Abstract
The c-fos gene is induced by a broad range of stimuli, and has been commonly used as a reliable marker for neural activity. Its induction mechanism and available reporter mouse lines are exclusively based on the c-fos promoter activity. Here, we demonstrate that multiple enhancers surrounding the c-fos gene are critical for ensuring robust c-fos response to various stimuli. Membrane depolarization, brain-derived neurotrophic factor (BDNF), and Forskolin activate distinct subsets of the enhancers to induce c-fos transcription in neurons, suggesting that stimulus-specific combinatorial activation of multiple enhancers underlies the broad inducibility of the c-fos gene. Accordingly, the functional requirement of key transcription factors varies depending on the type of stimulation. Combinatorial enhancer activation also occurs in the brain. Providing a comprehensive picture of the c-fos induction mechanism beyond the minimal promoter, our study should help in understanding the physiological nature of c-fos induction in relation to neural activity and plasticity.
INTRODUCTION
Neuronal activity generated spontaneously during early stages of brain development and by sensory experience throughout the lifetime plays an essential role in the proper development and function of neural circuits. Upon sensory experience, synaptic activity induces rapid calcium influx in postsynaptic neurons, which in turn mediates a multitude of intracellular events necessary for remodeling the synaptic connectivity of the circuit 1. A calcium rise within the postsynaptic compartments can promptly initiate strengthening or weakening of the synaptic connectivity through local biochemical actions such as mRNA translation, posttranslational modifications, and trafficking of synaptically localized proteins. In parallel, calcium influx can also induce a cell-wide adaptive response by activating nuclear gene expression through specific calcium-dependent signaling cascades. The timely synthesis and deployment of new gene products mediated by the activity-regulated gene expression program allows sustainable changes in the structure and function of individual synapses and the resulting behavioral plasticity.
A notable feature of the activity-induced transcription program is the biphasic nature of transcriptional induction. Many immediate early genes (IEGs) that are rapidly induced upon an increase in neural activity encode transcription factors (TFs) such as c-fos, Egr1, and Nr4a1. These IEG TFs can subsequently drive a second wave of transcription of delayed genes, many of which have been shown to be localized or act at synapses. Consistent with this biphasic arrangement of the transcription program, it has been shown that the application of transcription inhibitors can block the long-term synaptic plasticity and behavior in vertebrates only when they are administered during and/or immediately after stimulation 2. This result emphasizes the importance of a transcription mechanism that can ensure rapid IEG induction within the narrow temporal window following stimulation. Moreover, the diversity and specificity of the cellular response to different extracellular stimuli are mediated, at least in part, by distinct combinatorial induction of a relatively small number of IEG TFs 3. For example, in the rat pheochromocytoma cell line PC12, c-jun is induced by growth factors but not by membrane depolarization whereas c-fos is commonly induced by both agents 4.
The accessibility and assembly of transcription factor complexes at DNA regulatory regions, such as enhancers and promoters, are key regulatory steps in transcription and tightly governed by the status of epigenetic modifications. Unique combinations of epigenetic marks and nucleosome positioning provide information for the activity of the underlying DNA sequence. Enhancers can be defined as inter- and intragenic regions with an elevated level of mono-methylation at the lysine 4 residue of the histone H3 subunit (H3K4me1), whereas promoter regions of active genes are instead enriched by tri-methylation at the same residue (H3K4me3) 5. Even after establishment, the activity of enhancers can be suppressed (inactive), poised, or induced, depending on cell type, developmental stage, or extracellular signaling. We previously identified over 10,000 enhancers that control activity-dependent transcription in mouse cortical neurons, and also found that neuronal activity rapidly recruits RNA polymerase II (RNAPII) to a subset of neuronal enhancers (~2,500) and transcribes a novel class of lncRNAs, named “eRNAs” (enhancer RNAs) 6. Subsequent studies have established that eRNAs are expressed in a wide range of cell types and tissues in a manner that positively correlates with nearby mRNAs, which suggest that eRNA synthesis is an intrinsic regulatory mechanism of functionally active enhancers 7. Transcription activity at enhancers appears to be a functionally important process as the enhancer-specific H3K4me1/2 deposition at de novo enhancers was observed to occur in an enhancer transcription-dependent manner 8. In parallel, we and others have also found that eRNA transcripts play a functional role in target gene activation by various mechanisms depending on the cellular and/or genomic context 7. Therefore, enhancers have a more complex role in gene expression than previously appreciated.
Recent genome-scale studies of chromosomal organization have revealed that chromosomes are folded into topologically associated domains (TADs), which provide a three-dimensional (3D) structural barrier for enhancer sharing and allocation 9, 10. Within each TAD, multiple dispersed enhancers are often seen as physically associated with a common target gene via chromatin looping. In fact, about half of the active promoters in a mammalian cell show interactions with multiple enhancers, with an average number of 4.75 enhancers per active promoter 11, 12. However, how and why multiple enhancers coordinately regulate a common target gene is not well understood. Several transgenic studies in Drosophila and mice have suggested a few different modes of enhancer integration. Discrete enhancers might simply produce an additive effect on the expression level of a common target gene or be integrated, often with a functional hierarchy among the enhancer modules, to cause synergistic activation or to allow for fine-tuning (precision) and/or robustness of the transcriptional output in the face of dynamic environmental and genetic variability 13. So far, most studies concerning the functionality of enhancer clusters have been focused on developmentally regulated genes, however, the coordinated action of enhancer modules in stimulus-dependent transcriptional induction has not yet been explored. We noticed from our previous enhancer study in mouse cortical neurons that the c-fos gene is surrounded by five enhancers dispersed throughout a ~50 kb region 6. In primary neuronal cultures, c-fos transcription can be triggered by growth factors, cAMP signaling, and membrane depolarization induced by either neurotransmitters or KCl. Induction of c-fos expression has also been observed in the intact brain in response to a variety of physiological and pathological stimuli including seizure, traumatic injury, somatosensory stimulation, and long-term potentiation (LTP) 14
We reasoned that the several enhancers scattered around the c-fos gene might dynamically coordinate with each other to mediate robust c-fos induction in response to a wide range of stimuli. To test this idea, we have determined the functionality of the c-fos enhancer cluster by investigating the autonomous activity of individual enhancers as well as their functional integration during the induction of c-fos mRNA by three different stimuli (KCl-mediated membrane depolarization, BDNF, Forskolin to induce cAMP pathway). While all three stimuli efficiently induce c-fos transcription to a comparable level, each stimulus utilizes a distinctive subset of the enhancers and activity-regulated TFs to induce c-fos transcription. Characterizing individual c-fos enhancers by chromosome conformation capture (3C) analysis and an enhancer reporter assay, we have corroborated the recently emerging notion that the eRNA is a reliable marker for identifying activated enhancers. The functionality of individual enhancers in their native context was also evaluated by CRISPRi. Using eRNAs as a proxy for enhancer activity, that distinctive subsets of the c-fos enhancers are activated in the intact brain following chemically induced seizure or light stimulation. These results collectively demonstrate that the combinatorial activation of the c-fos enhancers enables the c-fos gene to be broadly responsive to various signaling pathways.
RESULTS
The enhancer cluster surrounding the c-fos gene
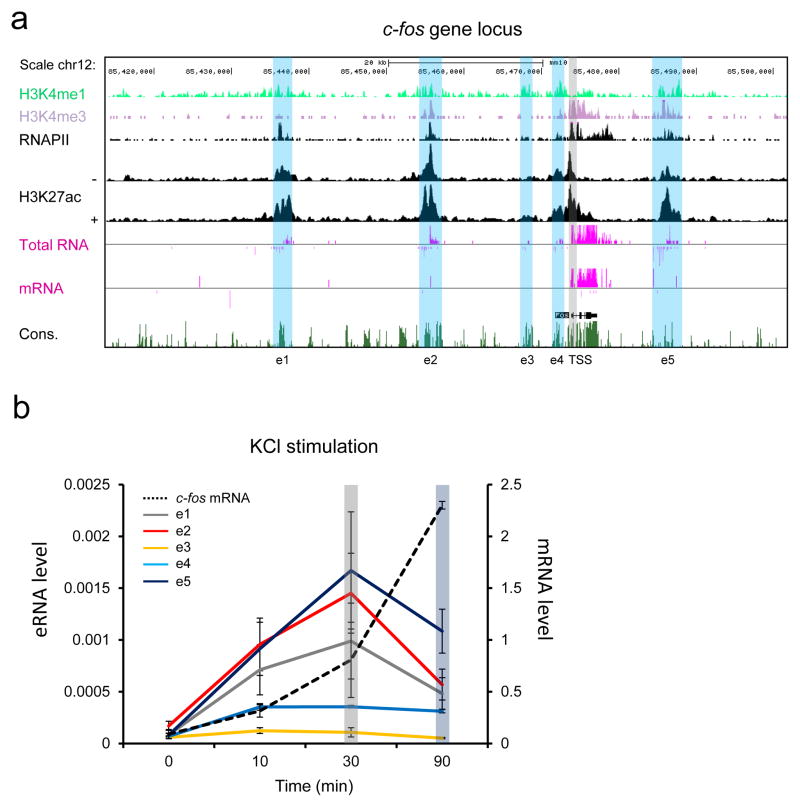
Our previous genome-wide study revealed that the c-fos gene is surrounded by five activity-regulated enhancers (Fig. 1a) 6. These enhancers exhibit a high level of the well-defined enhancer mark, H3K4me1 whereas the c-fos promoter region is enriched by H3K4me3. Upon membrane depolarization of mouse cortical neurons, all of the c-fos enhancers (e1 through e5) are inducibly bound by the general transcription coactivator, CBP, and several activity-regulated TFs in various combinations (Supplementary Fig. 1). RNAPII is also inducibly recruited to each of the c-fos enhancers and synthesizes eRNAs, which have been shown to positively correlate with the activity of the enhancers from which they are expressed 6, 12, 15, 16. These enhancers that we identified in primary culture of mouse cortical neurons appear to also be active in the intact brain throughout development, as all of them are tightly aligned with the peaks of an active enhancer marker, H3K27ac (Supplementary Fig. 1) 17. While all five enhancers show comparable levels of the H3K4me1 mark, the induction levels of RNAPII and eRNAs at individual enhancers are different, in that e1, e2, and e5 show much stronger transcription activity than e3 and e4 when neurons are depolarized by KCl. Notably we observed that the active promoter mark, H3K4me3 was also enriched at e2 and e5 enhancers, which transcribe eRNAs at much higher levels than the other enhancers. Several recent studies also reported the same observation that enhancers with very strong transcription activity are enriched by both H3K4me3 and H3K4me1 marks 18–20. It has been suggested that the H3K4me3/me1 ratio positively correlates with transcription level, independent of transcript stability. We have confirmed the induction of these eRNAs by RT-qPCR (Fig. 1b) and also observed that, just like the kinetic relationship we have previously shown between eRNA and mRNA from the Arc and Gadd45b genes 21, expression of all c-fos eRNAs peaked earlier than that of the c-fos mRNA. These results are consistent with a recent study of genome-scale CAGE (cap analysis of gene expression) analysis showing that enhancer transcription occurs most rapidly among the transcriptional changes during cellular differentiation or activation 22.
Figure 1.
Time course analysis of five c-fos eRNAs and mRNA. (a) A UCSC genome browser view of the c-fos genomic locus with RNA-seq and ChIP-Seq data (adapted from Kim et al. 10 and Malik et al. 48). + and − indicate the presence or absence of KCl. Blue and gray vertical bars indicate the locations of the c-fos enhancers and the promoter. (b) Expression levels of c-fos eRNA and mRNA in cortical neurons after KCl stimulation for various time points. Cortical neurons were depolarized with 55 mM KCl at DIV 6 and mRNA and eRNA levels were measured using RT-qPCR and normalized to Gapdh mRNA (n = 3 biological replicates). Error bars indicate SEM.
Combinatorial activation of the c-fos enhancers
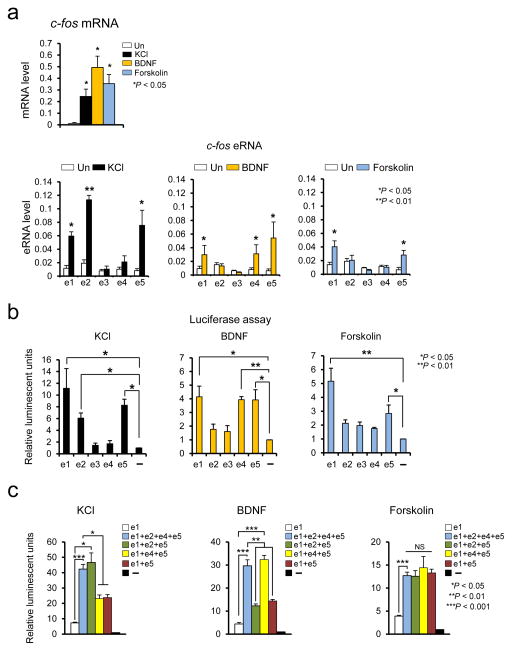
We reasoned that the previously observed broad inducibility of the c-fos gene might be enabled by synergistic and/or combinatorial actions of the surrounding enhancers. As the first step to test this idea, we treated neuronal cultures with three different stimuli – 55 mM KCl, Brain derived nerve growth factor (BDNF), and Forskolin which stimulates adenylate cyclase – to induce c-fos transcription, and then measured eRNA induction levels, which is an indicator of enhancer activity (Fig. 2a) 6, 12, 15, 16. All three stimuli effectively increased c-fos mRNA levels (Fig. 2a upper panel). Consistent with the RNA-seq profile, KCl-mediated membrane depolarization induced eRNA expression mainly from e1, e2, and e5. On the other hand, BDNF and Forskolin activated different subsets of the c-fos enhancers (e1, e4 and e5 by BDNF, and e1 and e5 by Forskolin) (Fig. 2a lower panel). In all cases, eRNA expression from the e3 enhancer was not reliably detected, which is consistent with the previous observation that the H3K27ac level is the lowest at e3 among the c-fos enhancers (Fig. 1 and Supplementary Fig. 1) 17, 23. This result suggests that different stimuli promote c-fos transcription through the activation of distinct subsets of the surrounding enhancers.
Figure 2.
The c-fos enhancer activities measured by eRNA analysis and luciferase reporter assay. (a) Expression of c-fos mRNA and eRNA in cortical neurons induced by KCl 30 min, BDNF 1 h, and Forskolin 1 h. The induction levels of the five c-fos eRNAs and mRNA were measured using RT-qPCR and normalized to Gapdh mRNA (c-fos mRNA, KCl: P = 0.0418, t(2) = 4.736, n = 3, BDNF: P = 0.0333, t(2) = 5.339, n = 3, Forskolin: P = 0.0448, t(2) = 4.566, n = 3, unpaired t-test with Welch correction; c-fos eRNA, KCl, e1: P = 0.0418, t(4) = 6.654, F = 10.187 [SDP = 0.0894], n = 3, e2: P = 0.008, t(4) = 11.103, F = 3.149 [SDP = 0.3267], n = 3, e5: P = 0.0264, t(4) = 3.436, F = 26.341 [SDP = 0.0566], n = 3, unpaired t-test, BDNF, e1: P = 0.0329, t(4) = 3.200, F = 16.876 [SDP = 0.0599], n = 3, e4: P = 0.0271, t(4) = 3.408, F = 17.578 [SDP = 0.0538], n = 3, unpaired t-test, e5 P = 0.0214, t(2) = 6.723, n = 3, unpaired t-test with Welch correction, Forskolin, e1: P = 0.0498, t(4) = 2.718, F = 6.333 [SDP = 0.1364], n = 3, e5: P = 0.0485, t(4) = 2.805, F = 4.048 [SDP = 0.1981], n = 3 biological replicates, unpaired t-test). (b,c) Combinational c-fos enhancer activities measured by the luciferase reporter assay. Each of the c-fos enhancers was fused directly upstream of the c-fos promoter, which then together drives luciferase expression. Stimulus-induced luciferase activities were measured after 6 h of KCl, BDNF, or Forskolin treatment in cortical neurons. (b, KCl, e1: P = 0.0242. t(3) = 6.306, n = 4, e2: P = 0.0103. t(3) = 5.784, n = 4, e5: P = 0.0191. t(2) = 7.140, n = 3, BDNF, e1: P = 0.0350. t(2) = 5.207, n = 3, e4: P = 0.0056. t(2) = 13.349, n = 3, e5: P = 0.0391. t(2) = 4.907, n = 3, Forskolin, e1 P = 0.0087. t(2) = 10.633, n = 3, e5: P = 0.0438. t(2) = 4.618, n = 3 biological replicates, unpaired t-test with Welch correction; c, KCl, e1+e2+e4+e5: P = 0.0001, t(5) = 12.256, n = 6, e1+e2+e5: P = 0.0246, t(2) = 6.259, n = 2, e1+e4+e5: P = 0.0240, t(2) = 6.576, n = 2, e1+e5: P = 0.0128, t(2) = 8.764, n = 2 biological replicates, unpaired t-test with Welch correction, BDNF, e1+e2+e4+e5: P = 0.0007, t(7) = 5.818, n = 6, unpaired t-test with Welch correction, e1+e2+e5: P = 0.0034, t(3) = 8.537, F = 1.411 [SDP = 0.3569], n = 3, e1+e4+e5: P = 0.0004, t(3) = 18.125, F = 6.346 [SDP = 0.1280], n = 3, e1+e5: P = 0.00303, t(3) = 2.822, F = 63.970 [SDP = 0.0946], n = 3, biological replicates, unpaired t-test, Forskolin, e1+e2+e4+e5: P = 0.0001, t(5) = 12.601, n = 6, unpaired t-test with Welch correction, e1+e2+e5: P = 0.0189, t(2) = 7.164, F = 126.28 [SDP = 0.0565], n = 3 biological replicates, unpaired t-test). Error bars indicate SEM; p values are from a two-tailed t test. NS, not significant. SDP is the P value from comparing the standard deviations for both groups. SDP > 0.05 means the two SDs are not significantly different and can be used for an unpaired t-test.
We then performed an enhancer reporter assay in order to examine whether individual enhancers can functionally regulate c-fos expression. We constructed a series of luciferase reporters, in which each enhancer region was fused to a common minimal c-fos promoter region. Luciferase activity was measured from neuronal cultures that had been transfected with each reporter and treated with three different stimuli (Fig. 2b). We found that the exact same subset of enhancers that was active in eRNA production upon each stimulus also promoted luciferase reporter expression. As enhancer loci and activities are widely varied in different cell or tissue types 24, we wondered whether the five c-fos enhancer regions we identified in our in vitro culture of mouse cortical neurons are preserved in other tissues. Examining publically available H3K27ac enrichment profiles in three different tissues (forebrain, heart, and liver) 17, we found that e2, e4, and e5 are active in all three tissues (Supplementary Fig. 1). The e1 enhancer was active in the heart and brain but not in the liver suggesting that its activity is tissue-specific. The H3K27ac level near the e3 enhancer was very low in all three tissues. We also measured the levels of eRNA induction from each enhancer in NIH 3T3 cells following serum stimulation and found that e5 and e4 are the only two enhancers that transcribe eRNAs upon serum stimulation (Supplementary Fig. 2a). The luciferase reporter assay further shows that e5 is the most responsive to serum stimulation in NIH 3T3 cells (Supplementary Fig. 2b). We then examine whether stimulus-specific combinations of enhancers contribute to the synergistic gene activation, by comparing the activities of the luciferase reporters driven by either a single enhancer or combinations of multiple enhancers (Fig. 2c). In this assay, we compared the activities of e1 alone, and combinations of KCl-, BDNF-, or Forskolin-activated enhancers (e1+e2+e5, e1+e4+e5, or e1+e5) and all four major enhancers (e1+e2+e4+e5). When compared to e1 enhancer alone, the combination of KCl-activated enhancers (e1+e2+e5) synergistically increased the luciferase reporter expression to the level comparable to the reporter expression driven by all four enhancers upon KCl stimulation. Likewise, the BDNF-specific enhancer combination (e1+e4+e5) synergistically increased the reporter expression to the level of four-enhancer combination when neurons were stimulated by BDNF. Forskolin stimulation of the same reporter constructs also yielded a similar result. Both e1+e2+e5 and e1+e5 combinations show very similar levels of reporter activity when stimulated by BDNF or Forskolin, confirming that neither stimulus induces e2 activity. Likewise, addition of e4 in KCl stimulation (e1+e4+e5) did not increase reporter activity more than the e1+e5 combination. These findings collectively demonstrate that distinct combinations of enhancers are utilized to induce c-fos expression depending on the types of stimuli or cells.
Interactions between enhancers and the c-fos promoter
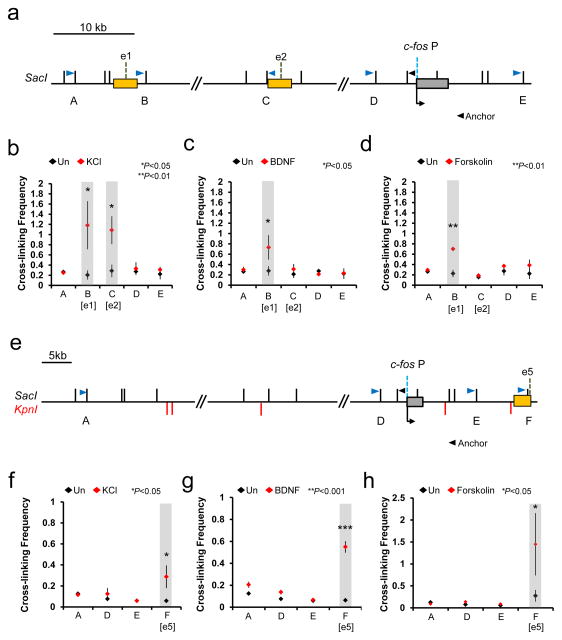
Chromosomal looping allows enhancers to be juxtaposed to their specific target promoters to regulate gene expression. We determined the chromosomal interactions between each of the c-fos enhancers and the promoter before and after each stimulus by Chromosome Conformation Capture (3C) analysis, to see whether the members of the stimulus-specific enhancer subset would selectively interact with the c-fos promoter, and also if the enhancer-promoter interaction occurs in a stimulus-dependent manner. In this 3C assay, we were only able to reliably analyze three enhancer regions (e1, e2, and e5) which are more remotely located from the c-fos promoter. The e3 and e4 enhancers were too close to the promoter (less than ~4 kb) and showed high levels of non-specific interaction with the c-fos promoter, as the rate of random collision and crosslinking efficiency become significantly higher as the genomic distance between the two restriction enzyme-digested genomic fragments decreases 25. Despite this limitation, the 3C result was consistent with the stimulus-specific eRNA induction profile and the enhancer reporter assay. Both e1 and e5 regions rapidly interact with the c-fos promoter upon stimulation by all three stimuli, whereas only KCl stimulation caused a detectable increase in the interaction between e2 and the promoter (Fig. 3). We previously found that the chromosomal interaction between the enhancer and the promoter for the Arc gene occurs inducibly upon KCl stimulation 21. Therefore, it appears that the induction of IEGs in neurons involves stimulus-induced chromosomal looping between the enhancer and promoter.
Figure 3.
Stimulus-dependent interactions between the c-fos enhancers and the promoter. (a,e) Schematic diagram of the loci analyzed by 3C. (b–d) 3C analysis for enhancer 1, 2 and the promoter before (black rhombus) and after (red rhombus) stimulation by KCl, BDNF, and Forskolin. (f–h) 3C analysis for enhancer 5 and the promoter before (black rhombus) and after (red rhombus) stimulation by KCl, BDNF, and Forskolin. Chromosomal interaction between the c-fos promoter and several genomic loci including the c-fos enhancers were measured by RT-qPCR using the primers indicated on the schematic diagram (blue and black arrowheads). The c-fos P represents the promoter, and e1, e2, and e5 represent enhancers 1, 2, and 5, respectively. The SacI and KpnI restriction enzyme sites (black and red vertical lines, respectively) are also shown (KCl stimulation, B [e1]: P = 0.0452, t(2) = 4.545, n = 3, unpaired t-test with Welch correction, C [e2]: P = 0.0109, t(4) = 4.492, F = 5.459 [SDP = 0.1548], n = 5, unpaired t-test, F [e5]: P = 0.0492, t(2) = 3.554, n = 3, unpaired t-test with Welch correction; BDNF stimulation, B [e1]: P = 0.0482, t(2) = 4.389, n = 3, unpaired t-test with Welch correction, F [e5]: P = 0.0001, t(4) = 14.393, F = 2.872 [SDP = 0.2583], n = 3, unpaired t-test; Forskolin stimulation, B [e1]: P = 0.0016, t(2) = 7.132, F = 49.091 [SDP = 0.0903], n = 3, F [e5]: P = 0.0489, t(2) = 3.481, F = 8.843 [SDP = 0.2065], n = 3 biological replicates, unpaired t-test, BDNF and Forskolin stimulation C [e2]: n = 5, KCl, BDNF, and Forskolin stimulation A, D, and E: n = 3 biological replicates). Note that e2 interactions were measured with templates that have been cut with either single enzyme (SacI) or double enzymes (SacI and KpnI). Error bars indicate SEM; p values are from a two-tailed t test. SDP is the P value from comparing the standard deviations for both groups. SDP > 0.05 means the two SDs are not significantly different and can be used for an unpaired t-test.
Transcription factors in KCl-mediated c-fos induction
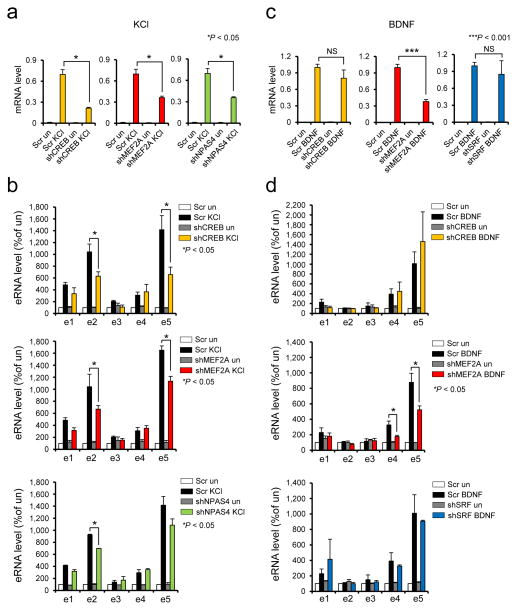
Having confirmed the activities of the c-fos enhancers in neurons, we next asked whether and how each of the activity-regulated TFs is functionally required for c-fos expression in response to different stimuli. Previous ChIP-seq analysis revealed that the c-fos promoter and enhancers are bound by different combinations of several activity-regulated TFs (Supplementary Fig. 1). SRF binding was shown to occur mainly at the promoter and e5 enhancer regardless of membrane depolarization. CREB binds to the promoter as well as e1 and e2 enhancers, and binding levels were slightly increased by KCl. On the other hand, binding of both CBP and NPAS4 occurs in a depolarization-dependent manner to all enhancers and weakly to the c-fos promoter. For comparison, we scanned each enhancer for the presence of canonical binding motifs of three activity-regulated TFs, CREB, MEF2, and SRF using Regulatory Sequence Analysis Tools (RSAT) software 26, and found the presence of cognate binding motifs for all of the experimentally determined TF binding sites although not all the motifs were bound by the corresponding TFs (Supplementary Fig. 3a,b). Notable from this analysis is that e1, e2, and e5 enhancers all have canonical binding motifs for all three TFs whereas the e3 and e4 enhancers only contain the MEF2 binding motif. To examine the functional requirement of each TF for different stimuli, we transduced neurons with lentiviruses expressing shRNAs against several activity-regulated TFs (CREB, SRF, MEF2A, MEF2D, MEF2C, NPAS4) and monitored how the knockdown of each TF affected the stimulus-dependent induction of c-fos mRNA and eRNAs. Knockdown of CREB, MEF2A, and NPAS4 significantly impaired c-fos induction by KCl-mediated membrane depolarization (Fig. 4a and Supplementary Fig. 4). Although knockdown of other MEF2 family member TFs, MEF2D and MEF2C, showed a trend toward a decrease in c-fos expression level, it was weaker than the impact of MEF2A knockdown, which was paralleled with little effect on eRNA transcription (Supplementary Figs. 5). This result was a surprise to us especially given that MEF2C is the most abundantly expressed isoform in mouse cortical neuron culture (Supplementary Fig. 6). It is unlikely that the result is due to the differences in their knockdown efficiency as the decreases in RNA levels of the TFs are quite comparable (Supplementary Fig. 5, 6, and 16). Knockdown of SRF did not affect the c-fos mRNA level at all. Consistently, knockdown of these TFs didn’t affect eRNA expression induced by KCl stimulation (Supplementary Fig. 5). This result is inconsistent with a previous study demonstrating that SRF contributes to KCl-mediated induction of c-fos transcription in PC12 cells 27. This discrepancy might result from some physiological difference between PC12 cells and primary neurons. Moreover, the analysis in PC12 cells was based on a transient expression assay with the plasmid containing the c-fos promoter region only, thus not accurately reflecting the endogenous genomic context where the surrounding enhancers are also active. It is also plausible that SRF activity might be efficiently compensated by other activity-regulated TFs in neurons. Cellular heterogeneity could have been a confounding factor in our analysis as the c-fos gene is known to be induced in many different cell types including glia cells and neurons. To investigate this issue, we counted the numbers of neurons and glia cells present in our typical embryonic culture of mouse cortical neurons (DIV 6) and found that GFAP positive glia cells are extremely outnumbered by Tuj-1 positive neurons (Supplementary Fig. 7a,b). 93% of DAPI stained cells were neurons whereas glia were only 1.2%, suggesting that the contribution from glia cell population would most likely be insignificant. We also prepared glia culture and measured induction levels of c-fos mRNAs as well as eRNAs in response to KCl, BDNF, and Forskolin (Supplementary Fig. 7a–c). Forskolin was the strongest inducer of the c-fos gene, then BDNF. KCl was the weakest in this glia culture. Importantly, eRNA activation patterns were dramatically different from those in neuronal cultures. The c-fos enhancers barely responded to KCl. Both BDNF and Forskolin activate all c-fos enhancers by ~2 fold. Therefore, the observed enhancer activation patterns in our in vitro culture most likely result from neuron-specific response to the stimulus.
Figure 4.
Function of CREB, MEF2A, NPAS4, and SRF transcription factors in KCl or BDNF-mediated induction of c-fos mRNA and eRNAs. (a,c) Effect of transcription factor knockdown in KCl or BNDF-mediated c-fos mRNA induction. Following knockdown of each transcription factor, cortical neurons were depolarized with KCl or stimulated with BDNF, and c-fos mRNA was measured. (b,d) Effect of transcription factor knockdown in KCl or BDNF-mediated c-fos eRNA induction (c-fos mRNA, KCl stimulation: shCREB P = 0.0222, t(2) = 6.600, F = 27.833 [SDP = 0.1192], n = 3; shMEF2A, P = 0.0417, t(2) = 4.741, F = 11.189 [SDP = 0.1849], n = 3; shNPAS4, P = 0.0455, t(2) = 4.524, F = 28.538 [SDP = 0.1178], n = 3; c-fos eRNA, KCl stimulation: e2, P = 0.0382, t(4) = 3.046, F = 1.078 [SDP = 0.4813], n = 3, e5, P = 0.0478, t(4) = 2.820, F = 3.517 [SDP = 0.2214], n = 3 for shCREB; e2, P = 0.0339, t(4) = 3.169, F = 2.995 [SDP = 0.2503], n = 3, e5, P = 0.0101, t(4) = 5.812, F = 2.441 [SDP = 0.2586], n = 3 for shMEF2A; e2, P = 0.0164, t(2) = 7.714, F = 9.324 [SDP = 0.2015], n = 2 for shNPAS4; c-fos mRNA, BDNF stimulation: shMEF2A, P = 0.0001, t(4) = 8.656, F = 2.157 [SDP = 0.3168], n = 3; c-fos eRNA, BDNF stimulation: e4, P = 0.0439, t(3) = 2.906, F = 8.631 [SDP = 0.1038], n = 3, e5, P = 0.0485, t(4) = 2.807, F = 5.381 [SDP = 0.1567], n = 3 for shMEF2A). All unpaired t-test. Error bars indicate SEM; p values are from a two-tailed t test. NS, not significant. SDP is the P value from comparing the standard deviations for both groups. SDP > 0.05 means the two SDs are not significantly different and can be used for an unpaired t-test.
Our study further demonstrated that the decrease in c-fos mRNA level is mainly caused by reduced activation of e2 and e5 enhancers, since the levels of eRNAs at those two enhancers were commonly decreased by the knockdown of CREB, MEF2A, and NPAS4 (Fig. 4b). The e1 enhancer is also active upon KCl stimulation as judged by eRNA expression, but knockdown of each of the three TFs had no considerable effect on e1 expression, suggesting that another unknown TF might play a redundant or more critical role at the e1 enhancer. However, since there was a trend toward a decrease in e1 eRNA level upon TF knockdown, although not significant (Fig. 4b), another possibility that we cannot rule out is that the lower level of e1 eRNAs might cause a difficulty in accurately measuring the dynamic change in e1 eRNA expression. A previous study of Npas4 KO neurons showed consistent results in that NPAS4 is functionally required for expression of several IEGs including the c-fos gene despite the fact that its own expression is regulated by activity 28. NPAS4 was unique among the activity-regulated TFs we investigated in that it binds to all c-fos enhancers but not to the promoter (Supplementary Fig. 1) 6. This result suggests that NPAS4 contributes to the KCl-mediated induction of the c-fos gene exclusively by regulating the activity of enhancers. Taken together, our study has revealed that e2 and e5 enhancers are particularly critical for KCl-induced c-fos expression where a subset of activity-regulated TFs, CREB, MEF2A, and NPAS4 play an important role.
Transcription factors in BDNF-mediated c-fos induction
We then performed a similar experiment with BDNF stimulation to test whether BDNF signaling requires a different set of TFs for c-fos induction. Unlike KCl stimulation, only knockdown of MEF2A but not CREB and SRF, significantly reduced the level of c-fos induction when neurons were stimulated by BDNF (Fig. 4c). It was previously shown that SRF is required for c-fos induction by growth factors in PC12 cells 29, however, in neurons we could not detect a significant impairment in BDNF-dependent induction of the endogenous c-fos gene by SRF knockdown. As neither of the stimuli show critical requirement of SRF for c-fos induction, we tested other IEGs, Arc and Egr1 for SRF dependency in their expression, and found that SRF depletion significantly impaired KCl-dependent induction of Arc and Egr1 (Supplementary Fig. 8). This result further demonstrates that the little or no effect of SRF knockdown on the c-fos induction is unlikely due to insufficient knockdown of SRF. We did not analyze NPAS4 for BDNF stimulation as the basal level of NPAS4 protein is very low in neurons, and its induction strictly requires membrane depolarization-mediated calcium signaling 30. We next examined the effect of TF knockdown on BDNF-dependent eRNA expression. In WT or scrambled control neurons, BDNF stimulation triggered rapid induction of eRNAs from e1, e4, and e5, whereas KCl-mediated membrane depolarization induced eRNA transcription from e1, e2, and e5 (Fig. 2a). Therefore e2 and e4 are uniquely responsive to KCl and BDNF, respectively. The decrease in BDNF-induced c-fos mRNA level by MEF2A depletion was paralleled by a decrease in eRNA levels at e4 and e5 (Fig. 4d). Consistently, the motif analysis shows that both e4 and e5 enhancers have multiple MEF2 binding sites (Supplementary Fig. 3a,b). Similar to the KCl-stimulation case, BDNF-dependent expression of e1 eRNAs was not significantly affected by the MEF2A knockdown. These results demonstrate that BDNF-induced expression of the c-fos gene is critically mediated by MEF2A, which mainly act on e4 and e5 enhancers. Since MEF2A is commonly important for both KCl and BDNF-induced c-fos induction and enhancer activation, we performed ChIP experiments to examine if MEF2A directly binds enhancers and/or the promoter to regulate the c-fos gene in mouse cortical neurons. We observed MEF2A binding at four enhancers (e1, e2, e4, e5) as well as the promoter, some of which also showed further increase in MEF2 binding in a stimulus-specific manner (Supplementary Fig. 9).
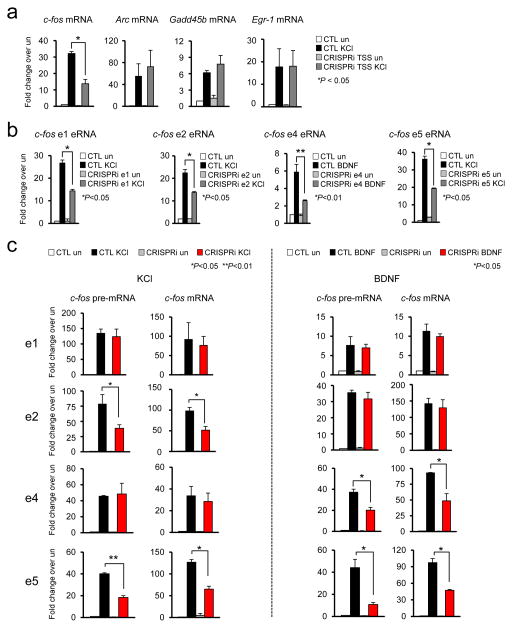
CRISPRi analysis of individual c-fos enhancers
The eRNA induction profile, enhancer reporter assay, and TF knockdown consistently suggest that e2 and e4 enhancers are selectively activated by KCl and BDNF, respectively. In order to further verify this intriguing observation, we utilized the CRISPRi system in which a catalytically inactivated mutant form of Cas9 (dCaps9) is fused to the KRAB (Krüppel-associated box) domain of Kox1 31. The KRAB domain protein can recruit KAP1 protein, which act as a scaffold for various heterochromatin-inducing factors such as heterochromatin protein 1 (HP1) and the histone methyltransferase SETDB1. The dCas9-KRAB fusion protein guided by sgRNA has been shown to efficiently silence expression of target genes 31, 32. We used CRISPRi to selectively silence the activity of individual c-fos enhancers and examined how that affected c-fos induction in response to KCl or BDNF. We first directed dCas9-KRAB to the c-fos transcription start site (TSS) and observed a decrease in the level of KCl-induced c-fos transcription, but not other IEGs, demonstrating the specificity of the system (Fig. 5a). The dCas9-KRAB caused a significant decrease in KCl- or BDNF-induced eRNA transcription (Fig. 5b). CRISPRi-mediated inhibition appears to specific to targeted enhancer as blocking e2 enhancer only reduced the activity of e2 enhancer but not other c-fos enhancers (Supplementary Fig 10). Based on the CRISPRi analysis with e5 enhancer, it appears that CRISPRi could also impair the physical interaction between the targeted enhancer and the promoter (Supplementary Fig 11). We then examined if the CRISPRi-mediated inhibition of individual enhancers affects the c-fos expression induced by either KCl or BDNF (Fig. 5c). Levels of pre-mRNA were measured in parallel with mRNAs in this analysis in order to evaluate the impact of CRISPRi more specifically on de novo transcription activity at the c-fos gene. CRISPRi at e2 and e5 caused a significant decrease in the induction levels of both RNA populations of the c-fos gene whereas suppression of e4 and e5 enhancer activities impaired BDNF-induced c-fos expression. The induction of other IEGs (Arc, Gadd45b, and Egr-1) was not affected by any CRISPRi (Supplementary Fig. 12). Together with eRNA induction profiles and luciferase reporter data, CRISPRi study demonstrates that e2 and e4 enhancers functionally contribute to the c-fos gene induction by specifically responding to KCl and BDNF stimulation, respectively. On the other hand, blocking e1 activity in the native context by CRISPRi did not have any impact on c-fos induction by either stimulus despite its inducible interaction with the c-fos promoter and eRNA transcription. We also showed by luciferase assay that e1 alone could enhance reporter expression in response to all three stimuli. Based on these results, we propose that e1 might act as a “shadow enhancer” for c-fos expression in native context, whose function is to ensure precise and robust transcription of target genes during extreme or adverse conditions that may occur throughout development and/or by dynamic environmental changes, where otherwise its function is dispensable by functionally redundant primary enhancers 33.
Figure 5.
Specific requirement of e2 and e4 enhancer in KCl or BDNF-mediated c-fos transcription. (a) Effect of c-fos promoter-targeted CRISPRi (dCas9-KRAB) (P = 0.0189, t(2) = 12.637, F = 4.830 [SDP = 0.2718], n = 3 biological replicates). (b) Effect of e1, e2, e4, and e5 enhancer-targeted CRISPRi on their respective eRNAs (c-fos e1 eRNA, P = 0.0151, t(2) = 8.037, F = 4.147 [SDP = 0.2906], n = 2; c-fos e2 eRNA, P = 0.0393, t(4) = 3.015, F = 3.461 [SDP = 0.2242], n = 3; c-fos e4 eRNA, P = 0.0206, t(2) = 6.864, F = 25.000 [SDP = 0.1257], n = 2; c-fos e5 eRNA, P = 0.0104, t(2) = 9.711, F = 32.398 [SDP = 0.1107], n = 2 biological replicates). (c) Effect of c-fos e1, e2, e4, and e5 enhancer-targeted CRISPRi. Following the suppression of e1, e2, e4, and e5 enhancer by CRISPRi, cortical neurons were stimulated by KCl or BDNF, and expression levels of c-fos pre-mRNA and mRNA were measured using RT-qPCR (KCl stimulation: e2 c-fos pre-mRNA, P = 0.0433, t(4) = 2.918, F = 1.388 [SDP = 0.4187], n = 3; c-fos mRNA, P = 0.0102, t(8) = 3.340, F = 1.114 [SDP = 0.459], n = 5; e5 c-fos pre-mRNA P = 0.0077, t(2) = 11.350, F = 3.550 [SDP = 0.3106], n = 2, c-fos mRNA, P = 0.0207, t(2) = 6.845, F = 1.122 [SDP = 0.4817], n = 2; BDNF stimulation: e4 c-fos pre-mRNA, P = 0.0485, t(2) = 4.373, F = 1.807 [SDP = 0.4868], n = 2, c-fos mRNA, P = 0.0322, t(2) = 5.440, F = 137.97 [SDP = 0.0541], n = 2; e5 c-fos pre-mRNA, P = 0.0489, t(2) = 4.353, F = 17.288 [SDP = 0.1503], n = 2, c-fos mRNA, P = 0.0223, t(2) = 6.582, F = 26.669 [SDP = 0.1218], n = 2 biological replicates). All unpaired t-test. Error bars indicate SEM; p values are from a two-tailed t test. SDP is the P value from comparing the standard deviations for both groups. SDP > 0.05 means the two SDs are not significantly different and can be used for an unpaired t-test.
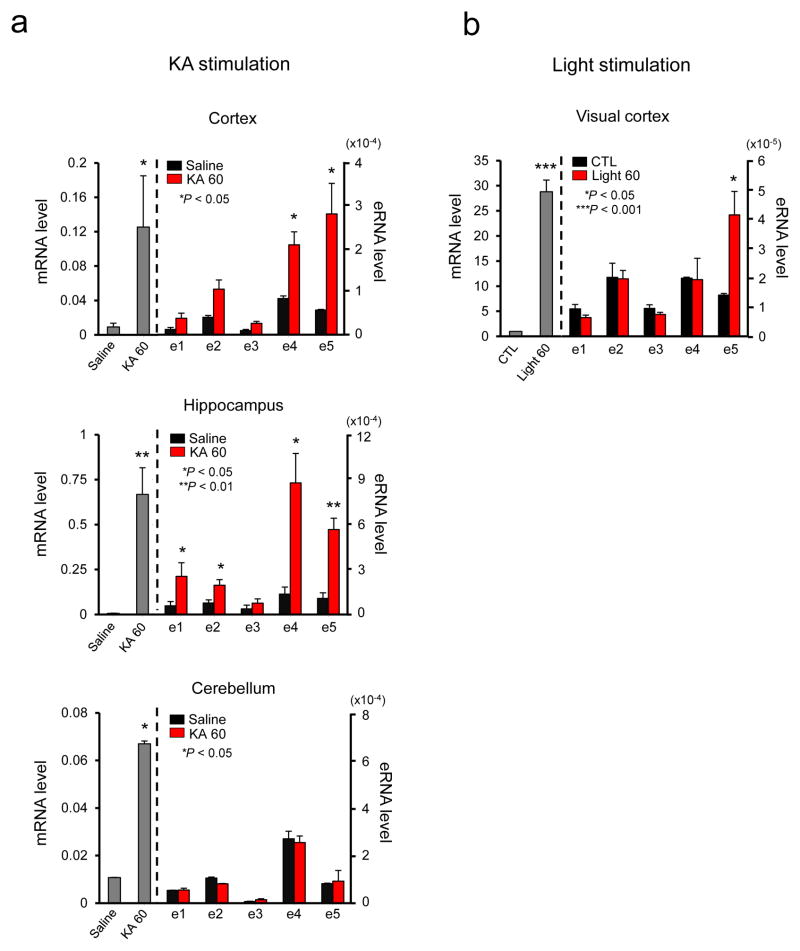
Combinatorial activation of c-fos enhancers in the brain
We next asked if combinatorial activation of c-fos enhancers also occurs in the brain. First we examined whether different brain regions activate distinctive sets of the c-fos enhancers to mediate c-fos induction upon chemically induced seizure. Young adult mice were injected intraperitoneally with Kainic acid (KA), which activates glutamate receptors to induce seizure. One hour after KA injection, a strong c-fos mRNA induction was detected in the cerebral cortex and hippocampus, and also weakly in the cerebellum (Fig. 6a) 34. We then measured individual eRNA levels in those brain regions. A single injection of KA led to induction of c-fos eRNAs from e4 and e5 in the cerebral cortex. On the other hand, eRNAs from all c-fos enhancers except e3 were induced in the hippocampus. We could not detect any induction of eRNA in the cerebellum, possibly because the KA-induced c-fos mRNA induction level was lower than other brain regions. We also investigated whether sensory stimulation can trigger eRNA transcription from the c-fos enhancers in the intact brain. We took dark-reared mice and stimulated their visual cortex by light for 1 h. The RT-qPCR analysis of the RNAs isolated from the visual cortex following visual stimulation detected eRNAs only from the e5 enhancer region (Fig. 6b). These results support our findings from in vitro neuronal culture that different combinations of enhancers are utilized to ensure c-fos induction in response to various stimuli, and also illustrate region-specific combinatorial activation of the c-fos enhancers.
Figure 6.
Characterization of c-fos enhancer activities in vivo. (a) c-fos mRNA expression in the cortex, hippocampus, and cerebellum following KA-evoked seizure. c-fos mRNA was measured using RT-qPCR 1 h after saline or KA administration (note that the y axes are scaled differently). c-fos eRNA induction in the cortex, hippocampus, and cerebellum following KA-evoked seizure (c-fos mRNA, Cortex: P = 0.0458, t(4) = 4.511, n = 5 mice, Hippocampus: P = 0.0067, t(5) = 4.455, n = 5 mice, Cerebellum: P = 0.0133, t(4) = 47.767, n = 5 mice; Cortex, c-fos e4, P = 0.0150, t(4) = 4.084, n = 5 mice, c-fos e5, P = 0.034, t(4) = 3.154, n = 5 mice; Hippocampus, c-fos e1, P = 0.0386, t(4) = 3.036, n = 5 mice, c-fos e2, P = 0.0419, t(5) = 2.717, n = 5 mice, c-fos e4, P = 0.0211, t(4) = 3.686, n = 5 mice, c-fos e5, P = 0.0028, t(5) = 5.454, n = 5 mice). (b) Expression of c-fos mRNA in the visual cortex of mice treated with light stimulation. c-fos eRNA induction in the visual cortex of mice following light stimulation. The levels of mRNA and eRNA were normalized to the level of Gapdh mRNA (c-fos mRNA, Visual cortex: P = 0.0069, t(2) = 11.966, n = 3 mice; c-fos e5, P = 0.0485, t(4) = 2.806, n = 3 mice). All unpaired t-test with Welch correction. Error bars indicate SEM; p values are from a two-tailed t test.
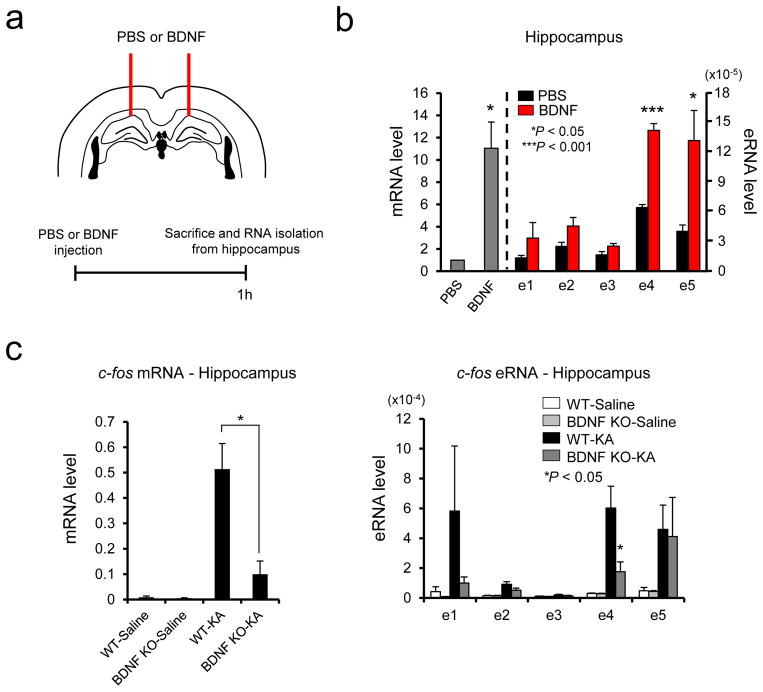
We wondered if our finding from the in vitro study that e2 and e4 are uniquely responsive to KCl and BDNF, respectively, could be observed in the intact brain in some way. However, unlike our in vitro culture system in which we could make a particular signaling dominate in the c-fos response, any sensory or chemical stimulation to the intact brain would involve multiple signaling pathways that would intricately intersect with each other. Despite this confounding issue, we attempted to investigate BDNF-induced activation of c-fos enhancers in the hippocampus. We directly injected recombinant BDNF proteins into the hippocampus and analyzed both c-fos mRNA and eRNAs 1 h later (Fig. 7a). A direct BDNF injection induced c-fos mRNA ~10 fold in the hippocampus compared to control PBS injection (Fig. 7b). In parallel, the e4 and e5 activities were mainly increased by BDNF. Notably, this pattern of enhancer activation is similar to the one from in vitro neuronal culture stimulated by BDNF except for the e1 enhancer despite the considerable differences present between the two experimental systems such as cell types and developmental stage. We next investigated whether any of BDNF-activated enhancers would be functionally important for BDNF-induced c-fos expression. We utilized Bdnf conditional KO mice in which the Bdnf gene has been selectively deleted in neurons during late embryogenesis 35. We then induced seizure by KA in both Bdnf KO and littermate control mice, and compared the expression levels of the c-fos mRNA and eRNAs between the two groups (Fig. 7c). While KA-induced seizure increased c-fos expression in the hippocampus, the induction level of the c-fos gene was dramatically diminished in the conditional Bdnf KO mice. This result implies that the downstream signaling pathways triggered by KA and BDNF significantly overlap or intersect with each other. Interestingly, KA-induced activation of e4 enhancer was severely impaired by Bdnf deletion whereas e5 enhancer activity was unaltered. These results strongly suggest that e4 enhancer is selectively responsive to BDNF-induced signaling pathways both in vitro and in vivo.
Figure 7.
Stimulus-specific combinatorial action of c-fos enhancers in vivo. (a) Schematic diagram of BDNF stereotaxic injection procedure. (b) c-fos mRNA and eRNA induction in the hippocampus following BDNF injection. c-fos mRNA and eRNA were measured using RT-qPCR 1 h after PBS or BDNF injection (c-fos mRNA, Hippocampus: P = 0.0236, t(3) = 4.267, n = 4 mice, unpaired t-test with Welch correction; c-fos e4, P = 0.0001, t(6) = 10.373, F = 5.082 [SDP = 0.1074], n = 4 mice, unpaired t-test, c-fos e5, P = 0.0402, t(3) = 3.475, n = 4 mice, unpaired t-test with Welch correction). (c) c-fos mRNA and eRNA induction in the hippocampus following KA-evoked seizure from wild-type and BDNF-KO mice. c-fos mRNA and eRNA were measured using RT-qPCR 1 h after saline or KA administration (c-fos mRNA, Hippocampus: P = 0.0216, t(4) = 3.660, F = 3.756 [SDP = 0.2103], n = 3 mice; c-fos e4, P = 0.0476, t(4) = 2.825, F = 10.113 [SDP = 0.0900], n = 3 mice, all unpaired t-test). The levels of mRNA and eRNA were normalized to the level of Gapdh mRNA. Error bars indicate SEM; p values are from a two-tailed t test. SDP is the P value from comparing the standard deviations for both groups. SDP > 0.05 means the two SDs are not significantly different and can be used for an unpaired t-test.
DISCUSSION
In this study, we demonstrate that multiple enhancers located near the c-fos gene can be activated in distinct combinations, in response to various stimuli or in different brain regions (Supplementary Fig. 15). We propose that such a combinatorial activation of enhancers present within a cluster could be a mechanism to allow the c-fos gene to be robustly responsive to a wide range of stimuli. As a consequence of such a mechanism, each stimulus exhibits a differential functional requirement of TFs, each of which also shows a distinct binding specificity in the c-fos gene area.
The c-fos gene is a prototypical IEG whose rapid and transient expression was originally observed in NIH 3T3 cells where the c-fos mRNA level rises within several minutes in response to the platelet-derived growth factor (PDGF) and subsides in ~ 30 minutes 36. In primary neuronal cultures, rapid c-fos transcription can be triggered by growth factors, a cAMP analog, or membrane depolarization induced by either neurotransmitters or KCl 14. Induction of c-fos expression has also been observed in the intact brain in response to a variety of physiological and pathological stimuli including seizure, traumatic injury, somatosensory stimulation, and long-term potentiation (LTP) 14. c-fos expression correlates very well with neuronal activity, and in some cases it has been thought to represent the synaptic plasticity response of neurons 37. In fact, c-fos has been the most popular marker for mapping functional neural circuits involved in sensory information processing due to its rapid and transient expression kinetics with very low background level, as well as its responsiveness to such a broad range of stimuli. Suppression of c-fos expression by antisense-mediated knockdown or a genetic deletion of the c-fos gene in mice caused a defect in various forms of learning behavior such as fear and spatial learning, and conditioned place preference 38–40. At the molecular level, FOS has recently been shown to regulate a wide range of activity-regulated genes that control synaptic function through its binding almost exclusively to neuronal enhancers 23.
The rationale of this study was two-fold. First, we wanted to determine the molecular mechanism that enables robust and reliable expression of the c-fos gene in response to various stimuli, which would help to understand the gene regulatory logic in multicellular organisms that has evolved to control gene expression with high precision and efficiency in the face of dynamic environmental changes and developmental cues. Secondly, we want to develop in vivo IEG reporters that are not only more potent in driving the expression of a fluorescent reporter protein but also tailored to be selectively responsive to a particular signal. The fos-based transgenic reporter mouse lines such as fosGFP and fosCreERT2 TRAP mice have been used to detect neuronal populations that are activated by behavioral or pharmacological stimuli 41, 42. However these mice utilize only the c-fos promoter region to induce the reporter fluorescent protein and do not include any of the c-fos enhancers we have characterized. Based on the findings presented in this study, the promoter-only reporters might not faithfully recapitulate the expression characteristics of the endogenous c-fos gene in vivo triggered by sensory or pharmacological stimuli. We have shown that combination of an enhancer with the c-fos minimal promoter can significantly increase the induction level of the luciferase reporter but in a stimulus-specific manner. Therefore, an array of reporters in which expression of a fluorescent protein gene is controlled by the c-fos promoter combined with a single or various combinations of the surrounding enhancers could serve as a versatile toolkit for the analysis of sensory stimulation-specific activation of particular neural circuits or populations in vivo and in acute brain slices.
Why are some genes regulated by multiple enhancers whereas others are not? While this intriguing question would require much work to be fully understood, several genetic studies of developmentally crucial genes suggest that enhancers with extensively overlapping activities help to ensure the robustness and precision of their expression in natural populations in the face of environmental and genetic variations 33. Evidence also suggests some other mechanisms of enhancer coordination 13. Multiple enhancers may simply decrease the failure rate in target gene expression by increasing the occurrence of productive interaction with their target gene. In some cases, different enhancers play non-redundant roles by acting sequentially during the developmental process of gene activation as shown in the regulation of the HoxD gene cluster during limb development 43. On the other hand, our study demonstrates another mode of enhancer coordination in which distinct subsets of enhancers dynamically respond to different types of stimuli to ensure the robust induction-capacity of the c-fos gene. An interesting question related to our study is how many and what types of genes are subject to the coordination of multiple enhancers like the c-fos gene. A recent three-dimensional chromatin interactome study in primary human fibroblast cells (IMR90) has shown that 46% of active genes do not interact with distal enhancers, and highly transcribed house keeping genes are enriched in this group 11. The rest of the active genes make extensive interactions with enhancers and are enriched for cell-type specific functions including signal transduction. In addition, recently defined super-enhancers, which refers to a set of clustered enhancers that exhibit an exceptionally high level of transcriptional activators or active enhancer chromatin marks such as H3K27ac, are also enriched near genes with a function in cell identity or cell-type specific pathways 44, 45. To the best of our knowledge, such a genome-wide analysis of chromosomal interactome has not been performed in the brain. Nonetheless, we provide here two additional inducible genes that appear to be regulated by multiple enhancers in a stimulus-dependent manner. The Peli1 gene is rapidly induced by either KCl or BDNF and multiple enhancers are located upstream of the gene (Supplementary Figs. 13). The eRNA expression analysis of four putative Peli1 enhancers revealed KCl and BDNF-specific activation patterns of the enhancers. The Igf-1 gene is not an IEG, but instead is induced more slowly, yet still appears to be regulated by interesting dynamics between multiple enhancers (Supplementary Figs. 14). Selective activation of e2 and e3 enhancers was observed by 6 h KCl stimulation. Interestingly, those two enhancers appear to be activated more rapidly by BDNF stimulation, which was then followed by slower activation of e1 enhancer at 6 h. While these are preliminary results, it might be that combinatorial action of multiple enhancers can be achieved spatially and/or temporally.
The c-fos enhancers can most likely be classified as one of the recently termed “super-enhancers”. A recent study showed that super-enhancers are the major platform to which multiple signaling pathways converge, to regulate genes that play prominent roles during development and tumorigenesis 46. The c-fos enhancers are clustered within a ~50 kb region flanking the c-fos gene and are highly enriched with an active enhancer mark, H3K27ac in the forebrain throughout development (Supplementary Fig. 1) 17. They also exhibit strong binding levels of key activity-regulated TFs in various combinations (Supplementary Fig. 1) 6. Although most of the activity-regulated TFs are broadly expressed throughout the brain, their downstream target genes and physiological functions in synapse development are not the same but rather distinct 14. Our study illustrates how the transcription output of each mammalian gene is controlled by the combinatorial interactions of multiple TFs that occur at the enhancers and the promoter. We also show that the functional requirement of individual TFs for the c-fos gene varies depending on the type of stimulation and possibly the brain regions, which is in part mediated by the combinatorial usage of multiple enhancers.
METHODS
All animal experiments performed in this study were reviewed and approved by the IACUC committee at UT Southwestern Medical Center.
Mouse cortical neuron, glial cell culture and stimulation
Embryonic day 16.5 (E16.5) mouse embryo cortices were dissected and then cultured in Neurobasal (NB) media supplemented with B-27 and Glutamax for cortical neuron culture and Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and Glutamax for glial cell culture. For KCl depolarization, days in vitro (DIV) 5 neurons were made quiescent overnight in 1 μM tetrodotoxin (TTX, Tocris) and then incubated for 10, 30, or 90 min in 55 mM KCl to stimulate gene expression. In the BDNF and Forskolin experiments, neurons were incubated with BDNF (10 ng/ml, R&D system) and, Forskolin (10 μM, Millipore) at DIV 6 for 1 h.
shRNA design, transfection, and retroviral infections
shRNAs against SRF, CREB, NPAS4, MEF2A, MEF2C, and MEF2D were designed as previously published 30, 47–50. The shRNAs were cloned into the HpaI/XhoI sites of the pLLX lentiviral vector. pLLX-shRNA-GFP, with the helper plasmids Δ8.9 and VsVg constructs, were transfected into HEK293T cells using Fugene-HD (Promega) for 48–72 h. The targeted sequences were as follows: Srf, 5′-AAGATGGAGTTCATCGACAAC-3′; Creb, 5′-GGAGTCTGTGGATAGTGTA-3′; Npas4, 5′-GGTTGACCCTGATAATTTA-3′; Mef2a, 5′-GTTATCTCAGGGTTCAAAT-3′; Mef2c, 5′-GTTCTTGCTGCCAGGTGGGAT-3′; Mef2d, 5′-GTAGCTCTCTGGTCACTCC-3′. Collected lentiviral supernatant was then added to DIV 3 neurons and the cells were harvested at DIV 6.
Chromosome Conformation Capture (3C)
3C analysis was performed as previously published 21, using the restriction enzymes SacI and KpnI. 3C RT-qPCR primer sequences were as follows: SacI c-fos pro-rev anchor primer, 5′-CTGACGGATAAACATTGTGC-3′; SacI c-fos-A primer, 5′-CATTCTTGGTTTCCACAGGC-3′; SacI c-fos-B primer, 5′-GATATTAGAGTTAAGGTGTGC-3′; SacI c-fos-C primer, 5′-CATCTTTGACCTATCAGCAC-3′; SacI c-fos-D primer, 5′-CAAGCTTGAACAGTTAGGTAG-3′; SacI c-fos-E primer, 5′-CTTACTGGGGCAAGCACTAG-3′; SacI c-fos-F primer, 5′-GTTCCATCTGCCATTTCAGCA-3′. A bacterial artificial chromosome (BAC) clone containing the c-fos gene was used as a control template. The BAC plasmid was digested with SacI or KpnI restriction enzymes overnight at 37 °C, and then ligated by T4 DNA ligase for 4 h at 16 °C. The DNA was subsequently purified by phenol/chloroform extraction and ethanol precipitation.
RT-qPCR
Total RNA was isolated from DIV 6 cultured cortical neurons using the reagent Trizol (Invitrogen). cDNA synthesis was performed as previously published 21. Primers employed were: c-fos coding forward, 5′-ATCCTTGGAGCCAGTCAAGA-3′, reverse, 5′-ATGATGCCGGAAACAAGAAG-3′; c-fos e1 forward, 5′-TCCGGTAAGGGCATTGTAAG-3′, reverse, 5′-CAAAGCCAGACCCTCATGTT-3′; c-fos e2 forward, 5′-TGCAGCTCTGCTCCTACTGA-3′, reverse, 5′-GAGGAGCAAGACTCCCACAG-3′; c-fos e3 forward, 5′-GGGGTGGTGCTAGTGGTAAA-3′, reverse, 5′-TCATCGTATGCCTCTGCTTG-3′; c-fos e4 forward, 5′-AGGGGGACACATGAGTTCTG-3′, reverse, 5′-GACAAGCCGGGTTAGACTGA-3′; c-fos e5 forward, 5′-CAACCCTGTCATCTATTTAGC-3′, reverse, 5′-TAAGAACTGCGGGGGTCTTC-3′; Arc coding forward, 5′-GTGAAGACAAGCCAGCATGA-3′, reverse, 5′-CCAAGAGGACCAAGGGTACA-3′; Gadd45b coding forward, 5′-GTTCTGCTGCGACAATGACA-3′, reverse, 5′-TTGGCTTTTCCAGGAATCTG-3′; Egr-1 coding forward, 5′-AACACTTTGTGGCCTGAACC-3′, reverse, 5′-AGGCAGAGGAAGACGATGAA-3′; c-fos pre-mRNA forward 5′-GCAGCAGTAGGATGGAGGAG-3′; c-fos pre-mRNA reverse 5′-TGTATGCACCTCATCGGAGA-3′; Arc pre-mRNA forward 5′-GGTGAGCTGGCTTAGCAAAT-3′; Arc pre-mRNA reverse 5′-AGTGGCATCAGAGTGGGTGT-3′; Gadd45b pre-mRNA forward 5′-GTGGAGGGCGACATTTCTCA-3′; Gadd45b pre-mRNA reverse 5′-AAATACTGATGGCGGTGGGG-3′; Egr-1 pre-mRNA forward 5′-ACAGCTCCAGGGTCTTGTGT-3′; Egr-1 pre-mRNA reverse 5′-ACGCAGCTTGAGTTCTCCAT-3′; Srf coding forward, 5′-GGTGCCAGGTAGTTGGTGAT-3′, reverse, 5′-TGAAGCCAGCATTCACAGTC-3′; Creb coding forward, 5′-CCGAGAAATGCTGCCTAAAC-3′, reverse, 5′-ACACAAGCCACCCAAAACTC-3′; Npas4 coding forward, 5′-CAGGGACAGGTTAGGGTTCA-3′, reverse, 5′-TTCAGCAGATCAGCCAGTTG-3′; Mef2a coding forward, 5′-AGGGCTGCTTGTCCTAGATG-3′, reverse, 5′-ACCAGCGCTGACCTGTCT-3′; Mef2c coding forward, 5′-GGTCTCCTGTTGACAGCTTGA-3′, reverse, 5′-CGAAGGTCTGGTGAGTCCA-3′; Mef2d coding forward, 5′-CTGGCACTAGGCAATGTCAC-3′, reverse, 5′-AGTGGGGCTGTTGCTGAG-3′; Gapdh forward, 5′-AGGTCGGTGTGAACGGATTTG-3′, reverse, 5′-TGTAGACCATGTAGTTGAGGTCA-3′; c-fos e1 for ChIP forward, 5′-GTGTTGAGTCTGTTCTGTCTC-3′, reverse, 5′-CATCTTCCCTGCTGCTCAGT-3′; c-fos e4 for ChIP forward, 5′-GTTTCTATATCGTGTTGTTCTC-3′, reverse, 5′-CTGTCACAGGCAGAGGTGAA-3′; Negative region for ChIP forward, 5′-GAGCATTTCTCCCTGGAAGG-3′, reverse, 5′-CAAAAAGGCTTTTTGCTGGCT-3′; Peli1 coding forward, 5′-GTAGTAACTGACACCGTTCC-3′, reverse, 5′-GGGACTGCGCTCACATATGA-3′; Peli1 e1 forward, 5′-CCCTCTGTAAAAGAAATTTGCA-3′, reverse, 5′-CATTACCAAGGGGTGGATGG-3′; Peli1 e2 forward, 5′-GGTTCAATTCACTGTCACCTC-3′, reverse, 5′-GGCCAAGCACTGGGTGTTTA-3′; Peli1 e3 forward, 5′-GACTTCACTGTGGTATCCTAG-3′, reverse, 5′-GAAGCATCCAGTCAGGCACT-3′; Peli1 e4 forward, 5′-CTAAATGAGCAACCAACCTGC-3′, reverse, 5′-CTCAGTCCTTGAGATATTGAC-3′; Peli1 e5 forward, 5′-GATGCTCCAAGGCAACCAAT-3′, reverse, 5′-CCTAACAGGAAGTGAATCTGG-3′. Igf-1 coding forward, 5′-CTATGGCTCCAGCATTCGGA-3′, reverse, 5′-GGATAGAGCGGGCTGCTTTT-3′; Igf-1 e1 forward, 5′-CAGCAGATTGCAGATATAATTC-3′, reverse, 5′-GGAATGACACCCAAGTTGTC-3′; Igf-1 e2 forward, 5′-GATCCTTTCTATCTCTGAGAAT-3′, reverse, 5′-GTGGTTTGGTCTGTTAAATGG-3′; Igf-1 e3 forward, 5′-GATAGCTCTTGCTAACTGGAA-3′, reverse, 5′-GATGTCTCGGGTAGAGGACT-3′; Igf-1 e4 forward, 5′-GAAGCTAATGACAAATGAGCTA-3′, reverse, 5′-CCTCAGACAGCCAGGGATTT-3′. PCR amplification conditions have been described 21. Statistical significance was evaluated by an unpaired two-tailed Student’s t-test.
Luciferase Reporter Assay
Cultured primary cortical neurons were transfected using the calcium phosphate precipitation method at DIV 3 and harvested at DIV 6. The NIH3T3 cells were transfected at 70% confluency using fugene HD (Promega). NIH3T3 cells are from American Type Culture Collection (ATCC Manassas, VA, USA), and were authenticated using the DNA Sanger Sequencing Mlutiplex PCR Assay. The cells have not been tested for mycoplasma contamination. The entire sequences of c-fos eRNA and promoter regions were amplified by PCR using a BAC clone (CHORI, RP24-233K8) and reporter constructs were generated with the firefly luciferase reporter plasmid pGL4.11[luc2P] (Promega). The pGL4.11-c-fosPromoter-luc2P plasmid, which contains 361 nucleotides from the start site and includes the first exon and intron sequence (212 nucleotides), was generated as the c-fos promoter region. Each of the full length c-fos eRNAs were subcloned into the KpnI/NheI or KpnI/SacI sites of the pGL4.11-c-fosPromoter-luc2P plasmid. Detailed c-fos eRNA genomic locations were as follows: pGL4.11-c-fose1Promoter-luc2P (+39.554 to +35.802 kb from start site); pGL4.11-c-fose2Promoter-luc2P (+20.034 to +16.454 kb from start site); pGL4.11-c-fose3Promoter-luc2P (+6.811 to +5.12 kb from start site); pGL4.11-c-fose4Promoter-luc2P (+3.432 to +1.167 kb from start site); pGL4.11-c-fose5Promoter-luc2P (−9.955 to −5.591 kb from start site). Genomic locations for combinational c-fos eRNA luciferase assay were as follows: pGL4.11-c-fose1+e2+e4+e5Promoter-luc2P (e1: +37.887 to +38.387 kb, e2: +18.482 to +17.990 kb, e4: +2.218 to 1.718 kb, e5: −12.160 to −12.648 kb from start site); pGL4.11-c-fose1+e2+e5Promoter-luc2P (e1: +37.887 to +38.387 kb, e2: +18.482 to +17.990 kb, e5: −12.160 to −12.648 kb from start site); pGL4.11-c-fose1+e4+e5Promoter-luc2P (e1: +37.887 to +38.387 kb, e4: +2.218 to 1.718 kb, e5: −12.160 to −12.648 kb from start site); pGL4.11-c-fose1+e5Promoter-luc2P (e1: +38.387 to +37.887 kb, e5: −12.160 to −12.648 kb from start site).
KA injection and visual stimulation
Adult C57BL/6 mice (2–4 months), wild type and BDNF knockout mice (4 – 6 months) 35 were injected intraperitoneally with Kainic acid (KA, Tocris) dissolved in 0.9% saline at 20 mg/kg, and control mice were injected with the same volume of 0.9% saline. Animals were sacrificed at 1 h after the injection. In the visual stimulation experiment, mice were placed in dark controlled environmental chambers for light deprivation. After 6 days, mice were taken out from the controlled environmental chambers and exposed to a lighted environment for 1 h, and then the visual cortex was isolated from the brain. Using Trizol reagent, total RNA was isolated from the cortex, hippocampus, cerebellum, and visual cortex, and then cDNA was synthesized.
Transcription repression by CRISPR-dCas9-KRAB (CRISPRi)
The c-fos promoter and enhancer target specific single guide RNAs (sgRNAs) were designed using the E-CRISPR design tool 51. Both of the lentiviral backbone plasmids, pHR-SFFV-dCas9-BFP-KRAB 31 and lentiGuide-Puro 52 were obtained from Addgene. The annealed DNA was cloned into the BsmBI site of the lentiGuide-Puro vector. The pHR-SFFV-dCas9-BFP-KRAB and lentiGuide-Puro-sgRNA, with the helper plasmids Δ8.9 and VsVg constructs, were transfected into HEK293T cells using Fugene-HD for 48–72 h. The targeted promoter, e1, e2, e4, and e5 sequences were as follows, promoter, 5′-AGAAGACTGGATAGAGCCGG-3′; e1, 5′-TGGGAAGCAGGACAGACTGG-3′; e2, 5′-ATTAGAAAGTGCTGAGGCGG-3; e4, 5′-GGATGGATCTTTAGGGGCGC-3; e5, 5′-ACCGCCCTGGCGCTCAGACC-3. Collected lentiviral supernatant was added to DIV 3 neurons, and cells were harvested at DIV 6.
BDNF stereotaxic injection
Two-month old male C57BL6 mice were anesthetized with tribromoeethanol (200 mg/kg, Sigma). 25 ng of BDNF was injected with a glass pipette in to the dorsal hippocampal CA1 region with the following coordinates, anteroposterior (AP) + 1.90 mm, medial lateral (ML) ± 1.25 mm, dorsal ventral (DV) – 1.20 mm. Animals were sacrificed at 1 h after the stereotaxic injection, and then the hippocampus was isolated from the brain.
Immunocytochemistry
DIV 6 primary cortical neurons and glial cells were fixed and permeabilized with 4% paraformaldehyde in PBS and 0.25% Tritron X-100 in PBS, respectively, and then incubated with primary antibody against Tuj-1 (Covance, 1:500) and GFAP (Sigma, 1:500). For secondary antibodies, Alexa Fluor 488- or 555-conjugated anti-IgG (Invitrogen, 1:600) antibodies were used. Cells were mounted in Vectashield mounting medium with DAPI (Vector laboratories) for fluorescence analysis, and imaged with a fluorescence microscope (Olympus LX51).
Chromatin Immunoprecipitation (ChIP)
ChIP assay was performed as previously published 21. Briefly, at DIV 6 cultured cortical neurons were incubated with KCl (55mM) for 30 min and BDNF (10 ng/ml) or Forskolin (10 μM) at DIV 6 for 1 h. Subsequently, cell lysates was incubated overnight with anti-Mef2a 53, and then the lysate was incubated with Protein A/G agarose beads (Santa Cruz). After post-immunoprecipitation, DNA was purified and then analyzed with RT-qPCR.
Western blot analysis
Western blot analysis was performed as previously published 21. Antibodies used for wetstern blot were anti-Mef2A (1:1000) 53, anti-Mef2C (Proteintech, 1:500), anti-Mef2D 53, or anti-β-actin (Sigma-Aldrich, 1:5000). Images were acquired by Odyssey (LI-COR).
Statistical analyses
The statistical analysis was performed using an unpaired Student’s t-test and unpaired t-test with Welch correction. No statistical methods were used to pre-determine sample size, but our sample sizes are similar to those used commonly in the field. Unpaired Student’s t-test and unpaired t-test with Welch correction were performed for comparison between two groups of samples. All detailed values were listed in each figure legend. Data distribution was assumed to be normal but this was not formally tested. Randomization and blinding were not used. We considered P < 0.05 to be statistically significant.
Supplementary Material
Acknowledgments
We would like to thank Dr. Lisa Monteggia and her lab members for providing us with the conditional Bdnf KO mice. We thank Dr. Michael Greenberg for providing us with MEF2A and MEF2D antibodies, which were used for ChIP experiments. We also thank Dr. Wei Xu for advice on stereotaxic injection, and Dr. Carla Green and J. Stubblefield for providing the dark controlled environment chamber. This work was supported by the National Science Foundation (NSF) BRAIN EAGER Award (IOS1451034) and the National Institute of Neurological Disorders and Stroke (NINDS) of the National Institutes of Health (NIH) under Award Number R01NS085418 (T.-K. Kim).
Footnotes
AUTHOR CONTRIBUTIONS
T.-K.K., J.-Y.J. designed the project, and J.-Y.J. performed the CRISPRi, 3C assays, luciferase reporter assay, ChIP, Immunocytochemistry, in vivo experiments, in vitro analysis of transcription factor knockdown, and eRNA and mRNA expression analysis. K.S. performed western blot and expression analysis for c-fos eRNA in NIH 3T3 cells. L.F performed primary cortical neuron culture and helped to make lentivirus, and CRISPRi and shRNA cloning. G.K performed bioinformatics analysis. J.-Y.J., K.S., and T.-K.K. wrote the manuscript. All authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Lyons MR, West AE. Mechanisms of specificity in neuronal activity-regulated gene transcription. Prog Neurobiol. 2011;94:259–295. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frey U, Frey S, Schollmeier F, Krug M. Influence of actinomycin D, a RNA synthesis inhibitor, on long-term potentiation in rat hippocampal neurons in vivo and in vitro. J Physiol. 1996;490(Pt 3):703–711. doi: 10.1113/jphysiol.1996.sp021179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP, Sheng M, Lau LF, Greenberg ME. Growth factors and membrane depolarization activate distinct programs of early response gene expression: dissociation of fos and jun induction. Genes Dev. 1989;3:304–313. doi: 10.1101/gad.3.3.304. [DOI] [PubMed] [Google Scholar]
- 5.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim TK, Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell. 2015;162:948–959. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaikkonen MU, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2013;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarzer W, Spitz F. The architecture of gene expression: integrating dispersed cis-regulatory modules into coherent regulatory domains. Curr Opin Genet Dev. 2014;27:74–82. doi: 10.1016/j.gde.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H, et al. Tissue-specific RNA expression marks distant-acting developmental enhancers. PLoS Genet. 2014;10:e1004610. doi: 10.1371/journal.pgen.1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nord AS, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Core LJ, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pekowska A, et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch F, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol. 2011;18:956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 21.Schaukowitch K, et al. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arner E, et al. Gene regulation. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik AN, et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci. 2014;17:1330–1339. doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 26.Thomas-Chollier M, et al. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39:W86–91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misra RP, et al. L-type voltage-sensitive calcium channel activation stimulates gene expression by a serum response factor-dependent pathway. J Biol Chem. 1994;269:25483–25493. [PubMed] [Google Scholar]
- 28.Ramamoorthi K, et al. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334:1669–1675. doi: 10.1126/science.1208049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheng M, Dougan ST, McFadden G, Greenberg ME. Calcium and growth factor pathways of c-fos transcriptional activation require distinct upstream regulatory sequences. Mol Cell Biol. 1988;8:2787–2796. doi: 10.1128/mcb.8.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Y, et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012;28:409–416. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan JI, Cohen DR, Hempstead JL, Curran T. Mapping patterns of c-fos expression in the central nervous system after seizure. Science. 1987;237:192–197. doi: 10.1126/science.3037702. [DOI] [PubMed] [Google Scholar]
- 35.Monteggia LM, et al. Essential role of brain-derived neurotrophic factor in adult hippocampal function. Proc Natl Acad Sci U S A. 2004;101:10827–10832. doi: 10.1073/pnas.0402141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 37.Kawashima T, Okuno H, Bito H. A new era for functional labeling of neurons: activity-dependent promoters have come of age. Front Neural Circuits. 2014;8:37. doi: 10.3389/fncir.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleischmann A, et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci. 2003;23:9116–9122. doi: 10.1523/JNEUROSCI.23-27-09116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamprecht R, Dudai Y. Transient expression of c-Fos in rat amygdala during training is required for encoding conditioned taste aversion memory. Learn Mem. 1996;3:31–41. doi: 10.1101/lm.3.1.31. [DOI] [PubMed] [Google Scholar]
- 40.Mileusnic R, Anokhin K, Rose SP. Antisense oligodeoxynucleotides to c-fos are amnestic for passive avoidance in the chick. Neuroreport. 1996;7:1269–1272. doi: 10.1097/00001756-199605170-00010. [DOI] [PubMed] [Google Scholar]
- 41.Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. J Neurosci. 2004;24:6466–6475. doi: 10.1523/JNEUROSCI.4737-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013;78:773–784. doi: 10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrey G, et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- 44.Chipumuro E, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hnisz D, et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell. 2015;58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinheiro EM, et al. Lpd depletion reveals that SRF specifies radial versus tangential migration of pyramidal neurons. Nat Cell Biol. 2011;13:989–995. doi: 10.1038/ncb2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rexach JE, et al. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat Chem Biol. 2012;8:253–261. doi: 10.1038/nchembio.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flavell SW, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 50.Pereira AH, et al. MEF2C silencing attenuates load-induced left ventricular hypertrophy by modulating mTOR/S6K pathway in mice. PLoS One. 2009;4:e8472. doi: 10.1371/journal.pone.0008472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 52.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.