Abstract
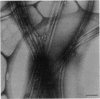
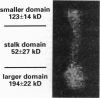
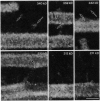
Kinesin is a cytoplasmic motor protein that moves along microtubules and can induce microtubule bundling and sliding in vitro. To determine how kinesin mediates microtubule interactions, we determined the shapes and mass distributions of squid brain kinesin, taxol-stabilized microtubules (squid and bovine), and adenosine 5'-[beta, gamma-imido]triphosphate-stabilized kinesin-microtubule complexes by high-resolution metal replication and by low-temperature, low-dose dark-field scanning transmission electron microscopy of unfixed, directly frozen preparations. Mass mapping by electron microscopy revealed kinesins loosely attached to the carbon support as asymmetrical dumbbell-shaped molecules, 40-52 nm long, with a mass of 379 +/- 15 kDa. The mass distribution and shape of these molecules suggest that these images represent kinesin in a shortened conformation. Kinesin-microtubule complexes were organized as bundles of linearly arrayed microtubules, stitched together at irregular intervals by cross-bridges typically < or = 25 nm long. The crossbridges had a mass of 360 +/- 15 kDa, consistent with one kinesin per crossbridge. These results suggest that kinesin has a second microtubule binding site in addition to the known site on the motor domain of the heavy chain; this second site may be located near the C terminus of the heavy chains or on the associated light chains. Thus, kinesin could play a role in either crosslinking or sliding microtubules.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baas P. W., Deitch J. S., Black M. M., Banker G. A. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom G. S., Wagner M. C., Pfister K. K., Brady S. T. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry. 1988 May 3;27(9):3409–3416. doi: 10.1021/bi00409a043. [DOI] [PubMed] [Google Scholar]
- Brady S. T., Pfister K. K., Bloom G. S. A monoclonal antibody against kinesin inhibits both anterograde and retrograde fast axonal transport in squid axoplasm. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1061–1065. doi: 10.1073/pnas.87.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Kanai Y., Cowan N. J., Hirokawa N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature. 1992 Dec 17;360(6405):674–677. doi: 10.1038/360674a0. [DOI] [PubMed] [Google Scholar]
- Dabora S. L., Sheetz M. P. The microtubule-dependent formation of a tubulovesicular network with characteristics of the ER from cultured cell extracts. Cell. 1988 Jul 1;54(1):27–35. doi: 10.1016/0092-8674(88)90176-6. [DOI] [PubMed] [Google Scholar]
- Goldstein L. S. The kinesin superfamily: tails of functional redundancy. Trends Cell Biol. 1991 Oct;1(4):93–98. doi: 10.1016/0962-8924(91)90036-9. [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Pfister K. K., Yorifuji H., Wagner M. C., Brady S. T., Bloom G. S. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 1989 Mar 10;56(5):867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- Hisanaga S., Murofushi H., Okuhara K., Sato R., Masuda Y., Sakai H., Hirokawa N. The molecular structure of adrenal medulla kinesin. Cell Motil Cytoskeleton. 1989;12(4):264–272. doi: 10.1002/cm.970120407. [DOI] [PubMed] [Google Scholar]
- Johnson K. A., Wall J. S. Structure and molecular weight of the dynein ATPase. J Cell Biol. 1983 Mar;96(3):669–678. doi: 10.1083/jcb.96.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik K. S., Orecchio L. D., Schnapp B., Inouye H., Neve R. L. The primary structure and analysis of the squid kinesin heavy chain. J Biol Chem. 1990 Feb 25;265(6):3278–3283. [PubMed] [Google Scholar]
- Mandelkow E. M., Rapp R., Mandelkow E. Microtubule structure studied by quick freezing: cryo-electron microscopy and freeze fracture. J Microsc. 1986 Mar;141(Pt 3):361–373. doi: 10.1111/j.1365-2818.1986.tb02729.x. [DOI] [PubMed] [Google Scholar]
- Navone F., Niclas J., Hom-Booher N., Sparks L., Bernstein H. D., McCaffrey G., Vale R. D. Cloning and expression of a human kinesin heavy chain gene: interaction of the COOH-terminal domain with cytoplasmic microtubules in transfected CV-1 cells. J Cell Biol. 1992 Jun;117(6):1263–1275. doi: 10.1083/jcb.117.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C., Lombillo V. A., Kuriyama R., McIntosh J. R. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992 Oct 8;359(6395):543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- Rodionov V. I., Gyoeva F. K., Kashina A. S., Kuznetsov S. A., Gelfand V. I. Microtubule-associated proteins and microtubule-based translocators have different binding sites on tubulin molecule. J Biol Chem. 1990 Apr 5;265(10):5702–5707. [PubMed] [Google Scholar]
- Roof D. M., Meluh P. B., Rose M. D. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992 Jul;118(1):95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W. S., Hoyt M. A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992 Aug 7;70(3):451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Schnapp B. J., Reese T. S., Bechtold R. Kinesin is bound with high affinity to squid axon organelles that move to the plus-end of microtubules. J Cell Biol. 1992 Oct;119(2):389–399. doi: 10.1083/jcb.119.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J. M., Heuser J., Yang J. T., Goldstein L. S. Identification of globular mechanochemical heads of kinesin. Nature. 1989 Mar 23;338(6213):355–357. doi: 10.1038/338355a0. [DOI] [PubMed] [Google Scholar]
- Schroer T. A., Sheetz M. P. Functions of microtubule-based motors. Annu Rev Physiol. 1991;53:629–652. doi: 10.1146/annurev.ph.53.030191.003213. [DOI] [PubMed] [Google Scholar]
- Urrutia R., McNiven M. A., Albanesi J. P., Murphy D. B., Kachar B. Purified kinesin promotes vesicle motility and induces active sliding between microtubules in vitro. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6701–6705. doi: 10.1073/pnas.88.15.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R. D., Schnapp B. J., Mitchison T., Steuer E., Reese T. S., Sheetz M. P. Different axoplasmic proteins generate movement in opposite directions along microtubules in vitro. Cell. 1985 Dec;43(3 Pt 2):623–632. doi: 10.1016/0092-8674(85)90234-x. [DOI] [PubMed] [Google Scholar]
- Wall J. S., Hainfeld J. F. Mass mapping with the scanning transmission electron microscope. Annu Rev Biophys Biophys Chem. 1986;15:355–376. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- Yang J. T., Laymon R. A., Goldstein L. S. A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding analyses. Cell. 1989 Mar 10;56(5):879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- Yang J. T., Saxton W. M., Stewart R. J., Raff E. C., Goldstein L. S. Evidence that the head of kinesin is sufficient for force generation and motility in vitro. Science. 1990 Jul 6;249(4964):42–47. doi: 10.1126/science.2142332. [DOI] [PubMed] [Google Scholar]