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 4.Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. 1995. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J Biol Chem 270:23619–23626. [DOI] [PubMed] [Google Scholar]
- 5.Potempa J, Pike R, Travis J. 1995. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect Immun 63:1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadowaki T, Nakayama K, Okamoto K, Abe N, Baba A, Shi Y, Ratnayake DB, Yamamoto K. 2000. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J Biochem 128:153–159. doi: 10.1093/oxfordjournals.jbchem.a022735. [DOI] [PubMed] [Google Scholar]
- 7.Andrian E, Mostefaoui Y, Rouabhia M, Grenier D. 2007. Regulation of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases by Porphyromonas gingivalis in an engineered human oral mucosa model. J Cell Physiol 211:56–62. doi: 10.1002/jcp.20894. [DOI] [PubMed] [Google Scholar]
- 8.Curtis MA, Kuramitsu HK, Lantz M, Macrina FL, Nakayama K, Potempa J, Reynolds EC, Aduse-Opoku J. 1999. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J Periodont Res 34:464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 9.Potempa J, Banbula A, Travis J. 2000. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol 2000 24:153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. [DOI] [PubMed] [Google Scholar]
- 10.O'Brien-Simpson NM, Paolini RA, Hoffmann B, Slakeski N, Dashper SG, Reynolds EC. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect Immun 69:7527–7534. doi: 10.1128/IAI.69.12.7527-7534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pathirana RD, O'Brien-Simpson NM, Brammar GC, Slakeski N, Reynolds EC. 2007. Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect Immun 75:1436–1442. doi: 10.1128/IAI.01627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtis MA, Aduse Opoku J, Rangarajan M, Gallagher A, Sterne JA, Reid CR, Evans HE, Samuelsson B. 2002. Attenuation of the virulence of Porphyromonas gingivalis by using a specific synthetic Kgp protease inhibitor. Infect Immun 70:6968–6975. doi: 10.1128/IAI.70.12.6968-6975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, Naito M, Nakayama K. 2011. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6:e21372. doi: 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A 107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayama K. 2015. Porphyromonas gingivalis and related bacteria: from colonial pigmentation to the type IX secretion system and gliding motility. J Periodont Res 50:1–8. doi: 10.1111/jre.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato K, Yukitake H, Narita Y, Shoji M, Naito M, Nakayama K. 2013. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol Lett 338:68–76. doi: 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- 17.McBride MJ, Zhu Y. 2013. Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J Bacteriol 195:270–278. doi: 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun TF, Khubbar MK, Saffarini DA, McBride MJ. 2005. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J Bacteriol 187:6943–6952. doi: 10.1128/JB.187.20.6943-6952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes RG, Samarasam MN, Shrivastava A, van Baaren JM, Pochiraju S, Bollampalli S, McBride MJ. 2010. Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB. J Bacteriol 192:1201–1211. doi: 10.1128/JB.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kharade SS, McBride MJ. 2015. Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J Bacteriol 197:147–158. doi: 10.1128/JB.02085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seers CA, Slakeski N, Veith PD, Nikolof T, Chen YY, Dashper SG, Reynolds EC. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J Bacteriol 188:6376–6386. doi: 10.1128/JB.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen KA, Travis J, Potempa J. 2007. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of gram-negative bacteria? J Bacteriol 189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomek MB, Neumann L, Nimeth I, Koerdt A, Andesner P, Messner P, Mach L, Potempa JS, Schaffer C. 2014. The S-layer proteins of Tannerella forsythia are secreted via a type IX secretion system that is decoupled from protein O-glycosylation. Mol Oral Microbiol 29:307–320. doi: 10.1111/omi.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narita Y, Sato K, Yukitake H, Shoji M, Nakane D, Nagano K, Yoshimura F, Naito M, Nakayama K. 2014. Lack of a surface layer in Tannerella forsythia mutants deficient in the type IX secretion system. Microbiology 160:2295–2303. doi: 10.1099/mic.0.080192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo Y, Ohara N, Sato K, Yoshimura M, Yukitake H, Naito M, Fujiwara T, Nakayama K. 2010. Tetratricopeptide repeat protein-associated proteins contribute to the virulence of Porphyromonas gingivalis. Infect Immun 78:2846–2856. doi: 10.1128/IAI.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds EC. 2012. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J Biol Chem 287:24605–24617. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen YY, Peng B, Yang Q, Glew MD, Veith PD, Cross KJ, Goldie KN, Chen D, O'Brien-Simpson N, Dashper SG, Reynolds EC. 2011. The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol Microbiol 79:1380–1401. doi: 10.1111/j.1365-2958.2010.07530.x. [DOI] [PubMed] [Google Scholar]
- 28.Pugsley AP. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev 57:50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricci DP, Silhavy TJ. 2012. The Bam machine: a molecular cooper. Biochim Biophys Acta 1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sklar JG, Wu T, Kahne D, Silhavy TJ. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi M, Sato K, Yukitake H, Noiri Y, Ebisu S, Nakayama K. 2010. A Porphyromonas gingivalis mutant defective in a putative glycosyltransferase exhibits defective biosynthesis of the polysaccharide portions of lipopolysaccharide, decreased gingipain activities, strong autoaggregation, and increased biofilm formation. Infect Immun 78:3801–3812. doi: 10.1128/IAI.00071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato K, Sakai E, Veith PD, Shoji M, Kikuchi Y, Yukitake H, Ohara N, Naito M, Okamoto K, Reynolds EC, Nakayama K. 2005. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J Biol Chem 280:8668–8677. doi: 10.1074/jbc.M413544200. [DOI] [PubMed] [Google Scholar]
- 33.Takii R, Kadowaki T, Baba A, Tsukuba T, Yamamoto K. 2005. A functional virulence complex composed of gingipains, adhesins, and lipopolysaccharide shows high affinity to host cells and matrix proteins and escapes recognition by host immune systems. Infect Immun 73:883–893. doi: 10.1128/IAI.73.2.883-893.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abiko Y, Hayakawa M, Aoki H, Kikuchi T, Shimatake H, Takiguchi H. 1990. Cloning of a Bacteroides gingivalis outer membrane protein gene in Escherichia coli. Arch Oral Biol 35:689–695. doi: 10.1016/0003-9969(90)90091-N. [DOI] [PubMed] [Google Scholar]
- 35.Curtis MA, Thickett A, Slaney JM, Rangarajan M, Aduse-Opoku J, Shepherd P, Paramonov N, Hounsell EF. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect Immun 67:3816–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rangarajan M, Aduse-Opoku J, Paramonov N, Hashim A, Bostanci N, Fraser OP, Tarelli E, Curtis MA. 2008. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J Bacteriol 190:2920–2932. doi: 10.1128/JB.01868-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Yoshimura M, Ohara N, Kondo Y, Shoji M, Okano S, Nakano Y, Abiko Y, Nakayama K. 2008. Proteome analysis of Porphyromonas gingivalis cells placed in a subcutaneous chamber of mice. Oral Microbiol Immunol 23:413–418. doi: 10.1111/j.1399-302X.2008.00444.x. [DOI] [PubMed] [Google Scholar]
- 39.Neiders ME, Chen PB, Suido H, Reynolds HS, Zambon JJ, Shlossman M, Genco RJ. 1989. Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodont Res 24:192–198. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto K, Nakayama K, Kadowaki T, Abe N, Ratnayake DB, Yamamoto K. 1998. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J Biol Chem 273:21225–21231. doi: 10.1074/jbc.273.33.21225. [DOI] [PubMed] [Google Scholar]
- 41.Shi Y, Ratnayake DB, Okamoto K, Abe N, Yamamoto K, Nakayama K. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem 274:17955–17960. [DOI] [PubMed] [Google Scholar]
- 42.Okamoto K, Kadowaki T, Nakayama K, Yamamoto K. 1996. Cloning and sequencing of the gene encoding a novel lysine-specific cysteine proteinase (Lys-gingipain) in Porphyromonas gingivalis: structural relationship with the arginine-specific cysteine proteinase (Arg-gingipain). J Biochem 120:398–406. doi: 10.1093/oxfordjournals.jbchem.a021426. [DOI] [PubMed] [Google Scholar]
- 43.Pavloff N, Potempa J, Pike RN, Prochazka V, Kiefer MC, Travis J, Barr PJ. 1995. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J Biol Chem 270:1007–1010. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama K, Ratnayake DB, Tsukuba T, Kadowaki T, Yamamoto K, Fujimura S. 1998. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol Microbiol 27:51–61. doi: 10.1046/j.1365-2958.1998.00656.x. [DOI] [PubMed] [Google Scholar]
- 45.Sakai E, Naito M, Sato K, Hotokezaka H, Kadowaki T, Kamaguchi A, Yamamoto K, Okamoto K, Nakayama K. 2007. Construction of recombinant hemagglutinin derived from the gingipain-encoding gene of Porphyromonas gingivalis, identification of its target protein on erythrocytes, and inhibition of hemagglutination by an interdomain regional peptide. J Bacteriol 189:3977–3986. doi: 10.1128/JB.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slakeski N, Seers CA, Ng K, Moore C, Cleal SM, Veith PD, Lo AW, Reynolds EC. 2011. C-terminal domain residues important for secretion and attachment of RgpB in Porphyromonas gingivalis. J Bacteriol 193:132–142. doi: 10.1128/JB.00773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakayama K. 1997. Domain-specific rearrangement between the two Arg-gingipain-encoding genes in Porphyromonas gingivalis: possible involvement of nonreciprocal recombination. Microbiol Immunol 41:185–196. doi: 10.1111/j.1348-0421.1997.tb01189.x. [DOI] [PubMed] [Google Scholar]
- 48.Veith PD, Nor Muhammad NA, Dashper SG, Likic VA, Gorasia DG, Chen D, Byrne SJ, Catmull DV, Reynolds EC. 2013. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J Proteome Res 12:4449–4461. doi: 10.1021/pr400487b. [DOI] [PubMed] [Google Scholar]
- 49.Smalley JW, Silver J, Marsh PJ, Birss AJ. 1998. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the μ-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem J 331(Pt 3):681–685. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen T, Dong H, Yong R, Duncan MJ. 2000. Pleiotropic pigmentation mutants of Porphyromonas gingivalis. Microb Pathog 28:235–247. doi: 10.1006/mpat.1999.0338. [DOI] [PubMed] [Google Scholar]
- 51.Hoover CI, Yoshimura F. 1994. Transposon-induced pigment-deficient mutants of Porphyromonas gingivalis. FEMS Microbiol Lett 124:43–48. doi: 10.1111/j.1574-6968.1994.tb07259.x. [DOI] [PubMed] [Google Scholar]
- 52.Sato K, Kido N, Murakami Y, Hoover CI, Nakayama K, Yoshimura F. 2009. Lipopolysaccharide biosynthesis-related genes are required for colony pigmentation of Porphyromonas gingivalis. Microbiology 155:1282–1293. doi: 10.1099/mic.0.025163-0. [DOI] [PubMed] [Google Scholar]
- 53.Shoji M, Ratnayake DB, Shi Y, Kadowaki T, Yamamoto K, Yoshimura F, Akamine A, Curtis MA, Nakayama K. 2002. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 148:1183–1191. doi: 10.1099/00221287-148-4-1183. [DOI] [PubMed] [Google Scholar]
- 54.Vanterpool E, Roy F, Fletcher HM. 2005. Inactivation of vimF, a putative glycosyltransferase gene downstream of vimE, alters glycosylation and activation of the gingipains in Porphyromonas gingivalis W83. Infect Immun 73:3971–3982. doi: 10.1128/IAI.73.7.3971-3982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vanterpool E, Roy F, Sandberg L, Fletcher HM. 2005. Altered gingipain maturation in vimA- and vimE-defective isogenic mutants of Porphyromonas gingivalis. Infect Immun 73:1357–1366. doi: 10.1128/IAI.73.3.1357-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vanterpool E, Roy F, Zhan W, Sheets SM, Sangberg L, Fletcher HM. 2006. VimA is part of the maturation pathway for the major gingipains of Porphyromonas gingivalis W83. Microbiology 152:3383–3389. doi: 10.1099/mic.0.29146-0. [DOI] [PubMed] [Google Scholar]
- 57.Shoji M, Yukitake H, Sato K, Shibata Y, Naito M, Aduse-Opoku J, Abiko Y, Curtis MA, Nakayama K. 2013. Identification of an O-antigen chain length regulator, WzzP, in Porphyromonas gingivalis. Microbiologyopen 2:383–401. doi: 10.1002/mbo3.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanterpool E, Roy F, Fletcher HM. 2004. The vimE gene downstream of vimA is independently expressed and is involved in modulating proteolytic activity in Porphyromonas gingivalis W83. Infect Immun 72:5555–5564. doi: 10.1128/IAI.72.10.5555-5564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shoji M, Sato K, Yukitake H, Naito M, Nakayama K. 2014. Involvement of the Wbp pathway in the biosynthesis of Porphyromonas gingivalis lipopolysaccharide with anionic polysaccharide. Sci Rep 4:5056. doi: 10.1038/srep05056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goemans C, Denoncin K, Collet JF. 2014. Folding mechanisms of periplasmic proteins. Biochim Biophys Acta 1843:1517–1528. doi: 10.1016/j.bbamcr.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 61.Walther DM, Rapaport D, Tommassen J. 2009. Biogenesis of β-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence. Cell Mol Life Sci 66:2789–2804. doi: 10.1007/s00018-009-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tommassen J. 2010. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology 156:2587–2596. doi: 10.1099/mic.0.042689-0. [DOI] [PubMed] [Google Scholar]
- 63.Rizzitello AE, Harper JR, Silhavy TJ. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol 183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dumetz F, Duchaud E, LaPatra SE, Le Marrec C, Claverol S, Urdaci MC, Le Henaff M. 2006. A protective immune response is generated in rainbow trout by an OmpH-like surface antigen (P18) of Flavobacterium psychrophilum. Appl Environ Microbiol 72:4845–4852. doi: 10.1128/AEM.00279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vuorio R, Hirvas L, Raybourne RB, Yu DT, Vaara M. 1991. The nucleotide and deduced amino acid sequence of the cationic 19 kDa outer membrane protein OmpH of Yersinia pseudotuberculosis. Biochim Biophys Acta 1129:124–126. doi: 10.1016/0167-4781(91)90226-C. [DOI] [PubMed] [Google Scholar]
- 66.Zhou XY, Gao JL, Hunter N, Potempa J, Nguyen KA. 2013. Sequence-independent processing site of the C-terminal domain (CTD) influences maturation of the RgpB protease from Porphyromonas gingivalis. Mol Microbiol 89:903–917. doi: 10.1111/mmi.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.