Significance
RNA-mediated heterochromatin assembly requires transcription by RNA polymerases and a plethora of RNA-processing factors. However, the mechanisms that differentiate productive gene transcription from transcriptional activity linked to the assembly of repressive heterochromatin have remained elusive. Here we find that factors promoting noncanonical termination of RNA polymerase II transcription trigger heterochromatin assembly. We propose that termination and 3′-end processing factors serve as molecular sensors that, in addition to determining the fate of target transcripts, mediate heterochromatin assembly. Importantly, these findings provide insights into the paradoxical interdependence of heterochromatin assembly and transcription of target loci and suggest a mechanism for specifying certain genomic regions, including developmental genes and repeat elements, as targets for heterochromatin assembly.
Keywords: heterochromatin, Dhp1/Xrn2, S. pombe, transcription termination
Abstract
Cotranscriptional RNA processing and surveillance factors mediate heterochromatin formation in diverse eukaryotes. In fission yeast, RNAi machinery and RNA elimination factors including the Mtl1–Red1 core and the exosome are involved in facultative heterochromatin assembly; however, the exact mechanisms remain unclear. Here we show that RNA elimination factors cooperate with the conserved exoribonuclease Dhp1/Rat1/Xrn2, which couples pre-mRNA 3′-end processing to transcription termination, to promote premature termination and facultative heterochromatin formation at meiotic genes. We also find that Dhp1 is critical for RNAi-mediated heterochromatin assembly at retroelements and regulated gene loci and facilitates the formation of constitutive heterochromatin at centromeric and mating-type loci. Remarkably, our results reveal that Dhp1 interacts with the Clr4/Suv39h methyltransferase complex and acts directly to nucleate heterochromatin. Our work uncovers a previously unidentified role for 3′-end processing and transcription termination machinery in gene silencing through premature termination and suggests that noncanonical transcription termination by Dhp1 and RNA elimination factors is linked to heterochromatin assembly. These findings have important implications for understanding silencing mechanisms targeting genes and repeat elements in higher eukaryotes.
Heterochromatin is a repressive form of chromatin that is critical for fundamental functions of eukaryotic genomes (1). Heterochromatin assembly pathways are highly conserved in eukaryotes and have been particularly well studied in the fission yeast Schizosaccharomyces pombe (1). Heterochromatin formation involves conserved histone-modifying enzymes. In addition to deacetylation of histones by histone deacetylases (HDACs), histone H3 is methylated on lysine 9 (H3K9me) by the Clr4/Suv39h family of methyltransferases (2). Methylated H3K9 is bound by members of the conserved family of HP1 proteins, which in turn associate with diverse effectors implicated in transcriptional and posttranscriptional silencing, proper segregation of chromosomes, and maintenance of genome stability (1, 3).
RNA polymerase II (RNAPII) transcriptional machinery and proteins involved in cotranscriptional processing of RNAs play important roles in targeting histone-modifying activities to nucleate heterochromatin at specific regions of the S. pombe genome (4–6). In particular, RNAi machinery is believed to function in the assembly of major constitutive heterochromatin domains coating centromeres, telomeres, and the mating type (mat) locus (7–9). RNAi-dependent mechanisms also assemble dynamically regulated facultative heterochromatin domains, hereafter referred to as “HOODs”, at discrete sites across the genome, including at developmental genes and retrotransposons (10). HOOD assembly is triggered by the specialized nuclear RNA processing and surveillance complex Mtl1–Red1 core (MTREC) and/or associated mRNA 3′-end processing activities, including the canonical poly(A) polymerase Pla1 and the poly(A)-binding protein Pab2, to direct transcripts into RNA degradation pathways and target heterochromatin assembly (10, 11). Notably, HOODs are detected under certain growth conditions or when the 3′-to-5′ exoribonuclease subunit Rrp6 of the nuclear exosome is compromised (10, 12).
RNA-processing factors such as MTREC also promote RNAi-independent assembly of facultative heterochromatin islands, which are modified in response to nutritional signals such as nitrogen starvation (11, 13–15). Heterochromatin islands preferentially target meiotic genes that are silenced during vegetative growth (15). Meiotic mRNAs that contain a determinant of selective removal (DSR) are recognized by the sequence-specific RNA-binding protein Mmi1 (16). Mmi1 in turn recruits MTREC and the Pir1/Iss10 protein to promote exosome-mediated elimination of transcripts and to recruit the Clr4 methyltransferase for heterochromatin island assembly (11, 13, 15, 17–20). MTREC also localizes to regions of the genome that do not assemble heterochromatin, suggesting that additional factors are needed to direct the formation of heterochromatin islands (11). Indeed, the exact mechanisms responsible for specifying MTREC-mediated heterochromatin assembly have remained unclear.
Here we report that RNA elimination factors promote premature transcription termination at facultative heterochromatin islands. This noncanonical transcription termination by RNA elimination factors requires the conserved nuclear exoribonuclease, Dhp1. The functional connection between Dhp1 and RNA elimination factors extends beyond meiotic heterochromatin islands and also is required for the production of small RNAs and RNAi-mediated assembly of HOODs at genes and retrotransposons. In addition, Dhp1 contributes to silencing and heterochromatin formation at constitutive heterochromatin domains such as at centromeres and the mat region. Our results show that Dhp1 associates with the Clr4 complex. These analyses suggest that the conserved termination factor Dhp1 plays a central role in transcription-dependent heterochromatin assembly pathways.
Results
DSR-Containing Meiotic Genes Are Targeted for Premature Transcription Termination.
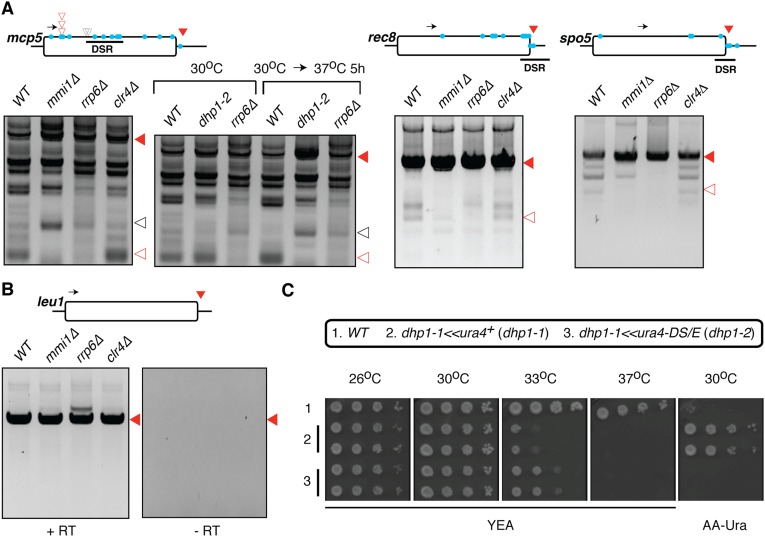
RNA elimination factors required for meiotic gene silencing have been shown to colocalize with 3′-end processing factors (18). To investigate whether meiotic gene silencing involves inappropriate 3′-end formation, we mapped the 3′ ends of ssm4 meiotic gene transcripts in vegetative cells using 3′ RACE. Our analyses of polyadenylated ssm4 transcripts revealed short, unstable RNA species in addition to the full-length transcript (Fig. 1A). Sequencing of the 3′ RACE products revealed that the 3′ ends of these abbreviated transcripts mapped adjacent to Mmi1-binding sites in and around the DSR (denoted by open red and gray triangles in Fig. 1A), indicating internal cleavage events leading to premature transcription termination. Importantly, deletion of the DSR in ssm4 abolished the premature termination that normally occurs upstream of the DSR (Fig. 1B). We also detected premature termination at other DSR-containing genes such as mcp5 and, to a lesser extent, at rec8 and spo5 loci (Fig. S1A). In contrast, the leu1 gene, which lacks a DSR, produced only a full-length transcript (Fig. S1B). These results suggest that DSR-containing meiotic genes are selectively targeted for premature termination.
Fig. 1.
Premature termination and silencing of DSR-containing meiotic gene transcripts requires RNA elimination factors and Dhp1. (A–C) Premature termination of the DSR-containing ssm4 transcript during vegetative growth was determined by 3′ RACE analysis using the indicated strains. Triangles indicate mapped 3′ ends of the RACE products as determined by sequencing: open red triangles, premature termination sites; open gray triangles, other internal cleavage sites; filled red triangles, putative 3′-end site of the full-length transcript. Blue circles indicate predicted Mmi1-binding sequences, and the black arrow indicates the gene-specific 3′ RACE forward oligonucleotide. The black square in B represents the 13xMYC epitope-tag. All positions are drawn to scale. Note that the 3′ RACE oligonucleotide, used for first-strand cDNA synthesis, adds 57 nt to the size of the PCR amplified products. (D) The ssm4 transcript is derepressed in the dhp1-2 mutant but not upon loss of Din1 or Rhn1. Strand-specific RT-PCR of ssm4 transcripts was performed using the indicated mutants. +RT and −RT indicate the presence or absence of reverse transcriptase enzyme in the reaction mixture. act1 mRNA was used as loading control. (E) The dhp1-2 mutation relieves the meiotic arrest in sme2∆ cells. Wild-type, sme2∆, and sme2∆ dhp1-2 strains were grown on Pombe minimal glutamate medium for 3 d, and the sporulation frequency (%) was measured (n > 225). Differential interference contrast (DIC) images are shown. (Scale bar, 10 microns.) (F and G) Dhp1 (F), but not Din1 or Rhn1 (G), promotes heterochromatin assembly at the ssm4 locus. Input and H3K9me2 ChIP DNA from the indicated strains were measured by real-time PCR amplification (qPCR) using gene-specific oligonucleotides, and H3K9me2 ChIP enrichment at ssm4 was calculated relative to the enrichment at act1. Error bars indicate the SD from two independent experiments. Here and in subsequent experiments involving dhp1-2, strains were grown first at 30 °C and then at 37 °C for 5 h.
Fig. S1.
Dhp1 and mRNA elimination factors promote premature termination of DSR-containing meiotic gene transcripts. (A) DSR-containing transcripts are prematurely terminated by factors involved in meiotic gene silencing. 3′ RACE was performed to analyze mcp5, rec8, and spo5 transcripts in wild-type and mmi1∆, rrp6∆, and clr4∆ mutant strains during vegetative growth. 3′ RACE analysis of mcp5 transcripts in wild-type and dhp1-2 mutant strains at the indicated temperature is shown also. Schematics of the mcp5, rec8, and spo5 genes are as described in Fig. 1A. (B) Mmi1 and the nuclear exosome do not prematurely terminate the non-DSR leu1 transcript. 3′ RACE was performed to analyze the leu1 transcript in wild-type, mmi1∆, rrp6∆, and clr4∆ strains during vegetative growth. +RT and −RT indicate the presence or absence of reverse-transcriptase enzyme in the reaction mixture. (C) Growth phenotype of dhp1-2 mutants. Serial dilutions of wild-type, dhp1-1<<ura4+ (dhp1-1), and dhp1-1<<ura4DS/E (dhp1-2) strains were spotted onto YEA medium or on AA medium without uracil. Plates were incubated at the indicated temperature for 2–3 d.
RNA Elimination Factors Promote Premature Transcription Termination.
Given the functional connection between 3′-end processing and degradation factors (18–21), we asked whether the elimination factors Mmi1 and Rrp6 are required for premature termination of DSR-containing gene transcripts. Remarkably, the loss of Mmi1 or Rrp6 prevented premature termination of ssm4 and mcp5 transcripts upstream of the DSR (Fig. 1A and Fig. S1A). Because RNA elimination factors promote the assembly of facultative heterochromatin on meiotic genes (13–15), premature termination could be mediated through heterochromatin formation. Strikingly, we found that the loss of Clr4, the sole H3K9 methyltransferase in S. pombe that is essential for formation of heterochromatin islands (15), did not affect premature transcription termination (Fig. 1A and Fig. S1A). These results reveal that premature termination likely precedes Clr4-mediated heterochromatin assembly and show that RNA elimination factors, which are involved in heterochromatin assembly, are also required for premature termination.
Dhp1 Promotes Premature Termination and Silencing of Meiotic Genes to Prevent Untimely Sexual Differentiation.
We identified Dhp1 as a factor that affects meiotic gene silencing (see below). Dhp1 is a conserved and essential protein related to Saccharomyces cerevisiae Rat1 and human Xrn2 that is implicated in coupling 3′-end processing to transcription termination (22–27). To test whether premature termination at meiotic genes requires Dhp1, we performed 3′ RACE of ssm4 in a mutant carrying the temperature-sensitive hypomorphic allele, dhp1-2 (Materials and Methods and Fig. S1C). Strikingly, at the restrictive temperature the dhp1-2 mutation prevented premature termination upstream of the ssm4 DSR, in a manner similar to mmi1∆ and rrp6∆ mutants (Fig. 1 A and C). We also detected Dhp1-dependent premature transcription termination in the mcp5 gene (Fig. S1A). Thus, premature termination at DSR-containing meiotic transcripts requires Dhp1.
We also observed stabilization of ssm4 meiotic gene transcripts specifically when dhp1-2 mutant cells were cultured at restrictive temperature (Fig. 1D). The accumulation of ssm4 mRNAs in dhp1-2 was comparable to that observed in rrp6∆ (Fig. 1D). In S. cerevisiae, the Dhp1 homolog collaborates with Rai1 and Rtt103 proteins, which are related to Din1 and Rhn1 in S. pombe, respectively (22). Rhn1 has been suggested to suppress the expression of certain meiotic loci (28). We found that, unlike dhp1-2, neither din1∆ nor rhn1∆ caused stabilization of the ssm4 transcript (Fig. 1D) (28). Together, our results clearly show that Dhp1, but not Din1 or Rhn1, is a key conserved factor required for silencing several DSR-containing meiotic transcripts.
To determine whether Dhp1-mediated silencing of meiotic genes is physiologically important, we used a read-out assay based on the rescue of the meiotic arrest caused by the deletion of sme2; sme2∆ zygotes arrest in meiotic prophase because of the retention of Mmi1 activity, which consequently prevents activation of the genes required for the completion of meiosis. A general up-regulation of meiotic gene expression can suppress the sme2∆ meiotic arrest, as has been observed in MTREC and exosome mutants (16, 17). Remarkably, we observed a significant amount of sporulation in dhp1-2 sme2∆ zygotes (40%) relative to the control (0%) (Fig. 1E), indicating a strong up-regulation of Mmi1-targeted genes in dhp1-2 cells. These results suggest an important role for Dhp1 in silencing meiotic loci to prevent untimely sexual differentiation.
Dhp1 Affects the Assembly of Heterochromatin Islands at Meiotic Genes.
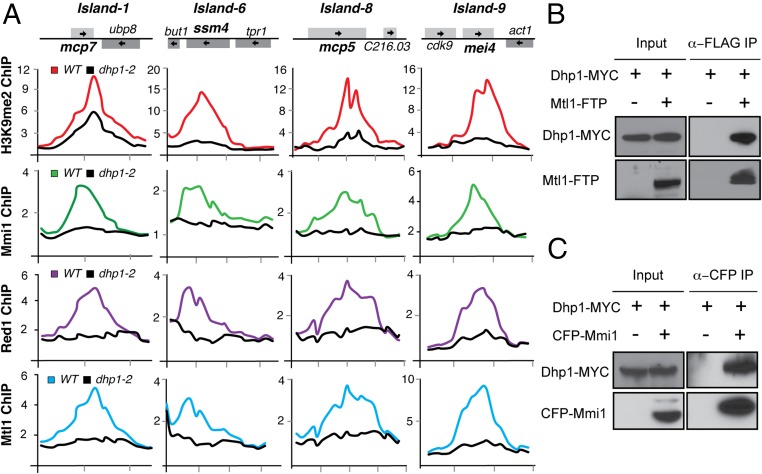
We wondered whether Dhp1, like RNA elimination factors, is also required for heterochromatin formation. Remarkably, ChIP analyses revealed that H3K9me levels were significantly reduced at the ssm4 locus in the dhp1-2 mutant (Fig. 1F). This effect was specific to the dhp1-2 mutant, because neither din1∆ nor rhn1∆ affected H3K9me levels (Fig. 1G). Moreover, we found that Dhp1 also is required for the assembly of other Mmi1- and exosome-dependent heterochromatin islands on meiotic genes akin to ssm4 (Fig. 2A). Together, these results clearly show that Dhp1 along with RNA elimination factors promotes premature transcription termination and also is required for heterochromatin assembly at meiotic gene loci during vegetative growth.
Fig. 2.
Dhp1 is required for the localization of mRNA elimination factors to heterochromatin islands containing meiotic genes. (A) Dhp1 affects the localization of mRNA elimination factors that are required for the assembly of heterochromatin islands. H3K9me2, Mmi1, Red1, and Mtl1 enrichments at the indicated heterochromatin islands in wild-type and dhp1-2 were determined by ChIP-chip analysis. The relative fold-enrichment of ChIP DNA (y axis) is plotted at the indicated chromosome position (x axis). (B) Coimmunoprecipitation analysis of Dhp1 and Mtl1 interaction. Flag-TEV-protein A (FTP) tagged Mtl1 was immunopurified using anti-FLAG M2 magnetic beads, and Dhp1-MYC was detected using anti-MYC antibody. (C) Coimmunoprecipitation analysis of Dhp1 and Mmi1 interaction. CFP-Mmi1 was immunopurified using anti-GFP agarose beads, and Dhp1-MYC was detected using anti-MYC antibody. IP, immunoprecipitation.
Dhp1 Associates with RNA Elimination Factors.
Because Dhp1 and RNA elimination machinery are both functionally required for premature transcription termination and for heterochromatin assembly, we wondered whether Dhp1 and RNA elimination machinery are physically interconnected. We noticed that a previous purification of the MTREC subunit Mtl1 detected several peptides corresponding to Dhp1 (29). We therefore asked whether Dhp1 interacts with factors involved in RNA elimination. Indeed, we found that Dhp1 coimmunoprecipitates with Mtl1, even with Benzonase treatment, indicating that the interactions with Dhp1 are not mediated by DNA or RNA (Fig. 2B and Fig. S2). We also detected an interaction between Dhp1 and Mmi1 (Fig. 2C). The interaction of Dhp1 with Mmi1 and the associated MTREC complex suggests that Dhp1 is part of a larger protein network that targets meiotic genes for premature transcription termination and heterochromatin-mediated silencing. Supporting this idea, we found that dhp1-2 showed severe defects in the localization of RNA elimination factors, including Mmi1, Red1, and Mtl1, at heterochromatin islands (Fig. 2A).
Fig. S2.
Dhp1 coimmunoprecipitates with Mtl1. Mtl1-FTP from cell lysates was immunopurified using anti-FLAG antibody-coupled magnetic beads and treated with or without Benzonase. Dhp1-MYC was detected using anti-MYC antibody.
RNAi-Dependent Heterochromatin Assembly at Genes and Retrotransposons Requires Dhp1.
MTREC along with associated RNA-processing factors also has been implicated in RNAi-dependent assembly of heterochromatin domains on developmental genes and retrotransposons (10, 11). To determine whether Dhp1 cooperates with MTREC to assemble RNAi-dependent HOODs, we introduced dhp1-2 into a strain carrying rrp6∆ that allows detection of HOODs under standard growth conditions (10). Remarkably, ChIP analysis of H3K9me distribution in the rrp6∆ dhp1-2 double mutant revealed defects in HOODs at several loci, including sexual differentiation genes such as myp2 and spcc1442.04c as well as Tf2 retrotransposons (Fig. 3A, Fig. S3, and Table S1).
Fig. 3.
Dhp1 is required for heterochromatin assembly and gene silencing at HOODs. (A) Dhp1 is crucial for small RNA production and heterochromatin assembly at HOODs. H3K9me2 distribution at the indicated HOODs in wild-type, rrp6∆, and rrp6∆ dhp1-2 strains was determined by ChIP-chip analysis. Normalized numbers of small RNA-sequencing (RNA-seq) reads mapping to the top and bottom DNA strands are represented by the signal above and below the line, respectively, and are plotted in alignment with HOOD loci. (B) Dhp1 is required for efficient silencing of target gene transcripts. The steady-state level of myp2 and spcc1442.04c transcripts from the indicated strains was determined by Northern blot analysis. Ethidium bromide (EtBr) staining of ribosomal RNA was used as a loading control. (C) Dhp1 promotes premature termination of a cryptic intron-containing meiotic gene transcript. The schematic is as described in Fig. 1 A–C. Introns 1 and 2 in spcc1442.04c are shown in orange.
Fig. S3.
Dhp1 affects heterochromatin formation and small RNA production at retrotransposons and developmental genes. (A) Dhp1 promotes RNAi-mediated heterochromatin silencing at retrotransposons. ChIP-chip analysis of H3K9me2 distribution is shown for HOOD-4 and HOOD-10 in wild-type, rrp6∆, and rrp6∆ dhp1-2 strains. Normalized numbers of small RNA-seq reads from retrotransposons are shown. (B) Dhp1 promotes assembly of HOODs. H3K9me2 ChIP enrichment relative to the act1 locus was calculated. Error bars indicate the SD from two independent experiments.
Table S1.
Dhp1 facilitates small RNA production and H3K9me2 at genes and retrotransposons
| Genomic regions | Location | Genomic features | Small RNAs | H3K9me2 | ||||
| Chromosome | Start | End | rrp6∆ | rrp6∆ dhp1-2 | rrp6∆ | rrp6∆ dhp1-2 | ||
| Subtelomeric regions | 1 | 1 | 9,289 | tlh1 | +++ | ++++ | +++ | +++ |
| 1 | 17,000 | 19,000 | SPAC212.06c | ++ | ++ | +++ | +++ | |
| 1 | 21,377 | 22,666 | SPAC212.04c meiosis↑ | + | – | +++ | ++ | |
| 1 | 28,579 | 32,648 | SPAC212.01c meiosis↑ | ++ | – | +++ | ++ | |
| SPAC977.01 memb | ||||||||
| 1 | 59,551 | 60,745 | SPAC977.14c memb | + | – | ++ | + | |
| 1 | 5,564,170 | 5,570,038 | SPAC750.04c memb | ++ | – | +++ | ++ | |
| SPAC750.05c meiosis↑, memb | ||||||||
| SPAC750.06c meiosis↑, memb (repeat region 5204, repeat region 5205, repeat region 5206, repeat region 5207) | ||||||||
| 1 | 5,572,541 | 5,574,430 | (Repeat region 5207, repeat region 5212) | + | + | +++ | +++ | |
| 2 | 5,244 | 9,353 | SPBC1348.01 meiosis↑, memb | + | – | +++ | ++ | |
| SPBC1348.02 meiosis↑, memb (repeat region 1a, repeat region 8b) | ||||||||
| 2 | 4,463,576 | 4,466,315 | SPBPB2B2.03c memb | + | – | + | – | |
| SPBPB2B2.04 meiosis↑ | ||||||||
| 2 | 4,501,916 | 4,507,716 | SPBPB2B2.17c memb | + | – | +++ | ++ | |
| SPBPB2B2.18 | ||||||||
| SPBPB2B2.19c meiosis↑, memb | ||||||||
| SPBCPT2R1.01 meiosis↑, memb (repeat region 4340, repeat region 4352) | ||||||||
| 2 | 4,512,962 | 4,514,857 | SPBCPT2R1.04c meiosis↑, memb | + | – | NP | NP | |
| (repeat region 4358) | ||||||||
| 2 | 4,515,585 | 4,515,702 | (intergenic region) | + | + | NP | NP | |
| 2 | 4,517,451 | 4,519,112 | SPBCPT2R1.09c | + | + | NP | NP | |
| 2 | 4,523,251 | 4,532,665 | SPBCPT2R1.07c | +++ | ++++ | NP | NP | |
| SPBCPT2R1.10 | ||||||||
| tlh2 | ||||||||
| HOOD-1 | 1 | 1,465,847 | 1,469,848 | tf2-1 | + | – | + | – |
| HOOD-2 | 1 | 1,564,163 | 1,568,414 | tf2-2 | + | – | + | – |
| HOOD-3 | 1 | 2,544,835 | 2,561,773 | isp6 meiosis↑ | ++++ | + | +++ | – |
| myp2 meiosis↑ | ||||||||
| SPAC4A8.06c | ||||||||
| SPAC4A8.07c | ||||||||
| vas1 | ||||||||
| HOOD-4 | 1 | 2,927,156 | 2,941,954 | tf2-3 | + | – | + | – |
| tf2-ORF (truncated) | ||||||||
| HOOD-5 | 1 | 2,977,013 | 2,988,899 | mfc1 memb | – | – | + | – |
| SPAPB1A11.02 | ||||||||
| SPAPB1A11.03 | ||||||||
| mca1 meiosis↑ | ||||||||
| HOOD-6 | 1 | 2,994,807 | 3,009,469 | lys12 | + | – | – | – |
| SPAC31G5.05c | ||||||||
| rgg8 | ||||||||
| dni1 meiosis↑, memb | ||||||||
| ups1 | ||||||||
| spk1 meiosis↑ | ||||||||
| eta2 meiosis↑ | ||||||||
| pac2 meiosis↑ | ||||||||
| maf1 | ||||||||
| HOOD-7 | 1 | 3,361,499 | 3,365,727 | tf2-4 | + | – | + | – |
| HOOD-8 | 1 | 3,736,902 | 3,743,575 | SPAP7G5.03 meiosis↑, memb | ++ | + | +* | +* |
| lys1 | ||||||||
| HOOD-9 | 1 | 3,791,825 | 3,796,114 | rad50 | ++ | ++ | ++* | ++* |
| HOOD-10 | 1 | 3,996,353 | 4,000,615 | tf2-5 | + | – | + | – |
| HOOD-11 | 1 | 4,022,326 | 4,0265,45 | tf2-6 | + | – | + | – |
| HOOD-12 | 1 | 5,069,824 | 5,083,205 | SPAC1006.02 meiosis↑ | ++ | – | + | – |
| SPAC1006.03c meiosis↑ | ||||||||
| mcp3 meiosis↑ | ||||||||
| och1 meiosis↑ | ||||||||
| rgf2 meiosis↑ | ||||||||
| HOOD-13 | 1 | 5,191,103 | 5,195,325 | tf2-7 | + | – | + | – |
| HOOD-14 | 1 | 5,195,656 | 5,199,909 | tf2-8 | + | – | + | – |
| HOOD-15 | 1 | 5,234,000 | 5,250,176 | SPAC14C4.04 meiosis↑ | ++++ | ++++ | +++ | ++ |
| man1 meiosis↑, memb | ||||||||
| SPAC14C4.06c | ||||||||
| SPAC14C4.07 memb | ||||||||
| mug5 meiosis↑ | ||||||||
| agn1 meiosis↑ | ||||||||
| SPAC14C4.10c meiosis↑, memb | ||||||||
| SPAC14C4.11 memb | ||||||||
| HOOD-16 | 2 | 91,600 | 101,684 | SPBPB10D8.04c memb | + | + | + | + |
| SPBPB10D8.05c memb | ||||||||
| SPBPB10D8.06c memb | ||||||||
| SPBPB10D8.07c memb | ||||||||
| HOOD-17 | 2 | 347,505 | 354,250 | SPBC1271.09 meiosis↑, memb | + | + | – | – |
| SPBC1271.10c memb (intergenic region) | ||||||||
| HOOD-18 | 2 | 898,107 | 902,233 | mcp5 meiosis↑ | ++ | + | + | – |
| HOOD-19 | 2 | 1,812,684 | 1,816,937 | tf2-9 | + | – | + | – |
| HOOD-20 | 2 | 1,965,175 | 1,969,519 | tf2-10 pseudo | + | – | + | – |
| HOOD-21 | 2 | 2,126,590 | 2,128,479 | SPBC23G7.14 meiosis↑ | + | – | +† | +† |
| rpp202 | ||||||||
| HOOD-22 | 2 | 4,414,469 | 4,418,768 | tf2-11 | + | – | + | – |
| HOOD-23 | 2 | 4,442,538 | 4,449,562 | SPBC8E4.02c meiosis↑ | ++++ | + | ++ | – |
| SPBC8E4.01c memb | ||||||||
| pho1 meiosis↑ | ||||||||
| HOOD-24 | 3 | 173,841 | 1,76,400 | SPCC1235.01 (internal repeats) | +++ | + | + | – |
| HOOD-25 | 3 | 254,411 | 256,353 | tf2-ORF (truncated) | + | – | + | – |
| HOOD-26 | 3 | 778,123 | 782,331 | tf2-12 | + | – | + | – |
| HOOD-27 | 3 | 1,047,657 | 1,056,145 | SPCC1259.08 meiosis↑ | + | + | +* | +* |
| SPCC1259.09c | ||||||||
| pgp1 | ||||||||
| gyp2 | ||||||||
| HOOD-28 | 3 | 1,168,500 | 1,176,000 | nte1 | ++ | + | +* | +* |
| SPCC4B3.03c meiosis↑, memb | ||||||||
| SPNCRNA.120 | ||||||||
| HOOD-29 | 3 | 1,179,500 | 1,182,650 | rhp26 | + | + | +* | +* |
| HOOD-30 | 3 | 1,196,050 | 1,196,500 | tf2-ORF (truncated) | + | – | + | – |
| HOOD-31 | 3 | 1,763,512 | 1,775,613 | SPCC1450.16c memb | ++++ | – | +++ | – |
| ste6 meiosis↑ | ||||||||
| SPCC1442.02 meiosis↑ | ||||||||
| SPCC1442.03 | ||||||||
| SPCC1442.04c meiosis↑ | ||||||||
| HOOD-32 | 3 | 2,320,230 | 2,324,503 | tf2-13-pseudo | + | – | + | – |
Meiosis↑, genes up-regulated during mating or meiosis; memb, genes encoding transmembrane domain proteins; NP, no probe on microarray; Tf2, considering high similarity among tf2 ORFs, it was not possible to define the exact origin of the signal; –, not detected or major reduction; +, low; ++, medium; +++, high; ++++, very high.
Near centromeric region.
Near mat locus.
We then examined small RNA production at HOODs in the dhp1-2 mutant. High-throughput sequencing analyses showed that the production of small RNAs mapping to HOODs was severely reduced in the dhp1-2 mutant (Fig. 3A, Fig. S3A, and Table S1). We also tested the effect of dhp1-2 on the silencing of loci within HOODs that require both RNAi and the exosome for efficient silencing (10). Consistent with the reduction in H3K9me and siRNA levels, we found that rrp6∆ dhp1-2 cells were severely defective in the silencing of genes within HOODs (Fig. 3B). Moreover, the up-regulation of transcripts observed in rrp6∆ dhp1-2 was comparable to that in rrp6∆ ago1∆ (Fig. 3B). Together, these results show that Dhp1 is required for RNAi-dependent heterochromatin assembly and silencing of loci within HOODs.
Dhp1 Promotes Premature Transcription Termination at a Gene Containing Cryptic Introns.
Because we found that Dhp1 along with RNA elimination machinery prematurely terminated meiotic mRNAs at heterochromatin islands, we wondered whether similar effects are observed at HOODs. To address this question, we performed 3′ RACE of the spcc1442.04c transcript, which contains cryptic introns that promote RNAi-mediated generation of small RNAs and heterochromatin assembly (11). We observed a premature transcription termination product in the wild-type strain; however, in the dhp1-2 mutant longer intermediate and full-length transcribed products were evident, indicating that Dhp1 is required for premature termination at HOODs, as it is at island loci (Fig. 3C). We also observed an accumulation of prematurely terminated transcripts in the absence of exosome activity, correlating with the H3K9me enrichment in rrp6∆ cells (Fig. 3C). Premature transcription termination again was dependent on Dhp1, as confirmed by a corresponding decrease in abbreviated transcripts in the rrp6∆ dhp1-2 double mutant (Fig. 3C). Taken together, these results indicate that Dhp1 promotes the premature termination of transcripts containing cryptic introns and is critical for channeling these RNAs into the RNAi pathway for small RNA production and heterochromatin assembly at developmental genes.
Heterochromatic Silencing at Centromeres Requires Dhp1.
We found that Dhp1 acts together with MTREC to promote RNAi-independent and -dependent facultative heterochromatin domains at various loci. However, MTREC and its associated factors are dispensable for the production of small RNAs and heterochromatin assembly at centromeres (11). We wondered whether Dhp1 also is dispensable for RNAi-dependent heterochromatin assembly at centromeres. To test this possibility, we examined the generation of small RNAs produced from dg/dh repeats at pericentromeric regions in the wild-type and dhp1-2 strains. We found that the level of small RNAs was higher in dhp1-2 than in wild-type cells (Fig. 4A), suggesting that small RNA production at centromeres can be triggered by other mechanisms (30).
Fig. 4.
Dhp1 promotes heterochromatin silencing at centromeres. (A, Upper) Schematic of the pericentromeric region showing the dg/dh inverted repeats and a ura4+ (otr1R::ura4+; red filled rectangle) reporter gene inserted within the outer centromeric repeat. (Lower) Northern blot analysis of dg small RNA levels in the indicated strains. (B) Northern blot analysis of dg and dh transcript levels in the indicated strains. Note the cumulative derepression in the ago1∆ dhp1-2 double mutant. (C) H3K9me2 enrichment at dg, otr1R::ura4+ loci in the indicated strains. Note that cumulative loss of pericentromeric heterochromatin occurs in the ago1∆ dhp1-2 double mutant. ChIP-qPCR and enrichment analysis at dg and otr1R::ura4+ relative to the act1 locus is as described in Fig. 1 F and G. Error bars indicate the SD from at least two independent experiments.
The increased production of siRNAs has been observed previously in mutants defective in either transcriptional silencing (clr3 HDAC mutant) or degradation (rrp6 exosome mutant) of transcripts in which RNAi processes extra primary dg/dh centromeric repeat transcripts into small RNAs (30, 31). Because we observed elevated levels of siRNAs in dhp1-2, we wondered whether mutant cells are defective in silencing. We observed that dhp1-2 alone caused little or no change in levels of dg/dh transcripts (Fig. 4B). However, we observed a cumulative increase in dg/dh transcript levels when dhp1-2 was combined with ago1∆ (Fig. 4B). Together with results showing elevated levels of dg/dh small RNAs in dhp1-2 (Fig. 4A), these data suggest that the centromeric repeats are derepressed in dhp1-2, but degradation by RNAi prevents transcript accumulation.
We then asked whether Dhp1 contributes to heterochromatin formation at centromeres. ChIP analyses showed that the levels of H3K9me at native centromeric repeats or at ura4+ inserted at a centromere were partially reduced in dhp1-2 relative to wild type (Fig. 4C). The reduction in H3K9me levels at centromeres in dhp1-2 was confirmed by ChIP-chip, which also revealed a reduction at subtelomere regions (Fig. S4 A and B). This result indicates that Dhp1 plays a more widespread role not only in facultative heterochromatin formation but also in the assembly of constitutive heterochromatin domains.
Fig. S4.
Dhp1 is required for heterochromatin assembly at centromeres and subtelomeres. (A) H3K9me2 enrichment at the centromere of chromosome 1 (cen1) in the wild-type and dhp1-2 strains was determined by ChIP-chip analysis. The relative fold-enrichment of ChIP DNA over the input (y axis) is plotted at the indicated chromosome position (x axis). (B) H3K9me2 enrichment at the left (tel1L) and right (tel1R) telomere ends of chromosome 1 in the wild-type and dhp1-2 strains was determined by ChIP-chip analysis. The relative fold-enrichment of ChIP DNA over the input (y axis) is plotted at the indicated chromosome position (x axis). (C) Dhp1 is epistatic to the nuclear exosome in pericentromeric heterochromatin assembly. H3K9me2 ChIP enrichment at the dg locus was determined relative to the act1 locus in the indicated strains by ChIP-qPCR. Error bars indicate the SD from two independent experiments.
We next explored the relationship between Dhp1 and RNAi at centromeres by comparing H3K9me levels in single and double mutants. In contrast to the partial effect observed in dhp1-2, severe reduction of H3K9me was observed in ago1∆ dhp1-2 (Fig. 4C). Because our previous work showed that rrp6∆ combined with ago1∆ causes a cumulative loss of heterochromatin at centromeres (32), we also assayed H3K9me levels in dhp1-2 rrp6∆. However, no major reduction in H3K9me was observed in the double mutant compared with either the rrp6∆ or dhp1-2 single mutant (Fig. S4C). Together, these results highlight the fact that Dhp1 does indeed affect heterochromatin assembly and silencing at centromeres, but its effects are masked by alternative mechanisms that trigger RNAi.
Dhp1 Is Required for the Restoration of Constitutive Heterochromatin in RNAi Mutants.
A transcription-dependent but RNAi-independent mechanism of heterochromatin assembly has been shown at centromeres (32). Deletion of factors such as Tfs1 encoding TFIIS, which promotes RNAPII processivity, can restore heterochromatin-mediated silencing in RNAi mutants (32), although restoration factors have not been identified. We found that the dhp1-2 mutation inhibited the restoration of silencing of the otr1R::ura4+ reporter gene in ago1∆ tfs1∆ (Fig. S5A). Although the loss of Tfs1 in the ago1∆ mutant restored heterochromatic silencing at pericentromeres, as previously reported (32), the dhp1-2 mutation combined with tfs1∆ and ago1∆ affected the formation of functional heterochromatin as indicated by defective silencing, increased thiabendazole (TBZ) sensitivity, and the drastic reduction in H3K9me at dg repeats (Fig. S5B). Together, these results underscore the functionally important role of Dhp1 in RNAi-independent heterochromatin assembly at centromeres.
Fig. S5.
Dhp1 promotes RNAi-independent heterochromatin formation. (A) H3K9me2 ChIP enrichment at the otr1R::ura4+ locus relative to the enrichment at the leu1 locus was determined by a competitive PCR method. (B) Dhp1 is required to maintain functional heterochromatin in the absence of RNAi. Serial dilution was used to determine ura4+ expression and TBZ sensitivity. The indicated strains were spotted onto medium with or without 5-FOA to counterselect for ura4+ expression. Strains also were spotted on YEA medium in the presence or absence of TBZ. Defective pericentromeric heterochromatin typically is associated with TBZ sensitivity.
Dhp1 and RNAi Function in Parallel to Assemble Heterochromatin at the mat Locus.
We further investigated whether Dhp1 affects heterochromatin assembly at the silent mat region. Heterochromatin formation at the mat locus involves redundant pathways (33, 34), which mask the effect of individual factors in this process. However, previous studies have described sensitized genetic backgrounds that can be used to study the effects of trans-acting factors on heterochromatin assembly. In particular, in a strain carrying a deletion of a local silencer adjacent to the mat2 locus, defects in heterochromatin formation cause derepression of the silent mat2P cassette and the ura4+ reporter (mat2P::ura4) located adjacent to this locus. Although ura4+derepression can be measured by plating cells on medium lacking uracil or on counterselective 5-fluoroorotic acid (FOA) medium, defects in mat2P silencing in nonswitching mat1M cells causes simultaneous expression of M and P mating-type information, resulting in haploid meiosis (34). Haploid meiosis can be detected by exposing colonies to iodine vapors, which stain the starch-like compound in cells undergoing haploid meiosis.
In this sensitized strain background, we found that dhp1-2 cells are defective in silencing at the mat locus. Moreover, ago1∆ dhp1-2 showed cumulative defects in the silencing of mat2P::ura4+ (Fig. 5 A and B). The double mutant also showed elevated levels of haploid meiosis. The dhp1-2 and ago1∆ single mutants stained yellow with exposure to iodine vapors, indicating little or no haploid meiosis, but the double mutant stained dark, indicating that haploid meiosis was induced as a result of a silencing defect (Fig. 5C). Microscopy analysis confirmed haploid meiosis in ago1∆ dhp1-2 (Fig. 5C). Consistently, ChIP analysis revealed a considerable reduction in H3K9me at the mat locus, and the loss of heterochromatin was even more severe in the ago1∆ dhp1-2 double mutant (Fig. 5D). The defects in heterochromatin formation and silencing also were detected in cells containing a ura4+ reporter (Kint2::ura4+) inserted at the normally silent mat2/3 interval (Fig. S6 A and B). Taken together, these results strongly suggest that Dhp1 plays an important role in heterochromatic silencing at the mat locus.
Fig. 5.
Dhp1 acts in parallel to RNAi for heterochromatin assembly at the silent mat locus. (A) Dhp1 affects heterochromatin silencing at the mat locus. A serial dilution assay was used to measure expression of the ura4+ reporter (mat2P::ura4+) inserted within the silent mat locus. The indicated strains were spotted onto medium with (nonselective; N/S) or without (selective; −URA) uracil or with counterselective medium containing FOA. (B) Dhp1 and RNAi simultaneously suppress a reporter gene in the silent mat locus. Strand-specific RT-qPCR amplification of ura4+ inserted within the silent mat locus (mat2P::ura4+) was performed using the indicated strains. The relative fold-change (ura4+/act1+) in RNA expression was calculated. Error bars indicate the SD from two independent experiments. (C) Dhp1 and RNAi are required to prevent haploid meiosis. Strains were grown and stained with iodine vapors. Dark iodine staining indicates haploid meiosis and reflects mat2P derepression. DIC images and the percentage of azygotic spores formed by the indicated mat1Msmt0 haploid strains are shown also (n > 492). (Scale bar, 10 microns.) The clr4∆ strain is shown as a control. (D) Dhp1 acts parallel to RNAi for heterochromatin assembly at the mat locus. Note that cumulative loss of pericentromeric heterochromatin occurs in the ago1∆ dhp1-2 double mutant. ChIP-qPCR and enrichment analysis at mat2P::ura4+ locus relative to act1 locus are as described in Fig. 1 F and G.
Fig. S6.
Dhp1 and RNAi act in parallel to mediate silencing of reporter genes inserted at the silent mat locus. (A) Dhp1 and RNAi are required for silencing at the mat locus. Serial dilution was used to measure the expression of the ura4+ reporter (Kint2::ura4+) inserted within the silent mat locus. The indicated strains were spotted onto medium without (nonselective; N/S) or with (+FOA) the counterselective medium containing 5-FOA. clr4∆ is shown as a control. (B) Dhp1 and RNAi act in parallel at the silent mat locus. RT-qPCR was used to measure transcript levels of ura4+ inserted within the silent mat locus (Kint2::ura4+). The relative fold-change (ura4+/act1+) in RNA levels was calculated. Error bars indicate the SD from two independent experiments.
Dhp1 Interacts with the Clr4 Methyltransferase Complex.
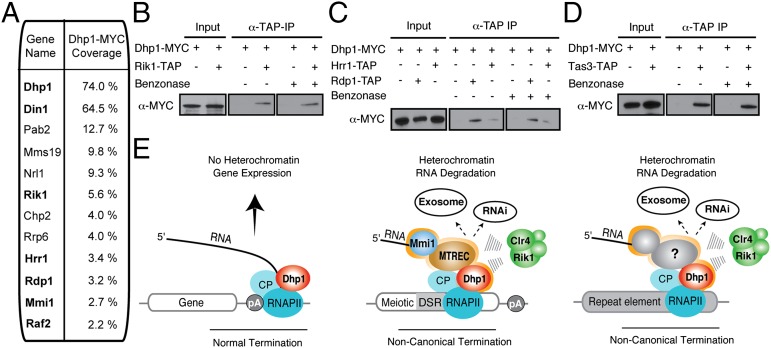
Our results indicate that Dhp1 is a central player in the assembly of all known transcription-dependent heterochromatin domains. To identify additional Dhp1-binding partners, we expressed Dhp1, which was tagged with a MYC epitope at its carboxyl terminus, under its native promoter. Mass spectrometry analysis specifically identified Dhp1, as well as the conserved Dhp1-interacting protein Din1, Mmi1, and many other heterochromatin assembly factors, in purified fractions from cells expressing tagged, but not untagged, Dhp1 (Fig. 6A and Fig. S7). In particular, we identified peptides derived from components of the Clr4-methyl transferase complex (ClrC), including Rik1 and Raf2, and confirmed the interactions (Fig. 6B and Fig. S8). We also detected other factors, including Rdp1 and Hrr1 of the RNA-dependent RNA polymerase complex (RDRC), Tas3 of the RNA-induced transcriptional silencing (RITS) complex, and Mms19, a transcription regulator (Fig. 6 A–D and Fig. S8) implicated in heterochromatin assembly (35, 36). These exciting findings indicate that, in addition to collaborating with RNA processing factors such as MTREC, a small fraction of Dhp1 likely participates in heterochromatin assembly directly through its association with the ClrC and RNAi factors.
Fig. 6.
Dhp1 associates with ClrC components to facilitate heterochromatin assembly directly. (A) Dhp1-MYC copurified with components of the core Clr4 methyl transferase complex. Relevant proteins associating with Dhp1 are shown. Proteins with a putative association or that have been verified by coimmunoprecipitation analysis are highlighted in bold. (B–D) Dhp1 interacts with components of the ClrC (Rik1), RDRC (Hrr1 and Rdp1), and RITS (Tas3) complexes. The indicated tandem affinity purification (TAP)-tagged proteins were immunopurified with or without Benzonase treatment, and Dhp1 was detected using anti-MYC antibody. (E) Model for Dhp1-mediated silencing and heterochromatin assembly. Dhp1 promotes normal transcriptional termination at actively expressed genes and does not trigger heterochromatin formation. At sites of facultative heterochromatin formation, Dhp1 collaborates with RNA-binding factors (e.g., Mmi1) and processing factors including MTREC to promote noncanonical termination, degradation of transcripts by the exosome and RNAi, and loading of Clr4 methyltransferase complex to promote heterochromatin assembly. In addition, Dhp1 also plays a role in constitutive heterochromatin assembly at repeat elements that does not require MTREC but likely involves additional factors.
Fig. S7.
Expression and affinity purification of MYC-tagged Dhp1. Extracts from an untagged and a MYC-tagged Dhp1 (Dhp1-MYC) strain were analyzed by SYPRO Ruby staining (Left) and by Western blotting (Right).
Fig. S8.
Dhp1 coimmunoprecipitates with Rik1 and Raf2. Rik1-TAP or Raf2-TAP from cell lysates was immunopurified, and Dhp1 was detected using anti-Dhp1 antibody (SI Materials and Methods).
Discussion
Dhp1 Function in Developmental Gene Control.
Eukaryotic cells use diverse, often overlapping, strategies to prevent inappropriate expression of genes (37–39). In addition to their role in transcriptional control, RNA surveillance factors closely monitor the transcriptome. In S. pombe, RNA elimination factors selectively degrade meiotic mRNAs in vegetative cells (11, 14–16, 40). In this study, we uncover a mode of gene silencing that requires close cooperation between RNA degradation and transcription termination factors. We demonstrate that, in addition to its canonical role in transcription termination, the essential 3′-end processing and termination factor Dhp1/Rat1/Xrn2 plays an important role in premature termination to promote silencing of meiotic genes and prevent untimely activation of the meiotic pathway. Interestingly, this function of Dhp1 is independent of its putative cofactors, Din1 and Rhn1, but requires RNA degradation factors including the nuclear exosome. Our analyses show that Dhp1 associates with RNA elimination factors, including MTREC, which are involved in the degradation of meiotic mRNAs by the exosome. In light of emerging evidence showing that the nuclear exosome is required for transcription termination at certain loci (12, 41–43) and that human Xrn2 is involved in nuclear mRNA decay (44, 45), we envision that Dhp1 acts together with elimination factors (e.g., Mmi1 and MTREC) and the exosome as part of a specialized protein assembly that couples premature transcription termination to degradation and silencing of meiotic mRNAs. This process might require the catalytic 5′–3′ exoribonuclease activity of Dhp1, in a manner similar to its role in termination of RNAPII transcripts at normal termination sites. However, another possibility is that Dhp1 serves as a scaffold for loading and/or stabilization of elimination factors that process meiotic mRNAs. Indeed, we find that mutant Dhp1 affects the localization of MTREC at meiotic genes. Regardless of the mechanism, this study implicates Dhp1 in the functional silencing of meiotic genes.
Dhp1 is also important for RNAi-mediated silencing of genes. Previously, we showed that Rrp6 and RNAi act in parallel to degrade transcripts containing cryptic introns via a mechanism that involves MTREC and its cofactors Pla1 and Pab2 (10, 11). In this study, we find that Dhp1 affects premature termination at a gene containing cryptic introns and is critical for processing transcripts into small RNAs. Consistent with its role in RNAi-mediated gene silencing, the rrp6∆ dhp1-2 double mutant showed cumulative defects in gene silencing in a manner similar to rrp6∆ ago1∆. These results highlighting the role of Dhp1 in small RNA production and gene silencing are highly relevant to gene-regulation studies in higher eukaryotes. In human cells, transcripts with defective splicing or impaired 3′-end formation are also prematurely terminated and cotranscriptionally degraded by the Dhp1 human homolog, Xrn2 (44). Given that splicing mutants affect the generation of small RNAs at loci containing cryptic introns in S. pombe (11, 46) and that splicing is coupled to 3′-end formation in many cases (47–51), it is possible that the stalled spliceosome engages Dhp1/Xrn2 to trigger transcription termination and RNA degradation (44, 52). This Dhp1-mediated early termination of cryptic intron-containing transcripts with its associated cotranscriptional degradation activity perhaps provides an early and effective way to suppress aberrant or untimely gene expression and supports the emerging view that the fate of the transcripts is determined during 3′-end formation (53).
Dhp1 and Heterochromatin Assembly.
RNA elimination machinery that participates in premature termination of meiotic gene transcripts also assembles heterochromatin at target gene loci (this study and ref. 15). We have previously reported that MTREC operates with both the nuclear exosome and RNAi and promotes the assembly of meiotic heterochromatin islands and HOODs (10, 11, 15). Here we show that Dhp1 also is required for assembly of meiotic islands and RNAi-dependent HOODs. Given the role of Dhp1 in transcription elongation (54), it is possible that Dhp1 might regulate the rate of RNAPII elongation and indirectly affect heterochromatin assembly. However, our results showing that Dhp1 interacts with ClrC components and RNAi proteins, in addition to MTREC, suggest a more direct role for Dhp1 in facultative heterochromatin assembly.
Although Dhp1 and MTREC mediate RNAi-mediated silencing at HOODs, both are dispensable for RNAi-mediated silencing at centromeres. Our analyses suggest that Dhp1 and RNAi act in parallel to assemble heterochromatin both at centromeres and at the mat locus. This result is reminiscent of previous work showing overlapping functions of the exosome and RNAi in centromeric heterochromatin assembly (32). Considering that Dhp1 and the exosome cooperate to assemble meiotic heterochromatin islands, it is conceivable that these factors also act together to assemble RNAi-independent heterochromatin at centromeres. Consistent with this possibility, we find that Dhp1 and the exosome have an epistatic relationship in constitutive heterochromatin formation. Together with data showing that Dhp1 is required for the restoration of centromeric heterochromatin in RNAi mutants, these observations suggest an RNAi-independent function for Dhp1 at constitutive heterochromatin loci. Nevertheless, we cannot rule out the possibility that other factors mask the effects of Dhp1 in the generation of centromeric siRNAs. Previous studies have implicated the Trf4/Air2/Mtr4p polyadenylation (TRAMP) complex in the production of centromeric siRNAs (30, 55), and another termination factor, Seb1 (Nrd1 in S. cerevisiae), affects heterochromatin assembly (56, 57).
How does Dhp1 affect MTREC- and RNAi-independent heterochromatin formation? Dhp1 may be a component of a specialized 3′ pre-mRNA processing and termination complex that directly engages the ClrC complex. In this regard, we note that the Rik1 subunit of ClrC shares sequence similarity with the cleavage and polyadenylation specificity factor (CPSF)-A protein, which functions in 3′-end processing (58). The significance of Rik1 homology to a 3′-end processing factor has remained a long-standing mystery. Given that Rik1 acts upstream of other ClrC components (59), the association of Dhp1 with Rik1 suggests a critical role for 3′-end processing machinery in the recruitment of Clr4. Indeed, Rik1 and Clr4 act at the 3′ end of convergent genes to suppress read-through transcription (60, 61). It is noteworthy, in this regard, that RNAi also has been implicated in transcription termination (62, 63).
In sum, this study reveals previously unrecognized direct cross-talk between 3′-end processing and heterochromatin machinery that hinges on the transcription termination factor Dhp1 (Fig. 6E). Dhp1 terminates RNAPII transcripts that do not target heterochromatin assembly, a function shown to require Dhp1 catalytic activity (22). Given the strong correlation between premature termination and heterochromatin assembly, it is possible that, in addition to the direct recruitment of assembly factors, the catalytic activity of Dhp1 also could be important in heterochromatic silencing. Current evidence suggests that Dhp1 forms a critical component of specialized machinery required for noncanonical termination and heterochromatin assembly (Fig. 6E). We envision that, in addition to determining the fate of coding and noncoding RNAs, 3′-end processing and termination factors serve to specify certain genomic regions including retroelements and repeat loci as preferential targets of heterochromatin assembly. Given that 3′-end processing factors leading to transcription termination also play a crucial role in the regulation of gene expression in Arabidopsis, Caenorhabditis elegans, and mammals (42, 44, 63–66), our results have important implications for understanding gene-regulatory mechanisms in metazoans.
Materials and Methods
All yeast strains and oligonucleotides used in this study are listed in Tables S2 and S3, respectively. Strains with gene deletions and strains expressing epitope-tagged proteins were constructed by standard procedures. The temperature-sensitive dhp1-1 (dhp1-1<<ura4+) strain, MP102, was kindly provided by Kazuo Tatebayashi, University of Tokyo, Tokyo. To facilitate the use of ura4+ as a reporter gene in our experiments, a modified version of the dhp1-1 mutant strain, dhp1-2 (dhp1-1<<ura4-DS/E), was generated in this work by converting the linked ura4+ gene into a ura4-DS/E minigene. The dhp1-2 mutant displayed a mild temperature-sensitive phenotype compared with the dhp1-1 mutant (Fig. S1C). In all experiments, unless otherwise noted, the dhp1-2 mutant phenotype was assayed after cells were first grown at 30 °C (permissive temperature) to early logarithmic phase and then were shifted to 37 °C (restrictive temperature) for 5 h. All strains analyzed in comparison with dhp1-2 were also subjected to the same growth conditions. All experimental methods carried out in this work, including 3′ RACE, reverse transcriptase PCR, ChIP, ChIP-chip, coimmunoprecipitation, Western blotting, Northern blotting, and small RNA sequencing, are described in SI Materials and Methods.
Table S2.
Strains used in this study
| Strain | Genotype |
| SPT999A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ |
| SY136 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ mei4∆C-9xmyc<<natN2 mmi1∆::kanMX |
| SPEN297 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ rrp6∆::kanMX |
| SPKZ569 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ clr4∆::kanMX |
| SPR826 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ ssm4-13xmyc::kanMX |
| SPR827 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ ssm4-DSR∆-13xmyc::kanMX |
| SPVC276A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ dhp1-1<<ura4+ |
| SPVC574A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ dhp1-1<<ura4DS/E |
| SPVC346A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ din1∆::kanMX |
| SPVC265A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ rhn1∆::natN2 |
| SPVC582A | h90 leu1-32 his2 otr1R(Sph1)::ura4+ sme2∆::ura4+ dhp1-1<<ura4DS/E |
| SPVC583A | h90 leu1-32 his2 otr1R(Sph1)::ura4+ sme2∆::ura4+ |
| SPVC584A | h90 leu1-32 his2 otr1R(Sph1)::ura4+ dhp1-1<<ura4DS/E |
| SPVC585A | h90 leu1-32 his2 otr1R(Sph1)::ura4+ |
| SPVC476A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ leu<<CFP-Mmi1 |
| SPVC474 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ dhp1-1<<ura4DS/E leu<<CFP-Mmi1 |
| SPF783A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ red1-13xmyc::kanMX |
| SPVC440A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ dhp1-1<<ura4DS/E red1-13xmyc::kanMX |
| SPF876 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ mtl1-13xmyc::kanMX |
| SPVC439A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ dhp1-1<<ura4DS/E mtl1-13xmyc::kanMX |
| SPVC344A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ dhp1-13xmyc::kanMX |
| SPVC601A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ dhp1-13xmyc::kanMX mtl1-FTP::kanMX |
| SPVC382A | mat1Msmt0 leu1-32 his2 rrp6∆::kanMX dhp1-1<<ura4DS/E |
| SPF599 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ rrp6∆::kanMX ago1∆::kanMX |
| SPR190 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ ago1∆::kanMX |
| SPVC462 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ ago1∆::kanMX dhp1-1<<ura4DS/E |
| SPF064 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ tfs1∆::kanMX |
| SPF222A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ ago1∆::kanMX tfs1∆::kanMX |
| SPVC533B | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ ago1∆::kanMX tfs1∆::kanMX dhp1-1<<ura4DS/E |
| SPVC565A | mat1Msmt0 leu1-32 his2 mat2P(∆Bg-Bs)::ura4+ |
| SPDF938 | mat1Msmt0 leu1-32 his2 mat2P(∆Bg-Bs)::ura4+ clr4∆::kanMX |
| SPVC565B | mat1Msmt0 leu1-32 his2 mat2P(∆Bg-Bs)::ura4+ dhp1-1<<ura4DS/E |
| SPVC566 | mat1Msmt0 leu1-32 his2 mat2P(∆Bg-Bs)::ura4+ ago1∆::kanMX |
| SPVC569A | mat1Msmt0 leu1-32 his2 mat2P(∆Bg-Bs)::ura4+ ago1∆::kanMX dhp1-1<<ura4DS/E |
| SPVC569C | mat1Msmt0 leu1-32 his2 mat2P(∆Bg-Bs)::ura4+ ago1∆::kanMX dhp1-1<<ura4DS/E |
| SPVC570A | h90 leu1-32 his2 kint2::ura4+ |
| SPK423 | h90 leu1-32 his2 kint2::ura4+ clr4∆::kanMX |
| SPVC571A | h90 leu1-32 his2 kint2::ura4+ dhp1-1<<ura4DS/E |
| SPVC572A | h90 leu1-32 his2 kint2::ura4+ ago1∆::kanMX |
| SPVC573A | h90 leu1-32 his2 kint2::ura4+ ago1∆::kanMX dhp1-1<<ura4DS/E |
| SPVC589A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ rik1-TAP::kanMX dhp1-13xmyc::kanMX |
| SPVC426B | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ hrr1-TAP::kanMX dhp1-13xmyc::kanMX |
| SPVC407A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ rdp1-TAP::kanMX dhp1-13xmyc::kanMX |
| SPVC406A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ tas3-TAP::kanMX dhp1-13xmyc::kanMX |
| SPR156 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ |
| SPKZ208A | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ rik1-TAP::kanMX |
| SPKZ189 | mat1Msmt0 leu1-32 his2 otr1R(Sph1)::ura4+ raf2-TAP::kanMX |
All strains used in this study contain ade6-210 or the ade6-216 mutation at the native ade6 gene locus. All strains used in this study contain ura4-DS/E or the ura4-D18 mutation at the native ura4 gene locus. N.B.: dhp1-1<<ura4+ is dhp1-1; dhp1-1<<ura4DS/E is dhp1-2.
Table S3.
Oligonucleotides used in this study
| Oligonucleotide | Sequence | Function |
| 3′ RACE | ||
| VC127 | AAGCAGTGGTATCAACGCAGAGTAC(T)30VN | 3′ RACE cDNA primer |
| VC128 | CTAATACGACTCACTATAGGGCAAGCAGTGGTAACAACGCAGAGT | UPM long |
| VC129 | CTAATACGACTCACTATAGGGC | UPM short |
| VC443 | TTCGTCTACTCCCACAAAGACAGCTAC | ssm4 FP |
| VC131 | AGCCAGGACAACTGGACTACACAAG | ssm4 FP |
| VC130 | AAGCAGTGGTAACAACGCAGAGT | 3′ RACE nested RP |
| VC491 | ATTTAGCGGAGGAGGAGGTGAGAATTAC | mcp5 FP |
| VC492 | TGAAAGAGAAGACTAAGGTGCAAGGAC | mcp5 FP |
| VC500 | AATGTTTCGAGCTTTGTTTACGAGGAC | rec8 FP |
| VC501 | GGAATTGACCAAGGGCACTATAA | rec8 FP |
| VC497 | GTTTCTCTCTCGATGCCTTCTT | spo5 FP |
| VC498 | AGAAATGAGTGTCGGTACAACAAAGGAG | spo5 FP |
| VC512 | GAGAAGAAGCGACCTGAGTTAAA | leu1 FP |
| VC513 | GTCGTCCTGAGCAAGGTTTAT | leu1 FP |
| VC531 | CCTGTGGTCAATCACCCTTATAC | spcc1442.04c FP |
| RT-PCR | ||
| VC453 | GTCCTCATCCGTGGAGAATTAG | ssm4 FP |
| VC456 | CAACAGTTGCCTTCTTGTCTTC | ssm4 RP |
| VC467 | AAATCGCAGCGTTGGTTATTG | act1 FP |
| VC470 | GTACGACCAGAGGCATACAAAG | act1 RP |
| qPCR | ||
| VC455 | TGTACCGGGAAGTTTGGATTTA | ssm4 FP |
| VC456 | CAACAGTTGCCTTCTTGTCTTC | ssm4 RP |
| VC467 | AAATCGCAGCGTTGGTTATTG | act1 FP |
| VC468 | TTTGTCCCATACCTACCATAATACC | act1 RP |
| VC519 | GTCAAATGCTAGTGGTCGAGATA | myp2 FP |
| VC520 | CGAGTTGGTGCATAAGACTAGAA | myp2 RP |
| VC517 | CCAAACTCTGTTGTTGCAGAAG | spcc1442.04c FP |
| VC518 | GATTCCTCAAGGTCGTTATCCC | spcc1442.04c FP |
| VC523 | CAGCATCTAATACACTTCCCTATCT | Tf2-3 FP |
| VC524 | GTAGGATTATCGAACCCGACAG | Tf2-3 RP |
| VC525 | AGGGCTGTAAGACAATAGTGAAG | Tf2-5 FP |
| VC526 | AGGTCGGTAGTCGATATACCAT | Tf2-5 RP |
| HDF-325 | AATTGTGGTGGTGTGGTAATAC | dg FP |
| HDF-326 | GGGTTCATCGTTTCCATTCAG | dg RP |
| VC441 | TTGGCTACTGGTTCCTACACAGAG | ura4 FP |
| VC381 | TCGCTACCGCAGTTTACAATCACTTC | ura4 RP |
| Northern blotting | ||
| myp2-fwd | TTTTGATGAACGGACATGGA | myp2 FP |
| myp2-T7-rev | TAATACGACTCACTATAgGGCCTCCAACACAGGATTCGTT | myp2 RP |
| SC1442.04c-fwd | TTGTATTCTCGACGCAACCA | spcc1442.04c FP |
| SC1442.04c-T7-rev | TAATACGACTCACTATAgGGTGGCAAGTTCATCTGACAGC | spcc1442.04c RP |
| dgT7-up-fwd | TAATACGACTCACTATAgGCTGCGGTTCACCCTTAAC | dg FP |
| dg-low-rev | CGGATCTAGCTTCGCCATC | dg RP |
| dhET7-up-fwdA | TAATACGACTCACTATAgGGAAAACACATCGTTGTC | dh FP |
| dhE-rev | GTCGTCTTGTAGCTGCATGTG | dh RP |
FP, forward primer; RP, reverse primer; UPM, universal primer mix.
SI Materials and Methods
Strains, Media, and Growth Conditions.
Strains with gene deletions and strains expressing epitope-tagged proteins were constructed by standard procedures (67). Strains SPR826 and SPR827 used for determining the requirement of the DSR for premature transcription termination in Fig. 1B were originally used and described in ref. 15. sme2∆ strains used in Fig. 1E were derived from the FY13308 strain provided by the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science, and Technology, Japan. SPVC474 and SPVC476A were derived by crossing with a strain expressing CFP-Mmi1 kindly provided by Masayuki Yamamoto, National Institute for Basic Biology, Okazaki, Japan. The allele used for expressing Mtl1-FTP was provided by Tomoyasu Sugiyama (National Cancer Institute, Bethesda). Standard cell culture methods and medium was used for all experiments.
3′ RACE.
Yeast cells grown to midlog phase were harvested, and total cellular RNA was extracted using the MasterPure Yeast RNA Purification Kit (Epicentre). 3′ RACE then was performed using the SMARTer RACE cDNA amplification kit (Clontech) with a few modifications to the manufacturer’s protocol. Briefly, DNase-treated total cellular RNA was used to synthesize cDNA using the 3′-RACE CDS Primer A oligonucleotide and SuperScript III Reverse-Transcriptase (Invitrogen) and was incubated at 55 °C for 30 min. 3′-RACE–ready cDNA then was amplified by touchdown PCR, using a gene-specific oligonucleotide and the universal primer mix, with the Advantage 2 Polymerase mix (Clontech). An additional nested PCR amplification was done to verify the specificity of the prematurely terminated unstable transcripts. 3′ RACE-amplified DNA was gel extracted and cloned into pCR4-TOPO plasmid (Invitrogen). Plasmids with the inserted 3′ RACE-amplified DNA then were sequenced using the flanking M13 oligonucleotides for identification of 3′ end of the transcript. Table S3 lists all oligonucleotides used for 3′ RACE mapping.
RT-PCR.
DNase-treated total cellular RNA was extracted as described above. Strand-specific RT-PCR reactions were performed using the OneStep RT-PCR Kit (Qiagen). Real-time RT-qPCR reactions were performed using the QuantiTect SYBR Green PCR Kit (Qiagen) on cDNA synthesized from DNase-treated total cellular RNA using a gene- and strand-specific oligonucleotide or oligo(dT)20. Table S3 lists all oligonucleotides used for RT-PCR and RT-qPCR.
Growth and Sporulation Assays.
To test ura4+ silencing and TBZ sensitivity, cells were spotted on Pombe minimal glutamate (PMG) plus supplements and yeast extract agar (YEA) media, respectively. 5-FOA (1 mg/mL) and TBZ (10 μg/mL) were used as indicated. Plates were incubated at 33 °C for 3 d unless otherwise stated. To assess sme2Δ rescue and haploid meiosis, cells were spotted in PMG plus supplements at 30 °C, were grown for 3 d, then were stained with iodine, and subsequently were mounted in a 2% agarose pad for live-imaging and sporulation counting. Images were acquired on a Delta Vision Elite microscope (GE Healthcare, Applied Precision) with a 100× 1.4 NA Plan Super Apochromat and 60× 1.42 NA Plan Apochromat oil lens (Olympus) and subsequently were processed with ImageJ (NIH).
ChIP and ChIP-chip.
ChIP and ChIP-chip experiments were performed as previously described (7). Anti-H3K9me2 (Ab1220; Abcam), anti–c-MYC 9E10 (MMS-150R; Covance), anti–c-MYC A-14 (Sc-789; Santa Cruz), or anti-GFP (Ab290; Abcam) antibody was used to immunoprecipitate H3K9me2, Red1- and Mtl1-MYC, and CFP-Mmi1 proteins, respectively. Immunoprecipitated DNA and DNA from whole-cell extracts were analyzed by real-time PCR (ChIP-qPCR) or multiplex PCR or were labeled with Cy5/Cy3 for microarray-based ChIP-chip using a custom 4 × 44K oligonucleotide array (Agilent).
Coimmunoprecipitation and Western Blotting.
Exponentially growing cells from 2L YEA cultures were harvested, washed, and flash-frozen in liquid nitrogen. Cell pellets then were ground with glass beads in Hepes buffer (pH 7.6) containing complete protease inhibitors (Roche) and 1 mM PMSF. Lysate was cleared by centrifugation at 27,143 × g for 1 h, and the supernatant was incubated with antibody-coupled beads [anti-FLAG M2 magnetic beads for immunoprecipitation of Flag-TEV-protein A (FTP) tagged Mtl1, IgG Sepharose 6 Fast Flow (GE) for immunoprecipitation of TAP-tagged proteins, and GFP-Trap A (ChromoTek) for CFP-Mmi1 immunoprecipitation] for 2 h. Beads were washed extensively, and TAP-tagged and CFP-tagged immunoprecipitated proteins were eluted by tobacco etch virus and glycine elution, respectively. For magnetic bead immunoprecipitation, proteins were eluted by incubation at 80 °C for 15 min. For Benzonase treatment, washed beads containing immunoprecipitated proteins were treated with 250 U of Benzonase (Sigma) for 30 min at room temperature. Then beads were washed again, and proteins were precipitated with 10% trichloroacetic acid and were resuspended in NuPAGE sample buffer. Samples were separated on a denaturing 4–12% Bis-Tris Gel (Invitrogen) for either mass spectrometry analysis as described elsewhere (11) or Western blotting. Mass spectrometry analysis was performed by Ming Zhou (Laboratory of Proteomics and Analytical Technologies, Frederick National Laboratory for Cancer Research, Frederick, MD). For Western analyses, protein from the denaturing gel was transferred onto a PVDF membrane (Immobilon-P; Millipore) and probed with anti–c-MYC 9E10 (MMS-150R; Covance), anti–c-MYC A-14 (Sc-789; Santa Cruz), anti-GFP (Ab290; Abcam), or anti-TAP (CAB1001; Pierce) primary antibody. For ECL, anti-rabbit or anti-mouse HRP-conjugated secondary antibody was used as a secondary antibody. FTP- or TAP-tagged proteins in the whole-cell extract were detected using peroxidase anti-peroxidase (PAP) antibody (P1291; Sigma). Anti-Dhp1 antibody (Pierce) was obtained by affinity purification from rabbit anti-Dhp1 antiserum, which was raised against a peptide antigen corresponding to the N terminus of Dhp1.
Small RNA and RNA-Seq.
Small RNA and RNA-seq library preparation, small RNA and RNA-seq library sequencing, and data analysis were performed as previously described (11). Briefly, the MasterPure Yeast RNA Purification Kit (Epicentre) was used to purify RNA for the construction of a small RNA or RNA-Seq library from exponentially growing cells. For RNA-seq, rRNA was removed using the Ribo-Zero rRNA Removal Magnetic Kit (Epicentre) before library construction using the ScriptSeq v2 RNA-Seq Library Preparation Kit (Epicentre). For small RNA-seq, 21- to 25-nt small RNAs were isolated by denaturing gel electrophoresis after ethanol precipitation. Libraries then were constructed using the NEBNext Small RNA Library Prep Set for Illumina (New England Biolabs). Amplified DNA of 140–150 bp was extracted from a 6% PAGE gel. Eluted DNA was ethanol precipitated overnight and resuspended in Tris-EDTA buffer. Libraries were analyzed using an Agilent 2100 BioAnalyzer and sequenced on the Illumina MiSeq platform.
Northern Blotting.
Total RNA (10 μg) was extracted using the MasterPure Yeast RNA Purification Kit (Epicentre). For Northern blotting of full-length transcripts, extracted RNA was resolved on a formaldehyde-agarose gel and blotted onto a BrightStar-Plus positively charged nylon membrane (Ambion). For Northern blotting of small RNA, small RNA was enriched using the mirVana microRNA Isolation Kit (Ambion), separated on a 6% PAGE gel, and blotted onto a Hybond N+ membrane (GE). RNA was UV-crosslinked to the membrane and subsequently hybridized with in vitro-transcribed α-32P–labeled oligonucleotide probes. The membrane then was exposed to the PhosphorImager screen overnight and imaged using ImageQuant software (GE Healthcare). Table S3 lists all oligonucleotides used for Northern blotting.
Acknowledgments
We are grateful to Dr. Ming Zhou for mass spectrometry analysis. FY13308 was provided by the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science, and Technology, Japan. We thank Dr. Kazuo Tatebayashi for providing the dhp1-1<<ura4+ strain (MP102), Dr. Masayuki Yamamoto for providing CFP-mmi1 allele, and Dr. Tomoyasu Sugiyama for mtl1-FTP allele. We also thank Jemima Barrowman for help in preparing and editing the manuscript, and members of the S.I.S.G. laboratory for helpful suggestions. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus database (accession no. GSE74741).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522127112/-/DCSupplemental.
References
- 1.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Eissenberg JC, Elgin SC. The HP1 protein family: Getting a grip on chromatin. Curr Opin Genet Dev. 2000;10(2):204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 4.Reyes-Turcu FE, Grewal SI. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev. 2012;22(2):156–163. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djupedal I, et al. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19(19):2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato H, et al. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309(5733):467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 7.Cam HP, et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37(8):809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 8.Hall IM, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297(5590):2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 9.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 10.Yamanaka S, et al. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2013;493(7433):557–560. doi: 10.1038/nature11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee NN, et al. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013;155(5):1061–1074. doi: 10.1016/j.cell.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah S, Wittmann S, Kilchert C, Vasiljeva L. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 2014;28(3):231–244. doi: 10.1101/gad.230177.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tashiro S, Asano T, Kanoh J, Ishikawa F. Transcription-induced chromatin association of RNA surveillance factors mediates facultative heterochromatin formation in fission yeast. Genes Cells. 2013;18(4):327–339. doi: 10.1111/gtc.12038. [DOI] [PubMed] [Google Scholar]
- 14.Hiriart E, et al. Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J. 2012;31(10):2296–2308. doi: 10.1038/emboj.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zofall M, et al. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335(6064):96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harigaya Y, et al. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442(7098):45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- 17.Yamashita A, Takayama T, Iwata R, Yamamoto M. A novel factor Iss10 regulates Mmi1-mediated selective elimination of meiotic transcripts. Nucleic Acids Res. 2013;41(21):9680–9687. doi: 10.1093/nar/gkt763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugiyama T, Sugioka-Sugiyama R. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J. 2011;30(6):1027–1039. doi: 10.1038/emboj.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HM, Futcher B, Leatherwood J. The fission yeast RNA binding protein Mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PLoS One. 2011;6(10):e26804. doi: 10.1371/journal.pone.0026804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.St-André O, et al. Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J Biol Chem. 2010;285(36):27859–27868. doi: 10.1074/jbc.M110.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 2010;29(13):2173–2181. doi: 10.1038/emboj.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432(7016):517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 23.Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: Implications for a unified allosteric-torpedo model. Genes Dev. 2006;20(8):954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23(11):1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugano S, Shobuike T, Takeda T, Sugino A, Ikeda H. Molecular analysis of the dhp1+ gene of Schizosaccharomyces pombe: An essential gene that has homology to the DST2 and RAT1 genes of Saccharomyces cerevisiae. Mol Gen Genet. 1994;243(1):1–8. doi: 10.1007/BF00283869. [DOI] [PubMed] [Google Scholar]
- 26.Shobuike T, Tatebayashi K, Tani T, Sugano S, Ikeda H. The dhp1(+) gene, encoding a putative nuclear 5′-->3′ exoribonuclease, is required for proper chromosome segregation in fission yeast. Nucleic Acids Res. 2001;29(6):1326–1333. doi: 10.1093/nar/29.6.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West S, Gromak N, Proudfoot NJ. Human 5′--> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432(7016):522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 28.Sugiyama T, Sugioka-Sugiyama R, Hada K, Niwa R. Rhn1, a nuclear protein, is required for suppression of meiotic mRNAs in mitotically dividing fission yeast. PLoS One. 2012;7(8):e42962. doi: 10.1371/journal.pone.0042962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egan ED, Braun CR, Gygi SP, Moazed D. Post-transcriptional regulation of meiotic genes by a nuclear RNA silencing complex. RNA. 2014;20(6):867–881. doi: 10.1261/rna.044479.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K, et al. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science. 2011;331(6024):1624–1627. doi: 10.1126/science.1198712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugiyama T, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128(3):491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 32.Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol. 2011;18(10):1132–1138. doi: 10.1038/nsmb.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304(5679):1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 34.Thon G, Cohen A, Klar AJ. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics. 1994;138(1):29–38. doi: 10.1093/genetics/138.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475(7355):244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303(5658):672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilchert C, Vasiljeva L. mRNA quality control goes transcriptional. Biochem Soc Trans. 2013;41(6):1666–1672. doi: 10.1042/BST20130202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Hoof A, Wagner EJ. A brief survey of mRNA surveillance. Trends Biochem Sci. 2011;36(11):585–592. doi: 10.1016/j.tibs.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y, et al. The fission yeast MTREC complex targets CUTs and unspliced pre-mRNAs to the nuclear exosome. Nat Commun. 2015;6:7050. doi: 10.1038/ncomms8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemay JF, et al. The RNA exosome promotes transcription termination of backtracked RNA polymerase II. Nat Struct Mol Biol. 2014;21(10):919–926. doi: 10.1038/nsmb.2893. [DOI] [PubMed] [Google Scholar]
- 42.Wagschal A, et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150(6):1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Almeida SF, García-Sacristán A, Custódio N, Carmo-Fonseca M. A link between nuclear RNA surveillance, the human exosome and RNA polymerase II transcriptional termination. Nucleic Acids Res. 2010;38(22):8015–8026. doi: 10.1093/nar/gkq703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davidson L, Kerr A, West S. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J. 2012;31(11):2566–2578. doi: 10.1038/emboj.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miki TS, Großhans H. The multifunctional RNase XRN2. Biochem Soc Trans. 2013;41(4):825–830. doi: 10.1042/BST20130001. [DOI] [PubMed] [Google Scholar]
- 46.Bayne EH, et al. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science. 2008;322(5901):602–606. doi: 10.1126/science.1164029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidson L, West S. Splicing-coupled 3′ end formation requires a terminal splice acceptor site, but not intron excision. Nucleic Acids Res. 2013;41(14):7101–7114. doi: 10.1093/nar/gkt446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell. 1999;3(3):371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 49.Fortes P, et al. Identification and characterization of RED120: A conserved PWI domain protein with links to splicing and 3′-end formation. FEBS Lett. 2007;581(16):3087–3097. doi: 10.1016/j.febslet.2007.05.066. [DOI] [PubMed] [Google Scholar]
- 50.Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell. 2006;23(2):195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 51.Millevoi S, et al. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′-end processing. EMBO Rep. 2002;3(9):869–874. doi: 10.1093/embo-reports/kvf173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dumesic PA, et al. Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell. 2013;152(5):957–968. doi: 10.1016/j.cell.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154(5):996–1009. doi: 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jimeno-González S, Haaning LL, Malagon F, Jensen TH. The yeast 5′-3′ exonuclease Rat1p functions during transcription elongation by RNA polymerase II. Mol Cell. 2010;37(4):580–587. doi: 10.1016/j.molcel.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 55.Bühler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15(10):1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marina DB, Shankar S, Natarajan P, Finn KJ, Madhani HD. A conserved ncRNA-binding protein recruits silencing factors to heterochromatin through an RNAi-independent mechanism. Genes Dev. 2013;27(17):1851–1856. doi: 10.1101/gad.226019.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell. 2008;29(3):313–323. doi: 10.1016/j.molcel.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 58.Neuwald AF, Poleksic A. PSI-BLAST searches using hidden markov models of structural repeats: Prediction of an unusual sliding DNA clamp and of beta-propellers in UV-damaged DNA-binding protein. Nucleic Acids Res. 2000;28(18):3570–3580. doi: 10.1093/nar/28.18.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jia S, Kobayashi R, Grewal SI. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005;7(10):1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 60.Zofall M, et al. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461(7262):419–422. doi: 10.1038/nature08321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132(6):983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 62.Zaratiegui M, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479(7371):135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guang S, et al. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465(7301):1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lai F, Gardini A, Zhang A, Shiekhattar R. Integrator mediates the biogenesis of enhancer RNAs. Nature. 2015;525(7569):399–403. doi: 10.1038/nature14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu F, Bakht S, Dean C. Cotranscriptional role for Arabidopsis DICER-LIKE 4 in transcription termination. Science. 2012;335(6076):1621–1623. doi: 10.1126/science.1214402. [DOI] [PubMed] [Google Scholar]
- 66.Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327(5961):94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 67.Bähler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14(10):943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]