Abstract
Purpose
EphA2, a member of the Eph receptor tyrosine kinases family, is an important regulator of tumour initiation, neo-vascularization and metastasis in a wide range of epithelial and mesenchymal cancers, however its role in colorectal cancer (CRC) recurrence and progression is unclear.
Experimental Design
EphA2 expression was determined by immunohistochemistry in stage II/III colorectal tumours (N=338), and findings correlated with clinical outcome. The correlation between EphA2 expression and stem cell markers CD44 and Lgr5 was examined. The role of EphA2 in migration/invasion was assessed using a panel of KRAS wild-type (WT) and mutant (MT) parental and invasive CRC cell line models.
Results
Colorectal tumours displayed significantly higher expression levels of EphA2 compared with matched normal tissue, which positively correlated with high CD44 and Lgr5 expression levels. Moreover, high EphA2 mRNA and protein expression were found to be associated with poor overall survival in stage II/III CRC tissues, in both univariate and multivariate analyses. Pre-clinically, we found that EphA2 was highly expressed in KRASMT CRC cells and that EphA2 levels are regulated by the KRAS-driven MAPK and RalGDS-RalA pathways. Moreover, EphA2 levels were elevated in several invasive daughter cell lines and down-regulation of EphA2 using RNAi or recombinant EFNA1, suppressed migration and invasion of KRASMT CRC cells.
Conclusions
These data show that EpHA2 is a poor prognostic marker in stage II/III CRC, which may be due to its ability to promote cell migration and invasion, providing support for the further investigation of EphA2 as a novel prognostic biomarker and therapeutic target.
Keywords: EphA2, invasion, biomarker, colorectal cancer
INTRODUCTION
Despite improvements in overall survival (OS) with the introduction of adjuvant 5-FU/folinic acid/oxaliplatin treatment for patients with locally advanced stage II and III colorectal cancer (CRC), the management of these patients remains an area of active clinical debate. Although the molecular targeted agents bevacizumab, cetuximab, panitumumab, aflibercept and ramucrirumab have each improved outcome of CRC patients with metastatic disease, no benefit from these agents have been seen in stage II/III CRC (1, 2). Understanding the molecular mechanisms of disease progression and resistance to available treatments, with the subsequent development and validation of novel therapeutic strategies, is therefore urgently required in order to develop stratified medicine approaches that individualise patient treatment in stage II/III disease.
EphA2 is a 130kDa glycoprotein receptor and belongs to the largest family of receptor tyrosine kinases (RTK), the Eph family. The Eph family can be divided into two subclasses, EphA and EphB, based on structural homology and affinity for binding either the GPI-anchored Ephrin-A or the transmembrane Ephrin-B ligands (3). Both EphA and EphB contain an extracellular region with an Ephrin binding domain, an epidermal growth factor-like motif and two fibronectin type-III domains as well as a cytoplasmic region including a juxtamembrane segment, the kinase domain, a sterile α-motif (SAM) and a PDZ binding motif (Post synaptic density protein, Drosophila disc large tumor suppressor, and Zonula occludens-1 protein). Eph-Ephrin complexes emanate bidirectional signaling into both Eph expressing cells (forward signaling) and Ephrin-expressing cells (reverse signaling) (4). In addition to ligand-dependent receptor activation, some studies have shown that even without ligands, Eph receptors, such as EphA2, can form Eph-Eph homodimers and oligomers thus facilitating the formation of signaling Ephrin-Eph heterotetramers (5, 6). A number of downstream signaling pathways have been linked to Ephrin-Eph complexes, including RAS/MAPK, FAK/SRC, ABL, RHO/RAC/CDC42 and PI3K/AKT/mTOR (7).
Among the different EphA receptors, the role of EphA2 in malignant transformation of normal cells, angiogenesis and metastasis has been studied extensively in cancers, particularly in breast, melanoma, glioblastoma and non-small cell lung carcinoma (NSCLC) (8-11). The role of EphA2 in invasion/migration and as a potential biomarker and therapeutic target in CRC is however unclear.
In this study, we demonstrate that EphA2 is overexpressed in invasive CRC models and correlates with increased expression of the stem cell marker CD44 in vitro and in clinical samples. We also show that EphA2 expression levels are regulated by KRAS through both the MAPK and RalGDS-RalA pathway and that treatment with EphA2-specific siRNA or recombinant human EFNA1 (rhEFNA1) abrogates migration/invasion of KRASMT CRC cells. In addition, EphA2 expression was found to be a prognostic biomarker of poor OS in stage II/III CRC. Taken together, our results indicate that EphA2-targeted therapies may represent a promising novel strategy to prevent disease recurrence and progression in stage II/III CRC.
MATERIALS AND METHODS
Materials
MG132 and cycloheximide were obtained from Sigma-Aldrich (Dorset, UK), Trametinib (GSK1120212) and Vemurafenib (PLX4032) from Selleck Chemicals LLC (Suffolk, UK), rhEFNA1 from R&D systems (Abingdon, UK). siRNAs targeting EphA2, EFNA1, KRAS, RalGDS, RalA, RalB, PLCε, TIAM1, PIK3CA were purchased from Qiagen (Crawley, UK). siRNAs targeting AKT1, AKT2, AKT3, p53, p63, p21, c-Src, YES, FYN were obtained from Dharmacon (Chicago, USA). The EphA2 plasmid was a kind gift from Dr. Koichi Miura (Osaka, Japan) (12).
Cell culture
Authentication and culture of HCT116, HKH-2, DLD-1, Dks-8, LoVo, LS174T, SW620, GP5D, RKO, WiDR, HT-29, LIM2405 and CACO-2 CRC cells have previously been described (13-15). COLO205 and CCD-18Co cells (2012) were obtained from the American Type Culture Collection (Authentication by short tandem repeat (STR) profiling/karyotyping/isoenzyme analysis) and maintained in RPMI and EMEM respectively. HCT116-p53 wild type, HCT116-p53 null (−/−), HCT116-p53 (+/−), HCT116-p53 (R248W/−) and HCT116-p53 (R248W/+) cells were provided by B. Vogelstein (Johns Hopkins University School of Medicine, Baltimore). The DiFi and OXCO-2 cells were received from Dr. Di Nicolantonio in March 2015 (University of Torino, Italy).
Western blotting
Western blot analysis has previously been described (14, 16). Anti-EphA2 (Invitrogen), anti-RalA (BD Transduction Laboratories, Oxfordshire, UK), anti-CBL (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-p53 (Santa Cruz Biotechnology) mouse monoclonal antibodies were used in conjunction with a HRP-conjugated sheep anti-mouse secondary antibody. Anti-pEphA2Y588, anti-pEphA2S897, anti-pEphA2Y772, anti-RalB, c-Src, YES, and FYN (Cell Signaling) rabbit polyclonal antibodies were used in conjunction with a HRP-conjugated anti-rabbit secondary antibody. Anti-EphA2 (Cell Signaling) and anti-pTyrosine (Cell Signaling) rabbit antibodies were used for co-immunoprecipitation (IP).
In vitro Migration and Invasion assays
Cell migration and invasion rates were performed as previously described (17).
Immunofluorescence
Immunofluorescence has previously been described (17). Anti-EphA2 (Invitrogen, 1:500) mouse monoclonal antibody was used.
Transwell indirect co-culture
CRC cell – fibroblast indirect co-culture was carried out using a Falcon® permeable support for 6 well plates with a 0.4μm transparent PET membrane and support companion plates.
Real-time reverse transcription-PCR analysis
RNA was isolated using the GeneJET RNA purification kit (Thermo Scientific, Leicestershire, UK) and reverse transcribed using the Moloney murine leukemia virus-based reverse transcriptase kit (Invitrogen). Q-PCR analysis was performed using the LightCycler® 480 probes master mix (LightCycler® 480II, Roche).
siRNA transfections
siRNA transfections were performed as previously described (13).
Generation of inducible EphA2-silenced CRC cell lines
Inducible EphA2-silenced HCT116 cells were generated as previously described (17). A pTRIPZ plasmid encoding Tet-inducible shRNA against EphA2 was used (Open Biosystems, Lafayette, United States).
Clinical-pathological data
The study cohort consisted of 509 stage I-IV CRC cases who received resection of the primary tumour at the National University Hospital of Singapore between 1990 and 1999 (18). The final dataset for survival analysis consisted of 335 stage II/III patients. The available clinical and pathological details (17), construction of the tissue microarray and methods of immunohistochemistry (IHC) for CD133 (18), Ki-67 (18), CD44 (19), LGR5 (20) and AXL (17) have previously been described and ethically approved for research (NUS-IRB 131–05-017). The TMA contained 1 core per colorectal tumour. In this study, we used anti-EphA2 antibody (Mouse monoclonal, Invitrogen, 1:100). Staining intensity was graded as 0 (no staining), 1 (weak staining), 2 (moderate staining) and 3 (strong staining); categories 0, 1 were classified EphA2-low, categories 2, 3 as EphA2-high. Scoring was done independently by Tingting Wang and Supriya Srivastava, who were both blinded to clinical outcome. In addition to EphA2 levels, OS and survival status (death by any cause was considered an event), age, gender, tumour size, ethnic group (Chinese/non-Chinese), invasion (either perineural and/or lymphatic and/or vascular), differentiation (1, 2 or 3), tumour site (rectal or colon), chemotherapy status and staging were available for each patient. Patients with an event occurring less than three months post-resection were excluded from the analysis, resulting in a revised stage II/III dataset of 313 patients (Supplementary Table S1A and S1B).
EphA2 expression in normal colonic epithelium and CRC was analysed using a tissue microarray (TMA) consisting of cores representing colorectal adenocarcinoma with matched normal colon tissue from 211 stage II/III CRC patients (21). This work was approved by the Office for Research Ethics Committees Northern Ireland (08/NIR02/105).
Validation cohort
An independent validation dataset was identified and the normalised, log-transformed data was obtained from the Gene Expression Omnibus (GEO) database, accession number GSE17536 (22, 23). The stage II/III patients were selected (n=114) and the probe set corresponding to EphA2 identified (203499_at). The distribution with respect to EphA2 from the main study (Stage II/III: Low: 49%; High: 51%) was applied to the remaining patients, a patient was therefore labelled as EphA2-low or EphA2-high. The factors age and stage were also extracted as was OS and survival status.
Statistical analysis
The unpaired two-tailed student’s t-test was used to determine statistically significant differences between treatment effects and calculated using GraphPad Prism version 5 for Windows (GraphPad Software, La Jolla California USA). Significance was defined as p<0.05. The non-parametric tau test was used to determine the Kendall rank correlation coefficient as a measure of association between EphA2 expression and other markers. Subsequently, the Fisher’s exact test and Bonferroni method (Statistical Package for the Social Sciences; SPSS) was used to determine the significance of the association. Methods on survival and univariate/multivariate analysis of Singapore data-set and validation cohort are further described in the supplementary methods.
RESULTS
Oncogenic KRAS is associated with high EphA2 expression levels in CRC cell lines
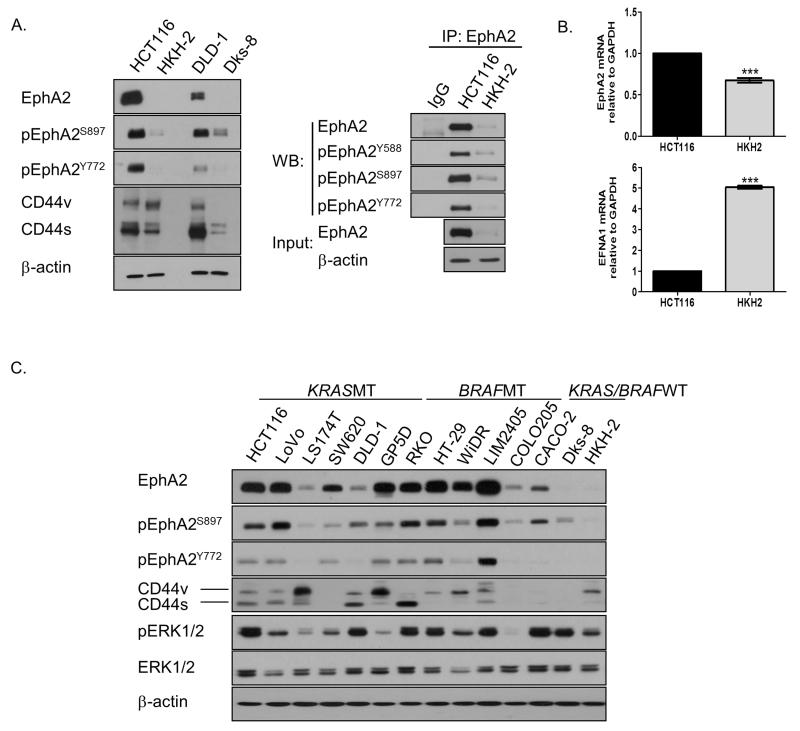
Previous KRAS siRNA screens from our lab have identified the RTK EphA2 as a potential KRAS target gene in KRASMT CRC cells (15). Based on these results, we analysed EphA2 expression and phosphorylation levels in the KRASMT HCT116 and DLD-1 and KRASWT HKH-2 and Dks-8 isogenic paired cell line models using Western blotting and co-immunoprecipitation (24). Constitutive EphA2 levels were significantly higher in the KRASMT HCT116 and DLD-1 cells compared to their KRASWT counterparts (Fig. 1A; Supplementary Fig. S1A). Phosphorylation levels of EphA2 at Y772 (tyrosine residue within the activation loop), S897 (ligand-independent serine residue) and Y588 (tyrosine residue located in the juxtamembrane region which controls its kinase activity) were likewise higher in KRASMT cells compared to their isogenic WT clones, suggesting that EphA2 is actively signaling in the KRASMT models (25-27). We also found increased EphA2 mRNA levels in the HCT116 cells compared to its WT clone (Fig. 1B). Moreover, significantly higher mRNA levels of EFNA1 (a major ligand for EphA2) were found in the KRASWT HKH-2 cell line, consistent with previous data showing an inverse correlation between expression of EphA2 and EFNA1 in breast cancer cells (Fig. 1B) (28). Silencing of EFNA1 markedly increased EphA2 levels in HCT116 and HKH-2 cells, indicating that EFNA1 negatively regulates EphA2 levels in CRC (Supplementary Fig. S1B). Furthermore, the half-life of EphA2 was markedly reduced in the HKH-2 cells, with no change and a 72% reduction in EphA2 levels 24 hours following treatment with the protein synthesis inhibitor cycloheximide in HCT116 and HKH-2 cells respectively (Supplementary Fig. S1C). These data would suggest that in addition to differences in transcriptional regulation, the increased EphA2 levels in KRASMT HCT116 are a result of decreased protein turn-over and increased protein stability.
Figure 1. EphA2 is highly expressed in KRASMT CRC cells.
A. Left panel: EphA2 expression and phosphorylation levels and CD44 expression in HCT116, HKH-2, DLD-1 and Dks-8 cells. Right panel: Lysates from HCT116 and HKH-2 cells were immunoprecipitated (IP) with anti-EphA2 antibody and then immunoblotted (WB) for EphA2, pEphA2Y588, pEphA2S897 and pEphA2Y772. B. Q-PCR analysis of EphA2 and EFNA1 in HCT116 and HKH-2 cells. Relative mRNA expression was calculated using the DDCt method with normalisation to GAPDH. Error bars represent mean ± SD of triplicate values from one of 3 independent experiments. C. EphA2, CD44, pERK1/2 and ERK1/2 levels in panel KRASMT, BRAFMT and KRAS/BRAFWT CRC cells.
Recent studies in lung cancer and glioma have suggested a role for EphA2 in regulating cancer stem-like properties (29, 30). Interestingly, we found that expression of CD44, a marker associated with CRC stem cells (31), was higher in KRASMT cells and correlated with high expression levels of EphA2 (Fig. 1A; Supplementary Fig. S1D). We also found high expression and phosphorylation levels of EphA2 and CD44 in a panel of non-matched KRASMT and BRAFMT CRC compared to KRASWT/BRAFWT CRC cells (Fig. 1C).
The RAS/MEK and RAS/RalGDS/RalA pathways regulate EphA2 expression levels
Given the differential expression of EphA2 in KRASMT, BRAFMT and KRASWT CRC cells, we hypothesized that KRAS may directly affect EphA2 expression. siRNA against KRAS or treatment with the MEK1/2 inhibitor GSK1120212 resulted in strong decreases in EphA2 mRNA and protein levels and significant increases in EFNA1 mRNA levels in KRASMT HCT116 cells (Fig. 2A). Similar results were also obtained in a panel of KRASMT and BRAFMT CRC cells using GSK1120212 (Supplementary Fig. S2A). Interestingly, treatment with the BRAF inhibitor Vemurafenib decreased EphA2 levels in BRAFMT cells, but not in KRASMT cells (Supplementary Fig. S2B); the sustained MAPK signaling following Vemurafenib or siBRAF in the KRASMT CRC cells is consistent with previous studies showing that inhibition of BRAF in the presence of oncogenic RAS induces BRAF binding to CRAF, leading to CRAF hyper-activation and sustained MEK1/2-ERK1/2 activation (32).
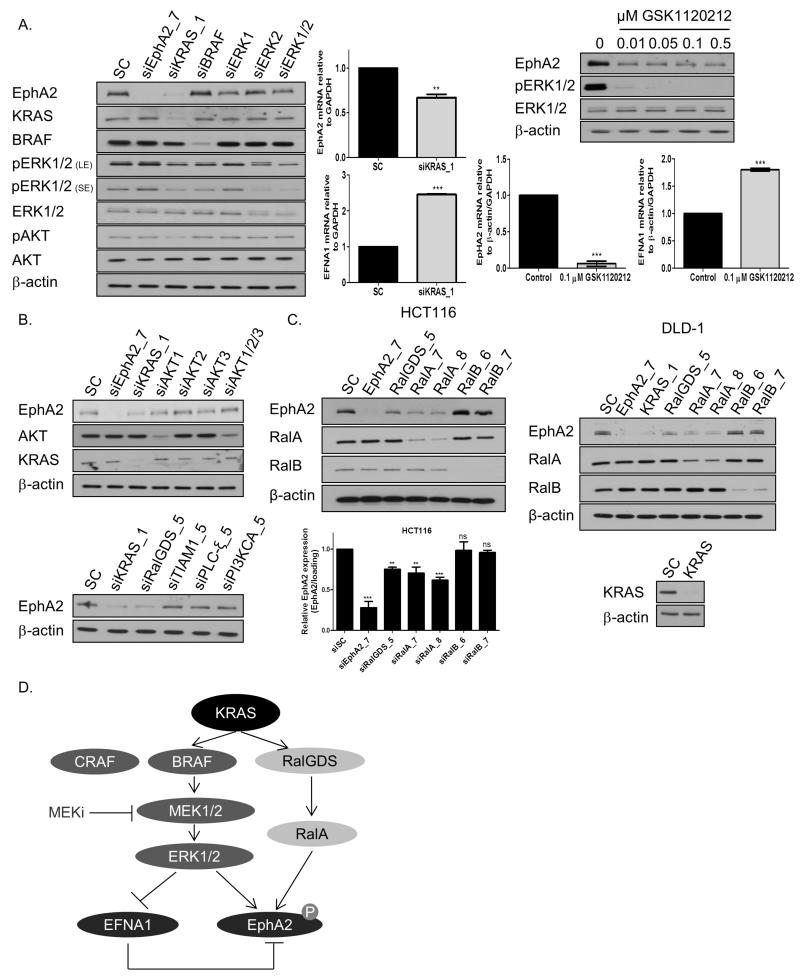
Figure 2. KRAS and downstream effectors MEK1/2 and RalGDS regulate EphA2 expression.
A. Left panel: HCT116 cells were transfected with 10nM SC or 10nM EphA2, KRAS, BRAF, ERK1, ERK2 or ERK1/ERK2 siRNA for 24h and levels of EphA2, KRAS, BRAF, pERK1/2, ERK1/2, pAKT and AKT determined. (LE=long exposure; SE=short exposure). Middle panel: HCT116 cells were transfected with 10nM SC or 10nM KRAS siRNA for 24h, levels of EphA2 and EFNA1 were determined by real-time PCR. Right upper panel: HCT116 cells were treated with GSK1120212 for 12h and EphA2, pERK1/2 and ERK1/2 levels determined by WB. Right lower panel: HCT116 cells were treated with GSK1120212 for 12h and EphA2 and EFNA1 mRNA levels determined by Real-Time PCR. B. Upper panel: HCT116 cells were transfected with 10nM SC or 10nM EphA2, KRAS, AKT1, AKT2, AKT3 or AKT1/2/3 siRNA for 24h. EphA2, KRAS, pAKT and AKT levels were determined by WB. Lower panel: Expression levels of EphA2 in HCT116 cells following transfection with 10nM SC or 10nM KRAS, RalGDS, TIAM1, PLCε or PI3KCA siRNA for 24h. C. Left panel: HCT116 cells were transfected with 10nM SC or 10nM EphA2, RalGDS, RalA or RalB siRNA for 24h, levels of EphA2 determined by WB and real-time PCR. Right panel: Expression levels of EphA2 in DLD-1 cells following transfection with 10nM SC or 10nM EphA2, KRAS, RalGDS, RalA or RalB siRNA for 24h. D. Schematic overview of pathways regulating EphA2/EFNA1 expression in KRASMT CRC. Error bars represent mean ± SD of triplicate values from one of 3 independent experiments.
Furthermore, RNAi against KRAS resulted in more potent inhibition of EphA2 expression compared to the effects of siERK1, siERK2 or combined siERK1/2, indicating that other KRAS effector pathways may regulate EphA2 expression (Fig. 2A). To further investigate the role of the additional effector pathways for RAS, we used RNAi against AKT1/2/3, PI3KCA, PLCε, TIAM1 and RalGDS and found that EphA2 levels were potently decreased by RalGDS gene silencing, whereas silencing of the other major RAS effectors did not significantly affect EphA2 levels (Fig. 2B; Supplementary Fig. S2C). RALGDS is a Guanine nucleotide Exchange Factor (GEF), coupling RAS to the GTPases RalA and RalB (33). Silencing of RalA, but not RalB, resulted in significant decreases in EphA2 mRNA and protein levels in HCT116 and DLD-1 cells (Fig. 2C; data not shown). There was no effect of siRalA on EFNA1 mRNA expression levels (Supplementary Fig. S2D). We also explored a potential regulation of EphA2 by p53 or the Src family kinases (SFK), using RNAi and the isogenic HCT116-p53wt, HCT116-p53 null (−/−), HCT116-p53 (+/−), HCT116-p53 (R248W/−) and HCT116-p53 (R248W/+) cells. These data showed that EphA2 is not regulated by p53 or the SFKs (Supplementary Fig. S2E and F). Taken together, these results would indicate that KRAS regulates EphA2 expression through both the MAPK and RalGDS-RalA pathways in KRASMT CRC models (Fig. 2D).
EphA2 regulates migration and invasion in CRC
Previous studies have shown a role for EphA2 in invasion and metastasis in breast, glioblastoma and NSCLC (9, 25, 30). To investigate a potential role for EphA2 in invasion and migration in CRC, we initially assessed expression and phosphorylation levels of EphA2 in the KRASMT, KRASWT (HKH-2) and KRASMT/+chr3 HCT116 parental and invasive sublines, previously generated in our lab using Matrigel Invasion Chambers (17). We found significant increases in EphA2 mRNA and protein expression and phosphorylation levels in the invasive HCT116, HKH-2 and HCT116+Chr3 sub-lines compared to their parental cells, and this was associated with markedly increased CD44 expression levels (Fig. 3A; Supplementary Fig. S3A). In addition, we also found marked decreases in EFNA1 mRNA levels in the HKH-2 invasive cells, compared to its parental cell line (Supplementary Fig. S3A). Silencing of EphA2 significantly reduced basal migration and invasion rates in KRASMT HCT116, DLD-1 and LoVo cells and in the invasive HCT116 and HKH-2 cells (Fig. 3A; Supplementary Fig. S3B; data not shown). We also established doxycycline-inducible EphA2 shRNA clones and assessed the effect of EphA2 targeting on migration. Treatment of two individual EphA2 shRNA clones with doxycycline for 72h resulted in potent decreases in migration rates similar to those observed with siRNA (Fig. 3A). Furthermore, the CD44+ subpopulation was significantly reduced in the corresponding EphA2 knockdown cells (Supplementary Fig. S3C). Importantly, transient over-expression of exogenous EphA2 resulted in increased invasion and migration of CRC cells (Fig. 3B; Supplementary Fig. S3D).
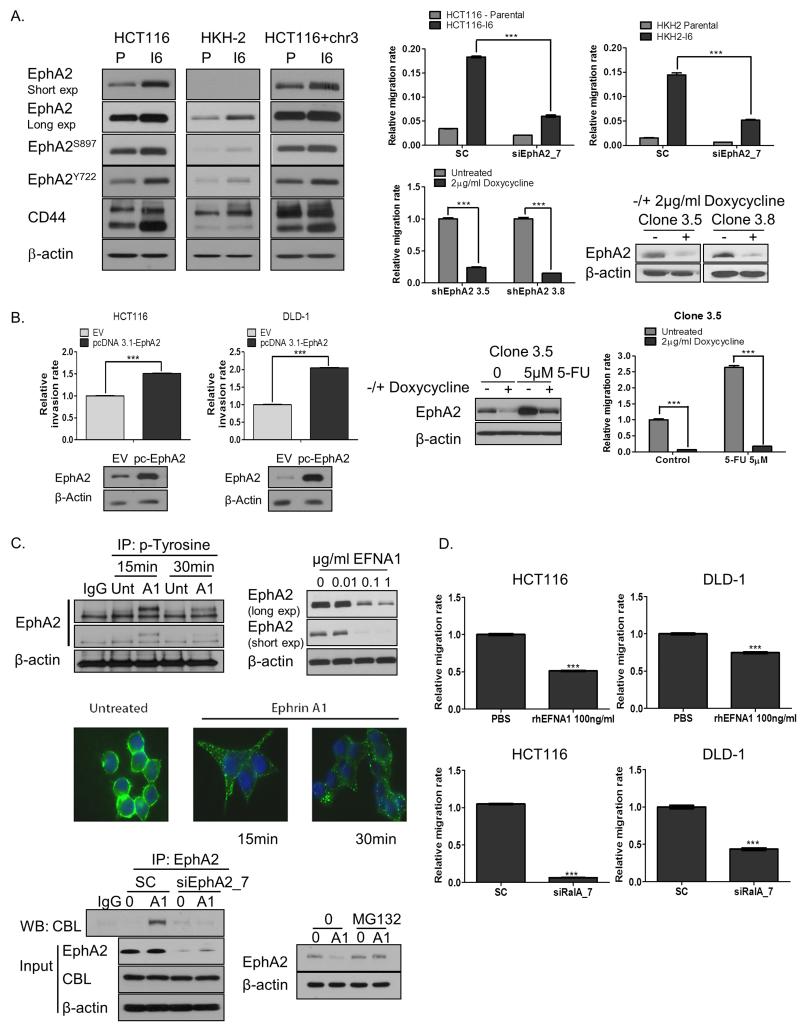
Figure 3. EphA2i reduces migration in KRASMT CRC cells.
A. Left panel: EphA2, EphA2S897, EphA2Y722 and CD44 levels in parental (P) and invasive CRC sublines (I6). Upper right panel: Migration of parental and invasive (I6) cells, transfected with 10nM SC or 10nM EphA2 siRNA. Lower right panel: Migration of HCT116 cells stably transfected with the EphA2 lentiviral pTRIPZ vector system in absence and presence of doxycycline for 24h. shEphA2 3.5 and 3.8 denote different clones. EphA2 expression was determined by WB. B. Left panel: CRC cells were transiently transfected with EV (PCDNA 3.1) or EphA2 expression construct for 24h and invasion rates determined using the xCELLigence system. EpHA2 levels were determined by WB. Right panel: Migration of HCT116 cells stably transfected with the EphA2 lentiviral pTRIPZ vector system in absence and presence of doxycycline for 72h and treated with 5-FU. EphA2 expression was determined by WB. C. Left upper panel: Lysates from HCT116 cells treated with vehicle or rhEFNA1 were immunoprecipitated (IP) with anti-phospho-tyrosine antibody and then immunoblotted (WB) for EphA2. Right upper panel: HCT116 cells were treated with rhEFNA1 for 24h and EphA2 expression measured by WB. Middle panel: HCT116 cells were treated with 0.1μg/ml rhEFNA1 for the indicated time. Immunofluorescent images with green staining EphA2 and DAPI staining (blue) indicating nuclei. Lower left panel: HCT116 cells were transfected with 10nM SC or 10nM EphA2 siRNA for 12h, followed by stimulation with 0.1μg/ml rhEFNA1 (A1) for 15min. Lysates were immunoprecipitated (IP) with anti-EphA2 antibody and then immunoblotted (WB) for CBL. Protein expression of EphA2 and CBL were also analysed. Lower right panel: HCT116 cells were treated with 0.1μM MG132 for 1h, followed by stimulation with 100ng/ml rhEFNA1 (A1) for 1h and EphA2 expression levels determined. D. Migration of CRC cells, incubated with 100ng/ml rhEFNA1 for 24h or transfected with 10nM SC or 10nM RalA siRNA. Error bars represent mean ± SD of quadruplicate values from one of 3 independent experiments.
Our group has previously shown that exposure to 5-FU significantly increases both migration and invasion of CRC cells (17). We now show that 5-FU treatment results in acute increases in EphA2 expression levels and that silencing of EphA2 abrogates the increased migratory potential following 5-FU treatment in CRC cells (Fig. 3B).
We also investigated the effect of the EphA2 ligand EFNA1 on CRC cell migration. Incubation of HCT116 cells with rhEFNA1 increased EphA2 tyrosine phosphorylation and resulted in EphA2 internalization and decreased protein expression after 15min and 30min respectively (Fig. 3C, upper-middle panel; Supplementary Fig. S3E). Immunoprecipitation experiments confirmed that EphA2 interacts with c-Cbl (an ubiquitin E3 ligase) in HCT116 following incubation with rhEFNA1 for 15 minutes (Fig. 3C, lower panel). Moreover, pre-treatment with the proteasome inhibitor MG132 attenuated rhEFNA1-induced EphA2 downregulation, providing further evidence that EphA2 downregulation is mediated by the ubiquitin proteasome system (Fig. 3C, lower panel). Importantly, incubation of CRC cells with rhEFNA1 significantly reduced basal migration rates in HCT116, DLD-1 and LoVo KRASMT cells, similar to those observed with RNAi against EphA2 (Fig. 3D; Supplementary Fig. S3F).
In view of our previous data showing that RalA regulates EphA2 expression levels, we also determined the effect of siRalA on migration of KRASMT CRC cells. Silencing of RalA significantly reduced basal migration rates in HCT116, DLD-1 and LoVo cells (Fig. 3D; Supplementary Fig. S3F). Collectively, all these data provide strong evidence that EphA2 is an important mediator of migration and invasion in CRC and that down-regulating EphA2 using RNAi or rhEFNA1 or by blocking upstream regulators of EphA2 expression, such as RalA, abrogates migration of KRASMT CRC cells.
EphA2 is differentially expressed in CRC versus matched normal tissue
To investigate the clinical importance of EphA2 in CRC, we assessed EphA2 expression in 211 (stage II-III) CRC samples by IHC (21). EphA2 was found to be highly expressed in colorectal adenocarcinoma compared to matched normal colon tissue (Fig. 4A). In contrast to our preclinical models, no correlation between KRAS mutational status and EphA2 expression levels was found within this dataset (data not shown). High EphA2 expression levels were also found in a second TMA including 509 stage I-IV CRC samples (Supplementary Fig. S4A) (18). In support of our preclinical data, a strong correlation was found between EphA2 expression and the stem-cell markers CD44 (p < 0.001) and Lgr5 (p < 0.001), both in the entire patient cohort and when the early stage II/III colorectal tumours were considered alone (Table 1; Supplementary table S2). In both surgery-only and surgery/chemotherapy cohorts, no significant statistical associations between pairs of clinical pathological factors were identified (Supplementary table S3; data not shown).
Figure 4. EphA2 is a strong negative prognostic factor in early stage CRC.
A. left panel: IHC scoring of EphA2 in CRC tissues. Right panel: EphA2 expression in matched normal colon tissue and carcinoma, as determined by IHC. B. Survival curve using Kaplan-Meier estimation comparing EphA2 levels in the entire stage II/III CRC group. C. Left panel: Survival curve using Kaplan-Meier estimation comparing EphA2 levels in the stage II/III surgery-only group. Right panel: Survival curves using Kaplan-Meier estimation comparing EphA2 levels in stage II/III validation group (GSE17536). Univariate (Cox proportional hazards regression) p-value reported. D. Left panel: HCT116 and HKH-2 cells were incubated with 25ng/ml TGF-α (T), EGF (E), HGF (H) or GAS6 (G) for 24h and EphA2, pERK1/2 and ERK1/2 expression determined by WB. Right upper panel: HKH-2 cells were co-cultured with CCD-18Co cells for the indicated time and EphA2 expression levels determined by WB. Right lower panel: HKH-2 cells were transfected with 10nM SC or KRAS siRNA for 12h and thereafter co-cultured with CCD-18Co cells for the indicated time and EphA2 expression levels determined by WB.
Table 1. EphA2 and clinical-pathological correlates in CRC.
Correlation between EphA2 and CD44, LGR5, CD133, Ki-67 and AXL in 338 stage II/III CRC cases of the Singapore dataset.
| Stage II/III Features | Low expression (0-1) (n=165) | High expression (2-3) (n=173) | Kendall’s tau | Fisher’s exact p (Bonferroni adjusted) |
|---|---|---|---|---|
| CD44 | 0.435 | <0.001* | ||
| Low expression | 125 | 56 | ||
| High expression | 40 | 117 | ||
| LGR5 | 0.205 | <0.001* | ||
| Low expression | 63 | 34 | ||
| High expression | 102 | 139 | ||
| CD133 expression | −0.003 | 1.000 | ||
| Low expression | 114 | 120 | ||
| High expression | 51 | 53 | ||
| Ki-67 expression | −0.019 | 0.740 | ||
| Low expression | 98 | 106 | ||
| High expression | 67 | 67 | ||
| AXL expression | 0.266 | <0.001* | ||
| Low expression | 79 | 39 | ||
| High expression | 86 | 134 |
EphA2 expression is a negative prognostic factor for survival in early stage CRC
Next, we assessed the prognostic value of EphA2 expression in the 313 stage II/III CRC (subgroup of 509 CRC patients) samples with mature survival data (18) (Supplementary table S1A). In univariate analysis, there was a significant correlation between high EphA2 expression and poor OS (p=0.0408) (Fig. 4B). When the stage II/III group was broken down into surgery-only and surgery/adjuvant chemotherapy cohorts, increased age (HR: 1.06, 95% CI: 1.03-1.09, p=0.00033), stage III (HR: 2.52, 95% CI: 1.51-4.19, p=0.00388) and high levels of EphA2 (HR: 1.97, 95% CI: 1.20-3.25, p=0.007890) were associated significantly with poorer prognosis when the surgery-only group was considered alone (Fig. 4C, left panel; Table 2A). Subsequently, we developed a full multivariate model and found that increased age (HR: 1.06, 95% CI: 1.03-1.09, p=0.00044), stage III (HR: 2.97, 95% CI: 1.66-5.32, p=0.00025) and high levels of EphA2 (HR: 1.97, 95% CI: 1.09-3.56, p=0.024) were correlated with an increased risk of death in the surgery-only group (Table 2A). The final multivariate model resulted in a concordance index (c-index) of 0.75. High EphA2 level, increased age and stage III were all associated with poorer prognosis in the surgery-only group (Table 2B). In contrast, there was no significant association between EphA2 expression and prognosis in the resection/adjuvant chemotherapy cohort, suggesting a treatment interaction effect (Supplementary Fig. S4B and Table S4). Further sub-analysis for patients with stage II or stage III disease alone, showed that high EphA2 expression was prognostic for poor OS in the stage II surgery-only cohort (p=0.000472) and the stage III surgery-only cohort (p=0.0375) but not in the stage II surgery-chemotherapy cohort (p=0.434) or the stage III surgery-chemotherapy subgroup (p=0.441) (Supplementary Fig. S4C and S5).
Table 2. EphA2 is an independent prognostic biomarker.
A. Univariate and multivariate analyses in stage II/III surgery-only CRC cohort (Singapore dataset) using the Cox Proportional Hazards Ratio method. B. Final multivariate model (Cox proportional hazards regression) of surgery-only patient group in Singapore dataset. C. Multivariate model (Cox proportional hazards regression) of GSE17536 patient group (Stage II/III).
| A. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate (Full) | |||||||
| FACTOR | N (n) | HR | 95% CI | p-value | HR | 95% CI | p-value | |
| Age (Ilinear term, per year increase) | 157 (67) | 1.06 | 1.03-1.09 | 0.000033 | 1.06 | 1.03-1.09 | 0.000440 | |
| Gender | Male | 68 (27) | 1.00 | 1.00 | ||||
| Female | 89 (40) | 1.45 | 0.89-2.36 | 0.140000 | 1.50 | 0.81-2.75 | 0.190000 | |
| Ethnic groud | Chinese | 137 (61) | 1.00 | 1.00 | ||||
| Non-Chinese | 20 (6) | 1.18 | 0.51-2.74 | 0.697000 | 1.56 | 0.63-3.87 | 0.340000 | |
| Tumour Site | Rectal | 24 (12) | 1.00 | 1.00 | ||||
| Non-Rectal | 133 (55) | 0.63 | 0.33-1.18 | 0.145000 | 0.73 | 0.37-1.46 | 0.380000 | |
| Stage | II | 114 (44) | 1.00 | 1.00 | ||||
| III | 43 (23) | 2.52 | 1.51-4.19 | 0.000388 | 2.97 | 1.66-5.32 | 0.000250 | |
| Differentiation Group | 1 | 4 (1) | 1.00 | 1.00 | ||||
| 2 | 140 (60) | 2.23 | 0.31-16.10 | 0.427000 | 5.62 | 0.70-45.17 | 0.100000 | |
| 3 | 13 (6) | 2.00 | 0.24-16.61 | 0.522000 | 5.98 | 0.64-56.32 | 0.120000 | |
| Tumour Size (linear term, per year increase) | 157 (67) | 0.99 | 0.89-1.10 | 0.890000 | 1.04 | 0.93-1.17 | 0.490000 | |
| Invasion | No Invasion | 18 (9) | 1.00 | 1.00 | ||||
| Invasion | 139 (58) | 1.81 | 0.89-3.67 | 0.099300 | 1.46 | 0.63-3.39 | 0.370000 | |
| EphA2 Level | Low | 76 (24) | 1.00 | 1.00 | ||||
| High | 81 (43) | 1.97 | 1.20-3.25 | 0.007890 | 1.97 | 1.09-3.56 | 0.024000 | |
| B. | |||||
|---|---|---|---|---|---|
| FACTOR | Multivariate (Final) | ||||
| N (n) | HR | 95% CI | p-value | ||
| Age (linear, per year increase) | 157 (67) | 1.06 | 1.03-1.09 | 0.000081 | |
| Stage | II | 114 (44) | 1 | ||
| III | 43 (23) | 2.59 | 1.51-4.44 | 0.000520 | |
| EphA2 Level | Low | 76 (24) | 1 | ||
| High | 81 (43) | 1.75 | 1.01-3.01 | 0.046000 | |
| C. | |||||
|---|---|---|---|---|---|
| FACTOR | Multivariate (Final) | ||||
| N (n) | HR | 95% CI | p-value | ||
| Age (linear, per year increase) | 114 (37) | 1.02 | 1.00-1.04 | 0.100000 | |
| Stage | II | 57 (12) | 1 | ||
| III | 57 (25) | 2.52 | 1.20-5.27 | 0.014000 | |
| EphA2 Level | Low | 56 (14 | 1 | ||
| High | 58 (23) | 2.31 | 1.15-4.67 | 0.019000 | |
Independent validation of EphA2 as poor prognostic biomarker in CRC
The prognostic role of EphA2 expression in CRC was validated using a publicly-available stage II/III (n=114) CRC microarray dataset (GSE17536) (17, 34). Analysis of EphA2 mRNA expression in this dataset revealed a significant association between high EphA2 expression and decreased OS (p=0.0277) (Fig. 4C, right panel). The multivariate model showed that stage III (HR: 2.52, 95% CI: 1.20-5.27, p=0.014000) and high EphA2 expression (HR: 2.31, 95% CI: 1.15-4.67, p=0.019) were associated with poorer OS (Table 2C). The model resulted in a c-index of 0.705. Taken together, these data indicate that EphA2 expression has the potential of predicting poor clinical outcome in early stage II/III CRC.
Growth factors regulate EphA2 levels in KRASWT CRC cells
Given the lack of correlation between EphA2 levels and KRAS status in our clinical samples, we determined whether the tumour microenvironment could influence EphA2 levels in KRASWT models. We found markedly increased EphA2 levels in the KRASWT HKH-2, DiFi, OXCO2 and Dks-8 cells following incubation with recombinant human EGF, TGF-α or HGF, thus supporting the idea that microenvironment-derived ligands can regulate EphA2 levels in KRASWT CRC (Fig. 4D, left panel; Supplementary Fig. S6A). In addition, incubation of HKH-2 cells with conditioned medium from CCD-18Co colon fibroblasts, or co-culture of HKH-2 cells with CCD-18Co cells, resulted in potent increases in EphA2 expression levels in the HKH-2 cell line (Fig. 4D, right panel; Supplementary Fig. S6B). Moreover, silencing of KRAS abrogated co-culture-induced EphA2 expression levels in HKH-2 cells, indicating that RAS activation is required for the EphA2 increases following co-culture in KRASWT CRC cells (Fig. 4D). Clinically, a significant positive association between TGF-α mRNA and EphA2 mRNA levels was observed in 4 independent CRC online datasets, further supporting the hypothesis that stromal-derived growth factors can regulate EphA2 expression levels in an in vivo setting (Supplementary Table S5).
DISCUSSION
Whilst there have been major developments in the treatment of metastatic CRC over the last two decades (e.g. the introduction of the VEGF and EGFR targeted agents bevacizumab, aflibercept, ramucirumab, cetuximab and panitumumab), translating these advances to stage II/III disease have shown no benefit in large phase III adjuvant clinical trials (1, 2). An improved understanding of the molecular mechanisms driving CRC progression and recurrence, will potentially lead to the identification of novel diagnostics and treatment strategies for stage II/III CRC. In this study, we provide evidence that EphA2 is an important mediator of CRC cell migration/invasion and could be a promising novel target for CRC, in particular for stage II/III CRC with high EphA2 expression levels.
High EphA2 expression has been reported in several tumours, including breast cancer (35), melanoma (36), NSCLC (11) and glioblastoma multiforme (37) and has been identified as a poor prognostic marker in these tumours. In CRC, genetic ablation of EphA2 in ApcMin/+ mice has been found to result in significant reduction in number and size of intestinal tumours, indicating that EphA2 plays a role in intestinal tumourigenesis (38). In this study, we have assessed the expression and prognostic relevance of EphA2 in tissues from patients with stage II and III CRC. In agreement with a previous study using RT-PCR to detect EphA2, the colorectal primary tumour tissue displayed marked upregulated expression of EphA2 when compared with matched normal tissue (39). The relative overexpression of EphA2 highlights the potential for exploitation of EphA2 as a therapeutic target in CRC. Previous studies in NSCLC and glioblastoma have shown a positive correlation between EphA2 and stem cell markers ALDH and DDEA-1a or CD44 respectively (10, 30). In addition, RNAi-mediated depletion of EphA2 or EFNA1-FC (a soluble EFNA1 dimer fused to Fc) resulted in a decreased cancer stem cell population and tumourigenicity in vivo. CD44 and Lgr5 have been identified as markers for CRC cancer cells with stem cell-like properties, and both markers have been associated with increased CRC tumourigenicity and metastasis (40, 41). Our study showed that high EphA2 expression significantly correlated with high expression levels of CD44 and Lgr5 in CRC tissues. Furthermore, shRNA mediated knockdown of EphA2 suppressed the CD44-high stem-like CRC cell population, indicating that EphA2 may be a promising target to prevent CRC recurrence and metastasis.
In order to model colorectal tumour cell invasion/metastasis, our group has previously generated invasive (KRASMT/KRASWT/+chr3) CRC daughter cells which displayed an epithelial-mesenchymal transition-like phenotype, high levels of CD44 and increased colony-forming ability (17). We now show that EphA2 is highly expressed in these invasive daughter cell lines and that si/shRNA mediated knockdown of EphA2 potently inhibited migration and invasion of parental and invasive CRC cells. In agreement with previous studies, we found that EphA2 was rapidly internalized and downregulated in a proteasome-dependent manner in response to EFNA1-ligand stimulation and this resulted in potent inhibition of migration in a panel of CRC cells (42). These data would indicate that EphA2 is an important regulator of migration and invasion in CRC.
A key finding of our study is that high EphA2 expression was significantly associated with poorer OS in stage II/III CRC. Importantly, further univariate and multivariate analyses revealed that the stage II/III surgery-only CRC patient cohort with high EphA2 expression levels have a shorter 5 years OS compared to stage II/III surgery-only CRC patients with low EphA2 expression. The results herein obtained from both public available datasets and our CRC TMA are the first to show that EphA2 may be a biomarker of poor prognosis in stage II/III CRC. A previous RNA sequencing study of 675 human cancer cells has suggested that EphA2 is strongly correlated with the expression of other oncogenes, such as c-MET and EGFR (43). It is therefore plausible that analysis of c-MET and/or EGFR together with EphA2 can further enhance the prognostic power of EphA2 in stage II/III CRC.
Given its potential role in CRC progression and metastasis, we further investigated how EphA2 expression levels are regulated in CRC. In contrast to previous studies, our data did not show a role for p53 or the Src Family Kinases in regulating EphA2 expression (44, 45). Using a systems biology approach, we previously identified EphA2 as a potential KRAS target in KRASMT CRC cells (15). In agreement with previous studies in breast cancer and melanoma, our examination of a large panel of CRC cells revealed high EphA2 expression in cells harbouring an activating mutation in KRAS or BRAF compared to KRAS/BRAFWT cells (28, 46). Inhibition of the MAPK pathway resulted in significant increased EFNA1 mRNA expression while reducing EphA2 mRNA and protein levels. These data and our results using RNAi-mediated knockdown of EFNA1 or recombinant EFNA1, would indicate that expression of EFNA1 contributes, at least in part, to EphA2 levels in CRC, consistent with previous findings in breast cancer (28).
Activated Ras-GTP can exert its function through multiple downstream effectors, such as the Raf kinases, the p110 catalytic subunits of class I PI3Ks, TIAM1, PLCε and the RalGDS-Ral effector pathway (47). This is the first study showing that KRAS also regulates mRNA and protein levels of EphA2 through the RalGDS-Ral pathway. The Ras-like small GTPases RalA and RalB interact with effectors such as Sec5, Filamin, RALBP1 and ZONAB and have been shown to regulate membrane trafficking, cell adhesion and transcription. A number of studies have shown that RalA and RalB are important drivers of survival, proliferation and metastasis in solid tumours, including CRC (48). Using RNAi against RalA and RalB, we found that RalA but not RalB regulated EphA2 expression levels and migration in KRASMT CRC cells. These results would indicate that anti-RalA selective therapies (49) may provide an effective therapeutic approach for KRASMT CRC with high expression levels of EphA2.
In contrast to the in vitro data, no correlation between the KRAS mutational status and EphA2 levels was found in our clinical samples. However, our data would suggest that microenvironment-derived ligands can regulate EphA2 levels in KRASWT tumours, which could explain why EphA2 levels were not significantly higher in the KRASMT CRC samples.
In conclusion, using preclinical CRC models and patient tissue samples, we have identified EphA2 as a key regulator of CRC cell migration and invasion. Moreover, EphA2 is highly expressed in CRC, associated with the CRC stem cell markers CD44 and Lgr5 and is a poor prognostic biomarker in CRC. A number of recent studies have identified the stem-like subtype as a dominant molecular subgroup with poor outcome in CRC (50). Our data would indicate that EphA2 targeted approaches may represent a promising treatment strategy for this stem-like subgroup with high EphA2 levels, in particular in the adjuvant disease setting where anti-EGFR agents have failed. A variety of therapeutic strategies have been developed to target EphA2, including activating monoclonal antibodies, ephrin ligands and selective kinase inhibitors (3). Finally, our data provide support for the further investigation of EphA2 as novel biomarker in early stage CRC.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Recent efforts to improve survival of patients with stage II/III colorectal cancer (CRC) by adding biological agents to 5-FU/oxaliplatin-based adjuvant therapies, have failed. Therefore, novel therapeutic approaches are needed to prevent recurrence and disease progression in these patients. In this study, we have analysed the expression of EphA2 in matched normal and tumour tissues from stage II/III CRC patients. Colorectal tumours expressed significantly higher levels of EphA2 which positively correlated with high levels of the stem cell markers CD44 and Lgr5, suggesting that increased EphA2 expression may be important in progression of this disease. Importantly, we found that high EphA2 expression was an independent adverse prognostic marker in stage II/III CRC. Moreover, down-regulating EphA2 using RNAi or rhEFNA1 decreased CRC cell migration/invasion, indicating that EphA2-targeted agents may be a novel treatment strategy for CRC, in particular for stage II/III CRC with high expression of EphA2.
ACKNOWLEDGMENTS
We thank the patients and staff who took part in the Northern Ireland Adjuvant Chemotherapy trial, trial funders Wyeth Ltd (now Pfizer Inc) and the Friends of the Cancer Centre. We are grateful to the Northern Ireland Biobank for its assistance in acquiring tissue for analysis. We thank Ms. Nicola Totton for technical assistance.
Financial support from Cancer Research UK (C212/A7402); Cancer Research UK fellowship (C13749/A7261).
Footnotes
Conflicts of interest: P.G. Johnston has an ownership interest in both Almac Diagnostics and Fusion Antibodies. He is a consultant/advisor for, and has received honoraria from, Chugai pharmaceuticals, Sanofi-Aventis and Pfizer. R. H. Wilson has received honoraria for advising Sanofi and Merck Serono. All other authors have no conflicts of interest to declare.
REFERENCES
- 1.Allegra CJ, Yothers G, O’Connell MJ, Sharif S, Petrelli NJ, Colangelo LH, et al. Phase III trial assessing bevacizumab in stages II and III carcinoma of the colon: results of NSABP protocol C-08. J Clin Oncol. 2011;29:11–6. doi: 10.1200/JCO.2010.30.0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts SR, Sargent DJ, Nair S, Mahoney MR, Mooney M, Thibodeau SN, et al. Effect of oxaliplatin, fluorouracil, and leucovorin with or without cetuximab on survival among patients with resected stage III colon cancer: a randomized trial. Jama. 2012;307:1383–93. doi: 10.1001/jama.2012.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–80. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6:462–75. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 5.Himanen JP. Ectodomain structures of Eph receptors. Seminars in cell & developmental biology. 2012;23:35–42. doi: 10.1016/j.semcdb.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 6.Janes PW, Nievergall E, Lackmann M. Concepts and consequences of Eph receptor clustering. Seminars in cell & developmental biology. 2012;23:43–50. doi: 10.1016/j.semcdb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Boyd AW, Bartlett PF, Lackmann M. Therapeutic targeting of EPH receptors and their ligands. Nat Rev Drug Discov. 2014;13:39–62. doi: 10.1038/nrd4175. [DOI] [PubMed] [Google Scholar]
- 8.Udayakumar D, Zhang G, Ji Z, Njauw CN, Mroz P, Tsao H. EphA2 is a critical oncogene in melanoma. Oncogene. 2011;30:4921–9. doi: 10.1038/onc.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brantley-Sieders DM, Zhuang G, Hicks D, Fang WB, Hwang Y, Cates JM, et al. The receptor tyrosine kinase EphA2 promotes mammary adenocarcinoma tumorigenesis and metastatic progression in mice by amplifying ErbB2 signaling. J Clin Invest. 2008;118:64–78. doi: 10.1172/JCI33154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binda E, Visioli A, Giani F, Lamorte G, Copetti M, Pitter KL, et al. The EphA2 receptor drives self-renewal and tumorigenicity in stem-like tumor-propagating cells from human glioblastomas. Cancer Cell. 2012;22:765–80. doi: 10.1016/j.ccr.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brannan JM, Dong W, Prudkin L, Behrens C, Lotan R, Bekele BN, et al. Expression of the receptor tyrosine kinase EphA2 is increased in smokers and predicts poor survival in non-small cell lung cancer. Clin Cancer Res. 2009;15:4423–30. doi: 10.1158/1078-0432.CCR-09-0473. [DOI] [PubMed] [Google Scholar]
- 12.Miura K, Wakayama Y, Tanino M, Orba Y, Sawa H, Hatakeyama M, et al. Involvement of EphA2-mediated tyrosine phosphorylation of Shp2 in Shp2-regulated activation of extracellular signal-regulated kinase. Oncogene. 2013;32:5292–301. doi: 10.1038/onc.2012.571. [DOI] [PubMed] [Google Scholar]
- 13.Kyula JN, Van Schaeybroeck S, Doherty J, Fenning CS, Longley DB, Johnston PG. Chemotherapy-Induced Activation of ADAM-17: A Novel Mechanism of Drug Resistance in Colorectal Cancer. Clin Cancer Res. 2010;16:3378–89. doi: 10.1158/1078-0432.CCR-10-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Schaeybroeck S, Kyula JN, Fenton A, Fenning CS, Sasazuki T, Shirasawa S, et al. Oncogenic Kras promotes chemotherapy-induced growth factor shedding via ADAM17. Cancer Res. 2011;71:1071–80. doi: 10.1158/0008-5472.CAN-10-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Schaeybroeck S, Kalimutho M, Dunne PD, Carson R, Allen W, Jithesh PV, et al. ADAM17-dependent c-MET-STAT3 signaling mediates resistance to MEK inhibitors in KRAS mutant colorectal cancer. Cell reports. 2014;7:1940–55. doi: 10.1016/j.celrep.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 16.Van Schaeybroeck S, Kelly DM, Kyula J, Stokesberry S, Fennell DA, Johnston PG, et al. Src and ADAM-17-mediated shedding of transforming growth factor-alpha is a mechanism of acute resistance to TRAIL. Cancer Res. 2008;68:8312–21. doi: 10.1158/0008-5472.CAN-07-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunne PD, McArt DG, Blayney JK, Kalimutho M, Greer S, Wang T, et al. AXL is a key regulator of inherent and chemotherapy-induced invasion and predicts a poor clinical outcome in early-stage colon cancer. Clin Cancer Res. 2014;20:164–75. doi: 10.1158/1078-0432.CCR-13-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong CW, Kim LG, Kong HH, Low LY, Iacopetta B, Soong R, et al. CD133 expression predicts for non-response to chemotherapy in colorectal cancer. Mod Pathol. 2010;23:450–7. doi: 10.1038/modpathol.2009.181. [DOI] [PubMed] [Google Scholar]
- 19.Wang T, Ong CW, Shi J, Srivastava S, Yan B, Cheng CL, et al. Sequential expression of putative stem cell markers in gastric carcinogenesis. Br J Cancer. 2011;105:658–65. doi: 10.1038/bjc.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Yeoh KG, Salto-Tellez M. Lgr5 expression is absent in human premalignant lesions of the stomach. Gut. 2012;61:1777–8. doi: 10.1136/gutjnl-2012-302372. [DOI] [PubMed] [Google Scholar]
- 21.Oladipo O, Conlon S, O’Grady A, Purcell C, Wilson C, Maxwell PJ, et al. The expression and prognostic impact of CXC-chemokines in stage II and III colorectal cancer epithelial and stromal tissue. Br J Cancer. 2011;104:480–7. doi: 10.1038/sj.bjc.6606055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, et al. NCBI GEO: archive for functional genomics data sets--10 years on. Nucleic acids research. 2011;39:D1005–10. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–8. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 25.Miao H, Li DQ, Mukherjee A, Guo H, Petty A, Cutter J, et al. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell. 2009;16:9–20. doi: 10.1016/j.ccr.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balasubramaniam D, Paul LN, Homan KT, Hall MC, Stauffacher CV. Specificity of HCPTP variants toward EphA2 tyrosines by quantitative selected reaction monitoring. Protein science : a publication of the Protein Society. 2011;20:1172–81. doi: 10.1002/pro.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang WB, Brantley-Sieders DM, Hwang Y, Ham AJ, Chen J. Identification and functional analysis of phosphorylated tyrosine residues within EphA2 receptor tyrosine kinase. J Biol Chem. 2008;283:16017–26. doi: 10.1074/jbc.M709934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macrae M, Neve RM, Rodriguez-Viciana P, Haqq C, Yeh J, Chen C, et al. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell. 2005;8:111–8. doi: 10.1016/j.ccr.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Miao H, Gale NW, Guo H, Qian J, Petty A, Kaspar J, et al. EphA2 promotes infiltrative invasion of glioma stem cells in vivo through cross-talk with Akt and regulates stem cell properties. Oncogene. 2015;34:558–67. doi: 10.1038/onc.2013.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song W, Ma Y, Wang J, Brantley-Sieders D, Chen J. JNK Signaling Mediates EPHA2-Dependent Tumor Cell Proliferation, Motility, and Cancer Stem Cell-like Properties in Non-Small Cell Lung Cancer. Cancer Res. 2014;74:2444–54. doi: 10.1158/0008-5472.CAN-13-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Todaro M, Francipane MG, Medema JP, Stassi G. Colon cancer stem cells: promise of targeted therapy. Gastroenterology. 2010;138:2151–62. doi: 10.1053/j.gastro.2009.12.063. [DOI] [PubMed] [Google Scholar]
- 32.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferro E, Trabalzini L. RalGDS family members couple Ras to Ral signalling and that’s not all. Cell Signal. 2010;22:1804–10. doi: 10.1016/j.cellsig.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 34.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–68. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zelinski DP, Zantek ND, Stewart JC, Irizarry AR, Kinch MS. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 2001;61:2301–6. [PubMed] [Google Scholar]
- 36.Easty DJ, Guthrie BA, Maung K, Farr CJ, Lindberg RA, Toso RJ, et al. Protein B61 as a new growth factor: expression of B61 and up-regulation of its receptor epithelial cell kinase during melanoma progression. Cancer Res. 1995;55:2528–32. [PubMed] [Google Scholar]
- 37.Wykosky J, Gibo DM, Stanton C, Debinski W. EphA2 as a novel molecular marker and target in glioblastoma multiforme. Molecular cancer research : MCR. 2005;3:541–51. doi: 10.1158/1541-7786.MCR-05-0056. [DOI] [PubMed] [Google Scholar]
- 38.Bogan C, Chen J, O’Sullivan MG, Cormier RT. Loss of EphA2 receptor tyrosine kinase reduces ApcMin/+ tumorigenesis. Int J Cancer. 2009;124:1366–71. doi: 10.1002/ijc.24083. [DOI] [PubMed] [Google Scholar]
- 39.Kataoka H, Igarashi H, Kanamori M, Ihara M, Wang JD, Wang YJ, et al. Correlation of EPHA2 overexpression with high microvessel count in human primary colorectal cancer. Cancer Sci. 2004;95:136–41. doi: 10.1111/j.1349-7006.2004.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du L, Rao G, Wang H, Li B, Tian W, Cui J, et al. CD44-positive cancer stem cells expressing cellular prion protein contribute to metastatic capacity in colorectal cancer. Cancer Res. 2013;73:2682–94. doi: 10.1158/0008-5472.CAN-12-3759. [DOI] [PubMed] [Google Scholar]
- 41.Tsuji S, Kawasaki Y, Furukawa S, Taniue K, Hayashi T, Okuno M, et al. The miR-363-GATA6-Lgr5 pathway is critical for colorectal tumourigenesis. Nature communications. 2014;5:3150. doi: 10.1038/ncomms4150. [DOI] [PubMed] [Google Scholar]
- 42.Walker-Daniels J, Riese DJ, 2nd, Kinch MS. c-Cbl-dependent EphA2 protein degradation is induced by ligand binding. Molecular cancer research : MCR. 2002;1:79–87. [PubMed] [Google Scholar]
- 43.Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306–12. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 44.Dohn M, Jiang J, Chen X. Receptor tyrosine kinase EphA2 is regulated by p53-family proteins and induces apoptosis. Oncogene. 2001;20:6503–15. doi: 10.1038/sj.onc.1204816. [DOI] [PubMed] [Google Scholar]
- 45.Du X, Baldwin C, Hooker E, Glorion P, Lemay S. Basal and Src kinase-mediated activation of the EphA2 promoter requires a cAMP-responsive element but is CREB-independent. J Cell Biochem. 2011;112:1268–76. doi: 10.1002/jcb.23018. [DOI] [PubMed] [Google Scholar]
- 46.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 47.Neel NF, Martin TD, Stratford JK, Zand TP, Reiner DJ, Der CJ. The RalGEF-Ral Effector Signaling Network: The Road Less Traveled for Anti-Ras Drug Discovery. Genes & cancer. 2011;2:275–87. doi: 10.1177/1947601911407329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin TD, Samuel JC, Routh ED, Der CJ, Yeh JJ. Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res. 2011;71:206–15. doi: 10.1158/0008-5472.CAN-10-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan C, Liu D, Li L, Wempe MF, Guin S, Khanna M, et al. Discovery and characterization of small molecules that target the GTPase Ral. Nature. 2014;515:443–7. doi: 10.1038/nature13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–25. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.