Abstract
Background
Differential Shannon entropy (DSE) and differential coefficient of variation (DCV) are effective metrics for the study of gene expression data. They can serve to augment differential expression (DE), and be applied in numerous settings whenever one seeks to measure differences in variability rather than mere differences in magnitude. A general purpose, easily accessible tool for DSE and DCV would help make these two metrics available to data scientists. Automated p value computations would additionally be useful, and are often easier to interpret than raw test statistic values alone.
Results
EntropyExplorer is an R package for calculating DSE, DCV and DE. It also computes corresponding p values for each metric. All features are available through a single R function call. Based on extensive investigations in the literature, the Fligner-Killeen test was chosen to compute DCV p values. No standard method was found to be appropriate for DSE, and so permutation testing is used to calculate DSE p values.
Conclusions
EntropyExplorer provides a convenient resource for calculating DSE, DCV, DE and associated p values. The package, along with its source code and reference manual, are freely available from the CRAN public repository at http://cran.r-project.org/web/packages/EntropyExplorer/index.html.
Electronic supplementary material
The online version of this article (doi:10.1186/s13104-015-1786-4) contains supplementary material, which is available to authorized users.
Keywords: Differential Shannon entropy, Differential coefficient of variation, Differential expression, Statistical tests
Background
Shannon entropy (SE) and coefficient of variation (CV) are used to measure the variability or dispersion of numerical data. Such variability has potential utility in numerous application domains, perhaps most notably in the analysis of high throughput biological data. Variability has been applied, for example, to study gene expression data in the context of human disease [1]. Increased entropy in particular, in both gene expression and protein interaction data, has been observed to be a characteristic of cancer [2]. Numerous other examples typify the utility of entropy [3–8] and coefficient of variation [9–12].
Shannon entropy is famously rooted in information theory [13]. To avoid confusion, we emphasize that we use the term “differential entropy” to denote a difference between two Shannon entropy values. This is distinct from information-theoretic terminology, in which “differential entropy” often means the entropy of a continuous, rather than a discrete, random variable [14].
We are particularly interested in differential analysis. In [15], we studied differential Shannon entropy (DSE) and differential coefficient of variation (DCV), and found them highly effective in identifying genes of potential interest not found by differential expression (DE) alone. DSE and DCV are applicable to other types of biological data as well, such as that produced by RNA-Seq technologies, although the usual caveats about careful interpretation apply. The usefulness of DSE and DCV is of course not limited to biological data. They may be applied to any numerical data for which normalized measures of differential variability are relevant.
Implementation
EntropyExplorer is implemented in R [16]. All features are wrapped into a single function call, which takes as input up to eight arguments. Two of these arguments are numerical matrices, with identical labels for each row. The output is a matrix with two, three or five columns that contains in each row two SE, CV or mean values; a DSE, DCV or DE value; and/or two p values, one raw and one adjusted. Output rows can be sorted by value, raw p value or adjusted p value, and can be filtered to show only the top-ranked rows.
Permutation testing for DSE is accomplished with the help of the R function sample.int. The default number of tests to be employed is set to 1000, which the user can override. The p value for DCV is calculated by applying the Fligner-Killeen test for homogeneity of variances, implemented via the R function fligner.test, to the log-transform of the input data. The R function t.test is used to find a p value for DE. Adjusted p values are calculated using the p.adjust function in R, which provides false discovery rate and multiple testing corrections. A more thorough explanation of p value calculations is provided in the discussion section.
EntropyExplorer checks that all matrix entries are positive. This is because calculations of a DSE value/p value and a DCV p value involve taking logarithms, which are undefined on data containing zeros or negative values. Also, CV becomes less meaningful when means approach zero or are negative. Experimental data may be noisy, however, and so EntropyExplorer provides mechanisms to handle non-positive values. An optional two-value argument permits the user to add a positive bias to all elements of one or both matrices prior to performing any other calculations. The argument can also be set to make this adjustment automatically, based on the least non-positive value in each matrix.
Metrics
Let represent a list of n positive numbers, and let denote their sum. The Shannon entropy of this list is
The coefficient of variation is
where is the sample mean and is the sample standard deviation. Given two such lists of positive numbers with Shannon entropies and , coefficients of variation and , and means and , , , and .
Shannon entropy falls in the range [0, 1]; DSE therefore also falls in the range [0, 1]. Lower (higher) SE corresponds to more (less) variability. CV falls in the range [0, ∞); DCV therefore also has a range of [0, ∞).
Application
EntropyExplorer is invoked as follows:
EntropyExplorer(expm1, expm2, dmetric, otype, ntop, nperm, shift, padjustmethod)
We refer the reader to the reference manual, included as Additional file 1 and available on the project webpage, for a detailed description of all arguments and options. Included with the package is a sample mRNA microarray dataset, consisting of a few rows from a dataset obtained from the Gene Expression Omnibus (GEO) [17]. This dataset, GSE10810, contains case/control data on breast cancer [18]. Figures 1 and 2 provide example uses of EntropyExplorer on the full data.
Fig. 1.
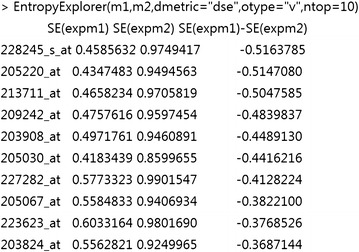
The output of EntropyExplorer on breast cancer data. The numerical matrices m1 and m2 have been read into R. The function call has specified “dse” for differential Shannon entropy, “v” for value, and 10 to return the top 10 values
Fig. 2.
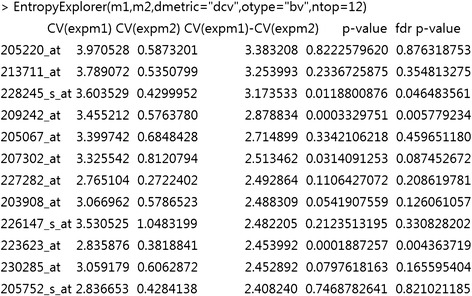
Another use of EntropyExplorer on breast cancer data. The function call has specified “dcv” for differential coefficient of variation, “bv” to specify both value and p value, and to sort by value, and 12 to return the top 12 rows
Discussion
In addition to calculating DSE, DCV and DE, EntropyExplorer can calculate both raw and adjusted p values for each. ANOVA-based tests are the standard way to obtain differential expression p values. We therefore use a t-test for this purpose. Certainly more sophisticated methods exist. See, for example, [19, 20]. Thus, we emphasize that EntropyExplorer includes DE only as a simple, convenient and straightforward point of comparison with the other two metrics. For DCV p values, we observe that 11 tests of equal relative variation were compared in [21], with the conclusion that the Fligner-Killeen test [22] is usually the most appropriate. It strikes a balance between type I and type II errors, and is robust to non-normal distributions.
Obtaining reliable p values for DSE proved much more challenging. We found no known method in the literature specific to DSE p values. We therefore investigated the extent to which SE is correlated to variance. A high correlation would suggest that they may be proxies for each other, in which case the p value of an F-test or some derivation thereof might serve as suitable estimate of the DSE p value. Unfortunately, correlations between SE and variance, or between SE and a function of variance, were not high enough to justify using one as a surrogate for the other. Table 1 shows the correlation between SE and variance V, and between SE and the function as an attempt to linearize the relationship, using the 16 datasets from [15]. The only notably high correlation is found in the obesity dataset. The obesity data, however, contains a large number of missing values, rendering the high correlation less reliable. We conclude that standard statistical tests related to variance do not appear suitable for testing DSE.
Table 1.
Correlations between SE and variance, and between SE and , on 16 microarray gene expression datasets
| Datasets | Correlation Between SE and Variance | Correlation between SE and | ||
|---|---|---|---|---|
| Case | Control | Case | Control | |
| Allergic Rhinitis | −0.5515 | −0.5769 | −0.9703 | −0.9658 |
| Asthma_GSE4302 | −0.4272 | −0.4677 | −0.1924 | −0.2004 |
| BreastCancer_GSE10810 | −0.3942 | −0.3378 | −0.1810 | −0.1265 |
| CLL_GSE8835 | 0.2251 | 0.2522 | −0.0806 | −0.0624 |
| ColorectalCancer_GSE9348 | 0.3122 | 0.4454 | −0.0086 | 0.0206 |
| CrohnsDisease_GSE6731 | −0.2826 | −0.2380 | −0.1664 | −0.4020 |
| LungAdenocarcinoma_GSE7670 | 0.0725 | 0.3360 | −0.0173 | 0.0105 |
| MS_GDS3920 | −0.3615 | −0.3320 | −0.0515 | −0.0559 |
| Obesity_GSE12050 | 0.9998 | 0.9990 | 0.1584 | 0.5420 |
| Pancreas_GDS4102 | −0.4137 | −0.4455 | −0.1331 | −0.0890 |
| ParkinsonsDisease_GSE20141 | −0.1732 | −0.2554 | −0.0024 | −0.0155 |
| ProstateCancer_GSE6919_GPL8300 | 0.2118 | 0.1552 | −0.0562 | −0.0699 |
| Psoriasis_GSE13355 | −0.6386 | −0.6554 | −0.5200 | −0.6779 |
| Schizophrenia_GSE17612 | 0.3632 | 0.3910 | 0.0170 | 0.0235 |
| T2D_GSE20966 | −0.6006 | −0.5550 | −0.4356 | −0.4663 |
| UlcerativeColitis_GSE6731 | −0.3112 | −0.2555 | −0.1799 | −0.1451 |
We also examined the distribution of DSE on the 16 datasets, with the goal of empirically determining a suitable reference distribution for DSE. From this, we could then estimate p values analytically. We applied the Kolmogorov–Smirnov (KS) test to compare the DSE distribution of each dataset to some of the more common reference distributions, such as normal, F, t, and Chi square. When performing a KS test, p values can be overly sensitive to deviations from the reference distribution [23], so a D-statistic value below 0.1 was used to identify matching distributions. In our experiments, only the Parkinson’s dataset produced a D-statistic below 0.1 when tested against a normal or standardized t distribution (Table 2). Figure 3 shows a sample distribution of DSE, in this case using prostate cancer data.
Table 2.
KS test D-statistic results comparing the DSE distribution against several common distributions
| Dataset | Distribution | ||||
|---|---|---|---|---|---|
| Normal | Chi-square | F | t | t (standardized DSE)* | |
| Allergic Rhinitis | 0.3109 | 1 | 1 | 0.4991 | 0.3526 |
| Asthma_GSE4302 | 0.2795 | 1 | 1 | 0.4895 | 0.3117 |
| BreastCancer_GSE10810 | 0.2115 | 1 | 1 | 0.4797 | 0.3944 |
| CLL_GSE8835 | 0.1506 | 1 | 0.9975 | 0.4519 | 0.1596 |
| ColorectalCancer_GSE9348 | 0.1232 | 1 | 0.9994 | 0.4514 | 0.2142 |
| CrohnsDisease_GSE6731 | 0.2131 | 1 | 0.987 | 0.4691 | 0.2392 |
| LungAdenocarcinoma_GSE7670 | 0.19 | 1 | 0.9999 | 0.4663 | 0.332 |
| MS_GDS3920 | 0.2703 | 1 | 0.9994 | 0.4813 | 0.3397 |
| Obesity_GSE12050 | 0.2352 | 1 | 0.9991 | 0.484 | 0.287 |
| Pancreas_GDS4102 | 0.2606 | 1 | 0.9937 | 0.4532 | 0.3254 |
| ParkinsonsDisease_GSE20141 | 0.0628 | 1 | 0.9361 | 0.3816 | 0.0582 |
| ProstateCancer_GSE6919_GPL8300 | 0.1575 | 1 | 1 | 0.4739 | 0.2522 |
| Psoriasis_GSE13355 | 0.3327 | 1 | 0.9999 | 0.4932 | 0.4195 |
| Schizophrenia_GSE17612 | 0.183 | 1 | 0.9998 | 0.4705 | 0.2138 |
| T2D_GSE20966 | 0.3271 | 1 | 0.9999 | 0.4936 | 0.3562 |
| UlcerativeColitis_GSE6731 | 0.2397 | 1 | 0.998 | 0.4831 | 0.3608 |
* The last column shows the results after first standardizing DSE by dividing each DSE by the standard deviation of all DSEs
Fig. 3.
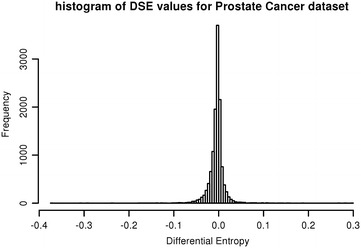
The distribution of differential Shannon entropy. The observed distribution of differential Shannon entropy in sample prostate cancer data is shown. Similar patterns were seen in all 16 data sets. None of the standard distributions tested matched the observed distributions closely enough to be considered as a reference distribution for obtaining p values
We conclude from this that none of the distributions tested are close enough approximations to the observed DSE distribution to be used as a proxy for obtaining p values. Thus, without a known distribution function or suitable surrogate, we resort to resampling in order to obtain reliable DSE p values. While computationally demanding, the following permutation test makes no assumptions about the underlying distribution of the data. Given two lists of numbers, containing n1 and n2 numerical elements respectively, we first calculate their DSE and then create a new list A containing all numbers from the two lists. Next we randomly permute the elements of A, then recalculate DSE, treating the first elements of A as one list and the last elements of A as a second list. The resultant p value is simply the proportion of all recalculated DSEs that are at least as extreme as the original DSE.
In addition to raw p values, EntropyExplorer also calculates p values adjusted for multiple testing. A user can choose to adjust based on FDR, Holm or another multiple-testing adjustment.
Conclusions
We have produced EntropyExplorer, an R package for calculating differential Shannon entropy, differential coefficient of variation and differential expression. This package also calculates raw and adjusted p values for each metric. These measures have been shown to complement one another [15], making this package an effective tool for users in search of more expansive suites of differential analysis methods.
Availability and requirements
Project name: EntropyExplorer.
Project home page: http://cran.r-project.org/web/packages/EntropyExplorer/index.html.
Operating system(s): Platform independent.
Programming language: R.
Other requirements: R version 3.0 or later is recommended.
License: GNU General Public License version 3.0 (GPLv3).
Any restrictions to use by non-academics: None.
Additional availability: EntropyExplorer is integrated into the GrAPPA toolkit at http://grappa.eecs.utk.edu/.
Authors’ contributions
KW implemented the package and performed numerous analytical tests. CAP led exhaustive software evaluations and isolated relevant data. AMS provided statistical expertise and assisted with human factors engineering. MAL directed the project and provided for its support. All authors participated in writing the paper. All authors read and approved the final manuscript.
Acknowledgements
This work has been supported in part by the National Institutes of Health under awards R01-AA-018776 and 3P20MD000516-07S1.
Competing interests
The authors declare that they have no competing interests.
Additional file
10.1186/s13104-015-1786-4 EntropyExplorer R package reference manual.
Contributor Information
Kai Wang, Email: kwang11@tennessee.edu.
Charles A. Phillips, Email: cphill25@tennessee.edu
Arnold M. Saxton, Email: asaxton@tennessee.edu
Michael A. Langston, Email: langston@tennessee.edu
References
- 1.Ho JW, Stefani M, dos Remedios CG, Charleston MA. Differential variability analysis of gene expression and its application to human diseases. Bioinformatics. 2008;24(13):i390–i398. doi: 10.1093/bioinformatics/btn142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.West J, Bianconi G, Severini S, Teschendorff AE. Differential network entropy reveals cancer system hallmarks. Scientific reports. 2012;2:802. doi: 10.1038/srep00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berretta R, Moscato P. Cancer biomarker discovery: the entropic hallmark. PLoS One. 2010;5(8):e12262. doi: 10.1371/journal.pone.0012262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherwin WB. Entropy and information approaches to genetic diversity and its expression: genomic geography. Entropy. 2010;12(7):1765–1798. doi: 10.3390/e12071765. [DOI] [Google Scholar]
- 5.Masisi L, Nelwamondo F, Marwala T. The use of entropy to measure structural diversity. In: IEEE 6th International Conference on Computational Cybernetics. Stará Lesná, Slovakia.
- 6.Chen B-S, Li C-W. On the interplay between entropy and robustness of gene regulatory networks. Entropy. 2010;12(5):1071–1101. doi: 10.3390/e12051071. [DOI] [Google Scholar]
- 7.Furlanello C, Serafini M, Merler S, Jurman G. Entropy-based gene ranking without selection bias for the predictive classification of microarray data. BMC Bioinform. 2003;4:54. doi: 10.1186/1471-2105-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohane IS, Kho AT, Butte AJ. Microarrays for an integrative genomics. Cambridge, Mass.: MIT Press; 2003.
- 9.Reed GF, Lynn F, Meade BD. Use of coefficient of variation in assessing variability of quantitative assays. Clin Diagn Lab Immunol. 2002;9(6):1235–1239. doi: 10.1128/CDLI.9.6.1235-1239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber EU, Shafir S, Blais AR. Predicting risk sensitivity in humans and lower animals: risk as variance or coefficient of variation. Psychol Rev. 2004;111(2):430–445. doi: 10.1037/0033-295X.111.2.430. [DOI] [PubMed] [Google Scholar]
- 11.Faber DS, Korn H. Applicability of the coefficient of variation method for analyzing synaptic plasticity. Biophys J. 1991;60(5):1288–1294. doi: 10.1016/S0006-3495(91)82162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedeian AG, Mossholder KW. On the use of the coefficient of variation as a measure of diversity. Organ Res Methods. 2000;3(3):285–297. doi: 10.1177/109442810033005. [DOI] [Google Scholar]
- 13.Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27(3):45. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 14.Cover TM, Thomas JA. Elements of information theory. 2. Hoboken: Wiley-Interscience; 2006. [Google Scholar]
- 15.Wang K, Phillips CA, Rogers GL, Barrenas F, Benson M, Langston MA. Differential Shannon entropy and differential coefficient of variation: alternatives and augmentations to differential expression in the search for disease-related genes. Int J Comput Biol Drug Des. 2014;7(2–3):183–194. doi: 10.1504/IJCBDD.2014.061656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R Core Team. R: A language and environment for statistical computing. In: R Foundation for Statistical Computing. Vienna, Austria; 2014.
- 17.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedraza V, Gomez-Capilla JA, Escaramis G, Gomez C, Torne P, Rivera JM, Gil A, Araque P, Olea N, Estivill X, et al. Gene expression signatures in breast cancer distinguish phenotype characteristics, histologic subtypes, and tumor invasiveness. Cancer. 2010;116(2):486–496. doi: 10.1002/cncr.24805. [DOI] [PubMed] [Google Scholar]
- 19.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donnelly SM, Kramer A. Testing for multiple species in fossil samples: an evaluation and comparison of tests for equal relative variation. Am J Phys Anthropol. 1999;108(4):507–529. doi: 10.1002/(SICI)1096-8644(199904)108:4<507::AID-AJPA8>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 22.Fligner MA, Killeen TJ. Distribution-free two-sample tests for scale. J Am Stat Assoc. 1976;71(353):210–213. doi: 10.1080/01621459.1976.10481517. [DOI] [Google Scholar]
- 23.Ghasemi A, Zahediasl S. Normality tests for statistical analysis: a guide for non-statisticians. Int J Endocrinol Metab. 2012;10(2):486–489. doi: 10.5812/ijem.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]


