Abstract
Obesity has been linked to breast cancer progression but the underlying mechanisms remain obscure. Here we report how leptin, an obesity-associated adipokine, regulates a transcriptional pathway to silence a genetic program of epithelial homeostasis in breast cancer stem-like cells (CSC) that promotes malignant progression. Using genome-wide ChIP-seq and RNA expression profiling, we defined a role for activated STAT3 and G9a histone methyltransferase in epigenetic silencing of miR-200c, which promotes the formation of breast CSCs defined by elevated cell surface levels of the leptin receptor (OBRhi). Inhibiting the STAT3/G9a pathway restored expression of miR-200c, which in turn reversed the CSC phenotype to a more differentiated epithelial phenotype. In a rat model of breast cancer driven by diet-induced obesity, STAT3 blockade suppressed the CSC-like OBRhi population and abrogated tumor progression. Together, our results show how targeting STAT3-G9a signaling regulates CSC plasticity during obesity-related breast cancer progression, suggesting a novel therapeutic paradigm to suppress CSC pools and limit breast malignancy.
Introduction
Obesity is an established risk factor for breast cancer incidence, progression, recurrence, and mortality (1, 2). The risk of breast cancer increases significantly in women who have an elevated body mass index (30 kg/m2). Being overweight or obese for a woman at the time she is diagnosed with breast cancer is linked to a high risk of recurrence regardless of treatment factors (3). In rodents, high body weight is also associated with increased incidence of spontaneous and chemically induced tumors (4). Obesity in both humans and rodents is characterized by increased leptin levels (4, 5). Accumulated evidence points to leptin signaling playing a significant role in mammary development and tumorigenesis (6). Interestingly, recent studies have further shown that leptin signaling may be involved in the promotion of the cancer stem cell (CSC) phenotype (7-9). It is worthy to note that CSCs, a subpopulation of cancer cells that have acquired the stem cell properties associated with normal stem cells, are considered to be the genesis of cancer and account for cancer initiation, progression, and recurrence. It has been shown that an enlarged CSC population is highly associated with tumor aggressiveness and recurrence (10, 11) and that in response to microenvironmental stimuli, the CSC population can expand through acquisition of epigenetic alterations that deregulate the balance of stemness versus differentiation and thereby drives the cancer progression (12). Nonetheless, the key epigenetic mechanisms governing the emergence or expansion of breast CSCs, particularly those in response to leptin signaling, still remain to be elucidated.
Using human mammary epithelial cells and human breast cancer cells (primary tumor cells and cell lines), this study not only delineates the critical epigenetic mechanism that leptin uses to promote the CSC traits but also provides molecular characterization of the leptin-induced breast CSC population. Using a diet-induced obesity animal model of breast cancer, our data further elucidate that therapeutic targeting of the leptin-induced CSC pool shows great promise to effectively prevent obesity-related breast cancer progression. Furthermore, correlation analysis of breast cancer patient specimens reveals that the molecular signature associated with leptin-induced stemness can be used to predict clinicopathologic features and will potentially benefit the stratification and evaluation of future therapeutic regimens.
Materials and Methods
Cell culture and treatment
The immortal normal mammary epithelial cells, MCF12A, and the breast cancer cell line MCF7 were purchased from ATCC. Cells lines have been tested for mycoplasma free and authenticated by ATCC using short tandem repeat (STR) profiling and have not been passaged for more than 6 months. Primary breast tumor cells [isolated from 12 low-grade tumors that include 10 luminal and 2 Her2+ tumors, and 12 high-grade tumors that include 11 triple-negative breast cancer (TNBC) and 1 luminal tumor] were purchased from Promab Inc. MCF12A cells were grown in DMEM/F12 medium supplemented with 5% horse serum, EGF (20 ng/mL), insulin (10 μg/mL), cholera toxin (1 ng/mL), hydrocortisone, and gentamycin (Sigma). MCF7 cells were cultured with DMEM medium supplemented with 10% FBS, penicillin (50 U/mL), and streptomycin (50 U/mL). Human recombinant leptin protein (50 μmol/L; R&D Systems) and 50 μmol/L STAT3-specific inhibitor, S3I-201, and 1 μmol/L G9a-specific inhibitor, BIX01294 (Selleck Chemicals) were used for the treatments in cells. Cells were serum-starved for 8 hours before leptin treatment.
Generation of stably expressed and knocked-down cell lines
Lentiviral infection was performed as described previously (13). Briefly, pCDH-puro or pCDH-miR-200c-puro was co-transfected with the third generation of lentiviral packaging plasmids: pMDLg-pRRE, PRSV-Rev, and PMD2G into 293T cells by Lipo-fectamine 2000 following the manufacturer’s instructions. At 24 and 48 hours posttransfection, culture medium was harvested and subsequently incubated with target cells in the presence of polybrene (5 μg/mL) for 24 hours. At 48 hours after infection, infected cells were harvested for gene and protein expression analysis or selected with puromycin (2 μg/mL) for 2 weeks to establish stably infected cells. Lentivirus particles expressing shRNAs, sh-STAT3, sh-G9a, and sh-OBR were purchased from Santa Cruz, and the infection procedure was performed according to the manufacturer’s instructions.
Mammary xenograft tumor
Isolation, infection, and culture of tumor spheres from human primary breast tumor cells (Promab Inc.) were performed as described previously (13). Mammary fat pads of 6-week-old female NOD/SCID mice were inoculated with indicated number of cells from the human primary breast tumor cells (grades II–III) for a total volume of 100 μL per injection site. At the end of the second week, before any palpable tumors, animals were given vehicle or S3I-201 intravenously at 5 mg/kg twice per week for 2 weeks (14). Four weeks after inoculation, tumor incidences were calculated and animals were sacrificed. All surgical procedures and animal manipulations were performed under regulations of Purdue Animal Care and Use Committee.
Establishment of the diet-induced obesity and breast cancer rat model
Twenty-day-old outbred Sprague-Dawley rats were purchased from Harlan Laboratories. Pups were weaned 24 hours after arrival and maintained in a temperature and light controlled animal facility. Animals were given ad libitum access to water and randomly assigned to either the standard regular rat chow diet (Harlan Teklad) or a Western diet (Research Diets). The regular rat chow diet contains 3.30 kcal/g with 18% protein, 5% fat, mainly from corn oil and 57% carbohydrates, primarily in the form of complex polysaccharides. The Western diet contains 4.47 kcal/g with 20% protein, 21% fat in the form of anhydrous milk fat, and 50% carbohydrates, 34% of which is sucrose. No significant difference was found in consumption among groups (data not shown). At 54 days old, rats were grouped according to their body fat composition determined by dual-energy x-ray absorptometry (DEXA, Lunar Corporation). Animals on the Western diet were placed into two groups with rats in the highest tertile being placed in the Western obese group and rats in the lowest tertile being placed in the Western lean group. At 65 days of age, half of the rats in Western diet each group were injected with 50 mg/kg of the chemical carcinogen N-methylnitrosourea (MNU; used in Fig. 5) and the remaining animals received 0.9% saline (used in Supplementary Fig. S5A and S5B). Rats were sacrificed by 168 days postinjection or when the animal reached a terminal state as determined by established guidelines (15). Intact mammary glands and tumors were collected and flash-frozen in liquid nitrogen or fixed in 10% neutral formalin for histologic evaluation. All animal experiments were conducted with approval of the Purdue Animal Care and Use Committee.
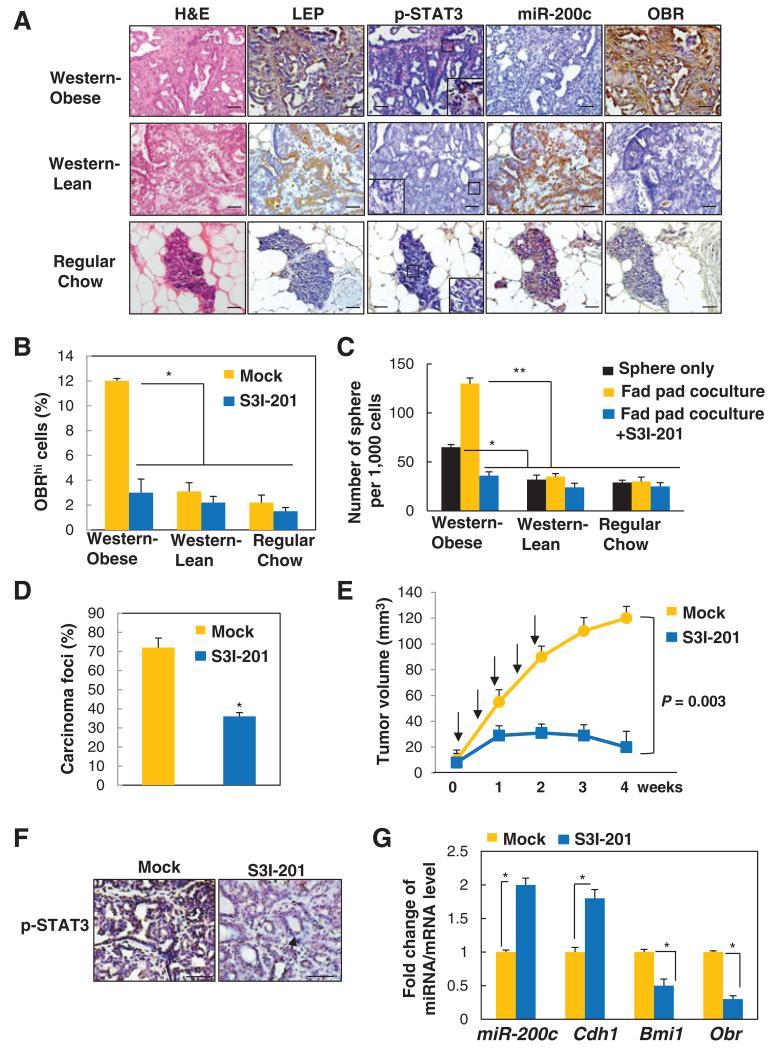
Figure 5.
STAT3 inhibitor suppresses leptin-induced CSCs and prevents obesity-related breast cancer progression. A, representative images showing expression levels of leptin (LEP), p-STAT3, miR-200c, and OBR (inset showing positive nuclear p-STAT3 in obsess rats; scale bar, 50 μm). B, percentage of OBRhi cells. C, number of tumor spheres (spheres only or spheres cocultured with the tumor-associated epithelia-free fat pads from individual diet groups) isolated from mammary tumors of the MNU-treated lean and obese rats with Western diet, and the MNU-treated control rats with regular diet, which were treated with S3I-201 (n = 5 per group; *, P < 0.05; **, P < 0.01). D, percentage of observed mammary carcinoma. E, mammary tumor growth (arrows, time points of treatment). F, representative p-STAT3 staining with distinct tumor morphologies: aggressive adenocarcinoma (left), benign adenoma (right; arrow, low cuboidal epithelial cells forming duct-like structures; scale bar, 100 μm). G, fold change of miRNA/mRNA expression in tumors isolated from MNU-treated obese rats with Western diet that had been treated with vehicle or S3I-201 (n = 5 per group; *, P < 0.05). Error bars, ±SD.
Sphere formation under coculture with epithelial-free mammary fad pad
Single tumor cells were isolated from the obese or lean rats and cultured in suspension in low-attachment culture wells to form spheres. The associated epithelial-free fat pads were isolated as described previously (16) and then washed in serum-free media and cut into explants of approximately 1 × 1 × 1 mm3 pieces. Explants were randomly distributed onto cell culture inserts (BD Biosciences; 3.0-μm pore size, high-density membrane), which were placed above the spheres cocultured in serum-free media.
Results
Leptin induces EMT and stem cell properties via activation of STAT3
To understand whether and how leptin influences mammary stem cell and CSC population, including the epithelial-mesenchymal transition (EMT) phenotype that was previously linked to CSC traits (10, 13), we first treated human mammary epithelial cells MCF12A and also breast cancer cells MCF7 with leptin for 3 days. We found that leptin treatment induced a prominent EMT phenotype from a cobblestonestone-like to a spindle-like morphology in both cell lines (Fig. 1A), accompanied by decreased expression of the epithelial marker, E-cadherin; increased expression of the mesenchymal marker, N-cadherin (Fig. 1B); and a substantial enhancement in the putative stem cell population marked by CD24−CD44+ (Fig. 1C).
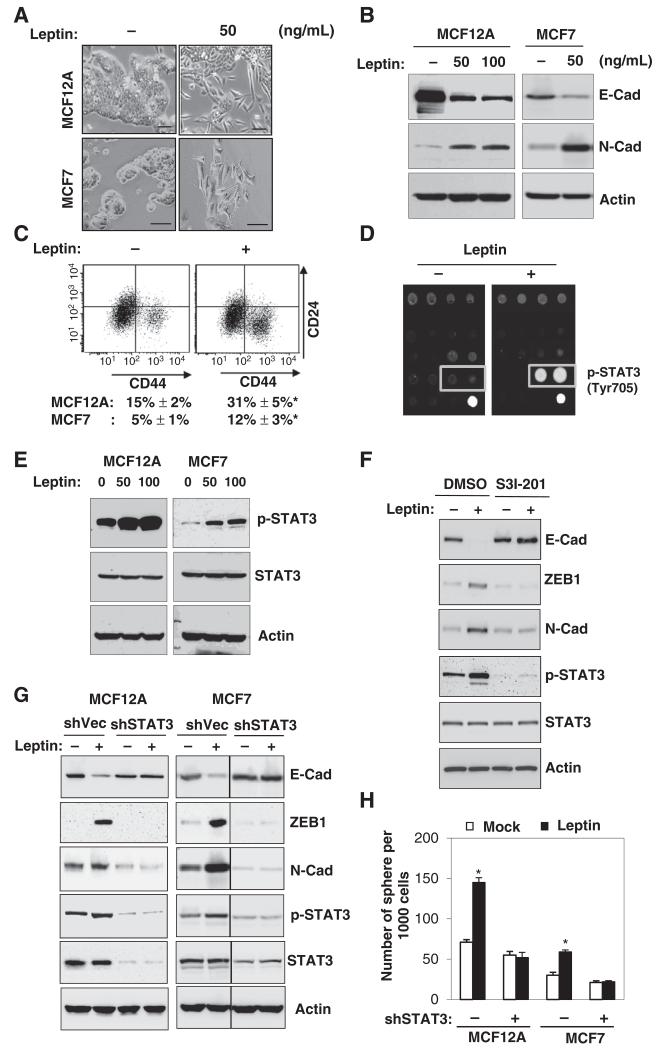
Figure 1.
Leptin induces EMT and stem cell properties via activation of STAT3. A-C, cell morphological change (A), protein expression of E-cadherin (epithelial marker) and N-cadherin (mesenchymal marker; B), and representative double staining CD24/CD44-FACS plots of MCF12A (top) and the percentages of CD24−CD44+ populations (bottom) in MCF12A and MCF7 cells treated with the indicated concentration of leptin for 3 days (C). Scale bar, 50 μm; n = 3; *, P < 0.05. D and E, representative antibody array image (bottom right, positive control) of MCF7 cells (D) and immunoblot (E) showing significant STAT3 phosphorylation (Y705) in MCF12A and MCF7 cells that were treated with leptin for 6 hours. F, EMT protein expression levels in MCF7 cells that were treated with leptin and S3I-201. G, EMT protein expression levels. H, number of spheres generated from MCF12A and MCF7 cells that stably expressed sh-STAT3 and was treated with leptin for 7 days. n = 3; *, P< 0.05. Leptin concentration. 50 ng/mL. Error bars, ±SD.
It is known that leptin, bound to leptin receptor (OBR/LEPR) on the cell surface, could activate a myriad of complex signaling cascades (2). To investigate the key mechanism downstream of leptin that could potentially impact EMT and stem cell properties in mammary epithelial cells, we next systematically examined the signaling pathways regulated by leptin treatment in MCF12A cells using a protein antibody array consisting of specific antibodies that detect the activation (phosphorylated or cleaved) of important intracellular signaling nodes involving a variety of pathways, such as AKT/mTOR/GSK3β, MAPK/ERK/p38/JNK, STAT1/3, p53, and caspase/PARP. Interestingly, leptin most significantly upregulated the phosphorylation of STAT3 at Tyr705 (a 10.5-fold increase, P < 0.05, Fig. 1D, Supplementary Table S1). This result can be validated by immunoblotting in both MCF12A and MCF7 cells (Fig. 1E). Furthermore, the blocking of STAT3 activity by S3I-201, a STAT3 inhibitor that blocks tyrosine phosphorylation, dimerization, and DNA binding of STAT3 in the nucleus (14) markedly increased E-cadherin expression, whereas it decreased the levels of N-cadherin and ZEB1 induced by leptin (Fig. 1F). Similarly, specific inhibition of STAT3 by knocking-down STAT3 using lentiviral delivered shRNA reversed leptin-promoted EMT phenotype and sphere formation in both MCF12A and MCF7 cells (Fig. 1G and H). Together, these data suggest that leptin promotes the EMT and stem cell properties via activation of STAT3.
Leptin induces STAT3-G9a interaction and their co-occupancy on targeted gene promoters involved in cell differentiation
Although STAT proteins primarily act as a transcription activator, evidence has suggested that they can mediate transcription repression of certain target genes by recruiting the corepressor complex instead of the coactivator complex under regulation of specific cellular contexts or chromatin features (17, 18). To determine mechanism(s) underlying the regulation of the stem cell properties by leptin-activated STAT3, a mass spectrometric analysis was used to identify potential chromatin proteins that specifically interact with STAT3 in response to leptin treatment in MCF7 cells. Interestingly, we found that a histone methyltransferase G9a was significantly associated with STAT3 in leptin-treated MCF12A cells, where endogenous STAT3 can be reciprocally coimmunoprecipitated with G9a in the nucleus (Supplementary Table S2, Fig. 2A). G9a, a H3K9 methyltransferase known to be highly expressed in a variety of human cancers (19, 20), is responsible for mono- and dimethylation of H3K9 to mediate epigenetic silencing. Nuclear STAT3 and G9a interaction appeared to be specific and highly responsive to leptin, as another known H3K9 methyltransferase Suv39h1 could not interact with STAT3 regardless of the presence or absence of leptin (Fig. 2A). GST pull-down data also validated the direct physical interaction between STAT3 and G9a proteins in vitro (Fig. 2B). To understand whether and how interaction of STAT3 and G9a impacts global gene expression, we performed genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) analysis targeting STAT3- and G9a-bound chromatin regions. We found that leptin most significantly enhanced STAT3-G9a co-occupancy on promoters of a cohort of genes involved in cell differentiation (Supplementary Fig. S1A and S1B), such as NUMB, GATAT3, FOXA2, HOXA5, by gene ontology enrichment analysis (FDR < 0.5, P < 0.01; Supplementary Table S3). Furthermore, STAT3-G9a also highly co-occupied promoter regions of several noncoding RNAs, such as MiR-200c-3p and MiR-34-5p (Supplementary Fig. S1C and S1D showing a representative MiR-200c ChIP-seq binding plot). Consistently, the expression levels of STAT3-G9a targeted genes/miRNAs were significantly downregulated by leptin treatment, among which miR-200c was the most repressed (Supplementary Fig. S1E). Furthermore, the gene/miRNA expression levels suppressed by leptin treatment were able to be restored by knocking down STAT3 or G9a (Supplementary Fig. S1E), suggesting that both STAT3 and G9a are required for leptin-mediated repression of these targeted genes/miRNAs.
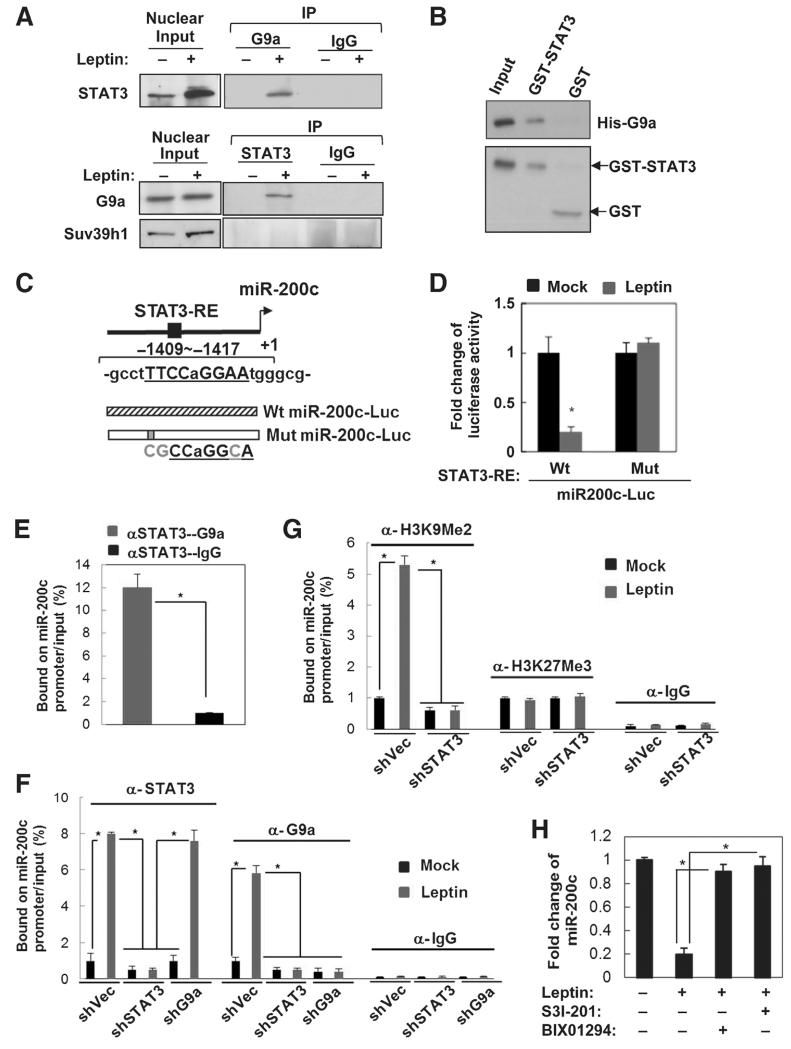
Figure 2.
Leptin-activated STAT3 recruits G9a to repress miR-200c via H3K9Me2 mediated gene silencing. A, reciprocal coimmunoprecipitation assay showing interaction between endogenous nuclear STAT3 and G9a in leptin-treated MCF7 cells (Input, 10% nuclear extract). B, GST pull-down assay showing direct association of GST-STAT3 and His-G9a in vitro. C, diagram showing MiR-200c promoter with the putative STAT3 response element (STAT3-RE, underline), and the structure of luciferase reporters driven by MiR-200c promoter with the wild-type (Wt) and mutated STAT3-RE (Mut, mutations are in gray). D, fold change of luciferase activity driven by MiR-200c promoter with the wild-type or mutated STAT3-RE under leptin treatment in MCF7 cells (n = 3; *, P < 0.05). E, sequential ChIP-PCR analysis showing the percentage of STAT3-G9a complex bound MiR200c promoter chromatin/input chromatin in leptin-treated MCF7 cells (n = 3; *, P < 0.05). IgG was used as a negative control. ChIP-PCR analysis showing the percentage of STAT3/G9a (F)- and H3K9Me2/H3K27Me3 (G)-bound MiR-200c promoter chromatin/input chromatin in MCF7 cells that expressed shSTAT3 along with leptin treatment. n = 3; *, P < 0.05. H, fold change of miR-200c expression in MCF7 cells under leptin and S3I-201/BIX01294 treatment. n = 3; *, P < 0.05. Error bars, ±SD.
Leptin-activated STAT3 recruits G9a to repress miR-200c via H3K9Me2-mediated epigenetic silencing and thereby promotes EMT stem cell phenotype
Notably, studies have shown that miR-200c is the most downregulated miRNA in the normal stem cell and neoplastic stem cell population (13, 21), where lost miR-200c expression de-repressed miR-200c targets, including ZEB1 and BMI1, to promote EMT and stem cell properties (13). Consistent with the ChIP-seq data, by analyzing the changes in the global miRNA expression profile in response to the leptin treatment using a genome-wide miRNA-PCR array consisting of 1,066 annotated miRNAs, we found that leptin significantly downregulated a subset of microRNAs (>2-fold change, P < 0.05) in MCF12A cells, among which microRNA-200c-3p (miR-200c) was the most significantly downregulated (a 6.2-fold decrease compared with the mock treatment, P < 0.01, Supplementary Fig. S2A). Despite the previous findings that link miR-200c to regulation of EMT/stem cell phenotype, the epigenetic mechanism that regulates miR-200c expression in response to the microenvironmental stimuli, such as leptin, is largely unknown, and the functional role of miR-200c in leptin-induced tumorigenesis remains unexplored.
Therefore, to investigate whether leptin indeed downregulates miR-200c through STAT3-G9a-mediated epigenetic silencing, we first analyzed, using promoter analysis, the transcription factor response elements located within the 2-kb region upstream of the transcription starting site of miR-200c (Genomatix MetInspector). We found a putative STAT3 response element located within the miR-200c promoter that had a high consensus score (Fig. 2C, the matrix similarity score > 0.95 as cutoff, score = 1 as perfect match). To validate the direct association of STAT3 with miR-200c in breast cancer cells, we performed ChIP analysis in MCF7 cells targeting the STAT3 consensus binding element using an antibody specifically against STAT3. Leptin treatment resulted in transcription repression of the luciferase driven by miR-200c, which was rescued by mutations of the STAT3 response element (Fig. 2D). The sequential ChIP results further revealed that STAT3 along with G9a were indeed bound to the STAT3 response element on miR-200c promoter (Fig. 2E). Leptin treatment significantly increased the association of both STAT3 and G9a with the miR-200c promoter where H3K9Me2 level was highly enhanced (Fig. 2F and G), whereas another major histone modification involved in gene silencing, H3K27Me3, remained unchanged (Fig. 2G). Interestingly, the association between G9a and miR-200c promoter as well as leptin-induced H3K9Me2 level was abolished upon knocking down of STAT3 (Fig. 2F and G), suggesting that STAT3 is required for recruitment of G9a to miR-200c promoter where it mediates silencing of MiR-200c via activation of H3K9Me2. Consistently, leptin treatment significantly downregulated miR-200c expression, which can be rescued by treatment of S3I-201 and also by a selective G9a inhibitor, BIX01294 (Fig. 2H). Together, these data suggest that nuclear STAT3 can recruit G9a in an miR-200c-bound complex to cooperatively regulate miR-200c expression.
Functionally, to understand whether downregulation of miR-200c is required for leptin-mediated modulation of EMT and stem cell phenotypes, we stably expressed miR-200c in MCF7 and MCF12A cells. We found that re-expression of miR-200c was able to reverse the mesenchymal phenotype that was previously induced by leptin to the epithelial phenotype (MET, Supplementary Fig. S2B and S2C). Restoration of miR-200c also significantly repressed the CD24−CD44+ population as well as the sphere formation that were promoted by the leptin treatment in both the MCF12A and MCF7 cells (Supplementary Fig. S2D and S2F). Together, these data suggest that downregulation of miR-200c is required for leptin-promoted EMT and stem cell phenotypes in both normal and neoplastic mammary epithelial cells.
miR-200c reciprocally regulates leptin signaling by targeting OBR
Next, we attempted to identify new miR-200c targets that may be involved in the regulation of leptin-promoted stemness. Using global gene expression profiling analysis of MCF7 cells that were under leptin treatment for 12 hours compared with that of the mock-treated cells, combined with the analysis result of miR-200c seed sequence matching targets from 3 miRNA target prediction databases (DIANSmT, miRand, TargetScan), 26 putative miR-200c targets were identified with expression levels significantly upregulated by leptin treatment (>2-fold change, n = 3, P < 0.05) and also with high context and conservation scores of miR-200c interaction (Fig. 3A, TargetScan 6.0). Interestingly, among these targets, leptin receptor (OBR/LEPR) showed a significant upregulation (4.8-fold) by knockdown of miR-200c, whereas 2 previously known miR-200c targets BMI1 and ZEB1 had only 2.1- and 3.7-fold of change, respectively (Fig. 3A). Immunoblot analysis further validated that OBR protein expression was markedly upregulated by knockdown of miR-200c, whereas it was downregulated by ectopic expression of miR-200c in MCF7 cells (Fig. 3B). To determine whether OBR is directly regulated by miR-200c through interaction of the complementary 3′-UTR region (conserved between human, mouse and rat, Fig. 3C), a luciferase reporter linked with 3′-untranslated region (UTR) of OBR was used. Indeed, expression of miR-200c significantly decreased OBR luciferase activity, whereas a reporter mutated in OBR 3′-UTR failed to respond to miR-200c expression (Fig. 3D). Furthermore, OBR along with p-STAT3 levels were elevated by the leptin, and leptin-induced OBR and p-STAT3 expression was abolished by re-expression of miR-200c, suggesting that leptin enhances OBR expression through downregulation of miR-200c (Fig. 3E). Consistently, knocking down either STAT3 or G9a, which restored miR-200c levels as shown in Supplementary Fig. S1E, effectively abolished the induced OBR expression by leptin (Supplementary Fig. S3A and S3B). Together, these data corroborate that OBR is a bona fide miR-200c target and may be involved in the self-reinforcing leptin–miR-200c signaling in regulation of stem cell properties, especially in the CSC population.
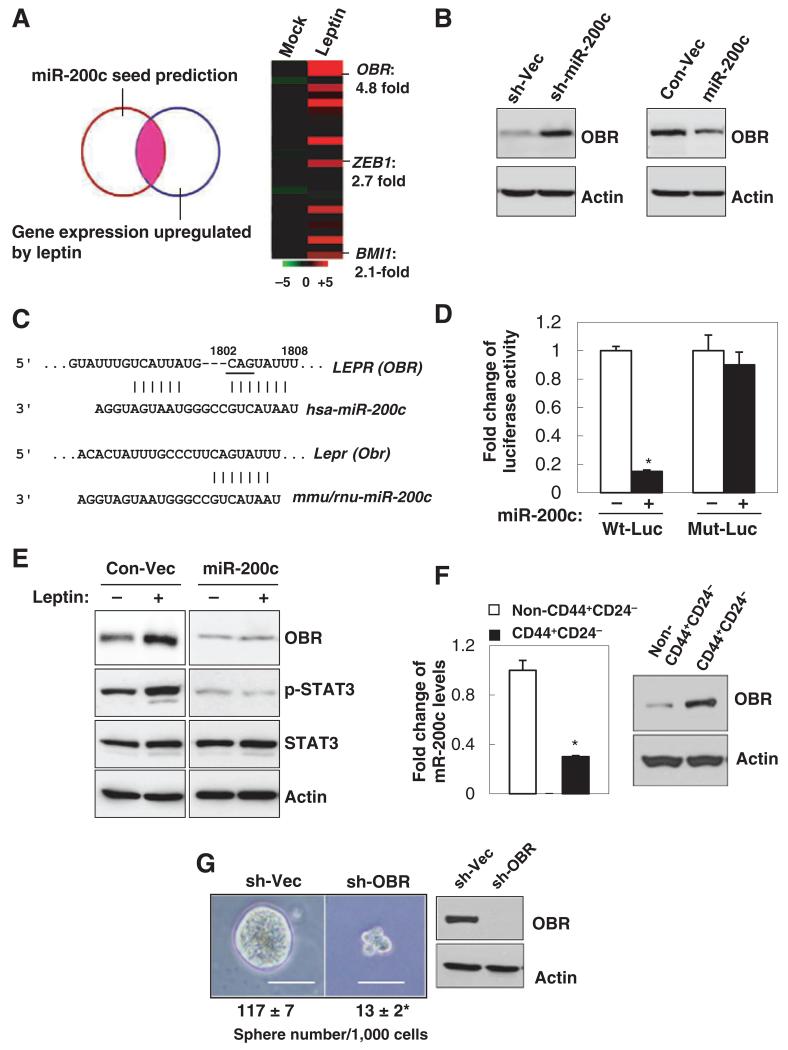
Figure 3.
miR-200c reciprocally antagonizes leptin signaling by targeting OBR. A, the overlapping genes were identified by miRNA seed sequence prediction and global gene expression profiles compared between vehicle and leptin-treated MCF7 cells (left). Heatmap showing mean expression values of these genes that were most significantly upregulated in leptin-treated MCF7 cells (right, n = 3 per each group; fold change > 2; P < 0.05). B, OBR expression levels in MCF7 cells that stably expressed miR-200c, sh-miR-200c, and the control vectors. C, diagram showing the putative miR-200c targeting seed sequence on OBR-3′UTR conserved between human, mouse, and rat (underline indicates mutation of CAG to ACC in Mut-Luc). D, fold change of luciferase activity driven by the wild-type or mutant OBR-3′UTR reporter. E, protein expression in MCF7 cells that stably expressed miR-200c under leptin treatment. F, miR-200c and OBR expression levels in the CD44+ CD24− population versus the non-CD44+CD24− population isolated from primary human breast tumor cells (PT2). G, number of tumor spheres (per 1,000 cells) generated from primary human breast tumor cells that stably expressed sh-Vector or sh-OBR (PT2, n = 3; scale bar, 100 μm; *, P < 0.05).
Thus, to determine whether OBR expression is functionally linked to the CSC traits, we first examined OBR expression in the CSC-enriched CD24−CD44+ cell population compared with the non-CD24−CD44+ cell population isolated from primary human breast tumor cells. Compared with the non-CD24−CD44+ cell population, OBR protein was found to be highly expressed in the CD24−CD44+ cell population, where miR-200c expression was reduced (Fig. 3F). Next, we stably knocked-down OBR using lentiviral delivered shRNA (sh-OBR) in primary human breast tumor cells. Suppression of OBR led to a significant decrease in tumor sphere formation (Fig. 3G), suggesting that OBR may play a role in maintenance of the CSC traits (no significant cell death or cell0cycle alteration was observed; data not shown).
OBRhi cell population is induced by leptin and enriched in CSC traits
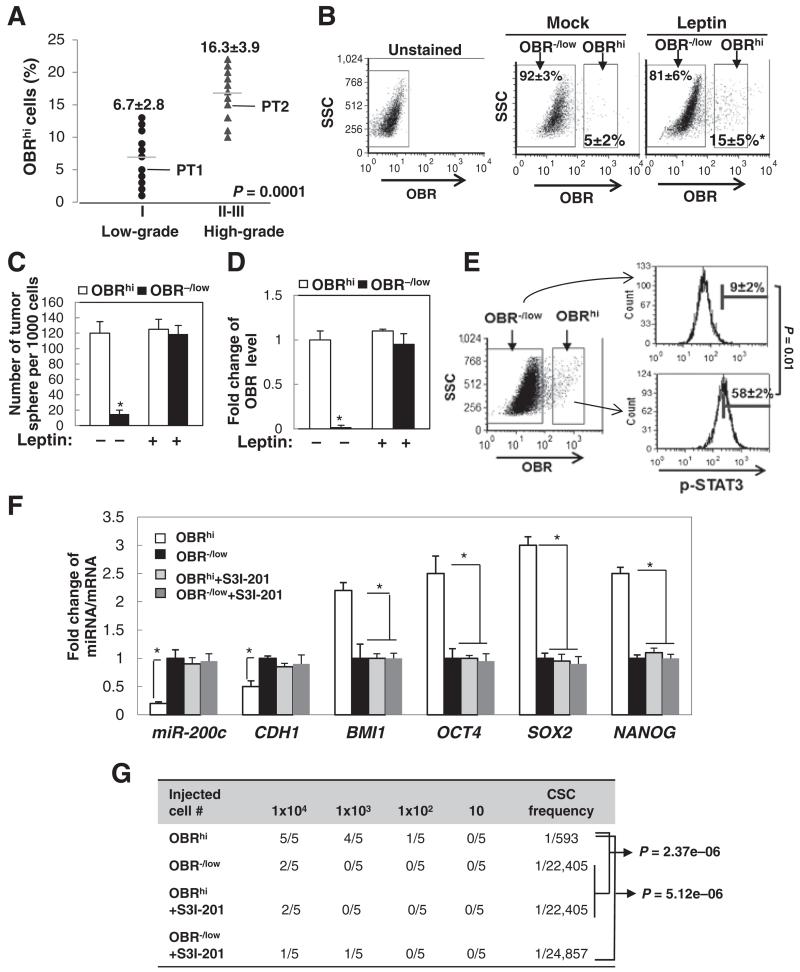
It is recognized that conventional CSC markers that lack a functional link and specificity create challenges for accurate characterization and treatment of CSCs. Previous studies using CD24−CD44+, the most widely used breast cancer stem cell marker, identified a population that may consist of not only CSCs but also other heterogeneous cells (22, 23). Because OBR is a cell surface protein whose expression is potentially associated with the CSC traits, we attempted to determine whether OBR could serve as an effective cell surface marker enriching the breast CSC population induced by leptin, based on the functional link (leptin–miR-200c–OBR) identified in Fig. 3. Using a fluorescence-conjugated antibody targeting OBR, primary human breast tumor cells were sorted for 2 populations with high (hi) or negative/low (−/low) endogenous OBR expression (fluorescence intensity OBR− : <101, OBRlow: 101–102, OBRhi > 102). We found that the majority of the tumor cells are OBR−/low, and the abundance of OBRhi population is correlated with high tumor grade (low grade I, OBRhi 6.7% ± 2.8%; high grade II–III, OBRhi 16.3% ± 3.9%, n = 12 patient samples/each group, P = 0.0001; Fig. 4A). Furthermore, leptin treatment to the primary tumor cells (PT1) for 3 days led to a significant increase of the OBRhi population (5% to 15%, P < 0.05; Fig. 4B). To understand how leptin influences OBRhi and OBR−/low populations in relation to the CSC traits, we FACS-sorted OBRhi and OBR−/low populations from the primary tumor cells, which were then maintained in sphere cultures individually under leptin or vehicle treatment for 10 days. We found that OBRhi cells showed superior sphere-forming capacity compared with OBR−/low cells in the absence of leptin. Interestingly, OBR−/low cells treated with leptin for 10 days started to exhibit sphere-forming capacity similar to OBRhi cells, accompanied by markedly elevated OBR expression, comparable to the OBR level in OBRhi cells (Fig. 4C and D).
Figure 4.
Leptin induces OBRhi cells, a population enriched in CSC traits. A, percentage of OBRhi cells in primary human tumor cells from low-grade (I) or high-grade breast tumors (II-III; n = 12 in each group). B, flow cytometric plots showing the changes in percentage of OBR−/low or OBRhi of primary human tumor cells (PT1) under leptin treatment for 3 days. C, number of tumor spheres. D, OBR expression level from primary human breast tumor cells (PT1) that had been FACS-sorted for high (hi) or negative/low (−/low) expression of OBR and then treated with leptin or vehicle for 10 days (sphere > 100 μm, n = 3; *, P < 0.05). E, horizontal bars showing the percentages of cells with high p-STAT3 levels in OBRhi or OBR−/low population that were isolated from primary human breast tumor cells (PT2) using intracellular flow cytometric staining (n = 3; *, P < 0.05). F, miR-200c and mRNA expression levels in OBRhi or OBR−/low cells that were treated with S3I-201 (n = 3; *, P < 0.05). G, NOD/SCID mice were inoculated with OBR−/low or OBRhi cells isolated from primary human breast tumor cells and treated with S3I-201. The frequency of the tumorigenic CSC was determined by extreme limiting dilution analysis (n = 5 per group). Error bars, ±SD.
We then investigated stemness-associated molecular signatures compared between the FACS-sorted OBRhi cells and OBR−/low cells. We found that, compared to OBR−/low cells, OBRhi cells exhibited substantially increased phospho-STAT3 (Y705), decreased expression of miR-200c and CDH1 (E-cadherin), and elevated levels of stemness-related genes, including NANOG, OCT4, SOX2, and BMI1 (known as a miR-200c target; Fig. 4E and F; ref. 13). In addition, OBRhi cells showed promoted mammary xenograft tumor formation with a highly enhanced CSC frequency (Fig. 4G). Blocking of STAT3 activation in OBRhi cells by S3I-201 treatment restored miR-200c and CDH1, repressed the levels of stemness-related genes, and significantly diminished the formation of mammary xenograft tumors (Fig. 4F and G). Together, these data suggest that a subset of breast tumor cells (OBR−/low) may acquire high OBR expression under leptin stimulation, through leptin-mediated silencing of miR-200c to de-repress OBR, and generate a CSC-like population (OBRhi) with substantially enhanced self-renewing and tumorigenic capacity.
Increased adiposity and tumor aggressiveness in diet-induced obesity and breast cancer rats
To study the complex interaction between the mammary epithelia and the microenvironment, with a focus on the mechanisms of leptin during the development of mammary cancer in vivo, we have established a diet-induced obesity (DIO) rat model of breast cancer (Supplementary Fig. S4A; ref. 24). Briefly, outbred female Sprague-Dawley (SD) rats were fed a “Western Diet” high in fat and simple sugars. Reflective of human obesity trends, approximately 30% of the rats on the Western Diet became obese, whereas 30% were diet resistant and remained lean. To study tumorigenesis, both obese and lean rats were injected with MNU carcinogen. Previous studies demonstrated that MNU-induced rat tumors had histologic and molecular features mimicking human breast cancer progression (25). As is characteristic in humans, we found that the Western-obese rats had substantially increased adiposity (a 2-fold increase of fat mass) and highly elevated leptin levels in their mammary tissues compared with lean rats (Supplementary Table S4, Fig. 5A). Compared with MNU-treated Western lean rats and regular diet-fed rats, which predominantly generated estrogen receptor–positive (ER+) benign adenomas and preneoplastic lesions, respectively, MNU-treated Western-obese rats developed ER-deficient adenocarcinomas along with significantly increased tumor incidences and aggressiveness (Supplementary Fig. S4B and S4C, Supplementary Table S5). The tumor sections from the Western-obese rats exhibited elevated expression of leptin (LEP), phospho-STAT3 Y705 (p-STAT3), and OBR, along with showing a significant reduction in miR-200c level compared with the Western lean and regular diet-fed rats (Fig. 5A, Supplementary Table S6). Interestingly, we observed that even in the absence of the carcinogen MNU, the histologically normal mammary tissues of the Western-obese rats exhibited elevated leptin (LEP) and p-STAT3 levels, along with decreased miR-200c and enhanced OBR expression, compared with the Western-lean rats and the regular diet-fed rats (Supplementary Fig. S5A and S5B), similar to the data shown in Fig. 5. Together, these data suggest that adiposity-induced leptin receptor overexpression and signaling dysregulation may serve as a cancer-predisposing condition, which likely further contributes to promoting tumor progression in the presence of a carcinogen.
Pharmacologic inhibition of STAT3 suppresses leptin-induced CSCs and cancer progression in diet-induced obesity and breast cancer rats
To further understand whether S3I-201 treatment could be therapeutically effective for obesity-promoted mammary tumorigenesis, we treated the Western-obese rats with S3I-201 twice per week for 2 weeks (i.v. 5 mg/kg, n = 5/group, Supplementary Fig. S5C-S5E) at 2 months after MNU injection, when the obese rats started to display mammary tumors. Compared with the Western lean and regular diet-fed rats, the percentage of OBRhi cells isolated from the primary tumors of the Western-obese rats was significantly elevated, which can be suppressed by the treatment of S3I-201 in vivo (Fig. 5B). To further understand the influence of leptin (secreted from the mammary adipose tissue) on formation of the tumor spheres (isolated from the mammary tumor), we used a coculture system that was physiologically parallel to the coexisting compartments of the mammary fat pad and the epithelial/tumor cells, which were separated by a basement membrane that allowed the passage of secreted molecules, such as leptin (16). Compared with the Western lean and regular diet-fed rats, the tumor sphere-forming capacity was markedly increased in the primary mammary tumor cells isolated from the Western-obese rats and was further enhanced by coculturing the spheres with the associated obese fat pads; the enhancement was completely abolished upon treatment of S3I-201 in the cultures (Fig. 5C).
Notably, S3I-201 treatment in the Western-obese rats significantly diminished the incidence of mammary carcinomas (Fig. 5D), decreased tumor growth (Fig. 5E), and reverted the aggressive adenocarcinoma phenotype to a phenotype similar to the benign adenoma observed in the lean rats (with low cuboidal epithelial cells forming duct-like structures, Fig. 5F and Supplementary Fig. S4B), accompanied by significantly suppressed p-STAT3 (Supplementary Fig. S5D-S5F), decreased Obr and Bmi1 expression, and increased levels of miR-200c and Cdh1 (E-cadherin; Fig. 5G). Together, these data suggest that diet-induced obesity, taking on a leptin–STAT3–miR-200c regulatory mechanism, promotes breast CSCs and the development of aggressive mammary carcinoma.
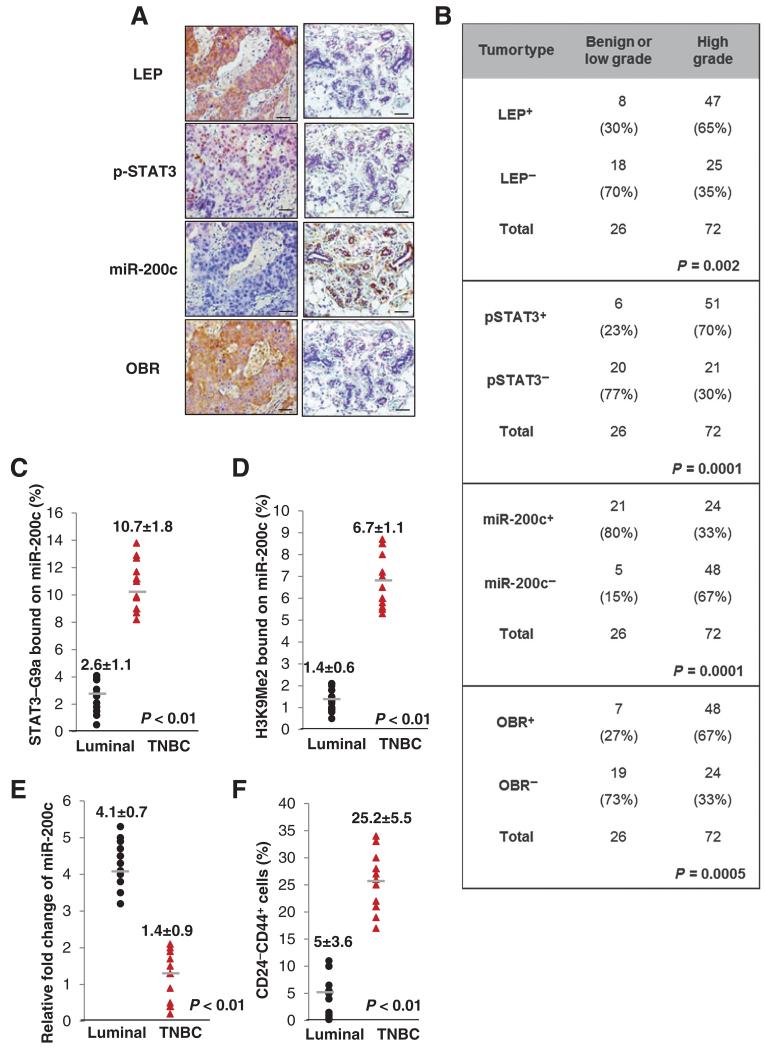
Overexpression of LEP, p-STAT3, and OBR is correlated with H3K9Me2-mediated silencing of miR-200c in poorly differentiated aggressive breast cancer
To further strengthen the pathologic correlation of leptin-regulated signaling cascade in human breast cancer, we performed a correlation analysis of LEP, p-STAT3, miR-200c, and OBR expression levels in human breast tissue microarrays consisting of a cohort of normal and malignant breast tissue samples. We found that miR-200c expression level was most abundantly expressed in benign lesions and well-differentiated low-grade breast tumors (grade I) and was negatively correlated with levels of LEP, p-STAT3, and OBR (Fig. 6A and B). In contrast, miR-200c expression level was significantly reduced in the poorly differentiated high-grade tumors (grades II-III), where LEP, p-STAT3, and OBR were most highly expressed (Fig. 6B, n = 98, P < 0.001). To further determine the correlation of leptin-regulated signaling cascade with the breast cancer subtypes, we performed sequential ChIP in primary human breast tumor samples (11 luminal breast cancer samples and 11 TNBC samples). Notably, TNBC, a breast cancer subtype deficient in expression of ER, progesterone receptor (PR), and HER2, tends to be more aggressive than the other subtypes and manifests high recurrence and poor survival (10). We found that, compared with luminal breast cancer, the association of STAT3-G9a to miR-200c promoter was markedly elevated in TNBC, where the level of H3K9Me2 on miR-200c promoter was also enhanced (Fig. 6C and D). Consistently, miR-200c expression was significantly reduced in TNBC, where the CD24−CD44+ population was highly enriched (Fig. 6E and F). Together, these data indicate that leptin–STAT3/G9a–miR-200c signaling is functionally linked to CSC traits and is correlated with aggressiveness and poor differentiation in human breast cancer.
Figure 6.
Overexpression of LEP, p-STAT3, and OBR is correlated with H3k9Me2-mediated silencing of miR-200c in poorly differentiated and triple-negative breast cancer. A, representative images showing expression levels of LEP, p-STAT3, miR-200c, and OBR in 98 human breast tissue specimens (scale bar, 100 μm). B, χ2 analysis of expression levels of LEP, p-STAT3, miR-200c, and OBR in 98 human breast tissue specimens, including benign and low-grade tumors (≤ tumor grade I) and high-grade tumors (tumor grades II-III). −, negative staining; +, positive staining. The association of STAT3-G9a (C) and H3K9Me2 (D) to MiR-200c promoter, and expression level of miR-200c (E) and the percentage of CD24−CD44+ cells (F) in luminal or TNBC (n = 11 in each group). Error bars, ±SD.
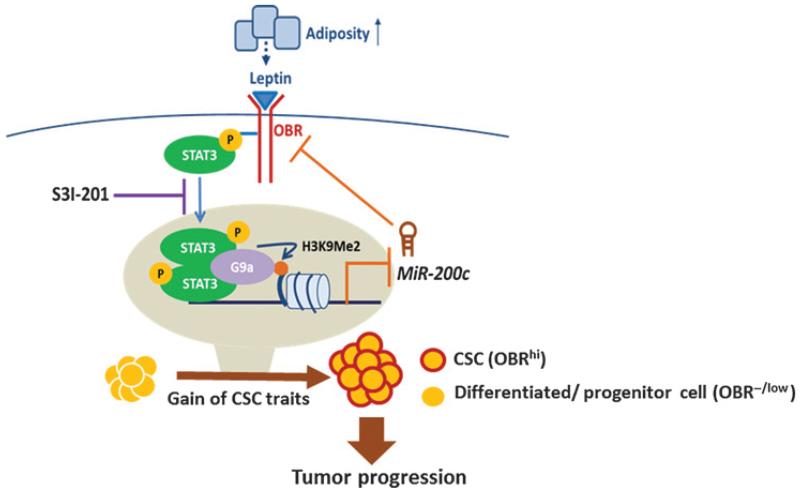
Together, our results reveal that leptin–STAT3–G9a–miR-200c regulatory axis plays a critical role in governing the stemness differentiation plasticity in breast cancer. During mammary cancer development, increased adiposity elevates leptin in the surrounding environment of the mammary epithelia to downregulate miR-200c via STAT3–G9a–mediated epigenetic silencing, which in turn allows a subset of OBR−/low cancer cell population to gain the EMT-CSC traits and high OBR expression, thereby inducing a highly self-renewing and tumorigenic OBRhi-CSC pool, contributing to mammary tumor progression. In contrast, blocking of STAT3 activation restores the expression of miR-200c, which can program OBRhi cells from the CSC phenotype to a differentiated epithelial phenotype, and thereby suppresses cancer progression (Fig. 7).
Figure 7.
A proposed model illustrating the leptin–STAT3–G9a–miR-200c regulatory axis plays a critical role in governing the CSC traits in response to microenvironmental adiposity, contributing to breast tumor progression.
Discussion
In response to various microenvironmental cues, STATs have been known as major transcription activators in regulation of genes and miRNAs involved in a myriad of cellular processes (26), and an integrated regulatory circuit of miRNAs, transcription factors, and chromatin-modifying activities is exploited to reinforce the breast cancer stem cell state (27). Echoing that work, our study provides the first evidence showing that STAT3 can recruit G9a to coordinately mediate epigenetic silencing of a specific cohort of gene targets involved in stemness differentiation plasticity. Interestingly, previous study suggests that a STAT family member, STAT5, represses Igk through a tetrameric binding pattern that involves a high-affinity STAT5-binding site adjacent to a low-affinity biding site on Igk promoter where tetrameric STAT5 likely recruits the corepressor complex instead of the coactivator complex (17). Further comprehensive analysis of the promoter sequences co-occupied by STAT3-G9a from our ChIP study will provide better understanding of how sequence features and chromatin contexts coordinately regulates the function of STAT3 (e.g., target selection for transcriptional activation vs. repression).
Furthermore, OBR has been shown to be overexpressed in aggressive epithelial cancers, including breast, colon, and ovarian cancers (6). In this study, we found that breast tumor cells can acquire high OBR expression (OBRhi) under the adiposity leptin enriched environment to generate a population with enhanced CSC properties and tumorigenic capacity. On the basis of the antagonistic regulation between OBR and miR-200c revealed by our data and the established role of lost miR-200c in CSCs from a broad spectrum of poorly differentiated epithelial cancers (21), OBR may serve as an effective cell surface marker for identification of CSC populations in multiple epithelial cancers. Furthermore, with the newly identified leptin–STAT3–miR-200c regulatory axis in this study, restoration of miR-200c is expected to be a therapeutic marker for determining the efficacy of STAT3 inhibitor for targeting the cancer stemness. This will be of great interest and importance to explore in future studies, given that several STAT3 inhibitors are currently tested under clinical trials for various cancer types (clinicaltrial.gov), and an effective therapeutic biomarker will not only guide the decision for personalized treatment but also prevent the adverse reactions from overtreatment.
It is also worth noting that existing obesity models do have limitations in applicability to both human obesity and breast cancer. Standard genetic models of obesity are the ob/ob and db/db mouse and the Zucker fa/fa rat, each of which contain a single mutation in the gene for leptin or its receptor (28). A drawback of these systems is that mutations in leptin signaling are very rare in humans and therefore do not represent the general cause, the complex polygenic human condition of obesity, or parallel the normal progression of human obesity (29, 30). It has been shown that DIO outbred Sprague-Dawley (SD) rats are an excellent model system given their mammary development, mammary tumorigenesis, and development of obesity are physiologically parallel to that of humans (31-33). Furthermore, MNU-induced mammary tumorigenesis in SD rats is a well-recognized model to study carcinogenesis, as it mimics the morphology and the global molecular phenotypes of human breast tumors (33). Indeed, Western-obese rats in our study manifested the development of aggressive, ER-negative mammary tumors with enhanced CSC-associated molecular phenotypes (Fig. 5 and Supplementary Fig. S4), which are accordingly enriched in the high-grade breast tumors and TNBC in patients with breast cancer (Figs. 5 and 6). This parallelism demonstrates that our DIO breast cancer rat model represents a valuable tool to better understand the association of obesity with breast cancer phenotypes and to be used for evaluation of therapeutics for treatment of obesity-related breast cancer.
Supplementary Material
Acknowledgments
The authors thank the assistance of Purdue Veterinary Medicine Histopathology Service Laboratory on histological sectioning and examination of the rat mammary tissue samples. They also thank National Taiwan University for technical assistance in animal experiments.
Grant Support
This study was supported by Women’s Global Health Institute-Mildred Elizabeth Edmundson Research Grant (C.-J. Chang), Walther Cancer Foundation-Obesity and CancerPilot Grant (I.G. Camarillo and C-J. Chang), American Cancer Society IRG Junior Investigator Award (#58-006-53 to C.-J. Chang), Showalter Research Scholar Grant (#206793 to C.-J. Chang), and P30 CA023168 to the Purdue University Center for Cancer Research in support of the use of facilities
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: C.-C. Chang, C.-J. Chang
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): I.G. Camarillo, C.-J. Chang
Writing, review, and/or revision of the manuscript: C.-C. Chang, C.-J. Chang
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): J.-Y. Yang
Study supervision: C.-C. Chang, C.-J. Chang
Other (experiment revision): M.-J. Wu
References
- 1.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348:1625–38. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 2.Cleary MP, Grossmann ME. Minireview: Obesity and breast cancer: the estrogen connection. Endocrinology. 2009;150:2537–42. doi: 10.1210/en.2009-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demark-Wahnefried W, Platz EA, Ligibel JA, Blair CK, Courneya KS, Meyerhardt JA, et al. The role of obesity in cancer survival and recurrence. Cancer Epidemiol Biomarkers Prev. 2012;21:1244–59. doi: 10.1158/1055-9965.EPI-12-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ligibel J. Obesity and breast cancer. Oncology (Williston Park) 2011;25:994–1000. [PubMed] [Google Scholar]
- 5.Grossmann ME, Cleary MP. The balance between leptin and adiponectin in the control of carcinogenesis - focus on mammary tumorigenesis. Biochimie. 2012;94:2164–71. doi: 10.1016/j.biochi.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu X, Juneja SC, Maihle NJ, Cleary MP. Leptin–a growth factor in normal and malignant breast cells and for normal mammary gland development. J Natl Cancer Inst. 2002;94:1704–11. doi: 10.1093/jnci/94.22.1704. [DOI] [PubMed] [Google Scholar]
- 7.Zheng Q, Dunlap SM, Zhu J, Downs-Kelly E, Rich J, Hursting SD, et al. Leptin deficiency suppresses MMTV-Wnt-1 mammary tumor growth in obese mice and abrogates tumor initiating cell survival. Endocr Relat Cancer. 2011;18:491–503. doi: 10.1530/ERC-11-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J, Scherer PE. Leptin and cancer: from cancer stem cells to metastasis. Endocr Relat Cancer. 2011;18:C25–29. doi: 10.1530/ERC-11-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan D, Avtanski D, Saxena NK, Sharma D. Leptin-induced epithelial-mesenchymal transition in breast cancer cells requires beta-catenin activation via Akt/GSK3- and MTA1/Wnt1 protein-dependent pathways. J Biol Chem. 2012;287:8598–612. doi: 10.1074/jbc.M111.322800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 11.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 12.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 13.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13:317–23. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci U S A. 2007;104:7391–6. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallace J. Humane endpoints and cancer research. Ilar J. 2000;41:87–93. doi: 10.1093/ilar.41.2.87. [DOI] [PubMed] [Google Scholar]
- 16.Salameh TS, Le TT, Nichols MB, Bauer E, Cheng J, Camarillo IG. An ex vivo co-culture model system to evaluate stromal-epithelial interactions in breast cancer. Int J Cancer. 2013;132:288–96. doi: 10.1002/ijc.27672. [DOI] [PubMed] [Google Scholar]
- 17.Mandal M, Powers SE, Maienschein-Cline M, Bartom ET, Hamel KM, Kee BL, et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat Immunol. 2011;12:1212–20. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov VN, Bhoumik A, Krasilnikov M, Raz R, Owen-Schaub LB, Levy D, et al. Cooperation between STAT3 and c-jun suppresses Fas transcription. Mol Cell. 2001;7:517–28. doi: 10.1016/s1097-2765(01)00199-x. [DOI] [PubMed] [Google Scholar]
- 19.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–8. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122:1469–86. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fillmore C, Kuperwasser C. Human breast cancer stem cell markers CD44 and CD24: enriching for cells with functional properties in mice or in man? Breast Cancer Res. 2007;9:303. doi: 10.1186/bcr1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le TT, Rehrer CW, Huff TB, Nichols MB, Camarillo IG, Cheng JX. Nonlinear optical imaging to evaluate the impact of obesity on mammary gland and tumor stroma. Mol Imaging. 2007;6:205–11. [PMC free article] [PubMed] [Google Scholar]
- 25.Camarillo IG, Thordarson G, Moffat JG, et al. Prolactin receptor expression in the epithelia and stroma of the rat mammary gland. J Endocrinol. 2001;171:85–95. doi: 10.1677/joe.0.1710085. [DOI] [PubMed] [Google Scholar]
- 26.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polytarchou C, Iliopoulos D, Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc Natl Acad Sci U S A. 2012;109:14470–5. doi: 10.1073/pnas.1212811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speakman J, Hambly C, Mitchell S, Krol E. Animal models of obesity. Obes Rev. 2007;8(Suppl 1):55–61. doi: 10.1111/j.1467-789X.2007.00319.x. [DOI] [PubMed] [Google Scholar]
- 29.Farooqi IS, O’Rahilly S. Monogenic obesity in humans. Annu Rev Med. 2005;56:443–58. doi: 10.1146/annurev.med.56.062904.144924. [DOI] [PubMed] [Google Scholar]
- 30.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, et al. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 31.Chan MM, Lu X, Merchant FM, Iglehart JD, Miron PL. Gene expression profiling of NMU-induced rat mammary tumors: cross species comparison with human breast cancer. Carcinogenesis. 2005;26:1343–53. doi: 10.1093/carcin/bgi100. [DOI] [PubMed] [Google Scholar]
- 32.Ghibaudi L, Cook J, Farley C, van Heek M, Hwa JJ. Fat intake affects adiposity, comorbidity factors, and energy metabolism of Sprague-Dawley rats. Obes Res. 2002;10:956–63. doi: 10.1038/oby.2002.130. [DOI] [PubMed] [Google Scholar]
- 33.Russo J, Gusterson BA, Rogers AE, Russo IH, Wellings SR, van Zwieten MJ. Comparative study of human and rat mammary tumorigenesis. Lab Invest. 1990;62:244–78. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.