Abstract
Biogenic amines are important signaling molecules, and the structural basis for their recognition by G Protein-Coupled Receptors (GPCRs) is well understood. Amines are also potent odors, with some activating olfactory trace amine-associated receptors (TAARs). Here, we report that teleost TAARs evolved a new way to recognize amines in a non-classical orientation. Chemical screens de-orphaned eleven zebrafish TAARs, with agonists including serotonin, histamine, tryptamine, 2-phenylethylamine, putrescine, and agmatine. Receptors from different clades contact ligands through aspartates on transmembrane α-helices III (canonical Asp3.32) or V (non-canonical Asp5.42), and diamine receptors contain both aspartates. Non-classical monoamine recognition evolved in two steps: an ancestral TAAR acquired Asp5.42, gaining diamine sensitivity, and subsequently lost Asp3.32. Through this transformation, the fish olfactory system dramatically expanded its capacity to detect amines, ecologically significant aquatic odors. The evolution of a second, alternative solution for amine detection by olfactory receptors highlights the tremendous structural versatility intrinsic to GPCRs.
DOI: http://dx.doi.org/10.7554/eLife.10441.001
Research Organism: Zebrafish
eLife digest
Many organisms make molecules called biogenic amines. These molecules, which include the human hormones adrenaline and histamine, have important roles in regulating the biology and behaviour of many animals. Some biogenic amines bind to receptor proteins called GPCRs on the surface of cells. Many drugs can affect the activity of GPCRs, so understanding how different GPCRs work is an important goal of the pharmaceutical industry. Like all proteins, GPCRs are made of chains of molecules called amino acids. The GPCRs that can detect biogenic amines use a particular amino acid named Asp3.32, and when this amino acid is mutated, these GPCRs become unable to bind to their target amine.
Trace amine-associated receptors (TAARs) are a type of GPCR that are found in many animals to detect odors. Most TAARs in mammals contain the Asp3.32 residue, and recognize amine odors. However, many fish TAARs do not contain Asp3.32, and it was not clear what molecules these fish receptors detect.
Here Li et al. find that these fish TAARs also recognize amines, and use a different amino acid called Asp5.42. Also, some TAARs contain both Asp3.32 and Asp5.42, and recognize chemicals with two amines named diamines. Some diamines that bind to TAARs are foul smelling odors; for example, cadaverine and putrescine are repulsive smells emitted by decomposing flesh. In total, the experiments identified amines that can bind to eleven zebrafish TAARs that previously had no odor partner.
Li et al. propose that some fish TAARs lost the Asp3.32 during the course of evolution to leave the Asp5.42 as the main interaction site for amines. This change dramatically altered how these TAARs interact with amines, which probably expanded the number of different amines that fish can detect. These findings open up new ways to study how the fish brain processes information about its surroundings.
Introduction
Biogenic amines are key intercellular signaling molecules that regulate physiology and behavior. In vertebrates, biogenic amines include adrenaline, serotonin, acetylcholine, histamine, and dopamine, and these biogenic amines can be detected by G Protein-Coupled Receptors (GPCRs) expressed on the plasma membrane of target cells. The biochemical and structural basis for biogenic amine recognition by GPCRs is well understood and involves an essential salt bridge between the ligand amino group and a highly conserved aspartic acid on the third transmembrane α-helix of the receptor (Asp3.32; Ballesteros-Weinstein indexing) (Katritch et al., 2013; Shi and Javitch, 2002). Mutation of Asp3.32 in adrenaline, serotonin, acetylcholine, histamine, and dopamine receptors eliminates amine recognition (Shi and Javitch, 2002; Surgand et al., 2006), and GPCRs that recognize amines through another motif have not been identified.
Trace amine-associated receptors (TAARs) are a family of vertebrate GPCRs distantly related to biogenic amine receptors (Liberles, 2015). There are 15 TAARs in mouse, 6 in human, and 112 in zebrafish, and in these or related species, all TAARs except TAAR1 function as olfactory receptors (Horowitz et al., 2014; Hussain et al., 2009; Liberles and Buck, 2006). All human TAARs (6/6), most mouse TAARs (13/15), and some zebrafish TAARs (27/112) retain Asp3.32, suggesting that many TAARs would recognize amines. High throughput chemical screens yielded ligands for TAAR1 (Borowsky et al., 2001; Bunzow et al., 2001; Revel et al., 2011) and many olfactory TAARs that retain Asp3.32, including TAAR3, TAAR4, TAAR5, TAAR7s, TAAR8s, TAAR9, and TAAR13c (Liberles and Buck, 2006; Ferrero et al., 2012; Hussain et al., 2013). Each of these receptors detects different amines, some of which evoke innate olfactory behaviors (Hussain et al., 2013; Dewan et al., 2013; Ferrero et al., 2011; Li et al., 2013; Li and Liberles, 2015). In mouse, TAAR4 detects the aversive carnivore odor 2-phenylethylamine (Ferrero et al., 2011), while TAAR5 detects the attractive and sexually dimorphic mouse odor trimethylamine (Liberles and Buck, 2006; Li et al., 2013). Knockout mice lacking TAAR4 and TAAR5 lose behavioral responses to TAAR ligands identified in cell culture studies (Dewan et al., 2013; Li et al., 2013).
The selectivity of olfactory TAARs for particular amine odors suggests structural variability within the ligand-binding pocket across the TAAR family. One clade of TAARs (including TAAR1, TAAR3, and TAAR4) detects primary amines derived by amino acid decarboxylation, while a second clade (including TAAR5, TAAR7s, TAAR8s, and TAAR9) prefers tertiary amines (Ferrero et al., 2012). Structural models and mutagenesis studies of TAAR1, TAAR7e, and TAAR7f predicted that ligand binding occurs in the membrane plane, Asp3.32 forms a salt bridge to the ligand amino group, and other transmembrane residues provide a scaffold that supports ligand binding and enables selectivity (Ferrero et al., 2012; Reese et al., 2014).
A third TAAR clade (clade III TAARs) arose in fish, and includes the majority (87/112) of zebrafish TAARs (Hussain et al., 2009). Most clade III TAARs lack Asp3.32 (85/87), and it has been proposed that these receptors may not detect amines (Hussain et al., 2009; Liberles, 2014). However, ligands have not been identified for any of these receptors. More generally, ligands are known for only one zebrafish TAAR, TAAR13c, which retains Asp3.32 and detects the carrion odor cadaverine (Hussain et al., 2013). Structure-activity analysis revealed that TAAR13c preferentially recognizes odd-chained, medium-sized diamines, and likely contains two distinct cation recognition sites within the ligand-binding pocket (Hussain et al., 2013). However, the structural basis for diamine recognition by TAAR13c was unknown.
Here, we find that TAAR13c recognizes cadaverine through two remote transmembrane aspartic acids, Asp3.32 and Asp5.42. A TAAR13c variant with a charge-neutralizing mutation at Asp5.42 has an altered ligand preference for monoamines rather than diamines. We propose that Asp3.32 and Asp5.42 together form a general di-cation recognition motif, and note this motif to be present in several zebrafish, mouse, and human TAARs. Surprisingly, Asp5.42 is retained in 84 out of 85 zebrafish TAARs that lack Asp3.32, raising the possibility that these receptors recognize amines positioned in a non-canonical inverted orientation with the amino group pointing towards transmembrane α-helix V. Using a high throughput chemical screening approach, we identified ligands for eleven additional zebrafish TAARs, and found that different receptors detect amines through salt bridge contacts at Asp3.32 and/or Asp5.42. TAAR10a, TAAR10b, TAAR12h, and TAAR12i are Asp3.32-containing clade I TAARs that detect serotonin, tryptamine, 2-phenylethylamine, and 3-methoxytyramine, while TAAR16c, TAAR16e, and TAAR16f are Asp5.42-containing clade III TAARs that detect N-methylpiperidine, N,N-dimethylcyclohexylamine, and isoamylamine. TAAR13a, TAAR13d, TAAR13e, and TAAR14d have both Asp3.32 and Asp5.42, and recognize the di-cationic molecules putrescine, histamine, and agmatine.
Based on these findings, we propose that a large clade of olfactory GPCRs evolved a novel way to detect amines that involves a non-classical salt bridge to transmembrane α-helix V. Furthermore, new TAAR ligands include several biogenic amines for which other receptors were identified, including serotonin, histamine, and 2-phenylethylamine. These findings illustrate that different members of the GPCR superfamily can evolve divergent ligand binding pockets for recognition of the same agonist.
Results
Two remote amine recognition sites in the cadaverine receptor
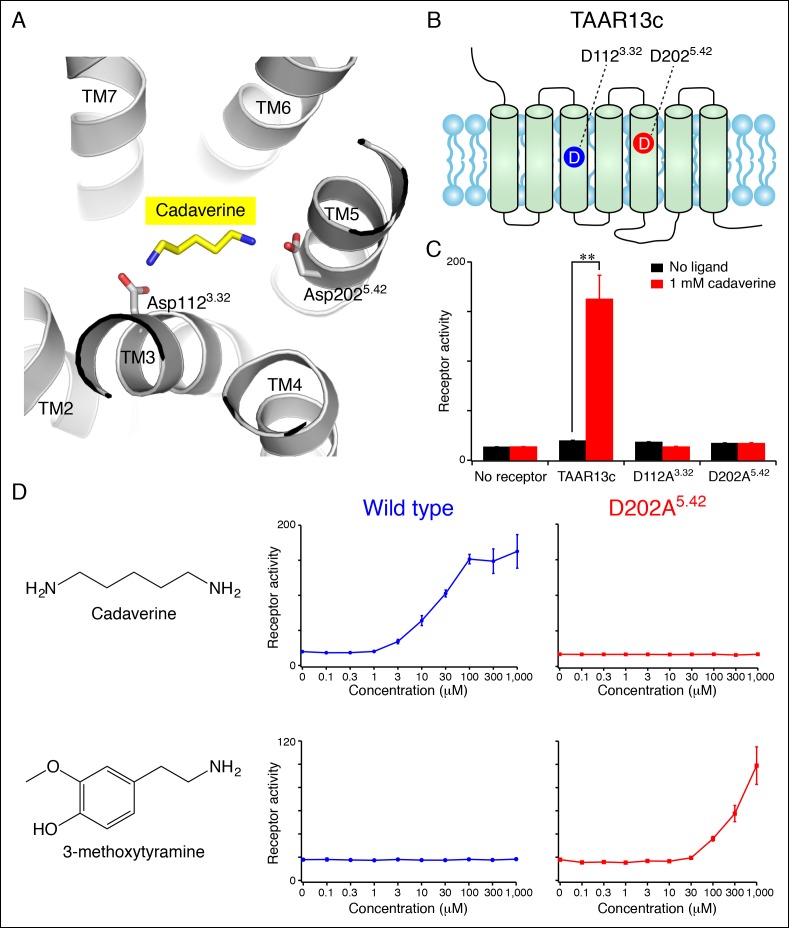
To gain structural insights into diamine recognition by the cadaverine receptor TAAR13c, we generated a TAAR13c homology model (Figure 1A) based on the X-ray crystal structure of agonist-bound β2 adrenergic receptor (Protein Data Bank Entry 4LDE) (Ring et al., 2013). The β2 adrenergic receptor is the closest homolog (33% amino acid identity) of TAAR13c for which an active-state structure is currently available. The homology model of TAAR13c indicated a canonical fold of seven transmembrane α-helices with ligand bound at the core. Using the homology model, we performed computational docking of cadaverine to the orthosteric binding site. One cadaverine amino group of the docked ligand was located near Asp1123.32, which corresponds to the highly conserved amine-contact site in biogenic amine receptors. The other cadaverine amino group was positioned at the opposing end of the ligand-binding pocket, 3 Å away from another aspartate, Asp2025.42. The second amino group was also within 5 Å of side chains from Thr2035.43 and Ser2766.55, Leu192 and Phe194 on extracellular loop 2, and the backbone of Ser1995.39 (additional nearby residues are depicted in Figure 1—figure supplement 1). Based on its proximity and charge, Asp2025.42 was deemed a prime candidate to function as a counterion that forms a salt bridge to the second cadaverine amino group. Position 5.42 in several other GPCRs directly contacts ligands through hydrogen bonds, salt bridges, or van der Waals interactions (Surgand et al., 2006), and our homology model suggests a role for this amino acid in ligand recognition by TAAR13c.
Figure 1. Diamine recognition sites in the cadaverine receptor TAAR13c.
(A) Structural modeling of zebrafish TAAR13c bound to cadaverine (yellow) reveals two aspartates (D1123.32 and D2025.42) with carboxylates (red) predicted to form salt bridges to ligand amino groups (blue). (B) Cartoon of zebrafish TAAR13c topology depicts the location of D1123.32 and D2025.42. (C) Charge-neutralizing mutation of D1123.32 and D2025.42 abrogates cadaverine responsiveness of zebrafish TAAR13c expressed in HEK-293 cells (mean ± sem, n = 3, **p<0.01 based on two-tailed unpaired Student's t-test). (D) D202A5.42 mutation transforms TAAR13c from a diamine receptor into a monoamine receptor (mean ± sem, n = 3).
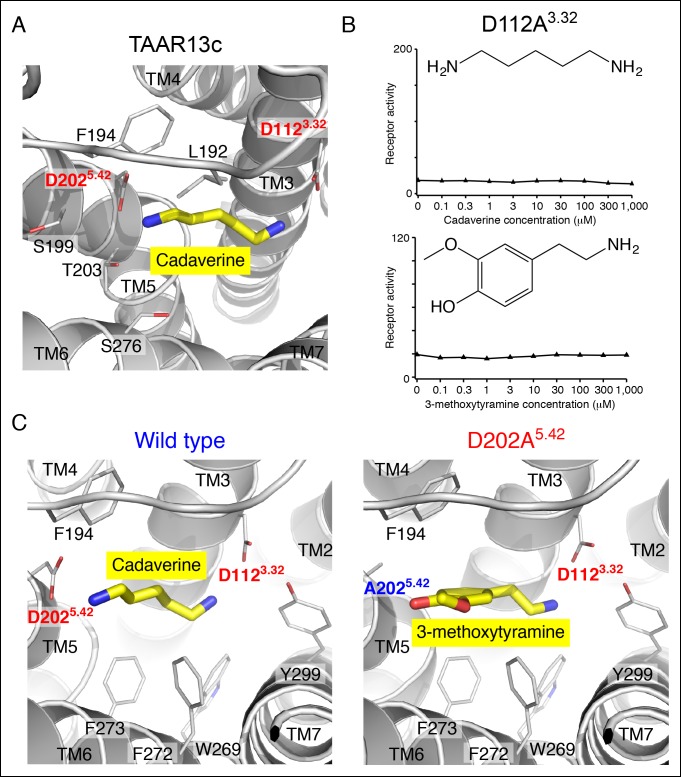
Figure 1—figure supplement 1. Functional analysis and structural modeling of TAAR13c mutants.
We used an established reporter gene assay (Liberles and Buck, 2006; Ferrero et al., 2012) to query the response properties of TAAR13c variants with charge-neutralizing mutations at Asp3.32 (TAAR13cD112A) and Asp5.42 (TAAR13cD202A). HEK-293 cells were co-transfected with plasmids encoding TAARs and a cAMP-dependent reporter gene encoding secreted alkaline phosphatase (Cre-Seap). Transfected cells were exposed to test ligands, and assayed for reporter activity using a fluorescent phosphatase substrate. As previously published with this paradigm (Hussain et al., 2013), cadaverine robustly activated TAAR13c, with a half maximal response occurring at ~10 μM. In contrast, cadaverine failed to activate either the TAAR13cD112A or TAAR13cD202A mutant at any concentration tested, up to 1 mM (Figure 1, Figure 1—figure supplement 1).
To ensure that mutation of Asp2025.42 did not impair protein folding or G protein coupling, we asked whether the mutant receptor could be activated by other ligands. We tested 50 chemicals for the ability to activate TAAR13cD202A, and identified the monoamine 3-methoxytyramine as an agonist (Figure 1D). 3-methoxytyramine failed to activate wild type TAAR13c at any concentration tested. Thus, charge-neutralizing mutation of Asp5.42 transformed TAAR13c from a diamine receptor into a monoamine receptor. The altered ligand-binding preference of the TAAR13cD202A mutant suggests that Asp5.42 contributes directly to the ligand-binding pocket, and homology modeling of TAAR13cD202A bound to 3-methoxytyramine (Figure 1—figure supplement 1) supports a direct interaction between this site and ligand. Based on modeling and mutagenesis studies, we conclude that Asp5.42 in TAAR13c forms a salt bridge to a cadaverine amino group.
Asp5.42 is widely conserved in clade III TAARs
We next asked whether Asp5.42 was present in other GPCRs, and might function more generally in amine detection. Biogenic amine receptors use Asp3.32 to contact primary amines (Shi and Javitch, 2002; Surgand et al., 2006), and Asp5.42 is not present in any of 12 serotonin, 5 dopamine, 5 acetylcholine, or 9 adrenergic receptors in mouse (Figure 2). Asp3.32 is also observed in all 4 mouse histamine receptors, while Asp5.42 is observed in one (histamine receptor H2). Histamine is di-cationic at low pH, and the polar imidazole group of the histamine ligand is predicted to contact a transmembrane α-helix 5 asparagine in histamine receptor H1 (Asn5.46) and transmembrane α-helix 5 anions in histamine receptors H2 (Asp5.42), H3 (Glu5.46), and H4 (Glu5.46) (Seifert et al., 2013).
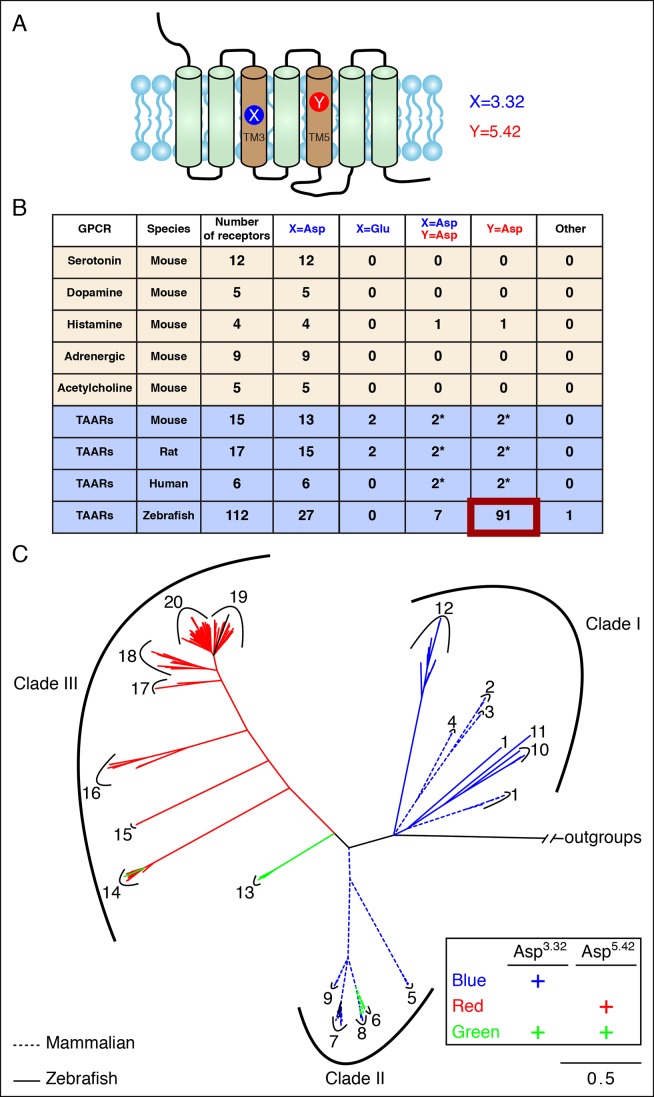
Figure 2. Asp5.42 is highly conserved in clade III TAARs.
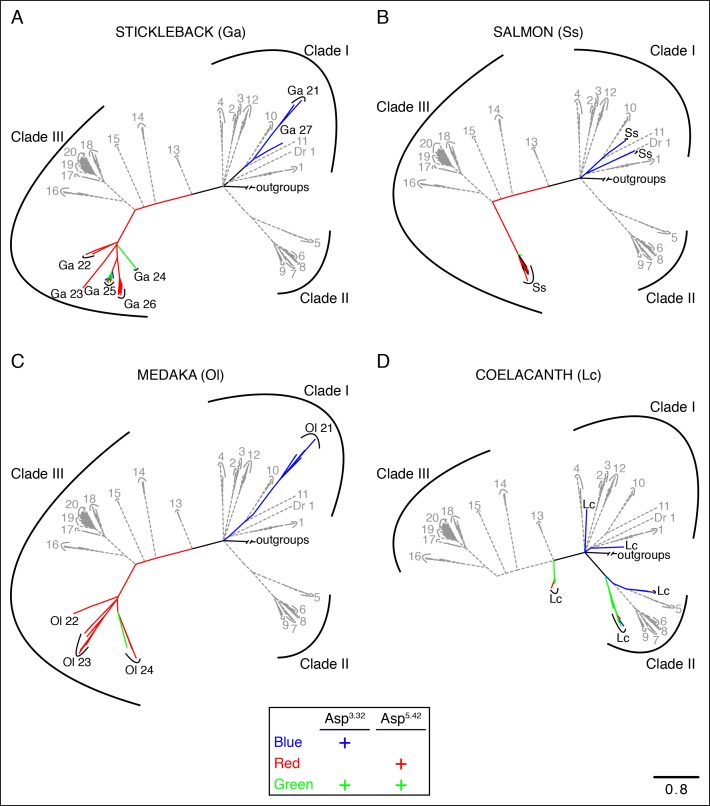
(A) Cartoon depiction indicates the location of positions 3.32 and 5.42 in GPCRs. (B) Bioinformatic analysis reveals the number of receptors with anions at positions 3.32 (X) or 5.42 (Y) across various biogenic receptor subfamilies in mouse, as well as TAARs in mouse, rat, humans, and zebrafish. Two mouse, rat, and human TAARs have Asp5.43 instead of Asp5.42, and these are included in columns marked Y = Asp (*). (C) Phylogenetic analysis of the TAAR family in zebrafish (solid lines), mice (dashed lines), rats (dashed lines), and humans (dashed lines); scale bar = 0.5 substitutions per site. TAARs containing only Asp3.32 (blue), only Asp5.42 (red), or both Asp3.32 and Asp5.42 (green) are depicted. Mammalian TAARs with Asp3.32 and Asp5.43 instead of Asp5.42 are green. Glu3.32-containing rodent TAARs and the one zebrafish TAAR, TAAR19f, that lacks both Asp3.32 and Asp5.42 are depicted in black.
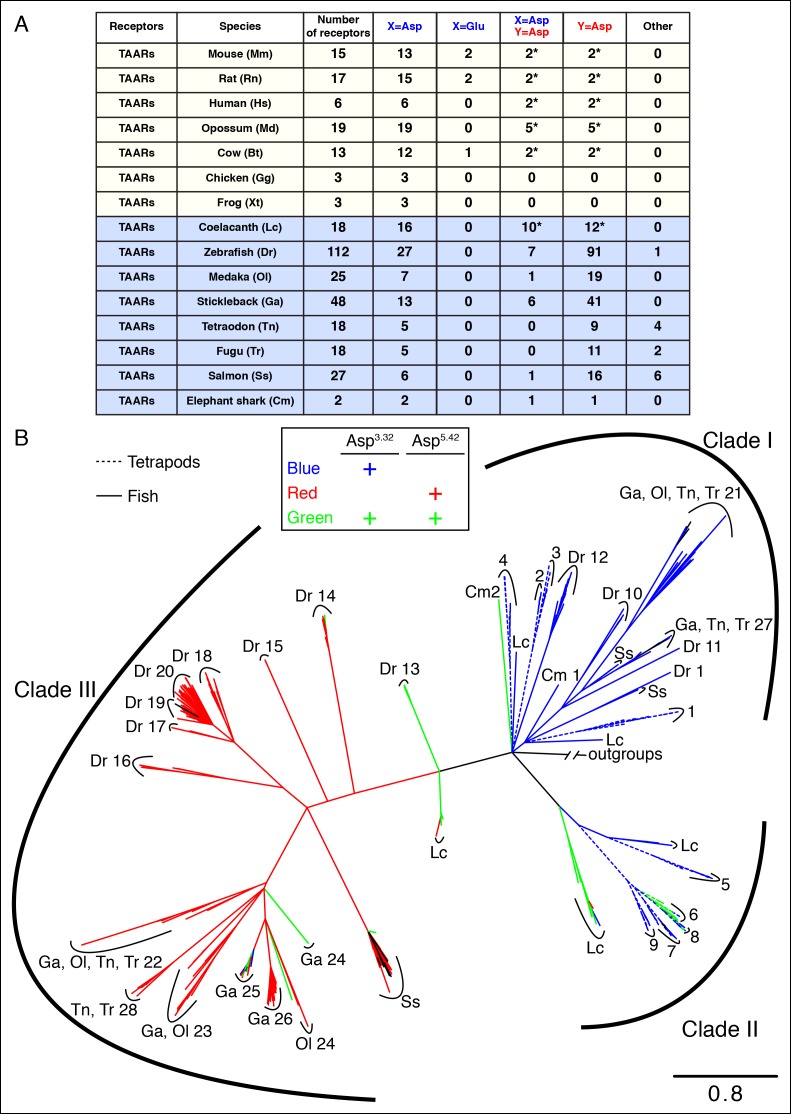
Figure 2—figure supplement 1. The phylogenetic tree of tetrapod and fish TAARs.
Figure 2—figure supplement 2. Phylogenetic analysis of TAARs from different fish species.
Most mammalian TAARs retain Asp3.32, including 6/6 human TAARs, 13/15 mouse TAARs, and 15/17 rat TAARs. In mouse and rat, two TAAR7s have conservative changes of Asp3.32 to Glu3.32; the effect of this substitution is unknown as ligands have not been found for Glu3.32-containing TAARs. A small number of mammalian TAARs have an aspartate at position 5.43 including two in humans (TAAR6, TAAR8), mice (TAAR6, TAAR8b), and rats (TAAR6, TAAR8a). Sequence alignments indicate that Asp5.43 of these receptors corresponds to Asp5.42 of TAAR13c, and Asp5.43 may be similarly positioned in the agonist-binding pocket. These mammalian TAARs are candidates to function as diamine receptors; however, ligands have not been identified for these receptors, perhaps due to difficulties in achieving their functional expression in heterologous cells. Mice lack a TAAR13c ortholog, yet a role for other TAARs in diamine recognition is supported by the finding that mice lacking all olfactory TAARs fail to avoid cadaverine odor (Dewan et al., 2013).
Analysis of the zebrafish TAAR repertoire surprisingly revealed that in contrast to mammalian TAARs and biogenic amine receptors, the vast majority of zebrafish TAARs (91/112) have an aspartate at position 5.42. Some of these receptors (7/91) also retain Asp3.32, including TAAR13c, while most (84/91) have lost Asp3.32. In several other fish species such as medaka, stickleback, fugu, and salmon, most TAARs also contain Asp5.42 but lack Asp3.32 (Figure 2—figure supplement 1). Phylogenetic analysis indicates that TAARs containing Asp5.42 but not Asp3.32 are clade III TAARs found in teleosts, and constitute a large branch of the TAAR family tree (Figure 2, Figure 2—figure supplement 2). Zebrafish lack clade II TAARs, and the teleost TAAR family evolved differently from mammalian TAARs by forming the large cohort of clade III TAARs. Several TAARs with both Asp5.42 and Asp3.32 reside between clade I TAARs and clade III TAARs phylogenetically (including TAAR13s in zebrafish), suggesting that clade III TAARs derived from clade I TAARs by first gaining Asp5.42 and then losing Asp3.32.
Ligands for clade III TAARs were unknown, and since they lack the canonical amine recognition site, were proposed to detect chemicals other than amines (Hussain et al., 2009; Liberles, 2014). However, the widespread retention of Asp5.42 in clade III TAARs lacking Asp3.32 raised the possibility that the corresponding 84 olfactory receptors in zebrafish indeed detect amines, but do so with ligands positioned in a non-classical orientation.
Identifying ligands for eleven 'orphan' zebrafish TAARs
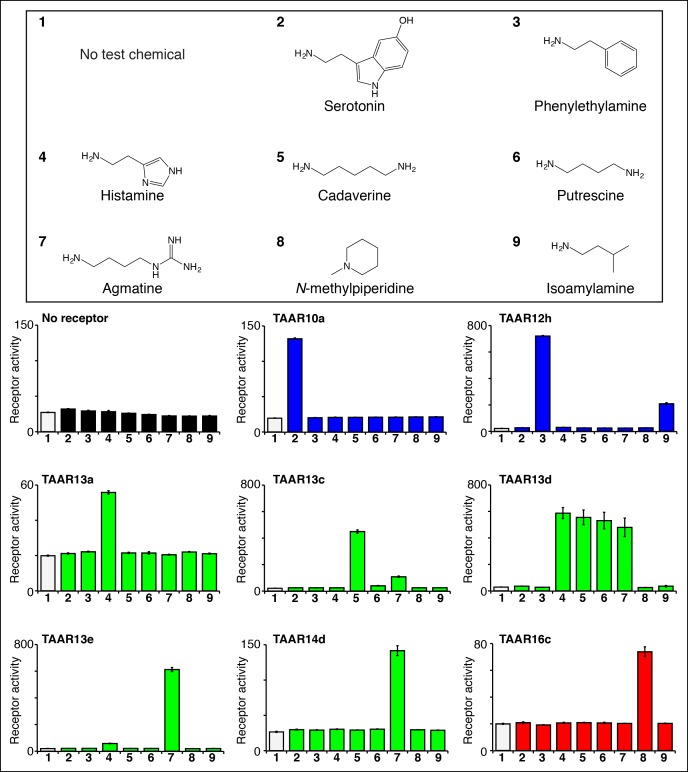
We used the high throughput reporter gene assay to identify agonists for additional zebrafish TAARs (Figure 3, Figure 3—figure supplement 1). Responses of 63 zebrafish TAARs were examined, including at least one representative from each TAAR subfamily in the zebrafish olfactory system (TAAR10 to TAAR20). Zebrafish TAARs were expressed as fusion proteins containing the N-terminal 20 amino acids of bovine rhodopsin, which enhances cell surface expression of some chemosensory receptors in heterologous cells (Krautwurst et al., 1998). As above, HEK-293 cells or Hana3A cells (Saito et al., 2004) (Hana3A cells express olfactory chaperones for promotion of receptor expression and were used for studies involving TAAR10b, TAAR12i, and TAAR16e) were transfected with plasmids encoding zebrafish TAARs and Cre-Seap, incubated with test chemicals, and assayed for reporter phosphatase activity. TAAR ligand identification in heterologous cells has been extensively validated; TAAR ligands identified by this technique also activate TAAR-containing neurons in vivo (Hussain et al., 2013; Dewan et al., 2013; Li et al., 2013;Zhang, et al., 2013 ), and behavioral responses to identified TAAR ligands are lost in TAAR knockout mice (Dewan et al., 2013; Li et al., 2013). Here, we used the heterologous expression approach to identify the first ligands for zebrafish TAAR10a, TAAR10b, TAAR12h, TAAR12i, TAAR13a, TAAR13d, TAAR13e, TAAR14d, TAAR16c, TAAR16e, and TAAR16f.
Figure 3. Identifying the first ligands for several zebrafish TAARs.
HEK-293 cells were cotransfected with Cre-Seap and plasmids encoding zebrafish TAARs, incubated with test chemicals (100 μM), and assayed for phosphatase activity using a fluorescent substrate (mean ± sem, n = 3). Zebrafish TAAR10a and TAAR12h are clade I TAARs containing Asp3.32 but not Asp5.42 (blue), zebrafish TAAR13a, TAAR13c, TAAR13d, and TAAR13e, and TAAR14d contain both Asp5.42 and Asp3.32 (green), and zebrafish TAAR16c is a clade III TAAR containing Asp5.42 but not Asp3.32 (red).
Figure 3—figure supplement 1. Functional expression of TAAR10b, TAAR12i, and TAAR16e in Hana3A cells.
Figure 3—figure supplement 2. Structure-function studies of zebrafish clade I TAARs: TAAR10a and TAAR12h.
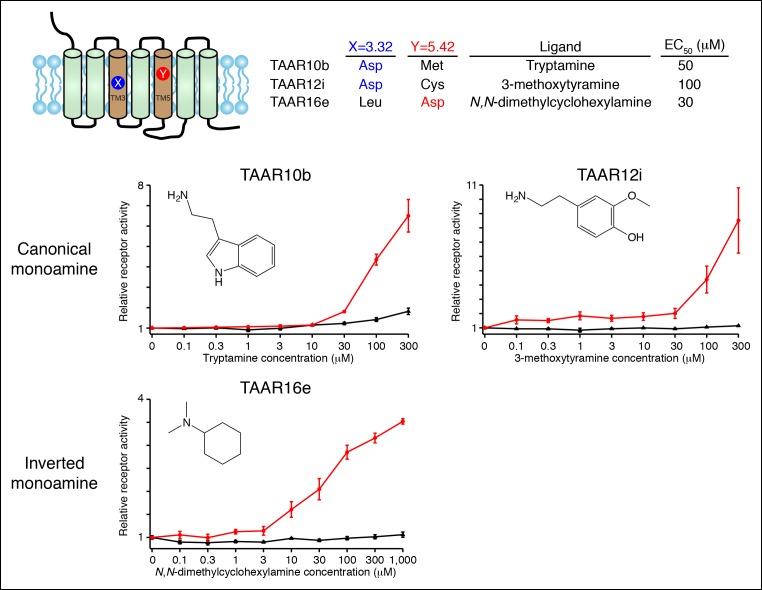
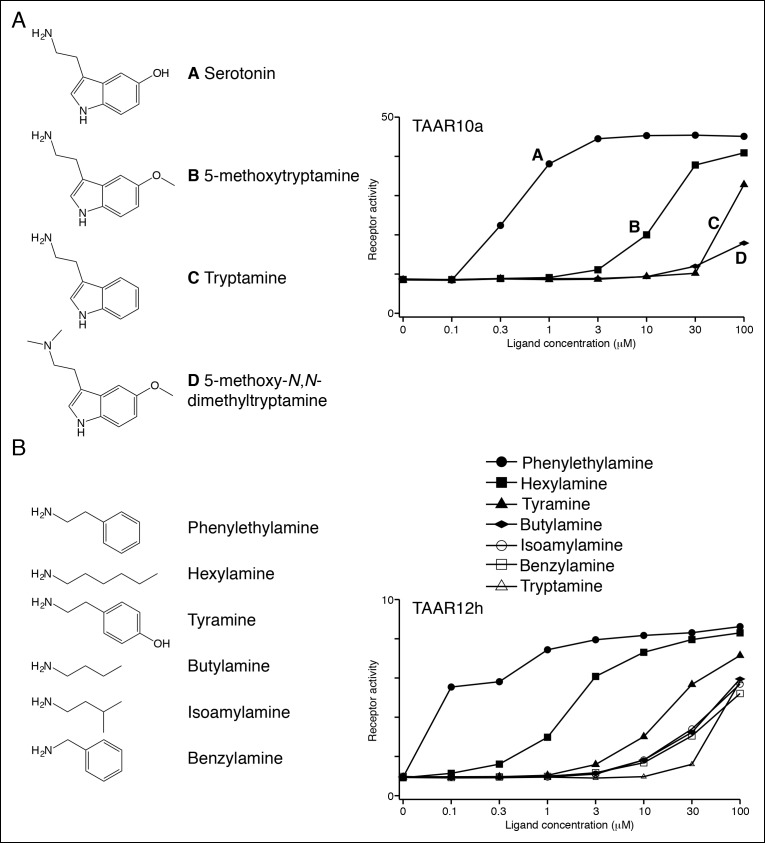
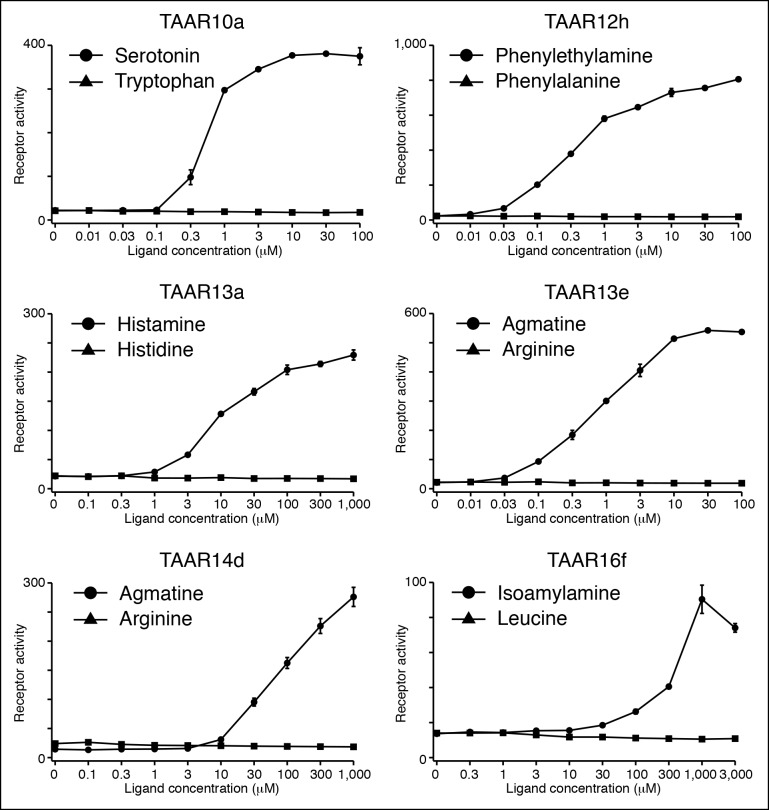
Ligands for clade I TAARs. TAAR10a, TAAR10b, TAAR12h, and TAAR12i are clade I TAARs that retain Asp3.32 but lack Asp5.42. TAAR10a sensitively detects serotonin (EC50 = ~0.5 μM), has significantly reduced affinity for the related indole amines 5-methoxytryptamine (EC50 = ~20 μM) and tryptamine (EC50 = ~70 μM), and does not detect other chemicals examined (Figure 3—figure supplement 2). The highest affinity ligand identified for TAAR12h was 2-phenylethylamine (EC50 = ~0.3 μM), and TAAR12h also detects other alkyl amines with reduced sensitivity (Figure 3—figure supplement 2). TAAR10b and TAAR12i detect the primary amines tryptamine and 3-methoxytyramine respectively (Figure 3—figure supplement 1).
Serotonin and 2-phenylethylamine are both biogenic amines that can be recognized by other GPCRs. Zebrafish lack an ortholog of mouse TAAR4, which detects 2-phenylethylamine (Ferrero et al., 2011), and mouse TAAR4 shares 41% identity with zebrafish TAAR12h. Likewise, zebrafish TAAR10a is distantly related to mouse serotonin receptors, sharing 27–35% identity. It is thought that the TAAR family derived from an ancestral duplicate of serotonin receptor subtype HTR4 (Lindemann and Hoener, 2005), and it is possible that either TAAR10a retains the ligand recognition properties of the ancestral TAAR or that serotonin responsiveness was lost and then re-gained in this lineage.
Ligands for putative di-cation receptors. Seven zebrafish TAARs retain both Asp3.32 and Asp5.42, including all five TAAR13s and two TAAR14s (TAAR14c and TAAR14d). We previously found that zebrafish TAAR13c detects the diamine cadaverine (Hussain et al., 2013), and now report that zebrafish TAAR13a, TAAR13d, TAAR13e, and TAAR14d also detect chemicals with two cations.
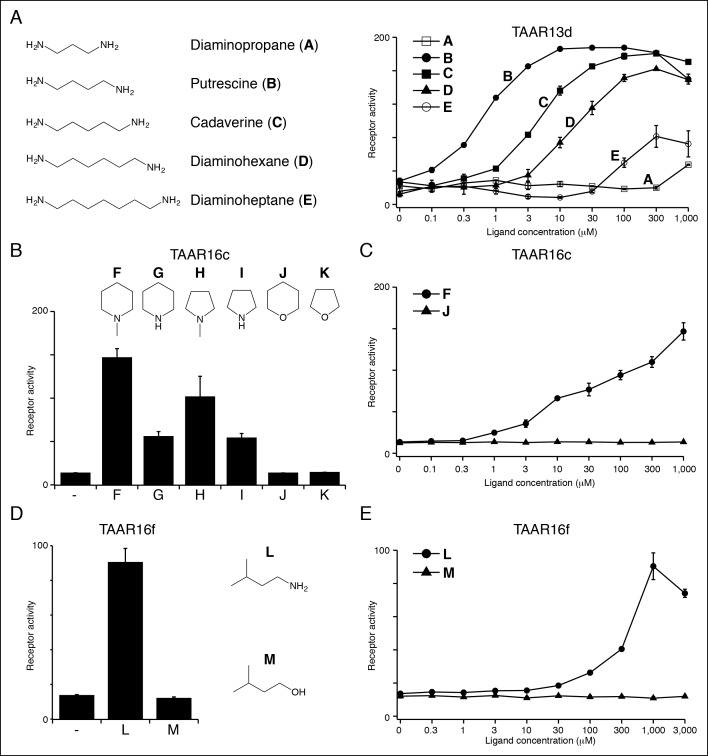
TAAR13d prefers medium-chained alkyl diamines with highest affinity for putrescine (EC50 = ~1 μM), and progressively reduced affinity for diamines with additional methylene groups (Figure 4A). TAAR13d has ~3,000-fold reduced sensitivity for diaminopropane, which is only one methylene group shorter than putrescine, indicating a strict minimal agonist length. The differing preference of TAAR13c and TAAR13d for cadaverine and putrescine respectively is consistent with cross-adaptation studies indicating that separate olfactory receptors detect these two diamines (Rolen, et al., 2003).
Figure 4. Structure-activity studies of zebrafish TAARs.
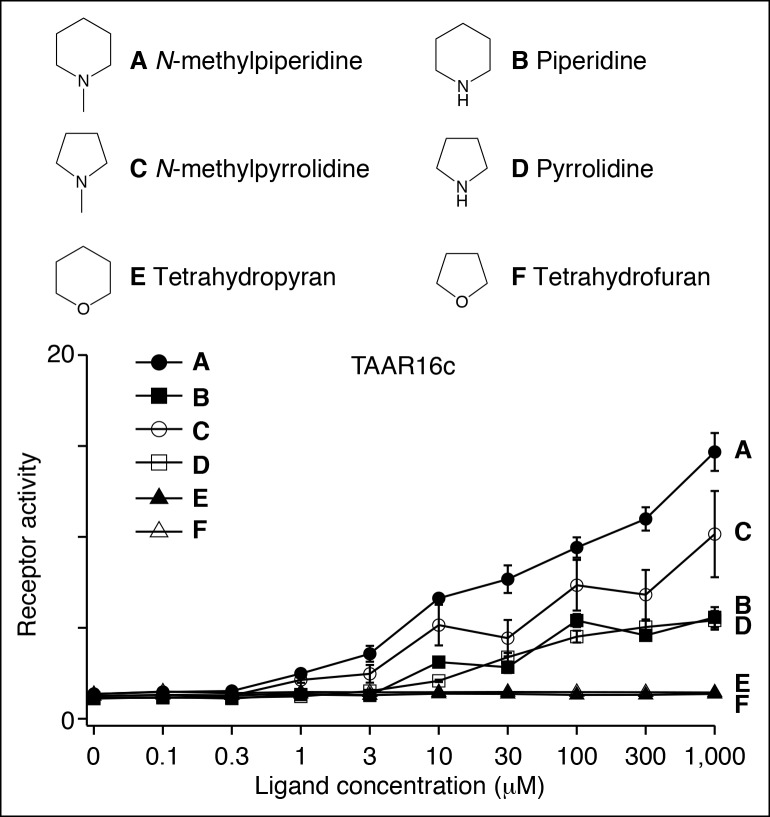
(A) Zebrafish TAAR13d displays highest affinity for putrescine among tested ligands, and reduced affinity for longer or shorter diamines. (B) Zebrafish TAAR16c recognizes several structurally related amines but not oxygen-containing analogs (1 mM). F: N-methylpiperidine; G: piperidine; H: N-methylpyrrolidine; I: pyrrolidine; J: tetrahydropyran; K: tetrahydrofuran. (C) Dose-dependent responses of TAAR16c for N-methylpiperidine and tetrahydropyran. (D) Zebrafish TAAR16f recognizes isoamylamine (L) but not isoamyl alcohol (M) at 1 mM. (E) Dose-dependent responses of TAAR16f for isoamylamine and isoamyl alcohol (mean ± sem, n = 3).
Figure 4—figure supplement 1. Several TAARs detect amines but not amino acids.
Figure 4—figure supplement 2. Structure-function studies of the zebrafish clade III TAAR, TAAR16c.
Histamine is an agonist for both TAAR13a (EC50 = ~20 μM) and TAAR13d (EC50 = ~7 μM). TAAR13d recognizes other alkyl diamines of medium length (see above), while TAAR13a is selective for histamine among tested ligands. TAAR13a and TAAR13d share only 18–28% identity with mouse histamine receptors, and the recognition of histamine by olfactory receptors is likely due to convergent evolution within the GPCR family. Agmatine also activates multiple TAARs, including TAAR13e with higher affinity (EC50 = ~1 μM) and TAAR14d with reduced affinity (EC50 = ~100 μM). Agmatine is derived from arginine by decarboxylation, reminiscent of several other TAAR ligands that are biogenic amines similarly derived from natural amino acids (Liberles, 2015). TAARs that detect agmatine, histamine, serotonin, 2-phenylethylamine, or isoamylamine (see below) are not activated by the natural amino acids from which their ligands were derived (Figure 4—figure supplement 1).
Ligands for clade III TAARs.Next, we found ligands for three clade III TAARs that lack Asp3.32. Despite the absence of the classical amine recognition motif present in all known amine-detecting GPCRs, zebrafish TAAR16c, TAAR16e, and TAAR16f detect amines. Zebrafish TAAR16c detects N-methylpiperidine (EC50 = ~10 μM), zebrafish TAAR16e detects N,N-dimethylcyclohexylamine (EC50 = ~30 μM), and zebrafish TAAR16f detects isoamylamine (EC50 = ~200 μM).
Removing the amino group of TAAR16c and TAAR16f ligands abolished responses (Figure 4, Figure 4—figure supplement 2). TAAR16c also detected N-methylpyrrolidine (EC50 = ~70 μM), and the corresponding secondary amines piperidine and pyrrolidine were partial agonists. In contrast, oxygen-containing analogs, tetrahydropyran and tetrahydrofuran did not activate TAAR16c at any concentration tested, indicating that the amine is essential for receptor agonism. Likewise, TAAR16f failed to detect isoamyl alcohol, and thus TAAR16f agonism also required a ligand amine. These studies indicate that these clade III TAARs are in fact amine receptors, and that the ligand amino group is an essential moiety recognized by the receptor.
Transmembrane aspartates are required for TAAR agonism
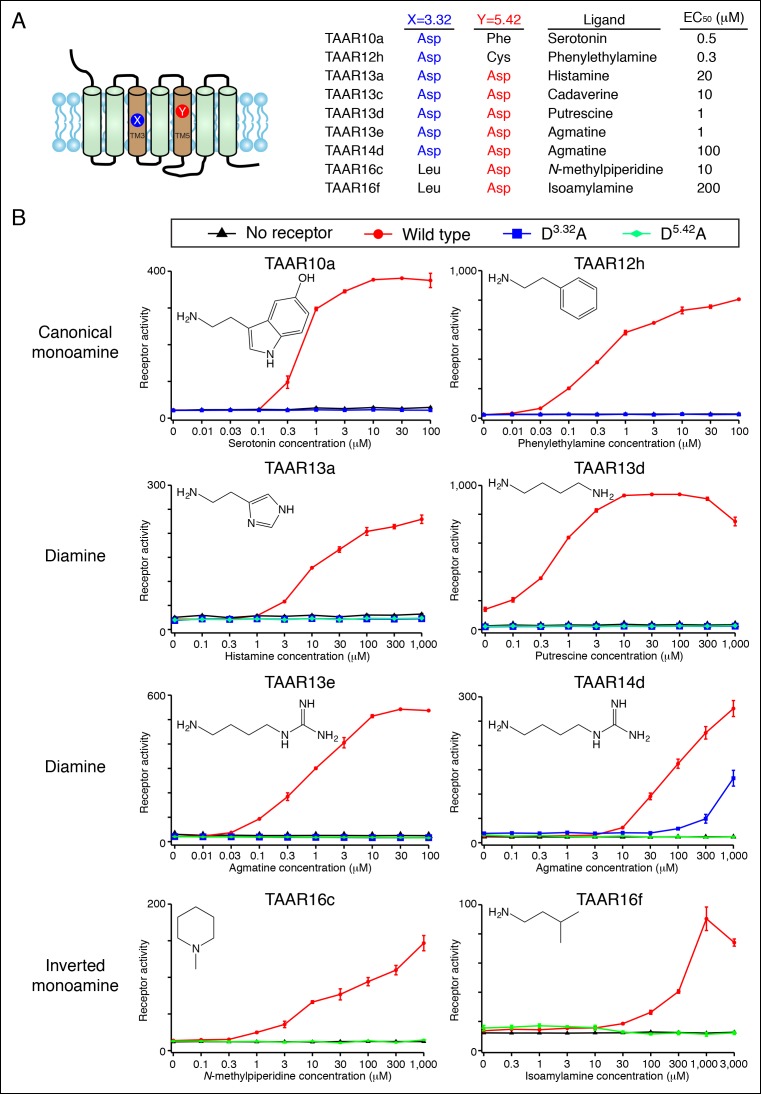
We showed that Asp3.32 and Asp5.42 are essential for cadaverine detection by TAAR13c (Figure 1), and here asked if the corresponding residues are required for activation of other fish TAARs (Figure 5). Charge-neutralizing mutation of Asp3.32 in clade I TAARs, TAAR10a and TAAR12h, completely eliminated responses to serotonin and 2-phenylethylamine respectively, while charge-neutralizing mutation of Asp5.42 in clade III TAARs, TAAR16c and TAAR16f, completely eliminated responses to N-methylpiperidine and isoamylamine respectively. Similarly, mutation of either Asp3.32 or Asp5.42 in TAAR13a, TAAR13d, TAAR13e, and TAAR14d reduced or abolished responses to all di-cationic ligands. The asymmetric diamines histamine and agmatine could be positioned in either orientation, with the primary amine contacting either Asp3.32 or Asp5.42. These mutagenesis studies reveal that clade I TAARs and clade III TAARs require different transmembrane aspartates for amine recognition.
Figure 5. Dose-dependent responses of zebrafish TAARs and charge-neutralizing TAAR mutants.
(A) The identities of amino acids 3.32 and 5.42 in nine 'de-orphaned' zebrafish TAARs, as well as preferred ligands and corresponding EC50s, are depicted. (B) Dose-dependent activation of TAARs and TAAR mutants by ligands indicated (mean ± sem, n = 3). Responses are shown in cells transfected with Cre-Seap alone (black) or together with wild type TAARs (red), D3.32A mutant TAARs (blue), and D5.42A mutant TAARs (green). D3.32 is lacking in clade III TAARs (TAAR16c, TAAR16f) and D5.42 is lacking in clade I TAARs (TAAR10a, TAAR12h), so the corresponding D3.32A and D5.42A mutants could not be generated.
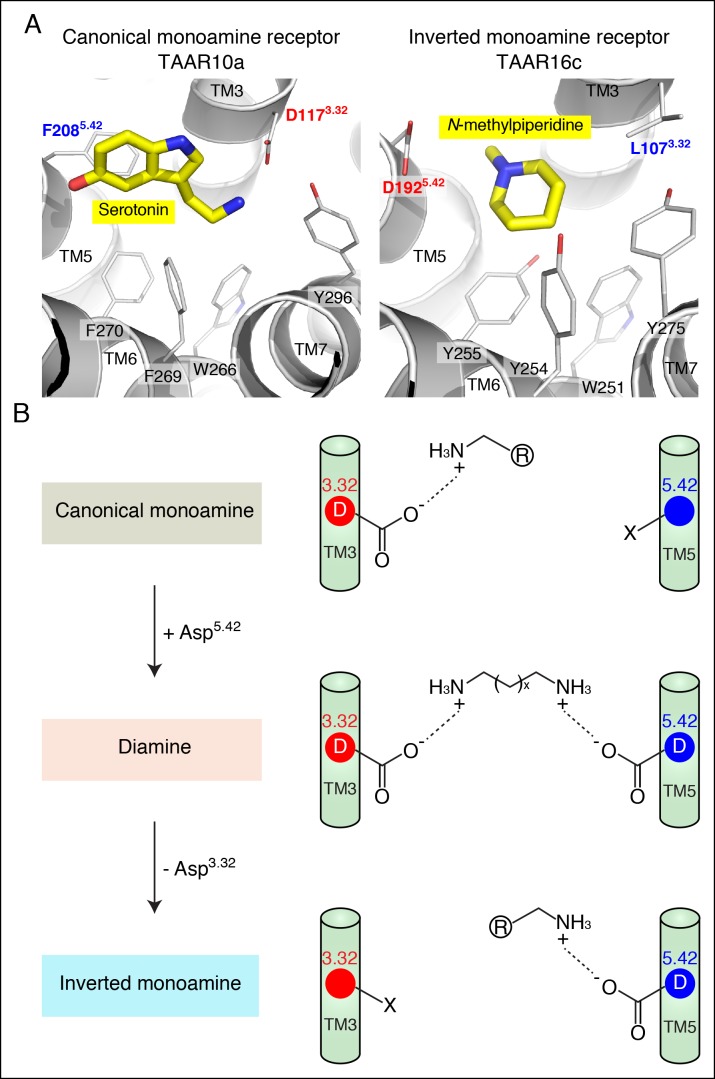
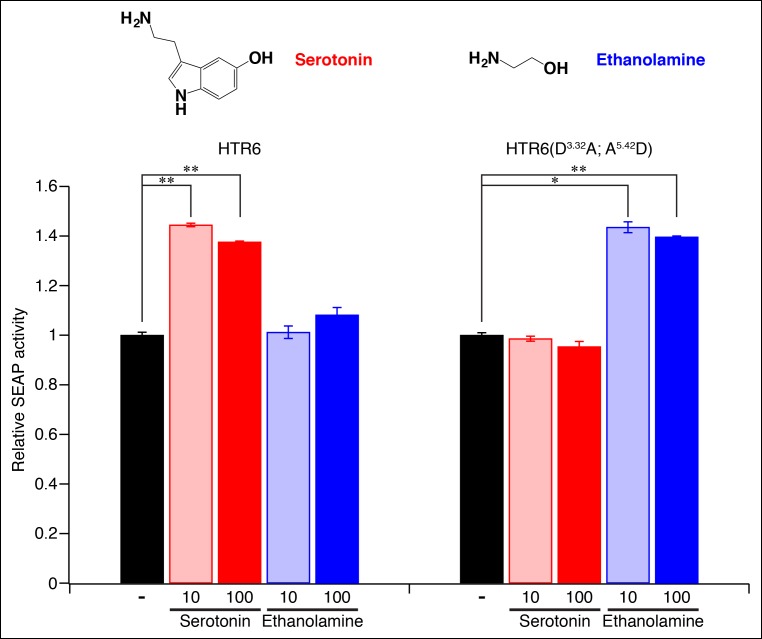
Homology models of serotonin-bound TAAR10a and N-methylpiperidine-bound TAAR16c were generated (Figure 6), as described previously for cadaverine-bound TAAR13c (Figure 1A). TAAR10a is a clade I TAAR and homology modeling indicated proximity of the serotonin amino group to Asp1173.32. In contrast, TAAR16c is a clade III TAAR, and homology modeling indicated ligand inversion with the N-methylpiperidine amino group near Asp1925.42. Inverting the ligand orientation in clade III TAARs would be expected to impact other ligand-receptor contacts; to test this model, we attempted to engineer such a ligand inversion in a canonical biogenic amine receptor, serotonin receptor 6 (HTR6). We analyzed the ligand preference of an HTR6 double mutant (D3.32A; A5.42D) in which the canonical amine recognition site at Asp3.32 was neutralized and a non-canonical amine recognition site at Asp5.42 was introduced. Ligand-identification studies revealed that the HTR6 double mutant no longer recognized serotonin, but instead recognized a different amine, ethanolamine (Figure 6—figure supplement 1). Re-orienting the ligand amine towards Asp5.42 presumably alters other receptor interactions dramatically, allowing Asp5.42-containing receptors to sample structurally distinct ligands. Together, these studies indicate that Asp5.42 forms a non-classical amine interaction site in GPCRs, and this site is present throughout a large clade of olfactory receptors.
Figure 6. Modeling the structure and evolution of non-classical amine recognition.
(A) Structural modeling of zebrafish TAAR10a and TAAR16c bound to serotonin and N-methylpiperidine (yellow) respectively. (B) A model for the birth of a large clade of olfactory receptors with non-classical amine recognition. We propose that clade III TAARs evolved a non-classical mode of amine recognition in two steps. First, an ancestral TAAR gained the ability to recognize diamines by acquiring Asp5.42. Subsequently, a diamine-detecting TAAR lost the canonical amine recognition site, Asp3.32, leading to non-classical amine recognition through a transmembrane α-helix V salt bridge. Extensive gene duplication and mutation expanded and diversified clade III TAARs, leading to a large clade of olfactory receptors with non-canonical amine-detection properties.
Figure 6—figure supplement 1. Re-engineering the amine contact site of HTR6.
Discussion
Sensory receptors define the capacity of an organism to perceive its external environment. Olfactory and taste receptor families can rapidly evolve, with dramatic species-specific variations in repertoire size and function (Nei et al., 2008; Shi and Zhang, 2007). In some lineages, receptor families can be reduced or die out, such as vomeronasal receptors in humans (Liman, 2006; Zhang and Webb, 2003) or sweet receptors in carnivores (Jiang et al., 2012), while in other lineages new receptor families can be born, such as formyl peptide receptors in rodents (Liberles et al., 2009; Rivière et al., 2009). Functional evolution of chemoreceptor families often involves a pattern of gene duplication and subsequent mutation (Ferrero et al., 2012), although isolated functional transformations can also occur, such as taste receptor re-purposing in hummingbirds that led to a novel receptor for sugars found in nectar (Baldwin et al., 2014).
Fish are aquatic animals, and efficient detection of water-soluble odors such as amines is important for their ecological niche. Zebrafish generally use the same families of olfactory receptors as other vertebrates, including odorant receptors, vomeronasal receptors, and TAARs (Kermen et al., 2013). The TAAR repertoire in mice and humans is relatively small, representing 1–2% of all main olfactory receptors. However, zebrafish have comparable numbers of TAARs (112) and odorant receptors (143), and expansion of the teleost TAAR family suggests a relatively expanded role for TAARs in fish chemosensation. However, prior to this study, ligands were known for only one fish TAAR (Hussain, et al., 2013). Moreover, the vast majority of zebrafish TAARs lack the canonical amine recognition motif, and the classes of ligands they might detect were unknown. Here, we show that almost all zebrafish TAARs (111/112) contain either a canonical amine-detection site on transmembrane α-helix III (Asp3.32), a non-canonical amine-detection site on transmembrane α-helix V (Asp5.42), or both. Thus, teleost TAARs likely represent a very large family of diverse amine receptors.
Several zebrafish TAAR ligands are biogenic amines produced during tissue decomposition (Shalaby, 1996). Cadaverine, histamine, agmatine, putrescine, tryptamine, 2-phenylethylamine, and isoamylamine are derived in one step from the natural amino acids lysine, histidine, arginine, ornithine, tryptophan, phenylalanine, and leucine by microbe-mediated decarboxylation. Levels of each of these amines are measured in commercially sold fish as indicators of food quality and freshness (Shalaby, 1996); for example, excessive histamine in consumed fish causes human scombroid poisoning (Morrow et al., 1991). While some of these biogenic amines are aversive to mammals (Dewan et al., 2013; Ferrero et al., 2011; Wisman and Shrira, 2015), they are reported to evoke variable behaviors in different fish species. For example, agmatine, putrescine, and cadaverine increase feeding-associated pecking behavior in goldfish (Rolen, et al., 2003; Hara, 2006) while cadaverine and putrescine evoke avoidance behavior in zebrafish (Hussain et al., 2013). Future studies are needed to examine the roles of each TAAR and its ligands in fish behavior.
The evolution of divergent solutions to the problem of amine detection by GPCRs reveals the extensive landscape of change possible within the GPCR superfamily. Here, we propose a two-step model for the functional evolution of clade III TAARs that is based on phylogenetic analysis, ligand identification studies, structural modeling, and mutagenesis (Figure 6). First, an ancestral clade I TAAR gained Asp5.42, and thus the ability to recognize di-cationic chemicals like diamines. Ligands were identified for five zebrafish TAARs that retain Asp3.32 and Asp5.42, and both aspartates are important for ligand recognition in each receptor. Second, a diamine receptor lost Asp3.32, leading to clade III TAARs that display non-classical monoamine recognition through a transmembrane α-helix V salt bridge to Asp5.42.
Flipping the amine orientation dramatically changes GPCR-ligand contacts, and presumably allows clade III TAARs to sample a distinct repertoire of structurally divergent amine odors. Once the new amine recognition motif was established, clade III TAARs dramatically expanded through a pattern of gene duplication and subsequent mutation. It seems likely that the massive expansion of this olfactory receptor clade increased the capacity of the fish olfactory system to detect amines, which are water-soluble and ecologically important stimuli for aquatic animals.
Materials and methods
Cloning of receptor genes
Zebrafish Taar genes were cloned from zebrafish genomic DNA (AB strain), and inserted into a modified pcDNA3.1- (Invitrogen, Waltham, MA, USA) vector containing a 5’ DNA extension encoding the first 20 amino acids of bovine rhodopsin followed by a cloning linker (GCGGCCGCC). Sequencing Taar genes revealed several AB strain polymorphisms as compared with Tübingen strain-derived genomic sequences deposited at NCBI. Amino acid-altering polymorphisms identified were as follows: Taar10a (NM_001082898): A467G (Ile to Val) and G545A (Val to Ile); Taar10b (NM_001082903): A338G (His to Arg), G496A (Ala to Thr), G544A (Val to Ile), and G775A (Val to Met); Taar12h (NM_001082909): A23G (Ile to Val), T26C (Tyr to His), C62T (Pro to Ser), C285A (Ser to Tyr), T743A (Phe to Ile), C874T and G875A (Gly to Ile); Taar12i (NM_001083085): T23C (Val to Ala), G313T, T314A (Val to Tyr), and T583G (Phe to Val); Taar13a (NM_001083102): C427G (Thr to Arg), T680A (Asn to Lys), T803G (Ile to Met), T882C (Phe to Leu), and T948C (Phe to Leu); Taar13d (NM_001083041): A112G (Ile to Val), A196C (Thr to Pro), G382A (Val to Ile), and G559A (Val to Ile); Taar13e (NM_001083043): A643G (Ile to Val) and G796A (Val to Ile); Taar16c (XM_009307036): A797C (Asn to Thr); Taar16e (XM_009307037): T103C (Val to Ala), G711A, C712T (Ala to Ile), and C844T (Ala to Val); Taar16f (XM_009305637): A23T (Asn to Ile), A59G (Asn to Ser), C190A (His to Lys), A202T (Thr to Ser), and A951C (Leu to Phe). Htr6 was cloned from mouse brain cDNA and inserted into pcDNA3.1-. Taar mutants were generated by overlap extension PCR and the Htr6 mutant was generated using TagMaster site-directed mutagenesis kit (GM Biosciences, Rockville, MD, USA).
TAAR functional assays
Reporter gene assays in HEK-293 cells or Hana3A cells (Saito et al., 2004) (for studies involving TAAR10b, TAAR12i, and TAAR16e) were performed as previously described (Liberles and Buck, 2006; Ferrero et al., 2012) with minor modifications. Modifications included use of tissue culture-treated 96 well plates (BD Biosciences, San Jose, CA, USA) pre-incubated with polylysine (250 ng per well); transfection using Lipofectamine 2000 (Invitrogen) reagent according to manufacturer's protocols, and introduction of test stimuli (see below) four to six hours after transfection.
Test stimuli for high throughput chemical screens
To identify TAAR agonists, chemicals were simultaneously tested in mixtures (83.3 μM per chemical, DMEM). Mix 1 contained GABA, β-alanine, 2-aminopentane, benzylamine, butylamine, and cystamine; mix 2 contained dibutylamine, dimethylamine, N,N-dimethylcyclohexylamine, ethylamine, ethyleneamine, and ethanolamine; mix 3 contained hexylamine, isoamylamine, isobutylamine, isopropylamine, indole, and cadaverine; mix 4 contained methylamine, N-methylindole, N-methylpyrrolidine, N-methylpiperidine, pyrrolidine, and putrescine; mix 5 contained hexanal, ethyl butyrate, tryptamine, histamine, 2-phenylethylamine, and trimethylamine; mix 6 contained 2-methylbutylamine, cysteamine, 1-amino propan-2-ol, 3-methylthiopropylamine, and agmatine; mix 7 contained tyramine, N,N-dimethylethanolamine, octopamine, N,N-dimethylglycine, 5-hydroxyindole-3-acetic acid, and 3-methoxytyramine; mix 8 contained 5-methoxytryptamine, 4-methoxy phenethylamine, N,N-dimethylphenethylamine, 5-methoxy-N,N-dimethyltryptamine, N,N-dimethylaniline, N,N-dimethylisopropylamine; mix 9 contained 1-dimethylamino propanol, 2-dimethylaminoethanethiol, and 1-(2-aminoethyl)-pyrrolidine; mix 10 contained 1,7-diaminoheptane, 1,8-diaminooctane, 1,6-diaminohexane, 1,10-diaminodecane, 1,3-diaminopropane; and mix 11 contained spermine, spermidine, dopamine, serotonin, and adenine. TAARs were tested with all 11 chemical mixtures, except TAAR14, TAAR15, and TAAR16 family members which were tested with mixes 1–10.
TAAR phylogenetic analysis
Amino acid sequences of 112 zebrafish TAARs, 15 mouse TAARs, 17 rat TAARs, 6 human TAARs, 12 biogenic amine receptors (zebrafish HTR2B, HRH2, and DRD2A; mouse HRH3, DRD3, HTR5, DRD1A, and ADRB1; and rat HTR2A, HRH2, DRD3, and ADRB2) and 5 odorant receptors (zebrafish OR103 and OR131, mouse OR121 and OR446, and rat ORI15) were aligned with MAFFT v7.017 (Katoh, 2002). For Figure 2—figure supplements 1, 2, published TAAR amino acid sequences from opossum, cow, chicken, frog, coelacanth, medaka, stickleback, tetraodon, fugu, Atlantic salmon, and elephant shark, as well as sea lamprey aminergic receptors, were analyzed (Hussain et al., 2009; Tessarolo et al., 2014). A phylogeny was inferred using a maximum likelihood approach implemented in RAxML (RAxML v7.7.1) using the JTT + Γ model of codon substitution, empirical base frequencies, and the rapid bootstrapping technique (100 replicates) (Stamatakis et al., 2008). Nodes with support values < 50 were collapsed in the program TreeGraph 2 and trees were visualized using FigTree v1.3.1 (Stöver and Müller, 2010).
TAAR structural modeling
The TAAR13c structural model was generated using SWISS-MODEL workspace (Biasini et al., 2014) (http://swissmodel.expasy.org/interactive) and based on the X-ray crystal structure of agonist-bound human β2 adrenergic receptor (Protein Data Bank entry 4LDE) (Ring et al., 2013). Modeling involved an active-state template, in which agonist binding is favored. The sequence alignment for homology modeling was verified by inspection to confirm proper alignment of sequence landmarks including conserved sequence motifs (DRY/DRH and NPxxY) and transmembrane proline residues. Homology models for TAAR10a (37% amino acid identity) and TAAR16c (30% amino acid identity) were prepared similarly using the same template. Models of receptor point mutants were prepared by side chain substitution in previously prepared homology models and then subjected to energy minimization prior to ligand docking. Docking of ligands into homology models was conducted with the Schrödinger suite (Schrodinger, 2015) using LigPrep version 3.4 and Glide version 6.7. First, the receptor model was prepared for docking by adding protons and assigning charges to ionizable side chains. Asp3.32 and Asp5.42 were modeled in the deprotonated state expected at physiological pH. The resulting model was then subjected to energy minimization using the Schrödinger impref utility prior to docking. Ligands were docked to the orthosteric binding pocket using Glide Extra Precision (Friesner et al., 2006), and figures were prepared in PyMol (Schrodinger, 2010).
Acknowledgments
We thank Xianchi Dong for assistance with GPCR modeling, Hiroaki Matsunami for Hana3A cells, and Sean Megason for zebrafish genomic DNA. This work was supported by a grant from the NIH (SDL, Award Number R01 DC013289).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grant:
National Institute for Health Research R01 DC013289 to Stephen D Liberles.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
QL, designed and interpreted experiments; wrote the manuscript; cloned TAARs and performed GPCR functional assays; generated and interpreted GPCR homology models; Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
YTB, designed and interpreted experiments; cloned TAARs and performed GPCR functional assays; Conception and design, Acquisition of data, Analysis and interpretation of data.
ZL, designed and interpreted experiments; cloned TAARs and performed GPCR functional assays; Conception and design, Acquisition of data, Analysis and interpretation of data.
MWB, designed and interpreted experiments; performed phylogenetic analysis, Conception and design, Acquisition of data, Analysis and interpretation of data.
ACK, designed and interpreted experiments;generated and interpreted GPCR homology models; Conception and design, Analysis and interpretation of data.
SDL, designed and interpreted experiments; wrote the manuscript; Conception and design, Analysis and interpretation of data, Drafting or revising the article.
References
- Baldwin MW, Toda Y, Nakagita T, O'Connell MJ, Klasing KC, Misaka T, Edwards SV, Liberles SD. Evolution of sweet taste perception in hummingbirds by transformation of the ancestral umami receptor. Science. 2014;345:929–933. doi: 10.1126/science.1255097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowsky B, Adham N, Jones KA, Raddatz R, Artymyshyn R, Ogozalek KL, Durkin MM, Lakhlani PP, Bonini JA, Pathirana S, Boyle N, Pu X, Kouranova E, Lichtblau H, Ochoa FY, Branchek TA, Gerald C. Trace amines: identification of a family of mammalian G protein-coupled receptors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8966–8971. doi: 10.1073/pnas.151105198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunzow JR, Sonders MS, Arttamangkul S, Harrison LM, Zhang G, Quigley DI, Darland T, Suchland KL, Pasumamula S, Kennedy JL, Olson SB, Magenis RE, Amara SG, Grandy DK. Amphetamine, 3,4-methylenedioxymethamphetamine, lysergic acid diethylamide, and metabolites of the catecholamine neurotransmitters are agonists of a rat trace amine receptor. Molecular Pharmacology. 2001;60:1181–1188. doi: 10.1124/mol.60.6.1181. [DOI] [PubMed] [Google Scholar]
- Dewan A, Pacifico R, Zhan R, Rinberg D, Bozza T. Non-redundant coding of aversive odours in the main olfactory pathway. Nature. 2013;497:486–489. doi: 10.1038/nature12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero DM, Lemon JK, Fluegge D, Pashkovski SL, Korzan WJ, Datta SR, Spehr M, Fendt M, Liberles SD. Detection and avoidance of a carnivore odor by prey. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11235–11240. doi: 10.1073/pnas.1103317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero DM, Wacker D, Roque MA, Baldwin MW, Stevens RC, Liberles SD. Agonists for 13 trace amine-associated receptors provide insight into the molecular basis of odor selectivity. ACS Chemical Biology. 2012;7:1184–1189. doi: 10.1021/cb300111e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Hara TJ. Feeding behaviour in some teleosts is triggered by single amino acids primarily through olfaction. Journal of Fish Biology. 2006;68:810–825. doi: 10.1111/j.0022-1112.2006.00967.x. [DOI] [Google Scholar]
- Horowitz LF, Saraiva LR, Kuang D, Yoon KH, Buck LB. Olfactory receptor patterning in a higher primate. The Journal of Neuroscience. 2014;34:12241–12252. doi: 10.1523/JNEUROSCI.1779-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A, Saraiva LR, Ferrero DM, Ahuja G, Krishna VS, Liberles SD, Korsching SI. High-affinity olfactory receptor for the death-associated odor cadaverine. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:19579–19584. doi: 10.1073/pnas.1318596110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A, Saraiva LR, Korsching SI. Positive darwinian selection and the birth of an olfactory receptor clade in teleosts. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4313–4318. doi: 10.1073/pnas.0803229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Josue J, Li X, Glaser D, Li W, Brand JG, Margolskee RF, Reed DR, Beauchamp GK. Major taste loss in carnivorous mammals. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4956–4961. doi: 10.1073/pnas.1118360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Research. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC. Structure-function of the g protein-coupled receptor superfamily. Annual Review of Pharmacology and Toxicology. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermen F, Franco LM, Wyatt C, Yaksi E. Neural circuits mediating olfactory-driven behavior in fish. Frontiers in Neural Circuits. 2013;7:62. doi: 10.3389/fncir.2013.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautwurst D, Yau KW, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/S0092-8674(00)81716-X. [DOI] [PubMed] [Google Scholar]
- Li Q, Korzan WJ, Ferrero DM, Chang RB, Roy DS, Buchi M, Lemon JK, Kaur AW, Stowers L, Fendt M, Liberles SD. Synchronous evolution of an odor biosynthesis pathway and behavioral response. Current Biology. 2013;23:11–20. doi: 10.1016/j.cub.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Liberles SD. Aversion and attraction through olfaction. Current Biology. 2015;25:R120–R129. doi: 10.1016/j.cub.2014.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- Liberles SD, Horowitz LF, Kuang D, Contos JJ, Wilson KL, Siltberg-Liberles J, Liberles DA, Buck LB. Formyl peptide receptors are candidate chemosensory receptors in the vomeronasal organ. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9842–9847. doi: 10.1073/pnas.0904464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD. Mammalian pheromones. Annual Review of Physiology. 2014;76:151–175. doi: 10.1146/annurev-physiol-021113-170334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD. Trace amine-associated receptors: ligands, neural circuits, and behaviors. Current Opinion in Neurobiology. 2015;34:1–7. doi: 10.1016/j.conb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER. Use it or lose it: molecular evolution of sensory signaling in primates. Pflügers Archiv : European Journal of Physiology. 2006;453:125–131. doi: 10.1007/s00424-006-0120-3. [DOI] [PubMed] [Google Scholar]
- Lindemann L, Hoener MC. A renaissance in trace amines inspired by a novel GPCR family. Trends in Pharmacological Sciences. 2005;26:274–281. doi: 10.1016/j.tips.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Morrow JD, Margolies GR, Rowland J, Roberts LJ. Evidence that histamine is the causative toxin of scombroid-fish poisoning. The New England Journal of Medicine. 1991;324:716–720. doi: 10.1056/NEJM199103143241102. [DOI] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nature Reviews. Genetics. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Reese EA, Norimatsu Y, Grandy MS, Suchland KL, Bunzow JR, Grandy DK. Exploring the determinants of trace amine-associated receptor 1's functional selectivity for the stereoisomers of amphetamine and methamphetamine. Journal of Medicinal Chemistry. 2014;57:378–390. doi: 10.1021/jm401316v. [DOI] [PubMed] [Google Scholar]
- Revel FG, Moreau J-L, Gainetdinov RR, Bradaia A, Sotnikova TD, Mory R, Durkin S, Zbinden KG, Norcross R, Meyer CA, Metzler V, Chaboz S, Ozmen L, Trube G, Pouzet B, Bettler B, Caron MG, Wettstein JG, Hoener MC. TAAR1 activation modulates monoaminergic neurotransmission, preventing hyperdopaminergic and hypoglutamatergic activity. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8485–8490. doi: 10.1073/pnas.1103029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring AM, Manglik A, Kruse AC, Enos MD, Weis WI, Garcia KC, Kobilka BK. Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature. 2009;459:574–577. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- Rolen SH, Sorensen PW, Mattson D, Caprio J. Polyamines as olfactory stimuli in the goldfish carassius auratus. Journal of Experimental Biology. 2003;206:1683–1696. doi: 10.1242/jeb.00338. [DOI] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC. The PyMOL molecular graphics system, version 1.3r1 2010.
- Schrodinger LLC. Schrodinger release 2015-2 2015.
- Seifert R, Strasser A, Schneider EH, Neumann D, Dove S, Buschauer A. Molecular and cellular analysis of human histamine receptor subtypes. Trends in Pharmacological Sciences. 2013;34:33–58. doi: 10.1016/j.tips.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby AR. Significance of biogenic amines to food safety and human health. Food Research International. 1996;29:675–690. doi: 10.1016/S0963-9969(96)00066-X. [DOI] [Google Scholar]
- Shi L, Javitch JA. The binding site of aminergic g protein-coupled receptors: the transmembrane segments and second extracellular loop. Annual Review of Pharmacology and Toxicology. 2002;42:437–467. doi: 10.1146/annurev.pharmtox.42.091101.144224. [DOI] [PubMed] [Google Scholar]
- Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Research. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Stöver BC, Müller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surgand J-S, Rodrigo J, Kellenberger E, Rognan D. A chemogenomic analysis of the transmembrane binding cavity of human g-protein-coupled receptors. Proteins. 2006;62:509–538. doi: 10.1002/prot.20768. [DOI] [PubMed] [Google Scholar]
- Tessarolo JA, Tabesh MJ, Nesbitt M, Davidson WS. Genomic organization and evolution of the trace amine-associated receptor (TAAR) repertoire in Atlantic salmon (Salmo salar) G3. 2014;4:1135–1141. doi: 10.1534/g3.114.010660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisman A, Shrira I. The smell of death: evidence that putrescine elicits threat management mechanisms. Frontiers in Psychology. 2015;6:1274. doi: 10.3389/fpsyg.2015.01274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Pacifico R, Cawley D, Feinstein P, Bozza T. Ultrasensitive detection of amines by a trace amine-associated receptor. Journal of Neuroscience. 2013;33:3228–3239. doi: 10.1523/JNEUROSCI.4299-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8337–8341. doi: 10.1073/pnas.1331721100. [DOI] [PMC free article] [PubMed] [Google Scholar]