Abstract
The initiation of DNA replication represents a committing step to cell proliferation. Appropriate replication onset depends on multiprotein complexes that help properly distinguish origin regions, generate nascent replication bubbles, and promote replisome formation. This review describes initiation systems employed by bacteria, archaea, and eukaryotes, with a focus on comparing and contrasting molecular mechanisms among organisms. Although commonalities can be found in the functional domains and strategies used to carry out and regulate initiation, many key participants have markedly different activities and appear to have evolved convergently. Despite significant advances in the field, major questions still persist in understanding how initiation programs are executed at the molecular level.
Keywords: DNA replication, AAA+ ATPases, initiators, helicases
INTRODUCTION
The initiation of DNA replication is a central event in the growth and division of all organisms. The assembly of replication machineries, or replisomes, is coordinated by dedicated initiation proteins, which ensure that DNA synthesis begins at the correct chromosomal locus in accordance with cell cycle cues. Faithful execution of initiation is crucial to viability; inappropriate replication onset is linked to changes in gene copy number, DNA damage, and genetic instability (1, 2).
Cellular initiation programs rely on proteinaceous, trans-acting factors that process chromosomal start sites (termed origins). The mechanics and control of these proteins are complex. Some factors and their activities are conserved across all three domains of life (bacteria, archaea, and eukaryotes); for example, all proteins responsible for recognizing origins (initiators) possess a set of evolutionarily related ATPase domains fused to a helix-turn-helix (HTH)-type DNA binding element (3-7). However, other aspects of initiation—such as DNA-unwinding and strand-synthesis enzymes (e.g., helicases and primases)—are distinct between bacterial and archaeal/eukaryal lineages (3, 8, 9). Hence, although common themes exist in how initiation is executed and controlled, the specific pathways and players that promote this process can vary dramatically.
The goal of this review is to highlight the prevalent mechanisms used to initiate DNA replication in cells. Given that a PubMed search for “DNA replication initiation” turns up >8,000 papers, it is impossible to be comprehensive. We apologize to those colleagues whose work could not be accommodated.
REPLICATION ORIGINS
Bacterial oriC
Bacterial chromosomes, which are typically (though not always) circular, generally bear a single origin distinguished by two classes of sequence elements: conserved repeats that are important for initiator recognition and an AT-rich DNA-unwinding element (DUE) where the replication bubble originates (10-12). The ~260 base-pair (bp) origin of Escherichia coli, oriC, has been a long-standing model system for studying initiation. E. coli oriC bears a complex array of conserved sequence motifs. Most of these are recognized by the ATP-dependent initiator, DnaA (13-15), whereas others are recognized by factors that modulate initiator/origin interactions (see Figure 1a). One prevalent motif is the DnaA box (sites R1–R5) (15, 16). DnaA boxes are recognized equivalently by ATP-bound and ADP-bound DnaA (ATP-DnaA and ADP-DnaA, respectively), with regions R1, R2, and R4 bound most tightly (17-20). I-sites and τ-sites constitute two other classes of DnaA binding sites that associate more weakly with the initiator and are preferentially recognized by ATP-DnaA (13, 18, 21). The DnaA boxes, I-sites, and τ-sites in oriC together form an organizing center that is critical for promoting the higher-order assembly of DnaA and origin activation (discussed below). Interestingly, the spacing between high- and low-affinity sites is important, as even two-base insertions can abrogate origin function (22).
Figure 1.
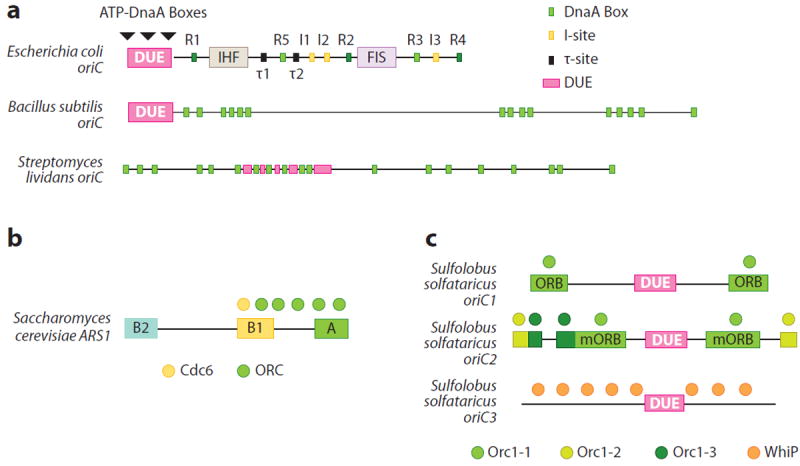
Overview of origin organization. (a) Bacterial oriCs from three representative species (Escherichia coli, Bacillus subtilis, Streptomyces lividans). Sites bound by DnaA and other factors, as well as the DNA-unwinding element (DUE), are labeled. (b) Saccharomyces cerevisiae ARS1 (autonomous replicating sequence 1). ARS consensus sequence elements (A, B1, B2) and ORC · Cdc6 binding sites are shown. (c) The three origins of Sulfolobus solfataricus. DUEs and binding sites for replication initiators are labeled. Abbreviations: ORB, origin recognition box; ORC, origin recognition complex; WhiP, winged-helix initiator protein.
The DUE contains several copies of a fourth type of DnaA binding element termed an ATP-DnaA box. These sites reside within three 13-bp AT-rich repeats historically designated as L, M, and R (10, 12). Similar to I-sites and τ-sites, ATP-DnaA boxes are bound preferentially by ATP-DnaA (14, 23). ATP-DnaA box recognition requires the cooperative assembly of multiple ATP-DnaA protomers and depends on initiator binding to the adjacent R1 element (23). As with the organizing center, the spacing between the DUE and other DnaA binding sites is critical for oriC function (24).
The direct, sequence-based readout of specific origin binding sites by DnaA appears preserved across bacteria (25-28). However, the replication origins of bacterial species display a unique number and arrangement of the consensus DnaA box sequence and its variants (Figure 1a) (29). DnaA orthologs also appear to differ in their relative affinity for DnaA box sequences (30). For example, Mycobacterium tuberculosis DnaA does not stably associate with a single DnaA box; instead, binding requires that multiple repeats be present in target substrates (31). This variation, coupled with the distinct arrangement of DnaA boxes among different oriCs, suggests that DnaA orthologs are fine-tuned to act only on their cognate origin. Consistent with this notion, Escherichia coli DnaA cannot initiate replication on Bacillus subtilis oriC or vice versa (32). Why origins are so diverse among bacteria is unclear but could reflect a speciation mechanism for preventing the replication of foreign origins acquired through phage infection or DNA uptake.
Eukaryotic Origins
Eukaryotic origins exhibit several differences from their bacterial counterparts. First, eukaryotic chromosomes invariably contain multiple origins. Second, eukaryotic origins frequently appear to be defined more by local DNA structure and chromatin environment than by sequence conservation (33). An exception is budding yeast, whose genome is punctuated by related DNA segments called autonomous replicating sequences (ARSs) (34). ARSs serve as recruitment sites for the origin recognition complex (ORC), a multiprotein assembly that functions as the initiator of replication in eukaryotes (35, 36).
The binding of ORC protects specific regions within certain ARSs known as ARS consensus sequences (ACSs). ACSs typically contain two conserved regions bound by ORC termed A and B1 (Figure 1b) (35-37). Interestingly, although ACSs can define a replication start site, they are not always sufficient to do so (38, 39). Initiation in Schizosaccharomyces pombe echoes this paradigm: Fission yeast origins frequently contain an AT-rich segment recognized by the Orc4 subunit of ORC (40, 41), but removal of these AT stretches affects origin activation only marginally in vivo (42, 43).
Metazoan replication start sites are more loosely encoded than those of yeast but exhibit some general similarities. For example, metazoan chromosomal regions occupied by ORC often (although not always) correlate with certain skewed patterns of DNA sequence bearing either AT- or GC-rich stretches (44-46). Nonreplicative proteins, such as transcription factors (47), can associate with ORC to help define particular origin loci. However, despite these generalities, a growing body of evidence points to contextual cues—e.g., proximity to promoter elements, DNA accessibility, nucleosome occupancy, and local structure—as major factors in specifying metazoan replication start sites (48-51). Indeed, in Xenopus egg extracts, replication can initiate at any nucleotide sequence in an exogenously introduced piece of DNA (52). Overall, the molecular logic that eukaryotic cells use to decide which region serves as a replication start site has yet to be fully defined (see Supplemental Material for more detail; follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org).
Archaeal Origins
Archaeal origins exhibit a mix of bacterial and eukaryotic features (53). As in eukaryotes, archaeal chromosomes frequently bear more than one origin (54-56). However, archaeal origins, similar to their bacterial counterparts, also contain conserved elements that function as sequence-specific binding sites for initiators (54).
Sulfolobus solfataricus, whose genome bears three loci termed oriC1, oriC2, and oriC3, exemplifies many of the attributes of archaeal origins (Figure 1c) (54). oriC1 contains three conserved sequence motifs: the origin recognition box (ORB) and the C2 and C3 loci. oriC2 contains similar elements, although its ORB repeats are smaller. Each type of site is bound by one of three distinct initiator paralogs (the Orc1-1, Orc1-2, and Orc1-3 proteins, respectively). Binding sites for archaeal Orc1 paralogs can overlap partially or fully (Figure 1c) (54), suggesting that cooperative or competitive interactions may occur among different initiator subfamilies.
As with different bacterial origins, the number, type, and order of Orc1 binding sites are not preserved among the three Sulfolobus origins. Nonetheless, the ORB-type sites within oriC1 and oriC2 are both arranged as inverted repeats separated by an AT-rich region of varying length that contains a DUE (54, 55). Surprisingly, oriC3 has no discernable Orc1 binding site and is instead protected by a winged-helix initiator protein (WhiP) that is unrelated to Orc1 and does not appear to bind ATP (55). These features suggest that origin multiplication in Sulfolobus likely occurred by the acquisition of extrachromosomal elements rather than by gene duplication (55).
Additional aspects of Sulfolobus replication origins are echoed in other archaeal species. One is origin multiplicity (55, 57). Others are GC sequence skew (58) and the existence of DUEs flanked by inverted ORB repeats (for review, see Reference 59). Whether contextual cues contribute to archaeal origin discrimination is still unclear.
INITIATORS AND ORIGIN PROCESSING/PRIMING/REMODELING
Bacterial DnaA
The DnaA initiator not only recognizes bacterial replication origins but also melts DNA to promote replication onset (12, 15). DnaA is modular, composed of four distinct domains (Figure 2a) (60, 61). The first (domain I) is a small, globular K-homology (KH)-type fold (62-64) that both homodimerizes and associates with many diverse regulatory proteins (for review, see References 65, 66). The second (domain II) is a variable and likely flexible linker element of unclear function (65). Domain III consists of an AAA+ (ATPases associated with various cellular activities) fold (3, 5, 6, 67, 68) that can form large homo-oligomeric arrays (61, 68-71), bind single-stranded (ss)DNA (Figure 2b) (72, 73), and promote oriC melting (71, 73, 74). The fourth region (domain IV) comprises a C-terminal HTH element (5, 75) that recognizes specific sites within the oriC organizing center (26, 28, 76). Domain III is the only functional fold conserved with eukaryotic and archaeal initiators (3, 5), indicating that this region was extant in an ancient initiator protein before bacteria and archaea split into different lineages.
Figure 2.
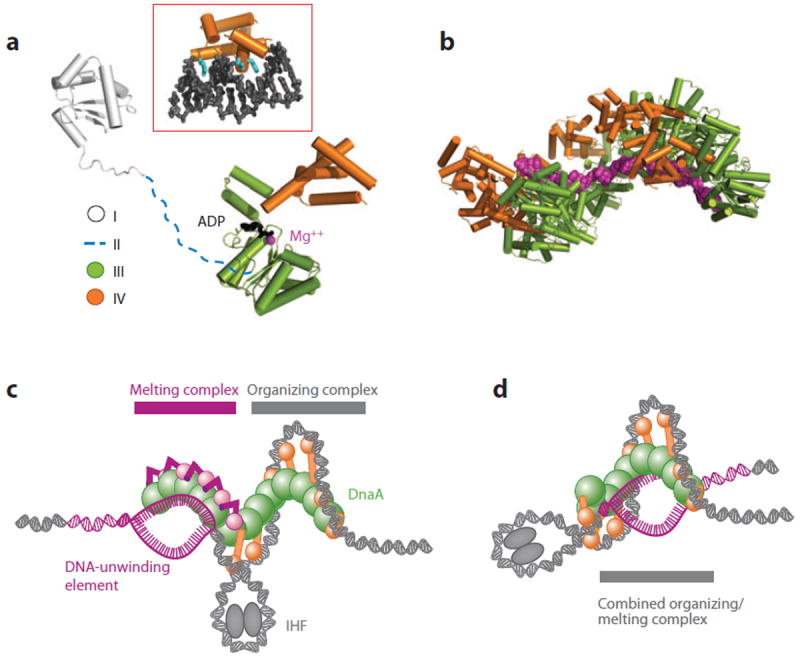
Structure and mechanism of DnaA. (a) Domain architecture of DnaA. Protein Data Bank models for domain I and an ADP state of domains III/IV of DnaA are shown (7, 62, 151). Inset: structure of domain IV bound to duplex DNA (28). (b) Structure of an AMPPCP-assembled DnaA helix (domains III and IV) bound to an extended single-stranded DNA substrate (magenta) (72). (c) Two-state model for IHF-assisted DnaA melting of oriC (7). This mechanism accounts for the existence of two different DnaA oligomers (12, 74), one of which wraps the organizing center of oriC and another that melts the DNA-unwinding element (DUE). (d) Loopback model for IHF-assisted melting, in which a single DnaA complex bound at the organizing center of oriC opens the DUE (296).
Prior to replication onset, strong DnaA boxes in oriC are bound by domain IV of ATP-DnaA or ADP-DnaA (77, 78). As initiation commences, additional copies of ATP-DnaA localize to oriC (78), filling weaker DnaA binding sites and coassociating into a large nucleoprotein complex (13, 22, 69, 79, 80). Domain III is the primary mediator of oligomerization (7, 21, 71, 72), although domains I and IV also participate in assembly (71, 74, 81). DnaA forms a helical oligomer that reconstitutes a hydrolysis-competent ATPase site through the joint action of several signature amino acid motifs (7, 70), including the Walker A, Walker B, Sensor I, and Sensor II elements from one DnaA subunit and a trans-acting arginine finger from a partner protomer (reviewed in Reference 65). Upon assembly, the DnaA helix appears to wrap duplex DNA around itself, stabilizing one or more positive DNA supercoils (Figure 2c) (7, 82).
The complex formed between DnaA and the organizing center of oriC serves as an essential prerequisite to DUE melting (10, 12, 23). Negative supercoiling is required for melting (12), suggesting that DNA opening might be aided by torsional strain (7). However, ATP-DnaA also associates directly with the DUE (23, 73) and can actively melt short DNA duplexes in an ATP-dependent manner by binding and stretching ssDNA along the helical axis of the DnaA oligomer (Figure 2b) (72). Although the precise mechanism is not fully understood, the available evidence indicates that either concomitant with or following DnaA self-assembly, the ATPase domains of the initiator deform and open the DUE directly (72). At least two models account for how DnaA organizes both ssDNA and double-stranded (ds)DNA to promote melting (Figure 2c,d), although the precise architecture of the nucleoprotein complex is not firmly established.
Eukaryotic Origin Recognition Complex
The eukaryotic initiator, ORC, was first discovered from biochemical studies using budding yeast (35, 36). Unlike the single-subunit DnaA protein, ORC comprises six protomers that transiently associate with a seventh factor known as Cdc6 (Cdc18 in fission yeast) (36, 83-85). Together, ORC and Cdc6 identify and mark chromosomal origins (35, 36) and promote the loading of the replicative eukaryotic helicase onto DNA (86-88).
As with DnaA, ORC is composed of many functional elements (Figure 3a). Orc1, Orc4, and Orc5, along with Cdc6, contain AAA+ ATPase domains closely related to that found in the bacterial initiator (3). Orc2 and Orc3 also appear predicated on an AAA+ scaffold, albeit a highly diverged one (84, 89), whereas Orc6 (at least in metazoans) is structurally related to the transcription factor TFIIB (90-92). Many of the AAA+ domains within ORC and Cdc6 are linked to other modules, in particular a predicted variant of an HTH fold termed a winged-helix domain (WHD), which may be involved in DNA binding (4, 84, 93). The N terminus of S. pombe Orc4 is further decorated with several AT-hook repeats that promote interactions between the initiator complex and origin DNA (40, 41, 94), whereas the N terminus of Orc1 universally bears a conserved bromo-adjacent homology (BAH) domain implicated in binding multiple factors (50, 95, 96), including transcriptional silencing factors such as Sir1 (in yeast) and HP1 (in human and Drosophila) (97-99). The Orc1 BAH domain binds to nucleosomes, specifically recognizing dimethylated histone H4 tails (100). BAH-dependent association of ORC with nucleosomes assists with sequence-independent recognition of replication origins (50, 96) and may have a role in the demonstrated ability of ORC to reposition nucleosomes (44, 51, 101). Contacts between the Orc1 BAH domain and histones are important physiologically, as their ablation has been linked to primordial dwarfism (100, 102).
Figure 3.
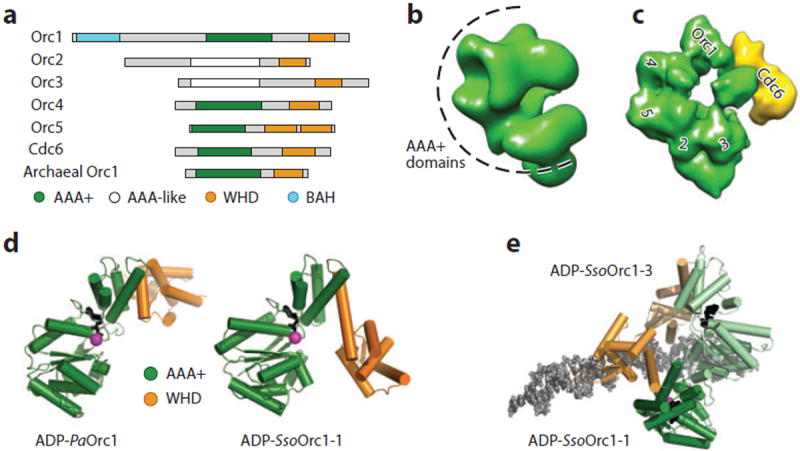
Structure of origin recognition complex (ORC)-type initiators. (a) Schematic of domains in Orc proteins. (b) Three-dimensional electron microscopy (EM) model of the Drosophila ORC indicating where the AAA+ (ATPases associated with various cellular activities) domains of the complex reside (89). (c) Three-dimensional EM model of the budding yeast ORC · Cdc6 complex. The positions of AAA+ subunits are labeled (103). (d) Structures of archaeal Orc1 proteins show variability in AAA+/WHD (winged-helix domain) position [Pyrobaculum aerophilum (4); Sulfolobus solfataricus (116)]. (e) Structure of S. solfataricus Orc1-1 and Orc1-3 paralogs complexed with overlapping sites in Sso-oriC2 (116). Abbreviation: BAH, bromo-adjacent homology.
As expected from the high degree of interspecies sequence conservation among Orc proteins, 3D electron microscopy (EM) reconstructions have revealed similar (although not identical) architectures for different Orc homologs. For example, Drosophila and Saccharomyces cerevisiae ORC form crescent-shaped particles (Figure 3b, c) (84, 89), both of which can accommodate five AAA+ initiator protomers (89, 103). The first five protomers of ORC appear to form the core of the crescent, associating in the order Orc1→Orc4→Orc5→Orc2→Orc3 (103-106). The location of Orc6 is less certain but likely maps near Orc2 or Orc3 (103, 105-107). Cdc6 binds directly to Orc1 and, in the yeast ORC model, contacts the open end of the crescent, converting ORC into a closed-ring-like particle (Figure 3c) (84).
Akin to DnaA, ORC relies on ATP to control several aspects of its activity. Some ATP-dependent responses observed in different ORC orthologs are not universal; for example, ATP promotes the ordered assembly of human ORC subunits (104, 105), whereas ORC from budding yeast and Drosophila can be purified as hexameric complexes even when nucleotide is not added (36, 108). Nonetheless, ATP frequently appears to control how ORC interacts with specific types of DNA substrates and certain classes of binding sites (109, 110). In S. cerevisiae, ATP also promotes the association of ORC with Cdc6 (111, 112), extending the footprint of the initiator on origins such as ARS1 (84, 103, 113). The ability of ATP to elicit specific structural and functional responses likely involves the pairwise coordination of AAA+ ATPase domains from adjoining subunits. Consistent with this idea, Orc4 of budding yeast bears a conserved arginine that maps to a secondary structure position typically occupied by AAA+ arginine fingers and that is critical for stimulating the ATPase activity of Orc1 in trans (111, 114, 115).
The action of ORC on DNA is not fully established. Although ORC binds readily to DNA (36) and is capable of altering local DNA structure and writhe (59), research has yet to show that the eukaryotic initiator can actively melt duplexes. Why ORC might have lost an ancestral ability to unwind replication origins (or why DnaA gained such a function) is unclear. The AAA+ domains of cellular initiators form a distinct subgroup (3) and possess a distinguishing α-helical appendage termed an initiator specific motif (ISM) (116). However, the specific architecture of the DnaA ISM differs from that of archaeal and eukaryotic initiators, as it forms a wedge-shaped structure that pushes adjoining subunits out of plane and into a helical oligomer (7). Differences in ISM configuration may cause adjoining subunits in ORC to adopt a less extreme helical organization, which in DnaA is necessary to support DNA stretching and melting (72, 74). Additional studies are needed to clarify how evolutionary divergence of the AAA+ scaffolds used by ORC and DnaA has led to distinct functions on DNA.
Archaeal Orc1
Archaea lack a DnaA ortholog, instead containing proteins more closely related to eukaryotic Orc1 and Cdc6. Archaeal Orc1 is the most streamlined of the cellular initiators, consisting only of an N-terminal AAA+ ATPase domain and a C-terminal WHD (4, 5, 116-118). Archaeal Orc1 proteins bind to origins (54, 119, 120), and the WHD contributes to both sequence-specific origin recognition and DNA affinity (121-123). Interestingly, the AAA+ domains of archaeal Orc1 proteins also bind to DNA, using their ISM to contact the phospho-diester backbone of target duplexes through either the major or the minor groove (116, 118). Although origin recognition relies on very few base-specific contacts (only one in Sulfolobus Orc1-1), the joint action of the Orc1 WHD and AAA+ domain creates an extended footprint that bends and underwinds the target DNA substrates (28, 116, 118). Biochemical data suggest that, as with eukaryotic replication start sites, local DNA structure is an important determinant for origin discrimination by the archaeal initiator (122).
Although many archaeal Orc1 proteins both possess hallmark AAA+ ATPase motifs and hydrolyze ATP (3, 4, 117, 124), the role of nucleotide in controlling initiator function is poorly understood. As with the HTH domain of DnaA (5, 7), the Orc1 WHD can adopt different orientations with respect to its adjoining AAA+ core in a manner influenced by both nucleotide and substrate DNA (Figure 3d) (4, 116-118). However, ATP has not been observed to foster the formation of higher-order Orc1 assemblies seen with DnaA. Moreover, although the binding of archaeal Orc1 to origin regions can alter the superhelical state of the DNA and promote melting in vitro, this effect is not nucleotide dependent and does not always result in duplex opening within the DUE (59, 120, 125). How Orc1 initiators respond to ATP binding/hydrolysis remains to be determined.
HELICASE LOADING AND ACTIVATION
Helicase Loading in Bacteria
The marking and processing of origins by initiator proteins facilitate the next phase of initiation, namely, the loading of replicative helicases onto DNA (reviewed in Reference 126). In bacteria, a single helicase—termed DnaB in E. coli—serves as the front end of the newly emerging replication fork. DnaB forms a twin-tiered, ring-shaped hexamer (Figure 4a) (127-129), in which a conserved N-terminal domain (NTD) structurally homologous to the helicase interaction domain of the DnaG primase forms one tier and the C-terminal ATPase domain (CTD) forms the other (128, 130-132). Adjoining NTDs in the DnaB hexamer self-assemble into a trimer-of-dimers configuration (127-129, 133), creating a collar that binds ssDNA (127). The CTD tier also interacts with nucleic acid substrates but serves as the site for DNA translocation (134-137). During replication, DnaB encircles the lagging template DNA strand (138), moving 5′→3′ with the ATPase domains oriented toward the duplex side of the fork (138-141). DNA unwinding likely occurs by a steric mechanism that excludes the leading template strand from the helicase interior during translocation (139, 142).
Figure 4.
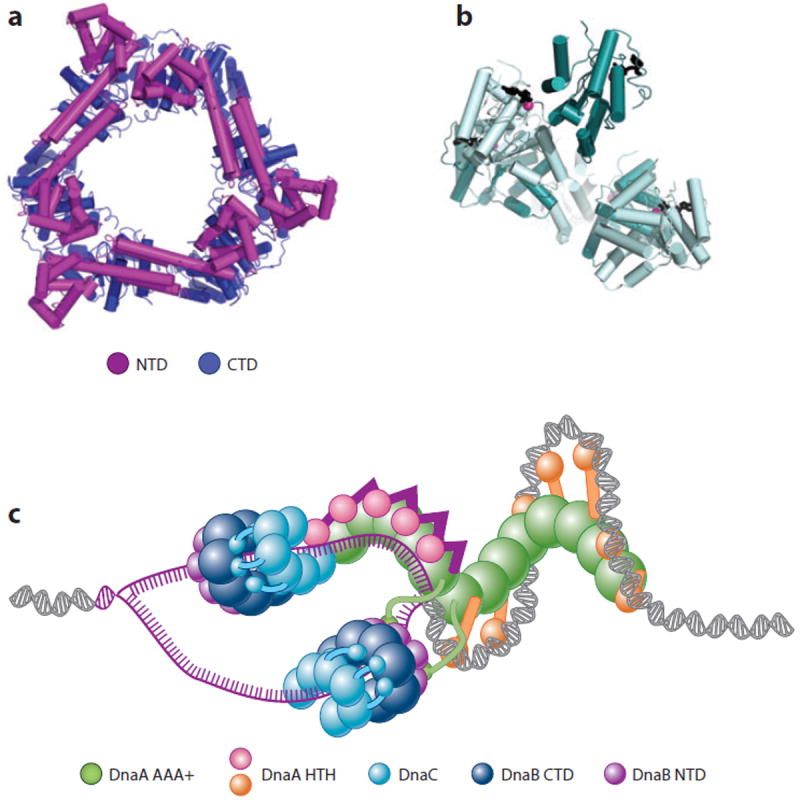
DnaB structure and loading. (a) Closed-ring DnaB hexamer from Bacillus stearothermophilus (128). (b) Structure of a spiral DnaC oligomer [AAA+ (ATPases associated with various cellular activities) domains] bound to nucleotide (147). (c) Model for loading of DnaB onto oriC following the two-state mechanism shown in Figure 1c (147). Abbreviations: CTD, C-terminal domain; HTH, helix-turn-helix; NTD, N-terminal domain.
E. coli DnaB is loaded onto the single-stranded DUE regions of oriC by the concerted action of both DnaA and a dedicated loading factor, DnaC (64, 143-146). DnaC is an AAA+ ATPase and paralog of DnaA (67, 147), but it lacks the C-terminal, duplex-DNA binding domain of the initiator. DnaC, like DnaA, bears a DnaB binding domain at its N terminus (148, 149); however, the folds of these regions are unrelated between the two protein families (62, 150, 151). DnaC also binds ssDNA in a cooperative and ATP-dependent manner (147, 152) by using its AAA+ domains to form a right-handed helical oligomer (Figure 4b) that exhibits a bipartite ATPase site similar to that of DnaA (147). When bound to ATP or poorly hydrolyzable ATP analogs, DnaC represses DnaB activity (146, 153, 154). Although ATP binding is not required for DnaC to load DnaB onto a ssDNA circle, productive association with nucleotide is necessary for DnaC to support the DnaA-dependent initiation of DNA replication on an oriC substrate in vivo and in vitro (155). Overall, the role of nucleotide in DnaC function remains enigmatic.
How DnaB is placed onto a melted origin is a central question in the field. DnaC forms a 6:6 complex with DnaB (156, 157) and has been proposed to crack open the helicase ring to allow ssDNA to engage the motor interior (126, 158, 159). Recruitment of DnaB to oriC is mediated, at least in part, by an interaction between domain I of DnaA and the helicase NTDs (63, 143). Coprecipitation studies using Aquifex aeolicus DnaC have further shown that the loader, whose N terminus binds to the C-terminal face of DnaB (149, 157), can use its ATPase domain to bind the AAA+ domain of DnaA in a nucleotide-dependent manner (147). Given that biochemical studies have shown that DnaA preferentially loads DnaB onto the bottom strand of the DUE in vitro (160) and that two DnaB hexamers are loaded onto opposite strands and in opposing orientations from one another at oriC (145), DnaA and DnaC may collaborate in orienting DnaB hexamers on the complementary strands of a melted DUE (Figure 4c). Recent studies have shown that the DnaG primase can bind to the DnaB · DnaC complex, stimulating ATP hydrolysis by the loader and causing DnaC to release the helicase (155).
Replicative helicase loading in other bacterial species exhibits both similarities to and differences from that of E. coli (126). For example, B. subtilis bears a DnaB helicase (named DnaC) as well as an AAA+-family loading protein (DnaI) that is an ortholog of E. coli DnaC. B. subtilis also utilizes two other proteins, DnaB (unrelated to E. coli DnaB) and DnaD, to aid helicase recruitment and deposition (161). During initiation, B. subtilis DnaB and DnaD, together with their cognate initiator DnaA, associate with oriC in the order DnaA→D→B (162, 163). DnaB and DnaD then interact with DNA, the DnaC helicase, and the DnaI · DnaC complex to facilitate helicase loading (161). As in E. coli, the DnaI loader can both bind ssDNA in an ATP-dependent manner and form a stable 6:6 complex with a replicative helicase target (148); however, data have also suggested that the B. subtilis helicase is assembled on the origin rather than loaded as a preformed hexamer (161).
The apparent lack of DnaC/DnaI loader orthologs in many species [e.g., Helicobacter pylori and M. tuberculosis (164, 165)] has confounded efforts to establish a general framework for helicase loading in bacteria. This absence raises the question of whether certain bacteria rely on as yet unrecognized loading factors, or whether their helicases are self-loading. In this respect, H. pylori DnaB has been observed to form a distinctive double hexamer, which appears predicated on a sixfold symmetric configuration of the N-terminal domains that differs from the trimer-of-dimers state seen in other DnaB orthologs (166). Moreover, expression of H. pylori DnaB in E. coli can complement a dnaC temperature-sensitive mutation (167). These data suggest that dedicated helicase-loading proteins may not be required in all bacterial species; DnaA may serve as the sole factor responsible for mediating the recruitment of the hexameric motor to replication origins in these instances (65). Overall, much remains to be discovered concerning the mechanisms by which initiation factors collaborate to facilitate helicase loading and the extent to which these strategies differ between species.
Helicase Loading and Activation in Eukaryotes
The replicative helicase of eukaryotes comprises an assembly of six distinct but evolutionarily related minichromosome maintenance (MCM) subunits termed Mcm2–7 (168). Mcm2–7 is superficially similar to DnaB, as it forms a hexameric ring composed of two tiers (169, 170)—one corresponding to an N-terminal oligomerization collar and the other to a set of C-terminal motor domains (6, 171)—that encircles and translocates along DNA (Figure 5a) (172-175). However, Mcm2–7 fundamentally differs from its bacterial counterpart in several ways. First, the NTDs of Mcm2–7 and DnaB are structurally unrelated (171, 176, 177). Second, the C-terminal motor domain of MCMs is an AAA+ protein (6, 178) and hence is evolutionarily and organizationally distinct from RecA-type motors such as DnaB (179-181). Third, Mcm2–7 translocates 3′→5′ along the leading strand template of the replication fork (182, 183), opposite to DnaB (140). Finally, whereas DnaB-type helicases exhibit closed-ring architectures in the absence of DNA and appear to switch into a lock washer configuration during DNA unwinding (127-129, 184-187), Mcm2–7 appears capable of spontaneously forming open-ring states when free of substrate and converts into a closed-ring form when bound to ATP and specific activating factors (Figure 5b) (170, 188). These differences support the idea that certain replication fork components evolved independently at least twice (189).
Figure 5.
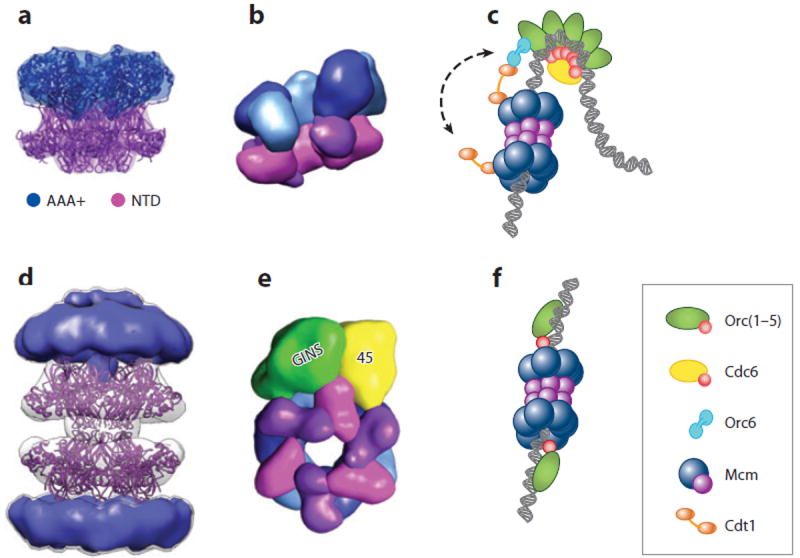
Minichromosome maintenance (MCM) structure and loading. (a) A closed-ring MCM homohexamer from Sulfolobus solfataricus (178). (b) Three-dimensional electron microscopy (EM) model for an open Mcm2–7 heterohexamer from Drosophila melanogaster (170). (c) Model for the loading of an Mcm2–7 double hexamer onto DNA by ORC · Cdc6 and Cdt1 (84, 175, 206). (d) Three-dimensional EM model for the Methanothermobacter thermautotrophicus MCM (235) showing a crystallographic model for an N-terminal domain (NTD) dodecamer (171) docked into the central region. (e) Three-dimensional EM model for the D. melanogaster CMG showing how GINS · Cdc45 latch onto and close an Mcm2–7 ring (170). (f) Model for the loading of archaeal MCMs onto DNA by Orc1 (54, 59).
Mcm2–7 subunits assemble in the order Mcm5→3→7→4→6→2(→5) (170, 190-192). Interactions between Mcm2 and Mcm5 are weak (190, 191) and appear to constitute a natural break point in the ring (170, 183). This discontinuity likely disrupts the processive coordination of ATPase activity among subunits and may explain why DNA unwinding by Mcm2–7 occurs in a narrow window of experimental conditions (183, 193). The Mcm2/5 gap may be a gate that topologically breaches the MCM motor domains (194). In S. cerevisiae, ATP binding appears capable of opening or closing the Mcm2/5 gate (174, 195); however, ATP does not exert such an effect with MCMs from Drosophila melanogaster (170) or the microsporidian parasite Encephalitozoon cuniculi (188). The functional implications of these differences on Mcm2–7 mechanism are unknown.
The loading of Mcm2–7 onto DNA [an event that marks the licensing of replication origins (196)] is accomplished by the joint action of ORC and Cdc6, together with a third protein called Cdt1 (reviewed in Reference 197). These factors form a distinct initiation intermediate known as the prereplicative complex (pre-RC) (198-200). As in bacteria, eukaryotes use an AAA+-type initiator to catalyze helicase deposition; however, the mechanics of Mcm2–7 loading differ from those used for DnaB. In budding yeast, Mcm2–7 is bound first by Cdt1, in part through an interaction with a WHD at the C terminus of Mcm6 (201-203). ORC and Cdc6 then collaborate with Cdt1 to load Mcm2–7 complexes onto DNA (204-206). ATP is required for loading, with ATP hydrolysis by Cdc6 assisting Mcm2–7 recruitment and nucleotide turnover by Orc1 promoting the stable association of the helicase with DNA (114, 207). Mcm2–7 loading appears highly cooperative, with two helicase rings deposited onto duplex DNA in a head-to-head (collar-to-collar) double hexamer for every round of action by ORC · Cdc6 (Figure 5c) (175, 208). How the Mcm2–7 double hexamer forms is not known, but the ability of Orc6 to bind two copies of Cdt1 may help ORC and Cdc6 count the number of MCMs to be used in each loading event (205, 206). Once loaded, Mcm2–7 complexes can passively slide along DNA in vitro but cannot unwind the duplex, suggesting that the double hexamer configuration corresponds to an inactive state (175).
The available structural and biochemical data on Mcm2–7 have several implications for how the helicase loads onto DNA. The break in the Mcm2/5 interface provides a way for DNA to enter the helicase interior (170, 183). Spontaneous adoption of a cracked-ring state suggests that the eukaryotic loading machinery may not use the energy of ATP to actively open the Mcm2–7 ring per se; instead, ATP may be involved in a switching mechanism that coordinates helicase recruitment with ring splitting and DNA entry (170). Because an open Mcm2–7 complex could in principle bind to any accessible DNA region in a chromosome, the initiation machinery, particularly Cdt1, may actually help prevent aberrant loading events as much as aid origin deposition. Curiously, multiple Mcm2–7 complexes can be loaded in vitro and during initiation in cells, even though only two are destined to become part of an active replication fork (209, 210). This so-called MCM paradox (211) differs from bacterial initiation, in which only two molecules of DnaB are deposited per round of origin firing (145, 212) and may have a role in helping to overcome replication fork stalling, for example, by the activation of dormant origins (212).
Although Mcm2–7 serves as the primary motor for unwinding DNA during replication, it lacks robust activity until two additional factors—Cdc45 and the heterotetrameric GINS assembly (comprising the Psf1, Psf2, Psf3, and Sld5 subunits)—associate with the helicase (193, 213-215). Both Cdc45 and GINS travel with Mcm2–7, forming a stable complex known as the CMG (216-219). The CMG exhibits dramatically higher ATPase and 3′→5′ DNA-unwinding activity than Mcm2–7 alone (193, 213). Activation of the CMG appears to occur through a conformational change in the helicase (193) promoted by GINS · Cdc45 binding to one edge of the Mcm2–7 ring and the concomitant closure of the Mcm2/5 interface (Figure 5e) (170). Several lines of evidence indicate that the CMG acts as a discrete, single particle during DNA unwinding and DNA replication (193, 210, 213, 214), implying that the Mcm2–7 double hexamers formed during loading separate once replication commences. How double hexamer separation occurs is not understood.
The observation that Mcm2–7 is initially loaded onto dsDNA fits with the apparent inability of ORC to melt DNA directly (see Eukaryotic Origin Replication Complex, above). However, this finding raises questions such as how origin DNA is melted and how Mcm2–7 associates with DNA in the context of the CMG to promote duplex unwinding. Although multiple models have been advanced for DNA unwinding by MCM proteins (reviewed in Reference 220), recent data appear to support a steric exclusion model akin to that of DnaB. For example, the CMG unwinds forked DNA substrates but not blunt-ended duplexes (193, 214). EM reconstructions of the CMG indicate that the GINS tetramer binds near the edge of the Mcm2–7 hexamer, precluding it from separating a DNA duplex that might emerge from the pore of the helicase ring (170). Perhaps most compellingly, functional replisomes formed in Xenopus egg extracts can bypass roadblocks on the lagging strand but not the leading strand, indicating that only one DNA strand enters and moves through the CMG during translocation (221).
How Mcm2–7 might transition from a ds-DNA to a ssDNA binding mode is unclear. One possibility is that as eukaryotic MCMs are activated upon entry into S phase (discussed below), they undergo a conformational rearrangement—possibly linked to a second ring-opening event—that couples the separation of the Mcm2–7 double hexamer with the concomitant melting of duplex DNA (222, 223). The disposition of DNA strands within the helicase once the CMG is formed is similarly unknown. Similar to the CMG, the GINS tetramer can bind to DNA substrates containing single-stranded regions but not to blunt-ended duplexes (193, 224). Cdc45, which appears to be a catalytically inactive homolog of the bacterial/archaeal 5′→3′ RecJ exonuclease (225), also associates with ssDNA but not ds-DNA (226). Within the CMG, two topologically segregated conduits exist, one formed by the Mcm2–7 ring and the other between the external face of Mcm2 · 5 · 3 and one side of GINS · Cdc45 (Figure 5e) (170). This configuration suggests that GINS and Cdc45 may help capture the lagging DNA strand as it is ejected from the Mcm2–7 complex during origin melting (170, 193, 223, 227). Alternatively, GINS and/or Cdc45 might prevent the leading strand from escaping the CMG during replication fork stalling or collapse; consistent with this idea, work in Xenopus egg extracts has shown that DNA damage causes part of the replisome (including GINS) to disassemble, whereas Cdc45 stays bound to Mcm2–7 at the fork (228). Overall, much remains to be discovered regarding how eukaryotic initiation machineries collaborate to open origin DNA and expand the resulting nascent replication bubble.
Helicase Loading in Archaea
Archaea do not utilize a bacterial DnaB-type helicase but rather retain one or more homologs of the eukaryotic MCM proteins (229-232). With the exception of certain MCM paralogs that appear catalytically defunct (233), archaeal MCMs typically form homohexameric rings that exhibit 3′ → 5′ helicase activity (230, 231, 234). Archaeal MCM protomers are composed of three elements: an N-terminal region that bears both a zinc binding domain and OB fold, a central AAA+ ATPase core, and a predicted C-terminal WHD of unknown function (reviewed in Reference 220). The NTD forms a singly or doubly hexameric collar with cyclic, sixfold symmetry that can bind ssDNA and dsDNA (171, 234, 235). The AAA+ motor elements assemble into a second ring coaxial with the N-terminal collar and may constitute the advancing end of the replisome during translocation (169, 234). A set of β-hairpin loops, present in both the NTD and AAA+ regions, forms a complex network that responds allosterically to ATP binding and hydrolysis to promote translocation and DNA unwinding (178, 234, 236, 237). A detailed understanding of the MCM-unwinding mechanism has yet to emerge.
As with eukaryotic Mcm2–7, full-length archaeal MCMs can assemble into both individual hexamers and NTD-to-NTD dodecamers (Figure 5d) (169, 235). Although the roles of the two oligomeric forms are not known, they may (as with eukaryotic Mcm2–7) reflect different functional states, with the double hexamer corresponding to a loaded configuration and the single hexamer corresponding to an active unwindase. Unlike eukaryotic Mcm2–7, open hexameric rings have not been observed in archaeal MCM structures; nonetheless, archaeal MCMs are both polymorphic and dynamic. For example, S. solfataricus MCMs undergo rapid subunit exchange (236), whereas the Methanothermobacter thermautotrophicus helicase forms heptameric assemblies that revert to hexamers upon binding duplex DNA (235). An open-end helical configuration, somewhat reminiscent of oligomerized DnaA, has even been observed by EM (238, 239). This latter observation suggests that archaeal MCMs might have a propensity to enter into a spiral state, possibly during helicase loading. Whether such a state engages ssDNA or aids in DNA melting is unresolved.
Although the establishment of an origin-dependent, archaeal MCM loading reaction has not been reported, the candidate helicase loader for archaea is Orc1. Interestingly, many archaeal origins contain inverted Orc1 binding sites (ORBs) separated by a distance sufficiently large to accommodate an MCM double hexamer (54, 118). The polar binding of Orc1 to these inverted ORBs is expected to orient the initiator’s C-terminal WHD toward this intervening region (116, 118), which often contains a DUE (59). In M. thermautotrophicus, the Orc1-1 C-terminal WHD associates with the MCM-NTD, whereas Orc1-2 destabilizes the helicase ring (240, 241). Together, these observations have led to a symmetrized model for the bidirectional loading of the archaeal replicative helicase, in which two Orc1 proteins chaperone an MCM double hexamer onto origin DNA (Figure 5f) (59). Future efforts are needed to establish whether helicase-loading factors exist in addition to Orc1.
The DNA-unwinding activity of the archaeal MCM is intrinsically more robust than that of the isolated eukaryotic Mcm2–7 (172, 183, 230, 234). Nevertheless, homologs of the eukaryotic helicase activators GINS and Cdc45 are present in archaea. Archaeal GINS is simpler than its eukaryotic counterpart, as it comprises a homotetramer of only one GINS subunit (GINS15) (242) or a heterotetramer of two subunits (GINS15 and GINS23) (243). Archaeal GINS interact with their respective MCM counterparts both in vitro and in vivo (243, 244), but a consistent effect of this interaction on helicase function has yet to emerge. GINS also forms a stable complex with archaeal RecJ-type proteins (243), although we do not know whether these prokaryotic Cdc45 homologs support MCM function.
REGULATION OF INITIATION FACTORS
Cellular organisms use an array of mechanisms for regulating initiation that are generally distinct among bacteria, archaea, and eukaryotes. Nonetheless, there exist common strategies that appear to have evolved convergently. One is the control of origin accessibility by DNA architecture factors [e.g., histones and nucleotide-associated proteins, which are discussed in the Supplemental Material and several recent reviews (18, 47, 245)]. Another strategy is the regulation of initiation proteins by posttranslational modifications and/or protein-protein interactions. In this section we compare and contrast a few of the many approaches used by cells to govern when and where initiation can occur.
Control of DnaA Function
A prevalent regulatory theme in bacteria is the control of DnaA’s nucleotide status and availability (Figure 6a). Although replication initiation depends on the binding of ATP to DnaA (see Bacterial oriC, above), once hydrolyzed, the initiator remains tightly bound to ADP, entering an inactive state that disfavors DnaA-oriC assembly (246). In E. coli, the intracellular concentrations of ATP-bound DnaA vary with the bacterial cell cycle, peaking at initiation (247). A portion of this active material derives from newly synthesized DnaA, which rapidly binds ATP (19). However, ADP-DnaA can be reactivated, both by acidic phospholipids [e.g., cardiolipin (248)] and by specific DNA loci [DnaA-reactivating sequences (DARSs)] (249). E. coli DnaA pools can also be adjusted transcriptionally (250) and by titrating free ATP-DnaA molecules to chromosomal loci such as datA (251), which contain multiple DnaA binding sites. In organisms such as E. coli and Caulobacter crescentus, Hda, a DnaA paralog, can stimulate initiator ATPase activity (246, 252). Hda functions by associating with polymerase sliding clamps that are loaded onto DNA following initiation and providing a catalytic arginine finger that activates the nucleotide binding site of DnaA in trans (253, 254). B. subtilis and other gram-positive bacteria appear to lack Hda but possess a functional analog termed YabA. As with Hda, YabA binds to both DnaA and the sliding clamp to block further DnaA binding to oriC, thereby preventing inappropriate reinitiation (255-257).
Figure 6.
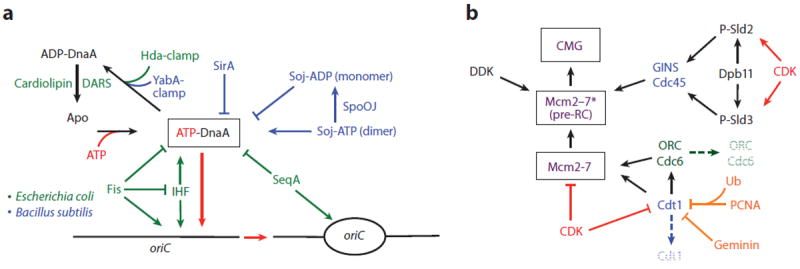
Overview of initiation factor regulation in cells. (a) Regulation of DnaA in bacteria, comparing and contrasting Escherichia coli and Bacillus subtilis systems. (b) Regulation of initiation proteins in eukaryotes. Arrows, activating interactions/events; inhibition lines, repressive interactions/events; hatched text, degradation. Abbreviations: CDK, cyclin-dependent kinase; DARSs, DnaA-reactivating sequences; DDK, Dbf4-dependent kinase; ORC, origin recognition complex; RC, replication complex.
Additional aspects of DnaA function are also subject to direct control. For example, the DnaA-binding protein DiaA is a positive regulator of initiation that, in E. coli, enhances DnaA-oriC assembly and aids DUE unwinding (258). DiaA and its homologs appear to function by forming homotetramers that bind to the N terminus of DnaA (259, 260), facilitating cooperative interactions between initiator protomers to maintain replication synchrony (260). Other proteins also bind to the N-terminal domain of E. coli DnaA; however, some of these interactions (such as the L2 ribosomal subunit) interfere with DnaA assembly to negatively regulate initiator activity (261). For its part, B. subtilis utilizes a different set of factors that binds to and controls DnaA, in particular SirA and Soj, which govern how the initiator associates with both itself and oriC (70, 262-264). Overall, a diverse array of mechanisms for regulating DnaA appears to exist among bacterial species, indicating that many new factors and pathways await discovery.
Regulation of Eukaryotic Initiation Proteins
Eukaryotes regulate initiation through a complex strategy that predominantly targets the assembly and activation of Mcm2–7 helicases through posttranslational modifications (197, 265). The cyclin-dependent kinases (CDKs) are a key set of players whose activities fluctuate in accordance with the phase of the cell cycle (1). In G1, CDKs are inactive, allowing ORC to recruit Mcm2–7 to replication origins and form the pre-RC. Upon entry into S phase, CDK activity increases, blocking pre-RC assembly but allowing for the recruitment of replisome components that activate Mcm2–7 and promote origin firing (Figure 6b) (266).
Mcm2–7 is activated by the action of two distinct cyclin-dependent kinases: CDK and a CDK variant termed the Dbf4-dependent kinase (DDK). DDK phosphorylates serine/threonine-rich sites—found predominantly in the unstructured N-terminal tails of Mcm2, 4, and 6 (reviewed in Reference 267)—that have been prephosphorylated by other kinases (268). Interestingly, two alterations in the Mcm2–7 NTDs overcome the requirement for DDK phosphorylation and either prematurely drive cells into S phase or bypass S phase checkpoints (269, 270). Although it is not known how DDK action and N-terminal mutations activate Mcm2–7, the available data suggest that these events may reshape the interface formed between newly loaded double hexamers (175), promoting the separation of the two helicase rings upon formation of the CMG.
Studies in budding yeast reveal that CDK also regulates Mcm2–7 function by controlling how two non-MCM factors, Sld2 and Sld3, chaperone CMG assembly. Modification of Sld2 and Sld3 by CDK promotes their association with Dpb11, a scaffolding protein that binds phosphorylated targets using BRCT repeats (Figure 6b). Formation of the CMG is completed by the arrival of Cdc45 via a (CDK-independent) interaction with Sld3 and of GINS through an interaction with phospho-Sld2 (265, 271, 272). Following entry into S phase and the initiation of DNA replication, GINS and Cdc45 continue to travel with Mcm2–7 as part of the CMG (273), whereas Sld2, Sld3, and Dpb11 are lost (218). The mechanism by which specific proteins are linked to or removed from Mcm2–7 is unclear but requires the transient association of another origin activator, Mcm10 (274, 275). At least part, if not all, of the yeast Sld2 · Dbp11 · Sld3 system appears present in metazoans (276-278). In vertebrates, this system is further embellished by additional regulatory factors [e.g., GemC1 (279)].
Metazoans employ several additional layers of control to ensure that helicase loading is tightly coordinated with cell cycle cues. For example, proteolysis can degrade Cdt1 following entry into S phase (280). In a strategy reminiscent of RIDA in bacteria (see Control of DnaA Function, above) (253, 254), proteolysis occurs only after Cdt1 binds to the PCNA sliding clamp and is ubiquitinated (280, 281). Mcm2–7 recruitment can also be prevented through Geminin, a Cdt1-sequestration factor that becomes active upon reaching a critical concentration threshold in the nucleus (282, 283). Structural and biochemical studies show that Geminin exerts this concentration-dependent effect by initially binding Cdt1 in a 2:1 complex (284), which subsequently dimerizes as protein levels increase to mask Cdt1 regions required for replication licensing (285).
Despite a focus on Mcm2–7, the helicase is not the sole nexus for regulating initiation in eukaryotes. For example, many organisms target Orc1 for ubiquitination and/or degradation to prevent re-replication (reviewed in Reference 1). CDK activity can stimulate the expression of Cdc6 and Mcm2–7 prior to origin firing (286, 287), as well as target pre-RC components to avoid aberrant secondary initiation events; interestingly, this latter activity can lead initiation factors to different destinies both within and between organisms. For instance, in budding yeast, phosphorylated Mcm2–7 and Cdt1 are routed out of the nucleus (288-290), whereas phosphorylated Cdc6 can be both sequestered and ubiquitinated for targeted proteolysis (291). Although the effect of CDK-dependent phosphorylation on yeast ORC is unclear, Cdk1 activity impairs the ability of ORC to bind DNA in Xenopus and mammalian systems (292, 293), precluding origin relicensing (294). Given the diverse actions seen in the regulation of eukaryotic initiator function, new investigations will likely reveal additional complexities.
Control of Initiation in Archaea
Compared with their bacterial and eukaryotic counterparts, the pathways and mechanisms by which archaea regulate initiation are far less well defined. Nonetheless, a growing body of evidence suggests that archaea likely possess cell cycle–dependent initiation control systems. For example, a strong consensus Orc1-1 binding site immediately precedes the orc1-1 open reading frame, which itself lies just downstream of S. solfataricus oriC1 (54), suggesting that archaeal initiators (like DnaA) may autoregulate their own expression. The whiP gene, which is adjacent to S. solfataricus oriC3, similarly can achieve extensive self-protection by the cooperative binding of WhiP protomers, at least in vitro (55). Genome-wide mapping of Sulfolobus acidocaldarius transcripts even shows a cell cycle–specific induction pattern for certain serine-threonine protein kinases belonging to the same family of kinases as eukaryotic CDKs (295). Overall, uncovering and defining the approaches used by archaea for regulating initiation is still very much a frontier area.
CONCLUDING REMARKS
All replicating systems must perform two fundamental tasks: (a) selecting the right time and place for initiating replication and (b) orchestrating DNA opening and replisome assembly. Cellular organisms have developed multiple strategies to overcome these challenges, often employing sets of proteins that appear to have evolved independently. The only common strategy universally adopted by cells is the use of an AAA+ ATPase to mark replication origins and to promote helicase recruitment; however, even these factors greatly differ in their specific molecular mechanisms. Future investigations will undoubtedly continue to reveal many unexpected surprises in this area.
Supplementary Material
SUMMARY POINTS.
The initiation of DNA replication in cells involves the timely recognition and melting of chromosomal origins and the appropriate loading of replicative helicases.
Initiation in cellular organisms relies on evolutionarily related AAA+ ATPase initiators and ring-shaped, hexameric helicases.
Despite certain commonalities, the participants, mechanisms, and regulation of initiation differ greatly among bacterial and archaeal/eukaryal lineages.
FUTURE ISSUES.
How do replication initiators bind and remodel origin DNA?
How are replicative helicases loaded onto origins?
How do different regulatory systems ensure that initiation of protein function accords with cell cycle cues?
Acknowledgments
The authors are indebted to members of the Berger lab for their help with the writing and editing of this review. This work was supported by Cancer Research UK (to A.C.), a Department of Energy Office of Science Graduate Fellowship (to I.V.H.), and the National Institutes of Health (GM071747, to J.M.B.).
Glossary
- Origin
chromosomal site for initiating DNA replication
- Initiator
an ATP-dependent protein or protein complex responsible for recognizing origins and recruiting replicative helicases to sites of replication
- Helix-turn-helix (HTH)
a type of DNA binding domain that includes specialized folds such as winged-helix domains
- Helicase
an enzyme that unwinds DNA; cellular replicative helicases form hexameric rings
- DUE
DNA-unwinding element
- ORC
origin recognition complex
- AAA+
ATPases associated with various cellular activities
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- WHD
winged-helix domain
- Loader
a protein or set of proteins that assists with reorganizing replicative helicases to promote their association with DNA
- MCM
minichromosome maintenance
- pre-RC
prereplicative complex
- CMG
Cdc45·Mcm2–7·GINS
- RIDA
regulatory inactivation of DnaA
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Alessandro Costa, Email: alessandro.costa@cancer.org.uk.
Iris V. Hood, Email: ihood@berkeley.edu.
James M. Berger, Email: jmberger@berkeley.edu.
LITERATURE CITED
- 1.Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21(5):497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 2.Sutera VA, Jr, Lovett ST. The role of replication initiation control in promoting survival of replication fork damage. Mol Microbiol. 2006;60(1):229–39. doi: 10.1111/j.1365-2958.2006.05093.x. [DOI] [PubMed] [Google Scholar]
- 3.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146(1–2):11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Smith CL, DeRyckere D, DeAngelis K, Martin GS, Berger JM. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol Cell. 2000;6(3):637–48. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 5.Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002;21(18):4763–73. doi: 10.1093/emboj/cdf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9(1):27–43. [PubMed] [Google Scholar]
- 7.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13(8):676–83. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 8.Aravind L, Leipe DD, Koonin EV. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26(18):4205–13. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leipe DD, Aravind L, Grishin NV, Koonin EV. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000;10(1):5–16. [PubMed] [Google Scholar]
- 10.Gille H, Messer W. Localized DNA melting and structural pertubations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J. 1991;10(6):1579–84. doi: 10.1002/j.1460-2075.1991.tb07678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowalski D, Eddy MJ. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989;8(13):4335–44. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52(5):743–55. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 13.McGarry KC, Ryan VT, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc Natl Acad Sci USA. 2004;101(9):2811–16. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Speck C, Weigel C, Messer W. ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 1999;18(21):6169–76. doi: 10.1093/emboj/18.21.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuller RS, Funnell BE, Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- 16.Matsui M, Oka A, Takanami M, Yasuda S, Hirota Y. Sites of dnaA protein-binding in the replication origin of the Escherichia coli K-12 chromosome. J Mol Biol. 1985;184(3):529–33. doi: 10.1016/0022-2836(85)90299-2. [DOI] [PubMed] [Google Scholar]
- 17.Margulies C, Kaguni JM. Ordered and sequential binding of DnaA protein to oriC, the chromosomal origin of Escherichia coli. J Biol Chem. 1996;271(29):17035–40. doi: 10.1074/jbc.271.29.17035. [DOI] [PubMed] [Google Scholar]
- 18.Leonard AC, Grimwade JE. Regulation of DnaA assembly and activity: taking directions from the genome. Annu Rev Microbiol. 2011;65:19–35. doi: 10.1146/annurev-micro-090110-102934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50(2):259–65. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- 20.Schaper S, Messer W. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J Biol Chem. 1995;270(29):17622–26. doi: 10.1074/jbc.270.29.17622. [DOI] [PubMed] [Google Scholar]
- 21.Kawakami H, Keyamura K, Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J Biol Chem. 2005;280(29):27420–30. doi: 10.1074/jbc.M502764200. [DOI] [PubMed] [Google Scholar]
- 22.Rozgaja TA, Grimwade JE, Iqbal M, Czerwonka C, Vora M, Leonard AC. Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol Microbiol. 2011;82(2):475–88. doi: 10.1111/j.1365-2958.2011.07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speck C, Messer W. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 2001;20(6):1469–76. doi: 10.1093/emboj/20.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu J, Bramhill D, Thompson CM. Open complex formation by DnaA initiation protein at the Escherichia coli chromosomal origin requires the 13-mers precisely spaced relative to the 9-mers. Mol Microbiol. 1994;11(5):903–11. doi: 10.1111/j.1365-2958.1994.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 25.Holz A, Schaefer C, Gille H, Jueterbock WR, Messer W. Mutations in the DnaA binding sites of the replication origin of Escherichia coli. Mol Gen Genet. 1992;233(1–2):81–88. doi: 10.1007/BF00587564. [DOI] [PubMed] [Google Scholar]
- 26.Sutton MD, Kaguni JM. Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J Biol Chem. 1997;272(37):23017–24. doi: 10.1074/jbc.272.37.23017. [DOI] [PubMed] [Google Scholar]
- 27.Tsodikov OV, Biswas T. Structural and thermodynamic signatures of DNA recognition by Mycobacterium tuberculosis DnaA. J Mol Biol. 2011;410:461–76. doi: 10.1016/j.jmb.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Fujikawa N, Kurumizaka H, Nureki O, Terada T, Shirouzu M, et al. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31(8):2077–86. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackiewicz P, Zakrzewska-Czerwinska J, Zawilak A, Dudek MR, Cebrat S. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 2004;32(13):3781–91. doi: 10.1093/nar/gkh699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zawilak-Pawlik A, Kois A, Majka J, Jakimowicz D, Smulczyk-Krawczyszyn A, et al. Architecture of bacterial replication initiation complexes: orisomes from four unrelated bacteria. Biochem J. 2005;389(Pt 2):471–81. doi: 10.1042/BJ20050143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zawilak A, Kois A, Konopa G, Smulczyk-Krawczyszyn A, Zakrzewska-Czerwińska J. Mycobacterium tuberculosis DnaA initiator protein: purification and DNA-binding requirements. Biochem J. 2004;382(Pt 1):247–52. doi: 10.1042/BJ20040338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krause M, Rückert B, Lurz R, Messer W. Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J Mol Biol. 1997;274(3):365–80. doi: 10.1006/jmbi.1997.1404. [DOI] [PubMed] [Google Scholar]
- 33.Mechali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol. 2010;11(10):728–38. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- 34.Stinchcomb DT, Struhl K, Davis RW. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979;282(5734):39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- 35.Diffley JF, Cocker JH. Protein-DNA interactions at a yeast replication origin. Nature. 1992;357(6374):169–72. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- 36.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357(6374):128–34. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 37.Diffley JF, Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci USA. 1988;85(7):2120–24. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Celniker SE, Sweder K, Srienc F, Bailey JE, Campbell JL. Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4(11):2455–66. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Houten JV, Newlon CS. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Mol Cell Biol. 1990;10(8):3917–25. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA. 1999;96(6):2656–61. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JK, Moon KY, Jiang Y, Hurwitz J. The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc Natl Acad Sci USA. 2001;98(24):13589–94. doi: 10.1073/pnas.251530398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okuno Y, Satoh H, Sekiguchi M, Masukata H. Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin. Mol Cell Biol. 1999;19(10):6699–709. doi: 10.1128/mcb.19.10.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heichinger C, Penkett CJ, Bähler J, Nurse P. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 2006;25(21):5171–79. doi: 10.1038/sj.emboj.7601390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20(2):201–11. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong D, Coleman TR, DePamphilis ML. Xenopus origin recognition complex (ORC) initiates DNA replication preferentially at sequences targeted by Schizosaccharomyces pombe ORC. EMBO J. 2003;22(13):3441–50. doi: 10.1093/emboj/cdg319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paixão S, Colaluca IN, Cubells M, Peverali FA, Destro A, et al. Modular structure of the human lamin B2 replicator. Mol Cell Biol. 2004;24(7):2958–67. doi: 10.1128/MCB.24.7.2958-2967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding Q, MacAlpine DM. Defining the replication program through the chromatin landscape. Crit Rev Biochem Mol Biol. 2011;46(2):165–79. doi: 10.3109/10409238.2011.560139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghosh M, Liu G, Randall G, Bevington J, Leffak M. Transcription factor binding and induced transcription alter chromosomal c-myc replicator activity. Mol Cell Biol. 2004;24(23):10193–207. doi: 10.1128/MCB.24.23.10193-10207.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23(4):897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller P, Park S, Shor E, Huebert DJ, Warren CL, et al. The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes Dev. 2010;24(13):1418–33. doi: 10.1101/gad.1906410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM, et al. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24(8):748–53. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harland RM, Laskey RA. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980;21(3):761–71. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- 53.Myllykallio H, Lopez P, López-García P, Heilig R, Saurin W, et al. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000;288(5474):2212–15. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- 54.Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116(1):25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 55.Robinson NP, Bell SD. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc Natl Acad Sci USA. 2007;104(14):5806–11. doi: 10.1073/pnas.0700206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc Natl Acad Sci USA. 2004;101(18):7046–51. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Norais C, Hawkins M, Hartman AL, Eisen JA, Myllykallio H, Allers T. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 2007;3(5):e77. doi: 10.1371/journal.pgen.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopez P, Philippe H, Myllykallio H, Forterre P. Identification of putative chromosomal origins of replication in Archaea. Mol Microbiol. 1999;32(4):883–86. doi: 10.1046/j.1365-2958.1999.01370.x. [DOI] [PubMed] [Google Scholar]
- 59.Wigley DB. ORC proteins: marking the start. Curr Opin Struct Biol. 2009;19(1):72–78. doi: 10.1016/j.sbi.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 60.Sutton MD, Kaguni JM. The Escherichia coli dnaA gene: four functional domains. J Mol Biol. 1997;274(4):546–61. doi: 10.1006/jmbi.1997.1425. [DOI] [PubMed] [Google Scholar]
- 61.Messer W, Blaesing F, Majka J, Nardmann J, Schaper S, et al. Functional domains of DnaA proteins. Biochimie. 1999;81(8–9):819–25. doi: 10.1016/s0300-9084(99)00215-1. [DOI] [PubMed] [Google Scholar]
- 62.Abe Y, Jo T, Matsuda Y, Matsunaga C, Katayama T, Ueda T. Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J Biol Chem. 2007;282(24):17816–27. doi: 10.1074/jbc.M701841200. [DOI] [PubMed] [Google Scholar]
- 63.Sutton MD, Carr KM, Vicente M, Kaguni JM. Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J Biol Chem. 1998;273(51):34255–62. doi: 10.1074/jbc.273.51.34255. [DOI] [PubMed] [Google Scholar]
- 64.Seitz H, Weigel C, Messer W. The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol Microbiol. 2000;37(5):1270–79. doi: 10.1046/j.1365-2958.2000.02096.x. [DOI] [PubMed] [Google Scholar]
- 65.Ozaki S, Katayama T. DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid. 2009;62(2):71–82. doi: 10.1016/j.plasmid.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 66.Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–75. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 67.Koonin EV. DnaC protein contains a modified ATP-binding motif and belongs to a novel family of ATPases including also DnaA. Nucleic Acids Res. 1992;20(8):1997. doi: 10.1093/nar/20.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 69.Funnell BE, Baker TA, Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem. 1987;262(21):10327–34. [PubMed] [Google Scholar]
- 70.Scholefield G, Errington J, Murray H. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 2012;31(6):1542–55. doi: 10.1038/emboj.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Felczak MM, Kaguni JM. The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J Biol Chem. 2004;279(49):51156–62. doi: 10.1074/jbc.M409695200. [DOI] [PubMed] [Google Scholar]
- 72.Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478(7368):209–13. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ozaki S, Kawakami H, Nakamura K, Fujikawa N, Kagawa W, et al. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem. 2008;283(13):8351–62. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 74.Duderstadt KE, Mott ML, Crisona NJ, Chuang K, Yang H, Berger JM. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J Biol Chem. 2010;285(36):28229–39. doi: 10.1074/jbc.M110.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roth A, Messer W. The DNA binding domain of the initiator protein DnaA. EMBO J. 1995;14(9):2106–11. doi: 10.1002/j.1460-2075.1995.tb07202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blaesing F, Weigel C, Welzeck M, Messer W. Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol Microbiol. 2000;36(3):557–69. doi: 10.1046/j.1365-2958.2000.01881.x. [DOI] [PubMed] [Google Scholar]
- 77.Samitt CE, Hansen FG, Miller JF, Schaechter M. In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 1989;8(3):989–93. doi: 10.1002/j.1460-2075.1989.tb03462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cassler MR, Grimwade JE, Leonard AC. Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 1995;14(23):5833–41. doi: 10.1002/j.1460-2075.1995.tb00271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grimwade JE, Torgue JJ, McGarry KC, Rozgaja T, Enloe ST, Leonard AC. Mutational analysis reveals Escherichia coli oriC interacts with both DnaA-ATP and DnaA-ADP during pre-RC assembly. Mol Microbiol. 2007;66(2):428–39. doi: 10.1111/j.1365-2958.2007.05930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Crooke E, Thresher R, Hwang DS, Griffith J, Kornberg A. Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. J Mol Biol. 1993;233(1):16–24. doi: 10.1006/jmbi.1993.1481. [DOI] [PubMed] [Google Scholar]
- 81.Felczak MM, Simmons LA, Kaguni JM. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J Biol Chem. 2005;280(26):24627–33. doi: 10.1074/jbc.M503684200. [DOI] [PubMed] [Google Scholar]
- 82.Zorman S, Seitz H, Sclavi B, Strick TR. Topological characterization of the DnaA-oriC complex using single-molecule nanomanipuation. Nucleic Acids Res. 2012;40:7375–83. doi: 10.1093/nar/gks371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81(5):667–76. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 84.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12(11):965–71. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379(6561):180–82. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 86.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404(6778):625–28. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 87.Maiorano D, Lemaitre JM, Mechali M. Stepwise regulated chromatin assembly of MCM2-7 proteins. J Biol Chem. 2000;275(12):8426–31. doi: 10.1074/jbc.275.12.8426. [DOI] [PubMed] [Google Scholar]
- 88.Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87(1):53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 89.Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, et al. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat Struct Mol Biol. 2006;13(8):684–90. doi: 10.1038/nsmb1121. [DOI] [PubMed] [Google Scholar]
- 90.Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci USA. 2003;100(16):9150–55. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu S, Balasov M, Wang H, Wu L, Chesnokov IN, Liu Y. Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proc Natl Acad Sci USA. 2011;108(18):7373–78. doi: 10.1073/pnas.1013676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Balasov M, Huijbregts RP, Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol Cell Biol. 2007;27(8):3143–53. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clarey MG, Botchan M, Nogales E. Single particle EM studies of the Drosophila melanogaster origin recognition complex and evidence for DNA wrapping. J Struct Biol. 2008;164(3):241–49. doi: 10.1016/j.jsb.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gaczynska M, Osmulski PA, Jiang Y, Lee JK, Bermudez V, Hurwitz J. Atomic force microscopic analysis of the binding of the Schizosaccharomyces pombe origin recognition complex and the spOrc4 protein with origin DNA. Proc Natl Acad Sci USA. 2004;101(52):17952–57. doi: 10.1073/pnas.0408369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Callebaut I, Courvalin JC, Mornon JP. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446(1):189–93. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- 96.Noguchi K, Vassiley A, Ghosh S, Yates JL, DePamphilis ML. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 2006;25(22):5372–82. doi: 10.1038/sj.emboj.7601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381(6579):251–53. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 98.Prasanth SG, Shen Z, Prasanth KV, Stillman B. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc Natl Acad Sci USA. 2010;107(34):15093–98. doi: 10.1073/pnas.1009945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, et al. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91(3):311–23. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 100.Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484(7392):115–19. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lubelsky Y, Sasaki T, Kuipers MA, Lucas I, Le Beau MM, et al. Pre-replication complex proteins assemble at regions of low nucleosome occupancy within the Chinese hamster dihydrofolate reductase initiation zone. Nucleic Acids Res. 2010;39(8):3141–55. doi: 10.1093/nar/gkq1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, et al. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet. 2011;43(4):350–55. doi: 10.1038/ng.776. [DOI] [PubMed] [Google Scholar]
- 103.Sun J, Kawakami H, Zech J, Speck C, Stillman B, Li H. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure. 2012;20(3):534–44. doi: 10.1016/j.str.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ranjan A, Gossen M. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc Natl Acad Sci USA. 2006;103(13):4864–69. doi: 10.1073/pnas.0510305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J Biol Chem. 2007;282(44):32370–83. doi: 10.1074/jbc.M705905200. [DOI] [PubMed] [Google Scholar]
- 106.Chen Z, Speck C, Wendel P, Tang C, Stillman B, Li H. The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2008;105(30):10326–31. doi: 10.1073/pnas.0803829105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dhar SK, Delmolino L, Dutta A. Architecture of the human origin recognition complex. J Biol Chem. 2001;276(31):29067–71. doi: 10.1074/jbc.M103078200. [DOI] [PubMed] [Google Scholar]
- 108.Gossen M, Pak DT, Hansen SK, Acharya JK, Botchan MR. A Drosophila homolog of the yeast origin recognition complex. Science. 1995;270(5242):1674–77. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- 109.Chesnokov I, Remus D, Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc Natl Acad Sci USA. 2001;98(21):11997–2002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88(4):493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 111.Klemm RD, Bell SP. ATP bound to the origin recognition complex is important for preRC formation. Proc Natl Acad Sci USA. 2001;98(15):8361–67. doi: 10.1073/pnas.131006898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mizushima T, Takahashi N, Stillman B. Cdc6p modulates the structure and DNA binding activity of the origin recognition complex in vitro. Genes Dev. 2000;14(13):1631–41. [PMC free article] [PubMed] [Google Scholar]
- 113.Santocanale C, Diffley JF. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15(23):6671–79. [PMC free article] [PubMed] [Google Scholar]
- 114.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16(6):967–78. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 115.Lee DG, Makhov AM, Klemm RD, Griffith JD, Bell SP. Regulation of origin recognition complex conformation and ATPase activity: differential effects of single-stranded and double-stranded DNA binding. EMBO J. 2000;19(17):4774–82. doi: 10.1093/emboj/19.17.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–13. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 117.Singleton MR, Morales R, Grainge I, Cook N, Isupov MN, Wigley DB. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J Mol Biol. 2004;343(3):547–57. doi: 10.1016/j.jmb.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 118.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–16. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 119.Duggin IG, McCallum SA, Bell SD. Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci USA. 2008;105(43):16737–42. doi: 10.1073/pnas.0806414105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grainge I, Gaudier M, Schuwirth BS, Westcott SL, Sandall J, et al. Biochemical analysis of a DNA replication origin in the archaeon Aeropyrum pernix. J Mol Biol. 2006;363(2):355–69. doi: 10.1016/j.jmb.2006.07.076. [DOI] [PubMed] [Google Scholar]
- 121.Capaldi SA, Berger JM. Biochemical characterization of Cdc6/Orc1 binding to the replication origin of the euryarchaeon Methanothermobacter thermoautotrophicus. Nucleic Acids Res. 2004;32(16):4821–32. doi: 10.1093/nar/gkh819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dueber EC, Costa A, Corn JE, Bell SD, Berger JM. Molecular determinants of origin discrimination by Orc1 initiators in archaea. Nucleic Acids Res. 2011;39(9):3621–31. doi: 10.1093/nar/gkq1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Majernik AI, Chong JP. A conserved mechanism for replication origin recognition and binding in archaea. Biochem J. 2008;409(2):511–18. doi: 10.1042/BJ20070213. [DOI] [PubMed] [Google Scholar]
- 124.Shin JH, Grabowski B, Kasiviswanathan R, Bell SD, Kelman Z. Regulation of minichromosome maintenance helicase activity by Cdc6. J Biol Chem. 2003;278(39):38059–67. doi: 10.1074/jbc.M305477200. [DOI] [PubMed] [Google Scholar]
- 125.Matsunaga F, Takemura K, Akita M, Adachi A, Yamagami T, Ishino Y. Localized melting of duplex DNA by Cdc6/Orc1 at the DNA replication origin in the hyperthermophilic archaeon Pyrococcus furiosus. Extremophiles. 2010;14(1):21–31. doi: 10.1007/s00792-009-0284-9. [DOI] [PubMed] [Google Scholar]
- 126.Soultanas P. Loading mechanisms of ring helicases at replication origins. Mol Microbiol. 2012;84(1):6–16. doi: 10.1111/j.1365-2958.2012.08012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lo YH, Tsai KL, Sun YJ, Chen WT, Huang CY, Hsiao CD. The crystal structure of a replicative hexameric helicase DnaC and its complex with single-stranded DNA. Nucleic Acids Res. 2009;37(3):804–14. doi: 10.1093/nar/gkn999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318(5849):459–63. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 129.Wang G, Klein MG, Tokonzaba E, Zhang Y, Holden LG, Chen XS. The structure of a DnaB-family replicative helicase and its interactions with primase. Nat Struct Mol Biol. 2008;15(1):94–100. doi: 10.1038/nsmb1356. [DOI] [PubMed] [Google Scholar]
- 130.Su XC, Schaeffer PM, Loscha KV, Gan PH, Dixon NE, Otting G. Monomeric solution structure of the helicase-binding domain of Escherichia coli DnaG primase. FEBS J. 2006;273(21):4997–5009. doi: 10.1111/j.1742-4658.2006.05495.x. [DOI] [PubMed] [Google Scholar]
- 131.Syson K, Thirlway J, Hounslow AM, Soultanas P, Waltho JP. Solution structure of the helicase-interaction domain of the primase DnaG: a model for helicase activation. Structure. 2005;13(4):609–16. doi: 10.1016/j.str.2005.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bailey S, Eliason WK, Steitz TA. The crystal structure of the Thermus aquaticus DnaB helicase monomer. Nucleic Acids Res. 2007;35(14):4728–36. doi: 10.1093/nar/gkm507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Biswas T, Tsodikov OV. Hexameric ring structure of the N-terminal domain of Mycobacterium tuberculosis DnaB helicase. FEBS J. 2008;275(12):3064–71. doi: 10.1111/j.1742-4658.2008.06460.x. [DOI] [PubMed] [Google Scholar]
- 134.Biswas EE, Biswas SB. Mechanism of DNA binding by the DnaB helicase of Escherichia coli: analysis of the roles of domain γ in DNA binding. Biochemistry. 1999;38(34):10929–39. doi: 10.1021/bi990049l. [DOI] [PubMed] [Google Scholar]
- 135.Lo YH, Liu SW, Sun YJ, Li HW, Hsiao CD. Mutations altering the interplay between GkDnaC helicase and DNA reveal an insight into helicase unwinding. PLoS ONE. 2011;6(12):e29016. doi: 10.1371/journal.pone.0029016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nitharwal RG, Verma V, Subbarao N, Dasgupta S, Choudhury NR, Dhar SK. DNA binding activity of Helicobacter pylori DnaB helicase: the role of the N-terminal domain in modulating DNA binding activities. FEBS J. 2012;279(2):234–50. doi: 10.1111/j.1742-4658.2011.08418.x. [DOI] [PubMed] [Google Scholar]
- 137.Nakayama N, Arai N, Kaziro Y, Arai K. Structural and functional studies of the dnaB protein using limited proteolysis. Characterization of domains for DNA-dependent ATP hydrolysis and for protein association in the primosome. J Biol Chem. 1984;259(1):88–96. [PubMed] [Google Scholar]
- 138.Jezewska MJ, Rajendran S, Bujalowska D, Bujalowski W. Does single-stranded DNA pass through the inner channel of the protein hexamer in the complex with the Escherichia coli DnaB helicase? Fluorescence energy transfer studies. J Biol Chem. 1998;273(17):10515–29. doi: 10.1074/jbc.273.17.10515. [DOI] [PubMed] [Google Scholar]
- 139.Kaplan DL. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J Mol Biol. 2000;301(2):285–99. doi: 10.1006/jmbi.2000.3965. [DOI] [PubMed] [Google Scholar]
- 140.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986;261(10):4738–48. [PubMed] [Google Scholar]
- 141.Lee EH, Kornberg A, Hidaka M, Kobayashi T, Horiuchi T. Escherichia coli replication termination protein impedes the action of helicases. Proc Natl Acad Sci USA. 1989;86(23):9104–8. doi: 10.1073/pnas.86.23.9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hacker KJ, Johnson KA. A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry. 1997;36(46):14080–87. doi: 10.1021/bi971644v. [DOI] [PubMed] [Google Scholar]
- 143.Marszalek J, Kaguni JM. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J Biol Chem. 1994;269(7):4883–90. [PubMed] [Google Scholar]
- 144.Wickner S, Hurwitz J. Interaction of Escherichia coli dnaB and dnaC(D) gene products in vitro. Proc Natl Acad Sci USA. 1975;72(3):921–25. doi: 10.1073/pnas.72.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fang L, Davey MJ, O’Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell. 1999;4(4):541–53. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- 146.Davey MJ, Fang L, McInerney P, Georgescu RE, O’Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 2002;21(12):3148–59. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135(4):623–34. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ioannou C, Schaeffer PM, Dixon NE, Soultanas P. Helicase binding to DnaI exposes a cryptic DNA-binding site during helicase loading in Bacillus subtilis. Nucleic Acids Res. 2006;34(18):5247–58. doi: 10.1093/nar/gkl690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ludlam AV, McNatt MW, Carr KM, Kaguni JM. Essential amino acids of Escherichia coli DnaC protein in an N-terminal domain interact with DnaB helicase. J Biol Chem. 2001;276(29):27345–53. doi: 10.1074/jbc.M101940200. [DOI] [PubMed] [Google Scholar]
- 150.Loscha KV, Jaudzems K, Ioannou C, Su XC, Hill FR, et al. A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader. Nucleic Acids Res. 2009;37(7):2395–404. doi: 10.1093/nar/gkp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lowery TJ, Pelton JG, Chandonia JM, Kim R, Yokota H, Wemmer DE. NMR structure of the N-terminal domain of the replication initiator protein DnaA. J Struct Funct Genomics. 2007;8(1):11–17. doi: 10.1007/s10969-007-9022-7. [DOI] [PubMed] [Google Scholar]
- 152.Biswas SB, Flowers S, Biswas-Fiss EE. Quantitative analysis of nucleotide modulation of DNA binding by DnaC protein of Escherichia coli. Biochem J. 2004;379(Pt 3):553–62. doi: 10.1042/BJ20031255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Allen GC, Jr, Kornberg A. Fine balance in the regulation of DnaB helicase by DnaC protein in replication in Escherichia coli. J Biol Chem. 1991;266(33):22096–101. [PubMed] [Google Scholar]
- 154.Wahle E, Lasken RS, Kornberg A. The dnaB-dnaC replication protein complex of Escherichia coli. II. Role of the complex in mobilizing dnaB functions. J Biol Chem. 1989;264(5):2469–75. [PubMed] [Google Scholar]
- 155.Makowska-Grzyska M, Kaguni JM. Primase directs the release of DnaC from DnaB. Mol Cell. 2010;37(1):90–101. doi: 10.1016/j.molcel.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kobori JA, Kornberg A. The Escherichia coli dnaC gene product. III. Properties of the dnaB-dnaC protein complex. J Biol Chem. 1982;257(22):13770–75. [PubMed] [Google Scholar]
- 157.Galletto R, Jezewska MJ, Bujalowski W. Interactions of the Escherichia coli DnaB helicase hexamer with the replication factor the DnaC protein. Effect of nucleotide cofactors and the ssDNA on protein-protein interactions and the topology of the complex. J Mol Biol. 2003;329(3):441–65. doi: 10.1016/s0022-2836(03)00435-2. [DOI] [PubMed] [Google Scholar]
- 158.Davey MJ, O’Donnell M. Replicative helicase loaders: ring breakers and ring makers. Curr Biol. 2003;13(15):R594–96. doi: 10.1016/s0960-9822(03)00523-2. [DOI] [PubMed] [Google Scholar]
- 159.Ahnert P, Picha KM, Patel SS. A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J. 2000;19(13):3418–27. doi: 10.1093/emboj/19.13.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weigel C, Seitz H. Strand-specific loading of DnaB helicase by DnaA to a substrate mimicking unwound oriC. Mol Microbiol. 2002;46(4):1149–56. doi: 10.1046/j.1365-2958.2002.03232.x. [DOI] [PubMed] [Google Scholar]
- 161.Velten M, McGovern S, Marsin S, Ehrlich SD, Noirot P, Polard P. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol Cell. 2003;11(4):1009–20. doi: 10.1016/s1097-2765(03)00130-8. [DOI] [PubMed] [Google Scholar]
- 162.Smits WK, Merrikh H, Bonilla CY, Grossman AD. Primosomal proteins DnaD and DnaB are recruited to chromosomal regions bound by DnaA in Bacillus subtilis. J Bacteriol. 2011;193(3):640–48. doi: 10.1128/JB.01253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Smits WK, Goranov AI, Grossman AD. Ordered association of helicase loader proteins with the Bacillus subtilis origin of replication in vivo. Mol Microbiol. 2010;75(2):452–61. doi: 10.1111/j.1365-2958.2009.06999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Alm RA, Trust TJ. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J Mol Med. 1999;77(12):834–46. doi: 10.1007/s001099900067. [DOI] [PubMed] [Google Scholar]
- 165.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 166.Stelter M, Gutsche I, Kapp U, Bazin A, Bajic G, et al. Architecture of a dodecameric bacterial replicative helicase. Structure. 2012;20(3):554–64. doi: 10.1016/j.str.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 167.Soni RK, Mehra P, Choudhury NR, Mukhopadhyay G, Dhar SK. Functional characterization of Helicobacter pylori DnaB helicase. Nucleic Acids Res. 2003;31(23):6828–40. doi: 10.1093/nar/gkg895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tye BK. MCM proteins in DNA replication. Annu Rev Biochem. 1999;68:649–86. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 169.Pape T, Meka H, Chen S, Vicentini G, van Heel M, Onesti S. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4(11):1079–83. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18(4):471–77. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat Struct Biol. 2003;10(3):160–67. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 172.Chong JP, Mahbubani HM, Khoo CY, Blow JJ. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375(6530):418–21. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- 173.Madine MA, Khoo CY, Mills AD, Laskey RA. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995;375(6530):421–24. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- 174.Bochman ML, Schwacha A. Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J Biol Chem. 2007;282(46):33795–804. doi: 10.1074/jbc.M703824200. [DOI] [PubMed] [Google Scholar]
- 175.Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139(4):719–30. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fass D, Bogden CE, Berger JM. Crystal structure of the N-terminal domain of the DnaB hexameric helicase. Structure. 1999;7(6):691–98. doi: 10.1016/s0969-2126(99)80090-2. [DOI] [PubMed] [Google Scholar]
- 177.Weigelt J, Brown SE, Miles CS, Dixon NE, Otting G. NMR structure of the N-terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer. Structure. 1999;7(6):681–90. doi: 10.1016/s0969-2126(99)80089-6. [DOI] [PubMed] [Google Scholar]
- 178.Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, et al. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci USA. 2008;105(51):20191–96. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Leipe DD, Koonin EV, Aravind L. Evolution and classification of P-loop kinases and related proteins. J Mol Biol. 2003;333(4):781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 180.Wang J. Nucleotide-dependent domain motions within rings of the RecA/AAA(+) superfamily. J Struct Biol. 2004;148(3):259–67. doi: 10.1016/j.jsb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 181.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32(17):5260–79. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272(39):24508–13. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 183.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31(2):287–93. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 184.San Martin MC, Stamford NP, Dammerova N, Dixon NE, Carazo JM. A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J Struct Biol. 1995;114(3):167–76. doi: 10.1006/jsbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- 185.Yu X, Jezewska MJ, Bujalowski W, Egelman EH. The hexameric E. coli DnaB helicase can exist in different Quaternary states. J Mol Biol. 1996;259(1):7–14. doi: 10.1006/jmbi.1996.0297. [DOI] [PubMed] [Google Scholar]
- 186.Yang S, Yu X, VanLoock MS, Jezewska MJ, Bujalowski W, Egelman EH. Flexibility of the rings: structural asymmetry in the DnaB hexameric helicase. J Mol Biol. 2002;321(5):839–49. doi: 10.1016/s0022-2836(02)00711-8. [DOI] [PubMed] [Google Scholar]
- 187.Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151(2):267–77. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lyubimov AY, Costa A, Bleichert F, Botchan MR, Berger JM. ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Proc Natl Acad Sci USA. 2012;109(30):11999–2004. doi: 10.1073/pnas.1209406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27(17):3389–401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Davey MJ, Indiani C, O’Donnell M. Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J Biol Chem. 2003;278(7):4491–99. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- 191.Crevel G, Ivetic A, Ohno K, Yamaguchi M, Cotterill S. Nearest neighbour analysis of MCM protein complexes in Drosophila melanogaster. Nucleic Acids Res. 2001;29(23):4834–42. doi: 10.1093/nar/29.23.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Schwacha A, Bell SP. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol Cell. 2001;8(5):1093–104. doi: 10.1016/s1097-2765(01)00389-6. [DOI] [PubMed] [Google Scholar]
- 193.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37(2):247–58. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bochman ML, Bell SP, Schwacha A. Subunit organization of Mcm2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol Cell Biol. 2008;28(19):5865–73. doi: 10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bochman ML, Schwacha A. The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2-7 ‘gate’. Nucleic Acids Res. 2010;38(18):6078–88. doi: 10.1093/nar/gkq422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Blow JJ. Preventing re-replication of DNA in a single cell cycle: evidence for a replication licensing factor. J Cell Biol. 1993;122(5):993–1002. doi: 10.1083/jcb.122.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol. 2009;21(6):771–77. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 198.Donovan S, Harwood J, Drury LS, Diffley JFX. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94(11):5611–16. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Maiorano D, Moreau J, Mechali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404(6778):622–25. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 200.Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78(2):303–16. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 201.Yanagi K, Mizuno T, You Z, Hanaoka F. Mouse geminin inhibits not only Cdt1-MCM6 interactions but also a novel intrinsic Cdt1 DNA binding activity. J Biol Chem. 2002;277(43):40871–80. doi: 10.1074/jbc.M206202200. [DOI] [PubMed] [Google Scholar]
- 202.Teer JK, Dutta A. Human Cdt1 lacking the evolutionarily conserved region that interacts with MCM2-7 is capable of inducing re-replication. J Biol Chem. 2008;283(11):6817–25. doi: 10.1074/jbc.M708767200. [DOI] [PubMed] [Google Scholar]
- 203.Wei Z, Liu C, Wu X, Xu N, Zhou B, et al. Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6. J Biol Chem. 2010;285(17):12469–73. doi: 10.1074/jbc.C109.094599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes Dev. 2007;21(22):2897–907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chen S, Bell SP. CDK prevents Mcm2-7 helicase loading by inhibiting Cdt1 interaction with Orc6. Genes Dev. 2011;25(4):363–72. doi: 10.1101/gad.2011511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Takara TJ, Bell SP. Multiple Cdt1 molecules act at each origin to load replication-competent Mcm2-7 helicases. EMBO J. 2011;30(24):4885–96. doi: 10.1038/emboj.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Randell JC, Bowers JL, Rodriguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol Cell. 2006;21(1):29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 208.Evrin C, Clarke P, Zech J, Lurz R, Sun J, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106(48):20240–45. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem. 2002;277(36):33049–57. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- 210.Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell. 2010;40(5):834–40. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Laskey RA, Madine MA. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 2003;4(1):26–30. doi: 10.1038/sj.embor.embor706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Klotz-Noack K, Blow JJ. A role for dormant origins in tumor suppression. Mol Cell. 2011;41(5):495–96. doi: 10.1016/j.molcel.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103(27):10236–41. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase ε in rolling circle DNA synthesis. Proc Natl Acad Sci USA. 2012;109(16):6042–47. doi: 10.1073/pnas.1203734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21(4):581–87. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 216.Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91(1):59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 217.Tercero JA, Labib K, Diffley JF. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 2000;19(9):2082–93. doi: 10.1093/emboj/19.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kanemaki M, Labib K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006;25(8):1753–63. doi: 10.1038/sj.emboj.7601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 2003;423(6941):720–24. doi: 10.1038/nature01692. [DOI] [PubMed] [Google Scholar]
- 220.Brewster AS, Chen XS. Insights into the MCM functional mechanism: lessons learned from the archaeal MCM complex. Crit Rev Biochem Mol Biol. 2010;45(3):243–56. doi: 10.3109/10409238.2010.484836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146(6):931–41. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146(1):80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Boos D, Frigola J, Diffley JF. Activation of the replicative DNA helicase: breaking up is hard to do. Curr Opin Cell Biol. 2012;24(3):423–30. doi: 10.1016/j.ceb.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 224.Boskovic J, Coloma J, Aparicio T, Zhou M, Robinson CV, et al. Molecular architecture of the human GINS complex. EMBO Rep. 2007;8(7):678–84. doi: 10.1038/sj.embor.7401002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sanchez-Pulido L, Ponting CP. Cdc45: The missing RecJ ortholog in eukaryotes? Bioinformatics. 2011;27:1885–88. doi: 10.1093/bioinformatics/btr332. [DOI] [PubMed] [Google Scholar]
- 226.Krastanova I, Sannino V, Amenitsch H, Gileadi O, Pisani FM, Onesti S. Structural and functional insights into the DNA replication factor Cdc45 reveal an evolutionary relationship to the DHH family of phosphoesterases. J Biol Chem. 2012;287(6):4121–28. doi: 10.1074/jbc.M111.285395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Botchan M, Berger J. DNA replication: making two forks from one prereplication complex. Mol Cell. 2010;40(6):860–61. doi: 10.1016/j.molcel.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hashimoto Y, Puddu F, Costanzo V. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol. 2012;19(1):17–24. doi: 10.1038/nsmb.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Barry ER, Bell SD. DNA replication in the archaea. Microbiol Mol Biol Rev. 2006;70(4):876–87. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kelman Z, Lee JK, Hurwitz J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum ΔH contains DNA helicase activity. Proc Natl Acad Sci USA. 1999;96(26):14783–88. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Chong JP, Hayashi MK, Simon MN, Xu RM, Stillman B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc Natl Acad Sci USA. 2000;97(4):1530–35. doi: 10.1073/pnas.030539597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Walters AD, Chong JP. An archaeal order with multiple minichromosome maintenance genes. Microbiology. 2010;156(Pt 5):1405–14. doi: 10.1099/mic.0.036707-0. [DOI] [PubMed] [Google Scholar]
- 233.Bae B, Chen YH, Costa A, Onesti S, Brunzelle JS, et al. Insights into the architecture of the replicative helicase from the structure of an archaeal MCM homolog. Structure. 2009;17(2):211–22. doi: 10.1016/j.str.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 234.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol. 2005;12(9):756–62. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 235.Costa A, Pape T, van Heel M, Brick P, Patwardhan A, Onesti S. Structural basis of the Methanothermobacter thermautotrophicus MCM helicase activity. Nucleic Acids Res. 2006;34(20):5829–38. doi: 10.1093/nar/gkl708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28(2):304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 237.Barry ER, Lovett JE, Costa A, Lea SM, Bell SD. Intersubunit allosteric communication mediated by a conserved loop in the MCM helicase. Proc Natl Acad Sci USA. 2009;106(4):1051–56. doi: 10.1073/pnas.0809192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Chen YJ, Yu X, Kasiviswanathan R, Shin JH, Kelman Z, Egelman EH. Structural polymorphism of Methanothermobacter thermautotrophicus MCM. J Mol Biol. 2005;346(2):389–94. doi: 10.1016/j.jmb.2004.11.076. [DOI] [PubMed] [Google Scholar]
- 239.Costa A, van Duinen G, Medagli B, Chong J, Sakakibara N, et al. Cryo-electron microscopy reveals a novel DNA-binding site on the MCM helicase. EMBO J. 2008;27(16):2250–58. doi: 10.1038/emboj.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Shin JH, Heo GY, Kelman Z. The Methanothermobacter thermautotrophicus Cdc6-2 protein, the putative helicase loader, dissociates the minichromosome maintenance helicase. J Bacteriol. 2008;190(11):4091–94. doi: 10.1128/JB.00233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kasiviswanathan R, Shin JH, Kelman Z. Interactions between the archaeal Cdc6 and MCM proteins modulate their biochemical properties. Nucleic Acids Res. 2005;33(15):4940–50. doi: 10.1093/nar/gki807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ogino H, Ishino S, Mayanagi K, Haugland GT, Birkeland NK, et al. The GINS complex from the thermophilic archaeon, Thermoplasma acidophilum may function as a homotetramer in DNA replication. Extremophiles. 2011;15(4):529–39. doi: 10.1007/s00792-011-0383-2. [DOI] [PubMed] [Google Scholar]
- 243.Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, Bell SD. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 2006;7(5):539–45. doi: 10.1038/sj.embor.7400649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yoshimochi T, Fujikane R, Kawanami M, Matsunaga F, Ishino Y. The GINS complex from Pyrococcus furiosus stimulates the MCM helicase activity. J Biol Chem. 2008;283(3):1601–9. doi: 10.1074/jbc.M707654200. [DOI] [PubMed] [Google Scholar]
- 245.Luijsterburg MS, White MF, van Driel R, Dame RT. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit Rev Biochem Mol Biol. 2008;43(6):393–418. doi: 10.1080/10409230802528488. [DOI] [PubMed] [Google Scholar]
- 246.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8(3):163–70. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 247.Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 1999;18(23):6642–52. doi: 10.1093/emboj/18.23.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sekimizu K, Kornberg A. Cardiolipin activation of dnaA protein, the initiation protein of replication in Escherichia coli. J Biol Chem. 1988;263(15):7131–35. [PubMed] [Google Scholar]
- 249.Fujimitsu K, Senriuchi T, Katayama T. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 2009;23(10):1221–33. doi: 10.1101/gad.1775809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Atlung T, Clausen ES, Hansen FG. Autoregulation of the dnaA gene of Escherichia coli K12. Mol Gen Genet. 1985;200(3):442–50. doi: 10.1007/BF00425729. [DOI] [PubMed] [Google Scholar]
- 251.Kitagawa R, Ozaki T, Moriya S, Ogawa T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 1998;12(19):3032–43. doi: 10.1101/gad.12.19.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Collier J, Shapiro L. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J Bacteriol. 2009;191(18):5706–16. doi: 10.1128/JB.00525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Nakamura K, Katayama T. Novel essential residues of Hda for interaction with DnaA in the regulatory inactivation of DnaA: unique roles for Hda AAA Box VI and VII motifs. Mol Microbiol. 2010;76(2):302–17. doi: 10.1111/j.1365-2958.2010.07074.x. [DOI] [PubMed] [Google Scholar]
- 254.Su’etsugu M, Shimuta TR, Ishida T, Kawakami H, Katayama T. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J Biol Chem. 2005;280(8):6528–36. doi: 10.1074/jbc.M412060200. [DOI] [PubMed] [Google Scholar]
- 255.Hayashi M, Ogura Y, Harry EJ, Ogasawara N, Moriya S. Bacillus subtilis YabA is involved in determining the timing and synchrony of replication initiation. FEMS Microbiol Lett. 2005;247(1):73–79. doi: 10.1016/j.femsle.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 256.Noirot-Gros MF, Dervyn E, Wu LJ, Mervelet P, Errington J, et al. An expanded view of bacterial DNA replication. Proc Natl Acad Sci USA. 2002;99(12):8342–47. doi: 10.1073/pnas.122040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Noirot-Gros MF, Velten M, Yoshimura M, McGovern S, Morimoto T, et al. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc Natl Acad Sci USA. 2006;103(7):2368–73. doi: 10.1073/pnas.0506914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Keyamura K, Fujikawa N, Ishida T, Ozaki S, Su’etsugu M, et al. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 2007;21(16):2083–99. doi: 10.1101/gad.1561207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Natrajan G, Noirot-Gros MF, Zawilak-Pawlik A, Kapp U, Terradot L. The structure of a DnaA/HobA complex from Helicobacter pylori provides insight into regulation of DNA replication in bacteria. Proc Natl Acad Sci USA. 2009;106(50):21115–20. doi: 10.1073/pnas.0908966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Keyamura K, Abe Y, Higashi M, Ueda T, Katayama T. DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J Biol Chem. 2009;284(37):25038–50. doi: 10.1074/jbc.M109.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Chodavarapu S, Felczak MM, Kaguni JM. Two forms of ribosomal protein L2 of Escherichia coli that inhibit DnaA in DNA replication. Nucleic Acids Res. 2011;39(10):4180–91. doi: 10.1093/nar/gkq1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Rahn-Lee L, Gorbatyuk B, Skovgaard O, Losick R. The conserved sporulation protein YneE inhibits DNA replication in Bacillus subtilis. J Bacteriol. 2009;191(11):3736–39. doi: 10.1128/JB.00216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wagner JK, Marquis KA, Rudner DZ. SirA enforces diploidy by inhibiting the replication initiator DnaA during spore formation in Bacillus subtilis. Mol Microbiol. 2009;73(5):963–74. doi: 10.1111/j.1365-2958.2009.06825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135(1):74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 265.Araki H. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol. 2010;22(6):766–71. doi: 10.1016/j.ceb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 266.Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14(18):R778–86. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 267.Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24(12):1208–19. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Randell JC, Fan A, Chan C, Francis LI, Heller RC, et al. Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell. 2010;40(3):353–63. doi: 10.1016/j.molcel.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Hardy CF, Dryga O, Seematter S, Pahl PMB, Sclafani RA. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci USA. 1997;94(7):3151–55. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463(7277):113–17. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445(7125):281–85. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 272.Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445(7125):328–32. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 273.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8(4):358–66. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 274.van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012;31(9):2195–206. doi: 10.1038/emboj.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 2012;31(9):2182–94. doi: 10.1038/emboj.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair. 2005;4(11):1227–39. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 277.Sanchez-Pulido L, Diffley JF, Ponting CP. Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr Biol. 2010;20(12):R509–10. doi: 10.1016/j.cub.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 278.Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, et al. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121(6):887–98. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 279.Balestrini A, Consentino C, Errico A, Garner E, Costanzo V. GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat Cell Biol. 2010;12(5):484–91. doi: 10.1038/ncb2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Arias EE, Walter JC. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 2005;19(1):114–26. doi: 10.1101/gad.1255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2006;8(1):84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- 282.Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290(5500):2309–12. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 283.Maiorano D, Lutzmann M, Mechali M. MCM proteins and DNA replication. Curr Opin Cell Biol. 2006;18(2):130–36. doi: 10.1016/j.ceb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 284.Lee C, Hong B, Choi JM, Kim Y, Watanabe S, et al. Structural basis for inhibition of the replication licensing factor Cdt1 by geminin. Nature. 2004;430(7002):913–17. doi: 10.1038/nature02813. [DOI] [PubMed] [Google Scholar]
- 285.De Marco V, Gillespie PJ, Li A, Karantzelis N, Christodoulou E, et al. Quaternary structure of the human Cdt1-Geminin complex regulates DNA replication licensing. Proc Natl Acad Sci USA. 2009;106(47):19807–12. doi: 10.1073/pnas.0905281106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Braun KA, Breeden LL. Nascent transcription of MCM2-7 is important for nuclear localization of the minichromosome maintenance complex in G1. Mol Biol Cell. 2007;18(4):1447–56. doi: 10.1091/mbc.E06-09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14(15):3788–99. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Labib K, Diffley JF, Kearsey SE. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat Cell Biol. 1999;1(7):415–22. doi: 10.1038/15649. [DOI] [PubMed] [Google Scholar]
- 289.Tanaka S, Diffley JF. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat Cell Biol. 2002;4(3):198–207. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- 290.Liku ME, Nguyen VQ, Rosales AW, Irie K, Li JJ. CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2-7 complex prevents chromosomal rereplication. Mol Biol Cell. 2005;16(10):5026–39. doi: 10.1091/mbc.E05-05-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Diffley JF. Quality control in the initiation of eukaryotic DNA replication. Philos Trans R Soc Lond B. 2011;366(1584):3545–53. doi: 10.1098/rstb.2011.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Li CJ, Vassilev A, DePamphilis ML. Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex’s largest subunit (Orc1) from binding to chromatin during mitosis. Mol Cell Biol. 2004;24(13):5875–86. doi: 10.1128/MCB.24.13.5875-5886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Mahbubani HM, Chong JP, Chevalier S, Thömmes P, Blow JJ. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J Cell Biol. 1997;136(1):125–35. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Tada S, Li A, Maiorano D, Méchali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3(2):107–13. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Lundgren M, Bernander R. Genome-wide transcription map of an archaeal cell cycle. Proc Natl Acad Sci USA. 2007;104(8):2939–44. doi: 10.1073/pnas.0611333104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Ozaki S, Katayama T. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012;40(4):1648–65. doi: 10.1093/nar/gkr832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


