Abstract
Molecular modeling and molecular dynamics simulations have been performed to elucidate feasible structures in the Y-family Dpo4 DNA polymerase for the 1S-(–)-trans-anti-B[c]Ph-N6-dA adduct, derived from the fjord region polycyclic aromatic hydrocarbon (PAH) benzo[c]-phenanthrene. Three types of models were delineated as follows: an intercalation model, a model with the aromatic ring system in the polymerase major groove open pocket, and a –1 deletion major groove model. All four 2′-deoxyribonucleoside 5′-triphosphates (dNTPs) were considered in the first two cases, and a normal Watson–Crick partner positioned to have skipped the modified template was employed as the incoming dNTP in the –1 deletion case. The trajectories derived from the dynamics simulations were analyzed in detail to evaluate the extents of distortion for each system. Overall, our results suggest that the major groove model is the least distorted, followed by the –1 deletion model, while the intercalation model is perturbed the most. The syn-dGTP and syn-dATP mismatches opposite the lesion are well-accommodated in the major groove model, as is the normal Watson–Crick partner dTTP. The intercalation model appears most likely to impede the polymerase. More broadly, these models look reasonable for other PAH metabolite-derived adducts to adenine with similar 1S stereochemistry. Furthermore, these models suggest how error-prone translesion synthesis by Y-family polymerases might produce mutations that may play a role in the initiation of cancer.
Introduction
Y-family DNA polymerases are believed to replace replicative DNA polymerases that are stalled at sites of bulky lesions such as those caused by the binding of polycyclic aromatic hydrocarbon (PAH)1 metabolites to DNA (1, 2). Such adducts, if not repaired, largely block primer extension by replicative DNA polymerases (3–9) but can be bypassed by the Y-family polymerases (9). In general, members of this latter family lack an intrinsic proofreading exonuclease, exhibit low processivity, and replicate undamaged DNA with low efficiency and fidelity (10). Translesion synthesis by Y-family DNA polymerases can be error-prone and may result in mutations, which, if present in critical cell cycle control genes such as oncogenes and tumor suppressor genes, can lead to cancer initiation (11, 12).
Structural characterizations of members of Y-family bypass DNA polymerases (13–17) reveal that they share common features with replicative polymerases. All possess a right-hand architecture containing thumb, palm, and finger subdomains in the polymerase domain. The core of the palm appears conserved between Y-family and replicative polymerases, while the thumb and finger are significantly smaller in the Y-family. Furthermore, Y-family polymerases contain a unique little finger subdomain, which is believed to play an important role in facilitating DNA binding (13–18). In addition, active sites of Y-family polymerases, in contrast to replicative polymerases, are solvent accessible and more spacious (13–16) and can in some cases accommodate two templating bases, and the nascent base pair is sterically less constrained than in replicative polymerases (13, 15).
A bulky DNA adduct at the active site is bypassed more easily by the Y-family DNA polymerases, and the spacious active site results in a relaxed geometric selection for the incoming 2′-deoxynucleotide 5′-triphosphate (dNTP) (9), thus compromising the efficiency and fidelity of DNA replication. The lesion bypass ability, accuracy, and efficiency of these polymerases vary significantly and depend on the type of DNA lesion (19–39).
The model Y-family Dpo4 DNA polymerase from the archaeon bacterium Sulfolobus solfataricus P2 is a member of the DinB family, which also includes eukaryotic Pol κ (18). This enzyme replicates an undamaged DNA template with an error rate of ~10−3 to 10−4 and frequently generates deletions (30, 40, 41). It is able to bypass cis–syn-cyclobutane dimers, 6-4 photoproducts, abasic sites, 1–2 cis-diamminedichloro-platinum adducted G–G, N2-acetylaminofluorene C8-modified guanine (30), and the benzo[a]pyrene (B[a]P)-derived 10S-(+)-trans-anti-B[a]P-N2-dG adduct (37). Dpo4 can insert the correct complementary nucleotide at each of the DNA lesions, but mispairs and frameshifts have also been observed (30).
Crystal structures of Dpo4 with unmodified DNA and the incoming nucleotide cocrystallized with the protein provide excellent models for investigating the structural features that determine lesion bypass efficiency and fidelity (13). The structures solved for unmodified Dpo4 show two modes of entry for a dNTP. In one (type I, PDB (42) ID: 1JX4), the entering nucleotide is paired with the normal, 3′ (proximal) template base. In the other crystal (type II, PDB ID: 1JXL), the entering nucleotide skipped this base and paired instead with the next 5′ (distal) base, its complement. This capacity of Dpo4 to accommodate two template residues within its active site suggests that this site may be sufficiently spacious to accommodate a bulky intercalated adduct and how a –1 deletion mutation might be engendered.

The nature of the geometric restraints imposed by the polymerase active site plays a key role in the selection of the incoming nucleotide (43, 44). A relatively large binding pocket might lower the nucleotide selectivity and not only allow for the incorporation of the incorrect dNTP but also facilitate the accommodation of bulky lesions within the active site. In replicative DNA polymerases, the dNTP binding pocket is tightly packed with protein residues on the minor groove side, and, to some extent, on the major groove side, and thereby strongly favor the incorporation of the correct dNTP because of these steric restraints (44). However, because of the small thumb and finger subdomains, the Dpo4 DNA polymerase is characterized by an active site with open pockets on both sides of the nascent base pair (13), suggesting that a bulky adduct may also readily fit in an extrahelical position.
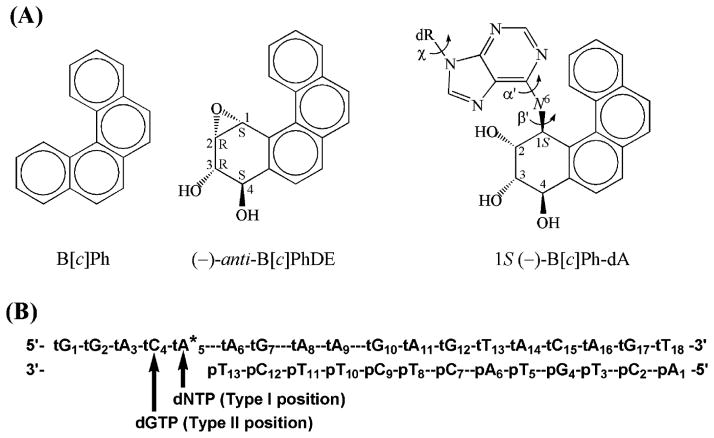
The fjord region PAH, benzo[c]phenanthrene (B[c]Ph), contains a sterically hindered, nonplanar aromatic region (45) (Figure 1A). In living cells, B[c]Ph is metabolically activated to a pair of highly reactive (+)- and (–)-anti-B[c]Ph diol epoxides (B[c]PhDE) (46, 47). The diol ep-oxides bind extensively to adenine residues in DNA (48, 49), forming the stereoisomeric 1R-(+)- and 1S-(–)-trans-anti-benzo[c]phenathrenyl-N6-deoxyadenosine [1R-(+)and 1S-(–)-B[c]Ph-dA] adducts (Figure 1A). These adducts are not efficiently removed by human nucleotide excision repair processes, at least in the sequence context of the human oncogene N-ras mutational hotspot codon 61 (CAA, Figure 1B) (50), which is mutated in a variety of human cancers (51–53). The 1S-(–)-B[c]Ph-dA adduct is of great interest, because it is more mutagenic than the corresponding 1R-(+) adduct in an SOS-induced Escherichia coli SMH77 system (54), where translesional bypass DNA polymerases are activated (55). However, cells containing the 1S-(–) adduct can survive with or without mutations (54). Therefore, translesion synthesis bypassing this adduct is likely to play an important role in the survival of cells. Primer extension studies have shown that the Y-family DNA polymerase Pol ι is capable of bypassing B[c]PhDE modified adenine adducts in vitro (33).
Figure 1.
(A) Structures of B[c]Ph, (–)-anti-B[c]PhDE, and 1S-(–)-B[c]Ph-dA. Torsion angles α′, β′, and χ are defined as follows: α′, N1–C6–N6–C1(B[c]Ph); β′, C6–N6–C1(B[c]Ph)-C2(B[c]Ph); and χ, O4′–C1′–N9–C4. (B) Human N-ras codon 61 sequence context employed in the MD simulation. * denotes the B[c]PhDE modification site. p and t denote the primer and template strand, respectively. Arrows denote the type I and type II positions for incoming dNTP. dR denotes 2′-deoxyribose.
High-resolution NMR solution structures reveal that the 1S-(–)-B[c]Ph-dA adduct in a duplex 11-mer has the B[c]Ph moiety intercalated into the B-DNA duplex with the aromatic ring system positioned on the 3′-side of the modified adenine and with Watson–Crick base pairs present, although distorted (56). Recent crystallographic studies of polycyclic aromatic lesions have revealed that the structures of the adducts in solution, as determined by high-resolution NMR, may well be preserved within a polymerase. Examples include a 10S-(+)-trans-anti-B[a]P-N2-dG adduct (57) and a N-(2′-deoxyguanosin-8-yl)-aminofluorene adduct (58) in the Bacillus fragment, BF. However, crystal structures resolved for a 10R-(+)-cis-anti-B[a]P-N6-dA adduct in Dpo4 (59) revealed two conformations of the B[a]P moiety. In one, the hydrocarbon is intercalated on the 5′-side of the modified base, with an orientation nearly identical to that found in a DNA duplex in solution (60). In the other conformation, however, the hydrocarbon moiety adducted to the base 3′ to the template is situated in the major groove open pocket of the Dpo4. Our recent modeling and molecular dynamics (MD) studies (37) of the 10S-(+)-trans-anti-B[a]P-N2-dG adduct have shown that a bulky PAH metabolite-derived lesion can be readily accommodated in the Dpo4 major groove as well as in the smaller minor groove pocket. However, a minor groove conformation does not apply in the current study because the B[c]Ph bulk would still be positioned in the major groove side open pocket when the N6-modified adenine adopts the syn glycosidic bond conformation.
The goal of our present work is to investigate possible structures of the 1S-(–)-B[c]Ph-dA adduct with the modified adenine residue as a templating base within a Dpo4 ternary complex, in an effort to understand the potential impact of the adduct on polymerase structure and functioning. We wished to make a comprehensive investigation of feasible locations for the B[c]Ph ring system within the polymerase. Accordingly, we considered three possible accommodation sites for the bulky aromatic ring system: (i) the NMR solution structure with B[c]Ph moiety intercalated on the 3′-side of the modified adenine, with a dNTP paired with the damaged template; (ii) a position in the major groove open pocket of the polymerase with the dNTP paired with the modified template adenine residue; and (iii) a position in the major groove polymerase open pocket with the dNTP paired with the base 5′ to the modified adenine. The Dpo4 type II crystal structure provided the initial models for cases (i) and (iii), while the type I crystal structure was utilized for case (ii) and the unmodified control (13). All four incoming dNTPs were considered for (i) and (ii) with purines both anti and syn, while the –1 deletion model was considered only with the normal Watson–Crick partner. MD simulations provided ensembles of structures for detailed analyses to evaluate the extent of distortion for each case as compared to the unmodified control. Our results suggest that the major groove orientation is least distorted among the three models and allows for accommodation of the normal partner dTTP, as well as the mismatched syn-dGTP and syn-dATP. The –1 deletion model is also feasible but with an intermediate degree of structural perturbation, whereas the intercalative structures represent the most significantly distorted conformations. This suggests that the latter might be the most blocking adduct conformation and impede the progress of the polymerase.
Computational Methods
Molecular Modeling
1. Unmodified Control and 1S-(–)-B[c]Ph-dA Major Groove Type I Model in Dpo4
The type I crystal structure of the ternary complex of Dpo4 polymerase with unmodified DNA and a correct incoming nucleotide (PDB ID: 1JX4) was employed as a starting model for the unmodified control. Hydrogen atoms not resolved in the crystal structure were added by the LEaP module in AMBER (61). Engineered polymerase residues 1, 76, 89, 157, 216, and 251 in the crystal structure were remodeled to natural methionine residues. The residue at the 5′-terminus of the template DNA strand, which was not located in the crystal structure, was modeled, guided by a canonical B-form conformation (62) with modifications to avoid collisions and optimize stacking (Table S1, Supporting Information). The missing C-terminal segment (residues 342–352) was not modeled because this part of the protein is far from the active site area. In the crystal structure, the incoming nucleotide is a dideoxydiphosphate nucleotide, ddADP. The Pα is in the position of the Pγ if it were dATP (13), which is out of the range for the phosphoryl transfer reaction. Additionally, the Dpo4 crystal structure contains only one Ca2+ in the active site. Using the dNTP and active site Mg2+ ions in the ternary complex of T7 DNA polymerase (63) as a guide, we remodeled ddADP to dTTP, Ca2+ to Mg2+ ion, and added the missing Mg2+ ion into the polymerase active site. The positions of these two Mg2+ ions were adjusted, and two water molecules were added to achieve proper octahedral coordination. The nucleotide binding Mg2+ ion was chelated with Oδ1 of Asp7, Oδ2 of Asp105, O of Phe8, O2α, O2β, and O1γ of dTTP. The catalytic Mg2+ ion was chelated with Oδ2 of Asp7, Oδ1 of Asp105, O2α of dTTP, O3′ of pT13, and the oxygen atoms of the two added water molecules. The DNA sequence was adjusted to match that of the human N-ras codon 61 sequence context (Figure 1B). The dideoxy-nucleotide of the 3′-terminus of the primer strand was remodeled as deoxynucleotides by adding the 3′-hydroxyl group.
This model was also employed in the creation of the major groove type I model. The templating adenine was covalently bonded to the B[c]Ph moiety, and the α′ and β′ torsion angles (Figure 1A) were varied over their 360° ranges at 30° intervals in combination to locate a collision-free position in the major groove open pocket. Linkage site torsional angles α′, β′, and χ of 160.0, −85.0, and −123.3°, respectively, were collision-free and represented a very narrow conformational domain that could accommodate the B[c]Ph moiety without modifying the polymerase; these are within an energetically favored domain for this adduct (64). Six starting structures were prepared by varying the base of the incoming dNTP: dTTP, dCTP, dATP, syn-dATP, dGTP, and syn-dGTP.
2. Intercalation and –1 Deletion Models of 1S-(–)-B[c]Ph-dA in Dpo4
The type II crystal structure of Dpo4, complexed with unmodified DNA and an incorrect incoming nucleotide (PDB ID: 1JXL) (13), was employed to model the 1S-(–)-B[c]Ph-dA intercalation and –1 deletion systems. This structure contains an unpaired residue on the template strand 3′ to the properly matched nascent base pair. Both this unpaired residue and the nascent base pair are in the active site. Again, hydrogen atoms were added with LEaP (61), and the C-terminal segment of the Dpo4 protein (residues 342–352) was not modeled. The nucleotide at the 5′-terminus of the template strand was not located in the crystal structure and was hence modeled. Furthermore, yet an additional residue had to be added to the 5′-end of the template so that the number of residues in the single-stranded overhang matched the unmodified control (which is based on the type I crystal), as the overhang was one residue shorter in the type II crystal. This modeling was guided by canonical B-DNA (62) with modifications to avoid collision with protein residues and optimize stacking interactions (Table S1, Supporting Information). In addition, the crystal structure contains a Mg2+ ion in the nucleotide binding position and a Ca2+ ion in the catalytic position. The Ca2+ ion was remodeled as a Mg2+ ion, the positions of the two Mg2+ ions were adjusted, and two water molecules were added to achieve proper octahedral coordination. The nucleotide binding Mg2+ ion was chelated with Oδ1 of Asp7, Oδ2 of Asp105, O of Phe8, O2δ and O2β of the dNTP, and O of one of the added water molecules. The catalytic Mg2+ ion was chelated with Oδ2 of Asp7, Oδ1 of Asp105, Oε2 of Glu106, O2α and O2δ of the dNTP, and O of the other added water molecule. The ethylene glycol molecule trapped between the primer terminus residue and the incoming nucleotide during crystallization was deleted. The DNA sequence was adjusted to match that of the N-ras codon 61 sequence context, as shown in Figure 1B.
To create the intercalation structures, we remodeled the 3′ (proximal) template base to a 3′-intercalated 1S-(–)-B[c]Ph-dA adduct in the active site, guided by the NMR solution structure (65), and the favored α′ and β′ domains computed for the adduct (64). The torsion angles of the modified adenine and its surrounding residues were adjusted (Table S1, Supporting Information) to relieve steric hindrance. Linkage site torsional angles α′, β′, and χ are 157.0, −82.0, and −35.5°, respectively. Six starting structures were created by varying the base of the dNTP: dTTP, dCTP, dATP, syn-dATP, dGTP, and syn-dGTP.
To create the –1 deletion structure where the 1S-(–)-B[c]Ph-dA adopts a major groove open pocket position with the incoming dGTP paired with the 5′ (distal) cytosine, the templating adenine was covalently bonded to the B[c]Ph moiety with α′ = 171.2° and β;′ = −89.8°, respectively, within an energetically favored domain for this adduct (64), and guided by steric feasibility. The glycosidic and backbone torsion angles of the two template bases at the active site were not changed from their crystal values. Only dGTP was considered for this structure. The Insight II 2000 software package from Accelrys Inc. was used for all modeling work.
Force Field Parametrization
Partial charges for 1S-(–)-B[c]Ph-dA were computed previously (66). Partial charges for dTTP, dATP, dGTP, and dCTP were those employed in previous work (67, 68). Parameters added to the AMBER 6.0 force field for the adduct were assigned by analogy to chemically similar atom types already available in the parm98 parameter set (69) and are given in the Supporting Information of our previous work (66).
MD Simulation Protocol
MD simulations were carried out using the AMBER 6.0 package (61) with the Cornell et al. force field (70) and the parm98 parameter set (69). Long-range electrostatic interactions were treated by using the particle mesh Ewald method (71, 72). A 9 Å cutoff was applied to the nonbonded Lennard–Jones interactions. The SHAKE algorithm (73) was applied to constrain all bonds involving hydrogen atoms with tolerance of 10−6 Å, and a 2 fs time step was used in the dynamics simulations. The translational motion of the center of mass was removed every 1 ps. Observation of trajectories showed no obvious overall rotation of the system; thus, energy leakage from internal motion to global rotation through the “flying ice cube effect” (74) did not happen in this case.
Eighteen Na+ ions were added to each system for neutralization using the LEaP module in AMBER 6.0. The systems were then solvated into a rectangular box of TIP3P water molecules (75), which extended ~9 Å from the polymerase–DNA ternary complex atoms in each direction. The Simulaid program (76) was employed to minimize the number of water molecules by optimizing the orientation of the rectangular water box, yielding a periodic water box of ~82 Å × 92 Å × 73 Å for the polymerase–DNA complex system. About 12000 water molecules were added to solvate the polymerase–DNA complex in each system.
All systems followed the same equilibration protocol: first, the water molecules and counterions were minimized for 5000 steps of steepest descent with solute atoms fixed, followed by 20 ps initial dynamics at 10 K with 5.0 kcal/mol restraints on the solute to allow the solvent to relax. The system was then heated from 10 to 300 K over 50 ps using the Berendsen coupling algorithm (77) with a coupling parameter of 1.0 ps, followed by dynamics at 300 K with 5.0 kcal/mol restraints on the solute for 50 ps. The restraints were then decreased to 1.0 kcal/mol for 25 ps and to 0 kcal/mol for another 50 ps. Production simulation was then performed at 1 atm, 300 K, with a coupling parameter of 1.0 ps, for 1 ns.
Trajectory Analysis
The PTRAJ and CARNAL modules of the AMBER package (61) were employed to analyze the trajectories.
Stability of the MD Simulations
Overall and linkage site structural fluctuations became reasonably stable after 200 ps of MD simulation for all systems. The root-mean-square deviation (RMSD) and linkage site torsion angles (α′, β′, χ) of the Dpo4–DNA–dNTP complex as a function of time are shown in Figures S1–S3 (Supporting Information).
Results
We carried out molecular modeling and dynamics simulations for an unmodified control, an intercalative model, a major groove model with incoming dNTP opposite the modified template in the type I crystal position, and another major groove model with incoming dGTP normal partner placed in the type II crystal position (Figure 1B), modeling a –1 deletion. For the intercalation and type I major groove structure, we investigated all four incoming dNTPs, and purines were studied in both syn and anti glycosidic bond conformations, since a Dpo4 structure with a damaged template opposite a syn incoming purine has been previously observed in a crystal structure (78). Glycosidic bond orientations for dNTPs are anti unless denoted syn.
Unmodified Control. Structural Features in the Active Site Are Consistent with Normal Nucleotide Incorporation
In the unmodified control simulation, the ternary complex contained an unmodified templating adenine with incoming dTTP. A stereoview of the ternary complex structure at the end of the simulation is shown in Figure 2A. We found that a number of structural features were maintained throughout the course of the 1 ns simulation. These include the Watson–Crick hydrogen bonds between the incoming dTTP and the unmodified template adenine that were highly occupied (>90%) (Table 1 and Figure S4, Supporting Information), the dNTP binding pocket (Figure 3A), the classical B-DNA base pair stacking (Figure S5A, Supporting Information), and chelation of the two active site Mg2+ ions by Asp7, Asp105, Glu106, and Phe8 (Figure 4A). Interestingly, Glu106, which was not chelated with metal ions in the crystal structure or in the starting model, became chelated with the catalytic Mg2+ ion during equilibration. The absence of the catalytic Mg2+ ion in the crystal structure was likely due to the presence of ddADP rather than ddATP in the crystal (13). However, our simulation contained the dNTP, and the simulation restored the chelation with Glu106 as in the type II crystal.
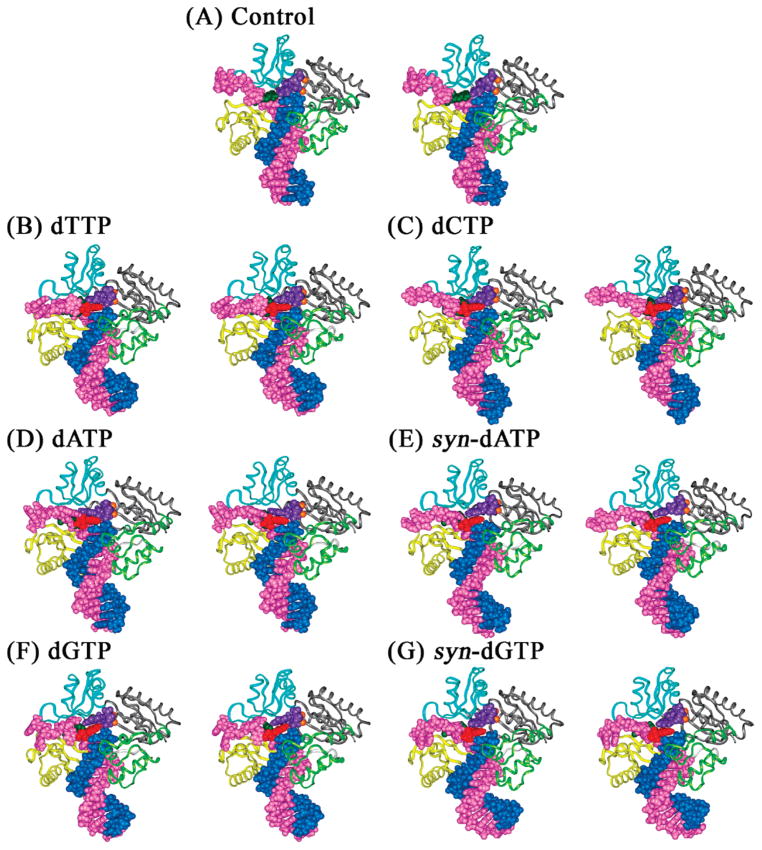
Figure 2.
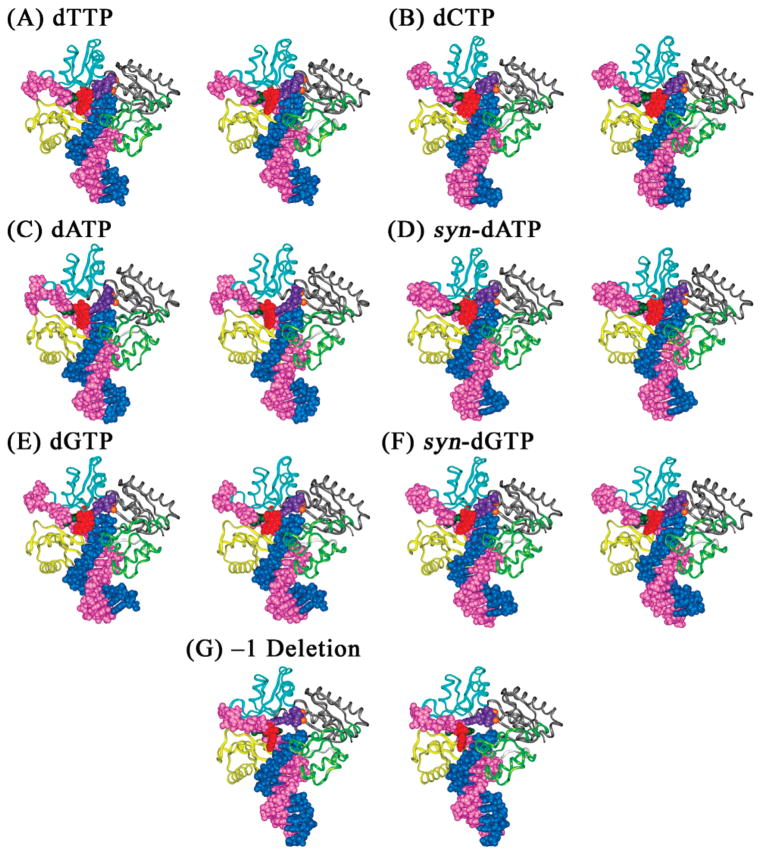
Structures of the unmodified control and 1S-(–)-B[c]Ph-dA intercalation models of the Dpo4 ternary complexes after 1 ns of MD. (A) Unmodified dA opposite dTTP; (B) 1S-(–)-B[c]Ph-dA opposite dTTP; (C) 1S-(–)-B[c]Ph-dA opposite dCTP; (D) 1S-(–)-B[c]Ph-dA opposite dATP; (E) 1S-(–)-B[c]Ph-dA opposite syn-dATP; (F) 1S-(–)-B[c]Ph-dA opposite dGTP; (G) 1S-(–)-B[c]Ph-dA opposite syn-dGTP. The DNA residues, Mg2+ ions, and dNTPs are shown in CPK and protein in ribbon. Color codes: palm, gray; thumb, light green; fingers, cyan; little finger, yellow; primer, blue; template, magenta; modified adenine base, dark green; B[c]Ph, red; incoming dNTP, purple; and Mg2+ ions, orange. All stereo images are constructed for viewing with a stereoviewer.
Table 1.
Hydrogen Bonds of >10% Occupancy between the Templating Residue and the Incoming dNTP
| model | hydrogen bonda | distance (Å)b | angle (°)b | occupancy (%)c |
| control | (tA5)N6–H61...O4(dTTP) | 2.9 ± 0.1 | 167.5 ± 7.1 | 98.1 |
| (dTTP)N3–H3...N1(tA5) | 3.0 ± 0.1 | 163.5 ± 9.6 | 94.2 | |
| intercalation | (tA5*)N6–H6...O4(dTTP) | 3.0 ± 0.2 | 162.7 ± 8.4 | 93.9 |
| (dTTP)N3–H3...N1(tA5*) | 3.1 ± 0.1 | 165.7 ± 7.2 | 84.9 | |
| (tA5*)O4Z-HO4Z...O4(dTTP) | 2.8 ± 0.2 | 170.5 ± 5.4 | 30.8 | |
| (dCTP)N4–H42...N3(tA5*) | 3.1 ± 0.1 | 165.9 ± 7.5 | 67.0 | |
| (dCTP)N4–H42...N1(tA5*) | 3.2 ± 0.1 | 147.0 ± 7.2 | 15.5 | |
| (dATP)N6–H61...N1(tA5*) | 3.1 ± 0.1 | 149.6 ± 9.2 | 31.7 | |
| (syn-dATP)N6–H62...N1(tA5*) | 3.1 ± 0.1 | 155.9 ± 8.1 | 95.2 | |
| (dGTP)N2–H21...N1(tA5*) | 3.1 ± 0.2 | 155.9 ± 10.3 | 59.4 | |
| (tA5*)N6–H6...O6(syn-dGTP) | 3.1 ± 0.2 | 158.0 ± 10.0 | 58.6 | |
| major groove type I | (tA5*)N6–H6...O4(dTTP) | 2.9 ± 0.1 | 168.9 ± 5.2 | 99.7 |
| (tA5*)O4Z-HO4Z...O4(dTTP) | 2.8 ± 0.1 | 169.2 ± 6.4 | 99.6 | |
| (dTTP)N3–H3...N1(tA5*) | 3.1 ± 0.1 | 165.2 ± 8.1 | 96.9 | |
| (dCTP)N4–H41...N1(tA5*) | 3.1 ± 0.2 | 150.6 ± 9.7 | 54.3 | |
| (dCTP)N4–H42...O4Z(tA5*) | 3.1 ± 0.2 | 157.7 ± 10.7 | 24.0 | |
| (tA5*)N6–H6...N1(dATP) | 3.2 ± 0.1 | 167.1 ± 7.1 | 35.3 | |
| (tA5*)O4Z-HO4Z...N1(dATP) | 2.9 ± 0.2 | 168.0 ± 8.1 | 27.0 | |
| (tA5*)O4Z-HO4Z...N7(dATP) | 3.0 ± 0.2 | 155.6 ± 9.6 | 19.2 | |
| (tA5*)O4Z-HO4Z...N1(syn-dATP) | 2.9 ± 0.1 | 162.8 ± 7.8 | 99.6 | |
| (syn-dATP)N6–H62...N1(tA5*) | 3.2 ± 0.1 | 144.6 ± 7.3 | 25.2 | |
| (tA5*)O4Z-HO4Z...O6(dGTP) | 2.8 ± 0.1 | 168.4 ± 6.5 | 100.0 | |
| (dGTP)N1–H1...N1(tA5*) | 3.0 ± 0.1 | 160.8 ± 9.6 | 98.5 | |
| (tA5*)N6–H6...O6(dGTP) | 3.1 ± 0.2 | 163.0 ± 7.9 | 93.3 | |
| (tA5*)O4Z-HO4Z...O6(syn-dGTP) | 2.7 ± 0.1 | 169.8 ± 5.7 | 99.6 | |
| (tA5*)N6–H6...O6(syn-dGTP) | 3.1 ± 0.2 | 167.2 ± 6.7 | 85.2 | |
| (syn-dGTP)N1–H1...O4Z(tA5*) | 3.2 ± 0.1 | 140.2 ± 3.9 | 11.4 | |
| –1 deletion | (dGTP)N1–H1...N3 (tC4) | 3.0 ± 0.1 | 168.2 ± 5.5 | 100.0 |
| (dGTP)N2–H21...O2(tC4) | 2.9 ± 0.1 | 170.0 ± 5.1 | 99.9 | |
| (tC4)N4–H41...O6(dGTP) | 2.9 ± 0.1 | 169.6 ± 6.0 | 99.6 | |
| (dGTP)N1–H1...O2(tC4) | 3.3 ± 0.1 | 149.1 ± 5.1 | 48.4 |
Hydrogen bonds are specified in the format (A)B–C...D(E), where A is the residue name of the donor atom, B is the name of the donor atom, C is the name of the hydrogen atom, D is the name of the acceptor atom, and E is the residue name of the acceptor atom.
denotes 1S-(–)-B[c]Ph-dA modification. Z denotes the B[c]Ph moiety on the adducted adenine.
Average values and standard deviations are provided.
The criterion for hydrogen bond occupancy is heavy atom–heavy atom distance ≤ 3.4 Å and heavy atom–hydrogen–heavy atom angle ≥ 135°.
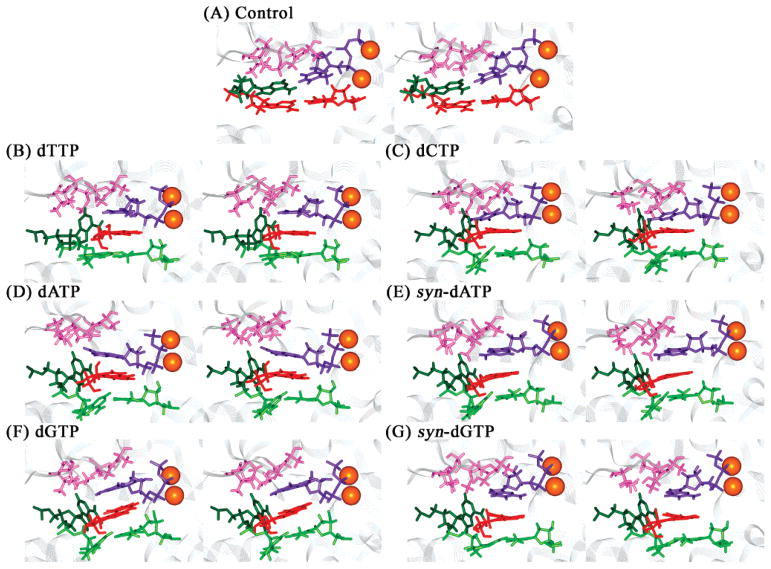
Figure 3.
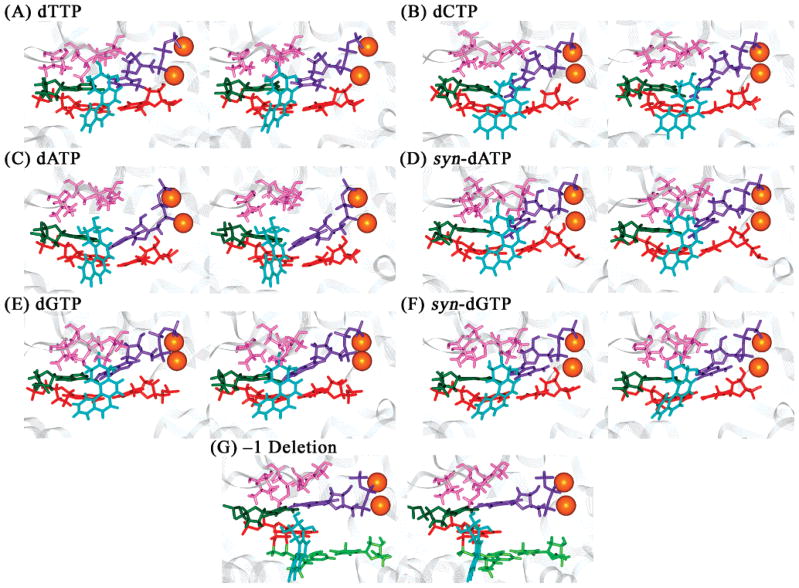
dNTP binding pockets of the unmodified control and 1S-(–)-B[c]Ph-dA intercalation structures of the Dpo4 ternary complexes after 1 ns of MD. Panels A–G are the same as in Figure 2. Color codes: ceiling, pink; floor, red; far wall, dark green; incoming dNTP, purple; tA6–pT13 base pair in B[c]PhDE modified systems, light green; and Mg2+ ions, orange spheres. All stereo images are constructed for viewing with a stereoviewer.
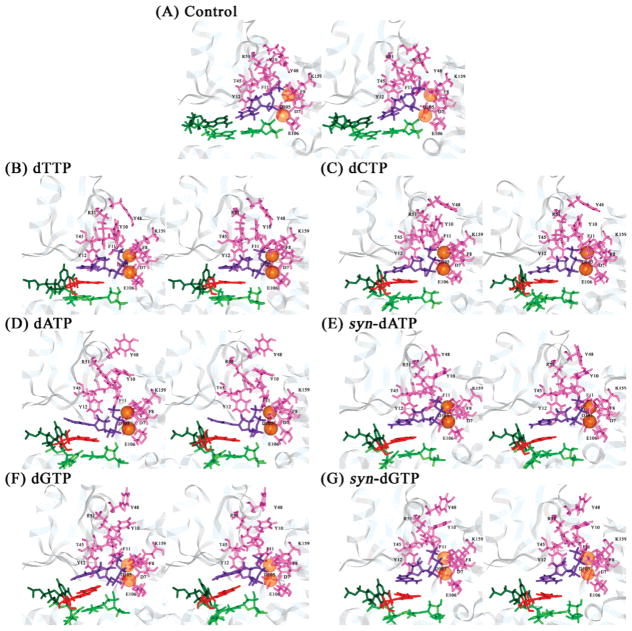
Figure 4.
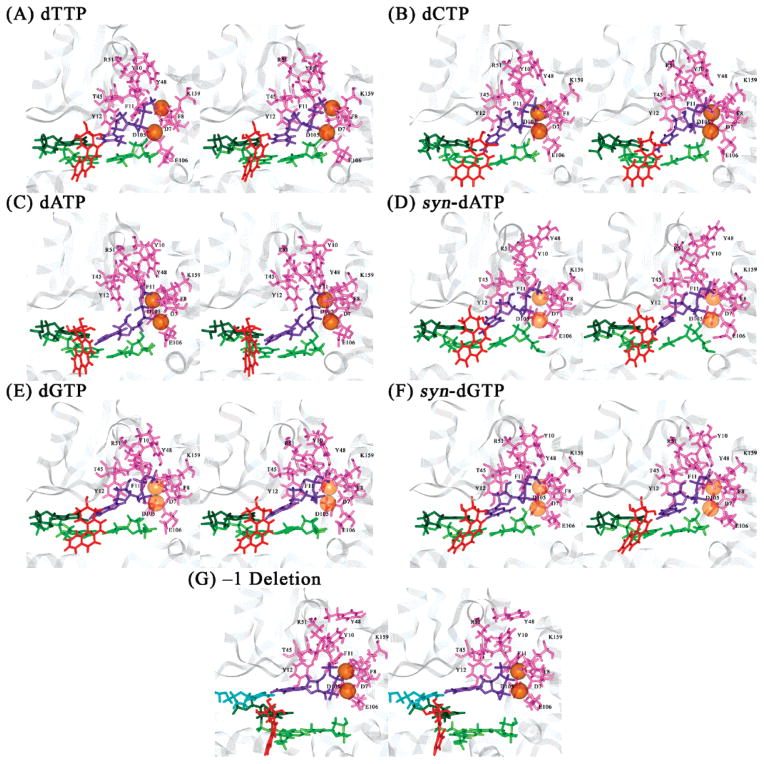
Interactions between polymerase residue and sugar-triphosphate of dNTP in the unmodified control and 1S-(–)-B[c]Ph-dA intercalation structures of the Dpo4 ternary complexes after 1 ns of MD. Panels A–G are the same as in Figure 2. Color codes: protein residues, pink with labels in black; dNTP, purple; Mg2+, transparent orange spheres; modified adenine base, dark green; B[c]Ph, red; and base pair tA6–pT13, light green. All stereo images are constructed for viewing with a stereoviewer.
An optimized A–T Watson–Crick base pair has a C1′–C1′ distance of 10.8 Å (79), as also observed in our unmodified control (10.8 ± 0.3 Å, 100% occupancy) (Table 2 and Figure S6A, Supporting Information). A combination of experimental and computational studies has shown that base pairs with C1′–C1′ distances up to ~12 Å can be accommodated by DNA polymerases (79). The efficiency of dNTP incorporation is significantly lowered when this distance is increased beyond ~12.5 Å. On the other hand, a decrease in the distance also penalizes the efficiency of dNTP incorporation.
Table 2.
Distances and Occupancies for C1′–C1′ and Pα–O3′
| model | dNTP | C1′–C1′ distance (Å)a | C1′–C1′ occupancy (%)b | Pα–O3′ distance (Å)a | Pα–O3′ occupancy (%)c |
|---|---|---|---|---|---|
| control | dTTP | 10.8 ± 0.3 | 100.0 | 3.5 ± 0.1 | 73.3 |
| intercalation | dTTP | 8.9 ± 0.3 | 1.1 | 5.7 ± 0.9 | 3.7 |
| dCTP | 8.7 ± 0.4 | 0.6 | 5.6 ± 0.6 | 0.05 | |
| dATP | 9.6 ± 0.3 | 40.7 | 4.0 ± 0.6 | 7.7 | |
| syn-dATP | 8.8 ± 0.4 | 3.0 | 6.5 ± 0.5 | 0.0 | |
| dGTP | 10.1 ± 0.4 | 88.9 | 4.1 ± 1.0 | 39.5 | |
| syn-dGTP | 9.9 ± 0.5 | 75.1 | 6.7 ± 0.6 | 0.0 | |
| major groove type I | dTTP | 10.8 ± 0.2 | 100.0 | 3.7 ± 0.1 | 9.4 |
| dCTP | 11.8 ± 0.4 | 71.5 | 4.8 ± 0.3 | 0.0 | |
| dATP | 11.5 ± 0.4 | 91.5 | 6.5 ± 0.7 | 0.0 | |
| syn-dATP | 11.1 ± 0.4 | 99.8 | 3.2 ± 0.1 | 99.8 | |
| dGTP | 12.3 ± 0.2 | 8.6 | 5.5 ± 0.5 | 0.8 | |
| syn-dGTP | 11.1 ± 0.3 | 99.8 | 3.1 ± 0.1 | 100.0 | |
| –1 deletion | dGTP | 10.7 ± 0.2 | 100.0 | 5.7 ± 0.8 | 2.8 |
Average values and standard deviations are provided. C1′–C1′ distances are for residues of the nascent base pair, and Pα–O3′ distances are between Pα of dNTP and O3′ of the primer terminus.
Criterion for C1′–C1′ distance occupancy: within 10.8 ± 1.2 Å.
Criterion for Pα–O3′ distance occupancy: ≤ 3.6 Å.
For the phosphoryl transfer reaction to take place, it is necessary for the distance between the Pα of the dNTP and the O3′ of the primer terminus (Pα–O3′ distance) to approach ~3.5 Å, which is close to the sum of their van der Waals radii (80). The Pα–O3′ distance is 3.5 ± 0.1 Å (Table 2 and Figure S6B, Supporting Information), with 73.3% occupancy of the near-reaction-ready distance (≤ ~3.6 Å) for the phosphoryl transfer reaction. Additionally, the HO3′ of pT13 is hydrogen bonded with Oδ1 of Asp105 for 72% of the simulation, stabilizing the position of O3′ of the pT13 for phosphoryl transfer.
Polymerase residues Lys159, Tyr10 in the palm subdomain, and Arg51, Thr45, Phe11, and Tyr12 in the finger subdomain form stable hydrogen bonds with atoms on the sugar-triphosphate of the incoming dTTP throughout the MD simulation (Figure 4A and Table S2, Supporting Information). Among these residues, Lys159, Arg51, and Phe11 are found to be highly conserved in Y-family DNA polymerases, namely, human Pol κ, Pol ι, and Pol η, yeast Pol η, as well as E. coli UmuC and DinB (81). The phenyl ring of Tyr12 also stacks with the sugar ring of the dTTP throughout the simulation. The hydroxyl group of Tyr48 is in the vicinity of the Pγ of the dTTP, interacting favorably with the oxygen atoms on Pγ and stabilizing the incoming nucleotide.
Intercalation Models Based on the Adduct NMR Solution Structure
In this series of simulations, the B[c]Ph moiety is intercalated on the 3′-side of the templating modified adenine with all four incoming dNTPs investigated. Stereoviews of the ternary structures after the 1 ns MD simulation are shown in Figure 2.
1. The dNTP Binding Pocket Is Enlarged to Accommodate the B[c]Ph Moiety
Employing the description devised by Kool (43), we define the incoming dNTP binding pocket as follows: the finger subdomain protein residues Val32, Ala42, Ala44, Ala57, Gly58, and Met76, which lie on top of the nascent base pair, form the “ceiling” of the pocket (Figure 3); base pair pT13–tA6, which is 3′ to the nascent base pair, comprises the “floor” of the binding pocket in the unmodified structure; however, in the B[c]PhDE modified systems, the floor is replaced by the B[c]Ph ring system; the DNA template residue tA5 in the active site forms the “far wall” in the unmodified structure. In the B[c]PhDE modified structures, the far wall is deepened and the pocket opening is enlarged due to the modification (Figure 3B–G). Thus, the lesion plays an important role in redefining the dNTP binding pocket.
To maximize stacking interactions between the B[c]-Ph ring system and the incoming nucleotide, the incoming dNTP is slid toward the modified template adenine (Figure 3B–G). This becomes feasible due to the deepening of the binding pocket, induced by the tilt of the modified base. The width of the nascent base pair is therefore reduced, as revealed by the distances between the C1′ of 1S-(–)-B[c]Ph-dA and the C1′ of the incoming dNTP (Table 2 and Figure S6A, Supporting Information), in comparison to the unmodified control. These widths are reduced by moderate to significant extents and reflect one component of the alterations to the active site induced by the intercalated B[c]Ph ring system.
2. Sampling of Near-Reaction-Ready Pα-O3′Distances Varies with Incoming dNTP
The Pα-O3′ distance was monitored during the course of the simulation for the modified structures with all dNTPs (Table 2 and Figure S6B, Supporting Information). We note that in the B[c]Ph intercalated starting structures modeled from the type II crystal structure of Dpo4 (13), the distance between O3′ of the primer terminus and Pα of the incoming dNTP is ~4.0 Å, due to an ethylene glycol molecule trapped between the primer terminus residue and the incoming nucleotide during crystallization (13), and that was, of course, removed from our starting models (see Computational Methods).
3. Mg2+ Ion Chelation Is Preserved in All Simulations
Conserved carboxylates, Asp7, Asp105, and Glu106, which play a key role in Y-family polymerase activity (13, 82), are chelated by the two positively charged Mg2+ ions in the active site of the Dpo4 DNA polymerase. Additionally, the carbonyl O of Phe8, dNTP O2β, and O2γ are chelated with the nucleotide binding Mg2+ ion, and dNTP O2α, O2β with the catalytic Mg2+ ion (Figure 4B–G). These interactions, and hence the positioning of the Mg2+ ions, are stably maintained throughout the MD simulations in all intercalation systems and are independent of the type of incoming dNTPs.
4. Hydrogen Bonding Interactions Stabilize the Nascent Base Pairs in the Active Site to a Lesser Extent Than in the Unmodified Control
Although the B[c]PhDE-modified adenine is severely buckled with respect to the incoming dNTP, there are distorted hydrogen bonding interactions in the nascent base pair. Such nonplanar base pairs have been observed in crystal and NMR solution structures (83–85), and quantum mechanical calculations also reveal that they are feasible (86). The hydrogen bonds between the nascent base pair with occupancy ≥ 10% are listed in Table 1. The time-dependent distances and angles of significantly occupied hydrogen bonds (occupancy ≥ 50%) between the nascent base pair are shown in Figure S4 (Supporting Information). It can be seen that either the number of hydrogen bonds between the nascent base pair or their occupancy is reduced or their geometry is suboptimal in the modified systems, in comparison to the unmodified control. Less significantly occupied hydrogen bonds between the nascent base pair may also aid in stabilizing the incoming dNTP in the modified systems.
The presence of the three hydroxyl groups on the B[c]-Ph benzylic ring and the tilt of the modified base permits formation of additional hydrogen bonds. These involve the B[c]Ph-adducted adenine and neighboring bases or protein residues other than the incoming dNTP. Those with occupancy ≥10% are listed in Table S3 (Supporting Information).
5. Stacking Interactions Stabilize the Incoming dNTP and the Intercalated B[c]Ph to Various Extents
The intercalated B[c]Ph moiety is partly “sandwiched” between the base of the incoming dNTP and the neighboring 3′-side base pair. However, the extent of stacking varies: the B[c]Ph moiety and the mismatched dNTP are sheared in the dATP and dCTP systems; thus, the bases of dATP and dCTP are not well-stacked with the B[c]Ph ring system (Figure S5, Supporting Information). The poorly occupied hydrogen bonds between the mismatched nascent base pairs involve N6 in dATP and N4 in dCTP (Table 1), shifting the dATP and dCTP toward the minor groove. The amino group of the protein residue Lys78 forms hydrogen bonds with N3 of dATP and O2 of dCTP to stabilize this shifted position. Consequently, only the five-membered ring is stacked in the dATP system, while in dCTP, the base is hardly stacked with the B[c]Ph ring system.
6. Protein Residue Interactions in the Active Site Vary with Different dNTPs
In comparison to the unmodified control, the hydrogen bond interactions involving the Lys159 and Arg51 are largely maintained throughout the simulation for all but the dATP and dCTP systems; those involving Tyr10 and Phe11 are reduced in all systems; those involving Thr45 are reduced in all but the dCTP systems; and those involving Tyr12 are reduced in all but the dCTP, dATP, and syn-dATP systems (Figure 4 and Table S2, Supporting Information). The phenyl ring of Tyr12 remains partially stacked with the sugar ring of the dNTP for all modified systems. Thus, the dATP system maintains the worst protein/sugar-triphosphate interactions among the six intercalated systems investigated (Table S2, Supporting Information).
The intercalation of the B[c]Ph moiety stretches and partly unwinds the DNA strands at the lesion site and alters the polymerase–DNA interactions. In the B[c]-PhDE-modified systems, the intercalation of the B[c]Ph moiety causes Asp105 to be too far from pT13, the primer terminus residue, to form hydrogen bonding interactions.
Major Groove Models with Incoming dNTP in the Type I Crystal Position
The presence of major and minor groove open pockets at the active site of Dpo4 allows for the accommodation of a bulky PAH metabolite-derived lesion on either side of the primer–template complex. However, a minor groove position cannot be obtained in an obvious way for an adduct to N6 of adenine. Our major groove starting models for dynamics, with incoming dNTP adopting the type I position (Figure 1B), place the B[c]Ph bulk in the major groove open pocket, with no collisions or disturbance to the polymerase crystal structure. The simulations show that this major groove position remained stable throughout the 1 ns time course, regardless of the identity of the incoming dNTP. Stereoviews of the ternary complex structures after 1 ns MD simulation are shown in Figure 5A–F.
Figure 5.
Structures of the 1S-(–)-B[c]Ph-dA major groove and –1 deletion models of the Dpo4 ternary complexes after 1 ns of MD. (A) 1S-(–)-B[c]Ph-dA opposite dTTP; (B) 1S-(–)-B[c]Ph-dA opposite dCTP; (C) 1S-(–)-B[c]Ph-dA opposite dATP; (D) 1S-(–)-B[c]Ph-dA opposite syn-dATP; (E) 1S-(–)-B[c]Ph-dA opposite dGTP; (F) 1S-(–)-B[c]Ph-dA opposite syn-dGTP; (G) 1S-(–)-B[c]Ph-dA 3′ to the templating cytosine base, which is paired with an incoming dGTP. Renderings and color codes are the same as in Figure 2. All stereo images are constructed for viewing with a stereoviewer.
We note that the accommodation of the B[c]Ph in the major groove pocket impairs the capability for selecting the complementary dNTP much less severely than the intercalation structures because the modified adenine remains in its normal anti conformation, as in the unmodified control (Figure 2A). However, to avoid steric collisions with the modified adenine, the bases of all incoming dNTPs, except for the complementary dTTP, are displaced toward the 3′-terminus of the primer and the width of the nascent base pair is increased (Figure 6A–F); this is revealed by the distance between the C1′ of 1S-(–)-B[c]Ph-dA and the C1′ of the incoming dNTP (Table 2 and Figure S7A, Supporting Information).
Figure 6.
dNTP binding pockets of the 1S-(–)-B[c]Ph-dA major groove and –1 deletion models of the Dpo4 ternary complexes after 1ns of MD. Panels A–G are the same as in Figure 5. Color codes are the same as in Figure 3 except that the B[c]Ph moiety is shown in cyan. All stereo images are constructed for viewing with a stereoviewer.
1. Sampling of Pα–O3′ Distances that Are Near-Reaction-Ready Varies with the Incoming dNTP
The Pα–O3′ distances for the major groove type I structures are shown in Table 2 and Figure S7B (Supporting Information). The near-reaction-ready distance domain (≤ ~3.6 Å) is achieved during about 9, 0.0, 0.0, 100, 0.8, and 100% of the simulation for dTTP, dCTP, dATP, syn-dATP, dGTP, and syn-dGTP systems, respectively, over the 1 ns time frame.
2. Mg2+ Ion Chelation Is Perturbed to Varying Extents
Chelation between the nucleotide binding Mg2+ ion and protein residues Asp7, Phe8, dNTP O2β, and O1γ, and between the catalytic Mg2+ ion and Asp7 and dNTP O2α is stably maintained in all major groove type I systems and is independent of the type of incoming dNTP (Figure 7A–F). However, chelations between the nucleotide binding Mg2+ ion and Asp105, and dNTP O2α and between the catalytic Mg2+ ion and Asp 105 and pT13 O3′ are all maintained only for the syn-dGTP system.
Figure 7.
Interactions between polymerase residue and sugar-triphosphate of dNTP in the 1S-(–)-B[c]Ph-dA major groove and –1 deletion models of the Dpo4 ternary complexes after 1 ns of MD. Panels A–G are the same as in Figure 5. Color codes are the same as in Figure 4 except that tC4 is shown in cyan in panel G.
3. Watson–Crick Hydrogen Bonding Geometry with the Incoming dTTP Is Well-Maintained
The time-dependent distances and angles of highly occupied hydrogen bonds (occupancy > 50%) in the nascent base pair are shown in Figure S8 (Supporting Information) and Table 1. It can be seen that the number of hydrogen bonds and their occupancies are highest in the dTTP and dGTP systems (with three hydrogen bonds of >90% occupancy) and lowest in the dATP system (all with <50% occupancy). However, the distance between the C1′ of 1S-(–)-B[c]Ph-dA and the C1′ of the incoming dGTP is 12.3 ± 0.2 Å, much greater than the optimal distance for base pair formation (79). The binding pockets in the major groove type I models are more selective for the correct incoming dNTP than the intercalation models, where the ability of the template base to form Watson–Crick hydrogen bonds with the complementary incoming dTTP is compromised.
The nascent base pair may also be stabilized to some extent by less significantly occupied hydrogen bonds (Table 1). Additional hydrogen bonds with ≥10% occupancy involving the B[c]Ph-adducted adenine and neighboring bases or protein residues other than the incoming dNTP are listed in Table S3 (Supporting Information).
4. Base Stacking Interactions Are Generally Maintained
In comparison to the unmodified control and intercalation models, the nascent base pair in the major groove type I models is generally well-stacked with the base pair 3′ to the modified adenine except for the dATP system; in this case, the base of the dATP is opened toward the major groove to facilitate hydrogen bond formation, so that it is no longer stacked with the 3′-terminal base of the primer strand (Figure S9C, Supporting Information).
5. Protein Residue Interactions in the Active Site Vary with Different dNTPs
In comparison to the unmodified control, hydrogen bond interactions involving the Tyr10, Lys159, and Arg51 are maintained throughout the simulation for all systems; those involving Phe11 are reduced in all but the dGTP and syn-dGTP systems; those involving Thr45 are reduced only in the dTTP system; and those involving Tyr12 are reduced in the dATP, syn-dATP, and syn-dGTP systems (Figure 7 and Table S2, Supporting Information); Tyr48, on the other hand, developed favorable interactions with the γ-phosphate in all but the syn-dATP system, as evidenced by the increased hydrogen bonding interactions between Tyr48 hydroxyl and oxygens on Pγ of the dNTPs (Tabel S2, Supporting Information).
Major Groove Model with the Incoming dGTP in the Type II Position: A –1 Deletion Model. The Ability of the Dpo4 Active Site to Accommodate Two Templating Bases Allows Pairing of an Incoming dGTP with the Cytosine 5′ to the Modified Adenine
The type II crystal structure of Dpo4 (13) revealed that the base 5′ to the modified adenine may also reside in the spacious active site of Dpo4 and is poised for Watson–Crick base pairing with an incoming dNTP. This structure provides a plausible model for –1 deletion when the proximal, 3′-template base is modified by a B[c]Ph-derived lesion positioned in the major groove open pocket. In our simulation, highly occupied (>99%) normal Watson–Crick hydrogen bonds are formed between the incoming dGTP and the cystosine 5′ to the modified adenine (Table 1 and Figure S8, Supporting Information).
To accommodate the nascent base pair 5′ to the modified adenine, the “roof” of the binding pocket is pushed upward to allow hydrogen bond formation, and the floor is replaced by the modified base (Figure 6G). This produces a binding pocket with a very large opening. Furthermore, the base of the incoming dGTP has to rotate more (become more twisted) relative to the 5′-base pair than if it were pairing with the 3′ (proximal) modified adenine (Figure S9G, Supporting Information); thus, it partially stacks with the modified adenine to form Watson–Crick hydrogen bonds with the 5′ (distal) cytosine. The base of the modified adenine is “pulled” toward the major groove by the B[c]Ph moiety to avoid steric collisions between the benzylic ring and the 5′ (distal) templating cytosine. Consequently, stacking between the modified adenine and its 3′-base is worse than in the unmodified control or in the major groove type I models.
The C1′–C1′ distance remained normal (10.7 ± 0.2 Å) for the nascent base pair (Table 2 and Figure S7A, Supporting Information); the near-reaction-ready Pα–O3′ distance, however, is sampled only during the first 100 ps of the simulation, for ~3% of the 1 ns time frame (Table 2 and Figure S7B, Supporting Information).
Chelation between the nucleotide binding Mg2+ ion and Asp7, Asp105, Phe8, dGTP O2β, and O2γand between the catalytic Mg2+ ion and Asp7, Asp105, Glu106, dGTP O2α, and O2β remained stable throughout the simulation (Figure 7G).
Hydrogen bonding interactions between the protein residues and the sugar-phosphate backbone of the incoming dGTP are seriously diminished as compared to the unmodified control and the type II crystal structure itself (13). Hydrogen bonding interactions involving Tyr10 and Phe11 (Figure 7G and Table S2, Supporting Information) are, however, maintained in the simulation, as in the unmodified control.
Discussion
We have employed molecular modeling and dynamics simulations to investigate three models of the 1S-(–)B[c]Ph-dA adduct in the Dpo4 active site. These are an intercalation model based on the NMR solution structure and type II crystal, a major groove model based on the type I crystal, and a –1 deletion model based on the type II crystal. All four incoming dNTPs were considered for the first two cases. The ensembles of structures from the dynamics trajectories were evaluated and compared to an unmodified control simulation, in relation to a number of structural criteria. These include occupancy and quality of hydrogen bonds between the nascent base pair, the distances between the C1′ of 1S-(–)-B[c]Ph-dA and the C1′ of the incoming dNTP and between Pα of dNTP and O3′ of the primer terminus, chelation of Mg2+ ions, stacking interactions at the active site, dNTP binding pocket geometry, and hydrogen bonding interactions between the polymerase and the incoming dNTP. While these structural criteria have been evaluated separately, undoubtedly there is an interdependence among them, and it is their composite quality for a given model that provides insights. For example, it is likely that the Pα–O3′ distance is tied to other elements. Table 3 summarizes our findings. These results permit us to consider the three models in terms of their plausibility for error-free or mutagenic nucleotide incorporation.
Table 3.
Composite Evaluation of Structural Distortion in the Dpo4 Ternary Complexes as Compared to Unmodified Control
| model | dNTP | HB geometrya | C1′–C1′ distanceb | Pα–O3′ distancec | Mg2+chelationd | stackinge | pocket geomtryf | protein interactiong | composite scoreh |
|---|---|---|---|---|---|---|---|---|---|
| control intercalation | dTTP | √ | √ | √ | √ | √ | √ | √ | 0 |
| dTTP | × | ×× | × | √ | √ | × | × | −6 | |
| dCTP | × | ×× | ×× | √ | ×× | × | × | −9 | |
| dATP | ×× | × | × | √ | × | × | ×× | −8 | |
| syn-dATP | × | ×× | ×× | √ | √ | × | × | −7 | |
| dGTP | × | × | × | √ | √ | × | × | −5 | |
| syn-dGTP | × | × | ×× | √ | √ | × | × | −6 | |
| major groove type I | dTTP | √ | √ | × | × | √ | √ | √ | −2 |
| dCTP | × | × | ×× | × | √ | √ | √ | −5 | |
| dATP | ×× | × | ×× | × | × | √ | × | −8 | |
| syn-dATP | × | √ | √ | × | √ | √ | × | −3 | |
| dGTP | √ | ×× | ×× | × | √ | √ | √ | −5 | |
| syn-dGTP | × | √ | √ | √ | √ | √ | √ | −1 | |
| –1 deletion | dGTP | √ | √ | × | √ | × | × | ×× | −5 |
Coplanarity, distance, angle, occupancy, and number of hydrogen bonds between the nascent base pair are considered. One cross denotes a decrease in one or more of these criteria relative to the control, and the number of hydrogen bonds of >50% occupancy ≥ 1; two crosses denote a decrease in one or more of these criteria and the number of hydrogen bonds of >50% occupancy = 0.
One cross denotes an average value for the C1′–C1′ distance 9.6–10.3 Å or 11.3–12.0 Å; two crosses denote an average value for the C1′–C1′ distance >12.0 or <9.6 Å.
One cross denotes a Pα–O3′ distance occupancy of ≤ 3.6 Å between 1 and 50% of the simulation time; two crosses denote this Pα–O3′ distance occupancy <1%.
Cross denotes disruption of 2–3 of the chelation interactions that involve the two Mg2+ ions.
One cross denotes decreased stacking interactions, but some stacking between the nascent base pair and the B[c]Ph moiety or the base pair 3′ to the modified base; two crosses denote no such stacking interactions.
One cross denotes distortion to the dNTP binding pocket geometry, as discussed in the text.
One cross denotes some disruption of hydrogen bonding interactions between the protein residues and the sugar-triphosphate backbone, but the disruption involves no more than three polymerase residues; two crosses denote disruption involving more than three polymerase residues.
Score is based on the unmodified control, which was assigned a base score of 0. Each cross penalizes the base score by –1.
The major groove type I model is generally the least disturbed, suited for dTTP incorporation opposite the damaged adenine and for accommodation of an A:G mismatch through incorporation of a syn-dGTP or an A:A mismatch through incorporation of a syn-dATP. The essential feature of this model is the situation of the B[c]-Ph aromatic ring system on the template major groove side in the polymerase open pocket. The nascent base pair and the polymerase residues comprising the active site region are not affected by the lesion, and normal Wat-son–Crick base pairing can be accommodated. Mismatches are accommodated to varying extents, depending on the geometric properties of the base pair itself.
The intercalation model is generally the most perturbed and overall appears most likely to impede the polymerase, but some nucleotide incorporation, particularly of dGTP, is not ruled out. The essential feature of this model, intercalation of the B[c]Ph ring system on the 3′-side of the modified adenine, offers the opportunity for favorable stacking interactions between incoming dNTP and the bulky aromatic rings. These interactions can serve to stabilize the position of the dNTP for possible reaction, while at the same time perturbing polymerase amino acid residues in the active site region. Hydrogen bonding in the nascent base pair is possible but severely distorted in every case.
The –1 deletion model for the 1S-(–)-B[c]Ph-dA adduct, while apparently an overall attractive model based on the type II crystal structure, has diminished hydrogen bonding interactions between the polymerase and the incoming dGTP, as compared to our control simulation and the crystal structure itself (13). This suggests that the incoming dGTP is less stably held in the –1 deletion position in the adduct than without modification. In addition, the pairing of the dGTP with the residue 5′ to the modified adenine in this model causes distortion to the binding pocket, together with displacement of the modified adenine toward the major groove, which adversely affects its stacking with its 3′-neighboring base. However, this model well supports Watson–Crick base pairing and is capable of achieving near-reaction-ready Pα–O3′ distances, since our simulation found such distances, although only during the first ~100 ps (following equilibration). All in all, this model appears feasible while less favored than the major groove type I model.
The accommodation of mismatched dNTPs in these models suggests that the insertions of G and A, in syn conformation, would be preferred over the insertion of C. However, the Dpo4 spacious active site with its diminished steric selectivity as compared to replicative polymerases does not entirely prohibit the possibility of some dCTP incorporation. For example, our intercalation model did sample the approach-to-reaction Pα–O3′ distance during 0.05% of the simulation, suggesting a small reaction capability.
Our 1 ns simulations for the 14 systems investigated in this study are of course limited by the sampling problem, whose solution requires an infinitely long trajectory in order to traverse all accessible states at the temperature of the simulation. This problem prevents observing conformational rearrangements that take place over a longer time frame, that is, with significant barriers between the states. Thus, we would not expect to observe dynamic interchange between the models that we investigated, nor can we be certain that the statistics for the various structural analyses are sufficient. Nonetheless, many important macromolecular motions do in fact occur in the nanosecond time frame (87), including torsional motions, sugar repuckering, bending, twisting, and hydrogen bond formation and breakage. Observing these kinds of motions provides a wealth of time-dependent structural information that is otherwise not accessible. Furthermore, the dynamics simulations enable us to investigate and evaluate rare structures that may be biologically important but experimentally difficult to capture.
In conclusion, we have studied three possible models for the 1S-(–)-B[c]Ph-dA adduct in the Dpo4 Y-family DNA polymerase employing molecular modeling and dynamics simulations. These include an intercalation model, based on the NMR solution structure and the type I crystal, a major groove model with dNTP in the type I crystal position, and a –1 deletion model based on the type II crystal. Our analyses of the dynamics simulations suggest that the major groove type I model is overall least distorted and would plausibly support both normal dTTP incorporation as well as mismatched syn-dGTP and syn-dATP. The –1 deletion model is intermediate in structural perturbation but looks feasible. Finally, the intercalation model is the most disturbed and seems most prone to impede the polymerase, although some capability for accommodating incoming dGTP is suggested. More broadly, similar types of models may also be reasonable for adenine adducts in DNA with 1S stereochemistry that are derived from other PAH metabolites.
Supplementary Material
Acknowledgments
We thank Professor Robert Sha-piro and Dr. Rebecca Perlow-Poehnelt for critical reading and helpful discussions of early versions of this manuscript. This work is supported by NIH Grants CA-28038 (S.B.), CA-46533 (D.J.P.), and CA-099194 (N.E.G.). Computations were carried out on our own Silicon Graphics Origin 300 server and Octane workstations and at NSF and DOE supercomputer centers.
Footnotes
Abbreviations: PAH, polycyclic aromatic hydrocarbons; dNTP, 2′-deoxyribonucleoside 5′-triphosphate; B[a]P, benzo[a]pyrene; B[c]Ph, benzo[c]phenanthrene; B[c]PhDE, benzo[c]phenanthrene diol epoxide; 1S-(–)-B[c]Ph-dA, 1S-(–)-trans-anti-benzo[c]phenanthrenyl-N6-deoxy-adenosine; MD, molecular dynamics; RMSD, root-mean-square deviation.
Supporting Information Available: Details of crystal structure remodeling (Table S1), additional hydrogen bonding interactions (Tables S2 and S3), stacking interactions (Figures S5 and S9), time dependences of RMSD (Figure S1), linkage site torsion angles (Figures S2 and S3), hydrogen bonds between the nascent base pair (Figures S4 and S8), and C1′–C1′ and Pα–O3′ distances (Figures S6 and S7) are provided. This material is available free of charge via the Internet at http//pubs.acs.org.
References
- 1.Cordonnier AM, Fuchs RPP. Replication of damaged DNA: Molecular defect in Xeroderma pigmentosum variant cells. Mutat Res. 1999;435:111–119. doi: 10.1016/s0921-8777(99)00047-6. [DOI] [PubMed] [Google Scholar]
- 2.Pages V, Fuchs RP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21:8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 3.Christner DF, Lakshman MK, Sayer JM, Jerina DM, Dipple A. Primer extension by various polymerases using oligonucleotide templates containing stereoisomeric benzo-[a]pyrene-deoxyadenosine adducts. Biochemistry. 1994;33:14297–14305. doi: 10.1021/bi00251a043. [DOI] [PubMed] [Google Scholar]
- 4.Chary P, Lloyd RS. In vitro replication by prokaryotic and eukaryotic polymerases on DNA templates containing site-specific and stereospecific benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide adducts. Nucleic Acids Res. 1995;23:1398–1405. doi: 10.1093/nar/23.8.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chary P, Lloyd RS. Impact of the stereochemistry of benzo[a]pyrene 7,8-dihydrodiol 9,10-epoxide-deoxyadenosine adducts on resistance to digestion by phosphodiesterases I and II and translesion synthesis with HIV-1 reverse transcriptase. Chem Res Toxicol. 1996;9:409–417. doi: 10.1021/tx950012b. [DOI] [PubMed] [Google Scholar]
- 6.Lipinski LJ, Ross HL, Zajc B, Sayer JM, Jerina DM, Dipple A. Effect of single benzo[a]pyrene diol epoxide-deoxyguanosine adducts on the action of DNA polymerases in vitro. Int J Oncol. 1998;13:269–273. doi: 10.3892/ijo.13.2.269. [DOI] [PubMed] [Google Scholar]
- 7.Alekseyev YO, Dzantiev L, Romano LJ. Effects of benzo[a]pyrene DNA adducts on Escherichia coli DNA polymerase I (Klenow fragment) primer-template interactions: Evidence for inhibition of the catalytically active ternary complex formation. Biochemistry. 2001;40:2282–2290. doi: 10.1021/bi002245u. [DOI] [PubMed] [Google Scholar]
- 8.Zhuang P, Kolbanovskiy A, Amin S, Geacintov NE. Base sequence dependence of in vitro translesional DNA replication past a bulky lesion catalyzed by the exo- Klenow fragment of Pol I. Biochemistry. 2001;40:6660–6669. doi: 10.1021/bi010005o. [DOI] [PubMed] [Google Scholar]
- 9.Goodman M. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg EC, Fischhaber PL, Kisker C. Error-prone DNA polymerases: Novel structures and the benefits of infidelity. Cell. 2001;107:9–12. doi: 10.1016/s0092-8674(01)00509-8. [DOI] [PubMed] [Google Scholar]
- 11.Luch A. Nature and nurture–lessons from chemical carcinogenesis. Nat Rev Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 12.Poirier MC. Chemical-induced DNA damage and human cancer risk. Nat Rev Cancer. 2004;4:630–637. doi: 10.1038/nrc1410. [DOI] [PubMed] [Google Scholar]
- 13.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: A mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 14.Silvian LF, Toth EA, Pham P, Goodman MF, Ellenberger T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat Struct Biol. 2001;8:984–989. doi: 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- 15.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase η: Implications for translesion DNA synthesis. Mol Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 16.Zhou BL, Pata JD, Steitz TA. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol Cell. 2001;8:427–437. doi: 10.1016/s1097-2765(01)00310-0. [DOI] [PubMed] [Google Scholar]
- 17.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-ι occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 18.Yang W. Damage repair DNA polymerase Y. Curr Opin Struct Biol. 2003;13:23–30. doi: 10.1016/s0959-440x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 19.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Polθ. Proc Natl Acad Sci USA. 2000;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerase ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 22.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 23.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 25.Tissier A, Frank EG, McDonald JP, Iwai S, Hanaoka F, Woodgate R. Misinsertion and bypass of thymine-thymine dimers by human DNA polymerase ι. EMBO J. 2000;19:5259–5266. doi: 10.1093/emboj/19.19.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaisman A, Masutani C, Hanaoka F, Chaney SG. Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η. Biochemistry. 2000;39:4575–4580. doi: 10.1021/bi000130k. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Yuan F, Wu X, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res. 2000;28:4717–4724. doi: 10.1093/nar/28.23.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase κ in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Yuan F, Xin H, Wu X, Rajpal DK, Yang D, Wang Z. Human DNA polymerase κ synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res. 2000;28:4147–4156. doi: 10.1093/nar/28.21.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): An archaeal DNA polymerase with lesion-bypass properties akin to eukaryotic pol η. Nucleic Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerlach VL, Feaver WJ, Fischhaber PL, Friedberg EC. Purification and characterization of pol κ, a DNA polymerase encoded by the human DINB1 gene. J Biol Chem. 2001;276:92–98. doi: 10.1074/jbc.M004413200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Yuan F, Wu X, Taylor JS, Wang Z. Response of human DNA polymerase ι to DNA lesions. Nucleic Acids Res. 2001;29:928–935. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank EG, Sayer JM, Kroth H, Ohashi E, Ohmori H, Jerina DM, Woodgate R. Translesion replication of benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyadenosine and deoxyguanosine by human DNA polymerase ι. Nucleic Acids Res. 2002;30:5284–5292. doi: 10.1093/nar/gkf643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehmann AR. Replication of damaged DNA in mammalian cells: New solutions to an old problem. Mutat Res. 2002;509:23–34. doi: 10.1016/s0027-5107(02)00227-0. [DOI] [PubMed] [Google Scholar]
- 35.Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. Translesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 36.Huang X, Kolbanovskiy A, Wu X, Zhang Y, Wang Z, Zhuang P, Amin S, Geacintov NE. Effects of base sequence context on translesion synthesis past a bulky (+)-trans-anti-B[a]P-N2-dG lesion catalyzed by the Y-family polymerase pol κ. Biochemistry. 2003;42:2456–2466. doi: 10.1021/bi026912q. [DOI] [PubMed] [Google Scholar]
- 37.Perlow-Poehnelt RA, Likhterov I, Scicchitano DA, Geacintov NE, Broyde S. The spacious active site of a Y-family DNA polymerase facilitates promiscuous nucleotide incorporation opposite a bulky carcinogen-DNA adduct: Elucidating the structure–function relationship through experimental and computational approaches. J Biol Chem. 2004;279:36951–36961. doi: 10.1074/jbc.M404332200. [DOI] [PubMed] [Google Scholar]
- 38.Washington MT, Minko IG, Johnson RE, Haracska L, Harris TM, Lloyd RS, Prakash L, Prakash S. Efficient and error-free replication past a minor-groove N2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase ζ. Mol Cell Biol. 2004;24:6900–6906. doi: 10.1128/MCB.24.16.6900-6906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash L, Prakash S. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol Cell Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kokoska RJ, Bebenek K, Boudsocq F, Woodgate R, Kunkel TA. Low fidelity DNA synthesis by a Y family DNA polymerase due to misalignment in the active site. J Biol Chem. 2002;277:19633–19638. doi: 10.1074/jbc.M202021200. [DOI] [PubMed] [Google Scholar]
- 41.Fiala KA, Suo Z. Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43:2106–2115. doi: 10.1021/bi0357457. [DOI] [PubMed] [Google Scholar]
- 42.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 44.Doublié S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Struct Fold Des. 1999;7:R31–R35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 45.Harvey RG. Polycyclic Aromatic Hydrocarbons. Wiley-VCH; New York: 1997. [Google Scholar]
- 46.Levin W, Wood AW, Chang RL, Ryan DE, Thomas PE, Yagi H, Thakker DR, Vyas K, Boyd C, Chu SY, Conney AH, Jerina DM. Oxidative metabolism of polycyclic aromatic hydrocarbons to ultimate carcinogens. Drug Metab Rev. 1982;13:555–580. doi: 10.3109/03602538209011087. [DOI] [PubMed] [Google Scholar]
- 47.Thakker DR, Levin W, Yagi H, Yeh HJ, Ryan DE, Thomas PE, Conney AH, Jerina DM. Stereo-selective metabolism of the (+)-(S,S)- and (–)-(R,R)-enantiomers of trans-3,4-dihydroxy-3,4-dihydrobenzo[c]-phenanthrene by rat and mouse liver microsomes and by a purified and reconstituted cytochrome P-450 system. J Biol Chem. 1986;261:5404–5413. [PubMed] [Google Scholar]
- 48.Agarwal SK, Sayer JM, Yeh HJC, Pannell LK, Hilton BD, Pigott MA, Dipple A, Yagi H, Jerina DM. Chemical characterization of DNA adducts derived from the configurationally isomeric benzo[c]phenanthrene-3,4-diol-1,2-epoxides. J Am Chem Soc. 1987;109:2497–2504. [Google Scholar]
- 49.Szeliga J, Dipple A. DNA adduct formation by polycyclic aromatic hydrocarbon dihydrodiol epoxides. Chem Res Toxicol. 1998;11:1–11. doi: 10.1021/tx970142f. [DOI] [PubMed] [Google Scholar]
- 50.Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H. Unrepaired fjord region polycyclic aromatic hydrocarbon-DNA adducts in ras codon 61 mutational hot spots. Cancer Res. 2000;60:1849–1856. [PubMed] [Google Scholar]
- 51.McKay IA, Paterson H, Brown BDT, Marshall CJ, Hall A. N-ras and Human Cancer. Anticancer Res. 1986;6:483–490. [PubMed] [Google Scholar]
- 52.Conti CJ. Mutations of genes of the ras family in human and experimental tumors. Prog Clin Biol Res. 1992;376:357–378. [PubMed] [Google Scholar]
- 53.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 54.Ponten I, Sayer JM, Pilcher AS, Yagi H, Kumar S, Jerina DM, Dipple A. Factors determining mutagenic potential for individual cis and trans opened benzo[c]phenanthrene diol epoxide-deoxyadenosine adducts. Biochemistry. 2000;39:4136–4144. doi: 10.1021/bi991719q. [DOI] [PubMed] [Google Scholar]
- 55.Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RP, Nohmi T. The dinB gene encodes an novel E. coli DNA polymerase (DNA pol IV) involved in mutagenesis. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 56.Cosman M, Fiala R, Hingerty BE, Laryea A, Lee H, Harvey RG, Amin S, Geacintov NE, Broyde S, Patel D. Solution conformation of the (+)-trans-anti-[BPh]dA adduct opposite dT in a DNA duplex: Intercalation of the covalently attached benzo[c]phenanthrene to the 5′-side of the adduct site without disruption of the modified base pair. Biochemistry. 1993;32:12488–12497. doi: 10.1021/bi00097a029. [DOI] [PubMed] [Google Scholar]
- 57.Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. Structure of a high-fidelity DNA polymerase bound to a benzo[a]pyrene adduct that blocks replication. J Biol Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 58.Hsu GW, Kiefer JR, Burnouf D, Becherel OJ, Fuchs RP, Beese LS. Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J Biol Chem. 2004;279:50280–50285. doi: 10.1074/jbc.M409224200. [DOI] [PubMed] [Google Scholar]
- 59.Ling H, Sayer JM, Plosky BS, Yagi H, Boudsocq F, Woodgate R, Jerina DM, Yang W. Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc Natl Acad Sci USA. 2004;101:2265–2269. doi: 10.1073/pnas.0308332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mao B, Gu Z, Gorin A, Chen J, Hingerty BE, Amin S, Broyde S, Geacintov NE, Patel DJ. Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex. Biochemistry. 1999;38:10831–10842. doi: 10.1021/bi991212f. [DOI] [PubMed] [Google Scholar]
- 61.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Ross W, Simmerling CL, Darden TA, Merz K, Stanton R, Cheng A, Vincent J, Crowley M, Ferguson D, Radner R, Seibel G, Singh UC, Weiner P, Kollman PA. AMBER 6. University of California; San Francisco, CA: 1999. [Google Scholar]
- 62.Arnott S, Campbell-Smith PJ, Chandrasekaran R. Atomic coordinates and molecular conformations for DNA-DNA, RNA-RNA, and DNA-RNA helices. In: Fasman G, editor. CRC Handbook of Biochemistry and Molecular Biology. Vol. 2. CRC Press; Cleveland: 1976. pp. 411–422. [Google Scholar]
- 63.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:231–232. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 64.Wu M, Yan SF, Tan J, Patel DJ, Geacintov NE, Broyde S. Conformational searches elucidate effects of stereochemistry on structures of deoxyadenosine covalently bound to tumorogenic metabolites of benzo[c]phenanthrene. Front Biosci. 2004;9:2807–2818. doi: 10.2741/1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cosman M, Laryea A, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (–)-trans-anti-benzo[c]phenanthrene-dA ([BPh]-dA) adduct opposite dT in a DNA duplex: Intercalation of the covalently attached benzo[c]phenanthrenyl ring to the 3′-side of the adduct site and comparison with the (+)-trans-anti-[BPh]dA opposite dT stereoisomer. Biochemistry. 1995;34:1295–1307. doi: 10.1021/bi00004a024. [DOI] [PubMed] [Google Scholar]
- 66.Wu M, Yan S, Patel DJ, Geacintov NE, Broyde S. Relating repair susceptibility of carcinogen-damaged DNA with structural distortion and thermodynamic stability. Nucleic Acids Res. 2002;30:3422–3432. doi: 10.1093/nar/gkf427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perlow RA, Broyde S. Evading the proofreading machinery of a replicative DNA polymerase: Induction of a mutation by an environmental carcinogen. J Biol Chem. 2001;309:519–536. doi: 10.1006/jmbi.2001.4674. [DOI] [PubMed] [Google Scholar]
- 68.Perlow RA, Broyde S. Toward understanding the mutagenicity of an environmental carcinogen: Structural insights into nucleotide incorporation preferences. J Mol Biol. 2002;322:291–309. doi: 10.1016/s0022-2836(02)00751-9. [DOI] [PubMed] [Google Scholar]
- 69.Cheatham TE, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J Biomol Struct Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 70.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins and nucleic acids. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 71.Darden T, York D, Pedersen L. Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 72.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pederson LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 73.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 74.Harvey SC, Tan RKZ, Cheatham TE. The flying ice cube: Velocity rescaling in molecular dynamics leads to violation of energy equipartition. J Comput Chem. 1998;19:726–740. [Google Scholar]
- 75.Jorgensen WL, Chandreskhar J, Madura JD, Imprey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 76.Mezei M. Optimal position of solute for simulations. J Comput Chem. 1997;18:812–815. [Google Scholar]
- 77.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 78.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis-syn thymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 79.Barsky D, Kool ET, Colvin ME. Interaction and solvation energies of nonpolar DNA base analogues and their role in polymerase insertion fidelity. J Biomol Struct Dyn. 1999;16:1119–1134. doi: 10.1080/07391102.1999.10508321. [DOI] [PubMed] [Google Scholar]
- 80.Jordan F. Lennard-Jones potential calculations of the barrier to rotation around the glycosidic C–N linkage in selected purine nucleosides and nucleotides. A direct comparison of the results of 6–12 potential calculations with results of semiempirical molecular orbital studies. J Theor Biol. 1973;41:375–395. doi: 10.1016/0022-5193(73)90125-2. [DOI] [PubMed] [Google Scholar]
- 81.Johnson RE, Trincao J, Aggarwal AK, Prakash S, Prakash L. Deoxynucleotide triphosphate binding mode conserved in Y family DNA polymerases. Mol Cell Biol. 2003;23:3008–3012. doi: 10.1128/MCB.23.8.3008-3012.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brautigam CA, Steitz TA. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr Opin Struct Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 83.Komarov VM, Polozov RV, Konoplev GG. Nonplanar structure of nitrous bases and noncoplanarity of Watson–Crick pairs. J Theor Biol. 1992;155:281–294. doi: 10.1016/s0022-5193(05)80600-9. [DOI] [PubMed] [Google Scholar]
- 84.El Amri C, Mauffret O, Monnot M, Tevanian G, Lescot E, Porumb H, Fermandjian S. A DNA hairpin with a single residue loop closed by a strongly distorted Watson–Crick G x C base-pair. J Mol Biol. 1999;294:427–442. doi: 10.1006/jmbi.1999.3270. [DOI] [PubMed] [Google Scholar]
- 85.Ennifar E, Yusupov M, Walter P, Marquet R, Ehresmann B, Ehresmann C, Dumas P. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges. Struct Fold Des. 1999;7:1439–1449. doi: 10.1016/s0969-2126(00)80033-7. [DOI] [PubMed] [Google Scholar]
- 86.Sponer J, Leszczynski J, Hobza P. Electronic properties, hydrogen bonding, stacking, and cation binding of DNA and RNA bases. Biopolymers. 2001–2002;61:3–31. doi: 10.1002/1097-0282(2001)61:1<3::AID-BIP10048>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 87.McCammon JA, Harvey SC. Dynamics of Proteins and Nucleic Acids. Cambridge University Press; New York: 1987. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.