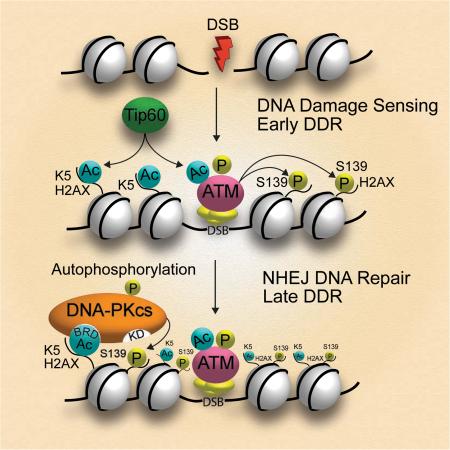
SUMMARY
Regulatory mechanisms underlying γH2AX induction and the associated cell fate decision during DNA damage response (DDR) remain obscure. Here we discover a bromodomain (BRD)-like module in DNA-PKcs (DNA-PKcs-BRD) that specifically recognize H2AX acetyl-lysine 5 (K5ac) for sequential induction of γH2AX and concurrent cell fate(s). First, top-down mass spectrometry of radiation-phenotypic, full-length H2AX revealed a radiation-inducible, K5ac-dependent induction of γH2AX. Combined approaches of sequence-structure modeling/docking, site-directed mutagenesis, and biochemical experiments illustrated that through docking on H2AX K5ac, this non-canonical BRD determines not only the H2AX-targeting activity of DNA-PKcs but also the over-activation of DNA-PKcs in radio-resistant tumor cells whereas a Kac antagonist JQ1 could bind to DNA-PKcs-BRD, leading to re-sensitization of tumor cells to radiation. This study elucidates the mechanism underlying the H2AX-dependent regulation of DNA-PKcs in IR-induced, differential DDR, and derives an unconventional, non-catalytic-domain target in DNA-PKs for overcoming resistance during cancer radiotherapy.
Graphical Abstract
INTRODUCTION
Based on the severity of DNA double-stranded breaks (DSBs) and the duration of stress exposure, cells take different decision-making pathways toward either apoptosis or survival(Lobrich and Jeggo, 2007). An acute ionizing radiation (IR) usually triggers pro-apoptotic signals in cells with irreparable DSBs or active DNA repair of survived cells, whereas cells constantly exposed to lower radiation doses can become tolerant or adapted to the frequent DNA damage caused by repeated irradiation(Mullenders et al., 2009). Cells with such an adaptive response are generally discerned by reduced sensitivity to stimuli as tumor cells can escape immunosurveillance under IR-adaptive conditions, contributing to an increased risk of chronic inflammation-associated carcinogenesis, and the acquired radio-resistance in tumor cells(Mullenders et al., 2009).
As one of the earliest cellular DDR, a replacement histone variant, H2AX, senses DSBs through rapid phosphorylation of the highly conserved Ser139(Bonner et al., 2008). This phosphorylation at Ser139, or γH2AX, then serves as a central scaffold that recruits protein factors associated with diverse functions including IR-induced cell-cycle arrest(Du et al., 2006), nucleosome dynamics(Heo et al., 2008), resulting in γH2AX foci over large chromatin domains surrounding DSBs(van Attikum and Gasser, 2009). Although evidences indicate the central role of DSB-inducible γH2AX in coordinating diverse processes of DSB repair and cell fate decision (Bonner et al., 2008), still obscure, however, is exactly how the phenotypic regulation of γH2AX is achieved, and its impact on either normal or abnormal cell fate decision.
As one of the two H2AX-targeting kinases that play redundant role in regulating γH2AX, DNA-PKcs not only promotes the H2AX-mediated apoptosis or DNA repair of damaged cells, but also, when over-activated, contributes to the resistance to DSB-induced apoptosis in human malignant cells(Deriano et al., 2005). These observations immediately raise the mechanistic questions as to how DNA-PKcs regulates these totally opposite DDRs? Based on a previous report that phosphorylation of H2AX by DNA-PK could be stimulated only in the context of acetylation-rich nucleosomes(Park et al., 2003), we reason there could be an acetylation-dependent mechanism underlying the activation of DNA-PKcs during H2AX-mediated DDR.
Given cross-regulations exist among different post-translational modifications (PTMs) on H2AX for either apoptosis/survival(Cook et al., 2009) or chromatin reorganization during DDR(Ikura et al., 2007), we first mapped the combinatorial PTM pattern on H2AX and its IR-induced changes by using a 12 Tesla FTICR mass spectrometry (MS) with ultrahigh mass accuracy and resolution that we have simultaneously identified multiple acetyl-lysine (Kac) in a full-length protein so that their relative abundances were quantified (Zhao et al., 2010). As a result, we observed an IR-inducible, concerted increase of both acetylated lysine 5 (K5ac) and γH2AX. Further, we found that, in the later phase of IR-induced DDR, in a K5ac-dependent manner DNA-PKcs was the primary kinase to phosphorylate H2AX Ser139. Combined approach utilizing molecular modeling/docking, site-directed mutagenesis, and biochemical/cell biology analyses revealed a novel BRD-like module in DNA-PKcs that not only specifically recognizes K5ac on H2AX but also tightly binds to JQ1, a small molecule antagonist of BET BRD and a Kac structure-mimic(Filippakopoulos et al., 2010). Further, we found that the DNA-PKcs activity for inducing γH2AX is K5ac/BRD-dependent, and this K5ac-depenent activity of DNA-PKcs acts as a double-edged sword, promoting either the DDR of acute-irradiated cells or the radio-resistance of chronic-irradiated cells. We mechanistically reveal that the K5ac induced on H2AX by prior irradiation is responsible for the early-phase over-activation of DNA-PKcs in radio-resistant leukemia cells(Deriano et al., 2005) where DNA-PKcs-BRD recognizes the H2AX K5ac during the activation of DNA-PKcs for nonhomologous end joining (NHEJ) repair. Unlike the mostly available drugs targeting the catalytic domain of DNA-PKcs(Bruce et al., 2012) our findings indicate a novel NHEJ pathway-specific, unconventional target of (+)-JQ1 that could readjust the off-balanced activity of DNA-PKcs in the H2AX-mediated NHEJ. As an immediate result of (+)-JQ1 pretreatment, the mitochondria-mediated apoptotic pathway was found reactivated in the radio-resistant leukemia cells that regained the sensitivity to IR.
RESULTS
The IR-inducible γ-Ser139 on H2AX Could be K5ac-Dependent
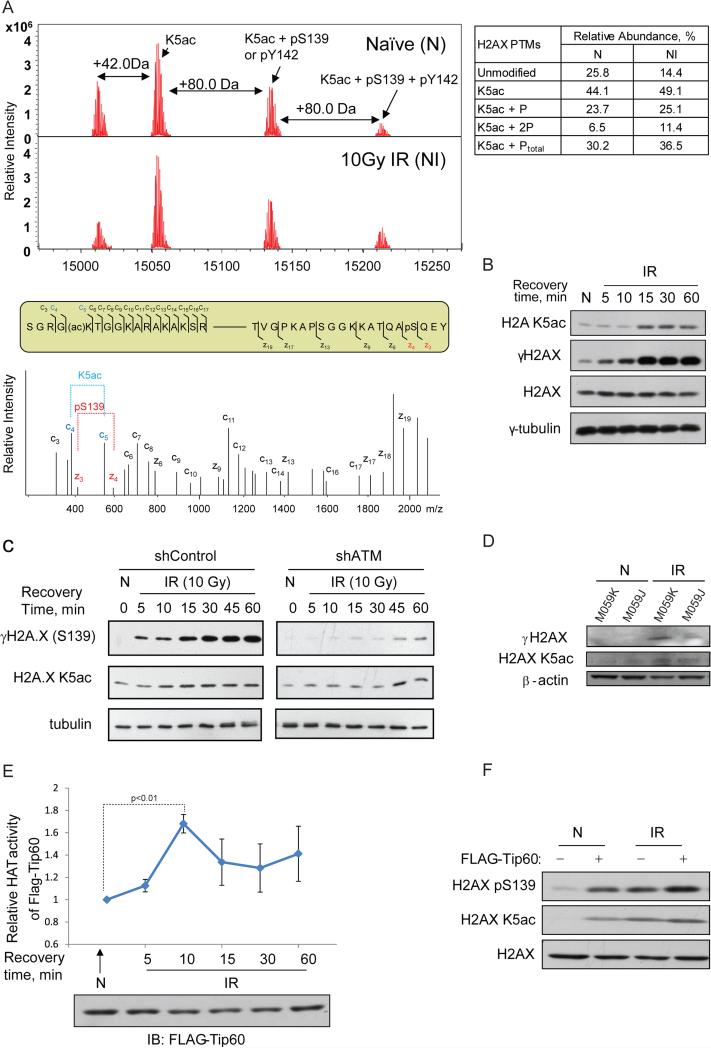
We first performed FTICR MS and electron capture dissociation (ECD) MS/MS (Zhao et al., 2010) to examine if 10 Gy IR could induce any concerted changes of different PTMs on H2AX. We purified by HPLC the histones extracted from 10 Gy-irradiated cells following a 1-hour post-radiation recovery, and used FTICR MS to analyze the single species in the fraction 19 (Figure S1A). From low to high m/z, three different forms of full-length H2AX were MS/MS-sequenced and identified in a combinatorial manner as the following: unmodified H2AX, H2AX K5ac, K5ac with γ-Ser139, and K5ac with both γ-Ser139 and γ-Tyr142 (Figure 1A and Figure S1B-1E).
Figure 1. IR-inducible phosphorylation of serine 139 (γ-Ser139) on H2AX could be K5ac-dependent.
(A), (Top) Top-down μESI-FTICR-MS analysis of full-length H2AX isolated from non-irradiated or 10 Gy-irradiated cells. +42 Da or +80 Da corresponds to addition of an acetyl or a phosphate group. Each mass spectrum was acquired in the ICR cell with 100 scans, (Middle) Sequence locations of ECD MS/MS fragment ions detected at either N-terminal (c ions) and C-terminal (z ions) on modified H2AX, and (Bottom) ECD MS/MS spectra of the H2AX precursor ions at m/z 1513.62 Da with 10+ charges, showing the fragment ions for K5ac and γ-Ser139. Both N-terminal (c ions) and C-terminal (z ions) fragment ions indicating the corresponding PTM are marked in red or blue, respectively. The ECD MS/MS spectrum shown with the low m/z fragment ions corresponds to N-terminal K5Ac and C-terminal γSer139. (B) Immunoblot analysis of 10 Gy IR-induced time-dependent changes of K5ac or γSer139. Non-irradiated or 10Gy-irradiated (100 rads/min for 10 min) Hela cells were collected at 5, 10, 15, 30, and 60 min after IR. Cells were lysed with 200 μl SDS sample buffer, and 5 μl was separated on 15% SDS-PAGE, transferred to PVDF and probed with the antibody against either γH2AX or H2A K5ac. (C) Immunoblot analysis of 10 Gy IR-induced time-dependent changes of K5ac or γSer139 in the wild-type F1-hTERT-shLacZ (control) versus ATM KD F1-hTERT-shATM cells. (D) DNA-PKcs directly contributes to the high-dose IR-induced γH2AX in the later phase of DDR. Both M059K and M059J DNA-PKcs-deficient human glioma cell lines were irradiated by a 10 Gy IR, allowed to recover for 60 mins. β-actin was used as the loading control. Quantitative analysis of the changes of K5ac or γSer139 was performed as described in Methods. One experimental point containing three technical duplicates representative of two is shown with the data precision indicated by the error bar based on mean ± SD of duplicate samples. (E) 10 Gy IR-induced time-dependent changes in the activity of Tip60. 293T cells were transfected with FLAG-Tip60, irradiated by 10 Gy IR, and harvested at each indicated time. Tip60 was immunoprecipitated using anti-flag antibody from the non-irradiated or irradiated cells collected at each recovery time and its time-dependent HAT activity was determined using a colorimetric assay (BioVision, Mountain view, CA) and normalized against the amount of FLAG-Tip60 determined by immunoblot. (F) Exogenous Tip60 triggered greater simultaneous increases of K5ac and γSer139 on H2AX in non-irradiated HeLa cells. HeLa cells without or with FLAG-Tip60 transfection were subjected to radiation and IB with indicated antibody. H2AX was used as the loading control.
Notably, in this particular HPLC fraction (Figure S1A) the acetylated forms of H2AX that have sufficient amounts for unambiguous MS/MS sequencing were found as the dominant species, while the population of non-acetylated H2AX was minor or could be suppressed during MS analysis so no γ-Ser139 was detected on the un-acetylated form of H2AX. Also, although there was a minor population of putative ubiquitinated H2AX was detected by MS, it was too weak to be sequenced by ECD MS/MS.
Because of simultaneous identification of both PTMs on full-length H2AX, we were able to quantitatively analyze IR-induced, concerted changes of multiply modified H2AX (Figure 1A): Following a high-dose IR and 1-hour recovery, the relative amount of unphosphorylated form in the total acetylated H2AX population was reduced by approximately 50% from 25.8% to 14.4% (table in Figure 1A). Meanwhile, the increases of both single and double phosphorylations (γ-Ser139 or and γ-Tyr142) occurred only on the K5ac form of H2AX, and both K5ac and γ-Ser139 or/and γ-Tyr142 were simultaneously increased compared to non-irradiated cells, indicating that both acetylation and phosphorylations on H2AX are coupled under a high-dose IR. Our discovery-driven, phenotype-specific, top-down MS analysis thus indicated that these H2AX PTMs are coordinately deposited during IR-induced DDR. Due to the m/z degeneracy between γ-Ser139 and γ-Tyr142, the proportion of these two phosphorylated forms was not distinguishable in MS spectra. We therefore used immunoblotting to determine which one of γ-Ser139 or γ-Tyr142 or both contributes to the IR-inducible abundance changes. First, we verified the K5ac-dependance of the DSB-induced site-specific phosphorylation(s) in different cell types after exposure to IR at different dose levels. In agreement with our FTICR MS results (Figure 1A), both K5ac and γSer139 showed the same trend of simultaneous increase with increasing doses of IR while γ-Tyr142 showed little IR-inducible change (Figure S2A), further indicating that the majority of the observed IR-inducible, K5ac-dependent increases of phosphorylation is from γ-Ser139. Results from IR-phenotypic screening of H2AX PTMs by using both top-down MS and immunoblotting have then generated the central hypothesis that γ-Ser139 may be induced by high-dose acute IR in the Kac-dependent way.
DNA-PKcs is the K5ac-dependent Kinase that Phosphorylates Serine 139
We then searched for a K5ac-dependent, H2AX-targeting kinase and clarified the biological implication of its K5ac-dependence. Previous work indicated that Ser139 phosphorylation was still inducible by a 10Gy IR on a K5R H2AX mutant(Xie et al., 2010), suggesting that not all PIKK family kinases for Ser139 phosphorylation, including ATM or DNA-PK(Bonner et al., 2008), function in a Kac-dependent manner. First, we investigated exactly how IR-induced changes of γ-Ser139 and K5ac are coordinated in 10 Gy-irradiated cells collected at different post-irradiation times. In Hela cells, within the first 10 min of recovery following a high-dose IR, γ-Ser139 increased whereas K5ac remained at its basal level (Figure 1B). After 15 min, however, both K5ac and γ-Ser139 increased significantly, indicating that the K5ac-dependence of Ser139 phosphorylation occurs only in irradiated cells with longer post-irradiation recovery. Further, to determine when ATM plays a dominant role in Ser139 phosphorylation, we compared the IR-induced, time-resolved changes of both PTMs in a pair of ATM knock-down (KD) cells, F1-hTERT-shATM (ATM KD), versus a wild-type control, F1-hTERT-shLacZ(Heffernan et al., 2002). While WT cells showed similar time-dependent changes as in Hela cells (Figure 1C, left), shRNA-mediated ATM KD led to diminished γ-Ser139 at an early stage of post-irradiation recovery. However, later simultaneous increases of both K5ac and γ-Ser139 were unaffected by ATM knock-down (Figure 1C, right), indicating that, during the early phase of DDR, ATM is the primary kinase targeting H2AX in a K5ac-independent manner.
DNA-PKcs, therefore, became the likely kinase responsible for the later increase of γ-Ser139, which is accompanied by an increase in K5ac. To conform this, we first examined the IR-induced changes of γ-Ser139 and K5ac during the late phase of DDR on a pair of human glioma cell lines M059K (WT DNA-PKcs) and M059J (DNA-PKcs-deficient)(Lees-Miller et al., 1995). Simultaneous increases of K5ac and γ-Ser139 in irradiated cells with a recovery longer than 15 min were observed only in WT cells but not in the DNA-PKcs-deficient cells (Figure 1D). Although a low-level of ATM expression was found in M059J cells(Fang et al., 2012), these DNA-PKcs-deficient cells clearly showed a lack of IR-inducible, coordinated increases of K5ac and γ-Ser139. Further, the composition of the 10 Gy IR-induced H2AX-interactome recently dissected by our multiplex quantitative proteomics indicated enhanced interactions between H2AX and the DNA-PK complex (Lee et al., 2012) coincident with the high-dose IR-induced increase of K5ac, also supporting that DNA-PKcs is the primary H2AX-targeting kinase at 1-hour post-IR recovery, which correlated with the defined H2AX PTM pattern (Figure 1A).
We then clarified the sequential order of depositing these PTMs on H2AX by examining the effect of a K5ac-targeting histone acetyltransferase (HAT) Tip60(Sun et al., 2007) on the level of γ-Ser139. First, the HAT activity of Tip60 showed a trend of changes similar to that of either K5ac or γ-Ser139 in the 10 Gy-irradiated cells during the same period of post-IR recovery (Figure 1E), implicating the IR-inducible Tip60 activity in depositing K5ac. Further, in non-irradiated cells with a low-level of K5ac, over-expression of Tip60 led to dramatic increases of both K5ac and γ-Ser139 (Figure 1F). Notably, over-expression of exogenous Tip60 contributed little to the already high level of K5ac in 10 Gy-irradiated cells, suggesting that, compared with non-irradiated cells, endogenous Tip60 in IR-responsive cells was fully activated by high-dose IR to acetylate H2AX. However, the low activity of endogenous Tip60 in non-irradiated was supplemented by over-expressed Tip60, resulting in higher K5 acetylation. These results indicated that the IR-inducible, Tip60-promoted K5 acetylation is a prerequisite for Ser139 phosphorylation, and recognition of H2AX by DNA-PK is enhanced due to more K5ac during the later DDR phase when the activity of DNA-PKcs depends on acetylated H2AX.
DNA-PKcs Contains a BRD-like Structural Module to Recognize H2AX K5ac
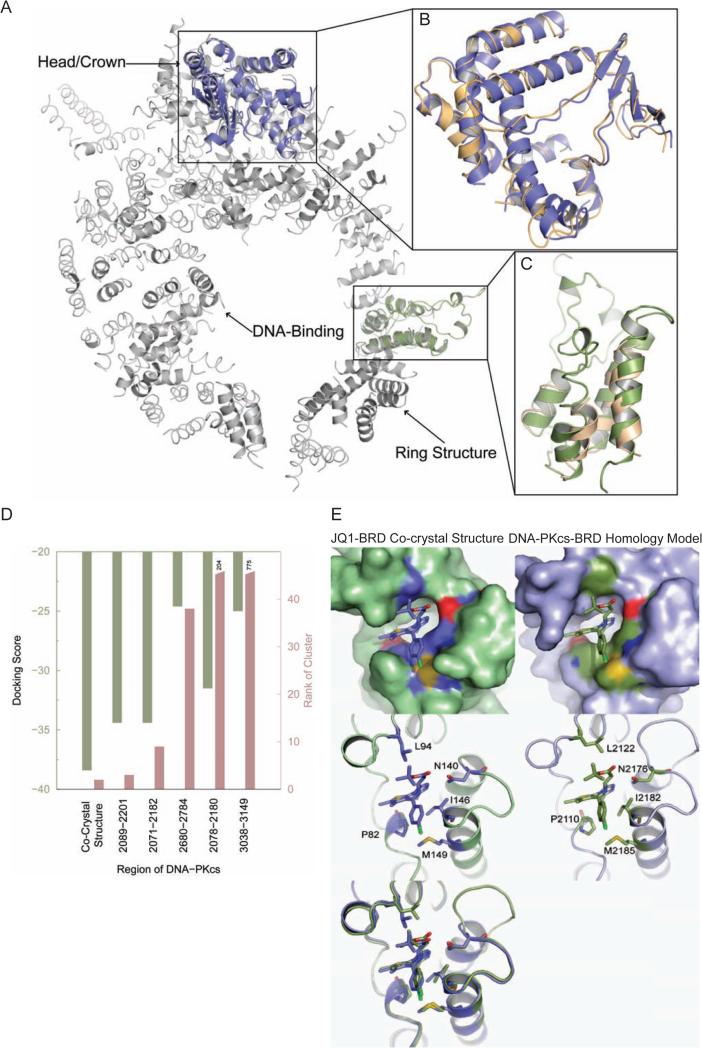
We next investigated the structural basis for the K5ac-dependent activation of DNA-PKcs. Based on the DNA-PK crystal structure(Sibanda et al., 2010), we used a sequence-structure modeling approach to explore whether any region in DNA-PKcs shares structural homology with BRDs. The crystal structure of DNA-PKcs features various regions named head, forehead, putative DNA-binding domain, and ring structure(Sibanda et al., 2010) (Protein Data Bank (PDB)(Berman et al., 2000) ID: 3KGV). However, due to low-resolution (6.6 Å), only the protein backbone forming different secondary structural elements was discernible, whereas the identities of the residues in the corresponding structural regions were not known. Thus, we first used Situs(Wriggers, 2009) to align the structure of the catalytic N- and C-lobes of phoshoinositide 3-kinase (PI3K, PDB ID: 1E8X) onto the crystal structure of DNA-PKcs (Figure S3). The PI3K catalytic subunit aligned well with the head/crown region of DNA-PKcs (Figure 2A and 2B). Further, using Situs, we examined alignments of the previously determined BRD structures with the DNA-PKcs structure and found significant structural homology between the BRD and a region of the HEAT repeats in the DNA-PKcs ring structure, which was confirmed using the Dali server(Holm and Rosenstrom, 2010) (Figure 2A and 2C, Table S1).
Figure 2. Molecular modeling/docking reveals a Kac-recognizing, BRD-like structure in DNA-PKcs.
(A) Structural alignment of DNA-PKcs (gray) with the catalytic subunit of PI3K (blue) and the BRD template (green). (B) Detailed structural alignment of PI3K catalytic subunit (blue) with DNA-PKcs (orange). (C) Detailed structural alignment of the BRD (green) with DNA-PKcs (orange). Here Dali reported a Ca root mean square deviation between BRD and the aligned region of DNA-PKcs to be 2.1 Å and a Z-score of 5.1, both of which indicate statistically significant structural homology (Table S1). The structures are rendered in cartoon representation using PyMOL. (D) Summary of docking results for different DNA-PKcs-BRD homology models. To select the best region that could form BRD in DNA-PKcs, docking studies were performed on all possible DNA-PKcs-BRD homology models. The docking score represents a combination of the stability of BRD-JQ1 complex and the strength of interaction of JQ1 with BRD. A negative docking score indicates better binding of JQ1 to BRD. The docking scores of the near-native poses are presented. The cluster rank illustrates the extent of sampling of the near-native poses. Top-ranked near-native clusters indicate better amenability of the BRD to bind JQ1. The co-crystal structure has the best docking score and cluster rank. Closest to the co-crystal structure is the putative BRD formed by residues 2089-2201 in DNA-PKcs. (E) Near-native docked pose of DNA-PKcs-BRD homology model. The surface representation of the pocket binding JQ1 (Top) in mouse BRD4 co-crystal structure (Left) and the DNA-PKcs-BRD homology model (Right) are shown with surface representation. The bound ligand (JQ1) is shown with sticks representation. The surface of residues in the pocket that are identical between mouse BRD4 and DNA-PKcs-BRD are colored in contrast to the rest of the binding pocket (blue, yellow and red in the co-crystal structure; green, yellow, dark blue and red in the BRD homology model). The binding poses are shown in the same view, but shown with the cartoon representation (Middle) to enable visualization of the positions of the residues in the binding pocket that are identical between mouse BRD4 and DNA-PKcs-BRD. These residues are shown as sticks and their positions in the corresponding sequences are labeled. The structural overlay of co-crystal structure and the BRD homology model (Bottom) further illustrates the similarities in the binding mode of JQ1 and the residues lining the binding pocket.
We then used a combination of sequence-structure alignment and molecular docking MedusaDock(Ding et al., 2010) to identify the exact sequence/location of this BRD in DNA-PKcs. Using FUGUE, we threaded the DNA-PKcs sequence through representative crystal-structures of BRDs to arrive at putative sequence-structure alignments that pinpointed BRD in several possible regions of DNA-PKcs (Table S2). To find the region that best represents a BRD, based on the co-crystal structure between BRD4(1) and an BET BRD-specific inhibitor (+)-JQ1(Filippakopoulos et al., 2010), we constructed homology models of BRD4 using sequence corresponding to all these regions and ranked the JQ1 binding ability of the homology models using MedusaDock. Considering both the docking score of the near-native pose and the rank of the near-native cluster (Figure 2D), we found residues 2089-2201 as the best possible region to form a putative BRD4 in DNA-PKcs as the alignment of DNA-PKcs with the crystal structure of mouse BRD4 (mBRD4, PDB ID 3JVK) indicated a large similarity (59%) with the BRD-characteristic left-handed bundle of four α-helices(Filippakopoulos et al., 2012), linked by loop regions of variable length (Figure S4). Strikingly, the structural model formed by DNA-PKcs-BRD and JQ1 with the published BRD4-JQ1 co-crystal structure (Figure 2E, top, and Figure S5) showed that many of the functionally significant residues in each Kac binding pocket were identical in their interactions with JQ1, e.g., the alignment recapitulates a conserved asparagine(Owen et al., 2000) involved in Kac binding (N140 in mBRD4 versus N2176 in DNA-PKcs) (Figure 2E, middle), indicating a good shape complementarity between both structures of BRDs in their JQ1-binding pockets (Figure 2E, bottom).
To experimentally validate our docking modeling that derives the JQ1-bound DNA-PKcs BRD, we expressed, purified, and refolded epitope-tagged DNA-PKcs fragments (aa1878-2700) comprising the putative BRD domain (aa2070-2200). Two mutants of this DNA-PKcs BRD fragment, a single mutant N2176A and a double mutant P2110A/N2176A, were also generated based on their locations in the JQ1/Kac binding pocket (Figure 2E). First, we used circular dichroism (CD) to confirm that WT or mutants of DNA-PK-BRD are properly refolded (Figure S6), and the WT DNA-PKcs BRD fragment showed unambiguously specific binding to JQ1 when incubated with Sepharose-immobilized JQ1 (Figure 3A). Compared with WT DNA-PKcs, the binding of JQ1 to both mutants was weakened (even with higher amounts of protein loaded), particularly for the N-depleted mutant(s) (Figure 3B), validating the accuracy of our modeling the JQ1 binding pocket in DNA-PKcs that was disrupted by substitution of a key pocket asparagine present in most BRDs(Owen et al., 2000). The binding constants were not accessible at this point due to the instability of refolded proteins.
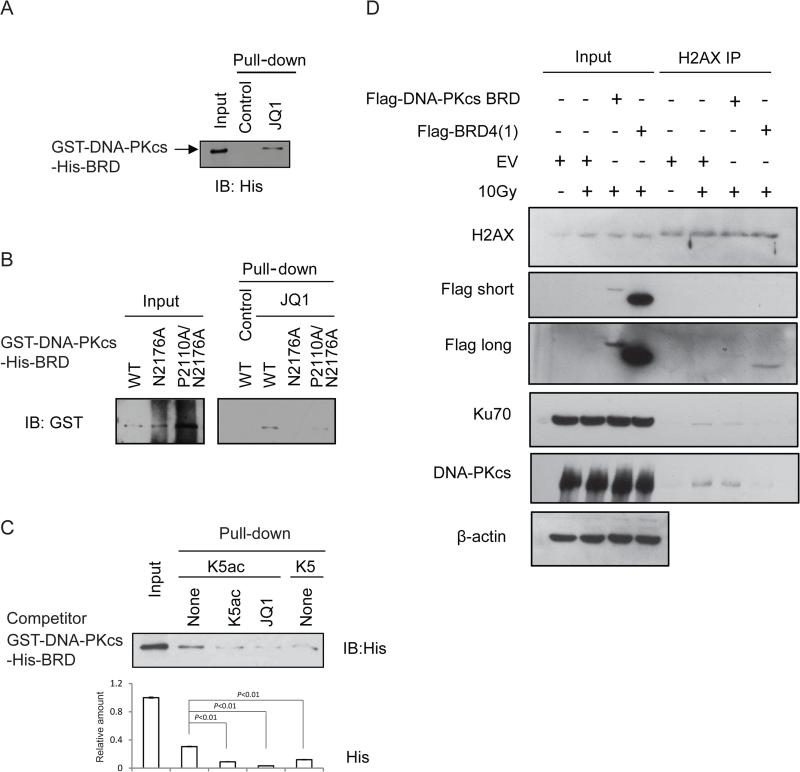
Figure 3. The DNA-PKcs-BRD specifically recognizes H2AX K5ac.
Immuoblot analysis of JQ1 binding to (A) the BRD-containing DNA-PKcs fragment (aa1878-2700), and (B) the mutants of this DNA-PKcs-BRD fragment including a single mutant N2176A and a double mutant P2110A/N2176A. These proteins were purified under denaturing conditions and refolded for the binding assay. (C) Analysis of comparative binding between the DNA-PKcs-BRD and the peptides containing either K5ac or non-acetylated K5 peptide without or with 10 μM JQ1. (D) Immunoblot analysis of the H2AX complexes pulled-down respectively from the U2OS cells transfected with either empty vector (EV) or the vector containing either gene expressing flag-tagged DNA-PKcs-BRD or flag-tagged BRD4(1). Some sets of the cells were irradiated by 10 Gy IR as indicated. The immunoprecipitates were blotted against the indicated antibody including DNA-PKcs or Ku70 or H2AX.
To prove the specificity of DNA-PKcs-BRD in recognizing K5ac we then compared binding between WT DNA-PKcs-BRD and H2AX peptides containing either un-acetylated K5 or K5ac. In line with the result from a peptide array study of BRD binding in which the highly conserved N-terminal sequences of both H2A and H2AX strongly bound to BRD4(Filippakopoulos et al., 2012), DNA-PKcs-BRD showed a stronger binding to the K5ac-containing peptide compared with its un-acetylated counterpart (Figure 3C). Further, addition of JQ1, the tighter binder of BRD4, completely abolished binding between DNA-PKcs-BRD and K5ac, indicating that the DNA-PKcs-BRD is a BRD4-like structure for Kac-recognizing with which JQ1 can compete.
Because JQ1 is a BET BRD-specific inhibitor that affects the functions of various Kac-recognizing, BET/BRD-containing proteins in cells, we investigated whether DNA-PKcs-BRD is indeed the functional module specifically involved in the IR-induced interaction between the DNA-PK complex and H2AX. We then generated a vector expressing only the BRD region of DNA-PKcs-BRD(aa2070-2200) with a size comparable to that of BRD4(1) and found its specific binding to Kac-mimic JQ1 was preserved (Figure S7A). In the 10Gy-IR-induced, H2AX-associated complexes pulled down by H2AX antibody respectively from the U2OS cells transfected with either empty vector (EV) or DNA-PKcs-BRD or BRD4(1) prior to 10Gy IR, we found that over-expression of either DNA-PKcs-BRD or BRD4(1) led to dissociation of endogenous DNA-PKcs from H2AX (Figure 3D). These results together indicate that the BRD of DNA-PKcs is the structural determinant for promoting the IR-induced interaction between the DNA-PK complex and H2AX, therefore, the exogenously expressed DNA-PKcs-BRD could disrupt IR-induced, BRD-dependent interaction between H2AX and the endogenous DNA-PKcs.
The DNA-PKcs-BRD Defines its activity for H2AX phosphorylation and the Associated Cell Fate Decision
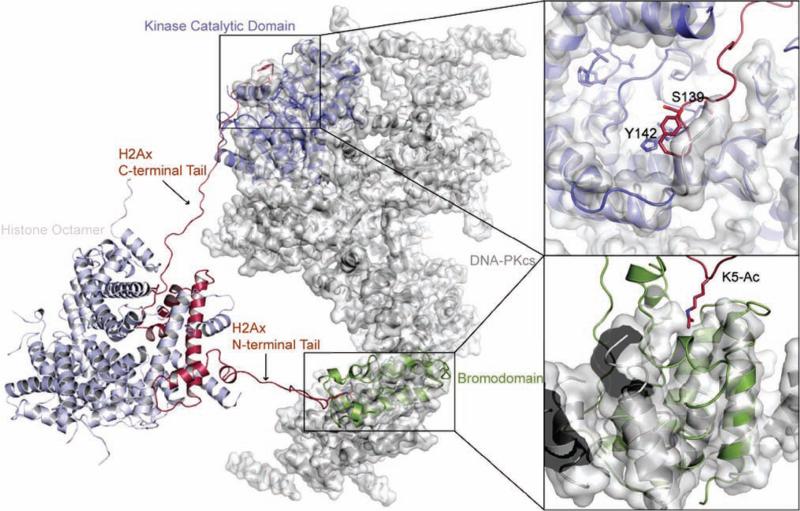
We next clarified structurally how the DNA-PKcs-BRD binding to the N-terminal K5ac facilitates phosphorylation of the C-terminal Ser139 on H2AX. Using the structure of the human nucleosome (PDB ID: 2CV5) as a template, we first constructed a homology model of an H2AX-containing nucleosome and then built its tail domains that are missing in the crystal structure. After manually placing the H2AX-containing nucleosome near DNA-PK, we set up constrained discrete molecular dynamics (DMD) simulations(Ding et al., 2008; Dokholyan et al., 1998), which showed a possibility for simultaneous binding of Kac and Ser139 to regions corresponding to the BRD and the catalytic domain of DNA-PKcs (Figure 4). Furthermore, the highly specific recognition of N6-Kac by BRD would fulfill a structural need for stabilization of the contact between its catalytic domain and the H2AX tail; this recognition would also enable DNA-PKcs to selectively bind K5ac-H2AX and target Ser139 for phosphorylation, an event that would be lost by substitution of lysine 5 with alanine, consequently making the kinase domain of DNA-PKcs less favorable to phosphorylate Ser139. The significant structural homology indicates that the binding of DNA-PKcs-BRD to K5ac could induce a conformational change favorable for DNA-PKcs to recognize H2AX Ser139.
Figure 4. Simultaneous binding of an H2AX-containing nucleosome to the BRD and the kinase domain of DNA-PKcs.
A structural model of an H2AX-containing nucleosome, with the N-terminal tail of H2AX bound to the putative BRD of DNA-PKcs (through K5ac) and the C-terminal tail positioned in the catalytic region of the kinase domain of DNA-PKcs (which would facilitate phosphorylation of Ser139). The detailed binding modes of Ser139 and K5ac are shown in the right. DNA-PKcs is rendered in the surface and cartoon representation, whereas the nucleosome is rendered using the cartoon representation. K5ac, Ser139, and Tyr142 are rendered using a stick representation. The PI3K catalytic subunit (blue) and the bromodomain template (green) are shown aligned to DNA-PKcs as in Fig. 2A-C. The structures are rendered using PyMOL.
As an immediate validation of the K5ac docking of DNA-PKcs, we determined how H2AX K5ac impacts on the co-localization between H2AX as well as the IR-induced DNA-PKcs activation by using the cells transfected with either FLAG-tagged WT H2AX or a non-acetylatable K5A mutant of H2AX that are correctly packed into chromatin(Du et al., 2006). In 10 Gy-irradiated cells, compared with the co-localized foci between more activated DNA-PKcs indicated by a DNA-PKcs activity marker, phospho-Thr2609(Chen et al., 2007) (in red) and WT H2AX, DNA-PKcs not only weakly co-localized with the non-acetylatable H2AX mutant K5A but also became less activated in the absence of Kac (Figure S7B). Notably, little γSer139 was detected on the FLAG-tagged H2AX K5A mutant from the 10 Gy-irradiated cells with a 1-hour post-IR recovery (Figure S7C), indicating that activation of DNA-PKcs requires prior acetylation at K5.
Further, to experimentally validate our modeling illustrating BRD/K5ac-dependent induction of γ-Ser139, we first studied how the exogenously expressed DNA-PKcs-BRD affects the γ-Ser139 level induced by an acute IR by transfecting U2OS cells respectively with either plasmid of EV or the one expressing BRD4 or DNA-PKcs-BRD or the single or triple mutant of DNA-PKcs-BRD prior to 10 Gy IR. As shown by Figure 5A-B, the IR-induced level of γSer139 was reduced with the over-expression of either DNA-PKcs-BRD or BRD4. Further, in correlation with the changes in the interaction between DNA-PKcs and H2AX, the significantly reduced level of IR-induced γSer139 in the U2OS cells expressing either DNA-PKcs-BRD (even with the lowest expressed amount) or BRD4 was not observed in those over-expressing either mutant of DNA-PKcs-BRDN2176A or DNA-PKcs-BRDP2110A/N2174A/M2185A (Figure 5A), supporting our structural model that DNA-PKcs phosphorylates H2AX Ser139 in a BRD-dependent way during acute IR wherein the exogenously expressed DNA-PKcs-BRD could compete off the endogenous DNA-PKcs from the same H2AX K5ac site and therefore disrupt the docking of endogenous DNA-Pkcs on K5ac for Ser139 phosphorylation. These loss-of-function experiments demonstrated that the specificity of the BRD of DNA-PKcs in defining its K5ac-dependent, H2AX-associated kinase activity in DDR as the DNA-PKcs-BRD structure plays a regulatory role in phosphorylating Ser139.
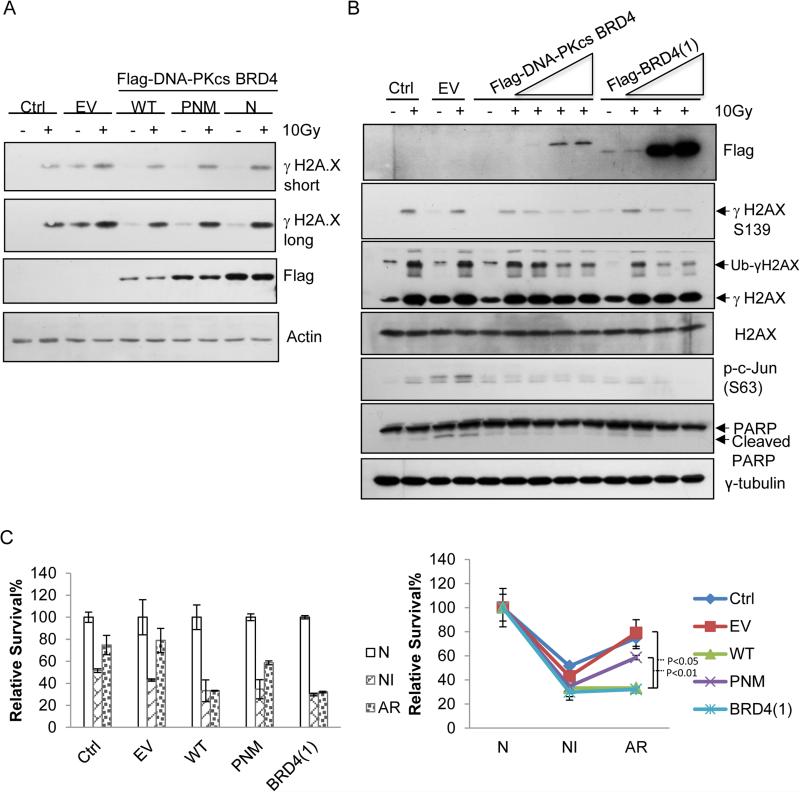
Figure 5. The non-canonical DNA-PKcs-BRD defines its kinase activity specifically for phosphorylating Ser139 and the associated cell fate decision.
(A) Immunoblot analysis of the effects of the exogenously expressed, empty vector (EV), wild-type DNA-PKcs-BRD (WT, aa2070-2200) or the mutants of single (N2174A or ‘N’) or triple (P2110A/N2174A/M2185A or ‘PNM’) site-substitution(s) within the BRD of DNA-PKcs on the 10Gy-induced level of γSer139 on H2AX. (B) Immunoblot analysis of the effects of the exogenously expressed, empty vector (EV), or wild-type DNA-PKcs-BRD (WT) or BRD4 on the propensity of the acute-IR-irradiated U2OS cells. Multiple apoptotic markers including phosphor-ser63 of c-Jun, cleaved PARP were assayed by the indicated antibodies. (C) Clonogenic survival analysis of the effects of the exogenously expressed, empty vector (EV), wild-type DNA-PKcs-BRD (WT) or the mutants of single (N) or triple (PNM) site-substitution(s) within the BRD of DNA-PKcs or BRD4 on the survival of differently irradiated cells. Following the transfection of each given vector the U2OS cells were treated respectively with either ‘non-irradiation (N), or 2 Gy acute-irradiation (NI), or 0.1Gy-priming for 24 hours then 2Gy-irradiation (AR).
We next investigated how the Kac-dependent activation of DNA-PKcs impacts on cellular sensitivity to radiation. Along with that the IR-induced level of γSer139 was reducing with the increasing expression of either DNA-PKcs-BRD or BRD4, we observed the decreasing amount of ubiquitinated γ-Ser139 H2AX that was observed in 10 Gy-irradiated cells transfected with EV (Figure 5B). Meanwhile, both phospho-Ser63 of c-Jun and cleaved PARP-1, the known apoptotic markers, were found diminished in the cells transfected by either DNA-PKcs-BRD or BRD4, indicating that by competing off the endogenous counterpart the expression of exogenous DNA-PKcs-BRD or BRD4 interferes the previously characterized DNA-PKcs function in the acute IR-induced, H2AX-mediated apoptosis(Sakasai et al., 2010). This result further indicated that specifically in IR-responsive cells the BRD of DNA-PKcs Defines the H2AX-mediated, Pro-apoptotic Activity of DNA-PKcs.
Dysregulated DNA-PKcs Activity in NHEJ Repair is BRD/K5ac-dependent that Contributes to Cellular Resistance to Radiation
Given the regulatory role of H2AX K5ac in DNA-PKcs activation as well as the previously observed radio-resistance that is associated with the early activation of DNA-PKcs(Deriano et al., 2005) we first determined whether DNA-PKcs-BRD impacts on the fate of chronically irradiated cells. Similarly, we respectively generated various radiation-phenotypic U2OS cells showing different sensitivities to IR, including ‘non-irradiated (N)’, 2 Gy acute-irradiated ‘IR-responsive (NI)’, and 2Gy-irradiated, 0.1 Gy-primed ‘IR-adaptive-resistance (AR)’ cells (Lee et al., 2012). Further, under defined IR-phenotypic conditions we comparatively analyzed the survival level of each phenotypic cell set with or without transfection of either WT or the mutant DNA-PKcs-BRD or BRD4 respectively by using clonogenic survival assay. As shown by Figure 5C, the increased percentages of the survived cells under AR with respect to that of the NI cells were reversed only by the exogenously expressed WT DNA-PKcs-BRD not by the mutant of DNA-PKcs-BRD. Note that similarly low percentages of the survived cells were observed for all cells following an acute 2Gy IR (NI), probably due to the acute, toxic effect of over-expressed DNA-PKcs-BRD or its mutant. These results indicated that, specifically in the IR-resistant cells, the BRD of DNA-PKcs may play the pro-survival role.
Because of the regulatory role of DNA-PKcs activity in NHEJ we then investigated the impact of DNA-PKcs-BRD on its NHEJ-associated activity. First, given that the integrity of the DNA PK complex involving other DNA end-joining components such as Ku70/80 is required for the activity of DNA-PKcs in NHEJ repair(Deriano et al., 2005), we asked whether the formation of the DNA-PKcs/Ku70 complex is mediated by the BRD-dependent H2AX code, K5ac. Accordingly, we evaluated the JQ1 effect of on the IR-induced association between H2AX and the DNA-PKcs/Ku70 complex by immunoprecipitating H2AX from the 10 Gy-irradiated cells respectively without or with a JQ1 pre-treatment. As shown by Figure 6A, the 10 Gy-induced association between H2AX and DNA-PK complex was completely disrupted in the JQ1 pre-treated cells similar to the effect of the exogenously expressed DNA-PKcs (Figure 3D), indicating that formation of the DSB-repairing DNA-PK complex was K5ac-dependent as JQ1 competed with K5ac for binding to DNA-PKcs-BRD due to its specificity in binding BRD4(Filippakopoulos et al., 2010), leading to destabilization of the DNA-PK complex involved in NHEJ.
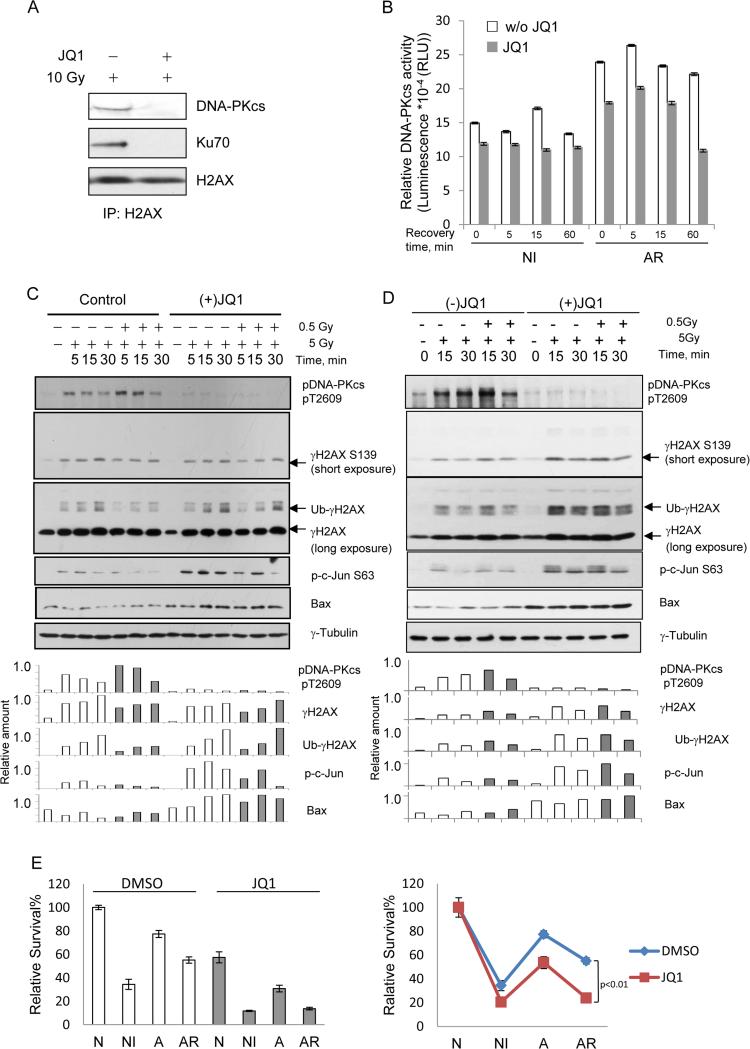
Figure 6. IR-induced, K5ac-dependent activation of DNA-PKcs contributes to the radio-resistance of tumor cells, which could be re-sensitized by the active form of (+)-JQ1.
(A) Formation of the DNA-PK complex in DSB end-repair is acetylation/BRD-dependent. Immunoblot analysis of the immunoprecipitates pulled down by anti-H2AX antibody immobilized on beads respectively from the nuclear fractions of the 10 Gy IR-treated HeLa cells pre-treated without or with JQ1. DNA-PKcs or Ku70 was visualized with the indicated antibodies with H2AX as the loading control. (B) Measurements of time-dependent DNA-PKcs activity in naïve or low dose IR-primed K562 leukemia cells without or with JQ1 pre-treatment using DNA-PK pull-down kinase assays. Each bar represents the mean ±SD of triplicates. (C) JQ1 re-sensitizes radio-resistant cancer cells through inhibiting the K5ac-initiated, over-active DNA-PKcs. Immunoblot analysis of time-dependent changes of DNA-PKcs-mediated NHEJ activity (indicated by pT2609) in naïve or low dose IR-primed K562 leukemia cells without or with JQ1 pre-treatment. Accordingly, the changes in γH2AX, ubiquitinated γH2AX, phosphor-ser63 on c-Jun, and expression of Bax were also measured. Immunoblot intensities were quantified by a densitometry. (D) The comparative effects of the active (+)-JQ1 vs. the inactive (−)-JQ1 on the cellular responsiveness to radiation and the cell fate decision. (E) Clonogenic survival analysis of the effect of (+)-JQ1 on the survival of U2OS cells under each of N, NI, AR condition.
We then investigated the impact of the acetyl-H2AX-dependent activation of DNA-PKcs in NHEJ repair and the associated cellular sensitivity to radiation. We first measured the time-dependent changes of DNA-PK activity in 5 Gy-irradiated leukemia K562 cells without or with a low-dose IR priming at 0.5 Gy, each phenotypically categorized as either NI or AR cells respectively(Lee et al., 2012). At the early time points of post-IR recovery, DNA-PKcs was more activated in AR cells than in NI cells (Figure 6B). Further, given that phosphorylation of Thr2609 on DNA-PKcs plays a critical role in the NHEJ pathway(Chen et al., 2007), the level of phospho (p)-Thr2609 should indicate the activation status of DNA-PKcs-mediated NHEJ repair. As measured with an anti-pThr2609 antibody, and similar to the change of DNA-PK activity (Figure 6B), the early activation of DNA-PKcs-mediated NHEJ repair was also observed in IR-resistant cells (Figure 6C, lanes 5, 6 versus 2, 3). Also, the IR-inducible apoptotic markers pSer63 c-Jun and Bax expression were suppressed in AR cells along with a reduced level of ubiquitinated γ-Ser139 H2AX. These observations agree with two other reports that showed: (1) similar early activation of DNA-PKcs in resistant B-cell chronic lymphocytic leukemia(Deriano et al., 2005), and (2) stimulated DDR through acetylation-dependent ubiquitination of H2AX(Ikura et al., 2007), together indicating that the over-activation of DNA-PKcs contributes to increased resistance to apoptosis and reduced sensitivity to radiation. Because compared with non-irradiated cells lysine 5 remained acetylated in the presence of prolonged IR (Figure S2B), we concluded that the early activation of DNA-PKcs in IR-resistant cells is caused by its BRD recognizing the residual K5Ac maintained by priming radiation.
More importantly, these results suggest that JQ1 could be a putative sensitizer of the tumor cells with radio-resistance. We therefore examined the effect of (+)-JQ1 treatment on the DNA-PKcs-mediated sensitivity to IR. While DNA-PK activity and the level of pThr2609 were significantly reduced in both IR-sensitive and -resistant cells pre-treated with (+)-JQ1 (Figure 6B and 6C), indicating the (+)-JQ1-mediated inhibition of the K5Ac-dependent activity of DNA-PKcs in NHEJ repair, both pSer63 c-Jun and Bax were more pronounced in the JQ1-treated cells under the IR-resistant condition along with increased level of ubiquitinated γ-Ser139 H2AX (30 min after the second radiation) (Figure 6C lanes 12, 13 versus 6,7 as well as lane 14 versus 7). Meanwhile, with measuring all markers of apoptosis and cell fate status mentioned above, we compared the effect of the pre-treatment with the active (+)-JQ1 vs. its inactive counterpart (−)-JQ1. As shown by Figure 6D, similar to non-treated cells under all conditions the pre-treatment using (−)-JQ1 caused little change in the responsiveness of the radio-resistant tumor cells to radiation, illustrating the specificity of the active form (+)-JQ1 in recognizing the BRD of DNA-PKcs. Also, we comparatively examined the effect of (+)-JQ1 on the survival of the cells respectively under N, NI, AR where the pre-treatment with (+)-JQ1 led to reduced survival under all different IR conditions with the more significant reduction of AR cells (Figure 6E).
Clearly, by inhibiting the over-heated, K5ac-dependent activity of DNA-PKcs, JQ1 could refractory cellular resistance to radiation and restore sensitivity of IR-resistant cells to DSB-induced apoptosis. Notably, although DNA-PKcs is over-activated in IR-resistant cells, little change at the level of γH2AX was observed. It is probably due to protein phosphatase 2A (PP2Ac) that we found was constitutively activated specifically in the cells under the chronic inflammatory state(Xie et al., 2013). The chronically active PP2Ac could dephosphorylate γH2AX and offset the increase of γH2AX caused by over-activated DNA-PKcs (unpublished data).
DiSCUSSION
Our discovery of the BRD-like domain module in DNA-PKcs was supported by other recent evidences from different groups including (1) BRD4 binding to H2A K5ac that is conserved in H2A and H2AX(Filippakopoulos et al., 2012), (2) with a DSB inducer (MNNG), ATM and DNA-PK play a sequential role in inducing γH2AX(Baritaud et al., 2012), and (3) the NHEJ pathway where DNA-PK is a major player could be the target pathway for re-sensitizing radio-resistant tumors(Srivastava et al., 2012).
Prior to our findings, H2AX PTMs was individually studied as little was found for the cooperative functions of multiple H2AX PTMs. The major technical barrier is that bottom-up MS proteomics (analysis of proteolytic digests) are capable only of identifying PTMs in MS-detectible peptides, so the connectivity among PTMs in disparate portions of a protein could be lost. Specifically for H2AX, although either γ-Ser139(Zhu et al., 2002) or H2AFZ K5ac(Choudhary et al., 2009) was individually identified by MS, possible connectivity among these PTMs was never observed because their remote locations separate them by >130 residues. In this regard, our top-down MS sequenced the full-length H2AX, simultaneously identifying different PTM sites within both N- and C-terminal tails. Without the enrichment step essential for low-abundance PTM analysis using bottom-up approaches, all of the phosphorylated/acetylated forms of H2AX were detected by FTICR MS at good signal-to-noise (S/N) with sufficient ion intensity for ECD MS/MS sequencing. By contrast, bottom-up analysis of the same set of samples gave no confident assignments of these modified peptides (data not shown). Meanwhile, unlike a bottom-up experiment in which most PTM-containing peptides, phosphopeptides in particular, are ionized much less efficiently than their unmodified counterparts, leading to poor S/N, different forms of an intact protein either modified or unmodified are equally ionized because either single or multiple PTM sites have little effect on the overall ionization efficiency of their parent proteins. As a result, relative intensities of the FTICR MS signals derived from either unmodified or modified H2AX were proportional to their relative abundances.
Mechanistically, we have revealed that, due to the unique BRD structure found only in DNA-PKcs, ATM and DNA-PK play the timing roles in H2AX phosphorylation during DDR: ATM is more involved in the γH2AX-mediated DSB recognition, whereas in a K5ac-dependent manner DNA-PKcs is more active in the γH2AX-mediated DNA repair or apoptosis of irreparable cells. Given that DSBs rapidly induce the Tip60-mediated acetylation of ATM for initiating its activity(Sun et al., 2007), the activated ATM therefore could immediately target H2AX Ser139 in a K5ac-independent way prior to the activation of DNA-PKcs. In parallel with the function of the K5ac depositor Tip60 in selective histone variant exchange at DNA lesions(Kusch et al., 2004), the timely K5ac-mediated docking of DNA-PKcs on H2AX promotes H2AX-mediated DSB repair (Figure 7).
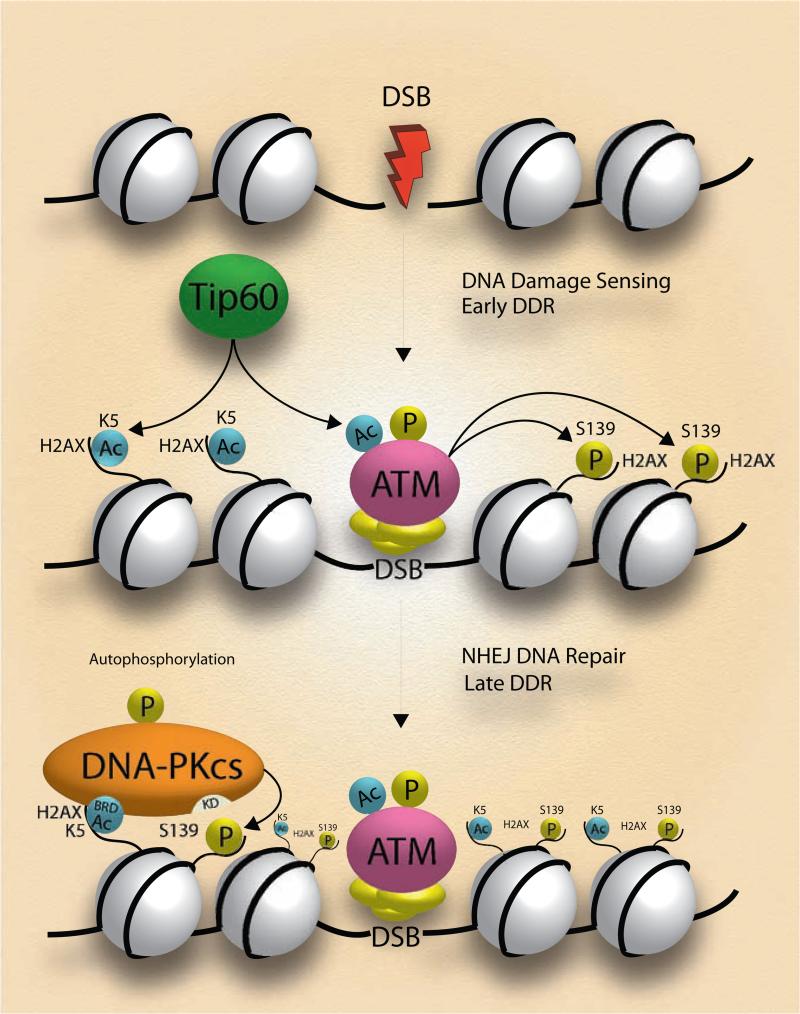
Figure 7. Mechanistic scheme of IR-induced, K5ac-dependent activation of DNA-PKcs for H2AX Ser139 phosphorylation.
Under an acute IR Tip60-mediated acetylation of K5 on H2AX facilitates the docking of DNA-PKcs that uses its BRD4-like domain to recognize K5ac. The K5ac-dependent activation of DNA-PKcs further phosphorylates H2AX Ser139. In the presence of prolonged low-dose IR, the residual K5ac is responsible for the early activation of DNA-PKcs that promotes the H2AX-mediated radioresistance.
Notably, in the IR phenotype-specific manner, this non-canonical BRD module in DNA-PKcs induces γH2AX and promotes apoptosis of acute-irradiated cells; in IR-responsive cells, introduction of an exogenous DNA-PKcs-BRD or BRD4 attenuates H2AX phosphorylation whereas expression of the DNA-PKcs constructs harboring mutations that abrogate key residues in DNA-PKcs-BRD had little effect on the γH2AX level (Figure 5A-B). This finding agrees with a reported role of BRD4 in inhibiting the radiation-induced γH2AX(Floyd et al., 2013). The level of γH2AX showed little change or a slight decrease in the radiation-responsive cells that were pre-treated with (+)-JQ1 whereas JQ1 treatment of the radiation-resistant cells led to increased levels of Ub-γH2AX (Figure 6C and 6D). This novel finding reveals that there is a differential effect of JQ1 depending upon either the radiation-responsive or radiation-resistant nature of the cancer cells: JQ1 inhibits only radiation-induced H2AX signaling in the radiation-responsive/sensitive cells and, conversely, JQ1 sensitizes the resistant cells through interrupting the DNA-PK BRD-mediated mechanism.
Although DNA-PKcs is a crucial enzyme in proper NHEJ repair, in a variety of carcinoma DNA-PKcs has been found not only over-expressed but also correlated with dysregulated NHEJ for radio-resistance. Recent report indicated that the therapeutic resistance of tumor cells could be overcome by the inhibition of NHEJ through interfering the DNA-binding domain of ligase IV(Srivastava et al., 2012). Here we showed that the DNA-PKcs-BRD4-interfered or JQ1-mediated inhibition of the dysregulated, NHEJ-specific activity of DNA-PKcs led to the activation of intrinsic apoptotic pathways and the re-sensitization of radio-resistant tumor cells similar to those induced by a ligase IV-targeting molecule SCR7, suggesting the NHEJ pathway is a reservoir of new drug targets for overcoming therapeutic resistance.
In summary we reveal the mechanism by which a DDR-phenotypic pattern of PTMs including γH2AX is sequentially established on H2AX, and how these H2AX PTMs impact in turn the activities of specific enzyme(s) involving in the H2AX-coordinated DDR and the corresponding cell fate decision. Although the precise mechanism underlying how over-activated, DNA-PKcs-mediated NHEJ repair inhibits apoptotic pathways remains to be further elucidated, we show that the BRD4-like domain of DNA-PKcs be a modulator of the acetylation-dependent NHEJ activity of DNA-PKcs. Because we revealed a novel intervention target of a non-catalytic BRD domain of DNA-PKcs that is different from its kinase catalytic domain most of conventional DNA-PKcs drugs are targeting at, co-administration of JQ1 with selected existing drug may represent a combination therapy strategy to increase the specificity and effectiveness of NHEJ-targeting treatments of cancer.
EXPERIMENTAL PROCEDURES
Cells, Reagents, and Antibodies
HEK 293T, HeLa, and U-2 OS cell lines were from ATCC. Human glioma cell lines M059K and M059J were a gift from Dr. Aziz Sancar's lab. Cells were maintained respectively in either DMEM or RPMI 1640 or McCoy's 5a medium (Invitrogen, CA). JQ1 (Structural Genomics Consortium) was re-suspended in DMSO to a final concentration at 5 mM. The Superfect transfection reagent was from Qiagen. Antibodies for DNA-PKcs, phospho-ser63 c-Jun, and Bax were from Cell Signaling. Antibodies of Tip60 and γTyr142H2AX were from Millipore. Antibodies for γH2AX (S139, ab22551), H2A K5ac (ab45152), H2AX (ab11175), and DNA-PKcs pT2609 (ab4194) were from Abcam. FLAG M2 was from Sigma. DNA-PKcs pT2609 for IB was a gift from Dr. Dale Ramsden. DNA-PKcs peptide substrate and Kinase-Glo Plus Luminescent Kinase Assay kit were from Promega.
Cell irradiation
Cells were grown to 60-70% confluence before IR. A γ-ray source (AECL Gammacell 40 Irradiator) irradiates cells at defined doses or for different lengths of time at a dose rate of 1 Gy/min. After IR the cells were allowed to recover for defined time and washed with PBS for further analysis.
Purification of full-length histone H2AX for top-down MS
Histone Purification Mini Kit (Active Motif) was used for histone extraction in the presence of protease (Roche Diagnostics) and phosphatase inhibitor cocktails (Sigma-Aldrich). Followed by acetone precipitation the resulting pellet containing histones was air-dried thoroughly, and then suspended in H2O. 50 μg histones were purified by RP HPLC with a ZORBAX C8 column (4.6 mm × 150 mm, 5 μm particle sizes). With solvent A (water premixed with 0.1% TFA) and solvent B (acetonitrile premixed with 0.1% TFA) core histones were eluted by using a multistep gradient with 0-5% B for 10 min, 5-24% B for 10 min, 25-64% B for 80 min, 65-99% B for 20 min, and isocratic gradient for 20 min at 100% with a flow rate of 200 μl/min. MS spectra was acquired by a hybrid Qe-FTICR MS, equipped with a 12.0 Tesla magnet (Apex 12.0 T AS, Bruker Daltonics) and an Apollo II microelectrospray (μESI) source. MS raw data acquired from ApexControl (Version 2.0, Build 36) were processed using DataAnalysis 3.4 Build 191 software.
Measurements of the bindings between DNA-PKcs-BRD and JQ1 or H2AX peptides
The JQ1-Sepharose or empty beads blocked by 3mg/ml BSA in PBS were incubated with the freshly refolded DNA-PKcs-BRD in a buffer containing 20 mM Tris (pH8.0), 150 mM NaCl, 1mM EDTA, 1% CA630, 1mg/ml BSA for 2 hrs and washed with the same buffer 3 times before eluted with SDS sample buffer. For the comparative studies of peptide binding and JQ1 competition, similar to the method (Fuchs et al., 2010) with minor modifications, 2 μg of wild-type H2AX peptide with the sequence SGRGKTGGKARAKAKSR-Peg-Biotin or the H2AX K5ac-containing peptide counterpart was incubated with streptavidin beads for 1 h at 4 °C and then washed twice with a peptide binding buffer (PBB; 50 mM Tris pH 8.0, 300 mM NaCl and 0.1% NP-40). The freshly refolded protein (80 pmol) was then incubated with each of the peptide on beads at 4 °C overnight and then washed three times with PBB. The DNA-PKcs-peptide complexes were eluted with 1X SDS gel sample buffer, separated in a NuPAGE 3-8% Tris-acetate gel (Invitrogen), and analyzed by IB against anti-His or anti-GST antibody. For JQ1 competition experiments, 10μM JQ1 was added.
JQ1 treatment of K562 cells
Prior to irradiation, K562 cells were pre-treated by 500nM (+)JQ1 or (−)JQ1 from a 5mM stock in DMSO) for 24hrs.
Supplementary Material
Significance.
Cells constantly exposed to ionizing radiation (IR) may acquire radioresistance. Through unknown mechanism this adaptive effect, if dysregulated, contributes to an increased risk of carcinogenesis. Here we discover a bromodomain (BRD)-like module in the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) or DNA-PKcs-BRD that specifically recognize H2AX acetyl-lysine 5 (K5ac) for sequential induction of phosphorylation of serine 139 on H2AX (γH2AX), an immediate indicator of DNA damage.
Although diverse BRD families are found presenting in different nuclear proteins including acetylases, ATP-dependent chromatin-remodeling complexes, helicases, methyltransferases, transcriptional activators/mediators, until now there has been no report of the presence of BRD(s) in a protein kinase directly functioning in chromatin-associated DSB recognition/repair. Our combined approaches of top-down mass spectrometry, sequence-structure modeling/docking, site-directed mutagenesis, and biochemical experiments illustrated that, through docking on H2AX K5ac, this non-canonical BRD determines not only the H2AX-targeting activity of DNA-PKcs but also the over-activation of DNA-PKcs in radio-resistant tumor cells whereas a Kac antagonist JQ1 could bind to DNA-PKcs-BRD, leading to re-sensitization of tumor cells to radiation.
To our knowledge this is the first report that a BRD occurs in a DNA repair protein that acts as both reader and writer of distinct histone codes for H2AX-mediated cell fate decision. Further, our discovery explained the nucleosome/chromatin-dependence of activation of DNA repair enzyme(s) and exactly how the active DNA-PK complex accesses damaged DNA in chromatin. We therefore reveal a new, Kac-dependent mechanism underlying the activation of DNA-PKcs wherein the DSB-inducible K5ac on H2AX is identified as a new epigenetic code to define the H2AX-mediated DDR and the associated cell fate decision.
Highlights.
Top-down mass spec reveals IR-induced concerted increases of K5ac and phospho-H2AX
A bromodomain module was found in DNA-PKcs to recognize the acetyl-lysine 5 on H2AX
The DNA-PKcs BRD defines the activity of DNA-PKcs to phosphorylate Ser139 on H2AX
DNA-PKcs activity in NHEJ repair is K5ac-dependent and contributes to radioresistance
ACKNOWLEDGMENTS
This work is primarily supported in part by multiple grants to X.C including Chinese 973 fund 2013CB910802, US NIH 1U24CA160035 from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC), NIAID 1U19AI109965, and was also supported by NIH R01GM103893 (to J.J.). We thank William K. Kaufmann for providing ATM/mutant cell lines, and Dale A. Ramsden for suggestions. We are grateful to Dr. James E. Bradner for providing JQ1 and to Dr. Howard Fried for proof-reading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
L. W., L. X., and Y.L. performed sample preparation including H2AX purification, cell culture, site-directed mutagenesis, and biochemical/immunoblot assays. L.Z. collected and analyzed the top-down MS data using FTICR. S.R. and N.V. D. performed modeling analysis. C.Z performed CD. J.J. and F.L. synthesized and provided immobilized JQ1. B.D.S and K.K. synthesized the H2AX peptides. X. C. conceived the project, designed the experiments, analyzed the data, and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Supplemental Information includes Methods, 7 figures, 2 tables.
REFERENCES
- Baritaud M, Cabon L, Delavallee L, Galan-Malo P, Gilles ME, Brunelle-Navas MN, Susin SA. AIF-mediated caspase-independent necroptosis requires ATM and DNAPK-induced histone H2AX Ser139 phosphorylation. Cell Death Dis. 2012;3:e390. doi: 10.1038/cddis.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. γH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce I, Akhlaq M, Bloomfield GC, Budd E, Cox B, Cuenoud B, Finan P, Gedeck P, Hatto J, Hayler JF, et al. Development of isoform selective PI3-kinase inhibitors as pharmacological tools for elucidating the PI3K pathway. Bioorg Med Chem Lett. 2012;22:5445–5450. doi: 10.1016/j.bmcl.2012.07.042. [DOI] [PubMed] [Google Scholar]
- Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. Ataxia telangiectasia mutated (ATM) is essential for DNAPKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science (New York, NY. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Guipaud O, Merle-Beral H, Binet JL, Ricoul M, Potocki-Veronese G, Favaudon V, Maciorowski Z, Muller C, Salles B, et al. Human chronic lymphocytic leukemia B cells can escape DNA damage-induced apoptosis through the nonhomologous end-joining DNA repair pathway. Blood. 2005;105:4776–4783. doi: 10.1182/blood-2004-07-2888. [DOI] [PubMed] [Google Scholar]
- Ding F, Tsao D, Nie H, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Yin S, Dokholyan NV. Rapid flexible docking using a stochastic rotamer library of ligands. J Chem Inf Model. 2010;50:1623–1632. doi: 10.1021/ci100218t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Discrete molecular dynamics studies of the folding of a protein-like model. Fold Des. 1998;3:577–587. doi: 10.1016/S1359-0278(98)00072-8. [DOI] [PubMed] [Google Scholar]
- Du YC, Gu S, Zhou J, Wang T, Cai H, Macinnes MA, Bradbury EM, Chen X. The dynamic alterations of H2AX complex during DNA repair detected by a proteomic approach reveal the critical roles of Ca(2+)/calmodulin in the ionizing radiation-induced cell cycle arrest. Mol Cell Proteomics. 2006;5:1033–1044. doi: 10.1074/mcp.M500327-MCP200. [DOI] [PubMed] [Google Scholar]
- Fang TC, Schaefer U, Mecklenbrauker I, Stienen A, Dewell S, Chen MS, Rioja I, Parravicini V, Prinjha RK, Chandwani R, et al. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. The Journal of experimental medicine. 2012;209:661–669. doi: 10.1084/jem.20112343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, Bryson BD, Rameseder J, Lee MJ, Blake EJ, et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498:246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of Combinatorial Histone Modifications on Antibody and Effector Protein Recognition. Current Biology. 2010;21:53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan TP, Simpson DA, Frank AR, Heinloth AN, Paules RS, Cordeiro-Stone M, Kaufmann WK. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Molecular and cellular biology. 2002;22:8552–8561. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-Mediated Exchange of Histone Variant H2AX Regulated by Phosphorylation of H2AX and ADP-Ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Suppl):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Molecular and cellular biology. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science (New York, NY. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Lee YY, Yu YB, Gunawardena HP, Xie L, Chen X. BCLAF1 is a radiation-induced H2AX-interacting partner involved in gammaH2AX-mediated regulation of apoptosis and DNA repair. Cell death & disease. 2012;3:e359. doi: 10.1038/cddis.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day RS, Barron GM, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science (New York, NY. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- Lobrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- Mullenders L, Atkinson M, Paretzke H, Sabatier L, Bouffler S. Assessing cancer risks of low-dose radiation. Nature reviews. 2009;9:596–604. doi: 10.1038/nrc2677. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. The EMBO journal. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Chan DW, Park JH, Oettinger MA, Kwon J. DNA-PK is activated by nucleosomes and phosphorylates H2AX within the nucleosomes in an acetylation-dependent manner. Nucleic Acids Res. 2003;31:6819–6827. doi: 10.1093/nar/gkg921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakasai R, Teraoka H, Takagi M, Tibbetts RS. Transcription-dependent activation of ataxia telangiectasia mutated prevents DNA-dependent protein kinase-mediated cell death in response to topoisomerase I poison. The Journal of biological chemistry. 2010;285:15201–15208. doi: 10.1074/jbc.M110.101808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, Kumar S, Pandey M, Singh RK, Ray P, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Molecular and cellular biology. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends in cell biology. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2009;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Odate S, Chandramouly G, Scully R. H2AX post-translational modifications in the ionizing radiation response and homologous recombination. Cell Cycle. 2010;9:3602–3610. doi: 10.4161/cc.9.17.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Liu C, Wang L, Gunawardena HP, Yu Y, Du R, Taxman DJ, Dai P, Yan Z, Yu J, et al. Protein phosphatase 2A catalytic subunit alpha plays a MyD88-dependent, central role in the gene-specific regulation of endotoxin tolerance. Cell Rep. 2013;3:678–688. doi: 10.1016/j.celrep.2013.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science (New York, NY. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Hunter TC, Pan S, Yau PM, Bradbury EM, Chen X. Residue-specific mass signatures for the efficient detection of protein modifications by mass spectrometry. Analytical chemistry. 2002;74:1687–1694. doi: 10.1021/ac010853p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.