ABSTRACT
Cadherin-11 (Cad11) cell adhesion molecule plays a role in prostate cancer cell migration. Because disassembly of adhesion complexes through endocytosis of adhesion proteins has been shown to play a role in cell migration, we examined whether Cad11 endocytosis plays a role in Cad11-mediated migration. The mechanism by which Cad11 is internalized is unknown. Using a GST pulldown assay, we found that clathrin binds to the Cad11 cytoplasmic domain but not to that of E-cadherin. Using deletion analysis, we identified a unique sequence motif, VFEEE, in the Cad11 membrane proximal region (amino acid residues 11–15) that binds to clathrin. Endocytosis assays using K+-depletion buffer showed that Cad11 internalization is clathrin dependent. Proximity ligation assays showed that Cad11 colocalizes with clathrin, and immunofluorescence assays showed that Cad11 localizes in vesicles that stain for the early endosomal marker Rab5. Deletion of the VFEEE sequence from the Cad11 cytoplasmic domain (Cad11-cla-Δ5) leads to inhibition of Cad11 internalization and reduces Cad11-mediated cell migration in C4-2B and PC3-mm2 prostate cancer cells. These observations suggest that clathrin-mediated internalization of Cad11 regulates surface trafficking of Cad11 and that dynamic turnover of Cad11 regulates the migratory function of Cad11 in prostate cancer cells.
KEY WORDS: Cadherin-11, Clathrin, Endocytosis, Migration, Prostate cancer
Summary: These findings show that recycling of cell adhesion molecules through endocytosis promotes their ability to control cell migration.
INTRODUCTION
The cadherin family adhesion molecules play important roles in cell–cell communication and cell sorting during development (Takeichi, 1990). In addition to mediating homophilic adhesion, cadherins have been shown to mediate cell polarity, junctional formation and cell migration (Gumbiner, 1996). The dynamic regulation of the surface levels of cadherin proteins is crucial for the coordination of these complex cellular functions.
Cadherins constitute a family of proteins (Hulpiau and van Roy, 2009). Different cadherins are expressed at various stages of development to mediate specific cellular activity (Angst et al., 2001). E-cadherin (E-Cad; also known as CDH1) is mainly expressed in epithelial cells and plays a role in maintaining cellular contact in polarized epithelial cells (Nelson et al., 2013). Cadherin-11 (Cad11; also known as osteoblast cadherin), by contrast, is a mesenchymal cadherin that is mainly expressed in osteoblasts (Cheng et al., 1998; Marie et al., 2014; Okazaki et al., 1994).
Aberrant expression of cell adhesion molecules occurs during cancer progression. Although E-Cad, which plays a role in maintaining cell polarity (Nelson et al., 2013), has been found to be downregulated in cancers, aberrant overexpression of mesenchymal cadherins, including N-cadherin (N-Cad; also known as CDH2) and Cad11 (Kosalková et al., 2015; Padmanabhan and Taneyhill, 2015; Tamura et al., 2008), are observed in some cancers. Such a cadherin switch has been shown to occur during the progression of prostate (Chu et al., 2008; Huang et al., 2010; Lee et al., 2010), breast (Tamura et al., 2008) and pancreatic (Martinez-Contreras et al., 2015; Xiumin et al., 2015) cancers. We have found previously that Cad11 expression in prostate cancer plays a role in the metastasis to bone, in part, through increased adhesion of prostate cancer cells to osteoblasts (Chu et al., 2008; Huang et al., 2010; Lee et al., 2013). A similar role of Cad11 has been observed in breast cancer bone metastasis (Tamura et al., 2008). We have also shown that expression of Cad11 increases the migration and invasion of prostate cancer cells (Huang et al., 2010). Interestingly, Cad11 expression is upregulated in castration-resistant prostate cancer (Lee et al., 2010), which frequently leads to prostate cancer metastasis in bone. These observations suggest that Cad11 might also be involved in prostate cancer bone metastasis by increasing cell migration. Taken together, these studies suggest that Cad11 plays important roles during both normal development and pathological conditions.
In migrating cells, the assembly and disassembly of adhesion complexes allow the cells to migrate (Broussard et al., 2008; Webb et al., 2002). Endocytosis of adhesion proteins has been shown to play a role in the disassembly of adhesion complexes (Alarcos et al., 2015; Bockus et al., 2015; Jin et al., 2015; Kowalczyk and Nanes, 2012; Mosesson et al., 2008; Xiao et al., 2005); however, the mechanism by which Cad11 is internalized is unknown. In this study, we identified proteins that interact with the cytoplasmic domain of Cad11 and found that clathrin is one of the Cad11-interacting proteins. We found that clathrin binds to the cytoplasmic domain of Cad11 but not to that of E-Cad, suggesting that there are differences in the regulatory mechanisms between these two cadherins. Our studies show that clathrin plays a role in Cad11 endocytosis and that the dynamic turnover of Cad11 promotes Cad11-mediated migration in prostate cancer cells.
RESULTS
Clathrin binds to the cytoplasmic domain of Cad11 but not to that of E-Cad
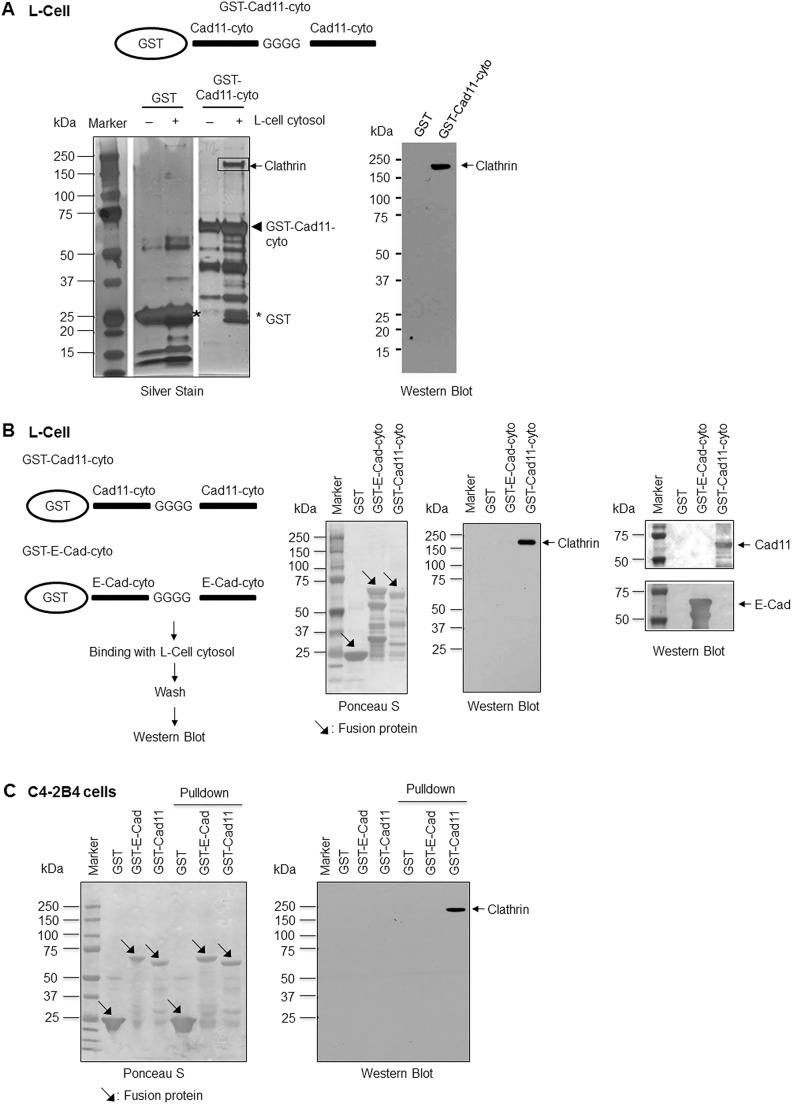
To search for proteins that interact with the cytoplasmic domain of Cad11, we used a GST fusion protein containing the Cad11 cytoplasmic (Cad11-cyto) domain in a GST pulldown assay. We constructed GST–Cad11-cyto, which contained two copies of the cytoplasmic domain in tandem to increase the likelihood of binding to interacting proteins (Fig. 1A, upper panel). To avoid the disruption of protein conformation of the individual cytoplasmic domain, four glycine residues were incorporated as a spacer in between the two copies of the cytoplasmic domain (Fig. 1A, upper panel). Recombinant GST–Cad11-cyto protein was used to pull down proteins from the cytosol of L-cells. Silver staining of an SDS-PAGE gel revealed a prominent band at approximately 180 kDa in the GST–Cad11-cyto, but not GST alone, samples (Fig. 1A, lower left panel). This protein was excised from the gel and identified as being clathrin by using mass spectrometry. The low molecular mass proteins in the GST–Cad11-cyto pulldown were found to be degradation products of GST–Cad11-cyto (data not shown). Immunoblotting using an antibody against clathrin further confirmed that the 180-kDa protein was clathrin (Fig. 1A, right panel). These results suggest that clathrin is one of the proteins that interact with the cytoplasmic domain of Cad11.
Fig. 1.
Clathrin binds to the Cad11 cytoplasmic domain. (A) A GST fusion protein containing two copies of the Cad11 cytoplasmic domain separated by four glycine residues was used in a pulldown assay (schematic). GST–Cad11-cyto-conjugated agarose or GST–agarose was incubated with or without the L-cell cytosolic fraction, and the proteins were separated by using SDS-PAGE and visualized with silver stain (left panel). The prominent protein at 180 kDa was identified to be clathrin by using mass spectrometry. Western blot showed that the 180 kDa protein pulled down by GST–Cad11-cyto agarose reacted with an antibody against clathrin (right panel). (B) Clathrin binds to the Cad11 cytoplasmic domain but not that of E-Cad. GST fusion proteins containing two copies of E-Cad (GST–E-Cad-cyto) or Cad11 (GST–Cad11-cyto) were incubated with L-cell cytosol in GST pulldown assays. The proteins were separated by using SDS-PAGE, transferred onto nitrocellulose membranes and visualized with Ponceau S. The membranes were immunoblotted with antibodies against clathrin, Cad11 or E-Cad. (C) The Cad11 cytoplasmic domain (GST–Cad11; construct GST–Cad11-cyto) but not that of E-Cad (GST–E-Cad; construct GST–E-Cad-cyto) pulled down clathrin in C4-2B4 cells. GST fusion proteins containing two copies of the E-cadherin or Cad11 were incubated with C4-2B4 cell cytosol in GST pulldown assays. The proteins were separated by SDS-PAGE, transferred onto nitrocellulose membranes and visualized with Ponceau S. The membranes were then immunoblotted with an antibody against clathrin.
Next, we examined whether clathrin also binds to the cytoplasmic domain of E-Cad. The cytoplasmic domain of E-Cad was expressed as a GST fusion protein (GST–E-Cad-cyto) using a strategy similar to that used with GST–Cad11-cyto (Fig. 1B, left panel). GST–E-Cad-cyto did not pull down clathrin from the L-cell cytosol, whereas GST–Cad11-cyto did (Fig. 1B, right panel). Immunoblotting using an antibody against the cytoplasmic domain of E-Cad or Cad11 confirmed the specificity of the GST–E-Cad-cyto and GST–Cad11-cyto constructs used in the pulldown assays (Fig. 1B, right panel). These observations suggest that clathrin binds to the cytoplasmic domain of Cad11 but not that of E-Cad under our experimental conditions. Similar results were obtained by using cell lysates that had been prepared from C4-2B4 cells (Fig. 1C), a cell line derived from the LNCaP prostate cancer cell line (Thalmann et al., 2000).
Clathrin binds to the juxtamembrane region of Cad11
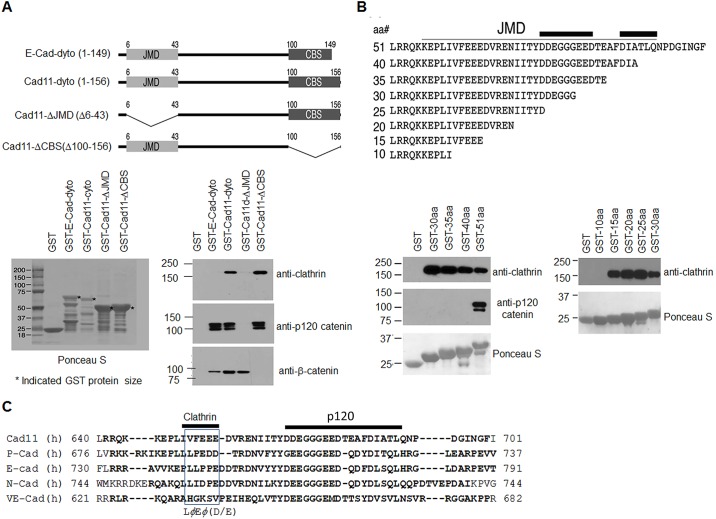
To map the clathrin-binding site on Cad11, we generated Cad11-cyto mutants that contained deletions in either the juxtamembrane domain (JMD) (GST–Cad11-ΔJMD) or the β-catenin-binding subdomain (CBS; β-catenin is also known as CTNNB1) (GST–Cad11-ΔCBS) (Fig. 2A, upper panel), and used them in pulldown assays. We found that clathrin bound to Cad11-cyto and Cad11-ΔCBS but not to Cad11-ΔJMD or E-Cad-cyto, suggesting that the clathrin-binding site is located within the JMD domain (Fig. 2A, lower right panel). It has previously been reported that p120-catenin (also known as CTNND1) binds to the JMD domain, whereas β-catenin binds to the CBS domain of cadherins (Nanes et al., 2012). As expected, p120-catenin was found to bind to the cytoplasmic domains of both E-Cad and Cad11, and also to Cad11-ΔCBS, but not to ΔJMD (Fig. 2A, lower right panel). β-catenin was found to bind to the cytoplasmic domains of E-Cad and Cad11, and also to Cad11-ΔJMD, but not to Cad11-ΔCBS (Fig. 2A, lower right panel). These results show that the clathrin-binding site is located within the JMD domain.
Fig. 2.
Clathrin binds to the juxtamembrane region of the Cad11 cytoplasmic domain. (A) Deletion constructs of the Cad11 cytoplasmic domain (top). Ponceau S staining of GST and the GST fusion proteins E-Cad-cyto, Cad11-cyto or Cad11-cyto with deletions in the juxtamembrane domain (Cad11-ΔJMD) or in the β-catenin binding domain (Cad11-ΔCBS) (lower left). Western blot with antibodies against clathrin, p120-catenin or β-catenin of the GST-pulldown products (lower right). (B) Sequences of Cad11 JMD-domain deletion mutants. Sequences associated with binding to p120-catenin in E-Cad are marked with bold lines. GST pulldown assays using GST and GST–Cad11-cyto proteins containing the N-terminal 51, 40, 35, 30, 25, 20, 15 or 10 amino acids of the Cad11 cytoplasmic domain (Ponceau S) were analyzed by immunoblotting with antibodies against clathrin or p120-catenin. p120-catenin only binds to the GST–Cad11-cyto51aa (GST-51aa) fusion protein, whereas clathrin binds to all GST–Cad11-cyto fusion proteins, except GST–Cad11-cyto10aa (GST-10aa). (C) Sequence alignment of the cytoplasmic domains of human (h) Cad11, E-Cad, P-Cad, N-Cad and VE-cadherin. The amino acid sequences that are essential for binding of Cad11-cyto to clathrin or p120-catenin (p120) are marked with thick lines. Blue indicates the clathrin-binding motif.
To further identify the clathrin-binding sequence within the JMD domain, we first generated GST fusion proteins containing the N-terminal 51, 40, 35 and 30 amino acid residues of the Cad11 cytoplasmic domain (Fig. 2B, upper panel). Clathrin was found to bind to all four deletion mutants, whereas p120-catenin only bound to the deletion mutant that contained the first 51 amino acids (Fig. 2B, lower left panel). These observations suggest that the sequence between amino acid residues 40 and 51 of Cad11 contains a motif that is crucial for the binding of p120-catenin. This observation is consistent with a previous report by Thoreson et al. (2000), showing that p120-catenin binds to amino acid residues 756–774 of the E-Cad cytoplasmic domain (Fig. 2B, upper panel, thick black lines). We further generated GST fusion proteins containing the N-terminal 25, 20, 15 and 10 amino acids of the Cad11 cytoplasmic domain (Fig. 2B, upper panel). We found that clathrin bound to all of these proteins except to the GST–Cad11-10aa construct (Fig. 2B, lower right panel). The difference in the amino acid sequences between the Cad11-10aa and Cad11-15aa constructs is VFEEE. Interestingly, this sequence is similar to the clathrin-binding box [LxEx(D/E)], which is located at the C-terminal tail of arrestin (Krupnick et al., 1997; Macedo et al., 2014). Sequence alignment analysis showed that the VFEEE sequence is unique to Cad11 and is absent in the corresponding region of E-Cad (Fig. 2C), consistent with the lack of clathrin binding to the E-Cad cytoplasmic domain (Fig. 1B). Sequence alignment of the cadherin cytoplasmic domains showed that the LxEx(D/E) motif might also be present in P-cadherin (P-Cad) and N-Cad, but whether clathrin binds to the cytoplasmic domain of P-Cad or N-Cad has not been determined.
Cad11 is internalized through a clathrin-dependent pathway in C4-2B cells
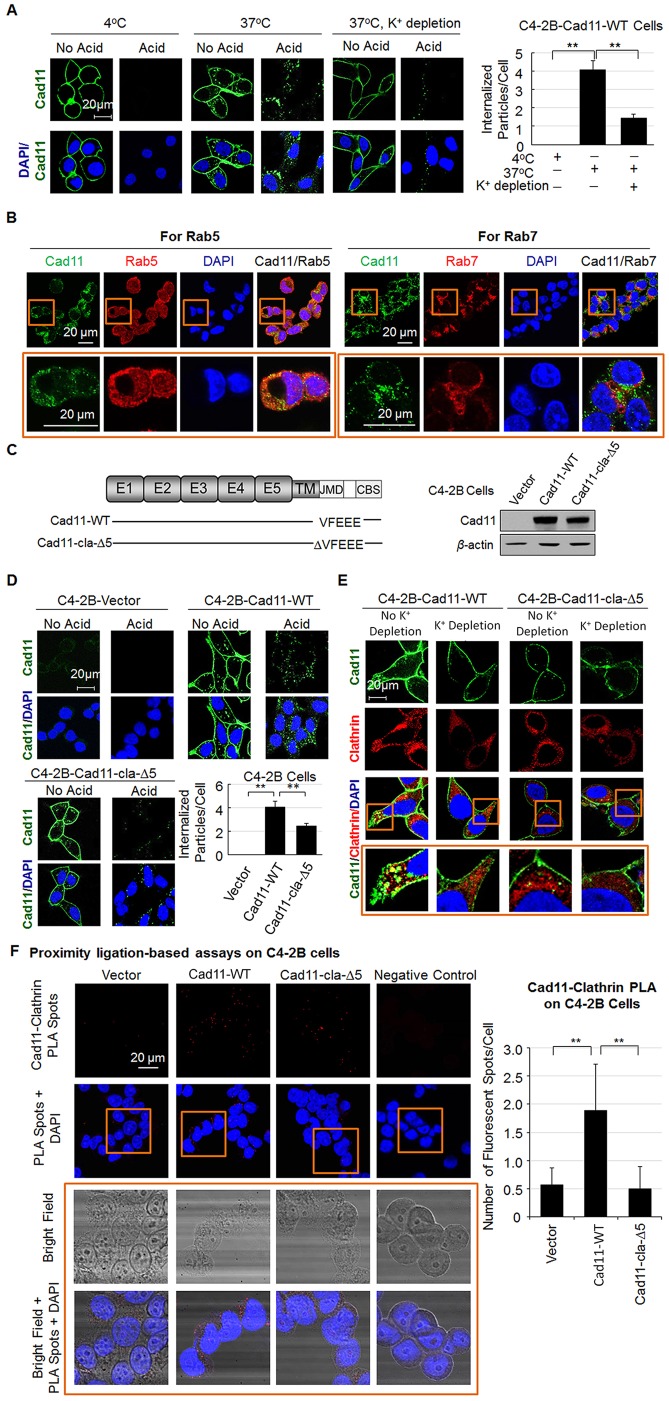
Clathrin is a major component of coated vesicles, which are known to play a role in the endocytosis of membrane proteins. To examine whether clathrin is involved in Cad11 endocytosis, we employed C4-2B prostate cancer cells that expressed low or undetectable endogenous Cad11. Cells were transfected with wild-type Cad11 (Cad11-WT), and Cad11 internalization was analyzed by using endocytosis assays. We found that at 4°C, a mouse antibody against Cad11 (clone 1A5) bound to Cad11 on the cell surface (Fig. 3A). When the temperature was shifted to 37°C to allow for endocytosis, the internalized Cad11–1A5-antibody complexes were detected as green dots in the cytosol (Fig. 3A). We next used a mild acid wash to remove antibodies associated with surface proteins but not those associated with internalized proteins. The loss of surface-bound antibodies against Cad11, but not those that had been internalized, after acid wash confirmed Cad11 internalization at 37° but not at 4°C (Fig. 3A). Depletion of K+ has been shown to prevent the assembly of clathrin into coated pits at the plasma membrane (Salazar and Gonzalez, 2002; Wang et al., 1993; Xiao et al., 2005). To test whether Cad11 internalization is mediated by clathrin, we conducted endocytosis assays under K+-deprived conditions. As shown in Fig. 3A, under 37°C assay conditions, depletion of intracellular K+ led to a 70% decrease of internalized Cad11 (green dots). These observations suggest that Cad11 internalization is mediated by clathrin-dependent endocytosis.
Fig. 3.
Clathrin mediates Cad11 endocytosis in C4-2B cells. (A) Endocytosis assay for Cad11. C4-2B cells expressing Cad11-WT were incubated with an antibody against Cad11 (mAb 1A5) at 4°C. Cad11 internalization was initiated by shifting the temperature to 37°C. A mild acid wash, which removes mAb 1A5 associated with surface Cad11, was used to distinguish the internalized Cad11 from surface Cad11. K+ depletion was used to block clathrin-mediated internalization of Cad11. After fixation, cells were incubated with fluorescence-labeled anti-mouse antibody, counterstained with DAPI for nuclei and imaged by using confocal microscopy. The number of internalized Cad11-protein-containing particles was quantified and expressed as mean±s.e.m. (n=10). **P<0.01 (Student's t-test). Magnification, ×400. (B) Colocalization of Cad11 (green) and Rab5 (red) or Rab7 (red) by immunofluorescent staining. Boxed areas are enlarged on the bottom row. (C) Expression of wild-type Cad11 (Cad11-WT) and mutant Cad11 with deletion of VFEEE in the JMD domain (Cad11-cla-Δ5) in C4-2B cells. (D) Endocytosis assay of C4-2B cells expressing Cad11-WT, Cad11-cla-Δ5 or vector alone. The data are expressed as mean±s.e.m. (n=10). **P<0.01 (Student's t-test). Magnification, ×400. (E) Colocalization of Cad11 and clathrin (enlarged in bottom row) in C4-2B cells expressing Cad11 or Cad11-cla-Δ5 after the internalization assay with or without K+ depletion. Cells were stained with the mAb 1A5 antibody against Cad11 and goat-anti-clathrin, and counterstained with DAPI. (F) Proximity ligation assay (PLA) to determine the interaction of Cad11 and clathrin in C4-2B cells. Red fluorescent spots indicate colocalization of Cad11 and clathrin. In the enlarged insets, bright-field images have been superimposed onto the fluorescence images to reveal the cell borders. Quantification of PLA spots per cell is shown in the right panel. mean±s.e.m. (n=8), **P<0.01 (Student's t-test). Assays lacking primary antibody were used as a negative control.
The endocytic pathway comprises a series of dynamic stages, including the transport of clathrin-coated vesicles to early endosomes, then the formation of late endosomes and multivesicular bodies, and finally fusion with lysosomes for degradation (Vanlandingham and Ceresa, 2009). We conducted immunofluorescence staining in Cad11-expressing C4-2B cells to determine whether internalized Cad11 colocalizes with clathrin in early endosomes or late endosomes. Immunofluorescence staining showed that internalized Cad11 colocalized in some vesicles with Rab5, an early endosome marker (Nielsen et al., 1999) (Fig. 3B, left; Fig. S1), but not in vesicles that were positively stained for Rab7 (Fig. 3B, right; Fig. S1), a late endosome marker (Vanlandingham and Ceresa, 2009). Similar results were obtained in PC3-mm2 cells (Fig. S2). These results indicate that internalized Cad11 is retained in early endosomes, which represent the recycling compartment.
To examine whether interactions between Cad11 and clathrin through the clathrin binding motif VFEEE are involved in Cad11 internalization, we generated a mutant Cad11 construct in which the VFEEE sequence had been deleted (Cad11-cla-Δ5) (Fig. 3C) and expressed it in C4-2B cells. Western blotting showed that the Cad11-WT and Cad11-cla-Δ5 constructs were expressed by cells at a similar level (Fig. 3C). Endocytosis assays performed at 37°C showed that more Cad11 was endocytosed in C4-2B cells expressing Cad11-WT cells than those expressing Cad11-cla-Δ5 cells (Fig. 3D). Quantification showed that Cad11 endocytosis was decreased by 40% in cells expressing Cad11-cla-Δ5 compared to that in cells expressing Cad11-WT (Fig. 3D). The C4-2B vector-expressing cells were used as a negative control (Fig. 3D). These observations indicate that Cad11 and clathrin interact through the clathrin binding motif VFEEE, regulating Cad11 endocytosis.
Next, we examined whether Cad11 and clathrin colocalize in endocytic vesicles. In C4-2B cells expressing Cad11-WT, internalized Cad11 was found to colocalize with clathrin in endocytic vesicles (Fig. 3E, see insets; Fig. S3). Incubation of cells in K+-depletion medium significantly reduced colocalization of Cad11 and clathrin in endocytic vesicles (Fig. 3E). In the C4-2B cells expressing Cad11-cla-Δ5, only a few clathrin-coated endocytic vesicles were co-stained with Cad11. Incubation of Cad11-cla-Δ5-expressing cells in K+-depletion medium did not further reduce the number of co-stained vesicles (Fig. 3E).
The interaction of clathrin with its substrates is usually transient and of low affinity. As a result, attempts to pull down Cad11 with endogenous clathrin in a co-immunoprecipitation assay were unsuccessful (data not shown). To further demonstrate an interaction between Cad11 and clathrin, we performed proximity-ligation-based assays (PLA) on C4-2B cells expressing the vector, Cad11-WT or Cad11-cla-Δ5. Cells were first incubated with both a mouse antibody against Cad11 and a goat antibody against clathrin, and then incubated with antibodies against mouse and goat antibodies that have a unique short DNA strand attached to each antibody. If the two epitopes on Cad11 and clathrin are sufficiently close, the attached oligonucleotides on the respective antibodies hybridize or become ligated, producing a template for a rolling circle DNA amplification, which can be probed efficiently with fluorescent oligonucleotide probes. The appearance of discrete fluorescent spots in the immunofluorescent images indicates that Cad11 and clathrin are present in close proximity. We found that the number of fluorescent spots in Cad11-WT-expressing C4-2B cells was significantly higher than that in C4-2B expressing the vector or Cad11-cla-Δ5 (Fig. 3F). These observations further support the notion that there is a direct interaction between Cad11 and clathrin in C4-2B cells expressing Cad11-WT.
Internalization of Cad11 in PC3mm2 cells
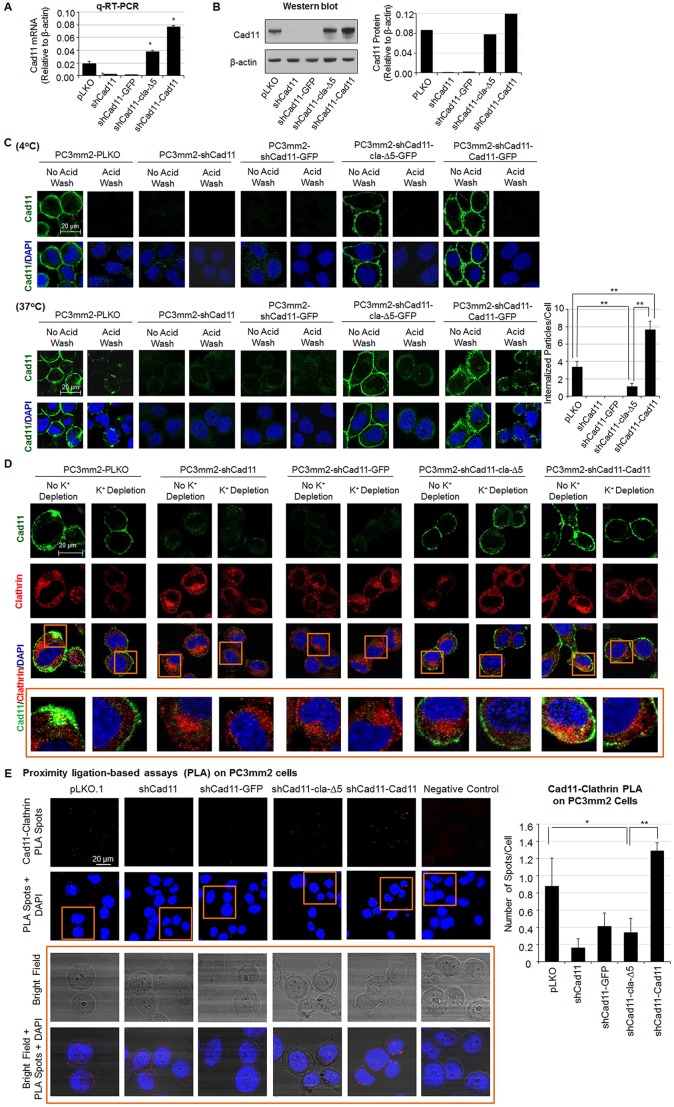
We further examined whether clathrin is also involved in Cad11 endocytosis in PC3mm2 cells, which endogenously express Cad11. To do so, we first depleted endogenous Cad11 and then reconstituted Cad11-deficient cells with either Cad11-WT or Cad11-cla-Δ5, which contains the deletion of the clathrin binding motif. Small hairpin RNA (shRNA) against Cad11 (shCad11), which targets the 3′ untranslated region of Cad11 in the pLKO.1 vector, was used to knock down endogenous Cad11 (PC3mm2-shCad11 cells). PC3mm2 cells that had been transfected with empty vector (PC3mm2-pLKO) were used as a control. PC3mm2-shCad11 cells were then transfected with cDNA encoding Cad11-cla-Δ5 in the pBMN-I-GFP vector (PC3mm2-shCad11-cla-Δ5-GFP). PC3mm2 cells transfected with empty pBMN-I-GFP vector (PC3mm2-shCad11-GFP) or Cad11 (PC3-mm2-shCad11-Cad11-GFP) were used as negative and positive controls, respectively. Quantitative real-time (qRT)-PCR and western blot analyses showed that the message levels of Cad11-cla-Δ5 in PC3mm2-shCad11-cla-Δ5-GFP cells were higher than the endogenous levels of Cad11 in the vector-transfected PC3mm2-pLKO cells, but the protein levels were comparable (Fig. 4A,B), whereas the levels of Cad11 in PC3mm2-shCad11-Cad11-GFP cells were higher than those in PC3mm2-pLKO cells (Fig. 4A,B). The lower levels of Cad11-cla-Δ5 compared to those for Cad11 in the reconstituted shCad11 cells is possibly due to differences in protein folding or stability because we have found that the introduction of a mutation and/or deletion to membrane proteins frequently leads to decreased levels of mutant protein production compared to that of the wild type.
Fig. 4.
Cad11 endocytosis in PC3mm2 cells. (A) Knockdown of endogenous Cad11 (shCad11) in PC3mm2 cells followed by reconstitution with a Cad11 cytoplasmic domain mutant (Cad11-cla-Δ5) or wild-type Cad11. Cad11 RNA levels relative to those of β-actin were determined by using qRT-PCR. *P<0.05 as compared to PC3mm2-shCad11-GFP vector control cells (Student's t-test). (B) Cad11 protein levels in the cells indicated were determined by western blotting and quantified relative to the levels of β-actin. (C) Cad11 internalization in PC3mm2 cells was determined at 4°C, followed by shifting the temperature to 37°C, with or without acid wash, and analyzed by using immunofluorescence and quantified as described in Fig. 3. **P<0.01 (Student's t-test). (D) Colocalization of Cad11 and clathrin in PC3mm2 cells. Cells were immunostained with Cad11 mAb 1A5 or an antibody against clathrin after the internalization assay with or without K+-depletion. Boxed areas are enlarged on the bottom row. (E) Proximity ligation assay (PLA) to determine the interaction of Cad11 and clathrin in PC3mm2 cells was performed as described in Fig. 3F. Quantification of PLA spots per cell is shown in the right panel. *P<0.05; **P<0.01 (Student's t-test).
When cells were incubated with the antibody against Cad11 at 4°C, Cad11 signals were readily detected at the cell surface in PC3mm2-pLKO, PC3mm2-shCad11-cla-Δ5-GFP and PC3mm2-shCad11-Cad11-GFP cells (Fig. 4C). After acid washing that removed only cell-surface-bound antibody, no Cad11 signals were detected in these cells (Fig. 4C, 4°C acid wash). These observations indicate that both the Cad11-cla-Δ5 mutant and wild type Cad11 proteins were expressed at the cell surface. No Cad11 signals were observed in PC3mm2-shCad11 and PC3mm2-shCad11-GFP cells at 4°C, indicating efficient knockdown of endogenous Cad11.
We next performed endocytosis assays on this panel of PC3mm2 cells. Upon shifting the temperature to 37°C, endocytic vesicles containing Cad11 (green dots) were detected in the cytosol of PC3mm2-pLKO and PC3mm2-shCad11-Cad11-GFP cells (Fig. 4C, 37°C, no acid wash). Further, these cytosolic Cad11 punctate signals persisted after acid wash (Fig. 4C, 37°C, acid wash). However, the number of internalized green dots decreased by 67% in PC3mm2-shCad11-cla-Δ5-GFP cells as compared to that on PC3mm2-pLKO cells (Fig. 4C). These observations suggest that clathrin is also involved in Cad11 endocytosis in PC3mm2 cells.
Immunostaining of Cad11 and clathrin in this panel of PC3mm2 cells showed colocalization of Cad11 and clathrin in the endocytic vesicles of PC3mm2-pLKO and PC3mm2-shCad11-Cad11 cells (Fig. 4D, see insets; Fig. S4), and the co-staining was diminished under K+-depleted conditions (Fig. 4D). In PC3mm2-shCad11-cla-Δ5-GFP cells, there was much less co-staining of Cad11 with clathrin compared to that observed in PC3mm2-pLKO and PC3mm2-shCad11-Cad11 cells (Fig. 4D). We further conducted PLA assays to detect the interaction of Cad11 and clathrin in PC3mm2 cells. We observed that there were fewer red fluorescent spots in PC3mm2-shCad11, PC3mm2-shCad11-GFP and PC3mm2-shCad11-cla-Δ5 cells as compared to PC3mm2-PLKO.1 cells, which endogenously express Cad11 (Fig. 4E). There were more red spots in PC3mm2-shCad11-Cad11 cells than PC3mm2-PLKO.1 cells (Fig. 4E), consistent with the overexpressed levels of wild-type Cad11 in the reconstituted shCad11 cells. Taken together, these observations indicate that the interaction of Cad11 and clathrin is one of the mechanisms responsible for Cad11 internalization in prostate cancer cells.
Effects of clathrin-dependent endocytosis on Cad11-mediated migration
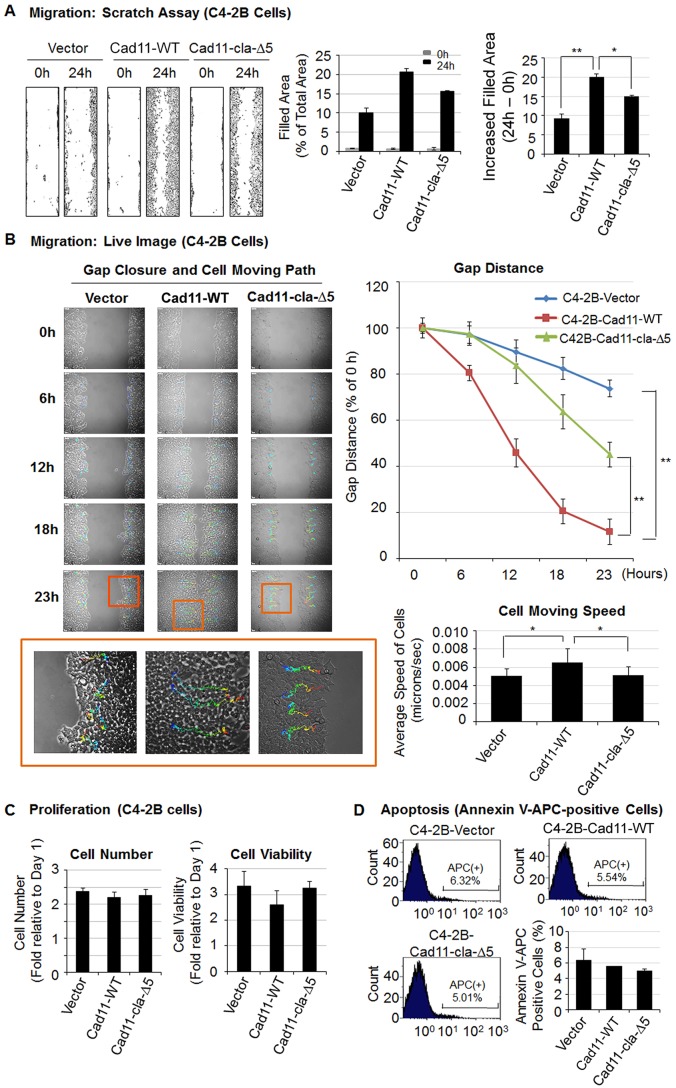
Previous studies have shown that Cad11 plays a role in the migration of prostate cancer cells (Huang et al., 2010; Lee et al., 2013, 2010). Because endocytosis plays a role in the turnover of Cad11 from the cell surface, which might have an effect on Cad11-mediated cellular activities, we examined whether clathrin-dependent endocytosis has an effect on Cad11-mediated migration. We performed wound healing migration assays on C4-2B cells expressing vector, Cad11-WT or Cad11-cla-Δ5 cells. Expression of Cad11-WT significantly increased C4-2B cell migration compared to C4-2B vector control cells (Fig. 5A). Expression of Cad11-cla-Δ5 also increased C4-2B cell migration, however, to a lesser extent than that in C4-2B cells expressing Cad11-WT (Fig. 5A). Moreover, we applied live-cell imaging to capture cell movement in ibidi migration chambers by using time-lapse recording. As shown in Fig. 5B, the gaps remained visible in C4-2B cells expressing the vector or Cad11-cla-Δ5, whereas the gap had almost closed in Cad-11-expressing C4-2B cells at 23 h. Quantification analysis revealed that the gap distance was significantly shorter in C4-2B cells expressing Cad11-WT than those expressing vector or Cad11-cla-Δ5 (Fig. 5B, upper right panel). Cad11-WT-expressing cells also moved further compared to C4-2B cells expressing vector or Cad11-cla-Δ5 (Fig. 5B, bottom left panel). Quantification of the average cell speed also showed significantly faster movement of C4-2B cells expressing Cad11-WT than of those expressing vector or Cad11-cla-Δ5 (Fig. 5B, lower right panel).
Fig. 5.
Effects of the Cad11 and clathrin interaction on Cad11-mediated cell migration, proliferation and apoptosis in C4-2B cells. (A) Cad11-mediated migration in C4-2B cells measured by using a wound healing scratch assay (left). The areas that were occupied by cells that migrated into the scratched gap before and after the 24-h incubation period was determined by using Image J and were expressed as a percentage (%) of the total area (middle). The increased areas that were occupied by migrated cells after a 24-h incubation were calculated by subtracting the background levels at 0 h (right). *P<0.05, **P<0.01 (Student's t-test). (B) Cell migration analyzed by live-cell imaging. Gap closure and the path of cell movement over 23 h (left). Boxed areas are enlarged in the bottom row. Quantification of gap distance (upper right panel) and speed of cell movement (lower right panel). *P<0.05 (Student's t-test). (C) Effect of Cad11-WT and Cad11-cla-Δ5 on proliferation of C4-2B cells, as measured by cell number (left) and viability (right). (n=2). (D) Flow cytometry analysis of C4-2B cells labeled with APC-conjugated annexin V (n=2). All results are shown as mean±s.e.m.
The possibility that changes in cell migration were due to an effect of Cad11-WT or Cad11-cla-Δ5 expression on cell proliferation was examined. We found that there was no significant difference in the cell numbers amongst the different C4-2B cell lines used in the migration assays at 24 h (Fig. 5C, left panel). Similarly, no significant difference in cell viability, as measured using a Presto Blue assay, was observed amongst the C4-2B cell lines used in the migration assays at 24 h (Fig. 5C, right panel). In addition, no significant difference was observed in the number of apoptotic cells, as measured using an annexin-V-binding assay, amongst the C4-2B cell lines (Fig. 5D).
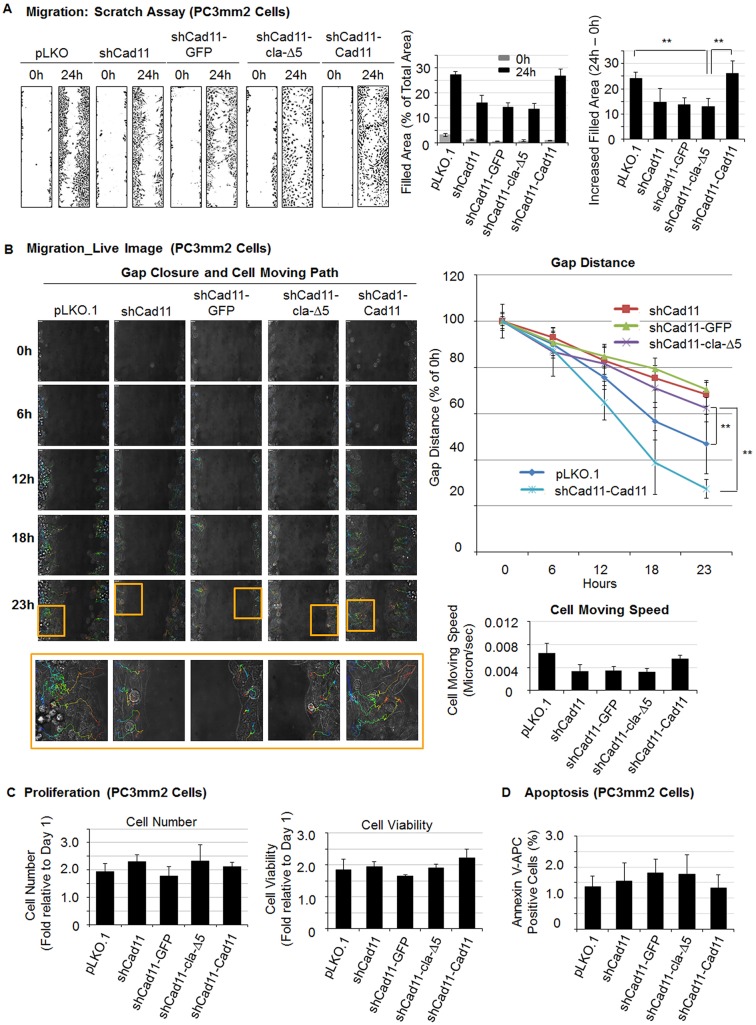
In addition to C4-2B cells, we also performed wound healing assays on the panel of PC3mm2 cells that is described in Fig. 4. Knockdown of endogenous Cad11 in PC3-mm2 cells (PC3mm2-shCad11 and PC3mm2-shCad11-GFP) significantly reduced cell migration relative to that in vector control cells (PC3mm2-pLKO) (Fig. 6A). The migratory activity was recovered by re-expression of wild-type Cad11 (PC3mm2-shCad11-Cad11) but not by Cad11-cla-Δ5 (PC3mm2-shCad11-Cad11-cla-Δ5) in PC3mm2 cells (Fig. 6A). Similarly, faster gap closure (Fig. 6B, upper panels) and longer paths of cell movement (Fig. 6B, lower panels) were observed in PC3mm2-pLKO.1 cells and in PC3mm2-shCad11-Cad11 cells compared to PC3mm2-shCad11-Cad11-cla-Δ5 cells with live-cell imaging. No significant differences in the cell numbers (Fig. 6C, left panel), cell viability (Fig. 6C, right panel) and the number of apoptotic cells (Fig. 6D) were observed amongst the different PC3mm2 cell lines used in the migration assays at 24 h. These observations indicate that the Cad11-mediated effects on cell migration were not due to cell proliferation. Taken together, these results suggest that turnover of Cad11 from the cell surface through clathrin-mediated endocytosis plays a role in Cad11-mediated migration in prostate cancer cells.
Fig. 6.
Effects of the Cad11 and clathrin interaction on Cad11-mediated cell migration, proliferation and apoptosis in PC3mm2 cells. The endogenous Cad11 in PC3mm2 cells was knocked down using Cad11 shRNA. The cells were then reconstituted with individual constructs. (A) Wound healing scratch assay (left). Quantification of scratch assay by using ImageJ, as described in Fig. 5. **P<0.01 (Student's t-test). (B) Cell migration analyzed by live-cell imaging. Gap closure and the path of cell movement over the time period of 23 h (left). Boxed areas are enlarged on the bottom row. Quantification of gap distance (upper right) and cell speed (lower right). (C) Effect of Cad11-WT and Cad11-cla-Δ5 on proliferation of PC3mm2 cells as measured by cell number (left) and viability (right). (D) Flow cytometry analysis of PC3mm2 cells labeled with APC-conjugated annexin V. All results are shown as mean±s.e.m. shCad11, shRNA against Cad11.
DISCUSSION
We showed that clathrin directly interacts with Cad11 and is involved in Cad11 endocytosis. We further showed that clathrin-mediated Cad11 endocytosis regulates Cad11-mediated migration. Although clathrin has been shown to mediate E-Cad endocytosis, we found that the E-Cad cytoplasmic domain does not contain the specific clathrin-binding motif VFEEE that is found in Cad11, and clathrin does not bind to the E-Cad cytoplasmic domain under our experimental conditions, suggesting that clathrin-mediated endocytosis of E-Cad and Cad11 are distinct.
The cadherin adhesion complex has been shown to play crucial roles during normal development as well as during malignancy (Kowalczyk and Nanes, 2012). Cad11 is a mesenchymal cadherin that is mainly expressed in osteoblasts, as well as in lung, testis and brain tissues at low levels (Okazaki et al., 1994). It is likely that Cad11 endocytosis provides dynamic changes that are necessary for Cad11-mediated cell migration, which plays a role in osteogenesis and neuronal cell migration during normal development. In Cad11-mediated neuronal migration during Xenopus development, it has been shown that excessive Cad11 expression prevents cell migration in the embryo and that cleavage of the Cad11 extracellular domain to reduce Cad11-mediated adhesion is important for Cad11-mediated neuronal migration (Borchers et al., 2001). Peglion et al. (2014) show that clathrin-mediated endocytosis of N-Cad plays a role in the cycling of adherens junction components for anterograde transport during collective cell movement. Gavard and Gutkind (2006) show that stimulation with VEGF promotes the rapid endocytosis of VE-cadherin, resulting in disruption of the endothelial barrier and increased vascular permeability. Recently, Padmanabhan and Taneyhill (2015) have shown that clathrin-mediated endocytosis of cadherin-6B plays a role in the neural crest cell epithelial–mesenchymal transition and migration. Thus, the dynamic changes in the surface levels of cadherin molecules through clathrin-mediated endocytosis affect cadherin-mediated adhesion and migratory activities.
Defects in the recycling of adhesion complexes have been shown to be involved in malignant transformation (Mosesson et al., 2008). Cad11 has been shown to promote the metastasis of prostate or breast cancer cells to bone (Chu et al., 2008; Tamura et al., 2008). Because acquisition of migration properties is a hallmark of metastatic cancer cells, it is possible that clathrin-mediated Cad11 endocytosis plays a role in the metastatic colonization of prostate cancer cells in bone. We have recently shown that Cad11 plays a role in cell migration through interaction with p80-angiomotin (Ortiz et al., 2015). Whether clathrin-mediated Cad11 endocytosis regulates Cad11–angiomotin complex formation and dissociation is unknown.
During classic clathrin-mediated endocytosis, membrane receptors are clustered in clathrin-coated pits, which is followed by membrane invagination and vesicle scission (Goldstein et al., 1979). In clathrin-coated pits, clathrin triskelia lattices recruit adaptor proteins – e.g. AP2, DAB and dynamin – to form endocytic vesicles (Hulpiau and van Roy, 2009). Thus, the binding of clathrin to cargo is typically indirect, and the adaptor protein interactions with cargo are also of low affinity. Although many reports describe clathrin-mediated endocytosis of E-cadherin (Ivanov et al., 2004; Kon et al., 2008; Le et al., 1999; Miyashita and Ozawa, 2007), it is likely that the interaction of clathrin with the E-Cad tail is indirect. Thus, clathrin does not bind to the E-Cad cytoplasmic domain under the experimental conditions used in the present study. We were able to pull down clathrin using the Cad11 cytoplasmic domain through the clathrin-binding motif in Cad11. However, we did not find AP-2 or DAB in the Cad11-cyto construct pulldown assay by western blotting (data not shown). In addition, we did not detect clathrin in PC3 cell extracts that had been immunoprecipitated with an antibody against Cad11 (data not shown). This might be due to the low affinity or the transient nature of the Cad11–clathrin interaction in vivo. It is also possible that the detergents used in the solubilization of Cad11 from membranes interfered with the Cad11–clathrin interaction.
Although clathrin-mediated endocytosis is a common mechanism for the turnover of cadherin-family proteins, the structural motifs utilized in this process appear to be distinct for different cadherins. In VE-cadherin, Nanes et al. (2012) have identified a dual-function motif comprising three highly conserved acidic residues (DEE) that alternatively serve as a p120-binding interface and an endocytic signal (Fig. 2C). In contrast, E-Cad contains a dileucine endocytic signal that is not present in VE-cadherin, and mutation of the DEE sequence in E-Cad only modestly inhibits its internalization (Nanes et al., 2012). Our studies revealed yet a third motif, the clathrin-binding motif VFEEE, in Cad11 that is distinct from the dileucine endocytic signal in E-Cad and the p120-binding site in VE-cadherin.
In conclusion, we have identified a unique structural motif that mediates Cad11 endocytosis. Because dynamic turnover of Cad11 is required for proper adhesion and migration activity, our studies provide a mechanism by which clathrin-mediated Cad11 endocytosis regulates the migratory function of Cad11 in prostate cancer cells.
MATERIALS AND METHODS
Cell lines and antibodies
Human kidney 293T (HEK293T), Phoenix cells and L-cells were purchased from American Culture Type Collection and were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS). Human prostate cancer cell lines C4-2B and PC3mm2 (Huang et al., 2010; Lira et al., 2008) were grown in RPMI 1640 (Invitrogen) containing 10% FBS. Goat anti-clathrin and mouse anti-Cad11 cyto (5B2H5) antibody were purchased from Santa Cruz Biotechnology and Invitrogen, respectively. Mouse anti-Cad11 monoclonal antibody mAb 1A5 was generated as described previously (Lee et al., 2013). Rabbit polyclonal antibody against Rab5 and rabbit recombinant monoclonal antibody against Rab7 were purchased from Abcam. Mouse anti-E-cad, anti-p120-catenin and anti-β-catenin antibodies were purchased from BD Transduction Lab. The oligonucleotides used were purchased from Sigma-Aldrich, and the sequences are listed in Table S1.
Construction and expression of GST–cyto fusion proteins
GST fusion proteins containing two copies of the Cad11 cytoplasmic domain (GST–Cad11-cyto) or the E-Cad cytoplasmic domain (GST–E-Cad-cyto) were constructed, as described in Ortiz et al. (2015). The construction of GST fusion proteins containing one copy of the cytoplasmic domains with deletions in the juxtamembrane domain (GST–ΔJMD-cyto) or β-catenin-binding domain (GST–ΔCBS-cyto), respectively, is described in Ortiz et al. (2015). GST fusion proteins containing the N-terminal 51, 40, 35, 30, 25, 20, 15 or 10 amino acids of the Cad11 cytoplasmic domain were generated using PCR with the forward primer and reverse primers listed in Table S1. The GST–cyto fusion proteins were expressed in Escherichia coli and purified using glutathione–agarose beads (GST beads, GE Healthcare Life Sciences).
GST pulldown assay for Cad11-cyto-associated proteins
L-cells, which do not express any of the major cadherins, were grown as a monolayer on tissue culture plates. Cells were then scraped from the plate in cold distilled water containing protease inhibitors, and homogenized with a Dounce homogenizer. After centrifugation of the cell lysates for 15 min at 15,700 g, the supernatant fraction was collected. The supernatant fraction was mixed with GST or GST–Cad11-cyto protein immobilized on glutathione–agarose beads on a rocker at 4°C overnight. The glutathione beads were spun down, washed three times with cold lysis buffer (50 mM phosphate, 100 mM NaCl and 0.01% Triton-X100) containing protease inhibitors and analyzed on 4–12% gradient NuPage gels (Novex). The gel was stained with silver stain. The specific protein band at 180 kDa was cut from the gel and analyzed by using mass spectrometry. For western blots, the gel was transferred to a nitrocellulose membrane (Schleicher & Schnell) and stained with Ponceau S, followed by immunoblotting with specific antibodies as indicated. Signals were detected with a chemiluminescent detection kit (Pierce Biotechnology).
Generation of cell lines expressing wild-type Cad11 or Cad11 mutants
Cad11 with deletion of VFEEE in the clathrin-binding site (Cad11-cla-Δ5) was generated using the QuikChange II XL site-directed mutagenesis kit (Stratagene) and verified with DNA sequencing. The sequences of the primers are listed in Table S1. Wild-type Cad11 (Cad11-WT) and mutant Cad11 (Cad11-cla-Δ5) in bicistronic pBMN-I-neo vectors were transfected into Phoenix cells, and the culture media were collected for infection of C4-2B cells, as described previously (Huang et al., 2010; Lira et al., 2008). C4-2B cells were infected with retrovirus and selected with G418 to generate C4-2B-Cad11-WT and C4-2B-Cad11-cla-Δ5 cell lines. C4-2B cells that had been infected with empty vector (C4-2B-vector) were used as a control.
PC3mm2 is a bone-metastasis-derived prostate cancer cell line expressing endogenous Cad11 (Chu et al., 2008). To express mutant Cad11 in PC3mm2 cells, we first knocked down endogenous Cad11 in PC3mm2 cells using lentiviral vector pLKO.1-puro containing Cad11 shRNA, as we have described previously (Ortiz et al., 2015). The lentiviral vector containing non-targeting shRNA (pLKO.1 puro, Sigma-Aldrich) was used as a control. Following the establishment of PC3mm2-shCad11 cells, we infected the cells with Cad11-cla-Δ5 or Cad11-WT retroviruses to generate PC3mm2-shCad11-Cad11-cla-Δ5-GFP, PC3mm2-shCad11-Cad11-GFP cells, as described above for C4-2B cells. PC3mm2 cells that had been infected with empty pBMN-I-GFP vector (PC3mm2-shCad11-GFP) were used as a control.
Endocytosis assay and colocalization with clathrin
Cells (5×104) were seeded onto coverslips in the well of a 24-well plate the day before the endocytosis assay was performed. Before starting the internalization assay, cells were incubated with anti-Cad11 mAb 1A5 (Lee et al., 2013), which recognizes the extracellular domain, at 4°C for 30 min to allow for antibody binding to cell surface Cad11. After washing cells with ice-cold PBS buffer containing 3% (w/v) BSA to remove unbound antibody, cells were changed to either fresh RPMI medium containing 10% FBS (v/v) or to K+-depletion buffer (20 mM Hepes, 140 mM NaCl, 1 mM CaCl2, 1 mM MgCl2), and incubated at 37°C for 30 min to allow internalization to occur. Then cells were washed with ice-cold 3% BSA in PBS wash buffer or washed with ice-cold acid wash buffer (0.5 M NaCl with 0.2 M acetic acid) for 4 min to remove the surface-bound antibody, followed by fixing cells with 100% methanol at −20°C for 6 min. Cells were immunostained with Alexa-Fluor-488-labeled anti-mouse secondary antibody (1:300) at room temperature for 1 h in the dark, followed by counterstaining with 4′, 6-diamidino-2-phenylindole (DAPI, 1:500 in PBS) for 10 min. The number of internalized vesicles with Cad11 was calculated using ImageJ software.
Colocalization by immunofluorescence
For the colocalization of Cad11 and clathrin, cells were incubated with anti-Cad11 antibody mAb 1A5 and goat anti-clathrin antibody (1:100). For the colocalization of Cad11 with endosome markers, cells were incubated with mouse anti-Cad11 antibody and rabbit anti-Rab5 antibody or anti-Rab7 antibody. After incubation at 4°C overnight, cells were incubated with anti-mouse secondary antibody conjugated to Alexa-Fluor-488 (1:400), anti-goat secondary antibody conjugated to Alexa-Fluor-594 (1:300) (Jackson ImmunoResearch) or anti-rabbit secondary antibody conjugated to Alexa-Fluor-594 (1:400) (Jackson ImmunoResearch) at room temperature for 1 h. Cells were then counterstained with DAPI for 10 min and mounted with anti-fading mounting medium.
Proximity ligation assay
The PLA was performed using Duolink PLA In Situ Red Starter Kit (Mouse and Goat, Sigma-Aldrich) as per the manufacturer's instruction. The primary antibodies were mouse anti-Cad11 antibody (Invitrogen, 1:150) and goat anti-clathrin antibody (1:100, Santa Cruz Biotechnology). Images were acquired using a FluoView 1000 IX2 confocal microscope (Olympus).
Wound healing migration assay
We employed a wound healing assay using both scratching and ibidi migration chambers. For scratching, cells were seeded onto 6-well plates and were grown to a confluent monolayer. Then ‘wounds’ were generated by scratching lines through the monolayer using 200-µl tips. The cells that moved into the created open gaps were then imaged over 24 h with a microscope at 4× magnification. The filled area was quantified using ImageJ software. When using µ-Slide 8-well ibiTreat microscopy chambers (Ibidi, Madison, Wisconsin), we used live-cell imaging to capture cell movement with time-lapse on an Olympus IX81 DSU spinning disk confocal microscope. The rate of gap closure and the speed of cell movement were analyzed from the time-lapse movies using 3I's Slidebook software.
Cell proliferation, viability and apoptosis assay
For cell proliferation assays, cells were seeded into 6-well plates (3×105/well) and allowed to attach onto the plate overnight. The number of cells was counted using a hemocytometer after digestion. The cell number counted the following day was set as day 1 and at 24 h later was set as day 2. The 24 h proliferation rate is expressed as folds of that at day 1.
For cell viability measurements, 3×104 cells in 200 µl of culture medium were seeded into wells of 96-well plates. The cell viability was determined on the following day (day 1) and another 24 h later (day 2). On the day of the assay, the medium was changed for 100 µl (for 96-well plate) of fresh medium. Presto Blue reagent (Life Technologies) was added to each well at 1:10 ratio, and the cells were further incubated at 37°C for 2 h. The medium (100 µl) was used for measuring absorbance optical density (OD) values at 570 nm and 600 nm. The wells containing culture medium without cells were used to take background readings.
To determine if cell apoptosis affects migration, we detected the early stage of cell apoptosis using flow cytometric analysis of cells labeled with APC-conjugated annexin V (BioLegend) on a FACScan™ model flow cytometer (Beckman Coulter ‘Gallios’).
Statistical analysis
Data from three or more independent experiments were used in analyses, and values are expressed as mean±s.e.m. Statistical significance was assessed by using Student's t-test. The level of significance was set at P<0.05.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
S.-H.L., L.-Y.Y.-L., A.P.K. and R.L.S. conceived and designed the experiments. T.P., M.A.B., X.L., Y.-C.L. and A.O. performed the experiments. R.L.S., T.P., L.-Y.Y.-L. and S.-H.L. analyzed the data. R.L.S., T.P., A.P.K., L.-Y.Y.-L. and S.-H.L. wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Institutes of Health [grant numbers P50 CA140388 to S.H.L., CA174798 to S.H.L. and CA16672 to M.D. Anderson Cancer Center]; the Prostate Cancer Foundation to S.H.L.; Cancer Prevention and Research Institute of Texas [grant numbers CPRIT RP110327 to S.H.L., RP150179 to S.H.L. and RP150282 to S.H.L. and G.E.G.]; by funds from the Sister Institute Network Fund to S.H.L.; by the Institutional Start-up fund to R.L.S.; and by the Institutional Research Grant (IRG) Program at the MD Anderson Cancer Center to R.L.S. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.176081/-/DC1
References
- Alarcos N., Gutiérrez M., Liras M., Sánchez F., Moreno M. and Douhal A. (2015). Direct observation of breaking of the intramolecular H-bond, and slowing down of the proton motion and tuning its mechanism in an HBO derivative. Phys. Chem. Chem. Phys. 17, 14569-14581. 10.1039/c5cp01437a [DOI] [PubMed] [Google Scholar]
- Angst B. D., Marcozzi C. and Magee A. I. (2001). The cadherin superfamily: diversity in form and function. J. Cell Sci. 114, 629-641. [DOI] [PubMed] [Google Scholar]
- Bockus A. T., Lexa K. W., Pye C. R., Kalgutkar A. S., Gardner J. W., Hund K. C. R., Hewitt W. M., Schwochert J. A., Glassey E., Price D. A. et al. (2015). Probing the physicochemical boundaries of cell permeability and oral bioavailability in lipophilic macrocycles inspired by natural products. J. Med. Chem. 58, 4581-4589. 10.1021/acs.jmedchem.5b00128 [DOI] [PubMed] [Google Scholar]
- Borchers A., David R. and Wedlich D. (2001). Xenopus cadherin-11 restrains cranial neural crest migration and influences neural crest specification. Development 128, 3049-3060. [DOI] [PubMed] [Google Scholar]
- Broussard J. A., Webb D. J. and Kaverina I. (2008). Asymmetric focal adhesion disassembly in motile cells. Curr. Opin. Cell Biol. 20, 85-90. 10.1016/j.ceb.2007.10.009 [DOI] [PubMed] [Google Scholar]
- Cheng S.-L., Lecanda F., Davidson M. K., Warlow P. M., Zhang S.-F., Zhang L., Suzuki S., St. John T. and Civitelli R. (1998). Human osteoblasts express a repertoire of cadherins, which are critical for BMP-2-induced osteogenic differentiation. J. Bone Miner. Res. 13, 633-644. 10.1359/jbmr.1998.13.4.633 [DOI] [PubMed] [Google Scholar]
- Chu K., Cheng C.-J., Ye X., Lee Y.-C., Zurita A. J., Chen D.-T., Yu-Lee L.-Y., Zhang S., Yeh E. T., Hu M. C.-T. et al. (2008). Cadherin-11 promotes the metastasis of prostate cancer cells to bone. Mol. Cancer Res. 6, 1259-1267. 10.1158/1541-7786.MCR-08-0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavard J. and Gutkind J. S. (2006). VEGF controls endothelial-cell permeability by promoting the beta-arrestin-dependent endocytosis of VE-cadherin. Nat. Cell Biol 8, 1223-1234. 10.1038/ncb1486 [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Anderson R. G. W. and Brown M. S. (1979). Coated pits, coated vesicles, and receptor-mediated endocytosis. Nature 279, 679-685. 10.1038/279679a0 [DOI] [PubMed] [Google Scholar]
- Gumbiner B. M. (1996). Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84, 345-357. 10.1016/S0092-8674(00)81279-9 [DOI] [PubMed] [Google Scholar]
- Huang C.-F., Lira C., Chu K., Bilen M. A., Lee Y.-C., Ye X., Kim S. M., Ortiz A., Wu F.-L. L., Logothetis C. J. et al. (2010). Cadherin-11 increases migration and invasion of prostate cancer cells and enhances their interaction with osteoblasts. Cancer Res. 70, 4580-4589. 10.1158/0008-5472.CAN-09-3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulpiau P. and van Roy F. (2009). Molecular evolution of the cadherin superfamily. Int. J. Biochem. Cell Biol. 41, 349-369. 10.1016/j.biocel.2008.09.027 [DOI] [PubMed] [Google Scholar]
- Ivanov A. I., Nusrat A. and Parkos C. A. (2004). Endocytosis of epithelial apical junctional proteins by a clathrin-mediated pathway into a unique storage compartment. Mol. Biol. Cell 15, 176-188. 10.1091/mbc.E03-05-0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J. K., Tien P. C., Cheng C. J., Song J. H., Huang C., Lin S. H. and Gallick G. E. (2015). Talin1 phosphorylation activates beta1 integrins: a novel mechanism to promote prostate cancer bone metastasis. Oncogene. 34, 1811-1821. 10.1038/onc.2014.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon S., Tanabe K., Watanabe T., Sabe H. and Satake M. (2008). Clathrin dependent endocytosis of E-cadherin is regulated by the Arf6GAP isoform SMAP1. Exp. Cell Res. 314, 1415-1428. 10.1016/j.yexcr.2007.11.006 [DOI] [PubMed] [Google Scholar]
- Kosalková K., Dominguez-Santos R., Coton M., Coton E., Garcia-Estrada C., Liras P. and Martín J. F. (2015). A natural short pathway synthesizes roquefortine C but not meleagrin in three different Penicillium roqueforti strains. Appl. Microbiol. Biotechnol. 99, 7601-7612. 10.1007/s00253-015-6676-0 [DOI] [PubMed] [Google Scholar]
- Kowalczyk A. P. and Nanes B. A. (2012). Adherens junction turnover: regulating adhesion through cadherin endocytosis, degradation, and recycling. Subcell. Biochem. 60, 197-222. 10.1007/978-94-007-4186-7_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupnick J. G., Goodman O. B. Jr., Keen J. H. and Benovic J. L. (1997). Arrestin/clathrin interaction. Localization of the clathrin binding domain of nonvisual arrestins to the carboxyl terminus. J. Biol. Chem. 272, 15011-15016. 10.1074/jbc.272.23.15011 [DOI] [PubMed] [Google Scholar]
- Le T. L., Yap A. S. and Stow J. L. (1999). Recycling of E-cadherin: a potential mechanism for regulating cadherin dynamics. J. Cell Biol. 146, 219-232. 10.1083/jcb.146.1.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-C., Cheng C.-J., Huang M., Bilen M. A., Ye X., Navone N. M., Chu K., Kao H.-H., Yu-Lee L.-Y., Wang Z. et al. (2010). Androgen depletion up-regulates cadherin-11 expression in prostate cancer. J. Pathol. 221, 68-76. 10.1002/path.2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-C., Bilen M. A., Yu G., Lin S.-C., Huang C.-F., Ortiz A., Cho H., Song J. H., Satcher R. L., Kuang J. et al. (2013). Inhibition of cell adhesion by a cadherin-11 antibody thwarts bone metastasis. Mol. Cancer Res. 11, 1401 10.1158/1541-7786.MCR-13-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira C. B. B., Chu K., Lee Y.-C., Hu M. C.-T. and Lin S.-H. (2008). Expression of the extracellular domain of OB-cadherin as an Fc fusion protein using bicistronic retroviral expression vector. Protein Expr. Purif. 61, 220-226. 10.1016/j.pep.2008.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo C. L., Vasconcelos L. H., Correia A. C., Martins I. R., Lira D. P., Santos B. V. and Silva B. A. (2014). Mechanisms underlying the relaxant effect of Galetin 3,6- dimethyl ether, from Piptadenia stipulacea (Benth.) Ducke, on guinea-pig trachea. Z. Naturforsch. C 69, 434-442. 10.5560/znc.2014-0017 [DOI] [PubMed] [Google Scholar]
- Marie P. J., Haÿ E., Modrowski D., Revollo L., Mbalaviele G. and Civitelli R. (2014). Cadherin-mediated cell–cell adhesion and signaling in the skeleton. Calcif. Tissue Int. 94, 46-54. 10.1007/s00223-013-9733-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras A., Lira R., Soria-Rodriguez C., Hori-Oshima S., Maldonado-Rodriguez A., Rojas-Montes O., Ayala-Figueroa R., Estrada-Guzman J. and Alvarez-Munoz M. T. (2015). [Cytomegalovirus: congenital infection and clinical presentation in infants with respiratory distress syndrome]. Rev. Med. Inst. Mex. Seguro Soc. 53, 286-293. [PubMed] [Google Scholar]
- Miyashita Y. and Ozawa M. (2007). Increased internalization of p120-uncoupled E-cadherin and a requirement for a dileucine motif in the cytoplasmic domain for endocytosis of the protein. J. Biol. Chem. 282, 11540-11548. 10.1074/jbc.M608351200 [DOI] [PubMed] [Google Scholar]
- Mosesson Y., Mills G. B. and Yarden Y. (2008). Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer 8, 835-850. 10.1038/nrc2521 [DOI] [PubMed] [Google Scholar]
- Nanes B. A., Chiasson-MacKenzie C., Lowery A. M., Ishiyama N., Faundez V., Ikura M., Vincent P. A. and Kowalczyk A. P. (2012). p120-catenin binding masks an endocytic signal conserved in classical cadherins. J. Cell Biol. 199, 365-380. 10.1083/jcb.201205029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W. J., Dickinson D. J. and Weis W. I. (2013). Roles of cadherins and catenins in cell-cell adhesion and epithelial cell polarity. Prog. Mol. Biol. Transl. Sci. 116, 3-23. 10.1016/B978-0-12-394311-8.00001-7 [DOI] [PubMed] [Google Scholar]
- Nielsen E., Severin F., Backer J. M., Hyman A. A. and Zerial M. (1999). Rab5 regulates motility of early endosomes on microtubules. Nat. Cell Biol. 1, 376-382. 10.1038/14075 [DOI] [PubMed] [Google Scholar]
- Okazaki M., Takeshita S., Kawai S., Kikuno R., Tsujimura A., Kudo A. and Amann E. (1994). Molecular cloning and characterization of OB-cadherin, a new member of cadherin family expressed in osteoblasts. J. Biol. Chem. 269, 12092-12098. [PubMed] [Google Scholar]
- Ortiz A., Lee Y. C., Yu G., Liu H. C., Lin S. C., Bilen M. A., Cho H., Yu-Lee L. Y. and Lin S. H. (2015). Angiomotin is a novel component of cadherin-11/beta-catenin/p120 complex and is critical for cadherin-11-mediated cell migration. FASEB J. 29, 1080-1091. 10.1096/fj.14-261594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan R. and Taneyhill L. A. (2015). Cadherin-6B undergoes macropinocytosis and clathrin-mediated endocytosis during cranial neural crest cell EMT. J. Cell Sci. 128, 1773-1786. 10.1242/jcs.164426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peglion F., Llense F. and Etienne-Manneville S. (2014). Adherens junction treadmilling during collective migration. Nat. Cell Biol. 16, 639-651. 10.1038/ncb2985 [DOI] [PubMed] [Google Scholar]
- Salazar G. and González A. (2002). Novel mechanism for regulation of epidermal growth factor receptor endocytosis revealed by protein kinase A inhibition. Mol. Biol. Cell 13, 1677-1693. 10.1091/mbc.01-08-0403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. (1990). Cadherins: a molecular family important in selective cell-cell adhesion. Annu. Rev. Biochem. 59, 237-252. 10.1146/annurev.bi.59.070190.001321 [DOI] [PubMed] [Google Scholar]
- Tamura D., Hiraga T., Myoui A., Yoshikawa H. and Yoneda T. (2008). Cadherin-11-mediated interactions with bone marrow stromal/osteoblastic cells support selective colonization of breast cancer cells in bone. Int. J. Oncol. 33, 17-24. 10.3892/ijo.33.1.17 [DOI] [PubMed] [Google Scholar]
- Thalmann G. N., Sikes R. A., Wu T. T., Degeorges A., Chang S.-M., Ozen M., Pathak S. and Chung L. W. K. (2000). LNCaP progression model of human prostate cancer: androgen-independence and osseous metastasis. Prostate 44, 91-103. [DOI] [PubMed] [Google Scholar]
- Thoreson M. A., Anastasiadis P. Z., Daniel J. M., Ireton R. C., Wheelock M. J., Johnson K. R., Hummingbird D. K. and Reynolds A. B. (2000). Selective uncoupling of P120(ctn) from E-cadherin disrupts strong adhesion. J. Cell Biol. 148, 189-202. 10.1083/jcb.148.1.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandingham P. A. and Ceresa B. P. (2009). Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J. Biol. Chem. 284, 12110-12124. 10.1074/jbc.M809277200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Rothberg K. G. and Anderson R. G. (1993). Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J. Cell Biol. 123, 1107-1117. 10.1083/jcb.123.5.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb D. J., Parsons J. T. and Horwitz A. F. (2002). Adhesion assembly, disassembly and turnover in migrating cells – over and over and over again. Nat. Cell Biol. 4, E97-100. 10.1038/ncb0402-e97 [DOI] [PubMed] [Google Scholar]
- Xiao K., Garner J., Buckley K. M., Vincent P. A., Chiasson C. M., Dejana E., Faundez V. and Kowalczyk A. P. (2005). p120-Catenin regulates clathrin-dependent endocytosis of VE-cadherin. Mol. Biol. Cell 16, 5141-5151. 10.1091/mbc.E05-05-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiumin C., Qingting Y., Zhibin L., Jiping Y., Liran X. and Huijun G. (2015). Asymptomatic stage of human immunodeficiency virus infection is the optimal timing for its management with Traditional Chinese Medicine. J. Tradit. Chin. Med. 35, 244-248. 10.1016/S0254-6272(15)30036-4 [DOI] [PubMed] [Google Scholar]