Abstract
Introduction
Multiple System Atrophy (MSA) is a neurodegenerative disease which presents heterogeneously with symptoms and signs of parkinsonism, ataxia and autonomic dysfunction. Although MSA typically occurs sporadically, rare pathology-proven MSA families following either autosomal recessive or autosomal dominant patterns have been described, indicating a heritable contribution to the pathogenesis.
Methods
We used Genome-Wide Complex Trait Analysis (GCTA) to estimate the heritable component of MSA due to common coding variability in imputed genotype data of 907 MSA cases and 3,866 population-matched controls. GCTA only assesses the effect of putative causal variants in linkage disequilibrium (LD) with all common SNPs on the genotyping platform.
Results
We estimate the heritability among common variants of MSA in pooled cases at 2.09–6.65%, with a wider range of values in geographic and diagnostic subgroups. Meta-analysis of our geographic cohorts reveals high between-group heterogeneity. Contributions of single chromosomes are generally negligible. We suggest that all calculated MSA heritability among common variants could be explained by the presence of misdiagnosed cases in the clinical subgroup based on a Bayesian estimate using literature-derived rates of misdiagnosis.
Discussion
MSA is a challenging disease to study due to high rates of misdiagnosis and low prevalence. Given our low estimates of heritability, common genetic variation appears to play a less prominent role in risk for MSA than in other complex neurodegenerative diseases such as Parkinson’s disease, Alzheimer’s disease, and Amyotrophic Lateral Sclerosis. The success of future gene discovery efforts rests on large pathologically-confirmed case series and an interrogation of both common and rare genetic variants.
Introduction
Genome Wide Association (GWA) studies have identified risk loci in several neurodegenerative diseases [1–5]. These loci, however only explain a relatively small proportion of the total heritable proportion of disease. Current conservative estimates of the heritable component of Parkinson’s Disease are ~30%, whereas the known GWA loci only account for 3–12% of the burden of disease. Clearly, an understanding of the known and unknown heritable component of disease has the power to inform the research community regarding the value of searching for additional genetic risk, and where to look for this risk.
A recently unpublished GWA study for MSA examined more than 5 million single nucleotide polymorphisms (SNPs) tagged to common genetic variants in 1,030 MSA samples. (Sailer et al, unpublished). The results, which included 918 MSA cases and 3884 controls after quality control, were unable to detect any genome-wide significant associations between tagged SNPs and MSA risk. Though sample size and power were limited in this study, this finding suggests that MSA etiology cannot be easily explained by common SNPs with moderate or large effects. This result, however, does not preclude the role of common variability in MSA; particularly for variants that may only have marginal effects, typical of those observed for GWA in complex disease. The genotype data generated as a part of this study afford an opportunity to look beyond the identification of individual risk loci, toward an estimate of the role and extent of common variants as a heritable component of disease risk.
We estimated the total heritability of MSA from common genetic variants (MAF > 0.01) with Genome-wide complex trait analysis (GCTA). Heritability is defined as phenotypic variation attributable to total genetic variation in all assessed loci. Total genetic variation is estimated in GCTA by generating a ‘genetic relatedness matrix’, which estimates overall genetic differences in each subject. If cases are more genetically similar to one another than they are to controls, we can quantify this higher relative similarity and use it to estimate the total heritability of the disease phenotype. Notably, this approach is possible only in unrelated populations and requires relatively large sample sizes for requisite statistical power. GCTA can examine only the effect of putative causal variants in LD with all common SNPs on the genotyping platform [6,7].
In this study, we estimated the total heritability of MSA with GCTA in order to guide future genetic research into the disease.
Methods
All participants provided written informed consent. Pre-imputation base calling quality control filters were applied using standard conditions. See Supplemental 1 for details. 907 MSA cases, 3,866 controls, and 107,447 SNPs passed initial quality control. All PCA and IBD analyses were carried out using linkage-pruned SNP sets.
Imputation was accomplished using MaCH to estimate subject haplotypes. In the process of imputation, sample genotype data is inferred using a reference haplotype database. Matching the genotypes to common haplotypes and ‘filling in’ (imputing) the missing bases based on typical haplotypes allow for millions of intervening nucleotide sequences not captured by the microarray to be incorporated into the overall genetic data for each sample. Further information can be found in Supplemental 1.
Genome-Wide Complex Trait Analysis
GCTA uses a REstricted Maximum Likelihood (REML) model to estimate the variance in phenotype explained by variance in genotype. Here, we used GCTA’s REML model to estimate the phenotypic variance of MSA, adjusting for population substructure using the top 20 European Ancestry population principal components. This heritability estimate was adjusted for actual population prevalence of MSA (estimated at 0.000046) [8,9]. We first ran GCTA using all samples in a pooled analysis, then divided MSA cases into several sample subsets based on geographic region of origin and whether cases had been pathologically confirmed or only clinically diagnosed (Table 1). Each of these groups were tested against matched controls to estimate total heritability of MSA both before and after imputation. We then meta-analyzed these subgroups under a random effects model in order to assess heterogeneity between the cohorts.
Table 1.
Summary statistics of all samples included within GCTA following stringent quality control analysis. Component numbers of control subjects do not sum to total due to incomplete annotation (i.e. unknown region of origin). Cases not explicitly labeled as pathologically confirmed were assumed to have only a clinical diagnosis.
| Cohort | Number of subjects | Path confirmed / Clinically Diagnosed |
|---|---|---|
| Cases | ||
| United Kingdom | 238 | 141/97 |
| United States | 127 | 108/19 |
| North European | 308 | 28/258 |
| South European | 234 | 14/220 |
| Total | 907 | 291/616 |
| Controls | ||
| United Kingdom | 945 | ---- |
| United States | 793 | ---- |
| North European | 944 | ---- |
| South European | 1184 | ---- |
| Total | 3877 | ---- |
GWAS
Please refer to Supplemental 1 for GWA study methods.
Bayesian Estimate of Parkinson’s Disease-Derived Heritability
We estimated rates of false clinical diagnosis from Osaki et. al 2009 [10]. Using values of 6–25% false positive rate, 70% of false positives as Parkinson’s cases, and heritability priors of 0.31 for PD and 0.1 for all other disorders, we calculated an expected rate of heritability to due misdiagnosis. Derivation of this formula may be found in Supplemental 1.
Results
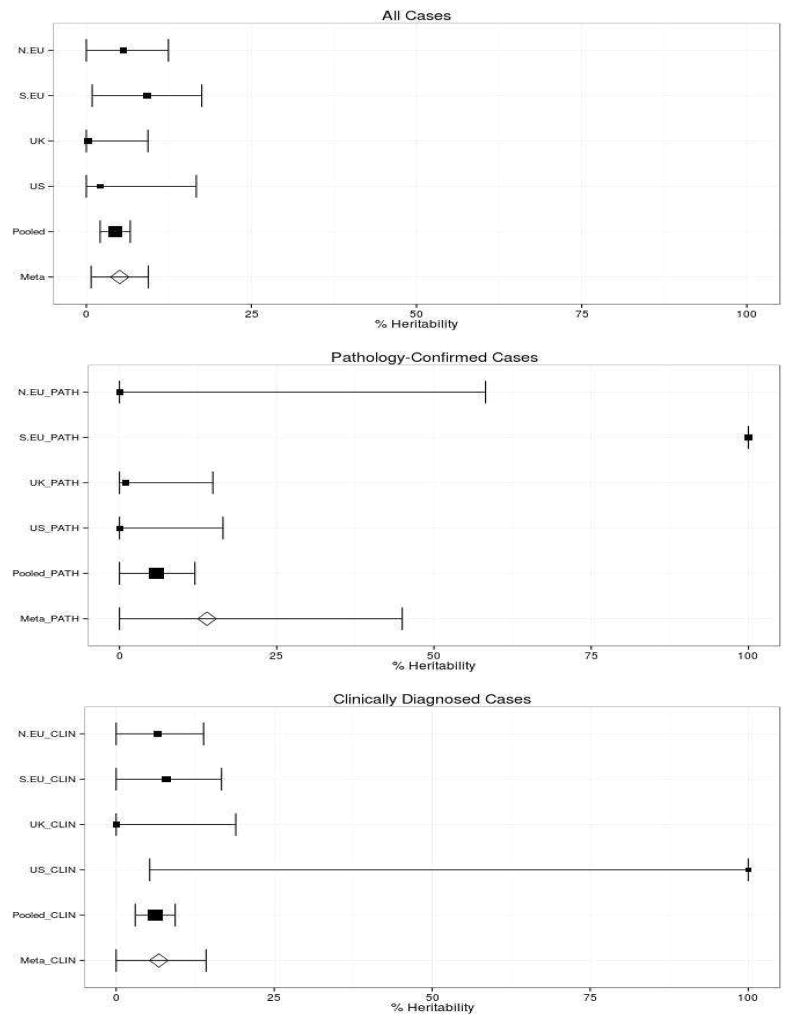
MSA cases and their respective geographic cohort controls show an insignificant amount of heterogeneity within each regional cohort (Supplemental 2). After initial quality control (QC) of genotyped data, we imputed the data using 1k Genomes reference haplotypes order to increase power and provide many more variants for assessment of total heritability. Standard errors of the estimates decreased uniformly post-imputation, likely reflecting improved statistical power and increased information content (Figure 1, Table 2).
Figure 1.
Heritability by Cohort in Diagnostic Subgroups
The size of the center point of these graphs is scaled to the sample size of each subgroup. Some of our cohorts have very high standard errors to due to low numbers of cases vs. controls.
Table 2A.
Heritability Estimates by Cohort and diagnostic subgroup
| Total: Cases & Controls (N) | Cases: Path (N) | Cases: Clinical (N) | Cohort | Imputation Status: 107,447 Genotyped SNPs / 11,138,628 Imputed SNPs | Heritability | Standard Error | 95% Confidence Interval (Percentage Heritability) | P-value |
|---|---|---|---|---|---|---|---|---|
| 1252 | 28 | 258 | N.EU All | Genotyped | 0.1067 | 0.0435 | 2.13–19.21% | 0.007724 |
| Imputed | 0.0563 | 0.0346 | 0–12.42% | 0.04445 | ||||
| 1418 | 14 | 220 | S.EU All | Genotyped | 0.1902 | 0.0506 | 9.09–28.95% | 7.903e–05 |
| Imputed | 0.0918 | 0.0423 | 0.88–17.48% | 0.01439 | ||||
| 920 | 108 | 19 | US All | Genotyped | 0.06652 | 0.0941 | 0–25.10% | 0.2399 |
| Imputed | 0.0210 | 0.0742 | 0–16.65% | 0.3858 | ||||
| 1183 | 141 | 97 | UK All | Genotyped | 0.0000 | 0.0545 | 0–9.33% | 0.5 |
| Imputed | 0.0026 | 0.0463 | 0–7.63% | 0.4793 | ||||
| 4784 | 291 | 616 | All Pooled | Genotyped | 0.0490 | 0.0139 | 2.1–7.63% | 0.0001796 |
| Imputed | 0.0437 | 0.0116 | 2.09–6.65% | 3.252e–05 | ||||
| Cohort Meta- Analysis Random Effects Model | Genotyped I^2 = 5.1% |
0.0962 | 0.0424 | 1.32–17.94% | 0.0232 | |||
| Imputed I^2 = 0% |
0.0506 | 0.0221 | 0.72–9.40% | 0.0223 |
We estimated heritability with GCTA using pooled samples, then divided analyses by population cohort and whether subjects were pathologically-confirmed or clinically diagnosed (Figure 1, Table 2). After estimating heritability of each of these subgroups, we also ran a random effects meta-analysis of all population cohorts in each diagnostic subset: all cases, pathologically-confirmed cases, and cases identified exclusively through clinical diagnosis.
We estimated heritability of pooled MSA samples to be about 4.37% in imputed data (95% CI 2.09–6.65%). Heritability estimates ranged widely in our geographic cohorts, from 0.26% in UK cases to 9.18% in Southern European cases. The UK and US cohorts were composed primarily of pathologically-confirmed cases (Table 1), while the Northern (Germany, Austria, Netherlands, Denmark) and Southern (Italy, Portugal, Spain) European samples were mostly identified by clinical means alone.
MSA, as with many parkinsonian disorders, is often misdiagnosed, making pathologically-confirmed cases significantly more reliable than clinical diagnoses. Therefore, we also estimated the heritability of pathologically confirmed cases alone with the intention of minimizing falsely attributed heritability stemming from genetic underpinnings of other neurodegenerative diseases (i.e. PD, PSP) (Table 2).
In pooled pathologically-confirmed samples, we found heritability to be near zero in genotyped data. However, this estimate increased to around 5.8% (95% CI 0–11.99%) in imputed data, suggesting that the imputed genotypes contribute a great deal to the heritability of MSA. Clinical-only estimates have a pooled heritability of 6.17%, slightly higher than the pooled estimate of all cases and pathologically-confirmed case in the imputed datasets. However, pathologically-confirmed and clinical estimates are characterized by a fairly large standard error, limiting conclusions that can be drawn (Table 2). Furthermore, our meta-analyses are characterized by rather high inter-sample heterogeneity in both the clinical and pathologically-confirmed subsets (30.2% and 78.6% respectively), suggesting that our geographic subpopulations may have some interpopulation differences that cannot be explained by random variation alone.
Curious to see which chromosomes contributed most to MSA heritability, we estimated the heritability from each chromosome. Chromosomal contributions tended to account for less than 1% of total heritability, and generally could not be said to contribute to heritability with 95% confidence. Chromosome 15 passed multiple-test corrections (p < 0.05/22 = 0.00227) in our clinical-only subgroup (0.25–1.70% heritability), but did not pass significance threshold in our pathologically-confirmed-only subgroup or our ‘all-cases’ subgroup. Chromosome 10 contributed significantly to heritability in our ‘all cases’ subgroup (0.43–1.05%) but neither in the clinical-only nor pathologically-confirmed only subgroups. For both chromosomes 10 and 15, the subgroups including clinical cases demonstrate higher heritability estimates than those in pathologically-confirmed cases alone (Supplemental 3).
PD Locus Exploration
Given our suspicion that MSA heritability may be driven in part by misdiagnosed PD cases, we replicated the GWAS performed by Sailer et al (unpublished) with our updated imputation and explored windows ±20 kilobases around Parkinson’s Disease GWA study loci from Nalls et al 2014 and from nearest genes associated with mRNA expression differences [1,11]. Of all variants included in this search, none demonstrated an association with MSA disease phenotype even at an extremely liberal significance cutoff of p < 0.05. Variants included in these windows without any p-value cutoff are included in the supplementary material (Supplemental 4).
Discussion
The usage of GCTA to estimate heritability of neurodegenerative diseases has yielded noteworthy results; heritability estimates obtained by simultaneously measuring all tagged SNPs have revealed values of 31% and 21% for PD and ALS, respectively [12,13]. Heritability estimates from GCTA are usually higher than those estimated from variance in GWA-significant loci alone, approaching heritability estimates reported from twin studies [14,15]. The identification of this “missing heritability”, which is attributed to the inability of GWA study hits to account for the full genetic variance of underlying phenotypic variance, suggests genetic discoveries yet to be made for diseases like PD and ALS without possible confounding factors of twin studies such as shared environment or similar treatment of twins. The strength of GCTA is that by analyzing SNPs simultaneously, it has the ability to combine the small effects of variants not passing significance thresholds in GWA studies, allowing for a much more comprehensive assessment of heritability of a particular phenotype.
Given that MSA shares several overlapping clinical features with both PD and ALS, with an estimated 14% of MSA cases misdiagnosed as other neurodegenerative diseases [15,16], it would be reasonable to hypothesize that MSA may manifest a similar heritability estimate as these diseases. However, MSA heritability estimates are markedly lower than those for PD or ALS: the average post-imputation MSA heritability was demonstrated to be <10% in all subgroups, with the 95% confidence interval in most subgroups overlapping 0% (Figure 1). Cohorts in which pathologically-confirmed cases make up a very small proportion of the total population (Northern and Southern European) have much higher estimates of heritability than those in which pathologically-confirmed cases constitute the majority (US and UK) (Table 2). Pathologically-confirmed samples are the gold standard for MSA cases due to the known problem of MSA misdiagnosis, but these cases comprise only a third of our already limited sample size (291 Pathologically-Confirmed / 907 Total). Our attempts to estimate heritability in pathologically-confirmed cases consequently suffer from very high standard error as a result of this lower sample size and limited power. Despite this reduction in power, our calculated Bayesian estimate of heritability due to misdiagnosed MSA suggests that the results of the pathologically-confirmed analysis may be more scientifically sound than our pooled meta-analysis results.
We acknowledge that our study sample size in conjunction with both the number and distribution of GWA study panel markers limit the lower boundary of variant frequency detected using GCTA, which improves with greater sample size. Thus, while GCTA is more comprehensive than other heritability assessments, we recognize that the entire heritability cannot be fully ascertained through GCTA. Indeed, the small sample sizes of some cohorts leads to unreasonable estimates of heritability after GCTA corrects for the very low prevalence of the disease (i.e. exceeding 100%). GCTA requires large sample sizes of at least several hundred cases to provide reliable estimates, and the very small (<100) case number in some of our subgroup analyses lead to highly unreliable estimates (e.g. 19 cases in US clinical, 14 cases in Southern European pathologically-confirmed). This is reflected in the very high heterogeneity of our cohort meta-analyses.
Individual chromosomes contribute a negligible fraction to MSA heritability, which is unsurprising given the overall very low heritability estimates. Few passed significance thresholds after multiple testing corrections (p<0.05/22). Intriguingly, while chromosome 15 seems to contribute to MSA heritability in the clinical-only subgroup, there is not even a trending relationship in the pathologically-confirmed subgroup (Supplemental 3). When these cases are pooled together, chromosome 15 exhibits a lower contribution to MSA heritability. Due to our limited power, it is challenging to say whether this difference is due to biological differences in pathologically-confirmed and clinical-only subgroups or simply by chance, but this finding does support the idea that some of our reported MSA heritability may be due to clinical misdiagnosis.
Initially, these findings may suggest that MSA is purely a disease of idiopathic origin, lacking any heritable component. However, this is not necessarily indicative of the absence of genetic risk genes. The low heritability estimate could be attributed to several factors: first, the SNPs used within standard GWA studies are limited to the common variants tagged by microarray-based genotyping methods. If putative causal variants associated with MSA are extremely rare (i.e. MAF<1%) and thus not tagged by genotyping platforms, they will go undetected. If a very rare variant only exists within a single case in the full cohort, it cannot be recognized as a genotypic variant shared among cases and thus would not contribute to the overall estimation of MSA heritability, again implicitly highlighting the essential role of sample size in this analysis. Since our cohort consists of all sporadic MSA cases, we are lacking the ability to detect rare variants that could explain MSA etiology within an affected family, which can be further scrutinized via segregation and linkage analyses. We were unable to obtain MSA familial cases for heritability analysis, as these are extremely rare. While one might expect a higher degree of heritability in such cases, the fact that these are so rare would suggest that this would be unrepresentative of the majority of MSA cases. In this case-control study design, one can infer that only modest genotypic variation in common SNPs exists between MSA cases and controls. Though this problem is somewhat ameliorated by imputation, imputation relies on already-demonstrated haplotypes, meaning that rarer genetic variants may be missed.
Secondly, even if causal variants are included within haplotype stretches of SNPs in the array, they may be in incomplete LD with the SNPs that have been genotyped [17]. This is also not mutually exclusive with the former, since causal variants in incomplete LD may also have MAF<1%, further compounding these effects.
Thirdly, rare variants are difficult to impute from array-based genotypes, as genotype platforms are generally based on common variants and imputation relies on linkage disequilibrium, it is also important to note that GCTA analysis does not account for non-additive genetic factors and possible environmental effects; thus, the heritability estimate given by GCTA represents a lower limit of MSA heritability that is likely to increase if such factors could be incorporated into the estimate.
Finally, the differences in diagnostic status (clinical vs. pathological) require further scrutiny. The Southern and Northern European cohorts exhibited the highest number of clinically diagnosed MSA cases and fewest pathological cases, while the US and UK together included 82% of all pathologically confirmed cases (Table 1) Interestingly, this corresponds with estimated heritability levels: the US and UK cohorts show lower heritability (between 0 and 3%), while the Northern and Southern European cohort estimates are much higher (between 0 and 17.48%) (Figure 1, Table 2) This is particularly noteworthy given the 6–25% rate of clinical misdiagnosis rate of MSA; many cases diagnosed as MSA are actually PD cases. PD exhibits a higher heritability estimate of about 31% [12,16]. Thus, it is possible that misdiagnosed cases within the clinical subpopulation are inflating the heritability estimate of MSA.
Indeed, our calculated Bayesian estimate of heritability due to misdiagnosed MSA is 1.00–4.19% based on a comprehensive literature overview of misdiagnosis rates (S1). Our GCTA estimate of MSA heritability using all cases falls in the range of 2.09–6.65%. These ranges overlap, suggesting that all MSA heritability estimated in this study could in principle be explained exclusively through heritability stemming from false positives.
The challenges in attempting to discover genetic risk factors for MSA are two-fold: the disease is extremely rare, so sample size is necessarily rather limited. Secondly, many cases that are clinically diagnosed are likely misdiagnosed, adding noise to any genetic differences that may underlie MSA. Attempting to select only pathologically-confirmed cases would decrease the already-small sample size, limiting statistical power further.
While our dataset represents the most comprehensive collection of MSA genotypes ever assembled (S5), our cohort numbers under 1000 cases and lacks adequate power required to detect uncommon variants and/or those with subtle effects. Future studies must rely on international collaboration to generate large, high-confidence (i.e. pathologically-confirmed), high-quality datasets. Furthermore, the use of next generation sequencing technology in pursuit of MSA genetic etiology could prove extremely valuable, as exomic variation could uncover novel rare variants with effects that have been missed by standard genotyping methods used in this study.
Supplementary Material
Table 2B.
Clinical Cohorts
| Total: Cases & Controls (N) | Cases: Path (N) | Cases: Clinical (N) | Cohort | Imputation Status: 107,447 Genotyped SNPs / 11,138,628 Imputed SNPs | Heritability | Standard Error | 95% Confidence Interval (Percentage Heritability) | P-value |
|---|---|---|---|---|---|---|---|---|
| 1224 | 0 | 258 | N.EU Clinical | Genotyped | 0.1184 | 0.0461 | 2.81–20.89% | 0.005081 |
| Imputed | 0.0658 | 0.0369 | 0–13.83% | 0.03057 | ||||
| 1404 | 0 | 220 | S.EU Clinical | Genotyped | 0.1977 | 0.0536 | 9.27–30.29% | 0.0001884 |
| Imputed | 0.0791 | 0.0446 | 0–16.67% | 0.03743 | ||||
| 812 | 0 | 19 | US Clinical | Genotyped | 1.146861 | 0.558051 | 0–100% | 0.01377 |
| Imputed | 0.5862 | 0.4435 | 5.31–100% | 0.089 | ||||
| 1042 | 0 | 97 | UK Clinical | Genotyped | 0.0000 | 0.1177 | 0–23.09% | 0.5 |
| Imputed | 0.0000 | 0.0965 | 0–18.93% | 0.5 | ||||
| 4482 | 0 | 616 | Clinical Pooled | Genotyped | 0.0854 | 0.0192 | 4.76–12.32% | 4.584e–06 |
| Imputed | 0.0617 | 0.0161 | 3.02–9.33% | 2.247e–05 | ||||
| 4482 | 0 | Clinical Cohort Meta- Analysis Random Effects Model | Genotyped I^2 = 21% |
0.1406 | 0.0420 | 5.82–22.31% | 0.284 | |
| Imputed I^2 = 30.2% |
0.0674 | 0.0382 | 0–14.23% | 0.078 |
Table 2C.
Pathologically confirmed cohorts
| Total: Cases & Controls (N) | Cases:P ath (N) | Cases: Clinical (N) | Cohort | Imputation Status: 107,447 Genotyped SNPs / 11,138,628 Imputed SNPs | Heritability | Standard Error | 95% Confidence Interval (Percentage Heritability) | P-value |
|---|---|---|---|---|---|---|---|---|
| 972 | 28 | 0 | N.EU Pathology Confirmed | Genotyped | 0.114253 | 0.385713 | 0–87.02% | 0.3866 |
| Imputed | 0.000001 | 0.296941 | 0–58.2% | 0.5 | ||||
| 1198 | 14 | 0 | S.EU Pathology Confirmed | Genotyped | 2.269129 | 0.726325 | 100% | 1.413e–06 |
| Imputed | 2.269129 | 0.601680 | 100% | 4.586e–07 | ||||
| 901 | 108 | 0 | US Pathology Confirmed | Genotyped | 0.0000 | 0.1076 | 0–21.09% | 0.5 |
| Imputed | 0.0000 | 0.0839 | 0–16.45% | 0.5 | ||||
| 1086 | 141 | 0 | UK Pathology Confirmed | Genotyped | 0.0545 | 0.0822 | 0–21.09% | 0.2514 |
| Imputed | 0.0100 | 0.0706 | 0–14.85% | 0.4467 | ||||
| 4168 | 291 | 0 | All Pathology Confirmed | Genotyped | 0.0050 | 0.0375 | 0–7.85% | 0.4467 |
| Imputed | 0.0580 | 0.0315 | 0–11.99% | 0.03102 | ||||
| 4168 | 291 | 0 | Path- Confirmed Cohort Meta- Analysis Random Effects Model | Genotyped I^2 = 68.7% |
0.1398 | 0.1615 | 0–45.62% | 0.3865 |
| Imputed I^2 = 78.6% |
0.1391 | 0.1584 | 0–44.95% | 0.3799 |
Highlights.
The heritable component of Multiple System Atrophy (MSA) is currently unknown.
We used Genome-Wide Complex Trait Analysis (GCTA) to estimate the heritable component of MSA due to common coding variability.
We estimate the heritability of MSA in pooled cases at 2.09–6.65%.
Common genetic variation appears to play a less prominent role in risk for MSA than in other complex neurodegenerative diseases.
Acknowledgments
DNA and brain samples: We used DNA samples and phenotype data from the NINDS Human Genetics Resource Center DNA and Cell Line Repository at Coriell (Newark, NJ, USA; http://ccr.coriell.org/ninds), and we would like to thank the patients and the submitters who contributed samples to this repository (S5). Human tissue was kindly obtained from the Queen Square Brain Bank (London, UK), the Institute of Psychiatry Brain Bank, King’s College (London, UK), the UK Parkinson’s disease tissue bank at Imperial College (London, UK), Newcastle Brain Tissue Resource at Newcastle University (Newcastle, UK), the Manchester Brain Bank at the University of Manchester (Manchester, UK), Jacksonville Brain Bank for Alzheimer’s, Parkinson’s and Related Disorders at the Mayo Clinic (Jacksonville, FL, USA), the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland (Baltimore, MD, USA), from the New York Brain Bank of the Taub Institute at Columbia University (New York, NY, USA), the Human Brain and Spinal Fluid Resource Center (Los Angeles, CA, USA), the Miami Brain Bank (Miami, FL, USA), the Center for Neurodegenerative Disease Research at the University of Pennsylvania (Philadelphia, PA, USA), the Harvard Brain Bank (Boston, MA, USA), the Emory University Alzheimer's Disease Research Center Brain Bank (Atlanta, GA, USA) , Neurobiobank München at the Ludwig-Maximillians-Universität (Munich, Germany), Brain Bank Center Würzburg (Würzburg, Germany), the Netherlands Brain Bank at the Netherlands Institute for Neuroscience (Amsterdam, Netherlands), and the Neurological Tissue Bank at the University of Barcelona (Barcelona, Spain).
This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services; project ZO1 AG000949
This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Md. (http://biowulf.nih.gov)
Footnotes
Conflicts of Interest: No relevant conflicts of interest.
- Conception and design of the study: M.F., T.R.P, M.N., A.B.S., H.H. Acquisition of data: A.S, S.S., D.H., T.R.P.; analysis of data: T.R.P., M.N., A.N.; interpretation of data: All authors
- Drafting the article or revising it critically for important intellectual content: All authors
- Final approval of the version to be submitted: All authors: M.F., T.R.P, M.N., A.B.S., H.H, A.S, S.S., D.H, A.N.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Höglinger GU, Melhem NM, Dickson DW, Sleiman PMA, Wang LS, Klei L, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrari R, Hernandez DG, Nalls MA, Rohrer JD, Ramasamy A, Kwok JBJ, et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol. 2014;13:686–699. doi: 10.1016/S1474-4422(14)70065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Deerlin VM, Sleiman PMA, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed Z, Asi YT, Sailer A, Lees AJ, Houlden H, Revesz T, et al. The neuropathology, pathophysiology and genetics of multiple system atrophy. Neuropathol Appl Neurobiol. 2012;38:4–24. doi: 10.1111/j.1365-2990.2011.01234.x. [DOI] [PubMed] [Google Scholar]
- 9.Bower JH, Maraganore DM, McDonnell SK, Rocca WA. Incidence of progressive supranuclear palsy and multiple system atrophy in Olmsted County, Minnesota, 1976 to 1990. Neurology. 1997;49:1284–1288. doi: 10.1212/wnl.49.5.1284. [DOI] [PubMed] [Google Scholar]
- 10.Osaki Y, Ben-Shlomo Y, Lees AJ, Wenning GK, Quinn NP. A validation exercise on the new consensus criteria for multiple system atrophy. Mov Disord Off J Mov Disord Soc. 2009;24:2272–2276. doi: 10.1002/mds.22826. [DOI] [PubMed] [Google Scholar]
- 11.Potashkin JA, Santiago JA, Ravina BM, Watts A, Leontovich AA. Biosignatures for Parkinson’s disease and atypical parkinsonian disorders patients. PloS One. 2012;7:e43595. doi: 10.1371/journal.pone.0043595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller MF, Saad M, Bras J, Bettella F, Nicolaou N, Simón-Sánchez J, et al. Using genome-wide complex trait analysis to quantify “missing heritability” in Parkinson’s disease. Hum Mol Genet. 2012;21:4996–5009. doi: 10.1093/hmg/dds335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller MF, Ferrucci L, Singleton AB, Tienari PJ, Laaksovirta H, Restagno G, et al. Genome-wide analysis of the heritability of amyotrophic lateral sclerosis. JAMA Neurol. 2014;71:1123–1134. doi: 10.1001/jamaneurol.2014.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Chalabi A, Fang F, Hanby MF, Leigh PN, Shaw CE, Ye W, et al. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81:1324–1326. doi: 10.1136/jnnp.2010.207464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirdefeldt K, Gatz M, Reynolds CA, Prescott CA, Pedersen NL. Heritability of Parkinson disease in Swedish twins: a longitudinal study. Neurobiol Aging. 2011;32:1923.e1–8. doi: 10.1016/j.neurobiolaging.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osaki Y, Wenning GK, Daniel SE, Hughes A, Lees AJ, Mathias CJ, et al. Do published criteria improve clinical diagnostic accuracy in multiple system atrophy? Neurology. 2002;59:1486–1491. doi: 10.1212/01.wnl.0000028690.15001.00. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poewe W, Wenning G. The differential diagnosis of Parkinson’s disease. Eur J Neurol Off J Eur Fed Neurol Soc. 2002;9(Suppl 3):23–30. doi: 10.1046/j.1468-1331.9.s3.3.x. [DOI] [PubMed] [Google Scholar]
- 24.Quinn NP. How to diagnose multiple system atrophy. Mov Disord Off J Mov Disord Soc. 2005;20(Suppl 12):S5–S10. doi: 10.1002/mds.20534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.