Abstract
The bile acid (BA)-sensing nuclear receptor, farnesoid X receptor (FXR), regulates postprandial metabolic responses, including inhibition of BA synthesis, by inducing the intestinal hormone, fibroblast growth factor (FGF)15 (FGF19 in human). In this study, we tested a novel hypothesis that FXR not only induces intestinal FGF15 but also primes the liver for effectively responding to the signal by transcriptional induction of the obligate coreceptor for FGF15, β-Klotho (βKL). Activation of FXR by a synthetic agonist, GW4064, in mice increased occupancy of FXR and its DNA-binding partner, retinoid X receptor-α, at FGF15-signaling component genes, particularly βKL, and induced expression of these genes. Interestingly, mRNA levels of Fgfr4, the FGF15 receptor, were not increased by GW4064, but protein levels increased as a result of βKL-dependent increased protein stability. Both FGF receptor 4 and βKL protein levels were substantially decreased in FXR-knockout (KO) mice, and FGF19 signaling, monitored by phosphorylated ERK, was blunted in FXR-KO mice, FXR-KO mouse hepatocytes, and FXR-down-regulated human hepatocytes. Overexpression of βKL in FXR-lacking hepatocytes partially restored FGF19 signaling and inhibition by FGF19 of Cyp7a1, which encodes the rate-limiting BA biosynthetic enzyme. In mice, transient inductions of intestinal Fgf15 and hepatic βKL were temporally correlated after GW4064 treatment, and pretreatment of hepatocytes with GW4064 before FGF19 treatment enhanced FGF19 signaling, which was abolished by transcriptional inhibition or βKL down-regulation. This study identifies FXR as a gut-liver metabolic coordinator for FGF15/19 action that orchestrates transient induction of hepatic βKL and intestinal Fgf15/19 in a temporally correlated manner.
Metabolic responses to a meal are regulated by an intricate interplay among pancreatic, intestinal, and hepatic hormones and factors. Although the roles of pancreatic and intestinal hormones, such as insulin, cholecystokinin, and glucagon-like peptide-1, in postprandial responses have been intensively studied, only recently has it been appreciated that bile acids (BAs) are important signaling molecules that control fed-state metabolism by activating nuclear and membrane BA receptors (1–6).
BAs are cholesterol metabolites that are synthesized in the liver, stored in the gall bladder, and are released into the small intestine in response to a meal, where they aid absorption and digestion of lipid nutrients. Nearly 95% of BAs are returned to the liver by portal circulation, resulting in transient increases in BA levels in the enterohepatic system (7). The nuclear receptor, farnesoid X receptor (FXR) (NR1H4), is the primary biosensor for BAs (2, 8). Postprandial activation of hepatic and intestinal FXR by transiently increased BA levels transcriptionally regulates its target genes in the liver and small intestine to control fed-state metabolism of cholesterol/BA, fatty acids, and glucose and to maintain metabolic homeostasis (1, 2, 6, 8, 9).
BA-activated intestinal FXR induces the synthesis of a late fed-state hormone, fibroblast growth factor (FGF)15 (FGF19 in human) (1, 10, 11). Secreted FGF15 then binds to its hepatic membrane receptor complex, FGF receptor 4 (FGFR4) and β-Klotho (βKL) (12, 13), and activates downstream cellular signaling pathways to mediate postprandial responses, including repression of BA synthesis through inhibition of Cyp7a1 gene expression (1, 10, 11, 14). It is largely unknown how the FGF15/19 signal is relayed to regulate Cyp7a1 and other FGF15/19 target genes, although a recent study has identified a nonreceptor tyrosine phosphatase and key FGF15/19-signaling component, Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2), as a novel regulator of BA homeostasis (15). Consistent with the in vivo role of the FXR/FGF15 regulatory pathway in inhibition of BA synthesis, Cyp7a1 expression was increased and BA levels elevated in mice lacking FGF15, FGFR4, βKL, Shp2, or FXR (10, 15–18).
In a recent genome-wide analysis in mice (19), FXR-binding peaks were detected at many hepatic genes involved in the FGF15 signaling pathway, including βKL. In this study, we, therefore, tested whether FXR might not only induce intestinal FGF15 but also prime the liver for effectively transmitting the signal. Here, we show that activation of the FXR pathway leads to temporally coordinated induction of intestinal Fgf15 and hepatic βKL, which results in enhanced responsiveness to FGF15.
Materials and Methods
Materials
Antibody against mouse βKL (2619-KB) was purchased from R&D Systems, for phospho (p)-Erk1/2 (9101) and total-Erk1/2 (4695) from Cell Signaling, and for human βKL (sc-74343), FXR (sc-1204), FGF15 (sc-27177), FGFR4 (sc-136988), and glyceraldehyde 3-phosphate dehydrogenase (sc-25778) from Santa Cruz Biotechnology, Inc. Antibodies for βKL (LS-B3568) and (ABS1113) used in immunohistochemistry (IHC) studies were purchased from LS Biology and EMD Millipore, respectively, small interfering RNA (siRNA) for mouse βKL (s96291 and s96292) from Applied Biosystems and human FXR siRNA (M-003414–01) and mouse FXR siRNA (M-045705–01) from Dharmacon. A mixture of βKL siRNA (s96291 and s96292) had been shown to be effective in previous studies (20, 21). For IHC studies, antibodies to βKL (goat and rabbit), FGFR4 (mouse), or FGF15 (goat) were detected with rabbit (ab64261) or mouse (ab64259)-specific horseradish peroxidase/3,3'-diaminobenzidine (HRP/DAP) IHC Detection kits (Abcam), or with the ImmunoCruz goat ABC Staining System (sc-2023; Santa Cruz Biotechnology, Inc).
Animal experiments
Twelve-week old male C57BL/6 wild-type (WT) mice or FXR-knockout (KO) mice (22) were fasted 12 hours and, then, injected ip with GW4064 (30 mg/kg in corn oil) at 10 am for different times (0.5, 1, 2, 3, 4, 6, and 8 h), and then, liver and intestine tissues were collected for further analyses. For FXR reconstitution experiments in FXR-KO mice, WT mice or FXR-KO mice were injected via the tail vein with adeno (Ad)-FXR (0.5–1 × 109 active particles/mouse in 200-μL PBS), and 1 week later, the mice were fasted overnight before killing. For in vivo FGF19 experiments, mice were fasted for 12 hours as previously reported (10, 14, 20) and then treated with purified recombinant human FGF19 (10 μg/kg) by tail vein injection for 15 minutes for FGF19 signaling studies or 2 hours for Cyp7a1 gene expression studies. All animal protocols were approved by the Institutional Animal Care and Use and Institutional Biosafety Committees at University of Illinois at Urbana-Champaign.
Primary hepatocyte experiments
Primary mouse hepatocytes (PMHs) were isolated from WT or FXR-KO mice as described previously (20). Primary human hepatocytes were isolated from 3 human donors (13–002, 14–017, and 15–006) and were obtained from the Liver Tissue Procurement and Distribution System of National Institute of Health. Hepatocytes were treated with a synthetic FXR agonist GW4064 (500nM) (23) for various times and with FGF19 (50 or 100 ng/mL) for 30 minutes as indicated in the figure legends. Mouse and human hepatocytes were in incubated in full media with 10% serum for 1 day and transfected with siRNA (mixture of s96291 and S96292) or pCMV-GFP-βKL (Addgene) DNA, and 36 hours (for plasmids) or 48 hours (for siRNA) later, cells were incubated with serum-free media overnight before treatment with GW4064 or FGF19. For protein stability studies, PMHs transfected with βKL siRNA were treated with GW4064 for 2 hours and then with 10μM cycloheximide (CHX) for the indicated times. For transcriptional inhibition studies, PMHs were treated with 5-μg/mL actinomycin D for 30 minutes, followed by GW4064 for 2 hours and FGF19 for the last 30 minutes.
IHC and immunofluorescence (IF) studies
For IHC studies, paraffin-embedded liver sections were prepared for analysis. Antibodies to βKL, FGFR4, or FGF15 were detected with HRP/DAB IHC Detection kits (Abcam), or the ImmunoCruz Staining System (Santa Cruz Biotechnology, Inc), and sections were imaged with a NanoZoomer Scanner (Hamamatsu). For IF studies, paraffin-embedded liver sections from WT and FXR-KO were incubated with βKL or FGFR4 antibody overnight and then Alexa Fluor 647 donkey antigoat or donkey antimouse IgG, for 1 hour. Nuclei were labeled with Hoechst 33258, and images were taken and analyzed by confocal microscopy (LSM700; Zeiss).
Plasmid construction and luciferase assay
A 700-bp fragment within intron 2 of the βKL gene, which contained the major FXR-binding site, and deletion fragments were cloned into pGL3-SV40-Luc. The inverted repeat 2 (IR2) site was mutated and the mutation was confirmed by sequencing. Luciferase activities were normalized to β-galactosidase activities.
Gel mobility shift assay
[γ-32P]-end labeled probe (31 mer) was incubated with partially purified FXR and retinoid X receptor (RXR), 100 ng of poly dIdC, and 20 μg of BSA. Antibodies or unlabeled competitor (1, 3, and 15 ng) were added 5 minutes before adding the probe.
Quantitative (q-) RT-PCR
Total RNA was isolated with TRIzol reagent, and q-RT-PCR was performed. The amount of mRNA was normalized to that of 36B4. Primer sequences used in q-RT-PCR and chromatin immunoprecipitation (ChIP) assays are presented in the Supplemental Figure 1.
Liver re-ChIP
Mice were fasted for 10 hours, treated with vehicle or GW4064 for 1 hour, and livers were collected for re-ChIP assays. Chromatin was first immunoprecipitated with FXR antibody, and the immunoprecipitate was washed, eluted, diluted 20-fold with 20mM Tris-HCl (pH 8.0), 150mM NaCl, 2mM EDTA, and 1% Triton X-100, and then reimmunoprecipitated. Precipitated genomic DNA was quantified by qPCR.
Results
Pharmacological activation of FXR increases occupancy of FXR and RXRα at FGF15-signaling component genes, particularly βKL, and induces transcriptional activation
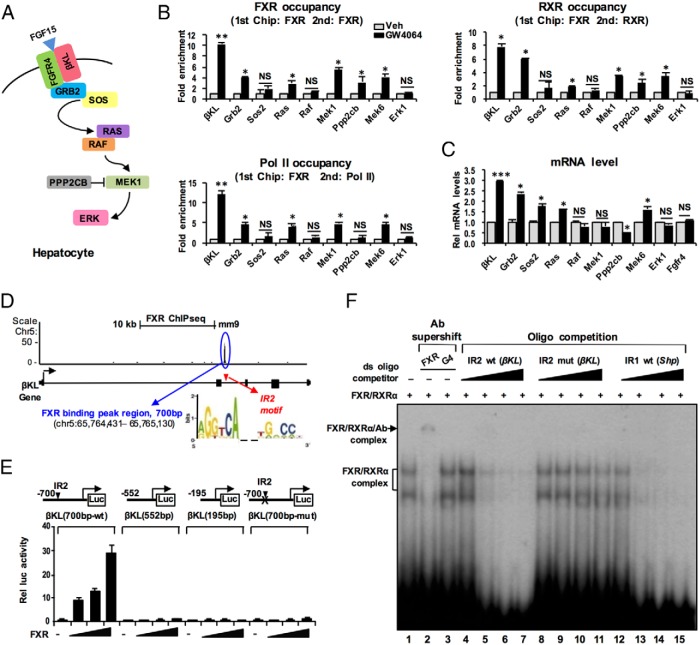
In liver ChIP-seq analysis in mice treated with GW4064 (19), FXR-binding peaks were detected at (or near) many genes in the FGF15 signaling pathway (Supplemental Figure 2), including βKL, Grb2, and Ras, but not at Fgfr4. To confirm these results, we examined the effects of activation of FXR on occupancy of FXR at FGF15 signaling pathway genes (Figure 1A) by ChIP analysis in mouse liver. A synthetic FXR agonist, GW4064, was used to specifically activate FXR because the natural agonists, BAs, activate multiple other signaling pathways, including the BA membrane receptors, TGR5 (2, 6, 24) and sphingosine 1-phosphate receptor-2 (S1P2) (25). In re-ChIP assays, occupancy of FXR and its DNA-binding partner, RXRα, was increased by GW4064 treatment in FXR-bound chromatin at the βKL, Grb2, Ras, Mek, and Ppp2cb genes, most prominently at βKL, whereas the occupancy at Sos2, Raf, and Mapk3 was not significantly increased (Figure 1B). Occupancy of RNA polymerase II, an indicator of gene activation, was increased over 10-fold at βKL and was also increased at most of the other genes with increased FXR occupancy (Figure 1B).
Figure 1.
Pharmacological activation of FXR increases its occupancy at FGF15 signaling pathway genes and increases their expression. A, FGF15/19-signaling components in hepatocytes. B and C, C57BL/6 mice were treated with GW4064 or vehicle for 1 hour, and liver re-ChIP assays (B) were performed to examine occupancy of FXR, RXRα, and RNA polymerase II at FXR-bound hepatic chromatin at the indicated genes, and (C) levels of the mRNA of the indicated genes were determined by q-RT-PCR; SEM, n = 3 mice; *, P < .05; **, P < .01; ***, P < .005; NS, not significant. D–F, βKL is a direct transcriptional target of FXR. D, A 700-bp fragment of βKL containing the WT or mutated IR2 motif or deletion mutations was cloned into pGL3-SV40-Luc as described in Materials and Methods. E, Luciferase reporter assays. Hepa1c1c7 cells were transfected with reporters and expression plasmids (0, 50, 100, and 200 ng) as indicated and treated with GW4064 (100nM) for 4 hours. Luciferase activity normalized to β-galactosidase activity was determined; SEM, n = 9. F, Gel mobility shift assays were performed using partially purified flag-FXR and flag-RXR, with a 32P-labeled oligonucleotide containing the βKL IR2 motif as probe. Antibody to FXR or the Gal4 DBD (G4), or competitor oligonucleotides containing the WT or mutated IR2 motif of βKL gene or the IR1 motif of the Shp promoter region were added.
Consistent with these results, βKL mRNA levels were increased 3-fold, levels of mRNA for other genes were increased by smaller amounts, and mRNA levels of the FGF15 receptor, Fgfr4, were not changed (Figure 1C). Similar results on mRNA levels of βKL and Fgfr4 were observed in hepatocytes isolated from WT or FXR-KO mice treated with GW4064 (Supplemental Figure 3).
βKL is the obligate coreceptor for FGF15/19 (11–13) and is most highly induced by FXR (Figure 1C), and importantly, liver-specific deletion of βKL in mice showed increased BA levels despite superphysiological serum FGF15 concentrations (11), indicating an essential role of βKL in mediating FGF15/19 signaling in liver hepatocytes in vivo. For these reasons, we focused further studies on βKL.
βKL is a direct transcriptional target of FXR
FXR occupancy was detected within intron 2 of βKL by liver ChIP-seq analysis in mice treated with GW4064 (Figure 1D) (19), and potential FXR-binding motifs were detected in 2 clustered regions within 700 bp of the βKL intron 2 sequence (Supplemental Figure 4). To identify the functional motif(s), the 700-bp sequence containing the FXR sites and deletion mutants were inserted into luciferase reporter plasmids (Figure 1E). Exogenous expression of FXR increased reporter activity with the full-length construct but not in deletion constructs lacking the IR2 motif or in the full-length construct with a mutated IR2 motif (Figure 1E). In gel mobility shift assays, addition of increasing amounts of partially purified flag-FXR and flag-RXRα resulted in the formation of FXR/RXRα DNA complexes with the βKL probe which were supershifted by FXR, but not Gal4 DNA binding domain, antibody (Figure 1F, lane 1–3). Unlabeled oligonucleotides containing the βKL IR2 or the inverted repeat-1 motif in the Shp promoter effectively competed for binding of FXR/RXRα (Figure 1F, lanes 4–7 and 12–15), and binding was not competed by oligonucleotides with the βKL IR2 motif mutated (Figure 1F, lanes 8–11). These results indicate that FXR/RXRα directly binds to the IR2 motif in the βKL gene, and the binding is required for FXR transactivation.
Activation of FXR increases protein levels of both FGFR4 and βKL
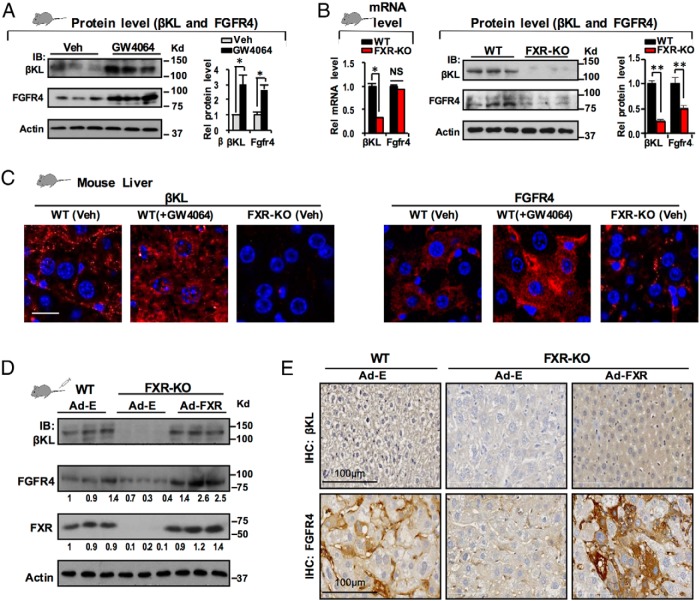
GW4064 treatment increased hepatic mRNA levels of βKL in mice (Figure 1C) and consistent with the increase, proteins levels increased 2- to 3-fold (Figure 2A). Although Fgfr4 mRNA levels did not increase (Figure 1C), protein levels increased a similar 2- to 3-fold (Figure 2A), suggesting a posttranscriptional mechanism for the increase.
Figure 2.
Activation of FXR increases protein levels of both FGFR4 and βKL. A, C57BL/6 WT mice (3 mice/group) were treated with vehicle or GW4064 for 4 hours and hepatic protein levels of βKL and FGFR4 were determined by IB. Levels were quantified with ImageJ at the right (SEM, n = 3; *, P < .05). B, WT and FXR-KO mice (3 mice/group) were used, and levels of βKL and FGFR4 mRNA were determined by q-RT-PCR (left) and protein levels (center, quantitation at right) by IB. C, Protein levels of βKL and FGFR4 in WT and FXR-KO mice treated with vehicle or GW4064 were detected by IF. D and E, WT or FXR-KO mice (3 mice/group) were tail vein injected with control Ad-E or Ad-FXR, and 1 week later, the mice were treated with GW4064 (30 mg/kg, ip) for 4 hours before killing. D, Protein levels of hepatic βKL, FGFR4, FXR, and actin as a loading control were determined by IB. The relative intensities of the bands for βKL and FGFR4 are shown. E, Protein levels of hepatic βKL and FGFR4 in WT and FXR-KO mice were determined by IHC.
Because GW4064 treatment increased βKL expression, to definitively show that the increase was mediated by FXR, we next examined expression in FXR-KO mice (22). The mRNA levels of βKL were diminished in FXR-KO mice, whereas those of Fgfr4 were not (Figure 2B, left), but protein levels of both βKL and FGFR4 were markedly decreased in FXR-KO mice as detected by immunoblotting (IB) (Figure 2B, center, quantitation in the right). Increased expression of βKL and FGFR4 in hepatocytes after GW4064 treatment and decreased basal protein levels in FXR-KO mice were also observed by IF (Figure 2C). Importantly, adenoviral-mediated expression of FXR in FXR-KO mice to levels similar to those in WT mice markedly increased protein levels of both βKL and FGFR4 as detected by IB (Figure 2D) and IHC (Figure 2E), although mRNA levels of FGFR4 were not changed (Supplemental Figure 5). These results demonstrate that pharmacological activation of FXR increased protein levels of both components of the FGF15/19 receptor complex, βKL and FGFR4.
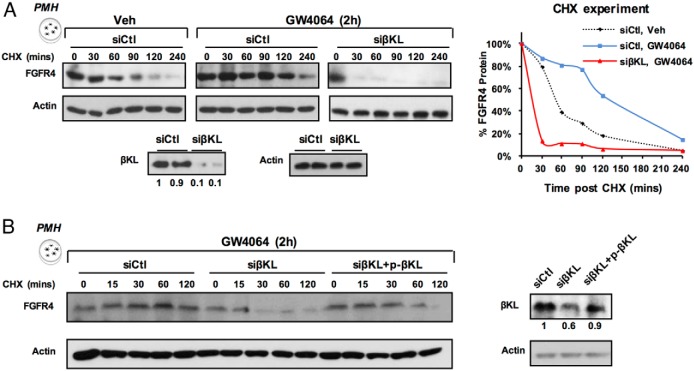
Activation of FXR by GW4064 treatment increased FGFR4 protein stability in a βKL-dependent manner
In βKL-KO mice, FGFR4 protein levels were markedly reduced (17, 18), which suggests protein levels of FGFR4 are dependent on βKL. To determine whether βKL stabilizes FGFR4, we examined FGFR4 stability by CHX studies in PMHs. FGFR4 was stabilized by GW4064 treatment (Figure 3A). The increased stability was reversed by down-regulation of βKL (Figure 3A) and, then, partially restored by overexpression of βKL (Figure 3B). These results indicate that FXR increases the amount of FGFR4 protein indirectly as a result of βKL-dependent increased protein stability.
Figure 3.
GW4064 increased FGFR4 protein stability in a βKL-dependent manner. A, PMHs were transfected with either control or βKL siRNA and/or (B) expression plasmid for βKL, and then, 48 hours later, cells were treated with vehicle or GW4064 for 2 hours, and further treated with CHX for indicated times. FGFR4 and actin protein levels were measured by IB. In A, the FGFR4 band intensities relative to the 0-minute time point were plotted at the right.
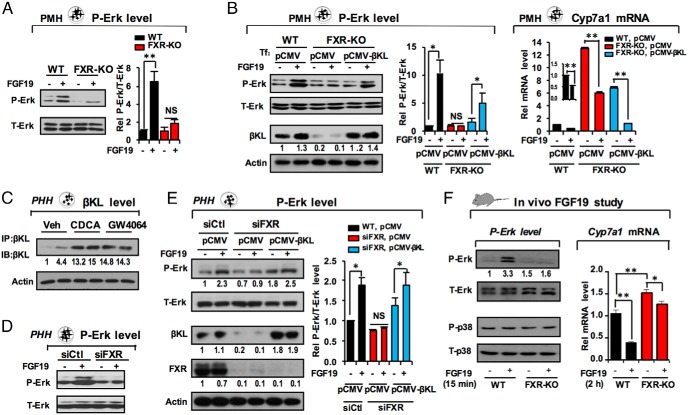
FGF19 signaling is impaired in FXR-KO mouse hepatocytes, in part due to diminished βKL expression
To determine whether decreased expression of the FGF15/19 receptor complex in FXR-KO mice inhibits FGF15 signaling, we examined levels of activated p-ERK as a measure of FGF15/19 signaling (14, 20, 26). Treatment with FGF19 resulted in a robust increase in p-ERK levels in hepatocytes from WT mice but not in those from FXR-KO mice (Figure 4A), suggesting that FXR has an important role in mediating FGF19 signaling.
Figure 4.
FGF19 signaling is impaired in FXR-lacking hepatocytes, in part due to diminished βKL expression. A and B, FGF19 signaling studies in PMHs. A, Hepatocytes were isolated from WT or FXR-KO mice and treated with 100 ng/mL of FGF19 for 30 minutes, and levels of total (T-Erk) and phosphorylated (P-Erk) ERK were determined by IB. Levels were quantified with ImageJ at the right (SEM, n = 6; **, P < .01; NS, not significant). B, Hepatocytes were transfected with plasmids, pCMV (control) or pCMV-βKL, for 36 hours and treated with FGF19 for 30 minutes, and T-Erk, P-Erk, and βKL levels were detected at the left (quantitation at the center; SEM, n = 5; *, P < .05; NS, not significant). Cyp7a1 mRNA levels were measured by q-RT-PCR (right; SEM, n = 3; *, P < .05; **, P < .01). C–E, FGF19 signaling studies in primary human hepatocytes. C, Human hepatocytes were treated with FXR agonists, chenodeoxycholic acid or GW4064, and βKL protein levels were detected (shown in duplicate). Consistent results were observed from 3 independent experiments (duplicate in each). D, Hepatocytes were transfected with control RNA or FXR siRNA, and 2–3 days later, hepatocytes were treated with FGF19 for 30 minutes and levels of total (T-Erk) and phosphorylated (P-Erk) ERK were determined by IB (left, quantitation at the right). E, Hepatocytes were transfected with pCMV or pCMV-βKL for 36 hours and treated with FGF19, and levels of T-Erk, P-Erk, and βKL were detected by IB (left). Protein levels quantified with ImageJ are shown at the right. In B and E, SEM, n = 3; *, P < .05; **, P < .01; NS, not significant. F, FGF19 signaling studies in vivo. WT and FXR-KO mice were tail vein injected as described in Materials and Methods with FGF19 (10 μg/kg) for 15 minutes for detecting P-Erk and P-38 levels and for 2 hours for measuring Cyp7a1 mRNA levels. Consistent results were obtained from 2 independent q-RT-PCR studies from a total of 7 mice/group (SEM, n = 7; *, P < .05; **, P < .01).
To determine whether decreased βKL expression in FXR-KO mice is the major determinant of impaired FGF15/19 signaling, βKL was expressed in hepatocytes from FXR-KO mice to a level similar to that in WT mice. Expression of βKL partially restored p-ERK levels (Figure 4B, left, quantitation in center). Further, the mRNA levels of Cyp7a1, a known FGF15/19 target (10, 27), were highly elevated in FXR-KO hepatocytes and the levels were partially but significantly reduced by overexpression of βKL (Figure 4B, right).
Because FXR induction of FGF19 was shown to occur in human hepatocytes (27), we also examined the FXR/βKL pathway in human hepatocytes. Treatment with FXR agonists, chenodeoxycholic acid or GW4064, resulted in marked increases in βKL protein levels (Figure 4C). Further, p-ERK levels were diminished in FXR-down-regulated human hepatocytes (Figure 4D) and exogenous expression of βKL partially restored p-ERK levels (Figure 4E, quantitation at the right).
These results suggest that impaired responsiveness to FGF19 signaling and consequent Cyp7a1 inhibition in FXR-lacking hepatocytes is due, at least in part, to decreased βKL expression, providing further evidence that FXR-mediated induction of βKL gene is important for transmitting the FGF15/19 signal.
Hepatic FGF19 signaling and repression of Cyp7a1 are also impaired in FXR-KO mice
Hepatic βKL and FGFR4 protein levels were markedly diminished in FXR-KO mice (Figure 2), and FGF19 signaling was impaired in FXR-lacking hepatocytes (Figure 4, A and D). To test whether FGF19 signaling is also compromised in FXR-KO mice in vivo, WT or FXR-KO mice were treated with FGF19, and then, p-ERK and Cyp7a1 mRNA levels were measured. FGF19 treatment increased p-ERK, but not p-p38, levels in WT mice, and FGF19-mediated increases in p-ERK levels were not detected in FXR-KO mice (Figure 4F, left), suggesting that FGF19 signaling is impaired in FXR-KO mice.
In gene expression studies, basal Cyp7a1 mRNA levels were significantly increased about 50% in FXR-KO mice (Figure 4F, right). Expression of Cyp7a1 was strongly inhibited about 65% in WT mice treated FGF19 for 2 hours, but only about 10% in FGF19-treated FXR-KO mice (Figure 4F, right), indicating that FGF19-mediated repression of Cyp7a1 is markedly blunted in these mice, which is consistent with impaired FGF19 signaling in FXR-KO mice (Figure 4F, left). Taken together with data presented above indicating that FXR induces βKL, these findings suggest that FGF19 signaling and FGF19-mediated repression of Cyp7a1 is impaired in liver hepatocytes lacking FXR, in part, due to decreased βKL expression.
Activation of FXR leads to temporally correlated transient induction of intestinal Fgf15 and hepatic βKL genes in mice
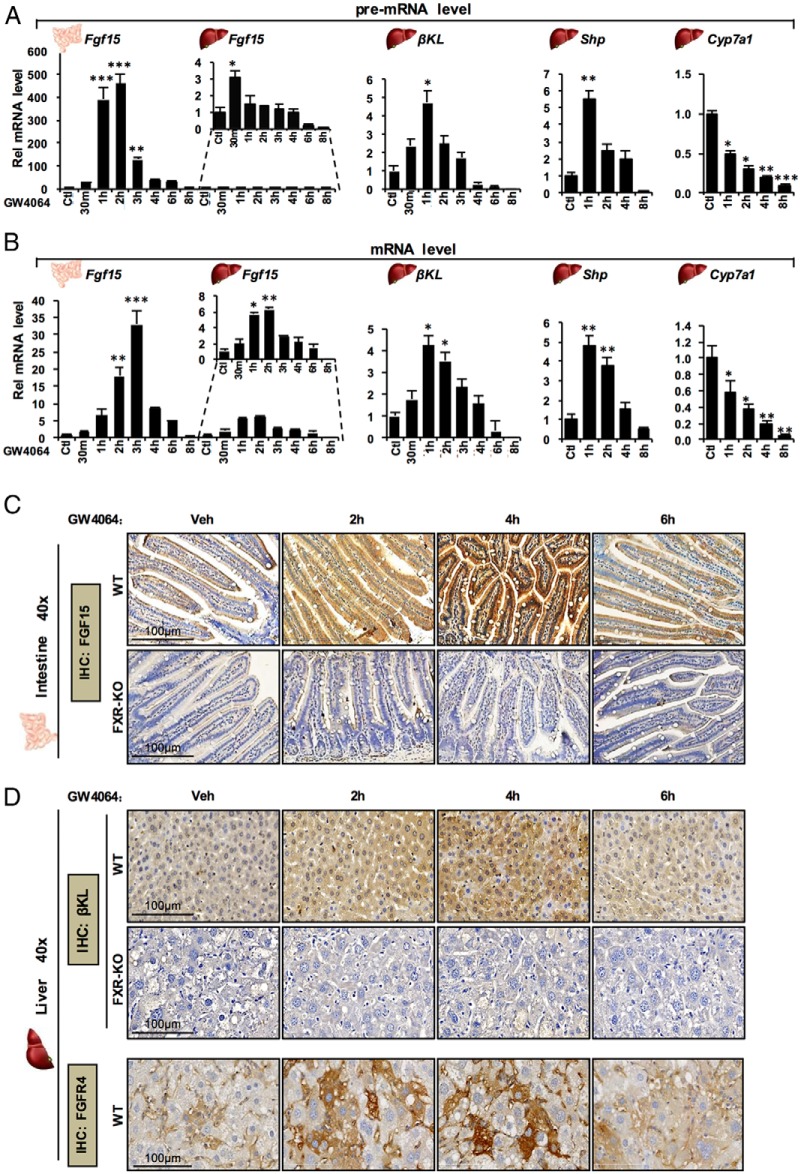
We further tested whether the transcriptional induction of βKL by FXR in the liver is temporally correlated with the induction of Fgf15 in the intestine in mice in vivo. Temporal changes in pre-mRNA and mRNA levels of βKL and Fgf15, and those of the known FXR direct and indirect targets, Shp and Cyp7a1, respectively, were measured after activation of FXR by GW4064 treatment. Pre-mRNA levels of these genes were also measured to monitor transcription (Figure 5A). Strikingly, intestinal Fgf15 pre-mRNA levels were increased over 400-fold, whereas only modest 3-fold increases were observed in the liver. Intestinal Fgf15 pre-mRNA levels peaked at 2 hours and decreased rapidly by 4–6 hours, returning to control levels by 8 hours. Hepatic βKL pre-mRNA levels were increased about 5-fold by 1 hour and then returned to levels below controls by 8 hours.
Figure 5.
Activation of FXR results in transient, temporally correlated induction of intestinal Fgf15 and hepatic βKL in mice in vivo. A and B, C57BL/6 WT mice were fasted for 12 hours and, then, ip injected with GW4064 for the indicated times before killing. Pre-mRNA (A) and mRNA levels (B) of Fgf15 in the intestine and liver, and of βKL, Shp, and Cyp7a1 in the liver were determined by q-RT-PCR (SEM, n = 3; *, P < .05; **, P < .01; ***, P < .005). C and D, WT and FXR-KO mice were injected with GW4064 for the times indicated and protein levels of intestinal FGF15 (C) and hepatic βKL and FGFR4 (D) were detected by IHC.
Intestinal Fgf15 mRNA levels were substantially increased by 2 hours and peaked at 3 hours, whereas hepatic βKL mRNA levels peaked at about 1–2 hours after GW4064 treatment (Figure 5B). A similar time course for pre-mRNA and mRNA levels for hepatic Shp and βKL was observed, but expression of Cyp7a1 was persistently decreased. These results indicate that FXR-mediated transcriptional induction of intestinal Fgf15 and hepatic βKL is transient and temporally correlated.
Temporal effects of GW4064 on protein levels of intestinal FGF15 and hepatic βKL in WT and FXR-KO mice were then determined by IHC analysis. FGF15 levels in the ileum were markedly increased at 2 hours, peaking at 4 hours, but decreased to near control levels by 6 hours (Figure 5C). Liver βKL levels also increased at 2 hours, peaking at 2–4 hours, and decreased to near basal levels 6 hours after treatment (Figure 5D and Supplemental Figure 6). In contrast, in FXR-KO mice, basal protein levels of FGF15 and βKL were markedly decreased, and protein levels were not increased by GW4064. Similar temporal expression patterns for FGFR4 were also observed in liver (Figure 5D). These results clearly demonstrate that basal expression of both βKL and FGF15 is FXR dependent and that the FXR induction of intestinal FGF15 and the hepatic FGF15 receptor complex, βKL and FGFR4, is transient and that their expression is temporally correlated in gut and liver.
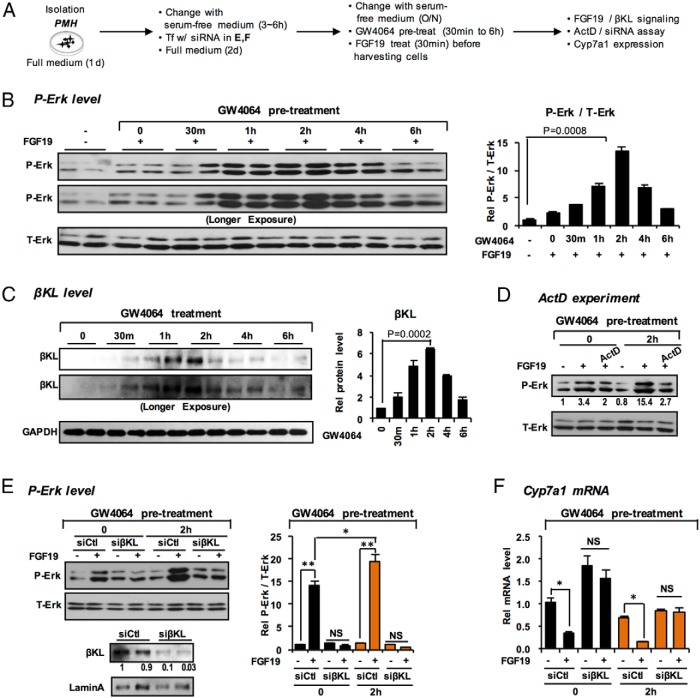
Pretreatment with GW4064 increases βKL expression and enhances responsiveness to FGF19
To test the idea that transcriptional induction of βKL by activated FXR “primes” the liver for FGF15/19 signaling, hepatocytes were pretreated with GW4064 before FGF19 treatment (Figure 6A). The p-ERK levels increased as early as 30 minutes after pretreatment with GW4064, reach a maximum 15-fold increase at 2 hours, and decreased to control levels by 6 hours (Figure 6B). Notably, the increases in βKL protein levels paralleled the increases in p-ERK (Figure 6C). Treatment with a transcriptional inhibitor, actinomycin D (Figure 6D), or down-regulation of βKL by siRNA (Figure 6E), blocked the increases in p-ERK levels, which is consistent with the increase being dependent on transcriptional activation of βKL gene (Figures 1 and 2). The mRNA levels of Cyp7a1 were significantly decreased in GW4064-pretreated hepatocytes and these decreases were abolished in βKL-down-regulated hepatocytes (Figure 6F). Pretreatment with GW4064 also enhanced responsiveness to FGF19 signaling in human hepatocytes (Supplemental Figure 7). These results indicate that treatment with GW4064 before FGF19 treatment enhanced responsiveness to FGF19 action in a manner that is dependent on transcriptional induction of βKL, providing further evidence that activation of FXR primes hepatocytes to respond to the FGF15/19 signal.
Figure 6.
Pretreatment with GW4064 enhances responsiveness to FGF19 signaling in hepatocytes by transcriptional induction of βKL. A, Experimental outline. B, Levels of T-Erk and P-Erk after GW4064 pretreatment for the times indicated before FGF19 treatment were determined by IB (quantitation at the right, n = 3). C, The protein levels of βKL were measured (quantitation at the right). D–F, Hepatocytes were pretreated with GW4064 for 2 hours before FGF19 treatment for 30 minutes (3 independent experiments). D, Hepatocytes were treated with actinomycin D (ActD) or vehicle followed by GW4064 for 2 hours and FGF19 for the last 30 minutes. Levels of T-Erk and P-Erk were determined by IB. E and F, Hepatocytes were transfected with either control or βKL siRNA for 48 hours before the pretreatment with GW4064 and FGF19 treatment. E, Levels of T-Erk and P-Erk were determined and quantified (right; SEM, n = 5 from 3 independent experiments; *, P < .05; **, P < .01; NS, not significant). F, The mRNA levels of Cyp7a1 were measured by q-RT-PCR (SEM, n = 3; *, P < .05; NS, not significant).
Discussion
In this study, we demonstrate that activated FXR primes the liver for intestinal FGF15/19 signaling through temporally correlated transient induction of intestinal FGF15 and hepatic βKL. βKL is the obligate coreceptor for FGF15 and thus, essential for regulation of metabolism mediated by FGF15 signaling in the liver. This conclusion was supported by our findings that: 1) βKL is a direct transcriptional target of FXR; 2) expression of both hepatic FGF15 receptor components, βKL and FGFR4, was substantially decreased in FXR-KO mice; 3) FGF15 signaling was impaired in FXR-KO mice, FXR-KO hepatocytes, and in FXR-down-regulated human hepatocytes, and the impaired FGF15 signaling was partially restored by overexpression of βKL; 4) FXR-mediated induction of intestinal FGF15 and hepatic βKL was temporally correlated in mice; and 5) pretreatment with GW4064 enhanced responsiveness to FGF19 signaling, which was abolished by transcriptional inhibition or down-regulation of βKL.
Interestingly, FGFR4 protein levels, but not mRNA levels, were increased in parallel with βKL by activation of FXR. Protein levels of both FGFR4 and βKL were substantially decreased in FXR-KO mice but were restored to nearly normal levels by adenoviral-mediated hepatic expression of FXR. FGFR4 levels are decreased in βKL-KO mice (17, 18), demonstrating that FGFR4 levels are dependent on βKL levels and suggesting that FGFR4 might be stabilized by forming a complex with βKL. We observed that FGFR4 stability was increased by GW4064 and the increased stability was reversed by down-regulation of βKL consistent with this possibility.
Transient, temporally correlated induction of intestinal Fgf15 and hepatic βKL after pharmacological activation of FXR provides a model for physiological effects after a meal. BA concentrations in the intestine are rapidly increased in response to food intake and most BAs are then returned to the liver through enterohepatic circulation. In humans, BA concentrations in the portal vein increase to a peak between 15 and 60 minutes after a meal (7), so that FXR is activated at roughly the similar time in both liver and intestine, whereas serum FGF19 levels peak relatively late, about 3 hours, after a meal (28). Utilizing a highly sensitive technique that combines immune-enrichment and mass spectrometry, Katafuchi et al detected serum levels of FGF15 and demonstrated that FXR-induced intestinal FGF15 is a bona-fide endocrine hormone that acts on the liver (11). In the current study, we show that substantial increases in intestinal FGF15 and hepatic βKL protein levels occurred in parallel, with detectable increases by 2 hours that peaked by 4 hours and decreased back to near basal levels by 6 hours after GW4064 treatment. Temporal changes in expression of intestinal FGF15 and hepatic βKL after activation of FXR support the intriguing concept that FXR both induces expression of intestinal FGF15 and the hepatic receptor complex for FGF15 in a temporally correlated manner.
Homeostatic mechanisms must terminate postprandial FGF15/19 signaling, in part because its chronic activation is associated with liver tumorigenesis (29). Indeed, FXR-mediated induction of βKL and Fgf15 after GW4064 treatment was transient, and mRNA and protein levels returned to basal levels about 6 hours after treatment (Figure 5). In contrast to these findings, treatment of mice with GW4064 did not increase mRNA levels of βKL in previous studies (26), but because FXR-mediated induction is transient, it might not have been observed in those studies with the treatment protocol used of 2 treatments by oral gavage at 14 and 2 hours before analysis of mRNA levels. The differences in administration of GW4064 by oral gavage compared with ip treatment in our study may also contribute to the conflicting results because GW4064 has poor bioavailability and may result in lower concentrations in the liver. In the present study, we clearly demonstrated that pre-mRNA and mRNA levels of FGF15 in the ileum and βKL in the liver, and importantly, protein levels of FGF15, βKL, and FGFR4 were transiently increased and returned to basal levels 6 hours after treatment from time-course experiments in vivo (Figure 5). Further, Cyp7a1 expression was substantially repressed at 2–4 hours when βKL protein levels were maximal, which is consistent with previous findings that BA synthesis and Cyp7a1 expression are increased in βKL-KO mice (17). Remarkably, strong repression of Cyp7a1 persisted when βKL, Fgf15, or Shp expression had returned to basal levels. Cyp7a1 regulation is complex and influenced by multiple factors, including the Jun N-terminal kinase pathway (4, 30–32), diurnal regulation, nutrient status, stress signaling, and hepatic inflammation, in addition to, cholesterol and BA levels (2–5, 33), some of which may underlie the prolonged repression of Cyp7a1, although the underlying mechanisms are not well understood.
The essential in vivo role of intestinal FXR in inhibiting hepatic Cyp7a1 expression by inducing intestinal FGF15 has been originally demonstrated in intestine- and liver-specific FXR-KO mouse studies (34). However, in liver-specific FXR-KO mice, although FGF15 induction was normal, Cyp7a1 repression was still partially attenuated and importantly, basal mRNA levels of Cyp7a1 were significantly increased 2-fold (34), which suggest that hepatic FXR also has a role in regulation of Cyp7a1 in vivo. Our observations that basal mRNA levels of Cyp7a1 are substantially increased and FGF19-mediated repression of Cyp7a1 is partially attenuated in FXR-KO mouse hepatocyte are qualitatively similar to these liver-specific FXR-KO studies, but quantitatively greater effects were observed in our studies. Nevertheless, this observation and the demonstration that hepatic FXR induces βKL in a temporally correlated manner and enhances FGF19 signaling are consistent with the previous finding in liver-specific FXR-KO mice (34), suggesting that hepatic FXR also contributes to the repression of Cyp7a1.
In previous studies, FGF15 treatment in vivo reduced mRNA levels of Cyp7a1 nearly 99% in WT mice but still showed robust (90%) repression in whole-body FXR-KO mice (26). In contrast, we observed that FGF19 signaling and FGF19 repression of Cyp7a1 were markedly attenuated in the whole-body FXR-KO mice (Figure 4F) and also in hepatocytes from FXR-KO mice (Figure 4B). Human FGF19 was used in our studies and FGF15 in the earlier study, which is unlikely to explain the difference because human FGF19 has been used in mouse studies (10, 14, 15, 20), and moreover, treatment with FGF19 or FGF15 did not show differences in Cyp7a1 mRNA levels in mice (35), so that the explanation for the different results is not clear. As noted above, Cyp7a1 regulation is complex and influenced by multiple factors, which could underlie the inconsistent results. Our findings from IB, IF, and IHC studies indicated that basal protein levels of both βKL and FGFR4 were substantially decreased and FGF19 signaling measured by p-ERK was impaired in FXR-KO mice, which support the conclusion that FGF19 signal-mediated repression of Cyp7a1 is attenuated in FXR-KO mice in part by decreased expression of the FGF15/19 receptor complex in the liver.
Mounting evidence indicates that FXR inhibits BA synthesis by transcriptional induction of many target genes, including hepatic Shp and intestinal Fgf15 (1, 2, 8, 9). Recent studies demonstrated that FXR also inhibits BA synthetic genes by inducing expression of a novel global transcriptional repressor, V-Maf avian musculoaponeurotic fibrosarcoma oncogene homolog G (MAFG) (36), and an epigenetic histone modifying corepressor, lysine-specific demethylase-1 (37). Thus, FXR inhibits BA synthesis and protects the liver from BA toxicity by multiple pathways, including induction of βKL identified in the current study, demonstrating the central role of FXR as a master gene regulator in maintaining BA homeostasis.
In conclusion, this study reveals a conceptually intriguing finding that FXR coordinates postprandial gut-liver FGF15 signaling by transient induction of hepatic βKL and intestinal Fgf15 genes in a temporally correlated manner. Pharmacological treatment of FGF19 or activation of FXR to induce endogenous FGF19 has been shown to have BA-lowering and insulin-sensitizing effects without promoting hepatic lipogenesis (1–3, 8, 14), providing an attractive therapeutic option for treatment of hepatobiliary diseases and diabetes, and in fact, both FXR agonists and FGF19 analogs are currently being tested in clinical trials (1–3, 6). The FXR/βKL/FGFR4 pathway identified in this study provides useful information for developing therapeutic strategies for treatment of FGF19-associated diseases.
Additional material
Supplementary data supplied by authors.
Acknowledgments
We thank the Liver Tissue Procurement and Distribution System of the NIH for providing human primary hepatocytes.
This work was supported by National Institutes of Health Grants DK062777 and DK095842 (to J.K.K.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- Ad
- adeno
- BA
- bile acid
- ChIP
- chromatin immunoprecipitation
- CHX
- cycloheximide
- FGF
- fibroblast growth factor
- FGFR4
- FGF receptor 4
- FXR
- farnesoid X receptor
- IB
- immunoblotting
- IF
- immunofluorescence
- IHC
- immunohistochemistry
- IR2
- inverted repeat 2
- βKL
- β-Klotho
- KO
- knockout
- p-
- phospho
- PMH
- primary mouse hepatocyte
- q-
- quantitative
- RXR
- retinoid X receptor
- Shp
- Src homology-2 domain containing protein tyrosine phosphatase-2
- siRNA
- small interfering RNA
- WT
- wild type.
References
- 1. Kliewer SA, Mangelsdorf DJ. Bile acids as hormones: the FXR-FGF15/19 pathway. Dig Dis. 2015;33:327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chiang JY. Bile acid metabolism and signaling. Compr Physiol. 2013;3:1191–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res. 2009;50:1509–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. [DOI] [PubMed] [Google Scholar]
- 7. Angelin B, Björkhem I, Einarsson K, Ewerth S. Hepatic uptake of bile acids in man. Fasting and postprandial concentrations of individual bile acids in portal venous and systemic blood serum. J Clin Invest. 1982;70:724–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13:213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matsubara T, Li F, Gonzalez FJ. FXR signaling in the enterohepatic system. Mol Cell Endocrinol. 2013;368:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Inagaki T, Choi M, Moschetta A, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. [DOI] [PubMed] [Google Scholar]
- 11. Katafuchi T, Esterházy D, Lemoff A, et al. Detection of FGF15 in plasma by stable isotope standards and capture by anti-peptide antibodies and targeted mass spectrometry. Cell Metab. 2015;21:898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin BC, Wang M, Blackmore C, Desnoyers LR. Liver-specific activities of FGF19 require Klotho β. J Biol Chem. 2007;282:27277–27284. [DOI] [PubMed] [Google Scholar]
- 13. Kurosu H, Choi M, Ogawa Y, et al. Tissue-specific expression of βKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J Biol Chem. 2007;282:26687–26695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kir S, Beddow SA, Samuel VT, et al. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science. 2011;331:1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li S, Hsu DD, Li B, et al. Cytoplasmic tyrosine phosphatase Shp2 coordinates hepatic regulation of bile acid and FGF15/19 signaling to repress bile acid synthesis. Cell Metab. 2014;20:320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu C, Wang F, Kan M, et al. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem. 2000;275:15482–15489. [DOI] [PubMed] [Google Scholar]
- 17. Ito S, Fujimori T, Furuya A, Satoh J, Nabeshima Y. Impaired negative feedback suppression of bile acid synthesis in mice lacking βKlotho. J Clin Invest. 2005;115:2202–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tomiyama K, Maeda R, Urakawa I, et al. Relevant use of Klotho in FGF19 subfamily signaling system in vivo. Proc Natl Acad Sci USA. 2010;107:1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee J, Seok SM, Yu P, et al. Genomic analysis of hepatic Farnesoid X Receptor (FXR) binding sites reveals altered binding in obesity and direct gene repression by FXR. Hepatol. 2012;56:108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fu T, Choi SE, Kim DH, et al. Aberrantly elevated microRNA-34a in obesity attenuates hepatic responses to FGF19 by targeting a membrane coreceptor β-Klotho. Proc Natl Acad Sci USA. 2012;109:16137–16142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fu T, Seok S, Choi S, et al. MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Mol Cell Biol. 2014;34:4130–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. [DOI] [PubMed] [Google Scholar]
- 23. Maloney PR, Parks DJ, Haffner CD, et al. Identification of a chemical tool for the orphan nuclear receptor FXR. J Med Chem. 2000;43:2971–2974. [DOI] [PubMed] [Google Scholar]
- 24. Maruyama T, Miyamoto Y, Nakamura T, et al. Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun. 2002;298:714–719. [DOI] [PubMed] [Google Scholar]
- 25. Studer E, Zhou X, Zhao R, et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. 2012;55:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56:1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7α-hydroxylase gene expression. Hepatology. 2009;49:297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lundåsen T, Gälman C, Angelin B, Rudling M. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J Intern Med. 2006;260:530–536. [DOI] [PubMed] [Google Scholar]
- 29. Lin BC, Desnoyers LR. FGF19 and cancer. Adv Exp Med Biol. 2012;728:183–194. [DOI] [PubMed] [Google Scholar]
- 30. Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7 a-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-jun N-terminal kinase pathway. J Biol Chem. 2001;276:15816–15822. [DOI] [PubMed] [Google Scholar]
- 31. Li T, Jahan A, Chiang JY. Bile acids and cytokines inhibit the human cholesterol 7 α-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 2006;43:1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu C, Wang F, Jin C, Huang X, McKeehan WL. Independent repression of bile acid synthesis and activation of c-Jun N-terminal kinase (JNK) by activated hepatocyte fibroblast growth factor receptor 4 (FGFR4) and bile acids. J Biol Chem. 2005;280:17707–17714. [DOI] [PubMed] [Google Scholar]
- 33. Ponugoti B, Fang S, Kemper JK. Functional interaction of HNF-4 and PGC-1α in CYP7A1 regulation is inhibited by a key lipogenic activator, SREBP-1c. Mol Endocrinol. 2007;21:2698–2712. [DOI] [PubMed] [Google Scholar]
- 34. Kim I, Ahn SH, Inagaki T, et al. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48:2664–2672. [DOI] [PubMed] [Google Scholar]
- 35. Potthoff MJ, Boney-Montoya J, Choi M, et al. FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1α pathway. Cell Metab. 2011;13:729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Aguiar Vallim TQ, Tarling EJ, Ahn H, et al. MAFG is a transcriptional repressor of bile acid synthesis and metabolism. Cell Metab. 2015;21:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim YC, Fang S, Byun S, Seok S, Kemper B, Kemper JK FX. R-induced lysine-specific histone demethylase, LSD1, reduces hepatic bile acid levels and protects the liver against bile acid toxicity. Hepatology. 2015;62:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.